Abstract
Cytogenetic and molecular abnormalities are known to influence post-transplant outcomes in acute myeloid leukemia (AML) but data assessing the prognostic value of combined genetic models in the HCT setting are limited. We developed an adapted European LeukemiaNet (aELN) risk classification based on available genetic data reported to the Center for International Blood and Marrow Transplant Research, to predict post-transplant outcomes in 2289 adult AML patients transplanted in first remission, between 2013 and 2017. Patients were stratified according to aELN into three groups: favorable (Fav, N=181), intermediate (IM, N=1185) and adverse (Adv, N=923). Univariate analysis demonstrated significant differences in 2-year overall survival (OS) (Fav: 67.7%, IM: 64.9% and Adv: 53.9%; p<0.001); disease-free survival (DFS) (Fav: 57.8%, IM: 55.5% and Adv: 45.3; p<0.001) and relapse (Fav: 28%, IM: 27.5% and Adv: 37.5%; p<0.001). Multivariate analysis (MVA) revealed no differences in outcomes between the Fav and IM groups, thus they were combined. On MVA, patients in the Adv risk group had the highest risk of relapse (HR 1.47 p=<0.001) and inferior DFS (HR 1.35 p<0.001) and OS (HR 1.39 p<0.001), even using myeloablative conditioning or in those without pre-HCT measurable-residual disease. Novel approaches to mitigate relapse in this high-risk group are urgently needed.
INTRODUCTION
Allogeneic hematopoietic cell transplantation (HCT) is an effective post-remission strategy for patients with acute myeloid leukemia (AML) in first complete remission (CR1). Due to considerable morbidity and mortality risk, careful consideration of patient (1) and disease-specific (2, 3) characteristics is required. Traditionally, biological stratification of AML risk has been based on cytogenetic findings at diagnosis (3, 4). HCT in CR1 is considered for eligible patients with intermediate and high-risk cytogenetics whereas chemotherapy is recommended for those with favorable cytogenetics (2, 5, 6). Approximately 45% of AML cases have normal karyotype and cannot be subdivided based on cytogenetics; these patients have been assigned intermediate risk (4). Subsequent identification of genomic mutations in AML, such as FLT3-ITD, NPM1, CEBPA, MLL/KMT2A, IDH1–2, DNMT3A, TET2, TP53 and BCOR amongst others, has proven valuable for subdividing cytogenetically normal (CN) AML into subsets with different outcomes (7).
In 2010, the European LeukemiaNet (ELN2010) proposed a standardized prognostic system incorporating both cytogenetic and select molecular abnormalities (CEBPA, NPM1 and FLT3-ITD) to distinguish 4 distinct genetic risk groups (8). Multiple studies confirmed the prognostic accuracy of ELN2010, applying it to patient cohorts receiving consolidation chemotherapy (9, 10) or HCT (11, 12). In 2017, the ELN updated and simplified their classification (ELN2017) into 3-groups of favorable, intermediate, and adverse risk (13). The new classification incorporated the addition of RUNX1, ASXL1 and TP53 mutations to the adverse risk group, the inclusion of biallelic (but not monoallelic) CEBPA mutations to the favorable group and stratification of FLT3-ITD on the basis of ITD-to-wild-type allelic ratio. Several groups have since applied ELN2017 to predict outcomes after chemotherapy (14–17) as well as compared the outcomes between ELN2010 and ELN2017 (14). Single institution studies have also evaluated the utility of ELN2017 in predicting post- HCT outcomes (18, 19), but large, multicenter studies evaluating the impact of a combined genetic model exclusively in the transplant setting are lacking.
We analyzed data from the Center for International Blood and Marrow Transplant Research (CIBMTR) to evaluate the prognostic ability of an adapted ELN2017 model to predict allogeneic HCT outcomes.
MATERIALS AND METHODS
Data Sources
Data was obtained from the CIBMTR which includes a voluntary network of over 500 transplantation centers worldwide that contribute detailed HCT data to a statistical center at the Medical College of Wisconsin in Milwaukee and the National Marrow Donor Program (NMDP®) Coordinating Center in Minneapolis. Participating centers are required to report all transplants consecutively; patients are followed longitudinally and compliance is monitored by on-site audits. Observational studies conducted by the CIBMTR are performed in compliance with all applicable federal regulations pertaining to the protection of human research subjects and approved by the National Marrow Donor Program® (NMDP)/Be The Match® Institutional Review Board. Protected Health Information used in the performance of such research is collected and maintained in CIBMTR’s capacity as a Public Health Authority under the Health Insurance Portability Accountability Privacy Rule.
Patient Selection
An initial cohort of 4777 adult patients (≥18 years) with a diagnosis of AML receiving their first allogeneic HCT between 2013 and 2017 and reported to the CIBMTR were included. Patients receiving a graft from a human leukocyte antigen (HLA)-matched sibling, fully matched unrelated donor, umbilical cord blood (UCB) graft and haploidentical donors were eligible for study. Patients with active disease (i.e., ≥5% bone marrow blasts prior to transplantation or those with evidence of extra medullary disease), remission status beyond CR1, a diagnosis of acute promyelocytic leukemia and recipients of syngeneic or mismatched unrelated donor grafts were excluded.
Cytogenetic and molecular abnormalities present at the time of diagnosis or prior to initiation of the conditioning regimen were reported to the CIBMTR. When required, additional review of reported genetic data was performed by three reviewers (AJ, DW and KC) to adjudicate any uncertainties in classification. Cases with incomplete genetic data (N=152) were excluded. Additionally, subjects with incomplete research consent forms and those from embargoed centers were also excluded. Ultimately, a cohort of 2289 transplant recipients from 163 centers was analyzed.
Cytogenetic and Molecular Classification
Patients were assessed for the presence of specific chromosomal and molecular abnormalities at diagnosis and prior to HCT. Cytogenetic data reported to the CIBMTR conformed to the International System of Cytogenetic Nomenclature (20). The definition of complex karyotype (≥ 3 abnormalities) and monosomal karyotype were made according to previously published criteria. (21, 22).
Comprehensive molecular information (i.e., FLT3-ITD, NPM1, CEBPA, ASXL1, RUNX1, TP53, DNMT3A, BCR-ABL1, IDH1 and IDH2 mutational status obtained by next-generation sequencing [NGS]) or PCR was reported to the CIBMTR beginning in 2013. Information regarding FLT3-ITD allelic ratio and CEBPA allelic status is not regularly reported to the CIBMTR, and was therefore not included in our classification. On the basis of available registry molecular and conventional cytogenetic data, we defined an adapted ELN genetic risk stratification (aELN) (Table 1) whereby patients were stratified into three distinct groups: favorable (Fav, N=181), intermediate (IM, N=1185) and adverse (Adv, N=923).
Table 1.
Adapted* ELN (aELN) risk stratification
| Favorable | t(8;21)(q22;q22.1); RUNX1-RUNX1T1 |
| inv(16)(p13.1q22) or t(16;16)(p13.1;q22); CBFB-MYH11 | |
| Mutated NPM1 without FLT3-ITD | |
| Mutated CEBPA | |
| Intermediate | Mutated NPM1 and FLT3-ITD |
| Wild-type NPM1 without FLT3-ITD | |
| t(9;11)(p21.3;q23.3); MLLT3-KMT2A | |
| Cytogenetic abnormalities not classified as favorable or adverse | |
| Adverse | t(6;9)(p23;q34.1); DEK-NUP214 |
| t(v;11q23.3); KMT2A rearranged | |
| t(9;22)(q34.1;q11.2); BCR-ABL1 | |
| inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2); GATA2,MECOM | |
| −5 or del(5q); −7; −17/abn(17p); mutated TP53 | |
| Complex karyotype (≥ 3 abnormalities) | |
| Monosomal karyotype | |
| Wild-type NPM1 and FLT3-ITD | |
| Mutated RUNX1 without good risk karyotype | |
| Mutated ASXL1 without good risk karyotype |
FLT3-ITD allelic ratio and CEBPA mono/bi-allelic status not available
The aELN classified patients with mutated NPM1 without FLT3-ITD and those with mutated CEBPA (without clarity of bi/mono allelic status) as favorable. Patients with mutated NPM1 with FLT3-ITD (irrespective of allelic ratio) and those with wild type NPM1 without FLT3-ITD were classified as intermediate. Patients with FLT3-ITD (irrespective of allelic ratio) and wild-type NPM1 were classified as adverse. Other chromosomal and molecular abnormalities were classified according to the 2017 ELN genetic risk criteria.
Study Endpoints and Variables
Disease-free survival (DFS, defined as time to relapse or death from any cause, with surviving patients in CR censored at last follow-up) was the study’s primary endpoint. Secondary endpoints were: overall survival (OS, defined as time to death from any cause with surviving patients censored at last follow-up), non-relapse mortality (NRM, defined as death without preceding disease relapse/progression, relapse being a competing event), relapse (any reported events of leukemia relapse with NRM as competing event), acute and chronic graft vs. host disease (GVHD, with death as competing risk (23). Patients were censored at subsequent HCT or last follow-up alive.
The study main effect was the influence of the aELN genetic risk group. Additional variables considered in the multivariate analysis (MVA) were: age at transplant, gender, race, Karnofsky performance score (KPS), HCT comorbidity index (HCT-CI), clinical onset of AML (de novo vs. transformed vs. therapy-related), time to achieve first remission, measurable residual disease (MRD) status at transplantation, conditioning intensity, graft source, donor type and in vivo T-cell depletion (anti-thymocyte globulin [ATG] or alemtuzumab). Given intrinsic biologic and treatment-related differences between younger (i.e., <60 years) and older (i.e., ≥60 years) patients and previous reports demonstrating age-related differences in the distribution and impact of specific genetic abnormalities (9, 24), post-transplant outcomes were also analyzed separately between differing age cohorts. We also identified the most commonly occurring abnormalities (i.e., reported in at least >100 patients) within the adverse aELN genetic group, and evaluated specific outcomes for these larger individual subsets.
Statistical Analysis
Univariate analysis was performed using the Kaplan-Meier Method and compared using the log-rank test for OS and DFS, while acute/chronic GVHD, NRM, and relapse used the cumulative incidence method considering competing risks, with comparisons performed using Gray’s competing hazard method (25). MVA was performed using the Cox proportional hazard model (26) for OS and DFS. The main effect of the aELN risk group was retained in all models. The assumption of proportional hazards for each factor in the Cox model was tested by adding time-dependent covariates as necessary. When the test indicated differential effects over time (non-proportional hazards), models were constructed breaking the post-transplant time course into two periods, using the maximized partial likelihood method to find the most appropriate breakpoint. A backward stepwise model selection approach was used to identify significant risk factors. Factors which were significant at a 5% level were kept in the final model. Potential interaction between the main effect and significant co-variates was also tested for all endpoints. Adjusted probabilities of DFS and OS and adjusted cumulative incidence functions of NRM and relapse were calculated using the MVA models, stratified on main effect and weighted by the pooled sample proportion value for each prognostic factor. These adjusted probabilities estimate likelihood of outcomes in populations with similar prognostic factors. All analyses were done using the statistical package SAS version 9.4 (Cary, NC).
RESULTS
Demographics
Baseline clinical features of all 2289 patients are presented in Table 2. The median age at HCT was 57.2 years (IQR 19.2–74.2 years). Importantly, 41% (936 patients) were ≥60 years, 76% (n=1743) had de novo AML and 49% (1115) received a myeloablative conditioning (MAC) regimen. More patients in the Adv group (53% vs. 41.8% for Fav/IM patients, p<0.001) had high HCT-CI. The frequency of secondary AML or therapy-related disease differed amongst groups (Fav 18.8%, IM 22.2% and Adv 26.9%; p=0.02). Pre-transplant measurable residual disease (MRD) was also significantly different between groups (Fav 21%, IM 11.5% and Adv 22.9%; p<0.001), however morphologic complete remission with incomplete blood count recovery (CRi) status was comparable for all cohorts (p=0.29).
Table 2.
Patient, disease and transplant characteristics
| Characteristic | Favorable | Intermediate | Adverse | P Value |
|---|---|---|---|---|
|
| ||||
| No. of patients | 181 | 1185 | 923 | |
| No. of centers | 66 | 144 | 125 | |
| Patient related | ||||
|
| ||||
| Age at HCT - no. (%) | 0.01a | |||
| Median (range) years | 57.3 (18.4–73.5) | 58.1 (19.6–74) | 56.2 (19–74.4) | |
| 18–29 | 12 (6.6) | 97 (8.2) | 104 (11.3) | |
| 30–59 | 88 (48.6) | 96 (48.9) | 473 (51.3) | |
| 60–69 | 63 (34.8) | 417 (35.2) | 287 (31.1) | |
| >=70 | 18 (9.9) | 92 (7.8) | 59 (6.4) | |
| Gender - no. (%) | 0.16a | |||
| Male | 95 (52.5) | 615 (51.9) | 517 (56) | |
| Female | 86 (47.5) | 570 (48.1) | 406 (44) | |
| Race - no. (%) | < 0.001a | |||
| Caucasian | 138 (76.2) | 942 (79.5) | 718 (77.8) | |
| African-American | 6 (3.3) | 70 (5.9) | 101 (10.9) | |
| Asian/Pacific Islander | 27 (14.9) | 110 (9.3) | 70 (7.6) | |
| Other/missing | 10 (5.6) | 63 (5.3) | 34 (3.7) | |
| Performance Score - no. (%) | 0.09a | |||
| <90 | 64 (35.4) | 434 (36.6) | 386 (41.8) | |
| >=90 | 115 (63.5) | 739 (62.4) | 524 (56.8) | |
| Missing | 2 (1.1) | 12 (1) | 13 (1.4) | |
| HCT-CI - no. (%) | < 0.001a | |||
| 0 | 39 (21.5) | 245 (20.7) | 171 (18.5) | |
| 1–2 | 62 (34.3) | 409 (34.5) | 249 (27) | |
| 3+ | 74 (40.9) | 497 (41.9) | 489 (53) | |
| Missing | 6 (3.3) | 34 (2.9) | 14 (1.5) | |
| Disease related | ||||
|
| ||||
| White blood count at diagnosis, x109/L | < 0.001a | |||
| Median (range) | 13.9 (0–270.5) | 7.2 (0–329.7) | 5 (0.4–290.9) | |
| <= 10 | 73 (40.3) | 597 (50.4) | 528 (57.2) | |
| 10 – 100 | 86 (47.5) | 396 (33.4) | 275 (29.8) | |
| > 100 | 14 (7.7) | 114 (9.6) | 57 (6.2) | |
| Missing | 8 (4.4) | 78 (6.6) | 63 (6.8) | |
| Clinical onset of AML - no. (%) | 0.02a | |||
| De novo | 147 (81.2) | 922 (77.8) | 674 (73) | |
| Transformed from MDS/MPN | 23 (12.7) | 188 (15.9) | 161 (17.4) | |
| Therapy related | 11 (6.1) | 75 (6.3) | 88 (9.5) | |
| CRi status pre-HCT - no. (%) | 0.29a | |||
| Yes | 49 (27.1) | 328 (27.7) | 269 (29.1) | |
| No | 132 (72.9) | 830 (70) | 636 (68.9) | |
| Missing | 0 | 27 (2.3) | 18 (2) | |
| MRD pre-HCT+ - no. (%) | < 0.001a | |||
| No | 128 (70.7) | 917 (77.4) | 666 (72.2) | |
| Yes | 38 (21) | 136 (11.5) | 211 (22.9) | |
| Missing | 15 (8.3) | 132 (11.1) | 46 (5) | |
| Transplant related | ||||
|
| ||||
| Conditioning intensity - no. (%) | 0.12a | |||
| MAC | 91 (50.3) | 547 (46.2) | 477 (51.7) | |
| RIC/NMA | 90 (49.7) | 637 (53.8) | 446 (48.3) | |
| Missing | 0 | 1 (0.1) | 0 | |
| Graft type - no. (%) | 0.62a | |||
| Bone marrow | 34 (18.8) | 173 (14.6) | 135 (14.6) | |
| Peripheral blood | 119 (65.7) | 827 (69.8) | 635 (68.8) | |
| Cord blood | 28 (15.5) | 185 (15.6) | 153 (16.6) | |
| Donor type - no. (%) | 0.68a | |||
| HLA-identical sibling | 49 (27.1) | 325 (27.4) | 223 (24.2) | |
| Other related | 14 (7.7) | 99 (8.4) | 69 (7.5) | |
| Haploidentical | 23 (12.7) | 124 (10.5) | 117 (12.7) | |
| HLA-Matched unrelated (8/8) | 67 (37) | 452 (38.1) | 361 (39.1) | |
| Cord blood | 28 (15.5) | 185 (15.6) | 153 (16.6) | |
| GVHD prophylaxis - no. (%) | 0.24a | |||
| Ex vivo T-cell depletion/CD34 selection | 10 (5.5) | 49 (4.1) | 55 (6) | |
| PTCy | 24 (13.3) | 179 (15.1) | 158 (17.1) | |
| TAC based | 111 (61.3) | 713 (60.2) | 554 (60) | |
| CSA based | 33 (18.2) | 230 (19.4) | 149 (16.1) | |
| Other | 3 (1.7) | 14 (1.2) | 7 (0.8) | |
| In vivo T-cell depletion - no. (%) | 0.46a | |||
| No | 137 (75.7) | 944 (79.7) | 733 (79.4) | |
| ATG/Alemtuzumab | 44 (24.3) | 241 (20.3) | 190 (20.6) | |
| Year of HCT - no. (%) | 0.15a | |||
| 2013 | 34 (18.8) | 220 (18.6) | 139 (15.1) | |
| 2014 | 36 (19.9) | 275 (23.2) | 237 (25.7) | |
| 2015 | 36 (19.9) | 278 (23.5) | 208 (22.5) | |
| 2016 | 47 (26) | 248 (20.9) | 188 (20.4) | |
| 2017 | 28 (15.5) | 164 (13.8) | 151 (16.4) | |
| Follow-up of survivors - median (range) months | 24.6 (0.4–62.8) | 35.3 (0.2–69.4) | 35.1 (0.4–63.3) | |
Hypothesis testing:
Pearson chi-square test
MRD was assessed by different methods including, but not limited to: next generation sequencing (NGS), polymerase chain reaction (PCR) testing, chromosomal / genomic microarray analysis, fluorescence in situ hybridization (FISH), karyotyping, flow cytometry.
Abbreviations: AML, acute myeloid leukemia; ATG, anti-thymocyte globulin; CMV, cytomegalovirus; CRi, complete remission with incomplete blood count recovery; CSA, cyclosporine; HCT, hematopoietic cell transplant; HiDAC, high-dose cytarabine; HMA, hypomethylating agent; IDAC; intermediate-dose cytarabine; MAC, myeloablative conditioning; MDS, myelodysplastic syndrome; aELN, adapted European LeukemiaNet; MMF, mycophenolate mofetil; MPN, myeloproliferative neoplasm; MRD, measurable residual disease; NMA, non-myeloablative; PTCy, post-transplant cyclophosphamide; RIC, reduced intensity conditioning; TAC, tacrolimus.
Of 923 patients with Adv risk, 380 (41.1%) had −5/del5q, or monosomy 7; 158 (17.1%) had wild-type NPM1 with mutated FLT3-ITD; 139 (15.06%) carried a t(v;11q23.3); KMT2A rearrangement and 137 patients (14.84%) had a complex karyotype (Table 3). Complex karyotype included all other Adv-risk patients (without del5/del7, wtNPM1 with FLT3-ITD or t(v;11q23.3) KMT2A rearrangement) having multiple (≥3) chromosomal abnormalities.
Table 3.
3A. Adverse-risk patient characteristics by genetic subgroups 3B. Multivariate Analysis of Outcomes. Comparison of abnormal 5/7 subgroup vs. Other Adverse aELN risk group*
| 3A. | |||
| Characteristic | Abnormal 5/7 | * Others | P Value |
|
| |||
| No. of patients | 380 | 543 | |
| Age at HCT - no. (%) | < 0.001a | ||
| Median (range) | 59.81 (18.98–75.84) | 51.92 (18.96–73.76) | |
| 18–59 | 195 (51.3) | 382 (70.3) | |
| >=60 | 185 (48.7) | 161 (29.7) | |
| Conditioning intensity - no. (%) | < 0.001a | ||
| MAC | 171 (45) | 306 (56.4) | |
| RIC/NMA | 209 (55) | 237 (43.6) | |
| MRD at time of HCT - no. (%) | 0.003a | ||
| No | 255 (67.1) | 411 (75.7) | |
| Yes | 108 (28.4) | 103 (19) | |
| Missing | 17 (4.5) | 29 (5.3) | |
| CRi status prior to conditioning - no. (%) | 0.06a | ||
| Yes | 127 (33.4) | 142 (26.2) | |
| No | 246 (64.7) | 390 (71.8) | |
| Missing | 7 (1.8) | 11 (2) | |
| 3B. | |||
| HR (95% CI) | p-value | ||
|
| |||
| Overall survival | 1.58 (1.32–1.90) | <0.0001 | |
| Disease-free survival | 1.54 (1.30–1.83) | <0.0001 | |
| Relapse | 1.59 (1.28–1.97) | <0.0001 | |
Reference group. Includes wtNPM1 with FLT3-ITD, t(v;11q23.3), DEK-NUP214, complex karyotype, BCR-ABL1, GATA2, MECOM, mutated TP53, non-del5/del7 monosomy, mutated RUNX1/ASXL1 without good-risk karyotype
Pearson chi-square test
Abbreviations: CRi, complete remission with incomplete blood count recovery; GVHD, graft-vs.-host disease; HCT, hematopoietic cell transplant; MAC, myeloablative conditioning; aELN, adapted European LeukemiaNet; MRD, measurable residual disease; NMA, non-myeloablative; RIC, reduced intensity conditioning.
Transplant outcomes by aELN classification
Overall, 1237 of 2289 patients (54%) were alive at last follow up with a median survival of 37 months (IQR, 24–51 months) and 1031 (83% of the survivors; 45% of the whole group) were alive and disease-free. Median follow up for survivors was 35 months.
Disease Relapse
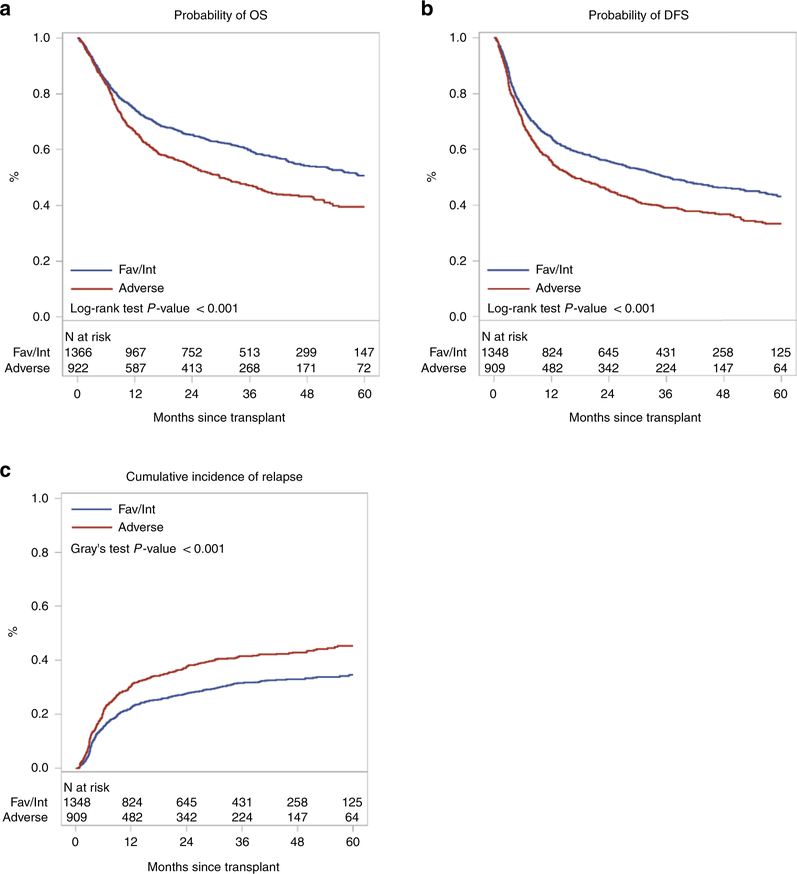
Cumulative incidence of relapse at two-years was 28% (95% confidence interval [CI], 21.5–35.1%), 27.5 % (24.9–30.1%) and 37.5% (34.3–40.7%) for Fav, IM and Adv cohorts, respectively (p<0.001) (Table 4, Figure 1C).
Table 4.
Univariate outcomes by aELN risk group
| Favorable (N = 181) | Intermediate (N = 1185) | Adverse (N = 923) | |||||
|---|---|---|---|---|---|---|---|
| Outcomes | N | Prob (95% CI) | N | Prob (95% CI) | N | Prob (95% CI) | P Value |
|
| |||||||
| Overall survival* | 181 | 1185 | 923 | <0.001 | |||
| 67.7 (60.5–74.6)% | 64.9 (62.1–67.6)% | 53.9 (50.6–57.2)% | |||||
| Disease free survival* | 178 | 1170 | 910 | <0.001 | |||
| 57.8 (50.3–65.2)% | 55.5 (52.6–58.4)% | 45.3 (42–48.6)% | |||||
| Relapse* | 178 | 1170 | 910 | <0.001 | |||
| 28 (21.5–35.1)% | 27.5 (24.9–30.1)% | 27.5 (24.9–30.1)% | |||||
| Non-relapse mortality* | 178 | 1170 | 910 | 0.467 | |||
| 14.2 (9.3–19.8)% | 17.1 (14.9–19.3)% | 17.3 (14.9–19.8)% | |||||
| Grade II-IV acute GVHD+ | 181 | 1171 | 913 | 0.423 | |||
| 30.9 (24.4–37.9)% | 30.9 (24.4–37.9)% | 35 (32–38.2)% | |||||
| Chronic GVHD* | 181 | 1177 | 918 | 0.442 | |||
| 40 (32.7–47.4)% | 43.6 (40.7–46.6)% | 40.2 (37–43.5)% | |||||
Outcomes at:
2 years
100 days.
Abbreviations: GVHD, graft-vs.-host disease; aELN, adapted European LeukemiaNet.
Figure 1.
OS (A), DFS (B) and cumulative incidence of disease relapse (C) for AML transplant recipients within favorable/intermediate vs. adverse aELN cohorts
Abbreviations: DFS, disease-free survival; aELN, adapted European LeukemiaNet; OS, overall survival.
MVA confirmed that Adv risk patients had higher relapse rates (HR for Adv vs. Fav/IM: 1.47, 95% CI 1.28–1.70, p<0.0001) with no differences observed between the Fav and IM groups; thus they were combined for subsequent analysis of other endpoints. Variables of interest considered in the MVA are listed in Supplemental Table S1.
Important variables independently associated with higher rates of relapse included: pre-transplant MRD (29.2% for MRD-negative [MRDneg] vs. 42% for MRD-positive [MRDpos]; HR: 1.47, 95% CI 1.23–1.76, p<0.0001); AML onset (29.9% for de novo vs. 40.1% for secondary and therapy-related disease; HR: 1.43, 95% CI 1.19–1.72, p=0.0001) and conditioning intensity (26.2% for MAC vs. 36.9% for non-myeloablative/reduced intensity regimens [NMA/RIC]; HR: 1.58, 95% CI 1.37–1.83, p<0.0001, Supplemental Table S2).
While patients receiving RIC/NMA conditioning experienced higher rates of relapse, the impact of aELN Adv risk on relapse was observed for patients receiving either MAC or RIC/NMA (p<0.001 for both).
Overall and Disease-Free Survival
There were significant differences in 2-year OS (Fav: 67.7% [95% CI, 60.5–74.6%], IM: 64.9% [62.1–67.6%] and Adv: 53.9% [50.6–57.2%]; p<0.001) and DFS (Fav: 57.8% [95% CI, 50.3–65.2%], IM: 55.5% [52.6–58.4%] and Adv: 45.3% [42–48.6%]; p<0.001) among aELN groups. (Table 4, Figure 1A, 1B). As above, Fav/IM were combined for further analysis.
The presence of pre-transplant MRD was associated with inferior two-year OS (63.7% for MRDneg vs. 46.6% for MRDpos, p<0.001) and DFS (54.4% for MRDneg vs. 37.4% for MRDpos, p<0.001). MAC led to superior two-year DFS (57.2 % vs. 46% for NMA/RIC regimens, p<0.001) and OS (66 % vs. 55.6% for MAC and NMA/RIC regimens, p<0.001).
Adv aELN was shown on MVA to be higher risk for both OS (HR for Adv vs. Fav/IM: 1.39, 95% CI 1.22–1.57, p<0.001) and DFS (HR for Adv vs. Fav/IM: 1.35, 95% CI 1.20–1.51, p<0.001, Table 5). Other variables independently associated with both OS and DFS included: age at HCT, pre-transplant MRD, donor type, clinical onset of AML and WBC at diagnosis (p≤0.01 for all, Supplemental Table S2).
Table 5:
Multivariate Analysis of outcomes. Comparison of Adverse vs. Favorable/Intermediate* aELN risk groups
| HR (95% CI) | p-value | |
|---|---|---|
|
| ||
| Overall survival | 1.39 (1.22–1.57) | <0.001 |
| Disease-free survival | 1.35 (1.20–1.51) | <0.001 |
| Relapse | 1.47 (1.28–1.70) | <0.001 |
| Non-relapse mortality | 1.01 (0.81–1.19) | 0.89 |
| Grade II-IV acute GVHD | 1.04 (0.91–1.19) | 0.53 |
| Chronic GVHD | 0.95 (0.83–1.08) | 0.42 |
Reference group
Abbreviations: GVHD, graft-vs.-host disease; aELN, adapted European LeukemiaNet.
Non-relapse Mortality and Graft-versus-Host Disease
There were no significant differences in NRM among aELN groups (two-year NRM: Fav: 14.2% [95% CI, 9.3–19.8%], IM: 17.1 % [95% CI, 14.9–19.2%] and Adv: 17.3 % [95% CI, 14.9–19.8%]; p=0.46). Similarly, aELN classification had no impact on the incidence of grade 2–4 acute GVHD (Fav: 30.9 % [95% CI, 24.4–37.9%], IM: 35.4 % [32.7–38.2%] and Adv: 35% [32–38.2%]; p=0.42) or chronic GVHD (Fav: 40% [32.7–47.4%], IM: 43.6 % [40.7–46.6%] and Adv: 40.2% [37–43.5%]; p=0.44).
Comparison of outcomes by aELN risk group and age
There were no significant differences in relapse rates among age groups (HR for Fav/IM ≥60 vs. <60: 1.19 [0.98–1.44] p=0.08; HR for Adv ≥60 vs. <60: 1.17 [0.95–1.44] p=0.15), but MVA confirmed higher NRM for older patients (≥60 y/o) in both Fav/IM (HR 1.40 [1.10–1.78] p=0.007) and Adv risk cohorts (HR 1.59 [1.18–2.13] p=0.002). Clinical outcomes were separately analyzed for two cohorts of younger patients (i.e., 18–39 vs. 40–59 years). MVA demonstrated inferior OS (HR 1.44 [1.18–1.75] p=0.0003) and DFS (HR 1.23 [1.03–1.46] p=0.02) for the 40–59 group. No significant differences in NRM or relapse were identified between these two younger cohorts. (Supplemental Table S2)
Outcomes by Genetic subsets within the aELN Adv Risk Cohort
Genetic subset comparisons within the Adv-risk group showed that patients harboring monosomy 5, del(5q) or monosomy 7 (abnormal 5/7 subgroup) had inferior 2-year OS (42.6%, p<0.001) and DFS (35.2%, p<0.001), as well as higher rates of relapse (45%, p= 0.002) when compared to other patients within the Adv risk cohort (OS 60%, DFS 50.8%, relapse 33.5%).
Patients in the abnormal 5/7 subgroup were older (median age 59.8 [IQR 18.92–75.84] vs. 51.9 [IQR 18.9–73.7] for other Adv-risk patients, p<0.001) and less often received MAC (45% MAC vs. 56.4% MAC for other Adv-risk patients p<0.001). Patients in the abnormal 5/7 subgroup also had a higher prevalence of pre-transplant MRD (28.4% vs. 19%, p=0.003) when compared to other Adv risk patients (p<0.001). MVA confirmed that abnormal 5/7 patients had higher relapse rates and inferior DFS and OS compared to other Adv risk patients (p<0.0001 for all. Table 3B).
DISCUSSION
This large registry analysis of patients with AML undergoing allogeneic HCT demonstrates robust prognostic stratification by aELN in regard to relapse, DFS and OS. Inferior survival in the Adv-risk cohort appears to be driven by higher relapse rates and ensuing inferior DFS. There were no significant interactions between the main effect of aELN risk group and any significant risk factors. This confirms that the aELN classification is applicable to all patients receiving HCT in CR1, both younger (<60 years) and older (≥60 years) and for patients with de novo and transformed/secondary AML. Additionally, the impact of adverse genetics is not overcome by greater conditioning intensity.
Previous reports examining the prognostic ability of ELN2017 as compared to ELN2010 support the new combined genetic prognostic model for treatment of newly diagnosed AML patients (14–17) and a recent single institution analysis also evaluated the prognostic impact of ELN2017 following HCT (18). Some of these studies are limited by patient heterogeneity, small sample sizes or different consolidation strategies. We assessed the impact of a combined genetic model exclusively in the transplant setting and restricted our analysis to patients receiving allogeneic HCT in CR1. While the effect of genetic abnormalities continues to be important beyond CR1, prior relapse, particularly early relapse, carries a heavier prognostic weight than most genetic abnormalities (27).
Fav and IM risk groups were combined in our study after pairwise multivariable analysis demonstrated no significant differences in any outcomes. This could, in part, be explained by a smaller number of Fav-risk AML patients proceeding to HCT in CR1, but perhaps most importantly by the presence of non-genetic, adverse clinical features in the Fav risk cohort (i.e., elevated WBC at diagnosis, failed induction, pre-transplant MRD, etc.) which could have resulted in early transplantation referral. Thus, due to inherent clinical differences between Fav patients referred to HCT and those with newly diagnosed disease, aELN distinguished only two (and not three) distinct prognostic groups in the HCT setting.
Despite inferior outcomes compared to the Fav/IM group, the overall survival for Adv patients was still 52% at 2 years, a modest difference from the Fav/IM group; but most importantly, significantly better than reported without allogeneic HCT. Herold and colleagues recently validated the ELN2017 risk stratification among 1116 adult AML patients enrolled on two multicenter phase III trials of the German AML Cooperative Group (16). Two-year survival for Adv patients was 22.1% (20.6% following chemotherapy or autoHCT consolidation for patients in CR1). Several cooperative and single-institution studies have also reported very poor outcomes for Adv risk patients receiving post-remission therapies other than allogeneic HCT (14, 15). While clear differences in outcomes were noted between Adv and Fav/IM groups, prognostic separation was clear when either were compared to patients receiving non-transplant consolidation. It is not unreasonable to suggest that allogeneic transplantation could have a partial ‘equalizing’ effect amongst different genetic subsets.
We identified the most commonly occurring genetic abnormalities (i.e., >100 cases) within the Adv aELN group. As reported, (18, 28) we confirmed that patients carrying monosomy 5, del(5q) or monosomy 7 (abnormal 5/7) had significantly higher rates of post-transplant relapse and inferior survival, even when transplanted in CR1. Monosomy 7 was the most common individual abnormality reported, with worse outcomes noted in the context of monosomal karyotype. Patients in the abnormal 5/7 subgroup were older and received less aggressive conditioning; however, the adverse impact of 5/7 was seen across all age groups and also in patients receiving MAC.
Unfortunately we could not assess in our analysis the clinical impact of post-transplant maintenance and preemptive strategies in our cohort or elucidate the indications for HCT in Fav/IM risk patients. Additional study limitations were intrinsic to a retrospective cohort analysis, including incomplete genetic data for some patients and lack of uniformity for MRD assessment methods among multiple institutions.
The impact of pre-HCT MRD has been previously reported by several groups (29–32). The presence of MRD correlates with increased post-transplant relapse, particularly in patients receiving RIC/NMA regimens. While we observed a higher proportion of pre-HCT MRD in the Adv aELN group, the presence of MRDpos was independently associated with higher post-HCT relapse across the aELN subsets. The patients in this study were transplanted prior to the publication of the ELN consensus guidelines for AML MRD assessment however, even without interlaboratory or detection technology standardization, residual disease pre-HCT was consistently associated with more relapse.
In conclusion, this adapted ELN classification had prognostic value for survival and relapse after allogeneic HCT. Novel peri-transplant pre-emptive and therapeutic strategies to reduce relapse remain necessary for all groups, but particularly those within the Adv cohort, those at highest-risk.
Supplementary Material
ACKNOWLEDGEMENTS
Data sharing:
CIBMTR supports accessibility of research in accord with the National Institutes of Health (NIH) Data Sharing Policy and the National Cancer Institute (NCI) Cancer Moonshot Public Access and Data Sharing Policy. The CIBMTR only releases de-identified datasets that comply with all relevant global regulations regarding privacy and confidentiality.
Additional contributing co-authors from the writing committee:
Kehinde Adekola, Ibrahim Ahmed, Talha Badar, Ashish Bajel, Qaiser Bashir, Chris Bredeson, Bruce Camitta, Jan Cerny, Stefan Ciurea, Corey S. Cutler, Shahinaz Gadalla, Sid Ganguly, Saar Gill, Nasheed Hossain, Luis Isola, Mark Juckett, Mary Laughlin, Hillard Lazarus, Jong Wook Lee, Richard Lin, Hongtao Liu, Vikram Matthews, Shahram Mori, Reinhold Munker, Ryotaro Nakamura, Miguel-Angel Perales, Marjolein van der Poel, Uday Popat, Jeffrey Pu, Marcie Riches, Olle Ringdén, David Rizzieri, Mitchell Sabloff, Lynn Savoie, Bart Scott, Anthony Stein, Masumi Ueda, Celalettin Ustun, Eric Wong.
The CIBMTR is supported primarily by Public Health Service U24CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); U24HL138660 from NHLBI and NCI; OT3HL147741, and U01HL128568 from the NHLBI; HHSH250201700006C, and HHSH250201700007C from the Health Resources and Services Administration (HRSA); and N00014–20-1–2705 and N00014–20-1–2832 from the Office of Naval Research; Additional federal support is provided by P01CA111412, R01CA152108, R01CA215134, R01CA218285, R01CA231141, R01AI128775, R01HL126589, R01HL129472, R01HL130388, R01HL131731, U01AI069197, U01AI126612, UG1HL06924, and BARDA. Support is also provided by Be the Match Foundation, Boston Children’s Hospital, Dana Farber, St. Baldrick’s Foundation, Stanford University, the Medical College of Wisconsin the National Marrow Donor Program, and from the following commercial entities: Actinium Pharmaceuticals, Inc.; Adienne SA; Allovir, Inc.; Amgen, Inc.; Angiocrine Bioscience; Astellas Pharma US; bluebird bio, Inc.; Bristol Myers Squibb Co.; Celgene Corp.; CSL Behring; CytoSen Therapeutics, Inc.; Daiichi Sankyo Co., Ltd.; ExcellThera; Fate Therapeutics; Gamida-Cell, Ltd.; Genentech Inc; Incyte Corporation; Janssen/Johnson & Johnson; Jazz Pharmaceuticals, Inc.; Kiadis Pharma; Kite, a Gilead Company; Kyowa Kirin; Legend Biotech; Magenta Therapeutics; Merck Sharp & Dohme Corp.; Millennium, the Takeda Oncology Co.; Miltenyi Biotec, Inc.; Novartis Pharmaceuticals Corporation; Omeros Corporation; Oncoimmune, Inc.; Orca Biosystems, Inc.; Pfizer, Inc.; Pharmacyclics, LLC; Sanofi Genzyme; Stemcyte; Takeda Pharma; Vor Biopharma; Xenikos BV. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration (HRSA) or any other agency of the U.S. Government.
COMPETING INTERESTS
Dr. Assal reports grants and personal fees from Incyte Corporation, personal fees from Boston Biomedical, personal fees from Alpha Insights, outside the submitted work.
Dr. Bejanyan reports personal fees from Magenta therapeutics, outside the submitted work.
Dr. Cahn reports Advisory Boards with Agios, AbbVie, Otsuka, Race Oncology.
Dr. Bhatt reports personal fees from Agios, grants and personal fees from Incyte, personal fees from Takeda, personal fees from Partner Therapeutics, personal fees from Omeros, grants and personal fees from Abbvie, grants from Jazz, grants from National Marrow Donor Program, other from Oncoceutics, personal fees from Partnership for health analytic research, LLC, grants and other from Pfizer, personal fees from CSL Behring, grants from Tolero Pharmaceuticals, personal fees from Rigel Pharmaceuticals, other from Novartis, personal fees from Genentech, outside the submitted work.
Dr. de Lima reports grants from Pfizer, grants from Celgene, personal fees from Kadmon, personal fees from Pfizer, personal fees from Incyte, personal fees from BMS, outside the submitted work.
Dr. Gale is a consultant to BeiGene Ltd., Kite Pharma Inc., Fusion Pharma LLC, LaJolla NanoMedical Inc., Mingsight Parmaceuticals Inc. and CStone Pharmaceuticals; Medical Director, FFF Enterprises Inc.; Partner, AZCA Inc.; Board of Directors, RakFond Foundation for Cancer Research Support; Scientific Advisory Board, Antegene Biotech LLC and StemRad Ltd.
Dr. Grunwald reports personal fees from Abbvie, personal fees from Agios, personal fees from Amgen, personal fees from Cardinal Health, personal fees from BMS/Celgene, personal fees from Daiichi Sankyo, personal fees and other from Incyte, personal fees from Merck, personal fees from Pfizer, personal fees from Premier, personal fees from Karius, other from Forma Therapeutics, other from Genentech/Roche, other from Janssen, personal fees from Astellas, personal fees from Trovagene, personal fees from Stemline, personal fees from Gilead, outside the submitted work.
Dr. Hildebrandt reports other from Pfizer, other from Kite Pharma, other from Incyte, other from Jazz Pharmaceuticals, other from Alexion Pharmaceutical, other from Karyopharm, other from Pharmacyclics, other from Takada, other from Jazz Pharmaceuticals, other from Kite Pharma, other from Incyte, other from Pfizer, other from Falk Foundation, other from Astellas Pharma, outside the submitted work.
Dr. Hourigan reports other from Sellas, outside the submitted work.
Dr. Kansagra reports other from Takeda, other from Jansen, other from Pfizer, other from Celgene/BMS, other from Sanofi, other from Alynylam, other from GSK, other from Karyopharm, other from Oncopeptide, outside the submitted work.
Dr. Kharfan-Dabaja reports other from Daiichi Sankyo, other from Pharmacyclics, outside the submitted work.
Dr. McGuirk reports grants and personal fees from AlloVir HCP, grants and personal fees from Juno Therapeutics, Inc, grants and personal fees from Gilead-Kite Pharmaceuticals, grants and other from Magenta Therapeutics, other from EcoR1 Cap, outside the submitted work.
Dr. Mishra reports grants from Novartis, from null, outside the submitted work.
Dr. Mussetti reports grants from Gilead, personal fees from Novartis, Brystol-Myers Squibb, outside the submitted work.
Dr. Patel reports personal fees from Kite Pharma, outside the submitted work.
Dr. Olsson reports personal fees from AstraZeneca, outside the submitted work.
Dr. Saad reports personal fees from Magenta Therapeutics, personal fees from Incyte Pharmaceuticals, personal fees from CareDx, outside the submitted work.
Dr. Sharma reports clinical trial salary support from Vertex Pharmaceuticals, CRISPR Therapeutics, Novartis paid to his institution, and personal consultancy fees from Spotlight Therapeutics, outside the submitted work.
All other authors declare no potential competing interests.
REFERENCES
- 1.Sorror ML, Maris MB, Storb R, Baron F, Sandmaier BM, Maloney DG, et al. Hematopoietic cell transplantation (HCT)-specific comorbidity index: a new tool for risk assessment before allogeneic HCT. Blood. 2005;106(8):2912–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koreth J, Schlenk R, Kopecky KJ, Honda S, Sierra J, Djulbegovic BJ, et al. Allogeneic stem cell transplantation for acute myeloid leukemia in first complete remission: systematic review and meta-analysis of prospective clinical trials. JAMA. 2009;301(22):2349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96(13):4075–83. [PubMed] [Google Scholar]
- 4.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–65. [DOI] [PubMed] [Google Scholar]
- 5.Dholaria B, Savani BN, Hamilton BK, Oran B, Liu HD, Tallman MS, et al. Hematopoietic Cell Transplantation in the Treatment of Newly Diagnosed Adult Acute Myeloid Leukemia: An Evidence-Based Review from the American Society of Transplantation and Cellular Therapy. Biol Blood Marrow Transplant. 2020. [DOI] [PubMed] [Google Scholar]
- 6.Oliansky DM, Appelbaum F, Cassileth PA, Keating A, Kerr J, Nieto Y, et al. The role of cytotoxic therapy with hematopoietic stem cell transplantation in the therapy of acute myelogenous leukemia in adults: an evidence-based review. Biol Blood Marrow Transplant. 2008;14(2):137–80. [DOI] [PubMed] [Google Scholar]
- 7.Grossmann V, Schnittger S, Kohlmann A, Eder C, Roller A, Dicker F, et al. A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood. 2012;120(15):2963–72. [DOI] [PubMed] [Google Scholar]
- 8.Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–74. [DOI] [PubMed] [Google Scholar]
- 9.Mrozek K, Marcucci G, Nicolet D, Maharry KS, Becker H, Whitman SP, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30(36):4515–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rollig C, Bornhauser M, Thiede C, Taube F, Kramer M, Mohr B, et al. Long-term prognosis of acute myeloid leukemia according to the new genetic risk classification of the European LeukemiaNet recommendations: evaluation of the proposed reporting system. J Clin Oncol. 2011;29(20):2758–65. [DOI] [PubMed] [Google Scholar]
- 11.Medeiros BC, Tian L, Robenson S, Laport GG, Johnston LJ, Shizuru JA, et al. European LeukemiaNet classification intermediate risk-1 cohort is associated with poor outcomes in adults with acute myeloid leukemia undergoing allogeneic hematopoietic cell transplantation. Blood Cancer J. 2014;4:e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oran B, Jimenez AM, De Lima M, Popat UR, Bassett R, Andersson B, et al. Age and Modified European LeukemiaNet Classification to Predict Transplant Outcomes: An Integrated Approach for Acute Myelogenous Leukemia Patients Undergoing Allogeneic Stem Cell Transplantation. Biol Blood Marrow Transplant. 2015;21(8):1405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boddu PC, Kadia TM, Garcia-Manero G, Cortes J, Alfayez M, Borthakur G, et al. Validation of the 2017 European LeukemiaNet classification for acute myeloid leukemia with NPM1 and FLT3-internal tandem duplication genotypes. Cancer. 2019;125(7):1091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada Y, Nagata Y, Kihara R, Ishikawa Y, Asou N, Ohtake S, et al. Prognostic analysis according to the 2017 ELN risk stratification by genetics in adult acute myeloid leukemia patients treated in the Japan Adult Leukemia Study Group (JALSG) AML201 study. Leuk Res. 2018;66:20–7. [DOI] [PubMed] [Google Scholar]
- 16.Herold T, Rothenberg-Thurley M, Grunwald VV, Janke H, Goerlich D, Sauerland MC, et al. Validation and refinement of the revised 2017 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia. 2020;34(12):3161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Straube J, Ling VY, Hill GR, Lane SW. The impact of age, NPM1(mut), and FLT3(ITD) allelic ratio in patients with acute myeloid leukemia. Blood. 2018;131(10):1148–53. [DOI] [PubMed] [Google Scholar]
- 18.Grimm J, Jentzsch M, Bill M, Goldmann K, Schulz J, Niederwieser D, et al. Prognostic impact of the ELN2017 risk classification in patients with AML receiving allogeneic transplantation. Blood Adv. 2020;4(16):3864–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen DK, Nieder ML, Thompson Z, Kim J, Pidala JA, Nishihori T, et al. ELN 2017 Risk Classification Predicts Survival of AML Patients Receiving Allogeneic Hematopoietic Stem Cell Transplantation. Biology of Blood and Marrow Transplantation. 2019;25(3):S112. [Google Scholar]
- 20.McGowan-Jordan J ISCN 2016: An International System for Human Cytogenomic Nomenclature (2016); Recommendations of the International Standing Human Committee on Human Cytogenomic Nomenclature Including New Sequence-based Cytogenomic: Karger; 2016.
- 21.Breems DA, Van Putten WL, De Greef GE, Van Zelderen-Bhola SL, Gerssen-Schoorl KB, Mellink CH, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J Clin Oncol. 2008;26(29):4791–7. [DOI] [PubMed] [Google Scholar]
- 22.Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002;100(13):4325–36. [DOI] [PubMed] [Google Scholar]
- 23.Przepiorka D, Weisdorf D, Martin P, Klingemann HG, Beatty P, Hows J, et al. 1994 Consensus Conference on Acute GVHD Grading. Bone Marrow Transplant. 1995;15(6):825–8. [PubMed] [Google Scholar]
- 24.Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, Schaich M, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood. 2006;107(10):4011–20. [DOI] [PubMed] [Google Scholar]
- 25.Fine JP, Gray RJ. A Proportional Hazards Model for the Subdistribution of a Competing Risk. Journal of the American Statistical Association. 1999;94(446):496–509. [Google Scholar]
- 26.Cox DR. Regression Models and Life-Tables. Journal of the Royal Statistical Society Series B (Methodological). 1972;34(2):187–220. [Google Scholar]
- 27.Breems DA, Van Putten WL, Huijgens PC, Ossenkoppele GJ, Verhoef GE, Verdonck LF, et al. Prognostic index for adult patients with acute myeloid leukemia in first relapse. J Clin Oncol. 2005;23(9):1969–78. [DOI] [PubMed] [Google Scholar]
- 28.Pasquini MC, Zhang MJ, Medeiros BC, Armand P, Hu ZH, Nishihori T, et al. Hematopoietic Cell Transplantation Outcomes in Monosomal Karyotype Myeloid Malignancies. Biol Blood Marrow Transplant. 2016;22(2):248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baron F, Labopin M, Ruggeri A, Sierra J, Robinson S, Labussiere-Wallet H, et al. Impact of detectable measurable residual disease on umbilical cord blood transplantation. Am J Hematol. 2020;95(9):1057–65. [DOI] [PubMed] [Google Scholar]
- 30.Buckley SA, Wood BL, Othus M, Hourigan CS, Ustun C, Linden MA, et al. Minimal residual disease prior to allogeneic hematopoietic cell transplantation in acute myeloid leukemia: a meta-analysis. Haematologica. 2017;102(5):865–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilleece MH, Labopin M, Yakoub-Agha I, Volin L, Socie G, Ljungman P, et al. Measurable residual disease, conditioning regimen intensity, and age predict outcome of allogeneic hematopoietic cell transplantation for acute myeloid leukemia in first remission: A registry analysis of 2292 patients by the Acute Leukemia Working Party European Society of Blood and Marrow Transplantation. Am J Hematol. 2018;93(9):1142–52. [DOI] [PubMed] [Google Scholar]
- 32.Hourigan CS, Dillon LW, Gui G, Logan BR, Fei M, Ghannam J, et al. Impact of Conditioning Intensity of Allogeneic Transplantation for Acute Myeloid Leukemia With Genomic Evidence of Residual Disease. J Clin Oncol. 2020;38(12):1273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.