ABSTRACT
Intestinal bacteria may influence lung homeostasis via the gut-lung axis. We conducted a single-center, quadruple-blinded, randomized trial in adult symptomatic Coronavirus Disease 2019 (Covid19) outpatients. Subjects were allocated 1:1 to probiotic formula (strains Lactiplantibacillus plantarum KABP022, KABP023, and KAPB033, plus strain Pediococcus acidilactici KABP021, totaling 2 × 109 colony-forming units (CFU)) or placebo, for 30 days. Co-primary endpoints included: i) proportion of patients in complete symptomatic and viral remission; ii) proportion progressing to moderate or severe disease with hospitalization, or death; and iii) days on Intensive Care Unit (ICU). Three hundred subjects were randomized (median age 37.0 years [range 18 to 60], 161 [53.7%] women, 126 [42.0%] having known metabolic risk factors), and 293 completed the study (97.7%). Complete remission was achieved by 78 of 147 (53.1%) in probiotic group compared to 41 of 146 (28.1%) in placebo (RR: 1.89 [95 CI 1.40–2.55]; P < .001), significant after multiplicity correction. No hospitalizations or deaths occurred during the study, precluding the assessment of remaining co-primary outcomes. Probiotic supplementation was well-tolerated and reduced nasopharyngeal viral load, lung infiltrates and duration of both digestive and non-digestive symptoms, compared to placebo. No significant compositional changes were detected in fecal microbiota between probiotic and placebo, but probiotic supplementation significantly increased specific IgM and IgG against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV2) compared to placebo. It is thus hypothesized this probiotic primarily acts by interacting with the host’s immune system rather than changing colonic microbiota composition. Future studies should replicate these findings and elucidate its mechanism of action (Registration: NCT04517422).
Abbreviations: AE: Adverse Event; BMI: Body Mass Index; CONSORT: CONsolidated Standards of Reporting Trials; CFU: Colony-Forming Units; eDRF: Electronic Daily Report Form; GLA: Gut-Lung Axis; GSRS: Gastrointestinal Symptoms Rating Scale; hsCRP: High-sensitivity C-Reactive Protein; HR: Hazard Ratio; ICU: Intensive Care Unit; OR: Odds Ratio; PCoA: Principal Coordinate Analysis; RR: Relative Risk; RT-qPCR: Real-Time Quantitative Polymerase Chain Reaction; SARS-CoV2: Severe acute respiratory syndrome coronavirus 2; SpO2: Peripheral Oxygen Saturation; WHO: World Health Organization
KEYWORDS: Covid19, SARS-CoV2, probiotic, viral load, chest x-ray, acquired immunity, gut-lung axis
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) is the causative agent of Coronavirus Disease 2019 (Covid19) global pandemic.1 SARS-CoV2 infection can range from asymptomatic to death, but most symptomatic patients typically display mild to moderate symptoms, even despite significant viral loads,2 and their condition can be managed on an outpatient basis. Symptoms can include dry cough, fever, shortness of breath, body aches, headache, fatigue, diarrhea and anosmia among others.3 However, no therapies have been approved for Covid19 outpatients to date.
Probiotics are defined as “live microorganisms that when administered in adequate amounts, confer a health benefit on the host”, and this definition entails the requirement of well-conducted studies in humans in the specified health indication.4 Recent evidence indicates a crosstalk between the gastro-intestinal tract and respiratory system, along with their respective microbiomes, referred to as the gut-lung axis (GLA).5,6 Meta-analyses have suggested oral probiotics may have a role in respiratory infections such as cold and influenza, but have also noted significant limitations, such as overreliance on subjective outcomes, small sample sizes and heterogeneity between individual trials.7,8 Particularly, heterogeneity between trials is not unexpected, as several probiotic effects are strain-specific, particularly immune-related effects.4,9–12 Based on said evidence, probiotics have been proposed for Covid19.13–15 At the time of writing, some observational, retrospective evidence has been reported,16 but no randomized, placebo-controlled trials.
The objective of this study was to test the efficacy and safety of the AB21© probiotic formula (Lactiplantibacillus plantarum stains KABP022, KABP023 and KABP033 plus Pediococcus acidilactici strain KABP021), in symptomatic Covid19 outpatients, by assessing clinical endpoints, nasopharyngeal and serum biomarkers, and its impact on the fecal microbiome.
Results
Participants
Of the 300 patients randomized, 293 completed the study between August 26th and December 10th 2020 and were available for primary analysis, while 7 were lost to follow-up (3 in probiotic and 4 in placebo, CONSORT Flowchart in Figure 1). Age ranged 18–60 years old, 126 (42.0%) had known metabolic risk factors for severe Covid19 (BMI ≥ 30, diabetes and/or hypertension) and median time from first symptom to study inclusion was 4 days (IQR 3–5). All patients were seropositive for SARS-CoV2-specific IgM, providing further confirmation of Covid19 diagnosis to RT-qPCR. In general, baseline characteristics were well balanced between groups (Table 1). Most common digestive complaints were diarrhea and nausea, followed by feeling of loose stools or incomplete evacuation and of abdominal pain. All remaining digestive symptoms (e.g. constipation, flatus, bloating, reflux) were reported by less than 10% of subjects in both study groups (Table S1), and not considered for further analysis. A few potential baseline imbalances were detected: i) higher incidence of lung infiltrates, of type II obesity, and lower SpO2 in probiotic group; ii) higher incidence of type I obesity and of shortness of breath in placebo group. Thus, said variables were considered for post-hoc sensitivity analyses.
Figure 1.
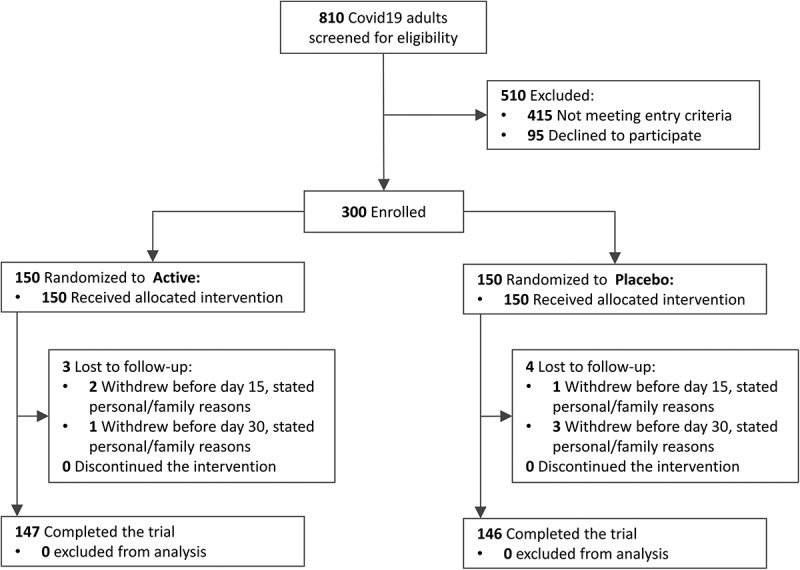
Patient enrollment and treatment assignment to active (≥2×109 CFU probiotic) or placebo among symptomatic Covid19 outpatients (CONSORT 2010 Flowchart).
Table 1.
Demographic and baseline characteristics of the randomized participants
| Characteristics | Probiotic (n = 150) | Placebo (n = 150) |
|---|---|---|
| Age (years) [median, IQR] | 34 (26–45) | 39 (27–49) |
| Sex (female) [n, %] | 82 (54.7%) | 79 (52.7%) |
| BMI (kg/m2) [median, IQR] | 27.5 (23.3–31.8) | 29.4 (27.1–32.9) |
|
31 (20.7%) | 72 (48.0%) |
|
16 (10.7%) | 0 (0.0%) |
| Smoker (yes) [n, %] | 22 (14.7%) | 20 (13.3%) |
| Diabetes (yes) [n, %] | 15 (10.0%) | 16 (10.7%) |
| Arterial hypertension (yes) [n, %] | 28 (18.7%) | 31 (20.7%) |
| Taking ≥2 medications daily (yes) [n, %] | 24 (16.0%) | 18 (12.0%) |
| Use of acetaminophen (yes) [n, %] | 83 (55.3%) | 70 (46.7%) |
| Days from symptom onset [median, IQR] | 4 (3–5) | 4 (3–5) |
| Fever (yes) [n, %] | 100 (66.7%) | 115 (76.7%) |
| Cough (yes) [n, %] | 138 (92.0%) | 133 (88.7%) |
| Headache (yes) [n, %] | 134 (89.3%) | 127 (84.7%) |
| Shortness of breath (yes) [n, %] | 42 (28.0%) | 64 (42.7%) |
| Body aches (yes) [n, %] | 94 (62.7%) | 97 (64.7%) |
| Diarrhea (yes) [n, %] | 41 (27.3%) | 54 (36.0%) |
| Loose stools (yes) [n, %] | 27 (18.9%) | 25 (16.7%) |
| Nausea (yes) [n, %] | 46 (30.7%) | 47 (31.3%) |
| Incomplete evacuation (yes) [n, %] | 27 (18.0%) | 30 (20.0%) |
| Abdominal pain (yes) [n, %] | 22 (14.7%) | 16 (10.7%) |
| Lung infiltrates (yes) [n, %] | 72 (48.0%) | 48 (32.0%) |
| SpO2 (%) [median, IQR] | 90 (90–91) | 91 (90–91) |
| SARS-CoV2 (log10 copies/mL) [median, IQR]a | 6.8 (6.7–6.9) | 6.8 (6.6–6.9) |
| SARS-CoV2 spike IgM (seropositive) [n, %]b | 150 (100%) | 150 (100%) |
| SARS-CoV2 spike IgG (seropositive) [n, %]b | 36 (24.0%) | 31 (20.7%) |
| hsCRP (mg/L) [median, IQR] | 3.2 (2.2–4.0) | 3.4 (2.8–3.9) |
| D-Dimer (mg/L) [median, IQR] | 2.0 (1.5–2.4) | 2.0 (1.3–2.8) |
BMI: Body Mass Index. hsCRP: High Sensitivity C-Reactive Protein. IQR: Interquartile range. SpO2: Peripheral Oxygen Saturation.a) As measured in nasopharyngeal swabs.b) As per test kit manufacturer instructions.
Primary clinical outcomes
Primary outcome of complete remission (i.e. complete symptomatic and viral clearance) on day 30 was achieved by 78 (53.1%) in the probiotic group compared to 41 (28.1%) in placebo (Table 2 and S2), the difference being significant at the multiplicity-corrected threshold of P = .01 (RR: 1.89 [95 CI 1.40–2.55], P < .001). No hospitalizations, ICU admissions or deaths occurred during the study, preventing the assessment of remaining primary outcomes (Table 2).
Table 2.
Primary outcomes and safety outcomes at the end of the 30-day intervention
| Probiotic | Placebo | RR (95 CI) | P-valuec | |
|---|---|---|---|---|
| Primary outcomes | ||||
| Complete remissiona [n, %] | 78/147 (53.1%) | 41/146 (28.1%) | 1.89 (1.40–2.55) | <0.001 |
| Hospitalized, moderateb [n, %] | 0/150 | 0/150 | - | 1.000 |
| Hospitalized, severeb [n, %] | 0/150 | 0/150 | - | 1.000 |
| Days of ICU stay [mean, SD] | 0 ± 0 | 0 ± 0 | - | 1.000 |
| Death [n, %] | 0/150 | 0/150 | - | 1.000 |
| Safety outcomes | ||||
| Patients with ≥ 1 AE [n, %] | 41/150 (27.3%) | 63/150 (42.0%) | 0.65 (0.47–0.90) | 0.008 |
|
7/24 (29.2%) | 8/18 (44.4%) | 0.66 (0.29–1.48) | 0.312 |
| Patients with ≥ 1 SAE [n, %] | 0/150 | 0/150 | - | 1.000 |
AE: Adverse Event. CI: Confidence Interval. ICU: Intensive Care Unit. SAE: Severe Adverse Event. SD: Standard Deviation. a) Requires negative RT-qPCR (viral clearance) plus complete resolution of all five Covid19 symptoms considered at study entry (symptomatic clearance). b) As per WHO Clinical Progression Scale.17 c) Calculated by Pearson Chi-squared test, Bonferroni-corrected threshold for significance is P = 0.01.
Secondary clinical outcomes
Patients in probiotic group reported significantly less days of fever, cough, headache, body aches (myalgia), shortness of breath (dyspnea), nausea, diarrhea and abdominal pain (Table 3). A significant effect was also observed on days with loose stools, although effect size was minimal. Importantly, only effects on fever were independent of their status at baseline, while incidence of other symptoms during the intervention was practically null in subjects who did not display them at study entry already. Patient compliance of electronic daily report form (eDRF) was high, with only 11 subjects in probiotic and 6 in placebo failing to report 100% complete diaries.
Table 3.
Days of each symptom after randomization, reported as median days (interquartile range), according to baseline status for each symptom (presence or absence at study entry). Number of subjects in each subgroup are indicated within parentheses below. P-values as calculated by Mann–Whitney non-parametric test. Number of subjects displaying each symptom at baseline within each treatment group can be found in Table 1
| Characteristic and baseline status | Probiotic | Placebo | P-value |
|---|---|---|---|
| Fever (temperature >37.5°C) | |||
|
2 (1–5) | 5 (4–8) | <0.001 |
|
2 (0–5) | 4 (4–5) | <0.001 |
| Cough | |||
|
10.5 (8–13) | 14 (12–17) | <0.001 |
|
0 (0–3.3) | 0 (0–0) | 0.238 |
| Headache | |||
|
7 (5–9) | 12 (9–14) | <0.001 |
|
0 (0–0) | 0 (0–0) | 0.404 |
| Shortness of breath | |||
|
2.5 (1–4) | 5 (2–6.3) | <0.001 |
|
0 (0–0) | 0 (0–0) | 1.000 |
| Body aches | |||
|
3 (2–6) | 7 (5–9) | <0.001 |
|
0 (0–0) | 0 (0–0) | 0.594 |
| Nausea | |||
|
2 (0–6) | 9 (0–14) | <0.001 |
|
0 (0–0) | 0 (0–0) | 0.479 |
| Diarrhea | |||
|
4 (0–6) | 8.5 (0–13.8) | 0.004 |
|
0 (0–0) | 0 (0–0) | 0.555 |
| Loose stools | |||
|
0 (0–0) | 0 (0–2) | 0.026 |
|
0 (0–0) | 0 (0–0) | 0.270 |
| Feeling of incomplete evacuation | |||
|
2 (0–3) | 0 (0–3.5) | 0.367 |
|
0 (0–0) | 0 (0–0) | 0.304 |
| Abdominal pain | |||
|
4 (0–6.5) | 10 (0–14) | 0.031 |
|
0 (0–0) | 0 (0–0) | 0.221 |
| Use of acetaminophen (post-hoc) | |||
|
1 (0–3) | 3 (3–6) | <0.001 |
|
1 (0–4) | 3 (3–7) | <0.001 |
Probiotic treatment was associated to lower nasopharyngeal viral load on days 15 and 30 compared to placebo (both P < .001; Figure 2(a)). Among subjects with lung infiltrates at baseline (n = 116), probiotic treatment was associated to lower radiographic scoring both on days 15 and 30 (both P < .001; Figure 2(b)). None of the subjects negative for lung infiltrates at baseline (n = 184) became positive for infiltrates on days 15 or 30. Compared to placebo, probiotic treatment was also associated to higher serum titers of SARS-CoV2-binding IgG and IgM on days 15 and 30 (all P < .001; Figure 2(c,d)) and lower serum levels of high-sensitivity C-reactive protein (hsCRP) and D-Dimer on day 15 (both P < .001), but not on day 30 (Figure 2(e,f)).
Figure 2.
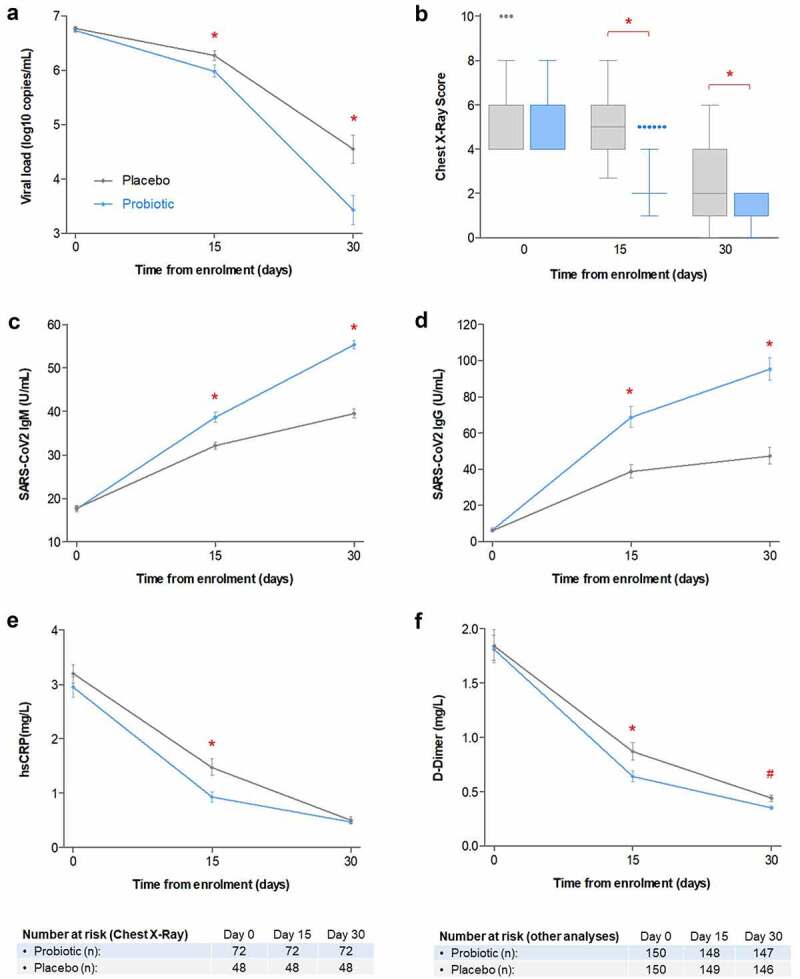
(a) Mean viral load (as base 10 logarithm of viral copies/mL), as measured by SARS-CoV2-specific RT-qPCR. (b) Box plot (median, quartiles, Tukey whiskers and individual outliers) of chest X-ray lug abnormality score, in subjects displaying lung infiltrates at baseline (n = 116). (c) Geometric mean serum titers of SARS-CoV2 spike-binding IgM. (d) Geometric mean serum titers of SARS-CoV2 spike-binding IgG. (e) Geometric means of serum levels of high-sensitivity hs-CRP. (f) Geometric means of serum levels of D-Dimer. Error bars denote 95%CI of the means. Probiotic treatment group is depicted in blue, while placebo is depicted in gray. Main effects of group, visit and group by visit were significant in all analyses ((a) to (f), P < .001). (*) Group by visit significance at specific timepoint (P < .001); (#) Group by visit statistical trend at specific timepoint (P < .10).
Compositional changes in gut microbiota
Fecal microbiome composition was characterized in a subset of probiotic and placebo patients (n = 100 each, Figure S2). A small but statistically significant increase in alpha diversity (Shannon index) was observed in both study groups on day 30 compared to day 0 (time effect P < .001; Figure 3(a)), but no significant differences were observed between groups. However, this time-dependent increase in alpha-diversity was not observed in the Chao1 abundance estimator (Figure 3(a)). Similarly, no significant compositional differences were observed between study groups, neither at baseline nor on day 30, based on beta diversity (Bray-Curtis index) (Figure 3(b)). In this regard, Principal Coordinate Analysis (PCoA) clustering was mostly driven by whether the microbiota was dominated by the Bacteroides genus, the Prevotella genus or the Firmicutes phylum (P < .0001, Figure 3(b)). The first coordinate (x-axis) separated Prevotella from Bacteroides and Firmicutes, while the second coordinate (y-axis) separated Bacteroides from Firmicutes. Noteworthy, no differences were observed in enterotype distribution between study groups (Figure S2).
Figure 3.
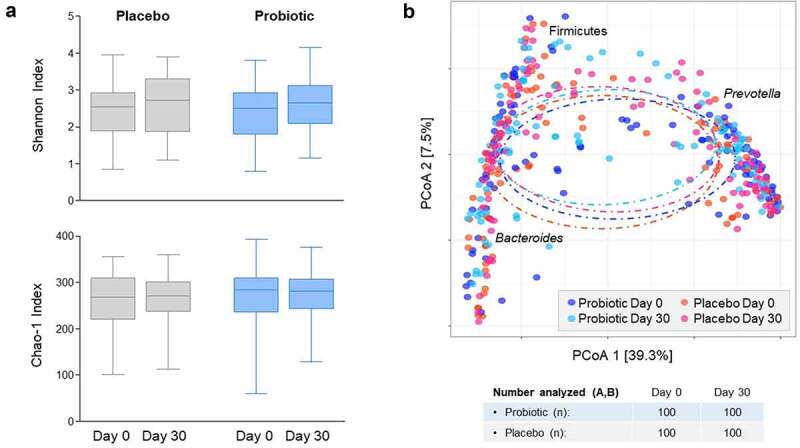
(a) Alpha diversity (Shannon diversity index and Chao1 abundance estimator). (b) Beta diversity (Bray-Curtiss index). Fecal microbiome analyses were performed by 16S rRNA sequencing in a random subset of study subjects (n = 100 from each group); obtained sequences were clustered into 97% similarity operational taxonomic unit (OTUs).
Post-hoc analyses
Several exploratory analyses were performed on the primary endpoint. Significance for complete remission was retained across all trial subpopulations assessed, defined by age, sex, presence of metabolic comorbidity, baseline viral load and days from symptom onset to randomization (Table S3). Sensitivity analyses were performed to assess the robustness of the primary endpoint to baseline imbalances, and the odds of association between probiotic treatment and complete remission remained statistically significant (unadjusted OR: 2.90 [95 CI 1.78–4.70], multivariate-adjusted OR: 2.98 [95 CI 1.77–5.03]; both P ≤ .001; Table S4). The effect of baseline enterotype on complete remission was also assessed in the subpopulation analyzed for fecal microbiome (n = 200) and remission was found to be independent of enterotype (Table S5).
Median time to overall symptom resolution (symptomatic clearance) was 5 days shorter in probiotic than placebo group (p < .001), the significance being robust to baseline imbalances as found by multivariate adjustment (Figure S1). Of note, higher BMI also produced a small but significant increase in time to symptom resolution. Days to symptom resolution were inversely correlated to IgM titers both on day 15 (rho = −0.25; P < .001) and day 30 (rho = −0.35; P < .001). A weak correlation was also observed with IgG titers on day 30 (rho = −0.14; P = .017), but not on day 15. Besides, days of use of acetaminophen were also significantly reduced in probiotic group (Table 3). Finally, age has been described as a key risk factor in Covid19 pathology, and the effect of age as a continuous covariate was further explored on SARS-CoV2-specific IgM and IgG, viral load and Brixia lung X-ray score. No significant effects were found for age, neither as independent factor nor its interaction with study group or time, for said variables (Table S6).
Safety
Serious adverse events (SAEs) were not reported for any of the 300 study subjects, while treatment-emergent adverse events (AEs) were reported in 41 (27.3%) and 63 (42.0%) subjects of probiotic and placebo groups, respectively (Table 2). The most frequent AEs were emergent fever, cough, body aches, pain when swallowing and conjunctivitis (Table S7), and no treatment-emergent hsCRP elevations occurred during the study. Incidence of AEs was generally higher in placebo than probiotic group, this trend being maintained in patients taking 2 or more medications daily. Therefore, many observed AEs could likely be natural symptom flares in Covid19.
Discussion
Few randomized, controlled trials have found effective therapies at reducing symptom duration and viral load in Covid19 outpatients so far.18–25 At the time of writing, only a few monoclonal antibodies have been recommended as treatments for Covid19 outpatients by FDA or EMA. Although effective, monoclonal antibodies are expensive, cannot be taken orally and the emergence of new SARS-CoV2 variants could jeopardize their efficacy.26 Therefore, an oral treatment helping reduce viral load, lung infiltrates and symptom duration could be a good addition to the therapeutic arsenal for Covid19 outpatients.
In this blinded, randomized study in Covid19 outpatients, the probiotic formula achieved a significant effect on improving remission rate against placebo (p < .001). No patients were hospitalized or died during the intervention, preventing assessment of remaining co-primary efficacy outcomes (frequency of progression to hospitalization, mortality ration, duration of ICU stay). Recent randomized trials in Covid19 outpatients also found a combined incidence of hospitalization plus emergency department visit of just 6% in placebo groups.18,19,23 Our entry criteria (e.g. maximum age limit of 60 years, SpO2 ≥ 90%) may have resulted in even lower risk of Covid19 worsening. However, the significance of the improvement in remission survived the Bonferroni correction for multiplicity of co-primary outcomes and was robust to multivariate adjustment for potential baseline imbalances, as well as to subject’s enterotype. Moreover, post-hoc analyses showed the effect was consistent across study subpopulations defined by age, sex, metabolic comorbidities, viral load at baseline and days from symptom initiation to randomization (all with p < .05). The positive effect in patients with metabolic comorbidities (i.e. obesity, diabetes and/or hypertension) could be of particular relevance, because of their higher risk of both severe Covid19 and long Covid19.27
Because most patients in the study had become symptom-free at the end of the study, complete remission mostly reflected whether patients achieved viral clearance. However, compared to placebo, probiotic intervention was also associated to shorter duration of both intestinal and non-intestinal Covid19 symptoms, shorter time to overall symptom resolution, and lower viral loads on day 15 and 30. Moderate-to-severe lung infiltrates in chest X-ray scans are frequent in hospitalized patients and related to worse outcomes,28 but as expected, they were absent or mild-to-moderate in our ambulatory study population. Nevertheless, probiotic intervention was associated to a significant reduction in severity of lung infiltrates in those patients displaying them, compared to placebo. Strikingly, effects on viral load on day 15 were significant but markedly less pronounced than on day 30, while benefits on symptoms and lung infiltrates seemed to occur earlier during the intervention. In this regard, recent reports suggest active viral particles correlate with RT-qPCR only during early symptom onset, high viral titers (~7 log and above) and low antibody levels.29
SARS-CoV2 spike-binding IgM were higher than IgG at baseline, as previously described,30 but this trend was reversed across the intervention. Probiotic intake was associated to higher titers of spike-binding IgM and IgG, compared to placebo. This effect was seemingly homogenous across age, but it must be pointed out that our study population was capped at 60 years old, thus studies in older subjects are warranted. In our study, higher spike-binding immunoglobulins correlated to shorter time to overall symptom resolution, especially IgM. Of note, neutralizing antibodies were not measured, but recent research indicates immunoglobulins to spike antigens provide a good correlate to both neutralization31,32 and efficacy,33 and spike-specific memory B-cells have been found to persist for months after infection.34,35 Our findings, together with the reduction of lung infiltrates and of nasopharyngeal viral load, suggest the specific probiotic strains used in this study can potentiate acquired humoral immunity against a respiratory pathogen, acting along the GLA.
Commensal gut microbiota has been found to influence immunity against viral lung infection in animal models.36,37 A previous randomized trial in more than 4,000 infants reported L. plantarum strain ATCC202195 significantly reduced lower respiratory tract infection – sepsis in infants (RR 0.66 [95 CI 0.51–0.88], P = .002).38 L. plantarum and related Lactobacillaceae species such as Pediococci, are common endophytes: bacteria living in wild vegetables and frequently ingested by herbivores and omnivores.39 Accordingly, the immune systems of the later evolved under repeated intestinal exposure to endophytes, regardless of whether these bacteria successfully colonize the intestine (i.e. become autochthonous) or are frequent nomadic commensals. Inspired by the success of the cited randomized trial38 but under the hypothesis that a cocktail of strains could better represent a natural ingestion of endophytic-type bacteria than a single strain, we chose a formula containing three different L. plantarum strains and one P. acidilactici (all of them originally isolated from humans on a vegetable-rich diet and not consuming probiotics). However, it must be stressed that existing evidence indicates probiotic immune effects are strain-specific,4,12 and effect from one strain cannot be directly extrapolated to other strains, even if from the same species (e.g. L. plantarum), until clinical trials with relevant endpoints are conducted.
Despite symptomatic clearance in the majority of patients, only a small increase in the alpha diversity in fecal microbiota (a proxy for distal colon microbiota) was noted across the 30-day study period. Furthermore, no changes in beta diversity were noted across the intervention, neither between groups nor between baseline and day 30. Principal coordinates analysis (PCoA) revealed that enterotype, not treatment or time, was the main driver in microbiota composition across the study. So far, Covid19-associated changes in fecal microbiome have been studied in hospitalized subjects and seem to be associated to the severity of the condition.40 In this regard, our results suggest microbiome changes could be minimal in Covid19 outpatients, but this observation warrant further studies.
In our view, the fact probiotic intervention succeeded at increasing acquired humoral immune response against SARS-CoV2 while not inducing detectable changes in fecal microbiota is noteworthy. The intestinal microbiota is a clear example of an ecological succession, where different bacterial groups bloom and dwindle following the availability and exhaustion of dietary nutrients, bacterial metabolites and oxygen, all under the modulation of transit time.41,42 This ecological succession is further influenced by the high disparity in bacterial densities between the small intestine (increasing from 104 to 108 cfu/mL) and the colon (1011–1012 cfu/mL).43,44 Accordingly, fecal microbiota is a proxy for the microbiota of the distal colon, but it becomes less and less representative of the microbial composition moving backwards toward the small intestine. Thus, the requirement for a probiotic to change fecal microbiota to be efficacious is a frequent misunderstanding.45 For instance, a probiotic dose of ~109 cfu could deliver a relevant microbial signal to the hundreds of Peyer patches and isolated lymphoid follicles in the ileum, regardless of compositional changes in the colon.46 Of note, this sensing of lactic acid bacteria by immune cells can require their capture by endocytosis.47,48 A probiotic could also trigger an adaptive reaction by the host’s microbiota trying to maintain its stato quo (i.e. ecological resilience),49 resulting in microbial proteome and metabolome changes which could in turn influence the host’s immunity. If successful, such adaptive response could prevent large compositional changes from extending across the colon. In this scenario, effects could be detectable only with high-resolution sequencing (e.g. single-nucleotide variants)50 or multi-omic approaches. Finally, a probiotic could overcome host’s microbiota adaptive response and ecological succession in significant numbers, producing clear compositional changes across the colon which could modulate the host’s immunity. In our study, our observations seem to rule out this last option, as no significant compositional changes were observed during the intervention, and baseline enterotype did not seem to influence the primary outcome. A graphical depiction of these possible mechanisms of action can be seen in Fig S3.
The apparent lack of changes in fecal microbiome leads us to hypothesize that observed clinical effects are mediated either by bacterial molecules produced by the probiotic strains or the host microbiome’s adaptation to probiotic intake. Specific bacterial signals to the host’s immune system might involve small molecules (e.g. short chain fatty acids, tryptophan metabolites, specific G-protein receptor ligands), which can act on mucosal immune cells but also permeate into circulation to tune immune cells in peripheral tissues.51–53 Some bacterial surface proteins in Lactobacilli are also recognized by antigen-presenting cells,10,11 which could result in systemic effects via migration of primed lymphocytes. Future studies should elucidate the mechanism of action of this probiotic on systemic immunity. Ileal microbiota sampling and multi-omic analyses could provide useful information.
Highly elevated serum hsCRP levels are a marker of poor prognosis in Covid19,3,54 yet hsCRP was only mildly elevated in our study population, as no subject displayed levels above normal range (i.e. >10 mg/L), and further declined during the intervention. Conversely, the majority of subjects in the study displayed abnormal serum levels of D-Dimer (i.e. >0.5 mg/L or μg/mL) at baseline. D-dimer serum levels declined in both probiotic and placebo groups as the study progressed, but probiotic achieved a faster decrease compared to placebo. Elevated D-Dimer levels are associated to higher risk of thrombotic events such as pulmonary embolism, and have been associated to Covid19 severity and mortality in meta-analyses.3,55 Therefore, the usefulness of this probiotic formula in helping prevent thrombotic complications in Covid19 warrants further investigations.
Treatment-emergent adverse events were characterized as in recent Covid19 trials19 and the results of this study highlight the safety of this probiotic formulation in Covid19 outpatients. Besides, no increases in hsCRP measurements were observed. Human supplementation with probiotics is generally considered as safe, based on the history of their use in foods, and is recognized as such for most probiotic strains by regulatory authorities.56,57 Conversely, probiotic use in patients with severe disease remains controversial due to concerns of bacteremia by lactic acid bacteria or microbial contaminants, especially immunosuppressed patients or those in intensive care units (ICU).57–59 Moreover, lymphopenia is frequent in severe Covid19 patients3 and could potentially interfere with the antibody-stimulating activity of this probiotic, reducing its efficacy. Therefore, additional studies must be conducted before the use of this probiotic can be recommended to patients with severe Covid19.
This study has some limitations that must be pointed out. First, all subjects in the study were of Hispanic ethnicity and were recruited in a single center. Hispanic ethnicity has been associated to higher mortality in Covid19.60 In our study, viral and symptomatic clearance in placebo group were markedly slower than in similar trials where Hispanic subjects accounted for 50% or less of the study population.18,19 Accordingly, although our study population could be regarded as particularly challenging, yet multicentric replication in other ethnicities is highly desirable. Second, no patients older than 60 years old were included in the study. The consistency of the effect across age subpopulations suggests the effects of this probiotic are not limited to young adults, yet additional studies in older populations are warranted. Third, no Covid19 aggravations requiring of hospitalization or ICU admission or resulting in death occurred in our study, probably owing to entry criteria preventing the entry of older patients or of those with lower SpO2. Thus, the usefulness of this probiotic on preventing Covid19 aggravation or death could not be directly assessed. Fourth, lenient entry criteria regarding the recent used of probiotics and antibiotics were used to facilitate patient recruitment, and dietary habits were not recorded in this study. These factors could have influenced microbiota composition. However, beta-diversity analysis indicated enterotypes explained a large fraction of the between-subject variability in microbiota composition in our study, these being markedly larger than the observed combined effect for anthropometric, dietary and medication factors in large cohort studies.61 Given the sample size, balanced distribution of enterotypes and lack of effect of enterotype on remission rate, it would seem unlikely that smaller random microbiota imbalances could explain the highly significant effects observed in this study.
In conclusion, this four-strain probiotic composition was associated with a significant increase in complete viral and symptomatic remission by day 30 in Covid19 outpatients, compared to placebo. Effect on hospitalization, ICU stay, and mortality could not be assessed because of lack of occurrences during the study. Significant effects were also observed in reducing symptom duration, viral load and lung infiltrates while increasing SARS-CoV2-specific IgM and IgG, and probiotic was well tolerated. No significant changes were detected in fecal microbiota (a proxy for distal colon microbiota), and probiotic efficacy on the primary endpoint was confirmed to be independent of the baseline enterotype. We thus hypothesize this probiotic may primarily act on the gut-lung axis (GLA) via crosstalk with the host’s immune system. Noteworthy, the observed stimulation of humoral immunity is unlikely to be dependent on a particular viral variant, an interesting trait given the emergence of new viral variants. Overall, consistency of effects across several objective endpoints and study subpopulations warrants replication studies. These studies should ensure the same strains and dose are used, while considering the immune-stimulating effects of this probiotic may require some days of buildup before beneficial effects can be observed.
Methods
Study design and participants
We conducted a randomized, parallel, quadruple-blinded, placebo-controlled study (prospectively registered as NCT04517422). Symptomatic Covid19 outpatients, 18–60 years old, with SARS-CoV2 confirmation by RT-qPCR62 within 72 h of enrollment were recruited at Hospital General Dr. Manuel Gea Gonzalez, a tertiary referral hospital in Mexico City (Mexico). Patients had to display one or more of the following Covid19 symptoms, with onset within 7 days of enrollment: fever (>37.5°C), cough, headache, muscle pain and shortness of breath. The choice of these symptoms was pragmatic, based on local experience with Covid19 at the time of study design. Full list of inclusion and exclusion criteria can be found in Supplementary Methods. The protocol was approved by the Research Ethics Committee of Hospital General Dr. Manuel Gea Conzalez (ref. 12–120-2020), complied with the Helsinki Declaration and followed the CONSORT reporting guideline (Annex 1 in Supplementary Information). All participants provided written, informed consent.
Randomization and blinding
Subjects were randomized 1:1 in blocks of six without stratification, using a randomization list generated with Sealed Envelope (https://www.sealedenvelope.com/) by an independent pharmacist. Enrollment and allocation were conducted by caregivers. Study products (probiotic or placebo) were given in coded, anonymous boxes, and were indistinguishable in form, color, and taste. All subjects, caregivers, investigators, and outcome assessors were unaware of the treatment allocation.
Study products
The active product (AB21© probiotic formula) consisted of capsules containing Lactiplantibacillus plantarum KABP033 (CECT30292), L. plantarum KABP022 (CECT7484), L. plantarum KABP023 (CECT7485) and Pediococcus acidilactici KABP021 (CECT7483), in a ratio of 3:1:1:1 colony-forming units (CFU), respectively, and a total dose of ≥2×109 total CFU, with a maltodextrin carrier. Placebo product consisted of capsules containing the maltodextrin carrier only. Identity of the four strains in the probiotic product and microbial quality of probiotic and placebo batches were verified (Supplementary Methods). Probiotic was also monitored for conformance to the specification of ≥2×109 CFU/capsule throughout the study in stability chamber (25 ± 2°C, 60 ± 5% relative humidity) by ISO17025-accredited company Silliker Iberica (Merieux Nutrisciences Group).
Patient procedures
The study was scheduled across three study site visits: day 0 (visit 1), day 15 (visit 2) and day 30 (visit 3). On day 0, study subjects were given the study product and were instructed to store it at room temperature and take one oral capsule daily, from day 1 to day 30, before breakfast. Subjects were also given access to a web-based electronic daily report form (eDRF) for symptoms recording. An infrared thermometer (Harbin Xiande Technology Development Co, Harbin, China) was provided to each subject for at home use during the study.
On all study visits, subjects were assessed for Covid19 severity using WHO Clinical Progression Scale17 and received chest pulmonary X-ray, which was rated according to Brixia score28 using the IA-Rx software (Annex 2 in Supplementary Information). Nasopharyngeal and venous blood samples were taken on each visit, and fecal samples were collected on the first and last visit with the GUT-OMR200 kit (DNAgenotek). Study subjects were also contacted by phone on days 5, 10, 20 and 25 (all ± 1 day) by a physician, as part of outpatient follow-up. Only acetaminophen (500 mg/dose, up to three times a day) was allowed as comedication for Covid19 symptoms (use was recorded in patient’s eDRF), to be used on-demand, and no other Covid19 therapies (e.g. corticosteroids) were allowed. All patients were recommended to rest as much as possible and not to change their diet.
Outcomes
Five co-primary efficacy outcomes were considered at the end of the 30 days intervention, using definitions from World Health Organization Clinical Progression Scale:17 i) fraction of subjects who progressed to remission (score of “0”); ii) fraction who progressed to hospitalization with moderate disease (scores of “4” or “5”); iii) fraction who progressed to hospitalization with severe disease (scores of “6” to “9”); iv) mortality rate (score of “10”); v) length of stay in Intensive Care Unit (ICU). Remission required a negative RT-qPCR (viral clearance) plus complete resolution of all five Covid19 symptoms considered at study entry (symptomatic clearance).
Prespecified secondary outcomes included: i) SARS-CoV2 viral load evaluated by RT-qPCR; ii) Plasma SARS-CoV2 spike-binding IgG and IgM titers; iii) lung infiltrates measured by chest X-ray and rated according to Brixia score;28 iv) duration from randomization of each of the five core Covid19 symptoms considered at baseline: fever (>37.5°C), cough, headache, body aches and shortness of breath; v) duration from randomization of gastrointestinal symptoms according to GSRS (Gastrointestinal Symptom Rating Scale);63 vi) serum high sensitivity C-reactive protein (hsCRP) and D-Dimer; and vii) fecal microbiome evaluated by 16S rRNA sequencing. These outcomes were analyzed in all subjects, except for microbiome which was analyzed in a random subset of 100 subjects (out of 150) per study arm.
Exploratory (post-hoc) outcomes included: i) robustness of the primary endpoint to baseline imbalances; ii) significance of the primary outcome in subpopulations split by age (less than 50 years old vs 50 years and older), sex (male vs female), metabolic comorbidity (diabetes, hypertension or obesity vs none), viral load at baseline (below vs above median value) and time from symptom onset to randomization (one to four days vs five or more days); iii) time to overall symptomatic resolution, defined as the disappearance of all five core Covid19 symptoms; iv) correlation between symptom duration and spike-binding IgM and IgG titers; v) number of days of use of acetaminophen; and vi) age-dependence of spike-binding IgM and IgG titers, viral load and chest X-ray score.
A treatment-emergent adverse event (AE) was defined as any event that first occurred or worsened in severity after the initiation of the intervention, akin to other trials in Covid19 outpatients.19 A serious AE (SAE) was defined when causing hospitalization, persistent disability or incapacitation, or death. Reporting of adverse events was monitored in phone calls (days 5, 10, 20, and 25) and study site visits (days 0, 15, and 30). Treatment-emergent serum levels of hsCRP >10 mg/L were also considered in safety analysis.
Because no hospitalizations were observed as the study progressed, the protocol was amended to include remission (defined as negative RT-qPCR plus symptomatic remission) to primary outcomes, as well as the duration of five specific Covid19 symptoms (fever, cough, headache, shortness of breath and body aches) as secondary outcomes, since these symptoms were already being recorded in patients eDRFs. All these changes were granted approval by the Research Ethics Committee before study unblinding by the independent pharmacist (on February 3, 2021).
Laboratory and microbiome analyses
SARS-CoV2 analysis was performed on nasopharyngeal samples using a validated RT-qPCR protocol (Supplementary Methods).62 Venous blood (stored at −70°C until the end of the study) was used to assay for SARS-CoV2 spike protein-specific IgG (DiaSorin SpA), SARS-CoV2 spike protein-specific IgM (Abbot Laboratories), D-dimer (Spinreact SA) and hsCRP (Abbot Laboratories), according to manufacturer’s instructions.
DNA was extracted from fecal samples obtained on days 0 and 30 from a random subset of patients from probiotic and placebo group (n = 100 each) with MoBio’s Soil DNA Isolation kit (Quiagen). Bacterial 16S rRNA genes were PCR-amplified with dual-barcoded primers targeting the V4 region (515 F and 806 R) and sequenced with Miseq (Illumina), obtaining an average of 29,400 quality-filtered reads per sample. Fastq files were quality-filtered and clustered into 97% similarity operational taxonomic unit (OTUs) using the Mothur software package64 and classified using the Silva database.65 Alpha-diversity (Shannon and Chao1 indexes) and beta-diversity (Bray-Curtiss index) were computed using the vegan R package. Changes in alpha diversity were assessed with a linear mixed-effect model for repeated measures (MMRMs) with visit and group as fixed factors, and a group by visit interaction. Beta diversity was assessed by Principal Coordinate Analysis (PCoA) and 2-way PERMANOVA (visit, group and group by visit interaction).
Sample size
No published data could be found regarding the risk of mild Covid19 progressing to hospitalization in Mexico, estimates based on local experience ranging 27% to 67%. Taking the average value (47%) and aiming at detecting a relative reduction of at least 35% with a two-sided alpha = 5% and power = 80% resulted in 150 subjects per study arm after accounting for dropouts.
Statistical analyses
All analyses were performed according to allocated randomization group, without any data exclusion or imputation for missing values. All statistical tests were two-tailed, and differences were considered significant at P< .05. For the five co-primary outcomes, a Bonferroni-type correction for multiplicity was applied post-hoc, resulting in a significance threshold of P < .01.
Co-primary outcomes were assessed by Pearson’s Chi-squared test. For secondary and exploratory outcomes, differences in days of symptoms were assessed with Mann–Whitney test. Differences between groups across days 0,15 and 30 in SARS-CoV2 viral load, SARS-CoV2-specific IgM and IgG, hsCRP and D-Dimer were assessed by linear mixed-effects models for repeated measures (with unstructured covariance matrix), while differences in Chest-X ray Brixia score were assessed by logit ordinal regression for repeated measures. These repeated measures analyses had study group and visit as fixed factors, and a group-by-visit interaction. Binomial logistic regression was used when adjusting primary outcome for baseline covariates in sensitivity analysis, and to calculate unadjusted and multivariate-adjusted odd-ratios (OR) and their 95% confidence interval. Time to overall symptom resolution was assessed by Kaplan-Meyer analysis, unadjusted and multivariate-adjusted hazard ratios (HR) and their 95% confidence intervals being calculated by Cox method. Finally, bivariate correlations were assessed by Spearman’s rank method. All statistical tests described in this section were performed with the SPSS program v.24 (IBS Corp.). Microbiota-specific analyses are described in the Laboratory analyses section.
Supplementary Material
Acknowledgments
Our sincere thanks to thank Paola Juarez-Valdez, Pamela Paez-Melendez, Yarumi Espinosa-Hernandez and Nidia Gómez-Ramirez for the nursing activities of the project.
Funding Statement
This work was fully sponsored by AB-Biotics SA (Kaneka Group).
Author contributions
PGC, JEM, and ATAA designed the study. TGM, CDNR, and IJE performed patient procedures. ELO and GLV performed laboratory analyses. CJG and PGC collected and curate the data. PGC conducted the statistical analyses. PGC and JEM drafted the manuscript. ATAA critically revised the manuscript.
Disclosure statement
ATAA reports receiving speaker fees from AB-Biotics SA (Kaneka Group). JEM is a staff scientist with no stock options or shares at AB-Biotics SA (Kaneka Group). Other authors report their institution was sponsored by AB-Biotics SA (Kaneka Group) for the submitted work.
Data availability statement
The data that support the findings of this study are available as deidentified individual patient data (IPD) in the ZENODO public repository (http://doi.org/10.5281/zenodo.5084711).
Supplementary material
Suppelemental data for this article can be accessed on the publisher’s website.
References
- 1.Boni MF, Lemey P, Jiang X, Lam TTY, Perry BW, Castoe TA, Rambaut A, Robertson DL.. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol. 2020;5:1408–16. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 2.Lavezzo E, Franchin E, Ciavarella C, Cuomo-Dannenburg G, Barzon L, Del Vecchio C, Rossi L, Manganelli R, Loregian A, Navarin N, et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature. 2020;584:425–429. doi: 10.1038/s41586-020-2488-1. [DOI] [PubMed] [Google Scholar]
- 3.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 4.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen SJ, et al. Expert consensus document: the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 5.Budden KF, Gellatly SL, Wood DLA, Cooper MA, Morrison M, Hugenholtz P, Hansbro PM. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 6.Dang AT, Marsland BJ. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019;12:843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 7.King S, Glanville J, Sanders ME, Fitzgerald A, Varley D. Effectiveness of probiotics on the duration of illness in healthy children and adults who develop common acute respiratory infectious conditions: a systematic review and meta-analysis. Br J Nutr. 2014;112:41–54. doi: 10.1017/S0007114514000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao Q, Dong BR, Wu T. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst Rev. 2015;2015:CD006895. doi: 10.1002/14651858.CD006895.pub3. [DOI] [PubMed] [Google Scholar]
- 9.Paineau D, Carcano D, Leyer G, Darquy S, Alyanakian MA, Simoneau G, Bergmann JF, Brassart D, Bornet F, Ouwehand AC. Effects of seven potential probiotic strains on specific immune responses in healthy adults: a double-blind, randomized, controlled trial. FEMS Immunol Med Microbiol. 2008;53:107–113. doi: 10.1111/j.1574-695X.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 10.van Hemert S, Meijerink M, Molenaar D, Bron PA, de Vos P, Kleerebezem M, Wells JM, Marco ML. Identification of Lactobacillus plantarum genes modulating the cytokine response of human peripheral blood mononuclear cells. BMC Microbiol. 2010;10:293. doi: 10.1186/1471-2180-10-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meijerink M, van Hemert S, Taverne N, Wels M, de Vos P, Bron PA, Savelkoul HF, van Bilsen J, Kleerebezem M, Wells JM. Identification of genetic loci in Lactobacillus plantarum that modulate the immune response of dendritic cells using comparative genome hybridization. PLoS One. 2010;5:e10632. doi: 10.1371/journal.pone.0010632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao W, Peng C, Sakandar HA, Kwok L-Y, Zhang W. Meta-analysis: randomized trials of Lactobacillus plantarum on immune regulation over the last decades. Front Immunol. 2021;12:728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baud D, Dimopoulou Agri V, Gibson GR, Reid G, Giannoni E. Using probiotics to flatten the curve of coronavirus disease COVID-2019 pandemic. Front Public Heal. 2020;8:186. doi: 10.3389/fpubh.2020.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mullish BH, Marchesi JR, McDonald JAK, Pass DA, Masetti G, Michael DR, Plummer S, Jack AA, Davies TS, Hughes TR, et al. Probiotics reduce self-reported symptoms of upper respiratory tract infection in overweight and obese adults: should we be considering probiotics during viral pandemics? Gut Microbes. 2021;13:1–9. doi: 10.1080/19490976.2021.1900997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng J, Zhang M, Yao G, Kwok L-Y, Zhang W. Probiotics as adjunctive treatment for patients contracted COVID-19: current understanding and future needs. Front Nutr. 2021;8:669808. doi: 10.3389/fnut.2021.669808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceccarelli G, Borrazzo C, Pinacchio C, Santinelli L, Pietro IG, Cavallari EN, Celani L, Marazzato M, Alessandri F, Ruberto F, et al. Oral bacteriotherapy in patients with COVID-19: a retrospective cohort study. Front Nutr. 2021;7:613928. doi: 10.3389/fnut.2020.613928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall JC, Murthy S, Diaz J, Adhikari N, Angus DC, Arabi YM, Baillie K, Bauer M, Berry S, Blackwood B, et al. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis. 2020;20:e192–7. doi: 10.1016/S1473-3099(20)30483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlieb RL, Nirula A, Chen P, Boscia J, Heller B, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V, et al. Effect of Bamlanivimab as monotherapy or in combination with Etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.López-Medina E, López P, Hurtado IC, Dávalos DM, Ramirez O, Martínez E, Díazgranados JA, Oñate JM, Chavarriaga H, Herrera S, et al. Effect of Ivermectin on time to resolution of symptoms among adults with mild COVID-19: a randomized clinical trial. JAMA. 2021;325:1426. doi: 10.1001/jama.2021.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feld JJ, Kandel C, Biondi MJ, Kozak RA, Zahoor MA, Lemieux C, Borgia SM, Boggild AK, Powis J, McCready J, et al. Peginterferon lambda for the treatment of outpatients with COVID-19: a phase 2, placebo-controlled randomised trial. Lancet Respir Med. 2021;9:498–510. doi: 10.1016/S2213-2600(20)30566-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jagannathan P, Andrews JR, Bonilla H, Hedlin H, Jacobson KB, Balasubramanian V, Purington N, Kamble S, de Vries CR, Quintero O, et al. Peginterferon Lambda-1a for treatment of outpatients with uncomplicated COVID-19: a randomized placebo-controlled trial. Nat Commun. 2021;12:12. doi: 10.1038/s41467-020-20168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reis G, Moreira Silva EADS, Medeiros Silva DC, Thabane L, Singh G, Park JJH, Forrest JI, Harari O, Quirino Dos Santos CV, Guimarães de Almeida APF, et al. Effect of early treatment with hydroxychloroquine or Lopinavir and Ritonavir on risk of hospitalization among patients with COVID-19: the TOGETHER randomized clinical trial. JAMA Netw open. 2021;4:e216468. doi: 10.1001/jamanetworkopen.2021.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas S, Patel D, Bittel B, Wolski K, Wang Q, Kumar A, Il’Giovine ZJ, Mehra R, McWilliams C, Nissen SE, et al. Effect of high-dose Zinc and Ascorbic acid supplementation vs usual care on symptom length and reduction among ambulatory patients with SARS-CoV-2 infection. JAMA Netw Open. 2021;4:e210369. doi: 10.1001/jamanetworkopen.2021.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ledford H. COVID antibody treatments show promise for preventing severe disease. Nature. 2021;591:513–514. doi: 10.1038/d41586-021-00650-7. [DOI] [PubMed] [Google Scholar]
- 26.Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nat. 2021 5937857 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 27.Khunti K, Davies MJ, Kosiborod MN, Nauck MA. Long COVID — metabolic risk factors and novel therapeutic management. Nat. Rev. Endocrinol. 2021;17:379–380. doi: 10.1038/s41574-021-00495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borghesi A, Zigliani A, Golemi S, Carapella N, Maculotti P, Farina D, Maroldi R. Chest X-ray severity index as a predictor of in-hospital mortality in coronavirus disease 2019: a study of 302 patients from Italy. Int J Infect Dis. 2020;96:291–293. doi: 10.1016/j.ijid.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh A, Stellrecht K, Arunachalam T, Barman T, Robek M, Waxman M, Elmendorf S, Metzger D. Lack of active SARS-CoV-2 virus in a subset of PCR-positive COVID-19 congregate care patients. J Clin Virol. 2021;141:104879. doi: 10.1016/j.jcv.2021.104879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, Hemmings O, O’Byrne A, Kouphou N, Galao RP, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dispinseri S, Secchi M, Pirillo MF, Tolazzi M, Borghi M, Brigatti C, De Angelis ML, Baratella M, Bazzigaluppi E, Venturi G, et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat Commun. 2021;12:2670. doi: 10.1038/s41467-021-22958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rockstroh A, Wolf J, Fertey J, Kalbitz S, Schroth S, Lübbert C, Ulbert S, Borte S. Correlation of humoral immune responses to different SARS-CoV-2 antigens with virus neutralizing antibodies and symptomatic severity in a German COVID-19 cohort Emerg Microbes Infect . 2021;10:774–781. doi: 10.1080/2222175120211913973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Earle K, Ambrosino D, Fiore-Gartland A, Goldblatt D, Gilbert P, Siber G, Dull P, Plotkin S. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39:4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anand SP, Prévost J, Nayrac M, Beaudoin-Bussières G, Benlarbi M, Gasser R, Brassard N, Laumaea A, Gong SY, Bourassa C, et al. Longitudinal analysis of humoral immunity against SARS-CoV-2 spike in convalescent individuals up to 8 months post-symptom onset. Cell Reports Med. 2021;2:100290. doi: 10.1016/j.xcrm.2021.100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, Grifoni A, Ramirez SI, Haupt S, Frazier A, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science . 2021;371:eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abt M, Osborne L, Monticelli L, Doering T, Alenghat T, Sonnenberg G, Paley M, Antenus M, Williams K, Erikson J, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradley KC, Finsterbusch K, Schnepf D, Crotta S, Llorian M, Davidson S, Fuchs SY, Staeheli P, Wack A. Microbiota-driven tonic interferon signals in lung stromal cells protect from influenza virus infection. Cell Rep. 2019;28:245–256.e4. doi: 10.1016/j.celrep.2019.05.105. [DOI] [PubMed] [Google Scholar]
- 38.Panigrahi P, Parida S, Nanda NC, Satpathy R, Pradhan L, Chandel D, Baccaglini L, Mohapatra A, Mohapatra SS, Misra PR, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017;548:407–412. doi: 10.1038/nature23480. [DOI] [PubMed] [Google Scholar]
- 39.Martínez-Romero E, Aguirre-Noyola JL, Bustamante-Brito R, González-Román P, Hernández-Oaxaca D, Higareda-Alvear V, Montes-Carreto LM, Martínez-Romero JC, Rosenblueth M, Servín-Garcidueñas LE. We and herbivores eat endophytes. Microb Biotechnol. 2020;1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeoh YK, Zuo T, Lui GC-Y-Y, Zhang F, Liu Q, Ayly L, Chung ACCK, Cheung CP, Tso EYYK, Fung KSCS, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falony G, Vieira-Silva S, Raes J. Richness and ecosystem development across faecal snapshots of the gut microbiota. Nat. Microbiol. 2018;3:526–528. doi: 10.1038/s41564-018-0143-5. [DOI] [PubMed] [Google Scholar]
- 42.Roager HM, Hansen LBS, Bahl MI, Frandsen HL, Carvalho V, Gøbel RJ, Dalgaard MD, Plichta DR, Sparholt MH, Vestergaard H, et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat Microbiol . 2016;1:16093. [DOI] [PubMed] [Google Scholar]
- 43.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2015;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kastl AJ, Terry NA, Wu GD, Albenberg LG. The structure and function of the human small intestinal microbiota. Curr Understanding Future Directions. 2020;9:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wieërs G, Belkhir L, Enaud R, Leclercq S, de Foy J-MP, Dequenne I, de Timary P, Cani PD, Feld JJ, Kandel C, et al. How probiotics affect the microbiota. Front Cell Infect Microbiol. 2019;9:454. doi: 10.3389/fcimb.2019.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mörbe UM, Jørgensen PB, Fenton TM, Von Burg N, Riis LB, Spencer J, Agace WW. Human gut-associated lymphoid tissues (GALT); diversity, structure, and function. Mucosal Immunol. 2021;14:793–802. doi: 10.1038/s41385-021-00389-4. [DOI] [PubMed] [Google Scholar]
- 47.Gutierrez-Merino J, Isla B, Combes T, Martinez-Estrada F, De Motes CM. Beneficial bacteria activate type-I interferon production via the intracellular cytosolic sensors STING and MAVS. Gut Microbes. 2020;11:771–788. doi: 10.1080/19490976.2019.1707015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss G, Rasmussen S, Zeuthen L, Nielsen B, Jarmer H, Jespersen L, Frøkiaer H. Lactobacillus acidophilus induces virus immune defence genes in murine dendritic cells by a Toll-like receptor-2-dependent mechanism. Immunology. 2010;131:268–281. doi: 10.1111/j.1365-2567.2010.03301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lozupone C, Stombaugh J, Gordon J, Jansson J, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma C, Zhang C, Chen D, Jiang S, Shen S, Huo D, Huang S, Zhai Q, Zhang J. Probiotic consumption influences universal adaptive mutations in indigenous human and mouse gut microbiota. Commun Biol. 2021;4:1–12. 2021 41. doi: 10.1038/s42003-021-02724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colosimo DA, Kohn JA, Luo PM, Piscotta FJ, Han SM, Pickard AJ, Rao A, Cross JR, Cohen LJ, Brady SF. Mapping interactions of microbial metabolites with Human G-protein-coupled receptors. Cell Host Microbe. 2019;26:273–282.e7. doi: 10.1016/j.chom.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. 2017;46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lynn DJ, Benson SC, Lynn MA, Pulendran B. Modulation of immune responses to vaccination by the microbiota: implications and potential mechanisms. Nat Rev Immunol. 2021;21:1–14. doi: 10.1038/s41577-020-00486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smilowitz NR, Kunichoff D, Garshick M, Shah B, Pillinger M, Hochman JS, Berger JS. C-reactive protein and clinical outcomes in patients with COVID-19. Eur Heart J. 2021;42:2270–2279. doi: 10.1093/eurheartj/ehaa1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nugroho J, Wardhana A, Maghfirah I, Mulia EPB, Rachmi DA, A’yun MQ, Septianda I. Relationship of D-dimer with severity and mortality in SARS-CoV-2 patients : a meta-analysis. Int J Lab Hematol. 2021;43:110–115. doi: 10.1111/ijlh.13336. [DOI] [PubMed] [Google Scholar]
- 56.Koutsoumanis K, Allende A, Alvarez-Ordóñez A, Bolton D, Bover-Cid S, Chemaly M, Davies R, De Cesare A, Hilbert F, Lindqvist R, et al. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 12: suitability of taxonomic units notified to EFSA until March 2020. EFSA J. 2020;18:e06174. doi: 10.2903/j.efsa.2020.6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat. Med. 2019;25:716–729. doi: 10.1038/s41591-019-0439-x. [DOI] [PubMed] [Google Scholar]
- 58.Yelin I, Flett KB, Merakou C, Mehrotra P, Stam J, Snesrud E, Hinkle M, Lesho E, McGann P, McAdam AJ, et al. Genomic and epidemiological evidence of bacterial transmission from probiotic capsule to blood in ICU patients. Nat Med. 2019;25:1728–1732. doi: 10.1038/s41591-019-0626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Didari T, Solki S, Mozaffari S, Nikfar S, Abdollahi M. A systematic review of the safety of probiotics. Expert Opin. Drug Saf. 2014;13:227–239. doi: 10.1517/14740338.2014.872627. [DOI] [PubMed] [Google Scholar]
- 60.Gold JAW, Rossen LM, Ahmad FB, Sutton P, Li Z, Salvatore PP, Coyle JP, DeCuir J, Baack BN, Durant TM, et al. Race, ethnicity, and age trends in persons who died from COVID-19 — United States, May–August 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1517–1521. doi: 10.15585/mmwr.mm6942e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al. Population-level analysis of gut microbiome variation. Science (80-). 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 62.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Svedlund J, Sjödin I, Dotevall G. GSRS-A clinical rating scale for gastrointestinal symptoms in patients with irritable bowel syndrome and peptic ulcer disease. Dig Dis Sci. 1988;33:129–134. doi: 10.1007/BF01535722. [DOI] [PubMed] [Google Scholar]
- 64.Schloss PD. Reintroducing mothur: 10 years later. Appl Environ Microbiol. 2020;86:e02343-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available as deidentified individual patient data (IPD) in the ZENODO public repository (http://doi.org/10.5281/zenodo.5084711).


