Abstract
We designed several new primers and modified previously described species- and type-specific primers targeting the Mycoplasma pneumoniae P1 adhesin gene. Optimized thermal profiles allowed one-step or nested PCR to be completed in less than 1 h. In 10 patients with pneumonia, M. pneumoniae type 1 was identified in 3 and type 2 in 7.
Mycoplasma pneumoniae is a causative agent of tracheobronchitis and primary atypical pneumonia in children (5) and one of the commonest causes of community-acquired pneumonia in adults, ranging in severity from mild to life-threatening (13, 25). M. pneumoniae also causes extrapulmonary complications and infections, involving the heart (16), central nervous system (3), and genitourinary tract (18).
Rapid confirmation of the diagnosis is important for clinical and epidemiological reasons (14), but culture is slow, technically demanding, and relatively insensitive (4, 18). Currently, diagnosis of M. pneumoniae infection usually relies on serology, which also has major limitations (4, 19). Recently, PCR has been accepted as a valuable method for diagnosis of M. pneumoniae infections (1, 4).
M. pneumoniae can be separated into two types on the basis of divergence of P1 gene sequence (21, 22, 23). PCR is the simplest and the most practical typing method (10). The relationship between these types and virulence, epidemic activity, reinfection, cross-protection or clinical severity, and complications of M. pneumoniae infection requires further investigation (10, 20).
In this study, we aimed to develop a faster and more practical PCR for detection and typing of M. pneumoniae.
Reference strains used were M. pneumoniae M129 (ATCC 29342), M. pneumoniae FH (ATCC 15531), and Mycoplasma genitalium (ATCC 33530), which were purchased directly from the American Type Culture Collection. Nasopharyngeal aspirates were obtained from 176 children admitted to the hospital with pneumonia during a 12-month period (April 1998 to March 1999). Specimens were stored at −70°C. Before processing, they were thawed and separated into two portions, each about 200 to 500 μl. One portion (the “original” specimen) was used directly for DNA extraction and PCR examination, and the other was inoculated into 2 ml of SP4 broth (“broth-enhanced” specimen). After 24 h of incubation at 37°C, 500 μl of broth was used for DNA extraction. DNA was prepared as previously described (6, 11, 26).
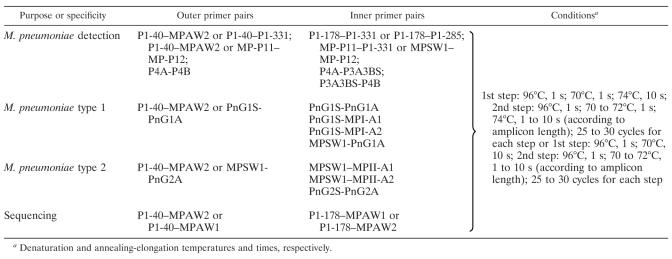
The following reference sequences were used to improve and design new primers: P1 adhesin gene sequences of M. pneumoniae type 1 (M129 [ATCC 29342]) (GenBank accession numbers M18639, M21519, and M20916); P1 adhesin gene sequences of M. pneumoniae type 2 (TW 7-5 and FH [ATCC 15531]) sequences as previously described (21); and the MgPa adhesin gene sequence of M. genitalium (G-37 [ATCC 33530]) (GenBank accession number M31431). The oligonucleotide primers used are listed in Table 1. The primer pairs used for initial screening for M. pneumoniae in clinical specimens and for optimizing thermal profiles in single-step and nested PCRs for detection and typing of M. pneumoniae, as well as the conditions used for PCRs, are shown in Tables 2 and 3.
TABLE 1.
Oligonucleotide primers used in this study
| Primer name | Tm (°C) (a/b)a | Sequenceb | Reference |
|---|---|---|---|
| P1-40 | 76.1/78 | 39TTGGc ATT CTC ATC CTC ACC GCC ACC63 | 25 |
| P1-178 | 77.0/78 | 177 CAA TGC CAT CAA CCC GCG CTT AAC C201 | 25 |
| P1-285 | 76.8/76 | 285GTT GTC GCG CAC TAA GGC CCA CG263 | 25 |
| P1-331 | 78.3/80 | 330CGT GGT TTG TTG ACT GCC ACT GCC G306 | 25 |
| MP-P11 | 77.0/78 | 176CCAA TGC CAT CAA CCC GCG CTT AAC200 | 2 |
| MP-P12 | 73.6/82 | 648CCC CCT/(G)dT/(C)TG CAA CTG CTC ATA GTA CACC621 | 2 |
| P4A | 78.9/88 | 3944/3968CTTC AGG CTC AGG TCA ATC TGG CGT GGA3971/3995 | 18 |
| P3A3BS | 73.4/84 | 4138/4162GAT GTT GAT GGT ATT GTA CGCACCCCAC4165/4189 | 18 |
| P3A3BA | 73.5/84 | 4162/4186GGGTGCG TAC AAT ACC ATC AAC ATC GTC4135/4159 | 18 |
| P4B | 76.7/84 | 4296/4320TACC GGA TCA AAC AGA TCG GTG ACT GGG T4268/4292 | 18 |
| MPSW1 (MP-1) | 78.1/90 | 459CGAC CAA ACC GGG CAG ATC ACC TTT AACC487 | 2 |
| MPSW2 | 53.0/66 | 526GTG AAA CGA GGT CAA AAA CAA GG550 | This study |
| MPAW | 55.0/64 | 1143/1158ATA AGG CGC ATC GTAC AGA A TC1122/1137 | This study |
| MPAW1 | 79.3/88 | 1149/1164GCG CGC ATA AGG CGC ATC GTAC AGA A TC1122/1137 | This study |
| MPAW2 | 83.4/90 | 1175/1190TCA ACG CGG TCA ATG GCG GTA CGG TTG C1148/1163 | This study |
| PnG1S | 76.2/88 | 96ACACTTCA CAA GTA CCA CGA CGC TCA A125 | 12 |
| PnG1A | 78.7/92 | 984TTGAGTTGG ACG GAC TGA CCC GAC TCC TC955 | 12 |
| PnG2S | 75.6/90 | 667GGGAGTT CGT CAG GCT CAG ACA GCA CTA A695 | 12 |
| PnG2A | 83.5/86 | 931CCACCTGT TCG GTG CCT TGG TCA CCG GAG903 | 12 |
| MPI-A1 | 72.2/84 | 700CGG TGG TGG AAG TAT TTT GAC CAC TCT C673 | This study |
| MPI-A2 | 75.1/86 | 702CCC GGT GGT GGA AGT ATT TTG ACC ACT C675 | This study |
| MPII-A1 | 73.6/86 | 701GTT TGG TTA GTG CTG TCT GAG CCT GAC G674 | This study |
| MPII-A2 | 73.5/86 | 704CCT GTT TGG TTA GTG CTG TCT GAG CCT G677 | This study |
The two primer Tm values are those provided by the primer synthesizer (Sigma-Aldrich) (a) and those calculated by the formula Tm = [4 × numbers of (G+C)] + [2 × numbers of (A+T)] (b).
Boldface numbers represent the numbered base positions at which primer sequences start and finish (numbering from the start codon of the P1 adhesin gene).
Underlined sequences show bases added to modify previously published primers and/or probes.
Letters in parentheses indicate alternative nucleotides in type 2.
TABLE 2.
Primer pairsa and conditions used for one-step PCRs for the detection and typing of M. pneumoniae
| Purpose or specificity | Primer pairs | Conditionsb |
|---|---|---|
| Screening for M. pneumoniae | P4A-P4B (unmodified) | 95°C, 60 s; 55°C, 60 s; 72°C, 60 s; 35 cycles |
| MP-P11–MP-P12 (unmodified) | ||
| M. pneumoniae detection | P1-40–P1-331 | 96°C, 1 s; 70°C, 1 s; 74°C, 1 to 10 s (according to amplicon length); 40 cycles |
| P1-178–P1-285 | ||
| MP-P11–MP-P12 | ||
| MP-P11–P1-331 | ||
| MPSW1–MP-P12 | ||
| P4A-P4B | ||
| P4A-P3A3BA | ||
| P3A3BS-P4B | ||
| M. pneumoniae type 1 | PnG1S-PnG1A | |
| PnG1S–MPI-A1 | ||
| PnG1S–MPI-A2 | ||
| MPSW1-PNG1A | ||
| M. pneumoniae type 2 | PnG2S-PnG2A | |
| MPSW1–MPII-A1 | ||
| MPSW1–MPII-A2 |
Primers are as shown in Table 1 (i.e., with modification, if applicable), unless otherwise stated.
Denaturation, annealing, and elongation temperatures and times, respectively.
TABLE 3.
Primer pairs and conditions used for nested PCRs for the detection and typing of M. pneumoniae
Denaturation and annealing-elongation temperatures and times, respectively.
The 25-μl amplification reaction mixtures were used as previously described (11). For nested PCR, 1 to 5 μl of the first-step PCR product was used as the template in the second-step PCR reaction systems. All PCRs were performed in a Perkin-Elmer thermocycler 9600. Twelve microliters of PCR products was analyzed by electrophoresis on 2% agarose gels, which were stained with 0.5-μg/ml ethidium bromide. If all controls were satisfactory, a visible band of the appropriate size on UV translumination was accepted as evidence of the presence of M. pneumoniae DNA.
Unmodified primer pairs P4A-P4B (18) and MP-P11–MP-P12 (2) were used in parallel to screen all original and broth-enhanced specimens for M. pneumoniae. The thermal profiles used were as previously described (2, 18). M. pneumoniae DNA was detected in 10 of 176 (6%) specimens from patients with pneumonia by using this single-round PCR—both specimen types were positive in six cases, while original or broth-enhanced specimens alone were positive in two cases each. Results were the same with both sets of detection primers for all but one specimen, which was positive only with primers P4A-P4B.
Nested PCR, including the use of type-specific inner primers, showed that three specimens contained M. pneumoniae type 1, while seven specimens contained type 2 P1 gene fragments. Amplicons (P1-40/MPAW2 or P1-40/MPAW1)—containing highly heterologous regions (partial Rep MP4) of the P1 adhesin gene—from all M. pneumoniae-positive specimens were sequenced to confirm the PCR typing results and the specificity of new type-specific primer pairs. Sequences of all amplicons were identical with those of the corresponding M. pneumoniae P1 adhesin gene sequences in GenBank (M129, type 1) or published previously (TW 7-5 and FH, type 2) (21).
It has been suggested that the times commonly used in PCR cycles are often unnecessarily long, which may reduce the specificity and sensitivity of reactions (8, 27). Rapid-cycle PCR can improve product specificity significantly (9). Newer types of thermocyclers have been used successfully for rapid-cycle PCR, with total reaction times between 90 s and 20 min (7, 12, 17, 24). However, because conventional heat-block thermocyclers are still most commonly used, our aim was to develop a faster PCR cycle that could be used with this type of equipment (e.g., Perkin-Elmer thermocycler 9600). To allow shorter ramp and/or incubation times and increase the specificity, we modified the most commonly used primer pairs targeting the P1 adhesin gene to increase their melting temperatures (Tm) (≥72°C), so that high annealing temperatures (≥70°C) could be used.
After clinical specimens containing M. pneumoniae (in one or both portions) had been identified by PCR as described above, both portions of those 10 specimens were used to optimize thermal profiles for the modified and new primers. The methods were adjusted to achieve sensitivities at least as high as those obtained by using unmodified primer pairs P4A-P4B (18) and MP-P11–MP-P12 (2), i.e., to give positive results in all known M. pneumoniae-positive specimens. This was achieved using denaturation and annealing and elongation at times and temperatures of 1 s at 96°C, 1 s at 70°C, and 1 to 10 s (depending on the amplicon length) at 70 to 74°C, respectively, in a Perkin-Elmer thermocycler 9600 (Tables 2 and 3).
With use of modified and new primers with optimized thermal profiles, the one-step PCR could be finished within 40 min and the nested PCR within 1 h (15). Thus, the whole procedure—DNA preparation (<2 h), detection of M. pneumoniae in clinical specimens (1 h), typing of M. pneumoniae for positive specimens (1 h), and electrophoresis (<2 h for both detection and typing)—could be completed in less than one working day. We believe that this is the fastest M. pneumoniae detection and typing method reported so far. In future, more rapid DNA preparation and automated amplicon detection systems (instead of traditional electrophoresis) will allow PCR detection and typing time to be further reduced.
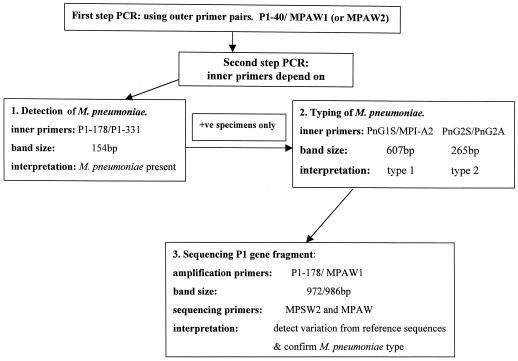
An algorithm was developed for clinical use of a rapid, nested PCR for detection and typing of M. pneumoniae (Fig. 1). We used a single outer primer pair (either P1-40–MPAW2 or P1-40–MPAW1) for the first round, followed by species- and type-specific primers for the second round, as required. The choice of primers was based on their sensitivity, band clarity, and suitable amplicon size. Our typing results showed that contrary to results in several other countries (20), type 2 was more commonly implicated than type 1 in a group of Australian children with pneumonia.
FIG. 1.
Algorithm for detection, typing, and sequencing of M. pneumoniae using nested PCR.
Acknowledgments
We thank Mark Wheeler for assistance with sequencing, Melanie Wong and Peter McIntyre for referring specimens and providing clinical data, Gregory James for helpful advice, and Zhengfang Ma for technical assistance and support.
REFERENCES
- 1.Abele-Horn M, Busch U, Nitschko H, Jacobs E, Bax R, Pfaff F, Schaffer B, Heesemann J. Molecular approaches to diagnosis of pulmonary diseases due to Mycoplasma pneumoniae. J Clin Microbiol. 1998;36:548–551. doi: 10.1128/jcm.36.2.548-551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Barbeyrac B, Bernet-Poggi C, Febrer F, Renaudin H, Dupon M, Bebear C. Detection of Mycoplasma pneumoniae and Mycoplasma genitalium in clinical samples by polymerase chain reaction. Clin Infect Dis. 1993;17(Suppl. 1):S83–S89. doi: 10.1093/clinids/17.supplement_1.s83. [DOI] [PubMed] [Google Scholar]
- 3.Dionisio D, Valassina M, Mata S, Rossetti R, Vivarelli A, Esperti F C, Benvenuti M, Catalani C, Uberti M. Encephalitis caused directly by Mycoplasma pneumoniae. Scand J Infect Dis. 1999;31:506–509. doi: 10.1080/00365549950164067. [DOI] [PubMed] [Google Scholar]
- 4.Dorigo-Zetsma J W, Zaat S A, Wertheim-van Dillen P M, Spanjaard L, Rijntjes J, van Waveren G, Jensen J S, Angulo A F, Dankert J. Comparison of PCR, culture, and serological tests for diagnosis of Mycoplasma pneumoniae respiratory tract infection in children. J Clin Microbiol. 1999;37:14–17. doi: 10.1128/jcm.37.1.14-17.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.File T M, Jr, Tan J S, Plouffe J F. The role of atypical pathogens: Mycoplasma pneumoniae, Chlamydia pneumoniae, and Legionella pneumophila in respiratory infection. Infect Dis Clin N Am. 1998;12:569–592. doi: 10.1016/s0891-5520(05)70199-9. [DOI] [PubMed] [Google Scholar]
- 6.Fink C G, Read S J, Sillis M. Direct sample polymerase chain reaction for the detection of Mycoplasma pneumoniae: a simple system for clinical application. Br J Biomed Sci. 1995;52:9–13. [PubMed] [Google Scholar]
- 7.Friedman N A, Meldrum D R. Capillary tube resistive thermal cycling. Anal Chem. 1998;70:2997–3002. doi: 10.1021/ac971303n. [DOI] [PubMed] [Google Scholar]
- 8.Gustafson C E, Alm R A, Trust T J. Effect of heat denaturation of target DNA on the PCR amplification. Gene. 1993;123:241–244. doi: 10.1016/0378-1119(93)90130-u. [DOI] [PubMed] [Google Scholar]
- 9.Harris S, Jones D B. Optimisation of the polymerase chain reaction. Br J Biomed Sci. 1997;54:166–173. [PubMed] [Google Scholar]
- 10.Jacobs E, Vonski M, Oberle K, Opitz O, Pietsch K. Are outbreaks and sporadic respiratory infections by Mycoplasma pneumoniae due to two distinct subtypes? Eur J Clin Microbiol Infect Dis. 1996;15:38–44. doi: 10.1007/BF01586183. [DOI] [PubMed] [Google Scholar]
- 11.Kong F, Ma Z, James G, Gordon S, Gilbert G L. Species identification and subtyping of Ureaplasma parvum and Ureaplasma urealyticum using PCR-based assays. J Clin Microbiol. 2000;38:1175–1179. doi: 10.1128/jcm.38.3.1175-1179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopp M U, Mello A J, Manz A. Chemical amplification: continuous-flow PCR on a chip. Science. 1998;280:1046–1048. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- 13.Lieberman D, Schlaeffer F, Lieberman D, Horowitz S, Horovitz O, Porath A. Mycoplasma pneumoniae community-acquired pneumonia: a review of 101 hospitalized adult patients. Respiration. 1996;63:261–266. doi: 10.1159/000196557. [DOI] [PubMed] [Google Scholar]
- 14.Lieberman D. Atypical pathogen pneumonia. Curr Opin Pulm Med. 1997;3:111–115. doi: 10.1097/00063198-199703000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Linz U, Delling U, Rubsamen-Waigmann H. Systematic studies on parameters influencing the performance of the polymerase chain reaction. J Clin Chem Clin Biochem. 1990;28:5–13. [PubMed] [Google Scholar]
- 16.Meseguer M A, Perez-Molina J A, Fernandez-Bustamante J, Gomez R, Martos I, Quero M C. Mycoplasma pneumoniae pericarditis and cardiac tamponade in a ten-year-old girl. Pediatr Infect Dis J. 1996;15:829–831. doi: 10.1097/00006454-199609000-00019. [DOI] [PubMed] [Google Scholar]
- 17.Oda R P, Strausbauch M A, Huhmer A F, Borson N, Jurrens S R, Craighead J, Wettstein P J, Eckloff B, Kline B, Landers J P. Infrared-mediated thermocycling for ultrafast polymerase chain reaction amplification of DNA. Anal Chem. 1998;70:4361–4368. doi: 10.1021/ac980452i. [DOI] [PubMed] [Google Scholar]
- 18.Sharma S, Brousseau R, Kasatiya S. Detection and confirmation of Mycoplasma pneumoniae in urogenital specimens by PCR. J Clin Microbiol. 1998;36:277–280. doi: 10.1128/jcm.36.1.277-280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sillis M. The limitations of IgM assays in the serological diagnosis of Mycoplasma pneumoniae infections. J Med Microbiol. 1990;33:253–258. doi: 10.1099/00222615-33-4-253. [DOI] [PubMed] [Google Scholar]
- 20.Su C J, Dallo S F, Baseman J B. Molecular distinctions among clinical isolates of Mycoplasma pneumoniae. J Clin Microbiol. 1990;28:1538–1540. doi: 10.1128/jcm.28.7.1538-1540.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su C J, Chavoya A, Dallo S F, Baseman J B. Sequence divergency of the cytadhesin gene of Mycoplasma pneumoniae. Infect Immun. 1990;58:2669–2674. doi: 10.1128/iai.58.8.2669-2674.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su C J, Dallo S F, Alderman H, Baseman J B. Distinctions in DNA and protein profiles among clinical isolates of Mycoplasma pneumoniae. J Gen Microbiol. 1991;137:2727–2732. doi: 10.1099/00221287-137-12-2727. [DOI] [PubMed] [Google Scholar]
- 23.Su C J, Dallo S F, Chavoya A, Baseman J B. Possible origin of sequence divergence in the P1 cytadhesin gene of Mycoplasma pneumoniae. Infect Immun. 1993;61:816–822. doi: 10.1128/iai.61.3.816-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swerdlow H, Dew-Jager K, Gesteland R F. Rapid cycle sequencing in an air thermal cycler. BioTechniques. 1993;15:512–519. [PubMed] [Google Scholar]
- 25.Talkington D F, Thacker W L, Keller D W, Jensen J S. Diagnosis of Mycoplasma pneumoniae infection in autopsy and open-lung biopsy tissues by nested PCR. J Clin Microbiol. 1998;36:1151–1153. doi: 10.1128/jcm.36.4.1151-1153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tjhie J H, van Kuppeveld F J, Roosendaal R, Melchers W J, Gordijn R, MacLaren D M, Walboomers J M, Meijer C J, van den Brule A J. Direct PCR enables detection of Mycoplasma pneumoniae in patients with respiratory tract infections. J Clin Microbiol. 1994;32:11–16. doi: 10.1128/jcm.32.1.11-16.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wittwer C T, Garling D J. Rapid cycle DNA amplification: time and temperature optimization. BioTechniques. 1991;10:76–83. [PubMed] [Google Scholar]