Abstract
This study proposed the diagnosis of COVID-19 by means of Raman spectroscopy. Samples of blood serum from 10 patients positive and 10 patients negative for COVID-19 by RT-PCR RNA and ELISA tests were analyzed. Raman spectra were obtained with a dispersive Raman spectrometer (830 nm, 350 mW) in triplicate, being submitted to exploratory analysis with principal component analysis (PCA) to identify the spectral differences and discriminant analysis with PCA (PCA-DA) and partial least squares (PLS-DA) for classification of the blood serum spectra into Control and COVID-19. The spectra of both groups positive and negative for COVID-19 showed peaks referred to the basal constitution of the serum (mainly albumin). The difference spectra showed decrease in the peaks referred to proteins and amino acids for the group positive. PCA variables showed more detailed spectral differences related to the biochemical alterations due to the COVID-19 such as increase in lipids, nitrogen compounds (urea and amines/amides) and nucleic acids, and decrease of proteins and amino acids (tryptophan) in the COVID-19 group. The discriminant analysis applied to the principal component loadings (PC2, PC4, PC5, and PC6) could classify spectra with 87% sensitivity and 100% specificity compared to 95% sensitivity and 100% specificity indicated in the RT-PCR kit leaflet, demonstrating the possibilities of a rapid, label-free, and costless technique for diagnosing COVID-19 infection.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10103-021-03488-7.
Keywords: Raman spectroscopy, SARS-CoV-2, COVID-19, Serum, Diagnosis, Principal component analysis
Introduction
The current COVID-19 pandemic, the disease caused by the new coronavirus SARS-CoV-2, has caused enormous loss of lives [1] and negative economical effects, with significant increase in costs of medical inputs and equipments for the public and private health sectors worldwide caused by the high demand [2]. In addition, in the countries where the virus circulates, local governments oriented by the World Health Organization (WHO) implemented spreading containment measures (closing borders and airports, closing trades and non-essential services, lockdown, social distance and massive testing) as well as the obligatory use of facial masks in public space [3, 4]. According to the WHO, a measure that has an important effect in controlling the transmission and consequently virus dissemination is the massive testing.
Vertical isolation or mitigation was initially adopted in the UK and the Netherlands, which later fell due to the large number of cases. Italy and Spain took a long time to determine horizontal social isolation, also called suppression, and account for the largest number of deaths. Suppression has been successfully adopted in China and South Korea [5]. In Brazil, a mixture of mitigation, suppression and lockdown has been adopted by most of the state governors and city mayors [6], while the country’s president was not able to address the pandemic in a satisfactory way due to ideological positioning [7].
Public infectious disease control agents are faced with the need for a fast and reliable diagnostic technique for carriers of the SARS-CoV-2 virus in order to better manage the isolation of infected patients during the COVID-19 pandemic. Early diagnosis of COVID-19 also contributes in reducing the chances of sustained transmission of the disease in a condition of flattening the contagion curve, in which the time to reach the “peak” and the herd immunity becomes longer [8, 9]. Another important fact is that in many countries like Brazil, testing is done only on symptomatic patients, leaving out a legion of asymptomatic individuals without diagnosis but with the virus [9, 10]. Also, there is a consensus that SARS-CoV-2 virus may become endemic with reinfections [11], and the COVID-19 disease may become a part of the “seasonal flu”.
In March 2020, WHO requested mass testing of the population in order to identify and isolate as many infected people as possible. One of the best examples of this testing came from South Korea, which did not quarantine, but tested millions of people who, together with others measures, reduced the number of cases and deaths. Along 2020, Brazil performed 19.54 tests for every thousand inhabitants, behind countries like Chile 110.57/thousand inhabitants, Argentina 19.94/thousand inhabitants, Italy 76.10/thousand inhabitants, USA 226.07/thousand inhabitants and UK 183.35/thousand inhabitants [9, 10]. Therefore, the rapid and accurate diagnosis of COVID-19 is extremely important for the management of positive patients with isolation and adaptation of available hospital beds to the demand [12, 13].
The gold standard technique for the detection of the COVID-19 is based on the identification of the virus genetic material (ribonucleic acid — RNA) by RT-PCR (reverse transcription–polymerase chain reaction) technique in nasopharyngeal and oropharyngeal samples collected using a swab [14, 15]. Bronchoalveolar and sputum lavage materials may also be used in hospitalized patients [16]. However, the PCR technique has some disadvantages such as the need for specific physical space, equipments, and trained personnel to perform the assay, higher costs compared to traditional serological tests for antibody detection, longer time to release the results (around three to four days) in some regions, and they can present a false negative if the collection is performed improperly [15, 17].
Rapid and accurate diagnoses are essential to reduce the impact caused by tests with false negative results, requesting the development of new diagnostic tests [15, 18]. Spectroscopic methods for the analysis of biological materials offer several advantages over molecular biochemical methods, including speed in obtaining diagnostic information, no need for sample preparation or for use of reagents (label-free) [19]. Raman spectroscopy is a non-destructive analytical technique that provides information about the molecular composition of the materials studied, using a minimum amount of sample without the need for preparation, in addition to quick diagnosis that can be done in a few minutes using statistical and computational tools [20]. In fact, Raman spectroscopy has been used to quantify biomarkers of kidney disease such as urea and creatinine in serum [21], to quantify glucose and lipid components of sera [22], and to quantify prostate-specific antigen (PSA) values in sera of prostate cancer patients [23] aiming diagnosis.
Up to date, several authors proposed the use of Raman spectroscopy for diagnosing viruses or virus diseases, particularly COVID-19. Desai et al. (2020) showed a preliminary report on the development of a statistical model to detect RNA viruses in saliva of infected patients by means of Raman spectroscopy, discussing possible applications and implications in the diagnosis of COVID-19 [24]. Masterson et al. (2020) proposed a method of “liquid biopsy” based on the evaluation of microRNA of cancers in plasma through surface-enhanced Raman spectroscopy (SERS) and plasmon-enhanced fluorescence (PEF), a method that, according to the authors, could be expanded to the diagnosis of COVID-19 [25]. Also, Dou et al. (2020) showed that viruses could be identified in a label-free way by tip-enhanced Raman spectroscopy (TERS), by revealing protein secondary structure and amino acid composition of the virus surface [26]. Carvalho and Nogueira (2020) suggested in a “letter to the editor” that the vibrational techniques such as Raman and Fourier-transform infrared (FT-IR) spectroscopies could address the challenges for early and fast diagnosis of COVID-19 [27]. Khan and Rehman (2020) discussed the possibilities and challenges of developing a spectroscopic test methodology based on FT-IR and Raman spectroscopies to analyze COVID-19 from samples of serum, blood, saliva, and urine, and compare spectral results with the current PCR method [28]. Goulart et al. (2021) employed dispersive Raman spectroscopy to unveil the spectral differences between subjects diagnosed with COVID-19 by positive serology (immunoglobulins M and G) versus control and developed discriminating models to classify the samples according to the immunoglobulin type [29].
The objective of this study is to show the results of a proposed methodology based on Raman spectroscopy of the serum and discriminant analysis to the diagnosis of serum samples of individuals with detected SARS-CoV-2 virus RNA by RT-PCR (COVID-19) versus individuals with not detected virus RNA by PCR (Control). Also, we present an exploratory analysis based on principal component analysis (PCA) as a tentative biochemical description of the spectral differences in serum from COVID-19 positive individuals with regard to the composition of proteins, amino acids, nucleic acids, lipids, and carotenoids.
Materials and methods
Serum samples
The study was approved by the Ethics and Research Committee of Universidade Anhembi Morumbi, under protocol No. 26691419.6.0000.5492. Among several patients, 20 individuals were selected, 10 of them presenting positive results for COVID-19 (COVID-19 group) and the other 10 individuals presenting negative results for COVID-19 (Control group). Confirmation of these results was obtained by subjecting both groups to RT-PCR and ELISA (enzyme-linked immunonosorbent assay) tests. From each individual a sample of nasopharynx secretion, a sample of oropharynx, and 5 mL of blood at the CIPAX Diagnostic Medicine (São José dos Campos, SP, Brazil) [12, 15–17, 30].
The samples of secretion and blood were collected between the 3rd and 10th day after the onset of the reported symptoms for the positive patients: fever (50%), cough (50%), sore throat (40%), anosmia (30%), malaise (30%), runny nose (30%), headache (20%), lack of appetite (10%), abdominal pain (10%), body pain (10%), and pneumonia (10%). To perform RT-PCR, the material from the nasopharynx and oropharynx was collected by means of two polyester swabs and immediately placed in a tube with screw cap containing 3 mL of sterile saline solution (swabs and tube by Kolplast, São Paulo, SP, Brazil). The tubes were transported in an upright position at a temperature of 2 to 8 °C until the sample was processed [16, 30, 31]. Whole blood was collected by means of venipuncture in a closed system using tubes with a separator gel (Vacuette® Z Serum Sep Clot Activator, Greiner Bio-One, Americana, SP, Brazil). The blood tubes were subjected to centrifugation for 10 min at 3000 RPM (model Elektra, Laborline, São Paulo, SP, Brazil) for serum extraction. Serum samples were used for serological tests [32].
The samples from nasopharynx and oropharynx were subjected to analyses performed using a RT-PCR assay (Abbott Real-Time SARS-CoV-2 kit, Abbott Laboratories, São Paulo, SP, Brazil), which characterizes the individuals as “detectable” and “undetectable” for COVID-19 disease [30]. The RT-PCR technique is considered the gold standard for the detection of COVID-19 disease by evidencing the RNA of the SARS-CoV-2 virus through the amplification of nucleic acid, with 95% sensitivity and 100% specificity as indicated in the leaflet for virus copies > 100 copies/mL [30]. The serum samples were submitted to the ELISA assay (Euroimmun Anti SARS-CoV-2 ELISA kit and Euroimmun Analyzer I, Euroimmun Brasil, São Caetano do Sul, SP, Brazil), for detection of IgA immunoglobulin. The results of ELISA tests were compared with RT-PCR and tests were in agreement for all patients. After the ELISA tests, the 20 serum samples were packed in thermal boxes, kept at 2 to 8 °C and then submitted to Raman spectroscopy.
Raman spectroscopy
At the time of spectroscopic analysis, 80 μL of serum was pipetted into an aluminum sample holder for reading the Raman spectra in triplicate. The spectra were obtained in a dispersive Raman spectrometer (model Dimension P1, Lambda Solutions Inc, MA, USA) with 830 nm excitation and 350 mW laser power. The spectrometer has a spectral resolution of 4 cm−1 in the spectral range between 400 and 1800 cm−1. The collection time for each spectrum was 30 s (3 s and 10 accumulations).
Before data analysis, Raman spectra were subjected to pre-processing to extract the fluorescence background (baseline) using a 7th order polynomial. Other interferences such as cosmic rays were removed manually and the spectra were then normalized by the area under the curve (1-norm). Spectra were then grouped in Control and COVID-19 according to the RT-PCR results. Each triplicate spectrum was considered an individual sample in the data analysis.
Principal component analysis and classification
The Raman spectra were submitted to PCA [33] in order to unveil the spectral differences between the two groups related to the changes in the biochemical constitution of the serum due to COVID-19. Also, the PCA variables (markedly the PCA loadings — PCs) were used in a discriminant analysis (DA) model. The DA model was also implemented via partial least squares (PLS) regression using the whole spectral information instead of the selected PCs. It has been used the software Chemoface, “Multivariate Calibration” menu (www.ufla.br/chemoface) [34] to develop the DA models using a cross-validation based on the “leave-one-out spectrum” approach. In these models, the groups Control and COVID-19 (numbered as 1 and 2, respectively, in the Supplementary Table 1) were used as independent variables (y), and the PCs or the entire spectra were used as dependent variables (x). Multivariate models such as PCA and PLS have been used for classification of Raman spectra of sera in normal and anemia [35] and for quantification of blood analytes in human serum for diagnosis [22, 23].
A Kolmogorov–Smirnov normality test was applied to check the normality of the PCs and Student’s t-test (with Welch correction whenever needed) or Mann–Whitney U-test were applied to the PCs in order to identify significant differences between Control and COVID-19 groups. The p-value was considered significant when p < 0.05.
Results and discussion
Raman spectra of serum samples of Control and COVID-19
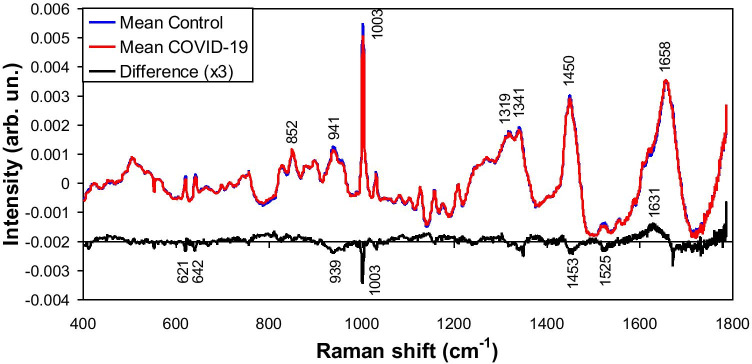
Figure 1 presents the mean Raman spectra of the Control group and COVID-19 group, showing the difference spectra between them. Both groups showed spectral features referred to human serum, with peaks at 852, 941, 1003, 1319, 1341, 1450, and 1658 cm−1 assigned to albumin [36–39]. The difference spectra (scale multiplied by 3) showed positive and negative peaks along the spectral range; this suggest that the composition of both samples (proteins, lipids, amino acids) differ in some extent. For instance, the labeled peaks at 621, 642, 939, 1003, and 1453 cm−1 (which will appear in the further exploratory analysis) can be assigned to proteins/amino acids [36–40]; the peak at 1525 cm−1 can be assigned to carotenoids [41, 42]. The following exploratory analysis by PCA has been used to understand the nature of this biochemical difference.
Fig. 1.
Mean Raman spectra of the blood serum of Control group and blood serum of patients with COVID-19 diagnosed by RT-PCR. Difference spectra (COVID-19 — Control) is also shown, with positive and negative peaks assigned to proteins, lipids, amino acids and nucleic acids as discussed in the text
Exploratory analysis
The exploratory analysis used the PCA variables (Scores — resembles Raman spectra and are related to the spectral variance presented in the data set and are in order of appearance, and PCs — intensities of each Score in the original data) and Fig. 2 shows the first 6 variables. These 6 Scores accounted for 99.4% of all spectral variation. Statistical significance of each PC in the group COVID-19 versus Control is also shown in Fig. 2. Also, Supplementary Fig. 1 shows the dispersion of the PCs along the samples in the dataset. The Score1 represented the spectral features of serum (mainly albumin and γ-globulin) [36–39], and both groups present similar constitution seen by the same intensity of PC1 (not significant difference). On the other hand, Score2 presented negative spectral features assigned to proteins (peaks at 939, 1003, 1454, and 1672 cm−1) [39, 42, 43] and carotenoids (peaks at 1159 and 1529 cm−1) [41, 42], higher (negative) for the Control group, and positive spectral features which could be assigned to amines/amides and nucleic acids (peaks at 794, 1142, 1366, and 1630 cm−1) [42, 44], higher for the COVID-19 group, as seen by PC2 (statistically significant difference, Welch-corrected t-test, p < 0.0001). Most of these spectral features are the same as the ones found in the difference spectrum in Fig. 1. A study showed that SARS-CoV-2 affects the 40S ribosomal subunit by preventing its binding with messenger RNA and decreasing host protein production [45]. COVID-19 increases chemical mediators of inflammation derived from granules that are inside the cells such as tissue macrophages, endothelial cells, and leukocytes [46–48]. These chemical mediators include vasoactive amines (histamine), serotonin, arachidonic acid, platelet-activating factor, cytokines, neuropeptins. The increase in amines seen in the Raman spectra of positive patients can also be explained by the increase in transaminases (TGO and TGP), which are enzymes responsible for transferring an amine group from one amino acid to a hydrocarbon to form a different amino acid and by increasing urea (the main functional compound of amides) and nitrogenous amine creatine synthesized in the liver, kidneys, and pancreas from amino acids glycine and arginine, also having methionine as a methyl donor [47, 49, 50]. Therefore, these amines could be responsible for the Raman features referred to amines/amides which were present in the sera of the positive COVID-19 group. Also, the presence of features assigned to nucleic acids could be due to the presence of SARS-CoV-2 RNA in sera of positive patients.
Fig. 2.
Plot of the first 6 principal component variables Scores and PCs. The features in Scores are assigned to serum constituents such as albumin, proteins, lipids, amino acids, nucleic acids, sulfur compounds and carotenoids. The intensities in PCs showed significant differences between groups for PC2, PC4 and PC6 (t-test and U-test, p < 0.05). NS = not significant
Score3 showed negative spectral features which are assigned to carotenoids (peaks at 1004, 1157, and 1525 cm−1) [41, 42], but without difference between groups, as seen by PC3 (not significant difference Mann–Whitney U-test, p = 0.387). Score4 presented negative features that may be assigned to amino acids — tryptophan (peaks at 759, 1003, and 1348 cm−1) [40, 42], higher (negative) for the Control group, and positive features assigned to lipids — unsaturated (1089, 1262, 1302, 1440, and 1657 cm−1) [39, 42, 43], higher for COVID-19, as seen by PC4 (statistically significant difference, Welch-corrected t-test, p < 0.01). Score5 presented spectral features which may be assigned to urea [51] and possible cosmic ray artifact; the PC5 showed no significant difference between groups (t-test, p = 0.125). Score6 showed negative spectral features which could be assigned to proteins and amino acids (peaks at 904, 942, and 1003 cm−1) [39, 42, 43], higher (negative) for the Control group, and positive spectral features which may be assigned to sulfur compounds (C–S stretching — sulfides?) (703 cm−1) [43, 52], remnant amines/nucleic acids (1256 and 1435 cm−1) [40, 42] and remnant carotenoids (1007, 1159 and 1522 cm−1) [41, 42], higher for COVID-19 group, as seen by the PC6 (statistically significant difference, t-test, p < 0.05). Therefore, the Control showed higher amount of amino acids and the COVID-19 group showed higher amounts of lipids, sulfur and carotenoids. Table 1 summarizes the assignments of the Raman bands found in the mean spectra, Fig. 1, and in the PCA analysis, Fig. 2 (Score plots).
Table 1.
Peak positions and assignments of the Raman bands seen in Figs. 1 and 2 according to the cited literature and reference spectra of pure compounds
| Biochemical group | Peak position (cm−1) | Assignment | References |
|---|---|---|---|
| Proteins (mainly albumin) and amino acids | 621 | Phenylalanine | [40] |
| 642 | Tyrosine | [40, 44] | |
| 759 | Tryptophan | [40, 42] | |
| 852 | Tyrosine; tryptophan – albumin | [36–40] | |
| 904 | C–C stretching | [39, 42, 43] | |
| 939/941/942 | C–C stretching – albumin | [36–39, 42, 43] | |
| 1003 | Ring breathing – phenylalanine; tryptophan (albumin) | [36–40, 42, 43] | |
| 1256 | Amide I – amines? | [40, 42] | |
| 1319 | Amide III – albumin | [36–40] | |
| 1341/1348 | Tryptophan – albumin | [36–39, 42] | |
| 1435 | CH2 deformation – amines? | [42] | |
| 1450/1453/1454 | CH2/CH3 deformations – albumin | [36–39, 42, 43] | |
| 1630/1631 | Amide I? | [40] | |
| 1658 | Amide I – albumin (C = O stretching) | [36–40] | |
| 1672 | Amide I | [39, 40, 42, 43] | |
| 1735 | ? | ||
| Nucleic acids | 794 | Ring breathing of bases (cytosine, thymine) (nucleoside conformation) | [42, 44] |
| 1142 | ? | ||
| 1256 | Ring mode of bases (cytosine, thymine) (nucleoside conformation) | [42, 44] | |
| 1366 | Thymine | [42, 44] | |
| 1630 | C–O stretching – thymine? | [44] | |
| Lipids (unsaturated) | 1089 | C − C stretching (gauche) | [39, 42, 43, 53] |
| 1262 | = C − H deformation | [39, 42, 43, 53] | |
| 1302 | CH2 twisting/scissoring | [39, 42, 43, 53] | |
| 1440 | CH3/CH2 deformations | [39, 42, 43, 53] | |
| 1657 | C = C stretching | [39, 42, 43, 53] | |
| Sulfur compounds? | 703 | C–S stretching | [43, 52] |
| Urea? | 1004 | C–N stretching | [51] |
| Carotenoids | 1004/1007 | C–CH3 rocking | [41, 42] |
| 1157/1159 | C–C stretching coupled with C–H in-plane bending | [41, 42] | |
| 1522/1525/1529 | C = C stretching of the conjugated chain | [38, 41] |
A study by Thomas et al. (2020) [54] showed a dysregulation of nitrogen metabolism in COVID-19 patients, with decreased circulating levels of most amino acids, except for tryptophan, and increased markers of oxidant stress (e.g. methionine sulfoxide, cystine), proteolysis, and kidney dysfunction (e.g. creatine, creatinine, polyamines). Increased circulating levels of glucose and free fatty acids were also observed; metabolite levels of these compounds correlated with clinical laboratory markers of inflammation and disease severity (i.e. IL-6 and C-reactive protein) and renal function (i.e. blood urea nitrogen) [54]. Higher levels of sulfides may be associated with the antiviral and anti-inflammatory action of hydrogen sulfide gas [55].
Phosphocholine has been found to be upregulated in positive COVID-19 patients, probably due to activated macrophage-mediated immunity, causing increase in arachidonic acid, which is responsible for the increase in nitric oxide that macrophages use as a cytotoxic metabolite in order to destroy the microorganism. In addition, there is an increase in protein C-reactive that binds to phosphocholine, activating the complement system and recruiting more phagocytes (inflammatory process) [46, 47, 49, 56, 57].
In a study of the proteomic and metabolomic characterization of serum from COVID-19 patients versus controls, Shen et al. (2020) [49] reported that more than 100 proteins were differentially expressed in COVID-19 positive patients compared to non-positive, where 50 of these proteins belonging to three pathways: activation of the complement system, macrophage function, and platelet degranulation. A number of metabolites (204) were correlated with disease severity and the most significantly changed metabolites were also involved in the same three biological processes. Therefore, the lower level of proteins and amino acids, and higher levels of lipids (including carotenoids), tryptophan, amines/amides and sulfur compounds seen in the Raman features are well aligned with the changes in the metabolism of positive patients.
Metabolites involved in arginine metabolism were also found to increase in COVID-19 patients [47, 50]. Therefore, an increase may be expected in the Raman features associated to compounds such as arginine, glutamine, glutamate and urea. The binding of SARS-CoV-2 to alveolar macrophages resulting in release of cytokines (interleukins) by macrophages [46]. These cytokines may appear as protein features, but these features were not seen in the positive samples probably due to low concentration.
Previous studies showed that Raman spectroscopy could be used to detect and quantify anti-Toxoplasma gondii IgG antibodies in sera of cats [58], to detect IgM antibody in dengue infection in human sera [59], and to detect IgM and IgG in sera of COVID-19 positive patients aiming diagnosis [29]. The Raman spectra were also used to detect hepatitis (B and C) virus infection in sera [60, 61] as well as dengue virus infection also in sera [62]. These studies of virus infections showed that hepatitis B presented difference in the metabolism of proteins, cholesterol, amino acids and nucleic acids due to liver infection [60], and hepatitis C presented difference related to sera components, but without specific biochemistry’s disease assignment [61]; the peaks at 1153 and 1510 cm−1 may have been erroneously assigned to carbohydrates and nucleic acids instead of carotenoids [60] and cytosine [61] in the control group. For the dengue virus patients, carotenoid peaks (1156 and 1516 cm−1) were decreased while peaks associated to immunoglobulins, adenosine diphosphate, and hemoglobin were increased in positive patients [62]. For the COVID-19 patients with positive IgM (IgM +) and IgG (IgG +), there were Raman features assigned to proteins including albumin in lower intensity in the group COVID-19 compared to Control, features referred to nucleic acids and tryptophan in the group COVID-19, particularly the IgG + /IgM + , and features assigned to lipids, phospholipids, and carotenoids in the group IgG + [29]. Therefore, different virus infection shows particular change in the biochemistry of the sera and consequently Raman features.
Discriminant analysis and classification
From the Raman spectra of sera and the first six PCs of both groups, classification models were developed using discriminant analysis in the Chemoface software. Each spectrum was considered a sample in the discriminant models (spectrum-wise analysis).
The models employed linear discriminant analysis (LDA) applied to the first six PCs (the ones with significant difference between Control and COVID-19), and employed PCA discriminant analysis (PCA-DA) and PLS discriminant analysis (PLS-DA) applied to the entire spectrum, where the number of loading vectors (for the PCA-DA) and latent variables (for the PLS-DA) used in the models were the ones that promoted the higher discrimination accuracy. Table 2 presents the results of the classification for each model in terms of sensitivity, specificity, and accuracy [63] using the individual classification seen in the Supplementary Table 1 (results of the classification for each sample spectrum). Better classification was achieved for the LDA using 4 PCs (PC2, PC4, PC5, and PC6), with 87% sensitivity, 100% specificity, and 93% accuracy. Despite not significant (Fig. 2), the PC5 (urea) was essential for the classification obtained by LDA; without it, the accuracy lowered to 88% (data not shown).
Table 2.
Sensitivity, specificity and percentage of correct classification (accuracy) for the classification of COVID-19 versus Control sera using the discriminations: LDA, PCA-DA and PLS-DA
| LDA | PCA-DA | PLS-DA | |
|---|---|---|---|
| No. of variables | 4 loadings* | 5 loadings | 5 latents |
| Sensitivity (%) | 87 | 77 | 80 |
| Specificity (%) | 100 | 100 | 100 |
| Accuracy (%) | 93 | 88 | 90 |
* Loadings 2, 4, 5 and 6 (PC2, PC4, PC5 and PC6)
The results of the discrimination accuracy are slightly better than the ones found in Goulart et al. (2021) [29]. In that study, the authors used 94 serum samples (54 control and 40 COVID-19) and the accuracy in classifying sera with positive IgM and IgG for COVID-19 versus control was as high as 90.3%. Therefore, the spectral model based on the PCR results presented in this study demonstrated accuracy comparable to the model based on immunoglobulin’s serology.
Future perspectives
It is extremely important that new technologies are used in the diagnosis of viral diseases, in order to make the diagnosis faster and more accurate, so that patients receive adequate and early treatment and minimize complications resulting from the disease. It is aligned with the need to develop a technique that has a lower cost per procedure for adoption as a screening technique in public health networks. The Raman technique can become a fast, accurate, and less expensive methodology for the diagnosis of viral infection, as well as a possible use in the differential diagnosis between COVID-19 and seasonal flu (A/H1N1), as well as associating spectral changes with the degree of infection with COVID-19 (viral load).
Studies by other research groups are needed to confirm the spectral findings and to better correlate the spectral signatures of lipids, nitrogen compounds (amines/amides and urea), and nucleic acids found in COVID-19 group with the actual difference in the composition of the sera from the groups due to the virus. Also, it is important to increase the number of positive subjects versus controls in order to check the reproducibility of the spectra from both control and COVID-19 aiming at the development of a technique for primary diagnosis as well as assessing the possibility of using the Raman technique for testing asymptomatic subjects on a large scale and assessing the presence of virus antibodies in serum, meeting the need for public health systems for the diagnosis of COVID-19 given the great need for identification and isolation of positive patients. The monitoring of positive patients and in resolution (cure) can also benefit from the advantages of the technique due to speed and lower cost.
The Raman spectral features reported for COVID-19 may be similar to other viral infections affecting the respiratory system such as seasonal influenza (A/H1N1 [64], and A/H3N2 [65]) employing surface enhanced Raman spectroscopy (SERS); therefore, studies are needed to identify the particular features associated only to COVID-19 and to promote a differential diagnosis from infections by the seasonal flu.
Conclusion
The Raman spectroscopy applied to diagnose COVID-19 in human serum showed biochemical alterations related to the presence of the SARS-CoV-2 such as increase in lipids, nitrogen compounds (urea and amines/amides) and nucleic acids, and decrease in proteins and amino acids (tryptophan). The models based on discriminant analysis applied to the principal component loadings (PC2, PC4, PC5 and PC6) could classify spectra with 87% sensitivity, 100% specificity, and 93% accuracy, demonstrating the possibilities of a rapid, label-free, and costless technique for diagnosing COVID-19 infection.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
Ethics approval and consent to participate
This study was performed in line with the principles of the Declaration of Helsinki and follows the ethical standards of scientific research involving human subjects edited by the Brazilian National Health Council – CNS, Resolution No. 466/12.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Word Health Organization (2020) WHO Coronavirus disease (COVID-19) dashboard. Word Health Organization – WHO. https://COVID19.who.int. Acessed 15 August 2021.
- 2.El Khatib AS (2020) Economy versus epidemiology: an analysis of trade-off between markets and lives in COVID 19 times. Contab Neg (En línea) 15 30 62 80 10.18800/contabilidad.202002.004
- 3.Kandel N, Chungong S, Omaar A, et al. Health security capacities in the context of COVID-19 outbreak: an analysis of International Health Regulations annual report data from 182 countries. Lancet. 2020;395(10229):P1047–1053. doi: 10.1016/S0140-6736(20)30553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auwaerter PG (2020) Coronavirus COVID-19 (SARS-COV-2). Johns Hopkins ABX Guide, Unbound Medicine. http://www.hopkinsguides.com/hopkins/view/Johns_Hopkins_ABX_Guide/540747/all/Coronavirus_COVID_19__SARS_CoV_2_. Acessed 05 August 2021.
- 5.Schuchmann AZ, Schnorrenberger BL, Chiquetti ME et al (2020) Vertical social isolation X horizontal social isolation: health and social dilemas in copping with the COVID-19 pandemic. Braz J Health Rev 3 2 3556 3576 10.34119/bjhrv3n2-185
- 6.Garcia Filho C, Vieira LJES, Silva RM. Internet searches for measures to address COVID-19 in Brazil: a description of searches in the first 100 days of 2020. Epidemiol Serv Saúde. 2020;29(3):e2020191. doi: 10.5123/s1679-49742020000300011. [DOI] [PubMed] [Google Scholar]
- 7.Barberia LG, Gómez EJ. Political and institutional perils of Brazil’s COVID-19 crisis. Lancet. 2020;396(10248):367–368. doi: 10.1016/S0140-6736(20)31681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson NM, Laydon D, Nedjati-Gilani G et al (2020) Impact of non-pharmaceutical interventions (NPIs) to reduce COVID-19 mortality and healthcare demand. Imperial College London. 10.25561/77482
- 9.Thunström L, Newbold SC, Finnoff D, et al. The benefits and costs of using social distancing to flatten the curve for COVID-19. J Benefit Cost Anal. 2020;11(2):179–195. doi: 10.1017/bca.2020.12. [DOI] [Google Scholar]
- 10.Marson FAL, Ortega MM. COVID-19 in Brazil. Pulmonology. 2020;26(4):241–244. doi: 10.1016/j.pulmoe.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kellam P, Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol. 2020;101(8):791–797. doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Word Health Organization (2020) Laboratory testing strategy recommendations for COVID-19. World Health Organization – WHO. https://apps.who.int/iris/bitstream/handle/10665/331509/WHO-COVID-19-lab_testing-2020.1-eng.pdf. Acessed 15 August 2021.
- 13.Tromberg BJ, Schwetz TA, Pérez-Stable EJ, et al. Rapid scaling up of Covid-19 diagnostic testing in the United States – The NIH RADx initiative. N Engl J Med. 2020;383(11):1071–1077. doi: 10.1056/NEJMsr2022263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long C, Xu H, Shen Q, et al. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol. 2020;126:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang YW, Schmitz JE, Persing DH, et al. Laboratory diagnosis of COVID 19: current issues and challenges. J Clin Microbiol. 2020;58(6):e00512–e520. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Instituto Adolfo Lutz (2020) Protocolo laboratorial para coleta, acondicionamento e transporte de amostras biológicas para investigação de SRAG e SG por SARS-CoV-2. Instituto Adolfo Lutz. http://www.ial.sp.gov.br/resources/insituto-adolfo-lutz/publicacoes/coronavirus/orientacoesdecoletaestrategiadeampliacao_sg.pdf. Acessed 15 August 2021.
- 17.Chan JF, Yip CC, To KK, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel Real Time Reverse Transcription PCR assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58(5):e00310–e320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen T, Bang DD, Wolff A. 2019 Novel Coronavirus Disease (COVID-19): paving the road for rapid detection and point-of-care diagnostics. Micromachines (Basel) 2020;11(3):306. doi: 10.3390/mi11030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S Giansante HE Giana AB Fernandes et al 2021 Analytical performance of Raman spectroscopy in assaying biochemical components in human serum Lasers Med Sci10.1007/s10103-021-03247-8 [DOI] [PubMed]
- 20.Krafft C, Popp J. The many facets of Raman spectroscopy for biomedical analysis. Anal Bioanal Chem. 2015;407(3):699–717. doi: 10.1007/s00216-014-8311-9. [DOI] [PubMed] [Google Scholar]
- 21.Almeida ML, Saatkamp CJ, Fernandes AB, et al. Estimating the concentration of urea and creatinine in the human serum of normal and dialysis patients through Raman spectroscopy. Lasers Med Sci. 2016;31(7):1415–1423. doi: 10.1007/s10103-016-2003-y. [DOI] [PubMed] [Google Scholar]
- 22.Silveira L, Borges RCF, Navarro RS, et al. Quantifying glucose and lipid components in human serum by Raman spectroscopy and multivariate statistics. Lasers Med Sci. 2017;32(4):787–795. doi: 10.1007/s10103-017-2173-2. [DOI] [PubMed] [Google Scholar]
- 23.Correia NA, Batista LTA, Nascimento RJM, et al. Detection of prostate cancer by Raman spectroscopy: a multivariate study on patients with normal and altered PSA values. J Photochem Photobiol B Biol. 2020;204:111801. doi: 10.1016/j.jphotobiol.2020.111801. [DOI] [PubMed] [Google Scholar]
- 24.Desai S, Mishra SV, Joshi A, et al. Raman spectroscopy based detection of RNA viruses in saliva: a preliminary report. J Biophotonics. 2020;13(10):e202000189. doi: 10.1002/jbio.202000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masterson AN, Liyanage T, Berman C, et al. A novel liquid biopsy-based approach for highly specific cancer diagnostics: mitigating false responses in assaying patient plasma-derived circulating microRNAs through combined SERS and plasmon-enhanced fluorescence analyses. Analyst. 2020;145(12):4173–4180. doi: 10.1039/d0an00538j. [DOI] [PubMed] [Google Scholar]
- 26.Dou T, Li Z, Zhang J, et al. Nanoscale structural characterization of individual viral particles using atomic force microscopy infrared spectroscopy (AFM-IR) and tip-enhanced Raman spectroscopy (TERS) Anal Chem. 2020;92(16):11297–11304. doi: 10.1021/acs.analchem.0c01971. [DOI] [PubMed] [Google Scholar]
- 27.Carvalho LFDCES, Nogueira MS. Optical techniques for fast screening — towards prevention of the coronavirus COVID-19 outbreak. Photodiagnosis Photodyn Ther. 2020;30:101765. doi: 10.1016/j.pdpdt.2020.101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan RS, Rehman IU. Spectroscopy as a tool for detection and monitoring of Coronavirus (COVID-19) Expert Rev Mol Diagn. 2020;20(7):647–649. doi: 10.1080/14737159.2020.1766968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ACC Goulart RA Zângaro HC Carvalho et al (2021) Diagnosing COVID-19 in human sera with detected immunoglobulinsIgM and IgG by means of Raman spectroscopy. J Raman Spectrosc. 10.1002/jrs.6235 [DOI] [PMC free article] [PubMed]
- 30.Abbott Molecular Inc (2020) Abbott RealTime SARS-CoV-2. Abbot. https://www.molecular.abbott/sal/Abbott%20RealTime%20SARS-CoV-2%20amp%20kit%20IFU%2051-608442%20R2.pdf. Acessed 15 August 2021.
- 31.Sociedade Brasileira de Patologia Clínica e Medicina Laboratorial (2020) Métodos laboratoriais para o diagnóstico de infecção capilar por SARS-CoV-2. Sociedade Brasileira de Patologia Clínica e Medicina Laboratorial – SBPC/ML. http://www.sbpc.org.br/wp-content/uploads/2020/04/MetodosLaboratoriaisDiagnosticoSARS-CoV-2.pdf. Acessed 15 August 2021.
- 32.Ejazi SA, Ghosh S, Ali N. Antibody detection assays for COVID-19 diagnosis: an early overview. Immunol Cell Biol. 2021;99(1):21–33. doi: 10.1111/imcb.12397. [DOI] [PubMed] [Google Scholar]
- 33.Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Phil Trans R Soc A. 2016;374(2065):20150202. doi: 10.1098/rsta.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nunes CA, Freitas MP, Pinheiro ACM, et al. Chemoface: a novel free user-friendly interface for chemometrics. J Braz Chem Soc. 2012;23(11):2003–2010. doi: 10.1590/S0103-50532012005000073. [DOI] [Google Scholar]
- 35.Silva AM, Oliveira FSS, de Brito PL, et al. Spectral model for diagnosis of acute leukemias in whole blood and plasma through Raman spectroscopy. J Biomed Opt. 2018;23(10):107002. doi: 10.1117/1.JBO.23.10.107002. [DOI] [PubMed] [Google Scholar]
- 36.Lin VJ, Koenig JL. Raman studies of bovine serum albumin. Biopolymers. 1976;15(1):203–218. doi: 10.1002/bip.1976.360150114. [DOI] [PubMed] [Google Scholar]
- 37.Lykina A, Artemyev D (2017) Analysis of albumin Raman scattering in visible and near-infrared ranges. In: Proceedings of the Saratov Fall Meeting 2017: Optical Technologies in Biophysics and Medicine XIX; Saratov, Russian Federation. 26–30 September 2017; p. 107160E. 10.1117/122317535
- 38.Parachalil DR, Bruno C, Bonnier F, et al. Raman spectroscopic screening of high and low molecular weight fractions of human serum. Analyst. 2019;144(14):4295–4311. doi: 10.1039/c9an00599d. [DOI] [PubMed] [Google Scholar]
- 39.Dingari NC, Horowitz GL, Kang JW, et al. Raman spectroscopy provides a powerful diagnostic tool for accurate determination of albumin glycation. PLoS ONE. 2012;7(2):e32406. doi: 10.1371/journal.pone.0032406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhar N, Sil S, Umapathy S. Potential of Raman spectroscopic techniques to study proteins. Spectrochim Acta A Mol Biomol Spectrosc. 2021;258:119712. doi: 10.1016/j.saa.2021.119712. [DOI] [PubMed] [Google Scholar]
- 41.Horiue H, Sasaki M, Yoshikawa Y, et al. Raman spectroscopic signatures of carotenoids and polyenes enable label-free visualization of microbial distributions within pink biofilms. Sci Rep. 2020;10(1):7704. doi: 10.1038/s41598-020-64737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Movasaghi Z, Rehman S, Rehman IU. Raman spectroscopy of biological tissues. Appl Spectrosc Rev. 2007;42(5):493–541. doi: 10.1080/05704920701551530. [DOI] [Google Scholar]
- 43.Lin-Vien D, Colthup N, Fateley W et al (1991) Appendix 3 — A summary of characteristic Raman and infrared frequencies. In: The handbook of infrared and Raman characteristic frequencies of organic molecules, 1st edn. Academic Press, San Diego, pp. 477 490 10.1016/B978-0-08-057116-4.50027-4
- 44.Kuhar N, Sil S, Verma T, et al. Challenges in application of Raman spectroscopy to biology and materials. RSC Adv. 2018;8:25888–25908. doi: 10.1039/C8RA04491K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thoms M, Buschauer R, Ameismeier M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369(6508):1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henry BM, Oliveira MHS, Benoit S, et al. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020;58(7):1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 47.Aloisio E, Chibireva M, Serafini L, et al. A comprehensive appraisal of laboratory biochemistry tests as major predictors of COVID-19 severity. Arch Pathol Lab Med. 2020;144(12):1457–1464. doi: 10.5858/arpa.2020-0389-SA. [DOI] [PubMed] [Google Scholar]
- 48.Wang D, Li R, Wang J, et al. Correlation analysis between disease severity and clinical and biochemical characteristics of 143 cases of COVID-19 in Wuhan, China: a descriptive study. BMC Infect Dis. 2020;20:519. doi: 10.1186/s12879-020-05242-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen B, Yi X, Sun Y, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xavier AR, Silva JS, Almeida JPCL, et al. COVID-19: clinical and laboratory manifestations in novel coronavirus infection. J Bras Patol Med Lab. 2020;56:e3232020. doi: 10.5935/1676-2444.20200049. [DOI] [Google Scholar]
- 51.Bispo JA, Vieira EES, Silveira L, et al. Correlating the amount of urea, creatinine, and glucose in urine from patients with diabetes mellitus and hypertension with the risk of developing renal lesions by means of Raman spectroscopy and principal component analysis. J Biomed Opt. 2013;18(8):87004. doi: 10.1117/1.JBO.18.8.087004. [DOI] [PubMed] [Google Scholar]
- 52.Nogami N, Sugta H, Miyazawa T. C-S stretching vibrations and molecular conformations of isobutyl methyl sulfide and related alkyl sulfides. Bull Chem Soc Japan. 1975;48(9):2417–2420. doi: 10.1246/bcsj.48.2417. [DOI] [Google Scholar]
- 53.Czamara K, Majzner K, Pacia MZ, et al. Raman spectroscopy of lipids: a review. J Raman Spectrosc. 2015;46(1):4–20. doi: 10.1002/jrs.4607. [DOI] [Google Scholar]
- 54.Thomas T, Stefanoni D, Reisz JA, et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5(14):e140327. doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ivanciuc T, Sbrana E, Ansar M et al (2016) Hydrogen sulfide is an antiviral and antiinflammatory endogenous gasotransmitter in the airways. Role in respiratory syncytial virus infection. Am J Resp Cell Mol Biol 55 5 684 696 10.1165/rcmb.2015-0385OC [DOI] [PMC free article] [PubMed]
- 56.Ciaccio M, Agnello L. Biochemical biomarkers alterations in Coronavirus Disease 2019 (COVID-19) Diagnosis. 2020;7(4):365–372. doi: 10.1515/dx-2020-0057. [DOI] [PubMed] [Google Scholar]
- 57.Chacolli MLA. Protein C reactive as inflammation marker. Rev Méd (Tacna) 2018;11(1):42–44. [Google Scholar]
- 58.Duarte J, Pacheco MT, Villaverde AB, et al. Near-infrared Raman spectroscopy to detect anti-Toxoplasma gondii antibody in blood sera of domestic cats: quantitative analysis based on partial least-squares multivariate statistics. J Biomed Opt. 2010;15(4):047002. doi: 10.1117/1.3463006. [DOI] [PubMed] [Google Scholar]
- 59.Bilal M, Saleem M, Bilal M, et al. Raman spectroscopy-based screening of IgM positive and negative sera for dengue virus infection. Laser Phys Lett. 2016;26(11):115602. doi: 10.1088/1054-660X/26/11/115602. [DOI] [Google Scholar]
- 60.Tong D, Chen C, Zhang J, et al. Application of Raman spectroscopy in the detection of hepatitis B virus infection. Photodiagnosis Photodyn Ther. 2019;28:248–252. doi: 10.1016/j.pdpdt.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 61.Cheng H, Xu C, Zhang D, et al. Multiclass identification of hepatitis C based on serum Raman spectroscopy. Photodiagnosis Photodyn Ther. 2020;30:101735. doi: 10.1016/j.pdpdt.2020.101735. [DOI] [PubMed] [Google Scholar]
- 62.Khan S, Ullah R, Saleem M, et al. Raman spectroscopic analysis of dengue virus infection in human blood sera. Optik. 2016;127(4):2086–2088. doi: 10.1016/j.ijleo.2015.11.060. [DOI] [Google Scholar]
- 63.Baratloo A, Hosseini M, Negida A, et al. Part 1: Simple definition and calculation of accuracy, sensitivity and specificity. Emerg (Tehran) 2015;3(2):48–49. [PMC free article] [PubMed] [Google Scholar]
- 64.Lim JY, Nam JS, Yang SE, et al. Identification of newly emerging influenza viruses by surface-enhanced Raman spectroscopy. Anal Chem. 2015;87(23):11652–11659. doi: 10.1021/acs.analchem.5b02661. [DOI] [PubMed] [Google Scholar]
- 65.Pezzotti G, Zhu W, Adachi T, et al. Metabolic machinery encrypted in the Raman spectrum of influenza A virus-inoculated mammalian cells. J Cell Physiol. 2019;235(6):146–5170. doi: 10.1002/jcp.29392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.