Abstract
Background
This study is aimed at constructing a risk signature to predict survival outcomes of ORCA patients.
Methods
We identified differentially expressed autophagy-related genes (DEARGs) based on the RNA sequencing data in the TCGA database; then, four independent survival-related ARGs were identified to construct an autophagy-associated signature for survival prediction of ORCA patients. The validity and robustness of the prognostic model were validated by clinicopathological data and survival data. Subsequently, four independent prognostic DEARGs that composed the model were evaluated individually.
Results
The expressions of 232 autophagy-related genes (ARGs) in 127 ORCA and 13 control tissues were compared, and 36 DEARGs were filtered out. We performed functional enrichment analysis and constructed protein–protein interaction network for 36 DEARGs. Univariate and multivariate Cox regression analyses were adopted for searching prognostic ARGs, and an autophagy-associated signature for ORCA patients was constructed. Eventually, 4 desirable independent survival-related ARGs (WDR45, MAPK9, VEGFA, and ATIC) were confirmed and comprised the prognostic model. We made use of multiple ways to verify the accuracy of the novel autophagy-related signature for survival evaluation, such as receiver-operator characteristic curve, Kaplan–Meier plotter, and clinicopathological correlational analyses. Four independent prognostic DEARGs that formed the model were also associated with the prognosis of ORCA patients.
Conclusions
The autophagy-related risk model can evaluate OS for ORCA patients independently since it is accurate and stable. Four prognostic ARGs that composed the model can be studied deeply for target treatment.
1. Background
Death rates are increasing for oropharyngeal cancer overall by 0.5% per year from 2009 to 2018. There is an estimated number of 54,010 new cases of oropharyngeal cancer (accounting for about 2.8% of new cancers), and 10,850 will die of it (accounting for about 1.8% of all deaths from cancer) in 2021 in the United States [1]. Among all oropharyngeal cancer patients, approximately half of those occur specifically in the oral cavity and were called oral cancer (ORCA) [2]. All over the world, 377,713 people were diagnosed with oral cancer in 2020, and the cancer caused 177,757 deaths; it means there was one person who died due to ORCA every 3 minutes [3]. The treatment and prognosis of ORCA patients relied on the staging system and conventional prognostic factors used in clinical practice, for example, the tumor-node-metastasis (TNM) staging system was the most important and well-known prognostic factor for ORCA patients [4], but the staging system was not accurate enough and prognostic factors were inadequate and nonspecific, leading to discouraging overall prognosis. Recent statistics show that the overall 5-year survival rate for ORCA patients was 52.0% in China [5]. Therefore, searching molecular prognostic markers which could be incorporated into the prognostic system is significant; they can benefit predicting clinical outcomes and targeted therapy. This study attempts to refine the prognostic signature of ORCA and used it in the clinical setting.
Autophagy is a process that can circulation and reuse cellular components; it is nonspecific. Autophagy-related genes (ARGs) are molecules that participate in autophagy. In recent studies, many researchers deemed that autophagy is an important process in ORCA [6–8]. Most of the molecular mechanisms which are involved in autophagy regulation deeply participate in signaling pathways that assumed significant roles in ORCA control [9, 10]. For example, autophagy is a significant AKT/mTOR pathway target, and the antioncogene that negatively regulates mTOR (such as AMPK and PTEN) will activate autophagy. In addition, many autophagy-inducing proteins are either oncoproteins or tumor suppressor proteins [11]. Some studies have proven that the autophagy inhibitor can enhance efficacy against aggressive ORCA [12, 13]. However, how to predict the prognosis of ORCA patients by several ARGs is still unclear. Therefore, our study utilized several screened ARGs to establish the prognostic risk model of ORCA. In this study, the relationship between prognostic ARGs and clinicopathological features in 127 ORCA patients were evaluated, and an autophagy-related signature was constructed to predict survival for ORCA patients. The risk score model had been verified from several aspects, and we proved its accuracy; we also verified the prognosis value of each ARG that comprised the autophagy-related risk model and proved them as prognostic biomarkers in ORCA; our report may shed light on evaluating the prognosis and targeting treatment of ORCA.
2. Methods
2.1. Data Acquisition
In total, 232 genes presently known as autophagy-related genes (ARGs) were downloaded from HADb (Human Autophagy Database, http://autophagy.lu/). RNA-seq data and clinical features of 127 ORCA patients and 13 nontumor samples were obtained from the TCGA website (The Cancer Genome Atlas database, https://tcga-data.nci.nih.gov/tcga/).
2.2. Identification of DEARGs and Functional Annotation
The “limma” package in R was applied to compare differential expressions between ORCA and nontumor counterparts; the criteria is false discovery rate (FDR) < 0.05 and ∣log2FoldChange | (∣log2FC∣) > 1. Then, we performed enrichment analyses of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) for DEARGs to find the major biological attributes. The R packages of ClusterProfiler and org.Hs.eg.db were used to perform GO and KEGG function enrichment analyses. The visualization of results was implemented with the R “ggplot2” package. STRING database (https://string-db.org/) was used to construct the protein–protein interaction (PPI) network of DEARGs. Cytoscape software visualized the interaction of the PPI network.
2.3. Construction and Verification of ARG-Based Prognostic Risk Model
Univariate Cox proportional risk regression analysis was performed to screen prognostic ARGs. All statistically significant prognostic ARGs were selected as candidates for multivariate Cox regression analysis and used to construct the prognostic risk model. The multivariate analysis provided the regression coefficient of prognostic ARGs in the risk score model.
| (1) |
The median risk score was determined as the threshold to divide the ORCA patients into the high- or low-risk group. Four risk genes comprising the risk model were used to construct the nomogram. We constructed a nomogram based on 4 independent prognosis-related ARGs to predict survival rates of patients at 1, 2, and 3 years via the rms package in R 4.0.1.
The predictive accuracy of the risk models was evaluated by the receiver-operator characteristic (ROC) analysis; besides, we compared survival probability of high-risk and low-risk groups using the “ggplot2” package in R. We also assessed the association between the prognostic risk model and classical clinical parameters (such as tumor grade, pathological stage, and T/N classification) by Cox proportional hazard regression analysis.
2.4. Validation for Prognostic Value of 4 Risk ARGs which Formed the Model
The relationships between expressions of 4 independent prognostic-related ARGs (WDR45, MAPK9, VEGFA, and ATIC) and OS of ORCA patients were evaluated individually by Kaplan–Meier survival curves. We also observed the mRNA expression pattern of the 4 genes in ORCA and normal tongue tissues via the Oncomine database (https://www.oncomine.org/).
To explore the roles of 4 risk ARGs in ORCA, a single-gene gene set enrichment analysis (GSEA) was performed. GSEA was conducted using a molecular signature database's (MSigDB's) c2.cp.kegg.v7.0.symbols.gmt gene sets in GSEA 3.0 software to identify gene sets that were significantly correlated with the expression of risk genes. We exhibited the top 10 biological processes most associated with the expression of the 4 risk genes.
3. Results
3.1. Identification of DEARGs
The expression profile of 232 ARGs in 127 ORCA tissue samples and 13 normal mouth samples were compared and analyzed. Finally, there were 36 differentially expressed ARGs including 29 upregulated ARGs and 7 downregulated ARGs with a threshold of ∣log2FoldChange | >1. The volcano plot, heat map, and box plot visualized the expression pattern of all DEARGs between ORCA and normal tissues (Figures 1(a)–1(c)). We listed all DEARGs in Table 1, including the log2FoldChange and statistical significance.
Figure 1.
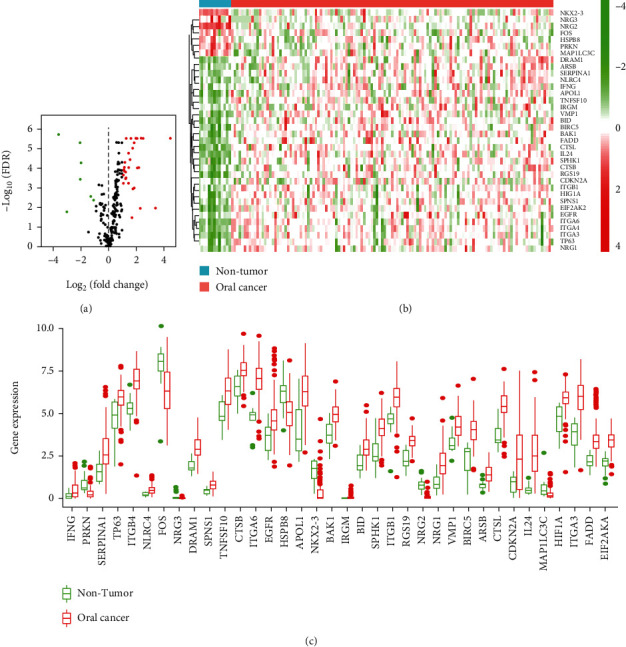
Differentially expressed autophagy-related genes (DEARGs) in ORCA tissues. (a) Volcano plot of autophagy-related genes (ARGs) in ORCA. Green: downregulated genes; red: upregulated genes. (b, c) Heat map and boxplot of the expression levels of 36 DEARGs in ORCA. ORCA: oral cancer.
Table 1.
All DEARGs, screened between normal mouth tissues and oral cancer tissues with criteria of FDR < 0.05 and ∣log2FoldChange | >1.
| Gene | Log2FC | p value | FDR |
|---|---|---|---|
| IFNG | 1.69529 | 0.02038 | 0.03289 |
| PRKN | -1.97667 | 5.69E-06 | 5.30E-05 |
| SERPINA1 | 1.87471 | 0.00030 | 0.00100 |
| TP63 | 1.07599 | 0.00015 | 0.00058 |
| ITGB4 | 1.73712 | 6.51E-07 | 9.53E-06 |
| NLRC4 | 1.25024 | 6.76E-05 | 0.00031 |
| FOS | -1.27976 | 0.00098 | 0.00276 |
| NRG3 | -3.00689 | 0.00871 | 0.01666 |
| DRAM1 | 1.46342 | 1.35E-06 | 1.69E-05 |
| SPNS1 | 1.13948 | 1.15E-05 | 9.07E-05 |
| TNFSF10 | 1.44512 | 0.00015 | 0.00059 |
| CTSB | 1.04712 | 4.40E-05 | 0.00021 |
| ITGA6 | 2.50098 | 1.04E-07 | 2.99E-06 |
| EGFR | 1.76767 | 0.00033 | 0.00109 |
| HSPB8 | -1.07785 | 0.00166 | 0.00420 |
| APOL1 | 2.08150 | 1.27E-05 | 9.30E-05 |
| NKX2-3 | -2.06465 | 3.12E-07 | 4.92E-06 |
| BAK1 | 1.13452 | 2.07E-05 | 0.00013 |
| IRGM | 2.29365 | 0.00536 | 0.01098 |
| BID | 1.23911 | 2.67E-05 | 0.00015 |
| SPHK1 | 1.54880 | 3.17E-06 | 3.61E-05 |
| ITGB1 | 1.46611 | 1.11E-05 | 9.07E-05 |
| RGS19 | 1.27753 | 9.97E-08 | 2.99E-06 |
| NRG2 | -3.60739 | 9.26E-09 | 1.90E-06 |
| NRG1 | 2.20708 | 1.23E-05 | 9.30E-05 |
| VMP1 | 1.13398 | 1.88E-05 | 0.00012 |
| BIRC5 | 2.00367 | 1.17E-07 | 2.99E-06 |
| ARSB | 1.22169 | 7.62E-05 | 0.00035 |
| CTSL | 1.85491 | 2.84E-07 | 4.92E-06 |
| CDKN2A | 3.39449 | 0.00511 | 0.01069 |
| IL24 | 4.48142 | 7.85E-08 | 2.99E-06 |
| MAP1LC3C | -2.03068 | 8.33E-05 | 0.00037 |
| HIF1A | 1.17363 | 4.80E-06 | 5.01E-05 |
| ITGA3 | 2.34824 | 1.04E-07 | 2.99E-06 |
| FADD | 2.08295 | 1.12E-07 | 2.99E-06 |
| EIF2AK2 | 1.61684 | 6.43E-08 | 2.99E-06 |
3.2. Functional Annotation for All DEARGs
In Figure2(a), all DEARGs are linked to form a protein-protein interaction network. The DEARGs are enriched in several biological processes (BP) such as autophagy and protein localization. In the molecular function (MF) term of GO analysis, some protein binding-related functions including “protease binding,” “integrin binding,” and “ubiquitin protein ligase binding” were significant roles these DEARGs played. For cellular components (CC), “focal adhesion,” “vacuolar membrane,” and “cell substrate” were significantly enriched (Figure2(b)). KEGG analysis exhibited that the DEARGs were significantly associated with apoptosis, platinum drug resistance, and hepatitis C (Figure2(c)).
Figure 2.
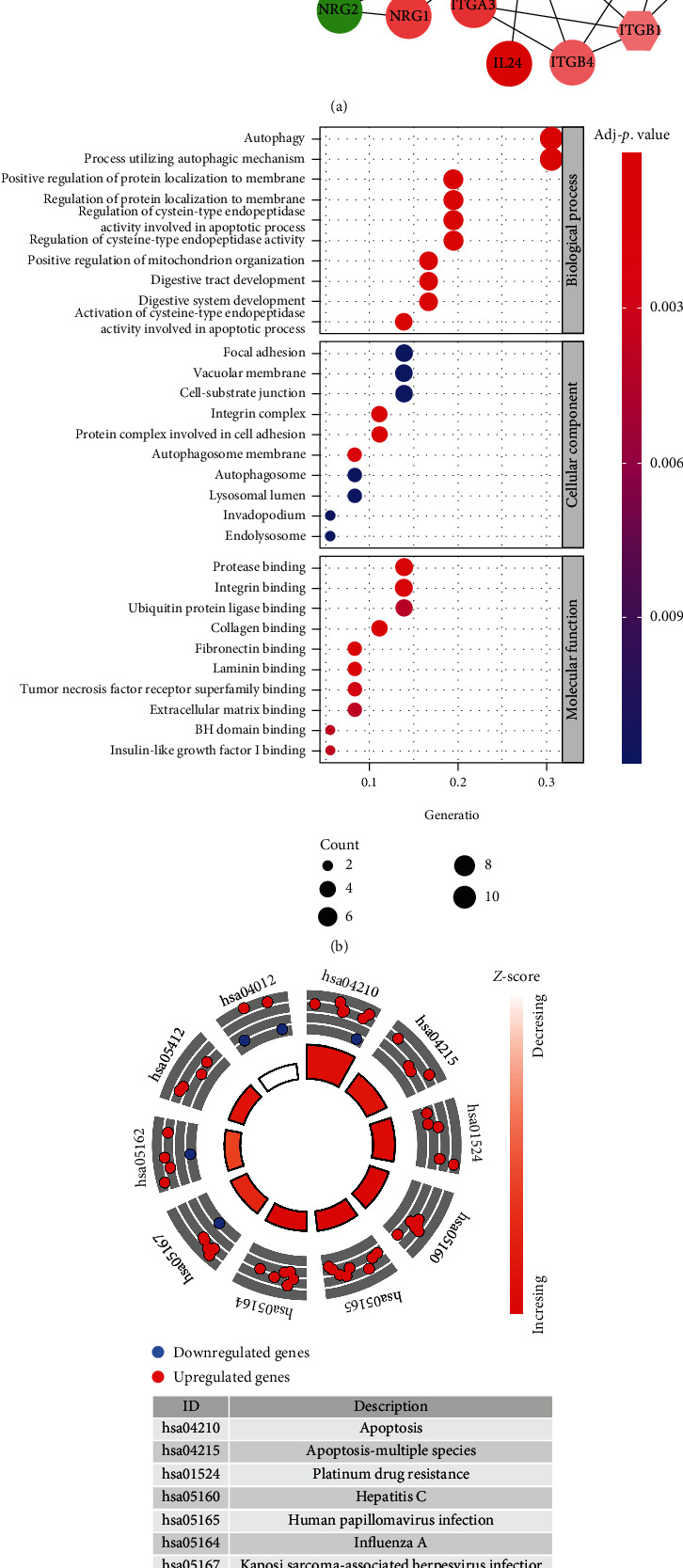
PPI network and functional enrichment analysis for 36 DEARGs. (a) PPI network of 36 DEARGs. Diamond nodes are hub genes which interact with more than 5 genes. Red nodes were upregulated DEARGs; green nodes were downregulated DEARGs. The color depth and size of nodes were related to log2FoldChange and p value, respectively. The width of links was significantly associated with combined score of protein interaction. (b) Gene Ontology analysis of DEARGs. (c) The circle plot of KEGG pathway analyses for DEARGs.
3.3. Establishment of Prognosis Prediction Model with ARGs
We performed the univariate Cox proportional hazard analysis on all ARGs; a total of 13 ARGs were significantly associated with the prognosis of ORCA patients (p < 0.05) (Figure 3(a)). Then, these prognostic ARGs entered the multivariate Cox regression analysis, and finally, 4 of them (WDR45, MAPK9, VEGFA, and ATIC) were screened as independent prognostic genes to construct the prognosis prediction model (Figure 3(b)). For each ORCA patient, the risk score = (−0.5801 × expression value of WDR45) + (0.8974 × expression value of MAPK9) + (0.2978 × expression value of VEGFA) + (0.6182 × expression value of ATIC). We divided the 126 ORCA cases into high- or low-risk groups according to the median values of the risk score. In Figure 3(c), the above 4 independent prognostic ARGs were incorporated into a nomogram model for predicting the individualized probability of survival times in clinical practice in ORCA patients. The score of every ARG can be found through the point scale located at the top of the nomogram. Then, the points of each ARG were summed, thereby estimating survival probability at 1, 2, and 3 years.
Figure 3.
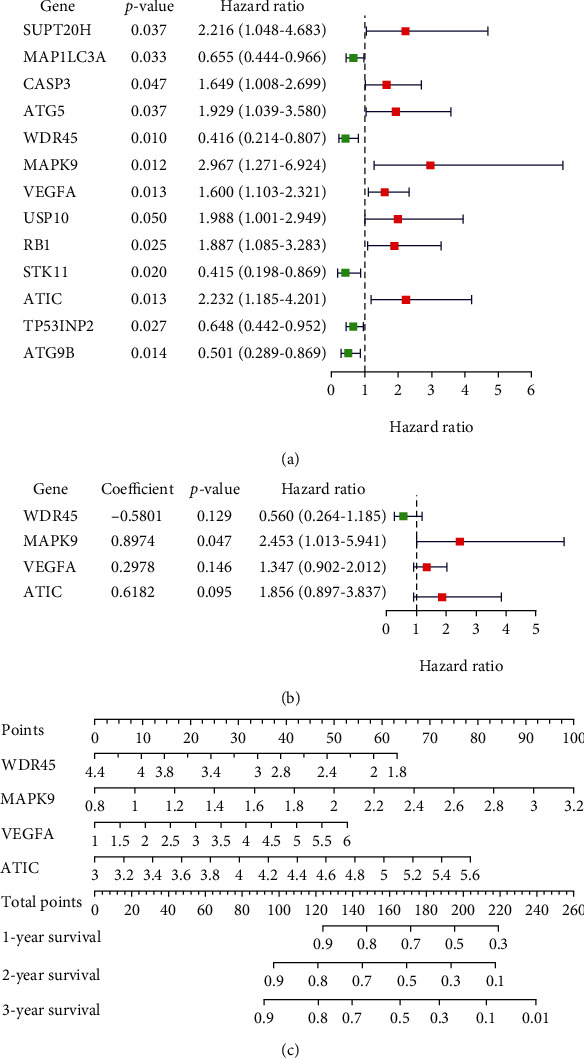
Establishment of a prognostic risk model making use of ARGs for ORCA patients. (a) 13 ARGs were prognostic genes based on univariate Cox analysis. (b) Regression coefficients, hazard ratios, and p value of 4 independent prognostic ARGs obtained from multivariate Cox model. (c) The nomogram prediction for overall survival probability at 1, 2, and 3 years for ORCA patients in TCGA cohort.
3.4. Testing the Risk Model
Patients with ORCA were stratified into high- and low-risk groups based on the median risk score, and Kaplan-Meier plots indicated that patients in the low-risk group had a significantly better prognosis (p = 4.425E − 05) (Figure 4(a)). The results of ROC curves revealed that this prognostic risk model had good predictive performance in predicting the overall survival of ORCA patients with areas under the ROC curve (AUCs) of 0.742 (Figure 4(b)). Figures 4(c)–4(e) indicate the risk distribution of patients, survival time of patients in high- and low-risk groups, and expression profile heat map of the 4 ARGs that formed the risk model.
Figure 4.
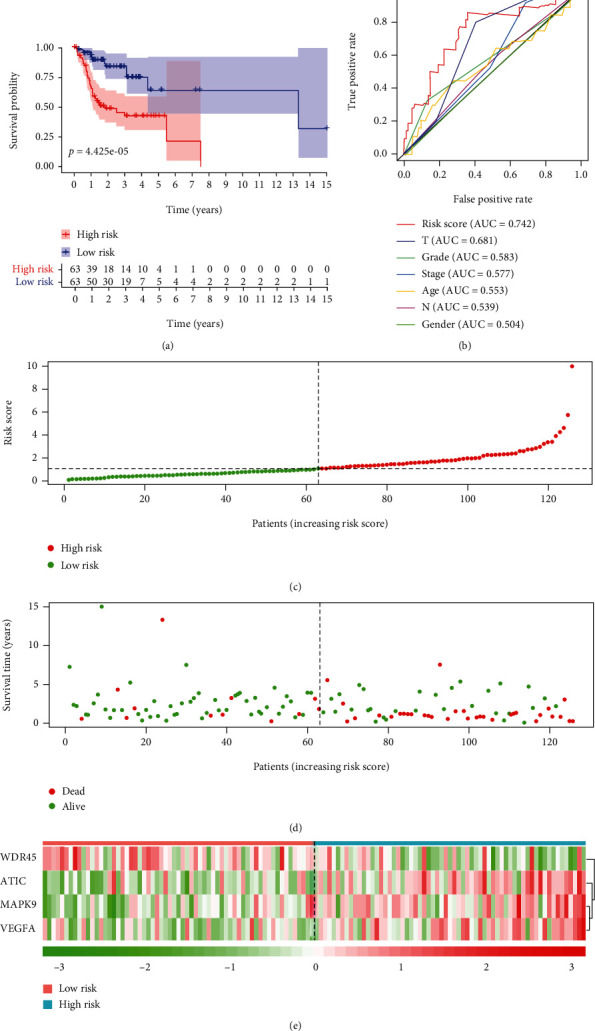
Validation of the prognostic risk model. (a) Kaplan-Meier curves of high-risk and low-risk ORCA patients. (b) ROC curves of risk score and traditional clinical parameters for prognosis predicting. (c) The distribution of risk scores. Each dot represents an ORCA patient. (d) The survival status of ORCA patients with different risk scores. (e) Heat map of 4 prognostic ARGs revealed the relationship between risk score and risk gene expression level.
We grouped the patients by age, sex, histological grade, clinical stage, and T/N classification to explore the association between the risk model and clinicopathological features. In Figure 5, we can find a trend of risk scores which were higher in patients classified with high grade, advanced pathologic stage, and terminal T/N stage (Figure 5(a)). The expressions of MAPK9, VEGFA, and ATIC all had a trend of being upregulated in patients with high grade, advanced pathologic stage, and terminal T/N stage; in addition, the expressions of WDR45 had an opposite trend (Figures 5(b)–5(e)). The expression regularity of 4 independent prognostic ARGs that comprised the risk model in patients with different clinicopathological features was in accordance with their expression trend in different risk groups. The expressions of MAPK9, VEGFA, and ATIC were higher in the high-risk group, and the expressions of WDR45 was higher in the low-risk group (Figure 4(e)). It proved that the risk prognostic model was better than any other clinicopathological features for predicting the prognosis for ORCA patients.
Figure 5.
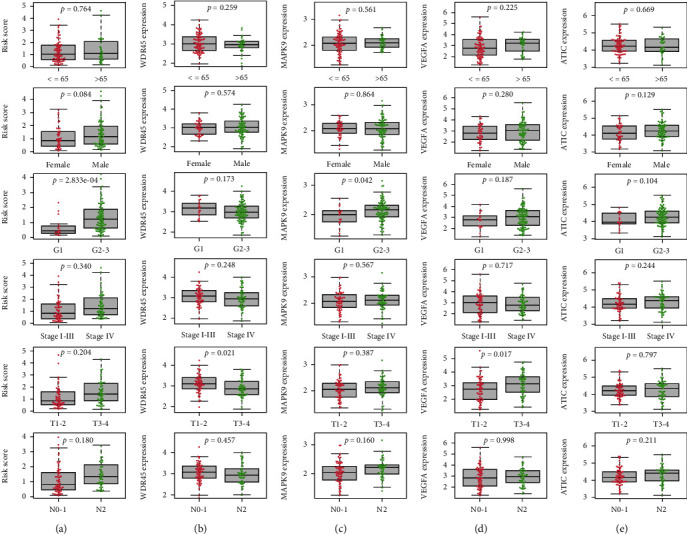
Clinical correlations between traditional clinicopathological indicators (age, gender, grade, pathological stage, T classification, and N classification) and the risk score (a). The association between expression of 4 prognostic ARGs [(b) WDR45, (c) MAPK9, (d) VEGFA, and (e) ATIC] and clinicopathological features.
The results in Table 2 suggested that our prognostic risk model could independently predict the prognosis of ORCA. Although T classification was also significantly associated with survival, ROC analysis demonstrated that T classification (AUC = 0.681) was not as reliable as the risk score (AUC = 0.742).
Table 2.
Univariate and multivariate Cox regression analyses of risk score and clinicopathologic features in the TCGA group oral cancer patients.
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Risk score | 1.622(1.354−1.942) | <0.001 | 1.628(1.327−1.997) | <0.001 |
| Age | 1.006(0.979−1.035) | 0.647 | 1.002(0.967−1.038) | 0.916 |
| Gender | 0.798(0.408−1.561) | 0.510 | 0.604(0.300−1.215) | 0.157 |
| Grade | 2.043(1.146−3.643) | 0.015 | 1.510(0.770−2.960) | 0.230 |
| Pathologic stage | 1.547(1.047−2.285) | 0.028 | 0.851(0.356−2.031) | 0.716 |
| T classification | 1.724(1.214−2.447) | 0.002 | 1.715(1.014−2.902) | 0.044 |
| N classification | 1.481(1.023−2.143) | 0.037 | 1.356(0.712−2.583) | 0.355 |
3.5. Validation of 4 Risk Genes that Comprised the Risk Model
WDR45, MAPK9, VEGFA, and ATIC were 4 prognostic-related ARGs which formed the prognostic risk model. As shown in Figure 6(a), upregulation of WDR45 was strongly correlated with longer patient survival, and MAPK9, VEGFA, and ATIC overexpression reduced OS in patients with ORCA. We also verified the expression of 4 risk genes in the Oncomine database (Figure 6(b)) and found that the expression trend of 4 prognostic ARGs is in accordance with their expression patterns in ORCA, and there are no tumor tissues in previous results in this paper in the TCGA database as shown in Figure 4(e) and Figures 5(b)–5(e). Single-gene GSEA of the 4 risk genes revealed their potential function in ORCA (Figure 7).
Figure 6.
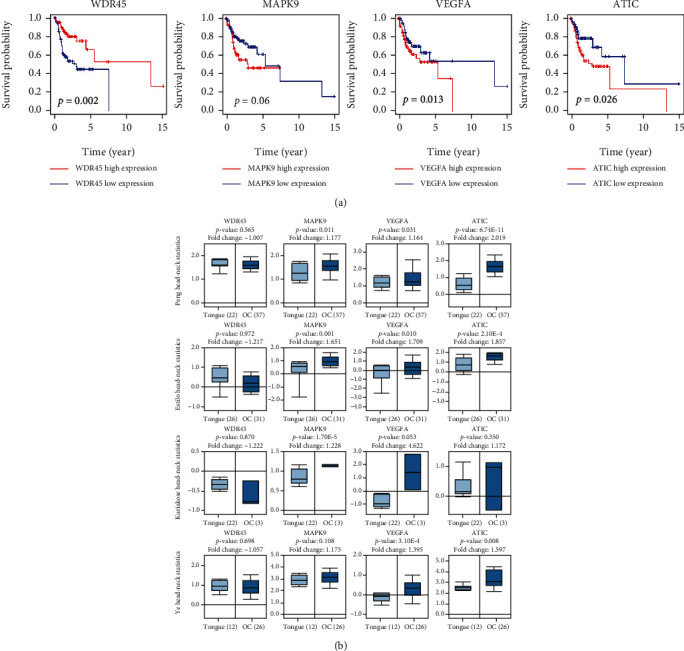
The correlation between 4 risk ARGs (WDR45, MAPK9, VEGFA, and ATIC) and overall survival of ORCA patients (a). The expressions of 4 risk genes (WDR45, MAPK9, VEGFA, and ATIC) between normal mouth and ORCA tissues were compared in Oncomine database (b).
Figure 7.
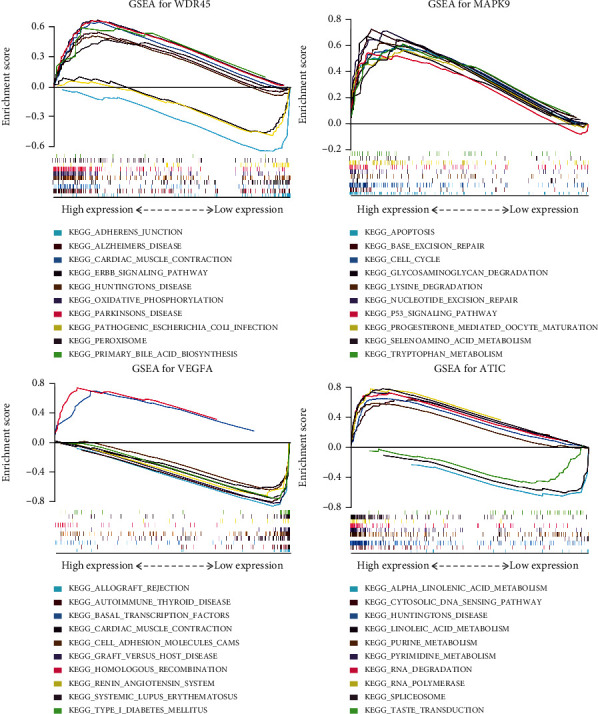
Single-gene GSEA analysis of 4 prognostic ARGs which formed the prognostic risk model in ORCA.
4. Discussion
Autophagy is a conserved and dynamic process whose function maintains cellular homeostasis [14]. Multiple studies had confirmed that autophagy played a significant role in many cancers [15–17]. Important experimental evidences had proven autophagy's potential as a therapeutic target for ORCA [18–20]. Hence, we constructed an autophagy-related prognostic risk model in this paper for prognosis prediction for ORCA patients.
The autophagy-related prognostic model is formed by 4 ARGs, including WDR45, MAPK9, VEGFA, and ATIC. According to our risk model, the expressions of MAPK9, VEGFA, and ATIC in the high-risk group were higher than those in the low risk-group; it is worth noting that their expressions in ORCA tissues were higher than those in the control, and their expressions all had a trend of upregulation in patients with advanced pathologic stage and high grade; in addition, high expressions of the 3 genes were significantly related to worse OS. What this means is that MAPK9, VEGFA, and ATIC were indeed high-risk factors in ORCA. The expression law of WDR45 was opposite to MAPK9, VEGFA, and ATIC and proved it was a low-risk factor in ORCA. The risk score model is significantly associated with clinicopathological indicators of ORCA patients. Our paper proved that the risk model comprised of WDR45, MAPK9, VEGFA, and ATIC for evaluating the prognosis of ORCA patients is clinically practicable.
MAPK9 is also called JNK2; recent studies have highlighted the oncogenic potential of MAPK9 in several human cancer cells, such as lung [21] and glioblastoma [22]. The role of MAPK9 in oral cancer is still controversial [23]. VEGFA encoded vascular endothelial growth factor A; many researchers reported that it was upregulated in oral squamous cell carcinomas [24, 25]. ATIC was considered an effective target for chemoradiosensitization [26]. Although there are some researches about independent prognostic ARGs in recent years, they are not incorporated into a prognostic evaluation system in the right proportions and ways, so exploring an effective prognostic risk model and novel targets for ORCA therapy is necessary.
Functional enrichment analysis showed that 36 DEARGs were mainly involved in apoptosis and inflammation-associated pathways. Dwivedi et al. reported that to regulate apoptosis may be the best way against ORCA [27]. A lot of reports deemed that oral inflammation promotes ORCA [28, 29]. Hence, clinical doctors maybe should pay more attention to the effect of inflammation on ORCA patients.
In this paper, we demonstrated that the novel prognostic risk model had excellent potential as a prognostic predictor which is even better than traditional clinicopathological indicators in ORCA. It can be incorporated into the clinical evaluation indicators to better predict clinical outcomes. Individual assessment of 4 risk genes that comprised the risk model in ORCA further proved that WDR45, MAPK9, VEGFA, and ATIC all play important parts in ORCA.
5. Conclusions
A novel autophagy-related model based on the expression levels of 4 risk genes was explored for survival prediction for ORCA patients. The 4 risk genes can benefit the underlying molecular mechanisms of ORCA and be utilized as potential therapeutic targets.
Acknowledgments
The study was supported by the Nosocomial Scientific Research Fund Projects from the International Peace Maternity and Child Health Hospital of Shanghai Jiao Tong University School of Medicine (No. GFY5801), Clinical Research Special Projects from the Shanghai Municipal Health Commission (No. 20204Y0230), and Shanghai Sailing Program from the Shanghai Science and Technology Committee (No. 19YF1452200). We also appreciate the support of the Youth Science and Technology Innovation Studio, Shanghai Jiao Tong University School of Medicine.
Abbreviations
- ARGs:
Autophagy- related genes
- AUC:
Area under the curve
- DEARGs:
Differentially expressed autophagy- related genes
- DEGs:
Differentially expressed genes
- FDR:
False discovery rate
- GO:
Gene Ontology
- GSEA:
Gene set enrichment analysis
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- KM plotter:
Kaplan-Meier plotter
- Log2FC:
Log2FoldChange/logarithm of fold change
- ORCA:
Oral cancer
- OS:
Overall survival
- PPI:
Protein-protein interaction
- ROC:
Receiver-operator characteristic
- TCGA:
The Cancer Genome Atlas.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Authors' Contributions
HF designed this study and drafted this paper. HF and XC conducted experiments and did statistical analysis. HF searched the references and participated in making the figure. HF revised the manuscript and supervised all the work.
References
- 1.Siegel R. L., Miller K. D., Fuchs H. E., Jemal A. Cancer statistics, 2021. CA: a Cancer Journal for Clinicians . 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Geum D. H., Roh Y. C., Yoon S. Y., et al. The impact factors on 5-year survival rate in patients operated with oral cancer. Journal of the Korean Association of Oral and Maxillofacial Surgeons . 2013;39(5):207–216. doi: 10.5125/jkaoms.2013.39.5.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung H., Ferlay J., Siegel R. L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians . 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Rikardsen O. G., Bjerkli I. H., Uhlin-Hansen L., Hadler-Olsen E., Steigen S. E. Clinicopathological characteristics of oral squamous cell carcinoma in northern Norway: a retrospective study. Bmc Oral Health . 2014;14(1) doi: 10.1186/1472-6831-14-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen S. W., Zhang Q., Guo Z. M., et al. Trends in clinical features and survival of oral cavity cancer: fifty years of experience with 3,362 consecutive cases from a single institution. Cancer Management and Research . 2018;Volume 10:4523–4535. doi: 10.2147/CMAR.S171251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandra T., Marina I. M., Daniela M., et al. Autophagy-a hidden but important actor on oral cancer scene. International Journal of Molecular Sciences . 2020;21(23):p. 9325. doi: 10.3390/ijms21239325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhury R. M., Singh G., Joshi A., Singh D. K., Bhatia S., Goswami S. Autophagy and oral cancers: a short review. Journal of Stomatology, Oral and Maxillofacial Surgery . 2018;119(1):37–39. doi: 10.1016/j.jormas.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Peña-Oyarzún D., Reyes M., Hernández-Cáceres M. P., et al. Role of autophagy in the microenvironment of oral squamous cell carcinoma. Frontiers in Oncology . 2020;10 doi: 10.3389/fonc.2020.602661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Wang C., Tang H., et al. Decrease of autophagy activity promotes malignant progression of tongue squamous cell carcinoma. Journal of Oral Pathology & Medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. . 2013;42(7):557–564. doi: 10.1111/jop.12049. [DOI] [PubMed] [Google Scholar]
- 10.Tang J. Y., Hsi E., Huang Y. C., Hsu N. C., Chu P. Y., Chai C. Y. High LC3 expression correlates with poor survival in patients with oral squamous cell carcinoma. Human Pathology . 2013;44(11):2558–2562. doi: 10.1016/j.humpath.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Ávalos Y., Canales J., Bravo-Sagua R., Criollo A., Lavandero S., Quest A. F. Tumor suppression and promotion by autophagy. BioMed Research International . 2014;2014:15. doi: 10.1155/2014/603980.603980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J. C., Yang D. Z., Wang W., et al. Inhibition of autophagy by 3-MA enhances IL-24-induced apoptosis in human oral squamous cell carcinoma cells. Journal of Experimental & Clinical Cancer Research . 2015;34(1) doi: 10.1186/s13046-015-0211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patil S., Rao R. S., Raj A. T. Dual role of autophagy in oral cancer. Journal of International Oral Health: JIOH . 2015;7(6):i–ii. [PMC free article] [PubMed] [Google Scholar]
- 14.Klionsky D. J., Emr S. D. Autophagy as a regulated pathway of cellular degradation. Science . 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fei H., Chen S., Xu C. Construction autophagy-related prognostic risk signature combined with clinicopathological validation analysis for survival prediction of kidney renal papillary cell carcinoma patients. BMC Cancer . 2021;21(1):p. 411. doi: 10.1186/s12885-021-08139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fei H., Chen S., Xu C. Construction autophagy-related prognostic risk signature to facilitate survival prediction, individual treatment and biomarker excavation of epithelial ovarian cancer patients. Journal of Ovarian Research . 2021;14(1):p. 41. doi: 10.1186/s13048-021-00791-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy J. M. M., Towers C. G., Thorburn A. Targeting autophagy in cancer. Nature Reviews. Cancer . 2017;17(9):528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi T. T., Yu X. X., Yan L. J., Xiao H. T. Research progress of hydroxychloroquine and autophagy inhibitors on cancer. Cancer Chemotherapy and Pharmacology . 2017;79(2):287–294. doi: 10.1007/s00280-016-3197-1. [DOI] [PubMed] [Google Scholar]
- 19.Gasiorkiewicz B. M., Koczurkiewicz-Adamczyk P., Piska K., Pekala E. Autophagy modulating agents as chemosensitizers for cisplatin therapy in cancer. Investigational New Drugs . 2021;39(2):538–563. doi: 10.1007/s10637-020-01032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng S., Shanmugam M. K., Kumar A. P., Yap C. T., Sethi G., Bishayee A. Targeting autophagy using natural compounds for cancer prevention and therapy. Cancer . 2019;125(8):1228–1246. doi: 10.1002/cncr.31978. [DOI] [PubMed] [Google Scholar]
- 21.Nitta R. T., Del Vecchio C. A., Chu A. H., Mitra S. S., Godwin A. K., Wong A. J. The role of the c-Jun N-terminal kinase 2-α-isoform in non-small cell lung carcinoma tumorigenesis. Oncogene . 2011;30(2):234–244. doi: 10.1038/onc.2010.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui J., Han S. Y., Wang C., et al. c-Jun NH2-Terminal kinase 2α2 promotes the tumorigenicity of human glioblastoma cells. Cancer Research . 2006;66(20):10024–10031. doi: 10.1158/0008-5472.CAN-06-0136. [DOI] [PubMed] [Google Scholar]
- 23.Gkouveris I., Nikitakis N. G. Role of JNK signaling in oral cancer: a mini review. Tumour Biology: the journal of the International Society for Oncodevelopmental Biology and Medicine . 2017;39(6, article 1010428317711659) doi: 10.1177/1010428317711659. [DOI] [PubMed] [Google Scholar]
- 24.Sales C. B., Buim M. E., de Souza R. O., et al. Elevated VEGFA mRNA levels in oral squamous cell carcinomas and tumor margins: a preliminary study. Journal of Oral Pathology & Medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology . 2016;45(7):481–485. doi: 10.1111/jop.12398. [DOI] [PubMed] [Google Scholar]
- 25.Naruse T., Kawasaki G., Yanamoto S., Mizuno A., Umeda M. Immunohistochemical study of VEGF expression in oral squamous cell carcinomas: correlation with the mTOR-HIF-1alpha pathway. Anticancer Research . 2011;31(12):4429–4437. [PubMed] [Google Scholar]
- 26.Liu X. F., Paila U. D., Teraoka S. N., et al. Identification of ATIC as a novel target for chemoradiosensitization. International Journal of Radiation Oncology • Biology • Physics . 2018;100(1):162–173. doi: 10.1016/j.ijrobp.2017.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dwivedi R., Pandey R., Chandra S., Mehrotra D. Apoptosis and genes involved in oral cancer - a comprehensive review. Oncology Reviews . 2020;14(2):108–120. doi: 10.4081/oncol.2020.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feller L., Altini M., Lemmer J. Inflammation in the context of oral cancer. Oral Oncology . 2013;49(9):887–892. doi: 10.1016/j.oraloncology.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Goertzen C., Mahdi H., Laliberte C., et al. Oral inflammation promotes oral squamous cell carcinoma invasion. Oncotarget . 2018;9(49):29047–29063. doi: 10.18632/oncotarget.25540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.


