Abstract
Erigeron annuus (L.) Pers. (annual, daisy or tall fleabane) is an annual herb native to North America but has been introduced and naturalized worldwide. In this study, its complete chloroplast (cp) genome was assembled from Illumina sequencing reads. The cp genome is 153,177 bp long with an A + T-biased base composition. It encodes a panel of 113 genes, including 80 protein-coding, 29 tRNA, and four rRNA genes. Nineteen genes are completely or partially duplicated, while 17 genes possess one or two introns. Phylogenetic analysis suggested E. annuus is mostly closely related to Erigeron canadensis L. and that the two genera Conyza and Erigeron are not mutually monophyletic.
Keywords: Erigeron annuus (L.) Pers, exotic plant high-throughput sequencing (HTS), invasive plant chloroplast genome
Erigeron annuus (L.) Pers., commonly known as annual, daisy or tall fleabane, is an annual herb species belonging to the family Asteraceae of the order Asterales (Chen et al. 2011). It is native to North America, but has been introduced and naturalized worldwide (Wu et al. 2004; Chen et al. 2011; Vuković 2015; Zimmermann et al. 2015; Seipel et al. 2016; Das et al. 2017; Shhagapsoev et al. 2018; Song et al. 2018; Sennikov and Kurtto 2019). To date, most studies of E. annuus have been focused upon its histochemistry (Kim et al. 2005; Yoo et al. 2008; Jeong et al. 2011; Kim and Choi 2015; Kim et al. 2018), invasive biology (Song et al. 2018; Sennikov and Kurtto 2019; Wei et al. 2020) and population genetics (Stratton 1991, 1992; Edwards et al. 2006; Tunaitienė et al. 2017). Little is known about its genomics (incl. chloroplast/cp genomics). In this study, we assembled the first complete cp genome for this invasive weed using high-throughput sequencing technology, and investigated its phylogenetic placement within the tribe Astereae (Asterales: Asteraceae).
Fresh leaves were sampled from an individual of E. annuus in Xunyangba Village, Ningshan County, Shaanxi Province, China (33°32'38′'N, 108°32'22′'E), and were used to isolate the total genomic DNA with the DNeasy Plant Mini Kit (Qiagen, CA, USA). A voucher specimen was held at herbarium of the College of Forestry, Northwest A&F University (https://en.nwsuaf.edu.cn/; Juanjuan Li, Email: wutong761014@163.com) under the accession number EANNU-2019-07-22. Library construction and Illumina PE150 sequencing (average insert size: 350 bp) were performed by Beijing Novogene Technology Co., Ltd. (Beijing, China) following the protocol of the manufacturer (Illumina, CA, USA). In all, 22.13 M of paired-end reads were retrieved, and were used to assemble the cp genome of E. annuus with the software NOVOPlasty v4.3.1 (Dierckxsens et al. 2017). The cp genome of Sonchus webbii Sch. Bip. (GenBank accession: MK033508) (Cho et al. 2019) was inputted as the initial seed sequence. Annotation of the cp genome was done in Geneious R11 (Biomatters Ltd., Auckland, New Zealand) by aligning with those of closely related taxa.
The cp genome of E. annuus is 153,177 bp in length, and comprises a pair of inverted repeat (IR) regions (25,019 bp) which were separated by a large single-copy (LSC) region (84,727 bp) and a small single-copy (SSC) region (18,412 bp). The base composition is asymmetric with the A + T contents of the entire genome, the SSC, LSC and IR regions being 63.0%, 69.1%, 65.1% and 57.0%, respectively. The cp genome encodes a panel of 113 genes [incl. 80 protein-coding (PCG), 29 tRNA and four rRNA genes]. In all, 17 genes are completely duplicated, including seven PCGs (ndhB, rpl2, rpl23, rps7, rps12, ycf2, and ycf15), six tRNAs (trnA-UGC, trnE-UUC, trnI-CAU, trnL-CAA, trnR-ACG, and trnV-GAC) and all four rRNAs (rrn4.5, rrn5, rrn16 and rrn23). Two PCGs (rps19 and ycf1) are partially duplicated. In addition, a single intron is presented in ten PCGs (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, and rps16) and five tRNA genes (trnA-UGC, trnE-UUC, trnG-ACC, trnK-UUU, and trnL-UAA), and a couple of introns are presented in two PCGs (clpP and ycf3).
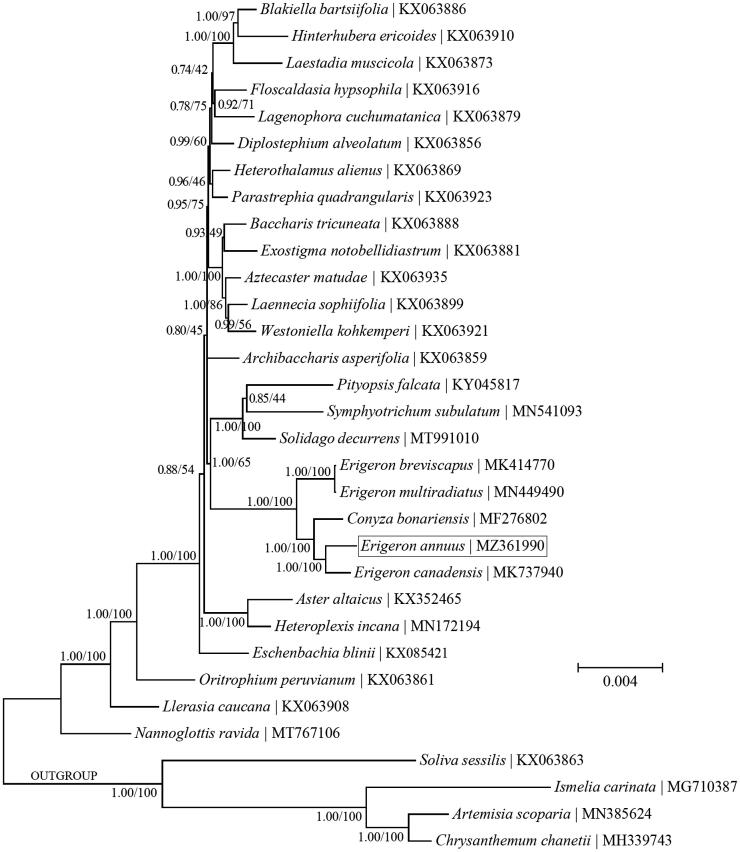
Phylogenetic analyses were conducted based on the coding sequences of cp PCGs to ascertain the phylogenetic placement of E. annuus within the tribe Astereae (Figure 1). Both Bayesian inference (BI) and maximum-likelihood (ML) methods were implemented using the software MrBayes v3.1.1 (Ronquist and Huelsenbeck 2003) as in TOPALi v2.5 (Milne et al. 2009) and the software MEGA11 (Tamura et al. 2021). The key parameters for BI analysis were set as follows: <Nucleotide substitution model: GTR±G ± I; Runs: 4; Generations: 200,000; Sample Freq.: 10; Burnin: 30%>, and those for ML analysis were set as follows: < Nucleotide substitution model: GTR±G ± I; Number of bootstrap replications: 500>. The nucleotide substitution models for BI and ML methods were inferred with the ‘Model Selection (MrBayes)’ function in TOPALi v2.5 and the ‘Find Best DNA/Protein Models (ML)’ function in MEGA11, respectively. The outgroup taxa used in this study are four species within the tribe Anthemideae (Asterales: Asteraceae), including Artemisia capillaris Thunb. (KU736963), Chrysanthemum boreale Makino (MG913594), Ismelia carinata (Schousb.) Sch. Bip. (MG710387) and Soliva sessilis Ruiz & Pav. (KX063863). The BI and ML analyses recovered the identical topology. E. annuus was found to be mostly closely related to the congener E. canadensis. In addition, the two genera Conyza and Erigeron were not mutually monophyletic but together formed a monophyletic clade. This finding appears to support the inclusion of Conyza within the genus Erigeron (Noyes 2000). However, further studies based on more extensive sampling are necessary to resolve this controversy.
Figure 1.
A combined phylogeny of the tribe Astereae based on the Bayesian inference (BI) and maximum-likelihood (ML) analysis of chloroplast protein-coding genes. The BI and ML analyses recovered the identical topology. The support values next to the nodes were Bayesian posterior probabilities according to the BI analysis (first value) and bootstrap percentages of 500 pseudoreplicates according to the ML analysis (second value). Four species within the tribe Anthemideae were included as the outgroup taxa.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession number MZ361990. The associated BioProject, SRA and Bio-Sample numbers are PRJNA736110, SRR14763427 and SAMN19609949, respectively.
References
- Chen Y, Chen Y, Brouillet L, Semple JC.. 2011. Tribe Astereae. Flora Chin. 20-21:545–652. [Google Scholar]
- Cho M-S, Yang JY, Yang T-J, Kim S-C.. 2019. Evolutionary comparison of the chloroplast genome in the woody Sonchus alliance (Asteraceae) on the Canary Islands. Genes. 10(3):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Rawat D, Shrivastava N, Kumar A, Sinha B, Singh P, Dash SS.. 2017. A contribution to the flora of Great Himalayan National Park, Himachal Pradesh, India. Nelumbo. 59(1):33–43. [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PJ, Frey D, Bailer H, Baltisberger M.. 2006. Genetic variation in native and invasive populations of Erigeron annuus as assessed by RAPD markers. Int J Plant Sci. 167(1):93–101. [Google Scholar]
- Jeong C-H, Jeong HR, Choi GN, Kim D-O, Lee U, Heo HJ.. 2011. Neuroprotective and anti-oxidant effects of caffeic acid isolated from Erigeron annuus leaf. Chin Med. 6(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Jung SJ, Chung I-S, Lee Y-H, Kim D-K, Kim S-H, Kwon B-M, Jeong T-S, Park M-H, Seoung N-S, et al. 2005. Ergosterol peroxide from flowers of Erigeron annuus L. as an anti-atherosclerosis agent. Arch Pharm Res. 28(5):541–545. [DOI] [PubMed] [Google Scholar]
- Kim D-Y, Won KJ, Hwang DI, Park SM, Kim HB, Lee HM.. 2018. Chemical composition of essential oil from Erigeron annuus (L.) Pers. flower and its effect on migration and proliferation in keratinocyte. J Essent Oil-Bear Plants. 21(5):1146–1154. [Google Scholar]
- Kim Y-H, Choi K-S.. 2015. Effect of the Erigeron annuus in vitro antioxidant properties and extract on serum lipid in mice. Korean J Food and Nutr. 28(3):387–395. [Google Scholar]
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F.. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25(1):126–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes RD. 2000. Biogeographical and evolutionary insights on Erigeron and allies (Asteraceae) from ITS sequence data. Pl Syst Evol. 220(1-2):93–114. [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. [DOI] [PubMed] [Google Scholar]
- Seipel T, Alexander JM, Edwards PJ, Kueffer C.. 2016. Range limits and population dynamics of non-native plants spreading along elevation gradients. Perspect Plant Ecol Evol Syst. 20:46–55. [Google Scholar]
- Sennikov AN, Kurtto A.. 2019. The taxonomy and invasion status assessment of Erigeron annuus s.l. (Asteraceae) in East Fennoscandia. Memo Soc Fauna Flora Fenn. 95:40–59. [Google Scholar]
- Shhagapsoev SH, Chadaeva VA, Tsepkova NL, Shhagapsoeva KA.. 2018. Materials for the blacklist of the central Caucasus flora (for the Kabardino-Balkar Republic). Russ J Biol Invasions. 9(4):384–391. [Google Scholar]
- Song U, Son D, Kang C, Lee EJ, Lee K, Park JS.. 2018. Mowing: a cause of invasion, but also a potential solution for management of the invasive, alien plant species Erigeron annuus (L.) Pers. J Environ Manage. 223:530–536. [DOI] [PubMed] [Google Scholar]
- Stratton DA. 1991. Life history variation within populations of an asexual plant, Erigeron annuus (Asteraceae). Am J Bot. 78(5):723–728. [Google Scholar]
- Stratton DA. 1992. Life-cycle components of selection in Erigeron annuus: II. Genetic variation. Evolution. 46(1):107–120. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S.. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunaitienė V, Patamsytė J, Naugžemys D, Kleizaitė V, Čėsnienė T, Rančelis V, Žvingila D.. 2017. Genetic and allelopathic differences between populations of daisy fleabane Erigeron annuus (L.) Pers. (Asteraceae) from disturbed and stable habitats. Biochem Syst Ecol. 70:294–303. [Google Scholar]
- Vuković N. 2015. Ecogeography of the invasive flora of Croatia. Zagreb (Croatia): University of Zagreb. [Google Scholar]
- Wei M, Wang S, Xiao H-G, Wu B-D, Jiang K, Wang C-Y.. 2020. Co-invasion of daisy fleabane and Canada goldenrod pose synergistic impacts on soil bacterial richness. J Cent South Univ. 27(6):1790–1801. [Google Scholar]
- Wu S-H, Hsieh C-F, Rejmánek M.. 2004. Catalogue of the naturalized flora of Taiwan. Taiwania. 49(1):16–31. [Google Scholar]
- Yoo NH, Jang DS, Yoo JL, Lee YM, Kim YS, Cho J-H, Kim JS.. 2008. Erigeroflavanone, a flavanone derivative from the flowers of Erigeron annuus with protein glycation and aldose reductase inhibitory activity. J Nat Prod. 71(4):713–715. [DOI] [PubMed] [Google Scholar]
- Zimmermann H, Loos J, von Wehrden H, Fischer J.. 2015. Aliens in Transylvania: risk maps of invasive alien plant species in Central Romania. NB. 24:55–65. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession number MZ361990. The associated BioProject, SRA and Bio-Sample numbers are PRJNA736110, SRR14763427 and SAMN19609949, respectively.