Abstract
Current methods to evaluate effects of kinase inhibitors in cancer are suboptimal. Analysis of changes in cancer metabolism in response to the inhibitors creates an opportunity for better understanding of the interplay between cell signaling and metabolism and, from the translational perspective, potential early evaluation of response to the inhibitors as well as treatment optimization. We performed genomic, metabolomic and fluxomic analyses to evaluate the mechanism of action of the Bruton’s tyrosine kinase (BTK) inhibitor ibrutinib (IBR) in mantle cell lymphoma (MCL) cells. Our comprehensive analysis of the data generated by these diverse technologies revealed that IBR profoundly affected key metabolic pathways in IBR-sensitive cells including glycolysis, pentose phosphate pathway, TCA cycle and glutaminolysis while having much less effects on IBR-poorly responsive cells. Changes in 1H MRS-detectable lactate and alanine concentrations emerged as promising biomarkers of response and resistance to IBR as demonstrated from experiments on various MCL cell lines. The metabolic network analysis on the 13C MRS and 13C LC-MS experimental data provided quantitative estimates of various intracellular fluxes and energy contributions. Glutaminolysis contributed over 50% of mitochondrial ATP production. Administration of the glutaminase inhibitor CB-839 induced growth suppression of the IBR-poorly responsive cells.
Keywords: kinase inhibition, cell signaling, cell metabolism, metabolic flux analysis, fluxomics, 13C MRS, 13C LC-MS, 13C metabolomics, molecular pathophysiology, lymphoma
INTRODUCTION
Small molecule kinase inhibitors have shown impressive clinical efficacy in large subsets of lymphomas and other malignancies. However, there are currently no reliable methods to detect response to kinase inhibitors at an early stage of therapy. While inhibitor’s effects on intracellular signaling pathways have been studied extensively, these agents also affect cell metabolism, opening the possibility that the inhibitor-induced metabolic changes may serve as biomarkers of response.
To begin to address this critical issue, we have used cells derived from mantle cell lymphoma (MCL), a currently non-curable type of lymphoma, and a Bruton’s tyrosine kinase (BTK) inhibitor ibrutinib (IBR) that has recently been approved by the FDA for treatment of this disease. IBR induces a response in 2/3 of MCL patients who are refractory to standard therapies (1, 2). However, the remaining 1/3 of MCL patients do not respond, and drug resistance also develops over time in a large subset of the responding patients. Therefore, the development of methods to accurately and promptly distinguish between responders and non-responders and the ability to precisely monitor responsiveness status to IBR and other kinase inhibitors would be of utmost importance to the management of patients with MCL and other malignancies.
We have performed a comprehensive metabolic analysis of IBR-sensitive and IBR-poorly responsive MCL cells following administration of IBR: RNA-Seq analysis focusing on metabolic genes, liquid chromatography mass spectrometry (LC-MS) and magnetic resonance spectroscopy (MRS) on intracellular metabolites and extracellular flux measurements. We have utilized 13C tracer experiments with MRS and LC-MS combined with comprehensive and compartmentalized metabolic network analysis methods (3–6) to determine intracellular fluxes through various metabolic pathways and measured contributions of these fluxes to overall tumor energy metabolism. We found high glycolysis and glutaminolysis activities and high oxidative pentose phosphate pathway (PPP) to glycolysis ratios in both IBR-sensitive MCL-RL and IBR-poorly responsive JeKo-1 cells under baseline conditions. We also found profound decreases in the activity of glycolysis, oxidative PPP, TCA cycle and glutaminolysis in MCL-RL cells after treatment with IBR. Much smaller changes of these pathways, in particular in glutaminolysis, were found in JeKo-1 cells in response to IBR. A glutaminase inhibitor CB-839 markedly inhibited cell growth of IBR-poorly responsive JeKo-1 cells but not IBR-sensitive MCL-RL cells.
This study provides detailed insight into metabolic pathway alterations induced by IBR in the IBR-sensitive and IBR–poorly responsive MCL cells. The applied methodologies and identified alterations enable better understanding of the pathophysiology of MCL cells. Furthermore, they open new horizons in evaluation of the effects of kinase inhibition and mechanisms of resistance to kinase inhibition with the related important implications for cancer therapy.
MATERIALS AND METHODS
Cell lines and drugs:
MCL-RL cells were derived from an MCL patient at the University of Pennsylvania. JeKo-1 and Rec-1 cells were purchased from the American Type Culture Collection (ATCC). SP49 cells were obtained from Dr. Raymond Lai, University of Alberta. Granta-519 cells were purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). The cell line identity was confirmed by diverse methods including flow cytometry, FISH, and short-tandem repeat (STR) studies. Flow cytometry six-color panel consisted of antibodies against CD45, CD20, CD5, CD23, and immunoglobulin kappa and lambda light chain. FISH detected fusion of immunoglobulin heavy chain (IgH) and cyclin D1 genes. STR test was done at University of Pennsylvania core facility. The cell lines were also found free of mycoplasma contamination using Mycoplasma detection kits from Thermo Fisher (Waltham, MA). All cells were grown in RPMI medium supplemented with 10% fetal bovine serum and 100 μg/ml penicillin/streptomycin under a humidified 37°C/5% CO2 incubator condition. Ibrutinib (PCI-32765) and CB-839 were purchased from Selleckchem (Houston, TX). [1,6-13C2] glucose, [1,2-13C2] glucose and [U-13C5, U-15N2] glutamine were acquired from Cambridge Isotope and Sigma Aldrich.
Cell count assay.
The MCL cells were placed in 96-well plates (Corning) at a concentration of 2×104 cells/well in RPMI medium supplemented with 10% FBS, cultured for 72 h in the absence or presence of various concentrations of BTK inhibitor IBR or glutaminase inhibitor CB-839. They were harvested, centrifuged, suspended in trypan blue-containing PBS and microscopically counted with a hemocytometer. The depicted results are representative of two separate counts. For kinetic studies, cells seeded in T-25 flasks at 105 cells/ml were grown and collected at 24 h, 48 h and 72 h of incubation with and without 500 nM IBR, and the viable cell numbers were measured using the Vi-CELL™ Cell Viability Analyzer (Beckman Coulter). Triplicate experimental data are presented.
Cell proliferation (BrdU incorporation) assay:
Cell proliferation was evaluated by detection of 5’-bromo-2’-deoxyuridine (BrdU) incorporation using the commercially available Cell Proliferation ELISA kit (Cell Signaling) according to the manufacturer’s protocol. In brief, the cells were seeded in 96-well plates at a concentration of 2×104 cells/well in RPMI medium supplemented with 10% FBS, cultured for 48 h in triplicates in the presence or absence of drug and labeled with BrdU for additional 24 h. After the culture plate centrifugation, supernatant removal and the plate drying, the cells were fixed and DNA denaturated by adding 200 ml FixDenat reagent. The amount of incorporated BrdU was determined by incubation with a specific antibody conjugated to peroxidase followed by colorimetric conversion of the substrate and optical density (OD) evaluation using an ELISA plate reader at 405 nm absorbance.
3-(4,5-dimethylthiazol-2-Yl)-2,5-diphenyltetrazolium bromide (MTT) enzymatic conversion assay:
Cells were plated in 96 well plates (3–5×103/well) and treated in triplicates with drug or vehicle alone for 72 h at 37°C. The treated cells were labeled with 10 μl of MTT (Promega) at 5 mg/ml for 4 h. Well contents were solubilized overnight with 10% SDS in 0.01 M HCl. Absorbance at 570 nm in each well was determined using a Titertek Multiskan reader.
Cell cycle assay:
MCL cells were stained with immunofluorescent dye showing DNA content distribution and analyzed by flow cytometry to identify G0/G1 and G2/M peaks separated by the S phase.
Cell division rate evaluation assay:
CFSE-labeled MCL cells were exposed in duplicate to IBR or its vehicle for 3 days and analyzed by FACS for the CFSE labeling pattern of responsive cells.
RNA-Seq analysis:
MCL cells were exposed to 500 nM IBR or its vehicle for 6 h. RNA was isolated, fragmented, adapter ligated and analyzed by high-throughput sequencing using HiSeq 2000/1000 Illumina. After assessing the sequencing quality by determining the error rate, the alignment of 100 bp paired-end reads was accomplished using the Tophat alignment program that allows splitting of individual reads across splice junctions. Multiple stages of alignment using trimmed reads were merged, PCR duplicates removed, and alignment statistics collected to determine if samples met required aligned read cutoffs. Variants were called from the transcriptome using SAMtools and filtered appropriately using depth greater than 10 and a phred-like quality score of greater than 20. Expression calculations were performed using the Cufflinks package which calculates the transcript-level expression by assembling aligned reads into transcripts, comparing them to known transcript annotations, and estimating their abundances, taking into account library size, biases in library preparation, and the statistical uncertainty associated with assigning reads to a particular isoform. The expressed genes were identified using Partek algorithms and assigned to the cell pathways and programs using KEGG, Ingenuity Pathway Analysis, and Gene Ontology databases using David as the search tool. The RNA-Seq data have been deposited at the Gene Expression Omnibus (GEO) website. The data record code is GSE126726.
Mass spectrometry (LC-MS):
For the no labeling experiment, cells were treated with either vehicle (DMSO) or 500 nM IBR for 48 h and harvested for LC-MS. Metabolite extraction and LC-MS procedures are as described (7). For the 13C labeling experiments, cells were first treated with either vehicle or 500 nM IBR and incubated for 48 h, then incubated for additional 8 h in the new medium with glutamine substituted with 2 mM [U-13C5, U-15N2] glutamine. Vehicle or IBR were added to the medium before incubation. After incubation, cells were quenched by adding a 5-fold excess of cold PBS to the medium, centrifuged at 4°C and rinsed 3 times with cold PBS and frozen. For dynamic analysis, aliquots of cells at 20 min, 40 min, 60 min, 90 min, 2 h and 3 h after incubation were harvested in the same way. Metabolite extraction and LC-MS followed as described (7). The labeling experiments were performed with cells in T-25 flasks with 1 million/ml cell density and 5–10 ml of medium.
Magnetic resonance spectroscopy (MRS):
Cell culture, treatment and harvest were performed similarly to the procedure used for LC-MS except that a larger number of cells was used. 1H MRS employed cells grown in T-75 flasks, and 13C MRS employed cells grown in T-150 flasks. Metabolites extraction was performed using either a perchloric acid extraction method (8) or a methanol-water extraction technique. MRS experiments utilized a Varian 9.4 T/8.9 cm vertical bore NMR spectrometer. 1H MRS employed a pulse-acquire sequence (8). 13C MRS experiments utilized the proton-observed carbon-edited (POCE) spectroscopy sequence (9) without 13C decoupling (4) and the 13C pulse-acquire sequence with 1H-decoupling (4). The former sequence enabled determination of 13C enrichment ratios of lactate, alanine and glutamate, and the latter sequence enabled determining multiplet ratios of 13C metabolite peaks (4).
Extracellular flux measurement:
Cells were treated for 48 h with vehicle or 500 nM IBR, re-suspended in fresh medium at 1–2 million/ml cell density, added with vehicle or IBR, and incubated for 6–8 h. The glucose, lactate and glutamine concentrations of the medium were measured by using YSI 2700 Biochemistry Analyzer (YSI Incorporated, Yellow Springs, Ohio) to calculate cellular uptake or export rates of these metabolites during the last 6–8 h.
Metabolic network analysis:
A three-compartment (extracellular medium, cell cytoplasm and mitochondria) non-linear metabolic model was used to fit experimental 13C MRS and LC-MS data for determination of various metabolic fluxes and metabolic parameters. Bonded cumomer and fragmented cumomer/mass isotopomer analysis methods (3, 4) were employed. See Supplementary Data for details.
Statistics:
All experiments were performed in triplicate. The analyzed data were presented as either mean and standard error (mean±SEM) or mean and standard deviation (mean±SD), indicated at each figure legend. A two-tailed student’s t-test was used to calculate p-values. P-values less than 0.05 were considered statistically significant. The fluxes determined from the metabolic network analysis were presented as the fitted flux and standard deviation calculated by Monte-Carlo simulations.
RESULTS
Differential metabolic effects of IBR on MCL cells
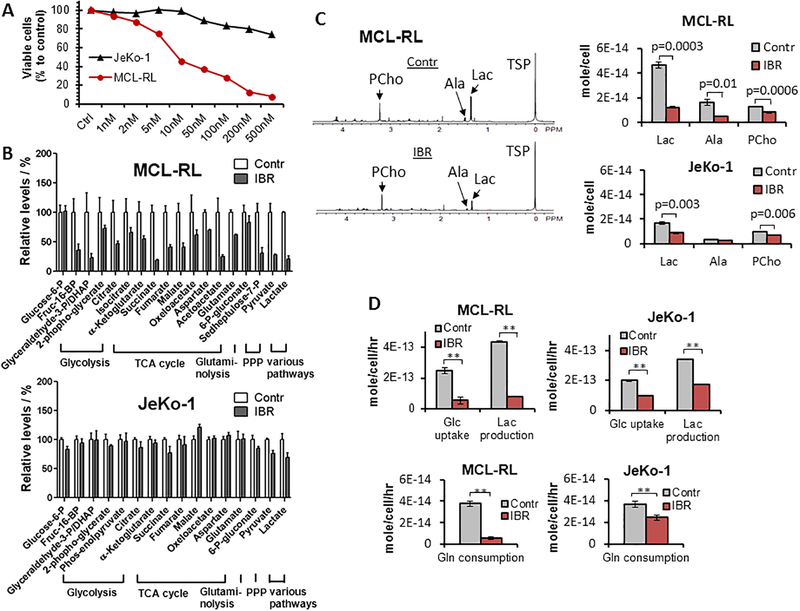
We first performed a basic metabolic assessment of two MCL lines, MCL-RL and JeKo-1, the former sensitive to IBR and the latter poorly responsive to IBR as determined by the viable cell count assay of the MCL cells after 72 h incubation in vehicle (DMSO) or various concentrations of IBR from 1 nM to 500 nM (Fig 1A). The LC-MS and 1H MRS experiments of the metabolites extracted from cells (Fig 1B and Fig 1C) and the extracellular flux measurements (i.e., import/export to/from the cell; Fig 1D) were conducted after 48 h incubation of the cells in vehicle or 500 nM IBR.
Figure 1. Effect of IBR on growth and metabolism of MCL cells.
(A) Impact of IBR on growth of the IBR-sensitive MCL-RL and IBR-poorly responsive JeKo-1 cell lines as determined by viable cell count. (B) Effect of IBR on intracellular concentration of key metabolites as measures by LC-MS. (C) Left; 1H MRS spectra of MCL-RL cells. The same number of cells (2.5×107 cells) was used for each spectrum. TSP, Trimethylsilylpropanoic acid; Lac, lactate; Ala, alanine; Pcho, phosphocholine. Right; intracellular metabolite concentrations of MCL-RL and JeKo-1 cells determined by 1H MRS. (D) Top; extracellular glucose and lactate fluxes. Bottom; extracellular glutamine fluxes after IBR treatment. **p<0.002. Data were presented as mean±SEM for Fig 1B while mean±SD for Fig 1C and 1D.
Figure 1B shows the LC-MS data of MCL-RL and JeKo-1 cells. The metabolites were grouped according to metabolic pathways: glycolysis, TCA cycle, glutaminolysis, and PPP with pyruvate and lactate categorized separately as the end products of various metabolic pathways (glycolysis and other pathways to be discussed later). In MCL-RL cells (Fig. 1B, upper panel), all glycolytic and TCA cycle metabolites decreased significantly in response to IBR except for glucose-6-P, which remained unchanged. In JeKo-1 cells (Fig. 1B, lower panel), all the metabolites in these pathways changed only minimally (<~10%). Of note, glutamate, a product of glutaminolysis, decreased by 40% in MCL-RL and remained unchanged in JeKo-1 cells. The changes of PPP metabolites were as follows: In MCL-RL cells, 6-P-gluconate did not significantly change, while sedoheptulose-7-P decreased by 70%; in JeKo-1 cells, 6-P-gluconate was unchanged and sedoheptulose-7-P was not detected. Finally, pyruvate and lactate decreased by 70% and 80%, respectively, in MCL-RL and by only 25% and 30%, respectively, in JeKo-1 cells. Overall, LC-MS analysis identified profound decreases in concentrations of key metabolites in response to IBR in IBR-sensitive MCL-RL but detected minimal or smaller changes in the IBR-poorly responsive Jeko-1 cells. Therefore, in addition to providing in-depth insight into metabolism of the malignant cells, these results indicate that changes in metabolite concentration reflect cell sensitivity to kinase inhibition and, hence, can serve as biomarkers of response to kinase inhibition.
Although LC-MS provides information about a large number of metabolites, it is an invasive technique requiring cell lysis. In search of a noninvasive method applicable to in vivo studies including the ones in cancer patients, we performed 1H MRS experiments. Three metabolites are most commonly detectable by in vivo 1H MRS of tumors: lactate, alanine and choline-containing compounds (10, 11). Thus, we focused on these metabolites. As shown in Fig. 1C, lactate decreased 73% in IBR-treated MCL-RL cells, while decreasing 48% in JeKo-1 cells. Noteworthy, alanine decreased 68% in MCL-RL after IBR, but no change occurred in JeKo-1 cells after this treatment. Phosphocholine decreased 36% in MCL-RL and 28% in JeKo-1 cells following IBR administration. These results indicate that lactate and alanine are the most promising biomarkers of response to IBR. Extension of the study to a larger panel of MCL cell lines and multiple time points after exposure of cells to IBR was performed and presented in a later section (Fig. 2).
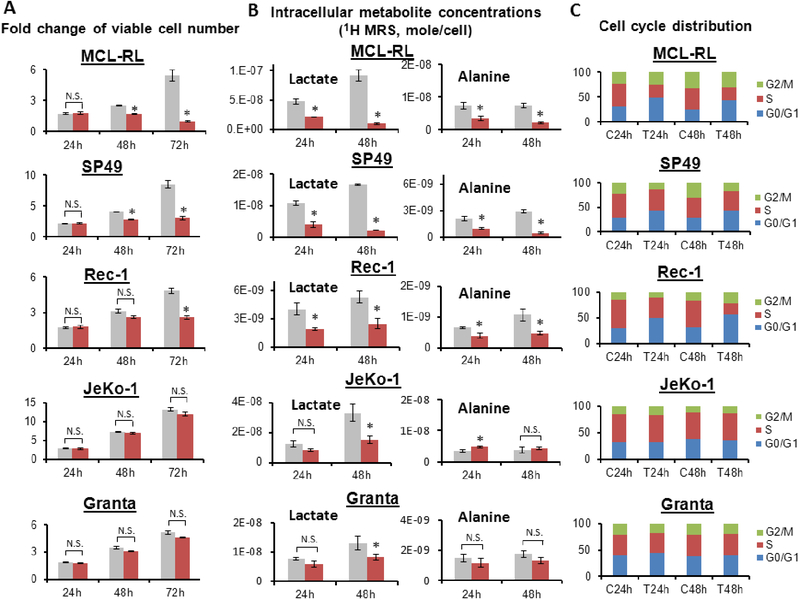
Figure 2. Kinetics of cell growth and metabolic changes upon IBR in various MCL cells.
(A) Viable cell number. (B) Intracellular 1H MRS metabolite concentrations. (C) Cell cycle distribution. *, p<0.05; N.S., statistically not significant. All data were presented as mean±SD. In Fig 2A and 2B, gray bar = control; red bar = IBR-treated.
Extracellular flux measurement also showed difference in IBR response between MCL-RL and JeKo-1 cells (Fig. 1D). In MCL-RL cells, glucose uptake and lactate secretion rates per cell decreased by 78±1% and 82±1% after IBR-treatment, respectively, compared to vehicle-treatment. In JeKo-1 cells, these rates decreased by 50±2% and 50±1%, respectively, after IBR. The glutamine uptake rate per cell decreased by 71±2% in MCL-RL after IBR and 41±3% in JeKo-1 cells.
IBR affects expression of metabolic genes
To determine if the differential cell-growth response of MCL cells to IBR (Fig. 1A) was associated with differential response of genes related to cell metabolism, we examined IBR-induced changes in the expression of metabolic genes in MCL-RL and JeKo-1 cells after 6 h incubation of the cells in either vehicle or 500 nM IBR (Table I). The study indicated that BTK inhibition had a broad suppressive effect on cell metabolism including glycolysis, pentose phosphate pathway, glutaminolysis, reductive carboxylation and lipid synthesis. While genes in some of the pathways were affected to a similar degree in both IBR-sensitive MCL-RL and IBR-poorly responsive JeKo-1 cell lines, others were suppressed predominantly, if not exclusively, in the MCL-RL cells, strongly suggesting that their products may contribute to differential drug response in these cells.
Table I.
IBR-induced changes in expression of selected metabolic genes in MCL cells
| Entrez ID | Gene Symbol | Enzyme/Protein Name | EC number | Fold Change MCL-RL | Fold Change JeKo-1 | Pathway/Products Affected |
|---|---|---|---|---|---|---|
| 92483 | LDHAL6B | NAD-L-lactate Dehydrogenase | EC 1.1.1.27 | −1.62 | * | |
| 22934 | RPIA | Ribose 5-phosphate isomerase A | EC 5.3.1.6 | −1.75 | 1.22 | Pentose phosphate |
| SLC38A5 | Solute Carrier Family 38 Member 5 | −1.75 | −1.02 | |||
| FASN | Fatty acid synthase | EC 2.3.1.85 | −1.70 | −1.25 | ||
| ALDH5A1 | EC 1.2.1.24 | 1.54 | 1.40 | |||
| 1543 | CYP1A1 | Monooxygenase | EC1.14.14.1 | −3.08 | −1.33 | |
| 9388 | LIPG | Triacylglycerol lipase | EC 3.1.1.3 | −3.04 | −1.46 |
data not shown due to very low gene expression of this LDH isoform in JeKo-1 cells
new enzymatic link for glutamine to glutamate inter-conversion through linked cytosolic catalysis of glutamine hydrolysis by phosphoribosyl pyrophosphate amidotransferase (PPAT)
Hexokinase 2 (HK2), the gatekeeper of the glycolytic pathway, was suppressed similarly in MCL-RL and JeKo-1 cells by IBR treatment. In contrast, expression of RPIA that encodes ribose 5-phosphate isomerase A in the pentose phosphate pathway (PPP) involved in nucleic acids synthesis was suppressed only in MCL-RL cells. Similarly, expressions of CAD, PPAT, GLS, MYC, SLC1A5 and SLC38A5 involved in glutaminolysis were suppressed in MCL-RL cells but remained essentially unchanged in JeKo-1 cells after IBR treatment. Expression of the MYC gene showed the largest change (over 5-fold) among the genes related to glutaminolysis. Expression of IDH1, which regulates conversion of α-KG to isocitrate in the reductive carboxylation pathway in the cytosol, decreased in MCL-RL but not in JeKo-1 cells. Finally, ATP citrate lyase (ACLY) and fatty acid synthase (FASN) as well as other genes related to steroid, cholesterol and lipid metabolism were more down-regulated in MCL-RL than in JeKo-1. These results indicate potential suitability of monitoring metabolic biomarkers of cell sensitivity to IBR, and strongly suggest a direct relationship between changes in metabolic status and inhibition of cell growth in response to this agent.
Kinetics of cell number, 1H MRS metabolite concentration and cell cycle distribution
We extended the cell growth and 1H MRS studies of Fig. 1A and 1C to elucidate the causal relationship between metabolic changes and cell growth. Several MCL cell lines with diverse sensitivity to IBR were used for generalization of the conclusion. The fold-changes of viable cell number at 24 h, 48 h and 72 h of vehicle or 500 nM IBR treatment were measured in MCL-RL, SP49, Rec-1, JeKo-1 and Granta-519 cell lines (Fig. 2A). The first three lines had viable cell number reduction by 50–80% at 72 h of IBR compared to vehicle control. We classified them as IBR-sensitive lines. Among them, MCL-RL was the most sensitive, SP49 was the next followed by Rec-1. JeKo-1 and Granta-519 lines had no significant difference in cell number between control and treatment at 72 h incubation with IBR. We classified these two lines as IBR-poorly responsive. There was no cell number difference between control and treatment at 24 h of IBR treatment in all five lines. At 48 h of IBR, MCL-RL and SP49 showed statistically significant difference in cell number between control and treatment while Rec-1, JeKo-1 and Granta-519 did not exhibit statistical difference in cell number between control and treatment.
Intracellular lactate and alanine concentrations at 24 h and 48 h exposure of the cells to either vehicle or 500 nM IBR were measured by 1H MRS experiments on the five cell lines (Fig. 2B). In three IBR-sensitive lines (MCL-RL, SP49 and Rec-1), both lactate and alanine decreased significantly (>50%) at 24 h of IBR treatment compared to control. The trend deepened at 48 h of IBR treatment in all three lines. In two IBR-poorly responsive lines (JeKo-1 and Granta-519), there was no statistically significant difference of lactate concentration between IBR treatment and vehicle treatment at 24 h. Alanine exhibited slight increase rather than decrease in the IBR-treated JeKo-1 cells compared to control at 24 h, while it did not exhibit difference between control and treatment in Granta-519 cells at that time. However, at 48 h both JeKo-1 and Granta-519 cells exhibited statistically significant decrease of lactate in IBR-treated cells compared to control while alanine remained unchanged. Combining the observations from Fig 2A and 2B, changes of both lactate and alanine concentrations in IBR-sensitive cells observed at 24 h of IBR treatment preceded cell number changes in these cells that were only observable at 48 h and later. Change of lactate with no change of alanine observed at 48 h IBR in both of the IBR-poorly responsive cell lines suggests that not lactate but alanine is the metabolite that correlates best with growth of these cell lines.
We analyzed the cell cycle distribution of the above MCL cell lines at 24 h and 48 h exposure to 500 nM IBR (Fig. 2C). The cell cycle arrest at G0/G1 phase was observed in all three IBR-sensitive lines (MCL-RL, SP49 and Rec-1) at both 24 h and 48 h of IBR treatment while no significant changes were observed in the IBR-poorly responsive lines. The timing of cell cycle arrest in IBR-sensitive lines matched timing of metabolic changes in these lines which occurred at 24 h (Fig. 2B) and preceded cell number changes observed at 48 h (Fig. 2A). No cell cycle change in IBR-poorly responsive lines (JeKo-1 and Granta-519) at 48 h of IBR matched no change in alanine but it did not correlate with lactate change in these lines. This is consistent with the finding of the preceding paragraph. It indicates that in the IBR-poorly responsive cell lines lactate is not a reliable biomarker of response but alanine is and suggests that metabolism related to alanine production plays an important role in the IBR resistance mechanism of these cells.
13C MRS metabolomics with [1,2-13C2]- and [1,6-13C2]-glucose tracer
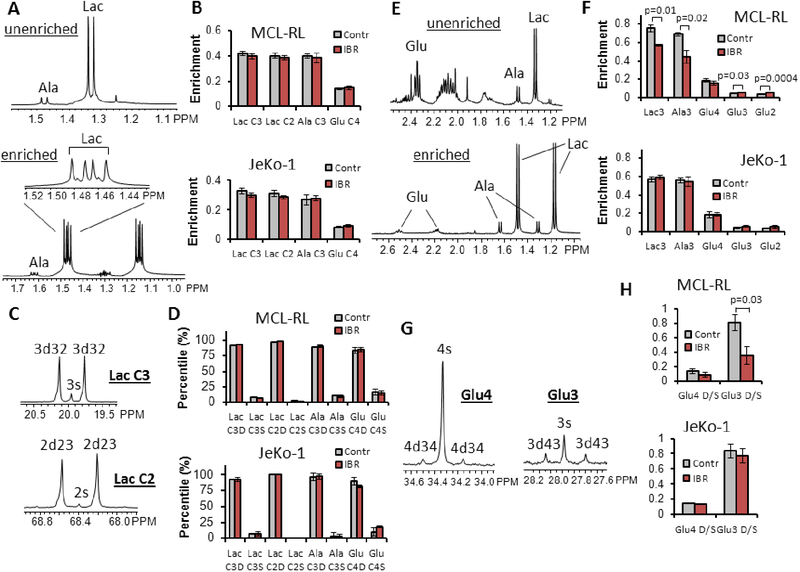
To study effects of IBR treatment on various metabolic pathways of the IBR-sensitive MCL-RL and IBR-poorly responsive JeKo-1 cells in detail, we performed 13C labeling experiments. [1,2-13C2] glucose labeling was employed to examine fluxes in the pentose phosphate pathway. Fig. 3A shows the spectral region around the lactate and alanine resonances obtained from an indirect 1H[13C] MRS experiment (i.e., POCE or proton observe carbon edited) of MCL-RL cells after 8 h incubation in the labeled medium; the upper panel is from 13C-unenriched molecules and the lower from the 13C-enriched molecules. Fig. 3B shows the calculated enrichment of lactate C3, lactate C2, alanine C3 and glutamate C4 from MCL-RL and JeKo-1 cells. Fig. 3C shows singlet and doublet peaks of lactate C3 and lactate C2 resonances of MCL-RL cells from a direct 13C MRS experiment (i.e., carbon observe, proton decouple). Fig. 3D shows percentages of doublet and singlet for lactate C3, lactate C2, alanine C3 and glutamate C4. These data were analyzed by the bonded cumomer analysis (4) to calculate fluxes in the PPP pathway (f2 through f5, Table II; see also a metabolic network diagram in the Supplementary Fig. S1). The fluxes were first calculated as relative fluxes normalized to glycolysis, then converted to absolute fluxes (Table II). We used the glucose uptake rate as a measure of glycolysis flux. The oxidative flux of the PPP, Fppp (G-6-P → Ru-5-P), and non-oxidative fluxes of the PPP, i.e., transketolase 1 and 2 and transaldolase fluxes Ftk1 (Ri-5-P + X-5-P →GA-3-P + S-7-P), Ftk2 (X-5-P + E-4-P → F-6-P + Ga-3-P) and Fta (GA-3-P + S-7-P → F-6-P + E-4-P), were determined. These fluxes decreased by 84% in MCL-RL and by 47% in JeKo-1 cells after 48 h IBR treatment, indicating that IBR affected the PPP pathway to a much larger extent in MCL-RL compared to JeKo-1 cells. The ratio of fluxes through the irreversible oxidative branch PPP to overall glycolysis was ~10% in both cell lines and remained relatively unchanged after IBR treatment, indicating that IBR has impacted the PPP and glycolysis similarly. The calculated Fppp corresponds to the NADPH production flux via the oxidative PPP branch.
Figure 3. 13C MRS metabolomics with [1,2-13C2]- and [1,6-13C2]-glucose tracer.
13C labeling experiments were performed by incubating MCL cells either in the [1,2-13C2] glucose containing medium (A-D) or [1,6-13C2] glucose containing medium (E-H) for 8 h, and the steady state 13C MRS data of MCL-RL and JeKo-1 cells were obtained. (A) Indirect 1H[13C] MRS spectra of vehicle-treated MCL-RL cells. Among the six peaks of the spectrum, the two small peaks are from lactate molecules labeled only at the C3 position (3s) while the four large peaks are from lactate molecules labeled at both C3 and C2 positions (3d32) (29). (B) 13C enrichment of each metabolite with vehicle or IBR treatment. (C) Direct 13C MRS spectra of vehicle-treated MCL-RL cells. (D) The doublet to singlet ratios of C3- and C2-lactate, C3-alanine and C4-glutamate of vehicle- or IBR-treated cells. (E) Proton Observe Carbon Edited (POCE) MRS spectra of MCL-RL cells. Top: Unlabeled metabolite spectrum. Bottom: Labeled metabolite spectrum. Enrichment ratios of glutamate C3 and C2 positions were calculated by comparing signal intensities of glutamate C3, C2 positions with the signal at C4 position in the direct 13C MRS spectrum. (F) 13C enrichment ratios of intracellular metabolites from MCL-RL and JeKo-1 cells with or without IBR treatment. The enrichment of each metabolite was calculated from (labeled metabolite area) / (unlabeled + labeled metabolite areas) of the POCE spectra. To determine enrichment of Glu3 and Glu2, relative intensities of Glu3, Glu2 and Glu4 from direct 13C MRS and correction of nuclear Overhauser enhancement (NOE) factors was used. (G) A 1H-decoupled 13C MRS spectrum of MCL-RL cells. Left: Glutamate C4 position peaks. 4s, C4 singlet; 4d34, C3–4 doublet. Right: Glutamate C3 position peaks. 3s, C3 singlet; 3d43, C4–3 doublet. (H) Doublet to singlet ratios of Glu4 and Glu3 peaks of MCL-RL and JeKo-1 cells with vehicle and IBR treatment. All data were presented as mean±SD.
Table II.
Metabolic fluxes determined from labeling experiments
| Flux | MCL-RL control | MCL-RL IBR | JeKo-1 control | JeKo-1 IBR | |
|---|---|---|---|---|---|
| f1 | Fmrg. metabolic rate of glucose. Glucose → G-6-P | 246±19 | 55±4 | 199±5 | 98±4 |
| f2* | Fppp. 6-P-G → Ru-5-P | 31.6±12.9 | 5.0±2.0 | 15.4±6.3 | 8.1±3.3 |
| f3* | Ftk1. Ri-5-P + X-5-P → GA-3-P + S-7-P | 10.5±4.3 | 1.7±0.7 | 5.2±2.1 | 2.7±1.1 |
| f4* | Ftk2. X-5-P + E-4-P → F-6-P + Ga-3-P | 10.5±4.3 | 1.7±0.7 | 5.2±2.1 | 2.7±1.1 |
| f5* | Fta. GA-3-P + S-7-P → F-6-P + E-4-P | 10.5±4.3 | 1.7±0.7 | 5.2±2.1 | 2.7±1.1 |
| f12** | Fpdh. pyruvate → acetyl-CoA | 31.5±6.0 | 12.9±2.4 | 19.4±3.7 | 15.8±3.0 |
| f13** | Fcs. acetyl-CoA → citrate | 31.5±6.0 | 12.9±2.4 | 19.4±3.7 | 15.8±3.0 |
| f16** | Fgls. glutaminei → glutamate | 17.0±2.9 | 4.6±0.8 | 5.9±1.0 | 5.8±1.0 |
| f12*** | Fpdh. pyruvate → acetyl-CoA | 33.3±5.0 | 17.5±2.7 | 40.6±6.1 | 31.1±5.1 |
| f13*** | Fcs. acetyl-CoA → citrate | 33.3±5.0 | 17.5±2.7 | 40.6±6.1 | 31.1±5.1 |
| f14*** | Fidh3. citrate → isocitrate (mitochondrial) | 6.4±0.9 | 4.7±0.8 | 4.4±0.8 | 3.5±0.5 |
| f16*** | Fgls. glutaminemito → glutamatemito | 27.0±5.0 | 12.7±2.5 | 36.2±6.8 | 27.6±5.0 |
| f22+f23*** | Frc. reductive carboxylation. αKG → isocitrate (cytosolic plus mitochondrial) | 4.3±0.7 | 3.1±0.4 | 5.5±0.8 | 3.6±0.6 |
| f30*** | Fkgdh. α-KG →succinate/SucCoA | 33.3±5.0 | 17.5±2.9 | 40.6±5.4 | 31.1±4.7 |
| f31*** | Fsdh. succinate/SucCoA → fumarate | 33.3±5.0 | 17.5±2.9 | 40.6±5.4 | 31.1±4.7 |
| f32*** | Ffuma. fumarate → malate | 33.3±5.0 | 17.5±2.9 | 40.6±5.4 | 31.1±4.7 |
| f33*** | Fmdh. malate →OAA | 33.3±5.0 | 17.5±2.9 | 40.6±5.4 | 31.1±4.7 |
| *** | ATPcyto | 468±51 | 105±12 | 379±42 | 187±21 |
| *** | ATPmito | 485±68 | 254±36 | 590±83 | 452±63 |
| *** | ATPq | 270±35 | 128±17 | 362±47 | 277±36 |
| *** | %LIPdnQ | 14±3 | 20±4 | 13±3 | 12±3 |
| *** | WE | 12.8±2.5 | 4.4±0.9 | 8.3±1.8 | 5.4±1.1 |
| *** | %ANAq | 98±21 | 96±21 | 99±21 | 98±20 |
Determined from the [1,2-13C2] glucose labeling experiment. Unit = nmole/106 cell/h.
Determined from the [1,6-13C2] glucose labeling experiment; relative to Fidh3 flux in the TCA cycle arbitrarily set to 10.
Determined from the [U-13C5] glutamine labeling experiment. Unit = nmole/106 cell/h for fluxes and ATP production rates.
ATPcyto, cytosolic ATP production rate
ATPmito, mitochondrial ATP production rate
ATPq, ATP production rate from glutamine
%LIPdnQ, glutamine contribution (%) to de novo lipid synthesis
WE,
%ANAq, glutamine contribution (%) to total anaplerosis
[1,6-13C2] glucose labeling was used to investigate mitochondrial and related metabolism. Fig. 3E shows spectra from indirect 1H[13C] MRS experiments, and Fig. 3G shows spectra obtained from direct 13C MRS (i.e., carbon observe proton decouple) after incubation of MCL-RL cells in [1,6-13C2] glucose-containing medium for 8 h. Fig. 3F shows the steady state 13C enrichments calculated from indirect 1H[13C] MRS data. C3 lactate enrichment decreased by 25% after IBR treatment, C3 alanine decreased 35%, C3 glutamate increased 29%, and C2 glutamate increased 42% in MCL-RL cells; changes were statistically significant (p-values shown). There was no significant change in the enrichment of C4 glutamate of MCL-RL cells or, importantly, of any of the C3 metabolites in JeKo-1 cells. The doublet to singlet (D/S) ratio of glutamate C4 and C3 resonances (Fig. 3H) was calculated from direct 13C MRS spectra (Fig. 3G). The D/S ratio at the glutamate C3 position decreased by 50% in MCL-RL after IBR treatment while there was no change in JeKo-1 cells. The D/S ratio at glutamate C4 did not change significantly in both cell lines. The enrichment and D/S ratio data were used to derive various metabolic fluxes at isotopic steady state (f12, f13 and f16; see Table II) using the bonded cumomer formalism (see Supplementary Data). The fluxes were presented as relative values to the flux from isocitrate to α-KG in the TCA cycle (f14; see Supplementary Fig. S1); the reference flux was arbitrarily set to 10. After treatment, Fpdh (the flux from pyruvate to acetyl-CoA) and Fcs (the flux from acetyl-CoA to citrate) decreased 60% in MCL-RL while decreasing only 19% in JeKo-1 cells. Strikingly, Fgls (the glutaminolysis flux, glutamine → glutamate) decreased by 73% in MCL-RL cells after treatment, while there was no substantial change in JeKo-1 cells. These results indicate that IBR profoundly inhibited glutaminolysis and TCA cycle in the IBR-sensitive MCL-RL cells but not in the IBR–poorly responsive JeKo-1 cells.
13C LC-MS metabolomics with [U-13C5, U-15N2] glutamine tracer
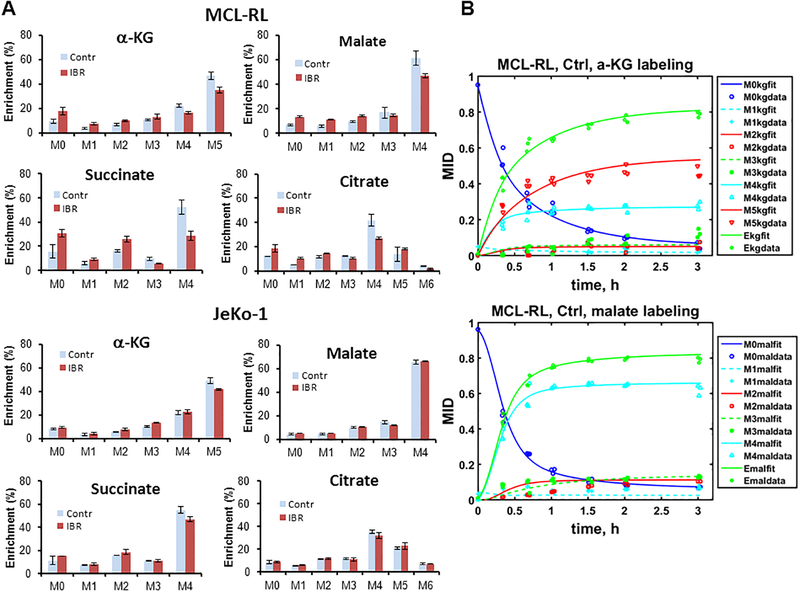
To study the effects of IBR on metabolic pathways in mitochondria in greater detail, we performed 13C LC-MS experiments. MCL-RL and JeKo-1 cells were treated with either vehicle or 500 nM IBR and cultured for 48 h, then incubated in the [U-13C5, U-15N2] glutamine-labeled medium. The labeled cells were analyzed 8 h after incubation (steady state labeling) or at multiple time points for a dynamic labeling analysis (See Materials and Methods.) Fig. 4A shows the steady state mass isotopologue distribution and labeling patterns of various metabolites from vehicle- and IBR-treated cells. After IBR treatment, significant changes were observed in M+5 α-ketoglutarate (α-KG), M+3 malate, M+4 succinate and M+4 citrate of MCL-RL cells but not of JeKo-1 cells. Glutaminolysis, TCA cycle, pyruvate cycling (both pyruvate-malate and pyruvate-citrate) and reductive carboxylation are involved in these labeling changes, which indicate alteration in these pathways after IBR treatment. The M+5 isotopologue of α-KG along with downstream M+4 isotopologues of malate, oxaloacetate and citrate are major biomarker isotopologues of glutaminolysis. Our data clearly indicate direct glutaminolysis contributions in both cell lines. MCL-RL cells showed a much greater change in these labeling patterns after IBR treatment than JeKo-1 cells. The presence of the tracer observed in the M+5 isotopologue of citrate and M+3 isotopologues of oxaloacetate and malate indicate carbon flow through reductive carboxylation. Labeling of M+3 isotopologues of oxaloacetate and malate also indicate pyruvate-citrate cycling along with labeling of M+3 and M+2 pyruvate and M+6 citrate. Fig. 4B shows time course labeling patterns of α-KG and malate in MCL-RL cells and fitting of the curve by fragmented cumomer/mass isotopomer analysis (See Supplementary Data.). Various metabolic fluxes and metabolic parameters were determined through this analysis and are presented in Table II.
Figure 4. 13C LC-MS metabolomics with [U-13C5, U-15N2] glutamine tracer.
(A) MCL-RL and JeKo-1 cells were labeled with [U-13C5, U-15N2] glutamine for 8 h. Mass isotopologues of α-ketoglutarate, malate, succinate and citrate are shown. Isotopologue information of other metabolites such as aspartate, glutamate, oxaloacetate, pyruvate and lactate was also obtained (data not shown). α-Ketoglutarate is initially labeled at M+5 by glutaminolysis and reductive carboxylation. Then it is labeled at all isotopologues of M+1 through M+5 after multiple scrambling in the TCA cycle and exchange of molecules between mitochondrion and cytosol. Succinate and malate are initially labeled as the M+4 isotopologue then labeled as other isotopologues as well after network scrambling. Citrate in the cytosol is initially labeled as the M+5 isotopologue after conversion from M+5 α-ketoglutarate. Citrate in the mitochondrion and cytosol are initially labeled as M+4 and M+5 isotopologues. The M+4 citrate is from the first turn of TCA cycle (clockwise), and the M+5 citrate is from the direct conversion from the α-ketoglutarate due to mitochondrial or cytosolic reductive carboxylation. Then the labeled carbons are distributed to other isotopologues after multiple turns of TCA cycle and scrambling through other pathways and exchange of the metabolites between mitochondrion and cytosol. For example, M+6 citrate in this figure suggests incorporation of labeled carbons from pyruvate produced in the multiple pathways including various pyruvate cycling routes (examples can be seen in Fig. S1). (B) MCL-RL and JeKo-1 cells were incubated in the [U-13C5, U-15N2] glutamine containing medium and collected at multiple time points for time course LC-MS analysis of the metabolite mass isotopologue fractions. The time course labeling of α-KG and malate along with fitting to the fragmented mass isotopomer model is presented. Ekg = (M1 + 2M2 + 3M3 + 4M4 + 5M5)/5; average enrichment of α-KG. Emal = (M1 + 2M2 + 3M3 + 4M4)/4; average enrichment of malate. Reversibility was assumed for all the reactions in the TCA cycle except for the reaction from oxaloacetate to citrate (Fig. S1). Data in Fig 4A were presented as mean±SD.
The dynamic labeling experiment and analysis enabled calculation of fluxes in absolute units (nmole/106 cell/h) rather than relative units (see Table II). Fpdh and Fcs decreased 50% in MCL-RL cells and 25% in JeKo-1 cells after IBR. In contrast, Fidh3 (citrate → isocitrate, mitochondria) decreased 25% in MCL-RL and 20% in JeKo-1 after IBR. Fidh3 was much smaller than the flux that entered into the TCA cycle (Fpdh; f12), being only 20% of Fpdh in MCL-RL and 12% of Fpdh in JeKo-1 cells (see Supplementary Fig. S1). This indicates a large transport flux from mitochondrial citrate to cytosolic citrate which is known to lead to pyruvate-citrate cycling and de novo lipid synthesis (5). This transport flux was reduced by 50% in MCL-RL after IBR treatment while diminishing by 25% in JeKo-1 cells. Fgls, the glutaminolysis flux (f16), decreased ~50% in MCL-RL cells after IBR treatment and ~20% in JeKo-1 cells. Reductive carboxylation flux (αKG → isocitrate, f22 + f23) decreased 30% in MCL-RL and 35% in JeKo-1 cells in response to IBR. The fluxes from α-KG to succinate/SucCoA (f30), from succinate/SucCoA to fumarate (f31), from fumarate to malate (f32), and from malate to oxaloacetate (f33) were very similar in both lines and all these fluxes decreased 50% in MCL-RL cells after IBR and by 25% in JeKo-1 cells. The flux through malic enzyme (ME2 and ME3) in mitochondria was near zero (not included in Table II), which indicates a very low production of pyruvate through malate-pyruvate cycling.
As alanine is produced through an aminotransferase reaction (GPT1 and GPT2; L-glutamate + pyruvate ⇌ α-ketoglutarate + L-alanine), the level of alanine will be affected by the availability of glutamate and pyruvate. Pyruvate is expected to be sufficient in both MCL-RL and JeKo-1 cells because a large amount of excess pyruvate is converted to lactate in both cell lines. Our flux analysis (Table II) indicates that glutaminolysis which leads to glutamate was profoundly suppressed in MCL-RL but not in JeKo-1 cells after IBR. This suggests differential glutamate production and resultant alanine levels after IBR treatment in MCL-RL and JeKo-1 cells, and is in full agreement with the differential IBR effect on alanine concentration in these cells, as shown in Fig. 1C and Fig. 2B.
Various metabolic parameters were also derived (Table II); see Supplementary Data for details. Cytosolic ATP production (ATPcyto) comprised ~50% of total ATP pool in MCL-RL and ~40% in JeKo-1 cells, which decreased by ~80% in MCL-RL cells and by ~50% in JeKo-1 in response to IBR. The overall mitochondrial ATP production (ATPmito) decreased after IBR by ~50% in MCL-RL cells and ~25% in JeKo-1 cells, with similar decreases seen in ATP derived from glutaminolysis. Glutaminolysis was responsible for ~60% of mitochondrial ATP synthesis in both MCL cell lines before drug treatment. Glutamine contribution to de novo lipid synthesis was quite modest at <15% in both MCL-RL and JeKo-1 cells before drug treatment. It increased to 20% after IBR in MCL-RL cells but not in JeKo-1 cells. The contribution of glutaminolysis to de novo lipid synthesis was negligible (not entered in Table II), which suggests that utilization of glutamine through reductive carboxylation is the key path of de novo lipid synthesis from glutamine in both of these cell lines.
The Warburg effect parameters (WE), defined as the ratio of the flux from pyruvate to lactate to the flux of pyruvate to mitochondria, were ~13 and ~8 for MCL-RL and JeKo-1 cells, respectively, with vehicle treatment. After IBR treatment, these parameters decreased to ~4 (~70% decrease) in MCL-RL and to ~5 (~38% decrease) in JeKo-1 cells. The high WE values of these cells at vehicle treatment indicate that most of the pyruvate derived from glycolysis was converted into lactate rather than entering into the TCA cycle. Reduction of this ratio after treatment implies that IBR primarily affected the pyruvate to lactate flux rather than the flux to mitochondria, in particular in MCL-RL cells. The glutamine contribution to total anaplerosis was nearly 100% in both cell lines, and was negligibly affected by IBR treatment.
Glutaminase inhibition suppressed growth of the IBR-poorly responsive cells
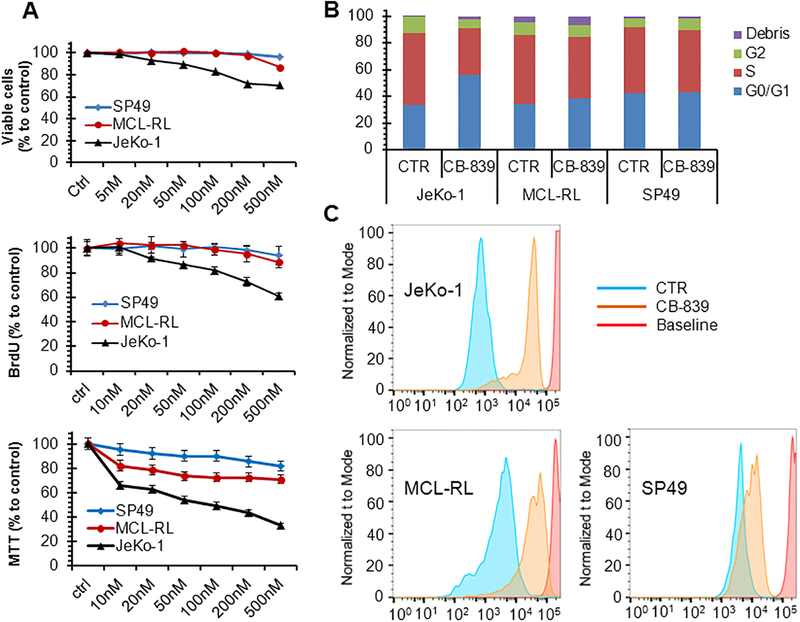
The importance of the glutamine metabolism in both MCL-RL and JeKo-1 cells and the limited changes in glutaminolysis in JeKo-1 cells compared to MCL-RL cells after IBR as indicated by multiple observations made in this study prompted us to examine the impact of a glutaminase inhibitor, CB-839, on various aspects of growth in these and other MCL cell lines. Fig. 5A shows the results after 72 h exposure of the cells to various concentrations of CB-839. Growth of JeKo-1 cells has been significantly impaired by the glutaminase inhibitor, as determined by decrease in cell number, diminished proliferation rate, and MTT conversion index. In contrast, MCL-RL and SP49 cells were much less affected. Similar results were obtained when the effect of CB-839 on cell cycle progression was measured (Fig. 5B). While JeKo-1 displayed a marked increase in the number of cells at the G0/G1 phase and a corresponding decrease in cells at the G2 phase in response to CB-839, these cell-cycle phase redistributions were much smaller in MCL-RL and essentially absent in SP49 cells. Finally, analysis of the cell division rate (Fig. 5C) also showed markedly diminished proliferative capacity in JeKo-1 cells in response to CB-839 that was much less pronounced in MCL-RL and SP49 cells. These results indicate that JeKo-1 cells are highly dependent on glutaminolysis for their growth and their resistance to IBR can be in a large part overcome by inhibition of this metabolic pathway.
Figure 5. Impact of glutaminolysis inhibition on growth of MCL cells.
(A) Growth inhibition of the depicted MCL cell lines in response to glutaminolysis inhibitor CB-839 as determined by viable cell count (top panel), BrdU uptake (middle panel), and MTT conversion index (bottom panel). (B) CB-839-mediated inhibition of cell cycle progression with cells exposed to the drug’s vehicle alone serving as control (CTR). (C) CB-839-mediated inhibition of cell division as determined by CSFE labeling pattern with cells exposed to the drug’s vehicle alone serving as control (CTR). Data were presented as mean±SEM.
DISCUSSION
While small-molecule kinase inhibitors continue to play an ever-increasing role in cancer research and therapy and have already become standards of care for a large number of malignancies, there are currently no methods to efficiently assess the activity of these inhibitors in vivo, including in cancer patients. By employing the BTK inhibitor IBR as an index inhibitor of kinase activity and MCL cells as a cancer model, we have demonstrated in this study that effective BTK inhibition profoundly affects cell metabolism and have identified metabolic biomarkers that detect response and resistance of MCL cells to IBR at an early stage. We have also shown that analysis of the expression of metabolic genes and 13C MRS and LC-MS based metabolomics permit comprehensive and detailed insight into metabolic pathway alterations upon IBR treatment in IBR-sensitive and IBR-poorly responsive MCL cells. While the BTK inhibitor potently inhibited most of the key metabolic pathways including glutaminolysis in the IBR-sensitive MCL-RL cells, it failed to do so in the IBR-poorly responsive JeKo-1 cells. This IBR-resistance could, however, be overcome in JeKo-1 by treating these cells with a glutaminase inhibitor CB-839.
While IBR has gained FDA approval and is widely used in therapy of MCL and other types of B-cell lymphoma (12), only a subset of the patients display response to this kinase inhibitor; the same is true for essentially all types of cancer treated with diverse kinase inhibitors. Despite considerable early hope, mutational genome analysis in MCL and many other malignancies failed to identify genetic biomarkers that predict response to IBR and other kinase inhibitors. Although diverse mutations have been identified in MCL, none has been linked to BTK activation (13–15). The MyD88 mutation which is highly predictive of IBR sensitivity, is frequently detected in lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia (16, 17), but is essentially absent in MCL. While resistance to IBR has been predicted based on the presence of mutations in the BTK and PLCγ genes (18, 19), this applies to only a fraction of IBR-resistant MCL patients. Furthermore, the mutations typically occur during therapy and require additional tissue biopsy to be detected. Moreover, resistance is not predicted on the basis of the ability of IBR to inhibit BTK autophosphorylation (20). The dominant mechanisms of IBR-resistance seem to emerge from MCL re-programing of cell signaling, enhanced or even enabled by lymphoma cell interactions with the microenvironment (21). These mechanisms, applicable to cancer in general, will likely prove difficult to evaluate in the clinical setting.
We have shown in this study that simultaneous decreases of both lactate and alanine are a strong metabolic indicator of response which was detectable as early as after 24 h exposure of three IBR-sensitive cell lines to the BTK inhibitor IBR while the effect on the cell number was observable only after 48 h exposure of these cell lines to IBR. The metabolic changes observed at 24 h after IBR treatment correlated with cell cycle changes at the same time. In the two IBR-poorly responsive cell lines, no significant metabolic changes were observable at 24 h post-exposure of these cells to IBR. At 48 h after exposure to IBR, substantial lactate changes were observed in both of the IBR-poorly responsive cell lines while alanine remained unchanged compared to vehicle control. This indicated that alanine is a promising marker of IBR resistance while lactate alone should not be used as a marker of response because no cell number or cell cycle changes were observable at 48 h post-IBR in these cell lines.
These findings carry considerable implications in the clinical imaging of MCL patients and maybe other types of cancer for monitoring the effects of kinase inhibitors. Current standard metabolic imaging tool for lymphoma patients, FDG-PET, has manifested its shortcoming in correctly monitoring response of tumor to kinase inhibitors. While FDG-PET images cancer by exploiting avid glucose uptake of cancer cells, interim FDG-PET results during the course of kinase inhibitor therapy did not correlate with the tumor volumetric response monitored at a later time (22, 23). The reason for this discrepancy is thought to be that even though the glycolytic pathway was suppressed by the kinase inhibitor, other active metabolic pathways may have provided sufficient energy and building blocks for survival and proliferation of the cancer cells (24). Our observation in the IBR-poorly responsive cells that lactate decreased even though cell number or cell cycle did not change compared to vehicle control exactly matches the observations in the clinical cancer patient setting with FDG-PET (22, 23). A unique attribute of 1H MRS is that it can simultaneously detect both lactate and alanine noninvasively from tumor animal models or tumor of patients in situ as we have demonstrated before (10, 25). Lactate mainly reflects glycolysis while alanine is responsive to glutaminolysis. Therefore, application of 1H MRS to MCL patients will have an important role for management of MCL treated with kinase inhibitors.
In this study, by using 13C MRS, 13C LC-MS and novel metabolic network analysis, we were able to determine many intracellular fluxes related to various metabolic pathways such as glycolysis, PPP, TCA cycle, glutaminolysis, reductive carboxylation, etc. From such fluxes, we were able to derive a number of critical metabolic parameters such as respective ATP productions from cytosol and from mitochondria, glutamine contribution to mitochondrial ATP production, glutamine and glutaminolysis contributions to de novo fatty acid synthesis, glutaminolysis contribution to total anaplerosis, the Warburg Effect parameter, etc. Such a wealth of information allowed us to understand the importance of glutaminolysis for energy production in the MCL cells. Minimal changes in glutaminolysis in JeKo-1 cells upon IBR treatment suggested that glutaminolysis provides a resistance mechanism to IBR in these cells, and led us to apply the glutaminase inhibitor CB-839 to effectively suppress growth of these cells. This ability of metabolic flux analysis to identify potentially effective alternative therapeutic approaches provides a remarkable opportunity for treatment protocol selection and adjustment of therapeutic strategies based on a comprehensive understanding of the metabolic characteristics of malignant cells.
The high oxidative PPP activity (e.g., up to 13% of glycolysis flux in MCL-RL at baseline) in the studied MCL cells indicates that PPP contributed to a large portion of NADPH production, the energetic source for key synthetic pathways including de novo lipogenesis. This high PPP activity may also account for the inhibition of reactive oxygen species (ROS) production in B-cells via glutathione recycling (26). Many investigators previously appreciated glutaminolysis contributions to cancer cell/tumor energy metabolism (26–28). Our study provided quantitative estimation of such contribution in MCL cells, i.e., over half of mitochondrial energy production from glutaminolysis.
In summary, our study provided a detailed metabolic understanding of MCL cells and their response to the BTK inhibitor IBR, suggested imaging biomarkers, lactate and alanine, to detect response and resistance to IBR in MCL, and proposed pathways to overcome IBR-resistance, foremost glutaminolysis, the major oxidative ATP producing pathway in these cells. From a broader perspective, our study and the experimental approaches used have far-reaching basic science and translational implications by paving the road to a better understanding of cancer cell signaling and metabolism cross-talk and molecular pathophysiology of MCL and cancer in general, which will impact management of patients with MCL and other malignancies.
Supplementary Material
Implications:
Our study demonstrates application of the advanced metabolomic/fluxomic techniques for comprehensive, precise and prompt evaluations of the effects of kinase inhibition in MCL cells and has strong translational implications by potentially permitting early evaluation of cancer patient response vs resistance to kinase inhibitors and on design of novel therapies for overcoming the resistance.
ACKNOWLEDGEMENT
This study was supported in part by NIH grants R01-CA172820 and R01-CA129544, Berman Family Fund and Daniel B. Allanoff Foundation.
List of Key Abbreviations
- BTK
Bruton’s tyrosine kinase
- IBR
BTK inhibitor ibrutinib
- MCL
mantle cell lymphoma
- LC-MS
liquid chromatography mass spectrometry
- MRS
magnetic resonance spectroscopy
- PPP
pentose phosphate pathway
- OxPhos
oxidative phosphorylation
- FDG-PET
fluorodeoxyglucose positron emission tomography
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Cheah CY, Seymour JF, and Wang ML Mantle Cell Lymphoma. Journal of Clinical Oncology, 34: 1256–1269, 2016. [DOI] [PubMed] [Google Scholar]
- 2.Wang ML, Rule S, Martin P, Goy A, Auer R, Kahl BS, Jurczak W, Advani RH, Romaguera JE, Williams ME, Barrientos JC, Chmielowska E, Radford J, Stilgenbauer S, Dreyling M, Jedrzejczak WW, Johnson P, Spurgeon SE, Li L, Zhang L, Newberry K, Ou ZS, Cheng N, Fang BL, McGreivy J, Clow F, Buggy JJ, Chang BY, Beaupre DM, Kunkel LA, and Blum KA Targeting BTK with Ibrutinib in Relapsed or Refractory Mantle-Cell Lymphoma. New England Journal of Medicine, 369: 507–516, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shestov AA, Lee SC, Nath K, Guo LL, Nelson DS, Roman JC, Leeper DB, Wasik MA, Blair IA, and Glickson JD C-13 MRS and LC-MS Flux Analysis of Tumor Intermediary Metabolism. Frontiers in Oncology, 6, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shestov AA, Mancuso A, Lee SC, Guo LL, Nelson DS, Roman JC, Henry PG, Leeper DB, Blair IA, and Glickson JD Bonded Cumomer Analysis of Human Melanoma Metabolism Monitored by C-13 NMR Spectroscopy of Perfused Tumor Cells. Journal of Biological Chemistry, 291: 5157–5171, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang L, Shestov AA, Swain P, Yang CD, Parker SJ, Wang QA, Terada LS, Adams ND, McCabe MT, Pietrak B, Schmidt S, Metallo CM, Dranka BP, Schwartz B, and DeBerardinis RJ Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature, 532: 255–258, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehrmohamadi M, Liu XJ, Shestov AA, and Locasale JW Characterization of the Usage of the Serine Metabolic Network in Human Cancer. Cell Reports, 9: 1507–1519, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo LL, Shestov AA, Worth AJ, Nath K, Nelson DS, Leeper DB, Glickson JD, and Blair IA Inhibition of Mitochondrial Complex II by the Anticancer Agent Lonidamine. Journal of Biological Chemistry, 291: 42–57, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SC, Marzec M, Liu XB, Wehrli S, Kantekure K, Ragunath PN, Nelson DS, Delikatny EJ, Glickson JD, and Wasik MA Decreased lactate concentration and glycolytic enzyme expression reflect inhibition of mTOR signal transduction pathway in B-cell lymphoma. Nmr in Biomedicine, 26: 106–114, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothman DL, Behar KL, Hetherington HP, Denhollander JA, Bendall MR, Petroff OAC, and Shulman RG H-1-Observe C-13-Decouple Spectroscopic Measurements of Lactate and Glutamate in the Rat-Brain Invivo. Proceedings of the National Academy of Sciences of the United States of America, 82: 1633–1637, 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SC, Delikatny EJ, Poptani H, Pickup S, and Glickson JD In vivo H-1 MRS of WSU-DLCL2 human non-Hodgkin’s lymphoma xenografts: response to rituximab and rituximab plus CHOP. Nmr in Biomedicine, 22: 259–265, 2009. [DOI] [PubMed] [Google Scholar]
- 11.Lee SC, Huang MQ, Nelson DS, Pickup S, Wehrli S, Adegbola O, Poptani H, Delikatny EJ, and Glickson JD In vivo MRS markers of response to CHOP chemotherapy in the WSU-DLCL2 human diffuse large B-cell lymphoma xenograft. Nmr in Biomedicine, 21: 723–733, 2008. [DOI] [PubMed] [Google Scholar]
- 12.Inamdar AA, Goy A, Ayoub NM, Attia C, Oton L, Taruvai V, Costales M, Lin YT, Pecora A, and Suh KS Mantle cell lymphoma in the era of precision medicine-diagnosis, biomarkers and therapeutic agents. Oncotarget, 7: 48692–48731, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bea S, Valdes-Mas R, Navarro A, Salaverria I, Martin-Garcia D, Jares P, Gine E, Pinyol M, Royo C, Nadeu F, Conde L, Juan M, Clot G, Vizan P, Di Croce L, Puente DA, Lopez-Guerra M, Moros A, Roue G, Aymerich M, Villamor N, Colomo L, Martinez A, Valera A, Martin-Subero JI, Amador V, Hernandez L, Rozman M, Enjuanes A, Forcada P, Muntanola A, Hartmann EM, Calasanz MJ, Rosenwald A, Ott G, Hernandez-Rivas JM, Klapper W, Siebert R, Wiestner A, Wilson WH, Colomer D, Lopez-Guillermo A, Lopez-Otin C, Puente XS, and Campo E Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America, 110: 18250–18255, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu CL, de Miranda NFCC, Chen LY, Wasik AM, Mansouri L, Jurczak W, Galazka K, Dlugosz-Danecka M, Machaczka M, Zhang HL, Peng RJ, Morin RD, Rosenquist R, Sander B, and Pan-Hammarstrom Q Genetic heterogeneity in primary and relapsed mantle cell lymphomas: Impact of recurrent CARD11 mutations. Oncotarget, 7: 38180–38190, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Jima D, Moffitt AB, Liu QQ, Czader M, Hsi ED, Fedoriw Y, Dunphy CH, Richards KL, Gill JI, Sun Z, Love C, Scotland P, Lock E, Levy S, Hsu DS, Dunson D, and Dave SS The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood, 123: 2988–2996, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treon SP, Xu L, and Hunter Z MYD88 Mutations and Response to Ibrutinib in Waldenstrom’s Macroglobulinemia. New England Journal of Medicine, 373: 584–586, 2015. [DOI] [PubMed] [Google Scholar]
- 17.Treon SP, Xu L, Yang G, Zhou YS, Liu X, Cao Y, Sheehy P, Manning RJ, Patterson CJ, Tripsas C, Arcaini L, Pinkus GS, Rodig SJ, Sohani AR, Harris NL, Laramie JM, Skifter DA, Lincoln SE, and Hunter ZR MYD88 L265P Somatic Mutation in Waldenstrom’s Macroglobulinemia. New England Journal of Medicine, 367: 826–833, 2012. [DOI] [PubMed] [Google Scholar]
- 18.Chiron D, Di Liberto M, Martin P, Huang XG, Sharman J, Blecua P, Mathew S, Vijay P, Eng K, Ali S, Johnson A, Chang B, Ely S, Elemento O, Mason CE, Leonard JP, and Chen-Kiang S Cell-Cycle Reprogramming for PI3K Inhibition Overrides a Relapse-Specific C481S BTK Mutation Revealed by Longitudinal Functional Genomics in Mantle Cell Lymphoma. Cancer Discovery, 4: 1022–1035, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woyach JA, Furman RR, Liu TM, Ozer HG, Zapatka M, Ruppert AS, Xue L, Li DHH, Steggerda SM, Versele M, Dave SS, Zhang J, Yilmaz AS, Jaglowski SM, Blum KA, Lozanski A, Lozanski G, James DF, Barrientos JC, Lichter P, Stilgenbauer S, Buggy JJ, Chang BY, Johnson AJ, and Byrd JC Resistance Mechanisms for the Bruton’s Tyrosine Kinase Inhibitor Ibrutinib. New England Journal of Medicine, 370: 2286–2294, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma J, Lu P, Guo AL, Cheng SH, Zong HL, Martin P, Coleman M, and Wang YL Characterization of ibrutinib-sensitive and -resistant mantle lymphoma cells. British Journal of Haematology, 166: 849–861, 2014. [DOI] [PubMed] [Google Scholar]
- 21.Zhao XH, Lwin T, Silva A, Shah B, Tao JC, Fang B, Zhang L, Fu K, Bi CF, Li JN, Jiang HJ, Meads MB, Jacobson T, Silva M, Distler A, Darville L, Zhang L, Han Y, Rebatchouk D, Di Liberto M, Moscinski LC, Koomen JM, Dalton WS, Shain KH, Wang M, Sotomayor E, and Tao JG Unification of de novo and acquired ibrutinib resistance in mantle cell lymphoma. Nature Communications, 8, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma WW, Jacene H, Song D, Vilardell F, Messersmith WA, Laheru D, Wahl R, Endres C, Jimeno A, Pomper MG, and Hidalgo M [F-18]Fluorodeoxyglucose Positron Emission Tomography Correlates With Akt Pathway Activity but Is Not Predictive of Clinical Outcome During mTOR Inhibitor Therapy. Journal of Clinical Oncology, 27: 2697–2704, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McArthur GA, Puzanov I, Amaravadi R, Ribas A, Chapman P, Kim KB, Sosman JA, Lee RJ, Nolop K, Flaherty KT, Callahan J, and Hicks RJ Marked, Homogeneous, and Early [F-18]Fluorodeoxyglucose-Positron Emission Tomography Responses to Vemurafenib in BRAF-Mutant Advanced Melanoma. Journal of Clinical Oncology, 30: 1628–1634, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deberardinis RJ, Sayed N, Ditsworth D, and Thompson CB Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev, 18: 54–61, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellon EA, Lee SC, Pickup S, Kim S, Goldstein SC, Floyd TF, Poptani H, Delikatny EJ, Reddy R, and Glickson JD Detection of Lactate With a Hadamard Slice Selected, Selective Multiple Quantum Coherence, Chemical Shift Imaging Sequence (HDMD-SelMQC-CSI) on a Clinical MRI Scanner: Application to Tumors and Muscle Ischemia. Magnetic Resonance in Medicine, 62: 1404–1413, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang HX, Zimmerman LJ, Liebler DC, Slebos RJC, Lorkiewicz PK, Higashi RM, Fan TWM, and Dang CV Glucose-Independent Glutamine Metabolism via TCA Cycling for Proliferation and Survival in B Cells. Cell Metabolism, 15: 110–121, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altman BJ, Stine ZE, and Dang CV From Krebs to clinic: glutamine metabolism to cancer therapy. Nature Reviews Cancer, 16: 619–634, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, and Thompson CB Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences of the United States of America, 104: 19345–19350, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd SG, Zeng HD, Wang PP, and Chatham JC Lactate isotopomer analysis by H-1 NMR spectroscopy: Consideration of long-range nuclear spin-spin interactions. Magnetic Resonance in Medicine, 51: 1279–1282, 2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.