Summary
Background
Mathematical modelling may aid in understanding the complex interactions between injury and immune response in critical illness.
Methods
We utilize a system biology model of COVID-19 to analyze the effect of altering baseline patient characteristics on the outcome of immunomodulatory therapies. We create example parameter sets meant to mimic diverse patient types. For each patient type, we define the optimal treatment, identify biologic programs responsible for clinical responses, and predict biomarkers of those programs.
Findings
Model states representing older and hyperinflamed patients respond better to immunomodulation than those representing obese and diabetic patients. The disparate clinical responses are driven by distinct biologic programs. Optimal treatment initiation time is determined by neutrophil recruitment, systemic cytokine expression, systemic microthrombosis and the renin-angiotensin system (RAS) in older patients, and by RAS, systemic microthrombosis and trans IL6 signalling for hyperinflamed patients. For older and hyperinflamed patients, IL6 modulating therapy is predicted to be optimal when initiated very early (<4th day of infection) and broad immunosuppression therapy (corticosteroids) is predicted to be optimally initiated later in the disease (7th – 9th day of infection). We show that markers of biologic programs identified by the model correspond to clinically identified markers of disease severity.
Interpretation
We demonstrate that modelling of COVID-19 pathobiology can suggest biomarkers that predict optimal response to a given immunomodulatory treatment. Mathematical modelling thus constitutes a novel adjunct to predictive enrichment and may aid in the reduction of heterogeneity in critical care trials.
Funding
C.V. received a Marie Skłodowska Curie Actions Individual Fellowship (MSCA-IF-GF-2020-101028945). R.K.J.'s research is supported by R01-CA208205, and U01-CA 224348, R35-CA197743 and grants from the National Foundation for Cancer Research, Jane's Trust Foundation, Advanced Medical Research Foundation and Harvard Ludwig Cancer Center. No funder had a role in production or approval of this manuscript.
Keywords: Systems biology, modelling, COVID-19, heterogeneity of treatment effect, clustering, principal component analysis, non-negative matrix factorization, NMF, treatment
Research in context.
Evidence before this study
Acute critical care syndromes are highly dynamic and multifactorial. Clinical trials in critical care may lump together patients at different stages of disease or with qualitatively different pathophysiology. This makes it challenging to assign a precise biological meaning to a given clinical observation.
Added value of this study
Here, we use mathematical modelling to demonstrate distinct biological pathways underlying differential response of COVID-19 to immunomodulatory therapies and outline an approach to prospective prediction of biomarkers of optimal treatment.
Implications of all the available evidence
Systems biology approaches could enable investigators to enrich a clinical trial with patients most likely to respond by using predicted biomarkers in inclusion criteria. Such ‘predictive enrichment’ based on prospectively predicted biomarkers is a potential strategy for speeding development of therapy.
Alt-text: Unlabelled box
Introduction
Major critical care diagnoses such as the Acute Respiratory Distress Syndrome (ARDS) are clinically defined1. However, broad clinical definitions result in the application of the same diagnosis to a population of patients who may be heterogeneous in terms of individual characteristics, aetiology of illness, severity of illness, pathophysiology and outcome.2,3 Even in incidences where initial aetiology is uniform, as in ARDS in the setting of COVID-19, persistent heterogeneity results in a broad spectrum of disease4 and response to therapy.5,6 Significant efforts7, 8, 9 have been directed towards reducing heterogeneity by defining ARDS subtypes, which might be expected to respond similarly to treatment. Retrospective analysis of clinical trial data may be used to define subsets based on markers which are statistically likely to be associated with common mechanisms - even if the details of these underlying mechanisms are not known. This effort has resulted in notable success.5,6,10, 11, 12 Applications of statistical inference of this sort, however, have several limitations. Retrospective analyses are limited to markers which have been reported in existing trials. Moreover, it is not always possible to know if identified markers reflect reproducible features of disease pathophysiology or are influenced by time of observation, timing of treatment, and/or confounded by other aspects of patient heterogeneity. Finally, given the complex and non-linear interactions between injury and immune response that characterizes critical illness such as ARDS, it may be difficult to relate biomarkers to underlying cellular mechanism.
Improved precision requires more accurate determination of the association between patient characteristics, biomarkers, and underlying biology. This may be achieved over the course of multiple trials. However, the number of possible associations/hypotheses is large, and the trial process is time consuming and expensive. In the physical sciences, mathematical models13 are widely employed to guide the interpretation and design of experiments. Mathematical modelling has a rich history in medicine as well,14, 15, 16, 17, 18 providing key insights into the pathophysiology of conditions ranging from HIV19 to solid tumours.15,20 In the setting of critical care, mathematical models can serve two related purposes. First, they can simulate the effect of multiple sources of heterogeneity on disease trajectory with a precision that is not practical in the context of a trial. Secondly, by identifying markers that are deterministically related to known mechanisms in the model, they can aid in hypothesizing the biologic meaning behind statistically identified biomarkers or even suggest novel marker/mechanism pairings which may be used to screen patients for inclusion in future therapeutic trials.
As a first test of the utility of mathematical modelling for understanding heterogeneity in ARDS here we employ our recently developed systems biology model of SARS-CoV-2 infection.16 COVID-19, the syndrome which results from SARS-CoV-2 infection, is a distinct illness while ARDS is a syndrome which encompasses diverse modes of injury. However, it is now well established that severe COVID-19 is physiologically similar to ARDS of other causes21,22 and is thus appropriate for this proof-of-principle study. We consider three immunomodulation strategies, applied at different times and to varying patient types. We find that the optimal treatment time differs between broad-based (corticosteroids) and targeted (anti-IL6 and anti-IL6R) immunosuppression and identify specific biologic pathways that account for these differences. We use this analysis to propose biomarkers that identify an optimal treatment strategy in a given patient and conclude by studying a subset of those markers in a clinical dataset of COVID-19 patients presenting to an emergency department (ED) within the Massachusetts General Brigham (MGB) healthcare system.
Immunomodulation in COVID-19
Since the start of the COVID-19 pandemic, it has been noted23 that elevated markers of innate immunity such as IL-6 are correlated with worse outcome.24,25 This observation led to the suggestion of Tocilizumab, a monoclonal antibody directed against the IL-6 receptor,26 as a potential therapy. However, clinical trials of anti-IL6 therapies are conflicting. Several smaller studies27, 28, 29, 30, 31 found no mortality benefit while two, large, studies5,32 did report a mortality benefit. Notably, the positive studies reported on somewhat different populations: Horby et al. found a benefit in patients on supplemental oxygen or more advanced respiratory support (in contrast to earlier reports), while the REMAP-CAP investigators reported a benefit in patients started on therapy within 24 hours of transfer to the ICU. Subsequently, it was noted that IL6 levels are not dramatically elevated in COVID-19 when compared to levels in pre-COVID-19 ARDS33,34 - calling into question the rationale for anti-IL6 therapy. Separate trials of the related anti-IL6R antibody Sarilumab showed no effect – even in the more severe patients who appeared to benefit from Tociluzimab.35 At the same time, the results of the RECOVERY trial6 demonstrated a substantial mortality benefit from the use of more broad-based immunosuppression with corticosteroids that mirrored results in pre-COVID-19 ARDS.36 We sought to understand these conflicting results by simulating both approaches in diverse patient types with multiple treatment initiation times.
Methods
System-Biology Modelling of COVID-19
The full details of the model are given in the supplementary material. The model used here is based on a previously validated version of our COVID-19 model.16 It expands on work by other groups by incorporating additional features of SARS-CoV-2 infection and ARDS pathology as well as novel methods of assessing patient outcome.37,38 Briefly, the original model included SARS-CoV-2 infection, the renin angiotensin system (RAS), dynamics of innate and adaptive immune cells, and the coagulation cascade. The model used here has been extended to explicitly incorporate antigen presenting cells, B cells and associated antibody production, viral antigen control of T-cell activation, and modulation of T-Cell activity by PD-L1/PD-1 inhibition. Values of model parameters, initial values and changes to model parameters made to simulate the effect of various treatments are given in Tables S1-S3. Sensitivity analysis, indicating robustness of current findings to changes in relevant parameter values is presented in Table S4 and Figure S2-S5. Disease severity was quantified both by viral load and oxygen saturation and followed until day 20.39
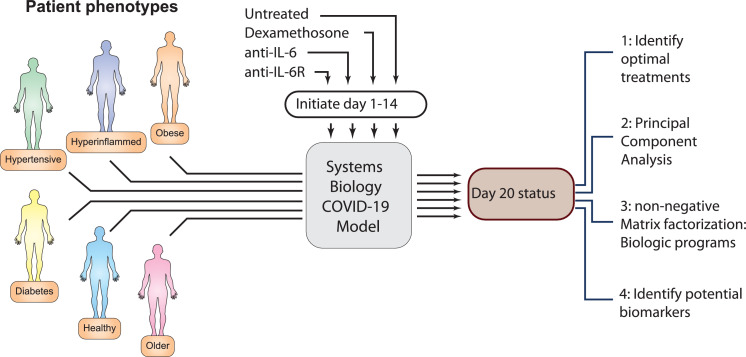
Patient types
In an attempt to mirror known risk factors for severe COVID-19,40 we modified model parameters in an attempt to simulate disease course in the following distinct patient types: young (healthy), diabetic (but not hyperinflamed), older, hyperinflamed, hypertensive and obese based on known risk factors for severe COVID-19.41 Alterations of model parameters to define each patient type are listed in Table 1. Herein we use the above specified clinical types to refer to the specific constellations of parameter values indicated in the Table, acknowledging that no single set of parameters will be fully representative of the heterogeneity of patients with the indicated co-morbidities, We employed the model to simulate the effect of three immunomodulatory treatments (corticosteroids, anti-IL6 and anti-IL6R), given on each possible day between days 1 and 14 (with day 1 indicating first viral exposure).
Table 1.
Alterations in Model Parameters to Represent Different Patient Phenotypes.16 Parameters increased and decreased are in green and red shading, respectively.
 |
Clustering and non-negative matrix factorization analysis
The model output consists of the value of each of the model variables, or features, on each of the 14 possible days following infection, plus day 20 which is taken as the endpoint. To gain insight into the 130 feature values obtained at each of the 15 timepoints for each of the six patient types, we performed unsupervised clustering, with principal component analysis (PCA), for each distinct patient type. We then sought to compare the untreated disease course with the day 20 outcome that resulted from initiating treatment on each of the first 14 days for each patient type, as illustrated in Figure 1. This large amount of data was then summarized by means of the top two principal components.
Figure 1.
Schematic representation of the systems biology approach to predict COVID-19 disease outcome in various patient phenotypes, treatments and treatment initiation times.
As the model consists of equations describing the time evolution of specific features of disease pathophysiology, model features can directly specify the underlying mechanism. We performed non-negative matrix factorization (NMF) analysis on the N x M matrix (where N represents features and M represent distinct patient phenotype).42 We used the Brunet algorithm for matrix factorization and selected the rank (k) based on cophenetic correlation.43 This results in the N x M matrix factorized into a N x k matrix (where k is the number of identified biologic programs), describing feature combinations which determine each clinical program, and a k x M matrix, describing clinical program usage per patient type.
In order to quantify patient outcomes, we developed a scoring system based on how closely the final model state (day 20) resembles either the state on day 1 or untreated day 20. Return to precisely the day 1 state would represent full recovery to baseline while, in the case of treatment, a model state more closely resembling the untreated day 20 state would represent no treatment effect. The score is defined as the ratio of the Euclidean distance between model state on day 20 of treated patients and untreated day 20, to the distance between day 20 of treated patients and day 1.
Where
and
Higher Treatment Score ratio indicates the specific treatment returned the patient towards baseline status.
Biomarker validation
In order to assess the utility of our predicted biomarkers, we developed a multinomial logistic regression model to predict ICU admission in a previously studied population of patients presenting to the ED with COVID-19.44 Patients from the Massachusetts General Brigham (MGB) healthcare system who were positive for SARS-CoV-2 between March and August of 2020 and had an ED encounter were included. We constructed separate models for older, obese, diabetic and hypertensive patients (Older defined as > 65), with diagnosis taken from the electronic medical record, and adjusted for age, sex, BMI, and race. We considered ICU admission as a surrogate for disease severity.
Ethics
This study was approved by the MGB Institutional Review Board (IRB protocol # 2020P000964).
Role of funding source
C.V. received a Marie Skłodowska Curie Actions Individual Fellowship (MSCA-IF-GF-2020-101028945). R.K.J.’s research is supported by R01-CA208205, and U01-CA 224348, R35-CA197743 and grants from the National Foundation for Cancer Research, Jane’s Trust Foundation, Advanced Medical Research Foundation and Harvard Ludwig Cancer Center. No funder had a role in production or approval of this manuscript.
Results
Treatments are more effective in older, hyperinflammed and hypertensive patients than other types
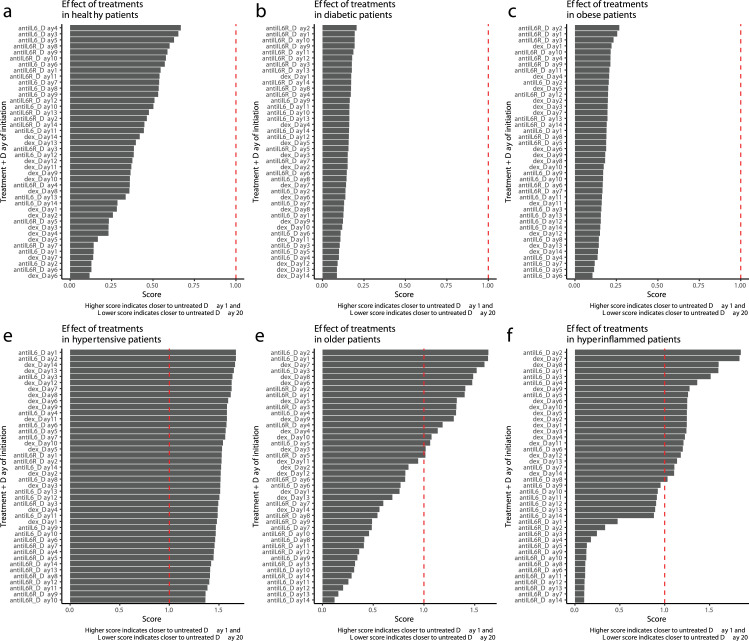
We first compared the treatment scores for each of the six patient types subjected to each of the three treatments on each of the 14 possible days of treatment initiation (Figure 2), where day 1 represents first virus exposure. Each plotted bar is the outcome score evaluated at day 20 in response to the listed treatment and timing. For the young, healthy patient, all three immunomodulatory treatments result in a day 20 state that is more similar to the untreated day 20 than to the baseline state - indicating that in these healthy patients, treatment had little effect on the course of disease (Figure 2a). Note that these patients recover without treatment, so their untreated day 20 status is closer to day 1 baseline than the other patient groups. Diabetic and obese patients (as defined in the model) also received little benefit from any of the treatments (Figures 2b,c). In contrast, the hypertensive type benefits from any of the three treatments, with little dependency on day of initiation (Figure 2d). More interestingly, older and hyper-inflamed patients range from no benefit to substantial benefit depending on the type of treatment and day of initiation (Figures 2e,f). Hyperinflammed patients benefit from corticosteroids or anti-IL6 therapy (but not anti-IL6 receptor therapy) anytime in the first 10 days after viral exposure, with most benefit coming from very early initiation of anti-IL6 therapy (between days 1-4 after virus infection) or somewhat later initiation of corticosteroids (between days 7-10). For older patients, the optimal window for treatment is somewhat shorter. Corticosteroids are most effective between days 7-9, and anti-IL6 and anti-IL6 receptor therapy is most effective before day 3.
Figure 2.
Optimal treatment type and timing varies by patient phenotype. Treatment score (see methods) for six distinct patient phenotypes. Each treatment is tested with initiation at each of the first 14 days following viral exposure. A score of 1.0 (shown by dashed vertical line) indicates treatment resulted in a patient status midway between day 20 and baseline (day 1). A score of less than one indicates treatment resulted in patient status being closer to untreated day 20 than day 1, while a score of greater than one indicates treatment moved the patient closer to baseline by day 20.
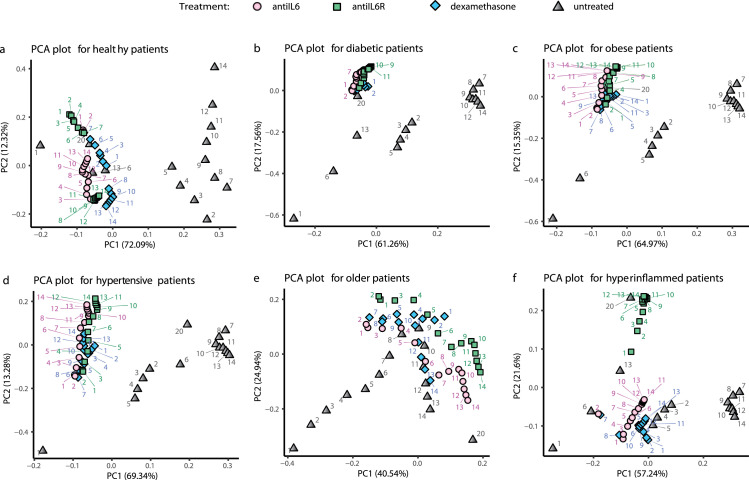
In order to examine a fuller disease trajectory, we next performed principal component analysis of the matrix of output variables. This is useful for visualizing how the important parameters are affected by various treatments and schedules. Figure 3 depicts the trajectory in principal component space for each of the timepoints of patient types with day 20 outcome of each of the treatments initiated at each of the fourteen possible days. In the case of the healthy, young patients (Figure 3a), the data for day 1 and day 20-untreated lie close to each other while other time points deviate, tracking the course of illness and return towards baseline over 20 days (gray triangles). For all except the young, healthy patient type, the untreated day 1 and day 20 are not in close proximity, suggesting persistent abnormalities on day 20 in the absence of treatment. For diabetic and obese patients (Figs 3b,c), all treatment outcomes at day 20 lie close to untreated day 20 status, suggesting little change in disease course with treatment irrespective of initiation day. Older and hyperinflamed patients differ substantially in principal component space when treatments are initiated on different days, confirming differential outcomes (Figs 3e,f). The hypertensive type moves away from day 20 untreated and closer to baseline, irrespective of treatment initiation day, suggesting most treatments are favourable (Fig 3d).
Figure 3.
Patient phenotype modifies treated and untreated disease trajectory. Model output for each of six patient phenotypes in the untreated (grey triangle), anti-IL6 treated (light pink circle), anti-IL6R treated (green square) and dexamethasone treated (blue diamond) cases depicted as principal component analysis (PCA) plots. The grey triangles show the trajectory of an untreated patient over 20 days (days indicated by blue text). The other data points represent the status achieved on day 20, when that treatment is given on the day indicated in the colored text. In this depiction, better treatment efficacy is indicated by less distance between the treated (colored) data point and the untreated patient at Day 1 (baseline condition). Less effective treatment is indicated by a treated (colored) data point that lies close to untreated day 20 data (grey) point. Percentages on the axis labels indicate the amount of variance of the feature matrix which is explained by each principal component.
Optimal treatment is determined by differential usage of clinical programs in distinct patient phenotypes
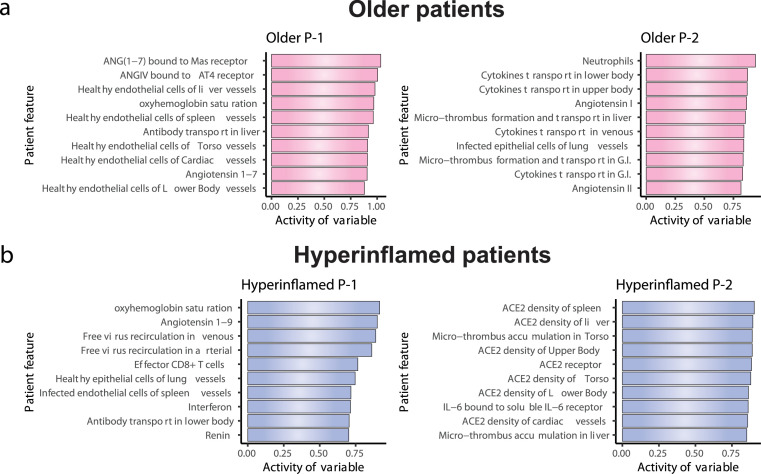
A key advantage of a modelling approach is the ability to move beyond the observation of differential outcomes and determine exactly why such an outcome occurred. To directly relate specific terms in the model to biological programs and clinical outcomes, we next performed non-negative matrix factorization and identified specific biologic programs responsible for treatment response in each patient type. Each output variable in the model represents a biological value that can be measured (IL-6 level in the lung, for example). The biologic programs defined here consist of combinations of such outputs from the model that are strongly associated with a given outcome. We sought to factor the N x M feature-patient matrix into two primary biologic programs (k = 2) for each patient type. We then separately identified dominant programs for patient type/treatment timing pairs that resulted in favourable outcomes versus those that did not using the treatment score defined above.
For older patients, we identified a program (P-1) that is low at baseline, becomes higher during disease progression, and then returns to baseline in untreated patients (Table 2). Another program for this patient type (P-2) is low at baseline, but increases over time, becoming high at day 20. Program-1 includes the number of healthy (uninfected) endothelial cells, the amount of Angiotensin1, 2, 3, 4, 5, 6, 7 bound to Mas receptor, and systemic antibody transport, while Program-2 includes extra-pulmonary cytokine production, systemic microthrombosis, and pulmonary neutrophil recruitment (Figure 4a). For hyperinflamed patients, Program-1 is defined by the level of circulating virus, number of CD8+ T-cells, interferon expression and the level of Angiotensin1, 2, 3, 4, 5, 6, 7. (Figure 4B, Table 2). This program also returns to baseline at day 20 in untreated patients, but Program-2 – composed of terms governing ACE2 density, systemic microthrombosis and trans IL6 signalling – remains elevated. Note that each program is specific for a given patient type (Figure 4).
Table 2.
Status and variation of clinical programs (P-1: Program-1, P-2: Program-2) based on usage by patient phenotypes (Information extracted from heatmap shown in Fig. S1).
| Program | State on day 1 (baseline) | State on day 20 (untreated) | Variation in program during untreated disease course | State after anti-IL6 initiation on optimal day | State after anti-IL6R initiation on optimal day | State after dexamethasone initiation on optimal day |
|---|---|---|---|---|---|---|
| Older | ||||||
| Older P-1 | Low | Low | High | Moderately high | Moderately high | Moderately high |
| Older P-2 | Low | High | Moderate | Low | Low | Low |
| Hyperinflamed | ||||||
| Hyperinflamed P-1 | Low | Low | High | Low | Low | Low |
| Hyperinflamed P-2 | Low | High | Moderate | Low | Moderately high | Low |
| Hypertensive | ||||||
| Hypertensive P-1 | Low | Low | Low | Moderately high | Moderately high | Moderately high |
| Hypertensive P-2 | Low | Moderately high | High | Low | Low | Low |
| Diabetics | ||||||
| Diabetic P-1 | Low | Moderately high | Low | Moderately high | Moderately high | Moderately high |
| Diabetic P-2 | Low | Low | High | Low | Low | Low |
| Obese | ||||||
| Obese P-1 | Low | Moderately high | Low | Moderately high | Moderately high | Moderately high |
| Obese P-2 | Low | Low | High | Low | Low | Low |
| Healthy | ||||||
| Healthy P-1 | Low | Low | Low | Low | Low | Low |
| Healthy P-2 | Low | Low | High | Low | Low | Low |
Figure 4.
Matrix factorization used to identify two biologic programs which determine outcome in older and hyperinflamed patients. For older patients and hyperinflamed patients, optimal treatment consists of very early anti-IL6 therapy (<4th day) or delayed (day 7th-9th) dexamethasone therapy. Matrix factorizations divides the patient-feature matrix into patient-program and program-feature matrices. The plot here depicts the feature contribution to 2 clinical programs that were generated during factorization.
We can next examine how treatments, applied at the optimal time, affect the identified programs. For example, for older patients, outcome with optimal treatment is largely determined by Program-2 status being reduced to baseline, which occurs for earlier timepoints of anti-IL6 and anti-IL6R therapy and later timepoints of corticosteroid (Figure S1e, Table 2). For hyperinflamed patients, Program-2 is reduced to baseline by earlier treatment initiation of anti-IL6 and later initiation of corticosteroid (anti-IL6R is not able to alter Program-2) (Figure S1f, Table 2).
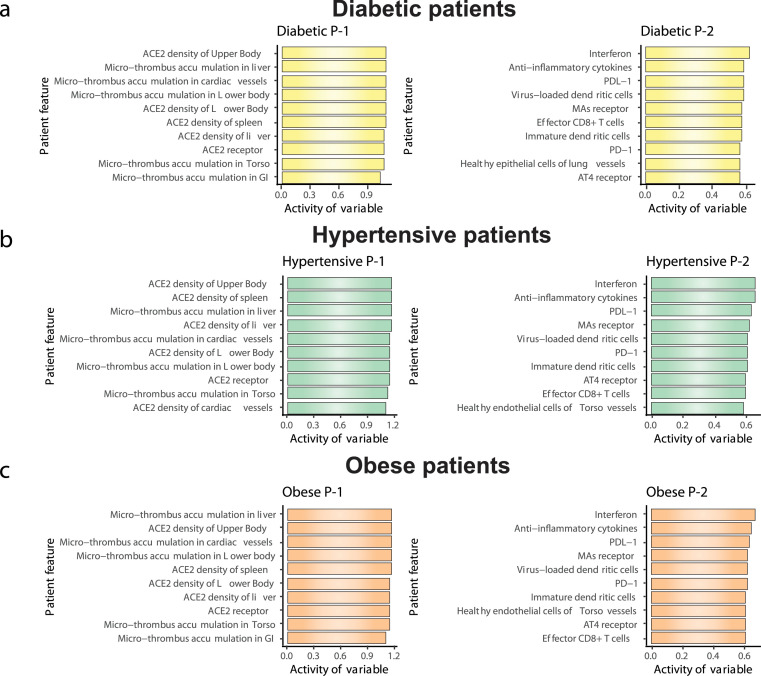
We conducted a similar analysis for the patient types that had little response to treatment (Figure 5). For diabetic and obese patients, Program-2 gets altered more during the untreated course of infection, but unlike Program-1, which remains moderately elevated, Program-2 returns to baseline on day 20 (Figure S1b and c, Table 2). On the other hand, for hypertensive patients, Program-2 does not come back to baseline at the end of untreated disease progression, indicating that it contains disease-driving parameters (Figure S1d, Table 2). Treatment initiated on any day changes the Program-2 state and brings it back toward baseline, but this change is accompanied by an increase in Program-1 as compared to untreated day 20. Irrespective of day of treatment initiation, the program states largely remain unchanged on day 20 as compared to untreated day 20. Program-2 in diabetic and obese individuals primarily consists of terms governing interferon expression, anti-inflammatory cytokines and PD-1/PDL-1 receptor signalling, whereas Program-1 consists of terms governing ACE2 density and systemic microthrombosis (Figure 5a and c). Based on components of Program-2, these data indicate that features, such as interferon activity, anti-inflammatory cytokine level and PD-1/PDL-1 receptor, can be altered by treatment of hypertensive patients, but Program-1 features, such as ACE2 density and systemic microthrombosis are less impacted by treatment (Figure 5b).
Figure 5.
Biologic programs determining outcome in Diabetic, Hypertensive and Obese patients. Depicted are the primary biologic programs (as determined by factorization of the feature matrix) responsible for outcome in the indicated patient phenotype. In the case of diabetic, hypertensive and obese patients the outcome was relatively insensitive to treatment protocol.
Clinical parameters corresponding to model features
As demonstrated in Figure 3 and Figure 4, model parameters correspond to defined biologic pathways, so it is possible to suggest clinically observable variables which may correspond to major features of the programs implicated by the analysis. In Table 3, we identify clinically observable biomarkers that may correspond to model programs that determine outcome in each patient type. In this way, the matrix factorization may be used to predict biomarkers for treatment response - even biomarkers which have not previously been measured in a clinical trial.
Table 3.
Clinically relevant biomarkers based on important features from clinical programs (P-1: Program-1, P-2: Program-2).
| Program | Feature name | Predicted biomarker |
|---|---|---|
| Older | ||
| Older P-1 | Ang(1 - 7) bound to Mas receptor | ACE2-Ang(1-7) ratios, ACE2 levels51,52 |
| Older P-1 | Oxygen saturation | Oxygen saturation |
| Older P-2 | Neutrophils | Neutrophil elastase myeloperoxidase53 |
| Hyperinflamed | ||
| Hyperinflamed P-1 | Oxygen saturation | Oxygen saturation |
| Hyperinflamed P-1 | Angiotensin 1-9 | Circulating Angiotensin 1-952 |
| Hyperinflamed P-1 | Free virus circulation | SARS-CoV-2 PCR cycle threshold |
| Obese, diabetic, hypertensive | ||
| Obese/Diabetic/Hypertensive P-1s | Micro thrombus in liver and cardiac vessels | D-dimer |
| Obese/Diabetic/Hypertensive P-1s | ACE2 density of upper body | ACE2 levels52 |
| Obese/Diabetic/Hypertensive P-2s | Interferon | IFN-γ levels |
| Obese/Diabetic/Hypertensive P-2s | Anti-inflammatory cytokines | IL-6, IL1β |
Finally, we sought to conduct a preliminary clinical analysis of our proposed biomarkers by constructing a multinomial logistic regression on two proposed biomarkers (IL6, D-dimer) in each of four phenotypes (older, obese, hypertensive, diabetic). Patient type in this case was determined form the electronic health record in a database of ∼5300 patients presenting to emergency departments within the Mass General Brigham healthcare system with COVID-19 and thus, may not entirely correspond to the parameter sets chosen in the model. Perhaps unsurprisingly (as both markers have previously been identified as prognostic in unselected populations of patients with COVID-19), both biomarkers are predictive of outcome in each of the clinical patient types (Table 4). However, this result suggests that model variables associated with prognosis are the same as those identified in clinical subpopulations, increasing confidence in the models ability to identify clinical programs responsible for outcome.
Table 4.
Logistic regression models showing the association of getting severe COVID-19 disease and comparable clinical variables (that correspond to some features of clinical programs) among patients from the Massachusetts General Brigham healthcare system who were positive for SARS-CoV-2 between March and August of 2020 and had an ED encounter. Logistic regression models developed using glm function in R. Sample size (n) for each model mentioned in patient phenotype column.
| Patient phenotype | Variable | Odds Ratio | 95% CI | p-value |
|---|---|---|---|---|
| Older (n = 2187) | First O2 saturation | 0.9074 | [0.8870;0.9283] | <0.001 |
| Neutrophils | 1.0547 | [1.0419;1.0677] | <0.001 | |
| Diabetes (n = 886) | D-dimer | 1.0002 | [1.0001;1.0003] | < 0.001 |
| IL-6 | 1.0085 | [1.0055;1.0115] | < 0.001 | |
| Obese (n = 2185) | D-dimer | 1.0001 | [1.0000;1.0002] | 0.00806 |
| IL-6 | 1.0177 | [1.0144;1.0210] | < 0.001 | |
| Hypertensive (1992) | D-dimer | 1.0001 | [1.0001;1.0002] | <0.001 |
| IL-6 | 1.017 | [1.0135;1.0205] | <0.001 |
Discussion
Efforts to deploy targeted treatments and personalized medicine have already seen substantial success in fields such as oncology, cardiology and rheumatology.45, 46, 47, 48 Compared to the largely chronic conditions with well-defined aetiology seen in these fields, acute critical care syndromes, such as ARDS, are heterogenous, dynamic and multifactorial. This makes it harder both to observe a significant treatment effect in a clinical trial and, crucially, to assign a precise biologic meaning to a given observation. As a first effort at deploying mathematical modelling to make sense of such an environment here, we report a newly developed framework for analysing patient trajectories and identifying key mechanistic programs that define patient subsets. We demonstrated the use of this approach using data from our mathematical model of COVID-19 (Figure 6), which predicts the response to different immunomodulatory therapies initiated at different times. Identification of key programs relies on a combination of principal component and non-negative matrix factorization analyses of model parameters, treatment modality, time of treatment, and patient factors to determine which features of the model determine treatment response in each type of patient. It is important to note that the model output features that characterize outcome are not the same as the parameters which are altered to create the phenotype in the model. In such a complex systems biology model, the few parameters we changed to specify each patient type have effects on many interacting biological pathways. This potentially non-linear behaviour is biologically relevant and not possible to account for without formal modelling of interactions as in the present work.
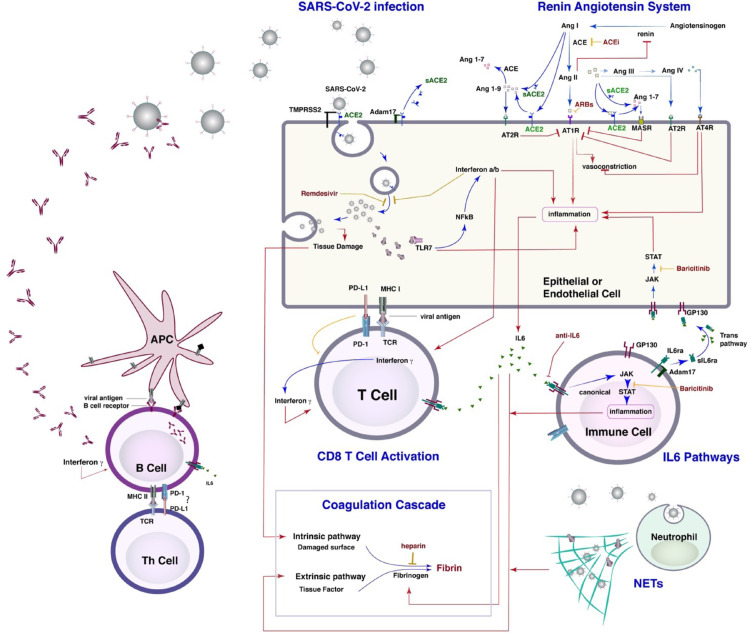
Figure 6.
Schematic of the model. The model consists of a lung model which incorporates viral infection of epithelial and endothelial cells and the innate immune response which includes classical and trans IL6 pathways, other inflammatory cytokines, activity of neutrophils, and macrophages, as well as the formation of NETs, and the coagulation cascade and activation of the renin-angiotensin system (RAS). In addition the model contains terms describing the adaptive immune response including conversion of dendritic cells to antigen presenting cells, formation of activated B-cells, antibody levels, T cell activation and immune checkpoints, The lung model is coupled with a PK/PD model for the virus and thrombi dissemination through the body.
The results suggest that targeted immunomodulatory treatment with anti-IL6 therapies is effective in certain patient types (hyperinflamed, older and hypertensive) but not others (obese, diabetic) and in all cases is most effective when given very early. Broad based immunosuppression (represented here by corticosteroids) is effective in a similar range of patients (and similarly ineffective in others) but is optimally provided later in the course of illness. For younger (healthy) individuals, immunomodulatory therapies have minimal impact on clinical outcome, which is characterized by return to baseline even in the absence of treatment.
In addition to the ability to test a broader range of treatment regimen/protocols than is practical in actual patients, modelling approaches have the advantage of providing information about the precise biologic origin for a given model outcome. Knowledge of the responsible program in the model may then be used to predict biomarkers that identify patients most likely to benefit from a given treatment protocol. Such prediction may be useful for enrichment of clinical trial populations. For example, the analysis predicts that immunomodulatory therapies provide the most benefit to patients corresponding to our type of older and hyperinflamed patients. In the case of the hyperinflamed type, the treatment is able to modulate the ACE2 density, microthrombosis formation and IL6 level, leading to better outcome. Based on these results, we speculate that IL-6, d-dimer and Angiotensin1, 2, 3, 4, 5, 6, 7 levels may be biomarkers of response to treatment in these phenotypes. IL-6 has previously been associated with outcome in severe COVID-19, but d-dimer has not been proposed as a biomarker specific to immunomodulatory treatment. In a clinical population, we find that both are associated with poor outcomes. While perhaps unsurprising, this result does indicate that the model is potentially able to identify biomarkers with real-world relevance. Indeed, using randomized clinical trial data, Sinha et al. showed that IL-6 was among the protein biomarkers that could lead to identification of clusters of patients with heterogeneity of treatment effects.12 Given the limitations of retrospective analysis, we were not able to investigate the predictive ability of infrequently measured markers such as Angiotensin,1, 2, 3, 4, 5, 6, 7 but we note that Angiotensin1, 2, 3, 4, 5, 6, 7 levels have recently been associated with poor prognosis in severe COVID-1949 and markers of the RAS have been associated with worse outcome in ARDS50 as well as having been studied in cardiovascular disease.51,52
Model Assumptions and Limitations
There are definite limitations to the approach described here. Chief among these is the fact that we have represented patient types with a single set of parameter choices for each type. The analysis, and the predicted response to treatment, is thus limited to those patients plausibly represented by our particular parameter choices. These particular parameter choices certainly do not represent the full diversity of a clinical population with COVID-19 ARDS, let alone non-COVID ARDS. Jenner et. al have reported on an innovative virtual patient cohort in which a computational model of COVID-19 is parameterized by selection from a normal distributions developed from clinical data38 (Table S5). Such an approach is readily applied to the model described here. Here we seek only to further demonstrate the utility and feasibility of modelling complex critical care syndromes such as COVID-19 ARDS by including simulations of specific treatments, developing a novel method of scoring outcomes and using the model outcomes to prospectively predict biomarkers. In this context, we point out that clinical trial simulations with more probabilistic parameter choices or iteration of the model based on emerging biologic data/insights and subsequent simulation is fast and inexpensive. Identification of predicted markers and biologic programs via a model is thus likely to be quite cost-effective when compared to retrospective analysis of clinical trial data. Moreover, while no doubt imperfect, generation of hypotheses regarding biologic mechanisms via a formally specified model is certainly more rigorous than unstructured speculation based on isolated clinical observations such as the elevation of a particular cytokine.
Here we have modelled COVID-19. The pathophysiology of ARDS or septic shock from other etiologies will certainly differ in important respects. However, there are important common pathways in critical illness and modelling can be a helpful framework for identifying them.37 Finally, we note that there are obstacles to experimentation based on model predictions, even if they are completely correct. The results discussed here suggest that anti-IL6 therapy is optimally provided very close to initial viral exposure - likely before a patient is symptomatic and certainly before they are likely to have presented for treatment. However, given the ease of testing new treatments in the model, it is imminently possible to conduct searches for optimized treatments that are more clinically-feasible.
In conclusion, we note that retrospective data are limited, and clinical trials are expensive. We believe that the use of mathematical modelling of critical illness is not only a powerful method for the generation and initial exploration of biologic hypotheses but holds great promise for decreasing heterogeneity and enhancing the precision of clinical trials in the ICU.
Contributions
S.S. developed the treatment score, performed the analysis of the model output, defined biological programs, and also validated model output using clinical data. C.V. ran model simulations for all patient types. C.C.H. interpreted the biological programs and wrote the initial draft. M.R.N developed the PK/PD model. M.R.N., A.B.P., A.V., M.J.K., S.D., T.S., R.K.J. and L.L.M. supervised the model simulations and output analysis. All authors were involved in reviewing the article.
Data sharing statement
The clinical data used in this study belongs to MGB healthcare and restrictions apply to the availability of this dataset. Researchers affiliated with the MGB may apply for access to these data through the MGB Institutional Review Board. The R code used in this study are available upon request addressed to the corresponding author: Lance L. Munn, PhD (munn@steele.mgh.harvard.edu).
Declaration of interests
LLM owns equity in Bayer AG and is a consultant for SimBiosys. R.K.J. received honorarium from Amgen; consultant fees from Chugai, Elpis, Merck, Ophthotech, Pfizer, SPARC, SynDevRx, XTuit; owns equity in Accurius, Enlight, Ophthotech, SynDevRx; and serves on the Boards of Trustees of Tekla Healthcare Investors, Tekla Life Sciences Investors, Tekla Healthcare Opportunities Fund, Tekla World Healthcare Fund; and received a grant from Boehringer Ingelheim. Neither any reagent nor any funding from these organizations was used in this study. Other coauthors have no conflict of interests to declare.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103809.
Appendix. Supplementary materials
References
- 1.Force ADT, Ranieri VM, Rubenfeld GD, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307(23):2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 2.Iwashyna TJ, Burke JF, Sussman JB, Prescott HC, Hayward RA, Angus DC. Implications of Heterogeneity of Treatment Effect for Reporting and Analysis of Randomized Trials in Critical Care. Am J Respir Crit Care Med. 2015;192(9):1045–1051. doi: 10.1164/rccm.201411-2125CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinha P, Calfee CS. Phenotypes in acute respiratory distress syndrome: moving towards precision medicine. Curr Opin Crit Care. 2019;25(1):12–20. doi: 10.1097/MCC.0000000000000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Driscoll M, Ribeiro Dos Santos G, Wang L, et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590(7844):140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 5.Investigators R-C, Gordon AC, Mouncey PR, et al. Interleukin-6 Receptor Antagonists in Critically Ill Patients with Covid-19. N Engl J Med. 2021 doi: 10.1056/NEJMoa2100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Group RC, Horby P, Lim WS, et al. Dexamethasone in Hospitalized Patients with Covid-19. N Engl J Med. 2021;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calfee CS, Janz DR, Bernard GR, et al. Distinct molecular phenotypes of direct vs indirect ARDS in single-center and multicenter studies. Chest. 2015;147(6):1539–1548. doi: 10.1378/chest.14-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha P, Delucchi KL, McAuley DF, O'Kane CM, Matthay MA, Calfee CS. Development and validation of parsimonious algorithms to classify acute respiratory distress syndrome phenotypes: a secondary analysis of randomised controlled trials. Lancet Respir Med. 2020;8(3):247–257. doi: 10.1016/S2213-2600(19)30369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prescott HC, Calfee CS, Thompson BT, Angus DC, Liu VX. Toward Smarter Lumping and Smarter Splitting: Rethinking Strategies for Sepsis and Acute Respiratory Distress Syndrome Clinical Trial Design. Am J Respir Crit Care Med. 2016;194(2):147–155. doi: 10.1164/rccm.201512-2544CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2(8):611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Famous KR, Delucchi K, Ware LB, et al. Acute Respiratory Distress Syndrome Subphenotypes Respond Differently to Randomized Fluid Management Strategy. Am J Respir Crit Care Med. 2017;195(3):331–338. doi: 10.1164/rccm.201603-0645OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinha P, Spicer A, Delucchi KL, McAuley DF, Calfee CS, Churpek MM. Comparison of machine learning clustering algorithms for detecting heterogeneity of treatment effect in acute respiratory distress syndrome: A secondary analysis of three randomised controlled trials. EBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rougier J, Sexton DMH, Murphy JM, Stainforth D. Analyzing the Climate Sensitivity of the HadSM3 Climate Model Using Ensembles from Different but Related Experiments. Journal of Climate. 2009;22(13):3540–3557. [Google Scholar]
- 14.Blair RH, Trichler DL, Gaille DP. Mathematical and statistical modeling in cancer systems biology. Front Physiol. 2012;3:227. doi: 10.3389/fphys.2012.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voutouri C, Kirkpatrick ND, Chung E, et al. Experimental and computational analyses reveal dynamics of tumor vessel cooption and optimal treatment strategies. Proc Natl Acad Sci U S A. 2019;116(7):2662–2671. doi: 10.1073/pnas.1818322116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voutouri C, Nikmaneshi MR, Hardin CC, et al. In silico dynamics of COVID-19 phenotypes for optimizing clinical management. Proc Natl Acad Sci U S A. 2021;118(3) doi: 10.1073/pnas.2021642118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perelson AS. Modelling viral and immune system dynamics. Nat Rev Immunol. 2002;2(1):28–36. doi: 10.1038/nri700. [DOI] [PubMed] [Google Scholar]
- 18.Perelson AS, Ke R. Mechanistic Modeling of SARS-CoV-2 and Other Infectious Diseases and the Effects of Therapeutics. Clin Pharmacol Ther. 2021;109(4):829–840. doi: 10.1002/cpt.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271(5255):1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 20.Stylianopoulos T, Munn LL, Jain RK. Reengineering the Physical Microenvironment of Tumors to Improve Drug Delivery and Efficacy: From Mathematical Modeling to Bench to Bedside. Trends Cancer. 2018;4(4):292–319. doi: 10.1016/j.trecan.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziehr DR, Alladina J, Petri CR, et al. Respiratory Pathophysiology of Mechanically Ventilated Patients with COVID-19: A Cohort Study. Am J Respir Crit Care Med. 2020;201(12):1560–1564. doi: 10.1164/rccm.202004-1163LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hariri LP, North CM, Shih AR, et al. Lung Histopathology in Coronavirus Disease 2019 as Compared With Severe Acute Respiratory Sydrome and H1N1 Influenza: A Systematic Review. Chest. 2021;159(1):73–84. doi: 10.1016/j.chest.2020.09.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C, Chen X, Cai Y, et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somers EC, Eschenauer GA, Troost JP, et al. Tocilizumab for treatment of mechanically ventilated patients with COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salama C, Han J, Yau L, et al. Tocilizumab in Patients Hospitalized with Covid-19 Pneumonia. N Engl J Med. 2021;384(1):20–30. doi: 10.1056/NEJMoa2030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stone JH, Frigault MJ, Serling-Boyd NJ, et al. Efficacy of Tocilizumab in Patients Hospitalized with Covid-19. N Engl J Med. 2020;383(24):2333–2344. doi: 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosas IO, Brau N, Waters M, et al. Tocilizumab in Hospitalized Patients with Severe Covid-19 Pneumonia. N Engl J Med. 2021;384(16):1503–1516. doi: 10.1056/NEJMoa2028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veiga VC, Prats J, Farias DLC, et al. Effect of tocilizumab on clinical outcomes at 15 days in patients with severe or critical coronavirus disease 2019: randomised controlled trial. BMJ. 2021;372:n84. doi: 10.1136/bmj.n84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Declercq J, Van Damme KFA, De Leeuw E, et al. Effect of anti-interleukin drugs in patients with COVID-19 and signs of cytokine release syndrome (COV-AID): a factorial, randomised, controlled trial. Lancet Respir Med. 2021 doi: 10.1016/S2213-2600(21)00377-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abani O, Abbas A, Abbas F, et al. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. The Lancet. 2021;397(10285):1637–1645. doi: 10.1016/S0140-6736(21)00676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson JG, Simpson LJ, Ferreira AM, et al. Cytokine profile in plasma of severe COVID-19 does not differ from ARDS and sepsis. JCI Insight. 2020;5(17) doi: 10.1172/jci.insight.140289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha P, Matthay MA, Calfee CS. Is a "Cytokine Storm" Relevant to COVID-19? JAMA Intern Med. 2020;180(9):1152–1154. doi: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 35.Lescure F-X, Honda H, Fowler RA, et al. Sarilumab in patients admitted to hospital with severe or critical COVID-19: a randomised, double-blind, placebo-controlled, phase 3 trial. The Lancet Respiratory Medicine. 2021;9(5):522–532. doi: 10.1016/S2213-2600(21)00099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villar J, Ferrando C, Martínez D, et al. Dexamethasone treatment for the acute respiratory distress syndrome: a multicentre, randomised controlled trial. Lancet Respir Med. 2020;8(3):267–276. doi: 10.1016/S2213-2600(19)30417-5. [DOI] [PubMed] [Google Scholar]
- 37.Thompson BT, Chambers RC, Liu KD. Acute Respiratory Distress Syndrome. N Engl J Med. 2017;377(6):562–572. doi: 10.1056/NEJMra1608077. [DOI] [PubMed] [Google Scholar]
- 38.Jenner AL, Aogo RA, Alfonso S, Crowe V, Deng X, Smith AP, et al. COVID-19 virtual patient cohort suggests immune mechanisms driving disease outcomes. PLoS Pathog. 2021;17(7) doi: 10.1371/journal.ppat.1009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy TK, Secomb TW. Theoretical analysis of the determinants of lung oxygen diffusing capacity. J Theor Biol. 2014;351:1–8. doi: 10.1016/j.jtbi.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horby PW, Pessoa-Amorim G, Peto L, et al. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): preliminary results of a randomised, controlled, open-label, platform trial. medRxiv. 2021 :2021.02.11.21249258. [Google Scholar]
- 41.Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 42.Naik GR, Ohio Library and Information Network. Non-negative matrix factorization techniques: advances in theory and applications. DOI of book: https://link.springer.com/book/10.1007%2F978-3-662-48331-2. 1 online resource. p.
- 43.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101(12):4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subudhi S, Verma A, Patel AB, et al. Comparing Machine Learning Algorithms for Predicting ICU Admission and Mortality in COVID-19. NPJ Digit Med. 2021 doi: 10.1038/s41746-021-00456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 46.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lotvall J, Akdis CA, Bacharier LB, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127(2):355–360. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 48.Leopold JA, Loscalzo J. Emerging Role of Precision Medicine in Cardiovascular Disease. Circ Res. 2018;122(9):1302–1315. doi: 10.1161/CIRCRESAHA.117.310782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reindl-Schwaighofer R, Hodlmoser S, Eskandary F, et al. ACE2 Elevation in Severe COVID-19. Am J Respir Crit Care Med. 2021;203(9):1191–1196. doi: 10.1164/rccm.202101-0142LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krenn K, Hobart P, Poglitsch M, Croize A, Ullrich R. Equilibrium Angiotensin Metabolite Profiling in Patients with Acute Respiratory Distress Syndrome Indicates Angiotensin-Converting Enzyme Inhibition. Am J Respir Crit Care Med. 2020;202(10):1468–1471. doi: 10.1164/rccm.201912-2504LE. [DOI] [PubMed] [Google Scholar]
- 51.Patel SK, Velkoska E, Burrell LM. Emerging markers in cardiovascular disease: where does angiotensin-converting enzyme 2 fit in? Clin Exp Pharmacol Physiol. 2013;40(8):551–559. doi: 10.1111/1440-1681.12069. [DOI] [PubMed] [Google Scholar]
- 52.Zhou X, Zhang P, Liang T, Chen Y, Liu D, Yu H. Relationship between circulating levels of angiotensin-converting enzyme 2-angiotensin-(1-7)-MAS axis and coronary heart disease. Heart Vessels. 2020;35(2):153–161. doi: 10.1007/s00380-019-01478-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boettcher M, Esser M, Trah J, et al. Markers of neutrophil activation and extracellular traps formation are predictive of appendicitis in mice and humans: a pilot study. Sci Rep. 2020;10(1):18240. doi: 10.1038/s41598-020-74370-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.