Abstract
The biophysical features of a cell can provide global insights into diverse molecular changes, especially in processes like the dedifferentiation of chondrocytes. Key biophysical markers of chondrocyte dedifferentiation include flattened cellular morphology and increased stress-fiber formation. During cartilage regeneration procedures, dedifferentiation of chondrocytes during in vitro expansion presents a critical limitation to the successful repair of cartilage tissue. Our study investigates how biophysical changes of chondrocytes during dedifferentiation influence the nuclear mechanics and gene expression of structural proteins located at the nuclear envelope. Through an experimental model of cell stretching and a detailed spatial intranuclear strain quantification, we identified that strain is amplified and the distribution of strain within the chromatin is altered under tensile loading in the dedifferentiated state. Further, using a confocal microscopy image-based finite element model and simulation of cell stretching, we found that the cell shape is the primary determinant of the strain amplification inside the chondrocyte nucleus in the dedifferentiated state. Additionally, we found that nuclear envelope proteins have lower gene expression in the dedifferentiated state. This study highlights the role of cell shape in nuclear mechanics and lays the groundwork to design biophysical strategies for the maintenance and enhancement of the chondrocyte phenotype during cell expansion with a goal of successful cartilage tissue engineering.
Significance
Chondrocytes dedifferentiate into a fibroblast-like phenotype in a non-native physical environment. Using high-resolution microscopy, intranuclear strain analysis, and finite element method-based computational modeling, we investigated how mechanical force causes abnormal intranuclear strain distribution in chondrocytes during the dedifferentiation process. Overall, our results suggest that the altered cell geometry is responsible for abnormal intranuclear strain during chondrocyte dedifferentiation, and the dedifferentiated state is associated with the lower gene expression of several nuclear envelope proteins.
Introduction
The mechanics of the cell nucleus and their biological implications, termed nuclear mechanobiology, have recently been shown to be important in numerous biological contexts: diseases (1), gene expression (2), differentiation (3, 4), developmental biology (5), and tissue homeostasis (6). The study of nuclear mechanobiology investigates how the mechanical microenvironment (i.e., stretch, compression, and shear) affects the spatiotemporal function of the nuclear envelope, the nucleoskeleton, the chromatin architecture, and hence the gene regulation. However, it is not well understood how mechanical stretch and differentiation-mediated cell morphology changes affect the intranuclear mechanics. In this study, dedifferentiation of chondrocytes into fibroblast-like cells in culture is used as a model system to investigate the effect of the differentiation process in the nuclear mechanics. The dedifferentiation of chondrocytes into a fibroblast-like phenotype during the expansion process in two-dimensional (2D) monolayer culture provides an ideal system to investigate the relationship between the cell mechanical environment and nuclear mechanics.
Chondrocyte-based tissue engineering has received specific attention for treating osteoarthritis, a debilitating disease affecting a large worldwide population (7). Pharmaceutical therapies to treat this degenerative disease are limited, and current disease management protocols rely on lifestyle changes and the use of medications to ameliorate pain or to reduce inflammation. Chondrocyte-based tissue engineering is a promising approach with the goal to rebuild the damaged articular cartilage in order to restore the functions of the knee joint (8). Autologous chondrocyte implantation (ACI) is an emerging technology for cartilage tissue engineering, where chondrocytes extracted from the patient are expanded in vitro and then implanted back in the knee joint to regenerate functional articular cartilage (8, 9, 10). However, there is a critical challenge that limits the potential widespread application of this technology, which involves the expansion of the chondrocytes in vitro. During the expansion process and passaging in 2D monolayer culture, the chondrocytes dedifferentiate into fibroblast-like cells. Consequently, the scalability of the ACI technology is limited because growing enough cells for a large defect requires extensive time, which can further dedifferentiate chondrocytes in 2D culture. From clinical follow-up studies, it is now apparent that the success of the ACI treatment is limited because of post-operative fibrosis that leads to inferior biomechanical and functional response of the knee joint (11). Those clinical studies also indicate the consequence of the in vitro dedifferentiation before the implantation process.
Chondrocytes are known to be mechanosensitive. It is well known that mechanical loading on chondrocytes in the native cartilage environment induces several pathophysiological responses mediated by several mechanobiological pathways (12, 13, 14). In vitro, the monolayer culture of chondrocytes on a dish imparts a passive 2D mechanical stretch on the cell, aided by the flat cell shape and contractile forces imparted by F-actin stress fibers. Dedifferentiated chondrocytes in monolayer culture have a higher cell stiffness mediated by a cell membrane to F-actin adhesion mechanism (15), indicating higher contractile forces within the cell. Interestingly, several studies suggest that, if the chondrocytes are instead encapsulated in a three-dimensional (3D) environment, the chondrocyte phenotype prevails. Even the late-passage chondroblasts (a term used to define a cell showing the phenotype of both chondrocyte and fibroblast) redifferentiate into chondrocytes (16, 17) when cultured in 3D, suggesting that release of the cellular contraction experienced in 2D ameliorates the dedifferentiation phenotype.
Mechanical loading affects not only the chondrocyte cytoskeletal mechanics but also the cell nucleus as well (18, 19, 20), which might have unknown implications in the chondrocyte dedifferentiation process. A recent study found that the loss of extracellular matrix stiffness and the lack of 3D chondrocyte matrix decreases the cell and nuclear stiffness in chondrocytes, suggesting a critical role for the cell's mechanical environment in the maintenance of the chondrocyte phenotype and nuclear mechanics (21). Another recent study discovered that, under shear stress, the chondrocyte nucleus experiences complex strain patterns resulting from heterogeneous intranuclear chromatin architecture, further highlighting the importance of chondrocyte nuclear mechanics (22). However, in the context of chondrocyte dedifferentiation, it is not understood how the mechanical stretch and gradual dedifferentiation-mediated alteration in the chondrocyte's biophysical features affect the intranuclear mechanics and the structural integrity of the nucleus. Such knowledge could be potentially harnessed for improving cartilage tissue engineering as intranuclear mechanics can directly affect the genomic expression stability of cells (23).
In this study, we hypothesized that cell spreading of chondrocytes during the expansion processes results in altered cell mechanics leading to an abnormal strain burden in the nucleus via flat cell morphology and weaker nuclear envelope. To test this hypothesis, we mechanically stretched the chondrocytes in a controlled manner to visualize the strain patterns in early-passage (population doubling 0 [PD0]) and late-passage chondrocyte (population doubling 16 [PD16]) nuclei using a technique already established in the context of studying chondrocyte dedifferentiation and pathology (24, 25, 26). We found an amplified strain magnitude and an abnormal distribution of strain inside the chromatin of the chondrocyte that might be associated with further degeneration of the chondrocyte phenotype, as indicated by changes in gene expression of the dedifferentiated chondrocytes compared with early-passage chondrocytes. Next, using image-based computational modeling of cell stretching, we found that the flattened cellular morphology is a key mediator of higher strain inside the cell nucleus. Gene expression analysis of chondrocytes suggested lower expression of several structural proteins in the cell nucleus, the implication of which could be further investigated in the context of chondrocyte dedifferentiation and maintenance.
Materials and methods
Chondrocyte extraction and culture
Primary bovine chondrocytes were harvested from the load-bearing region of the medial condyle from young bovine joints less than 6 months old (27, 28). The cells were seeded directly on the substrate after harvesting and these cells were described as PD0 cells, corresponding to passage 0. Harvested cells were cultured in DMEM-F12 (Gibco) with 10% fetal bovine serum (FBS) and passaged at around 80% confluency on petri dishes (Corning) until PD16, corresponding to passage 4.
Imaging and geometrical characterization of chondrocytes
PD0 and PD16 chondrocytes were cultured on tissue culture petri dishes (Corning), and StageFlexer type I collagen-treated membranes (FlexCell International Corporation) for 24 h prior to fixation to allow complete adhesion. Chondrocytes were fixed with cold 4% paraformaldehyde (Electron Microscopy Sciences) in 1× PBS, washed with 1× PBS three times, permeabilized in 0.1% Triton X-100 (Sigma-Aldrich), and again washed with 1× PBS three times. Next, cells were stained with DAPI (Thermo Fisher Scientific) for nuclear DNA staining (1:500) and Alexa Fluor 488 phalloidin (Thermo Fisher Scientific) for cytoplasmic F-actin staining (1:500). Fluorescent images of the PD0 and PD16 chondrocytes were captured using an inverted confocal microscope (Nikon Eclipse Ti AIR, Nikon, Tokyo, Japan) with a 10× air objective and a 60× oil objective. Cells to image were chosen at random and distributed across the substrate. The area, the length, and the aspect ratio of both the cell and the nucleus for imaged cells were measured using the image analysis software, Fiji.
Gene expression analysis on PD0 and PD16 chondrocytes
Total RNA was extracted from the PD0 and PD16 chondrocytes on type I collagen-treated membranes (FlexCell International Corporation) using Aurum Total RNA Mini Kit, reverse transcribed into cDNA via iScript Reverse Transcription Supermix. Real-time quantitative PCR was performed with SsoAdvanced Universal SYBR Green Supermix in a CFX96 Touch thermocycler (all kits and devices from Bio-Rad Laboratories) using 10 ng of cDNA per reaction. All data were normalized to the reference genes HPRT1 and RPL10A. Primers were custom designed and validated. Primer sequences are listed in Table S1.
Experimental setup for chondrocyte mechanical stretching
The FlexCell (FlexCell International Corporation) cell stretching device was used to apply controlled biaxial mechanical stretching on the chondrocytes. StageFlexer type I collagen-treated membranes (FlexCell International Corporation) were used for the chondrocyte culture and stretching. The device was modified for custom imaging specific to the current application using a two-photon microscope (Olympus with a tunable Mai-Tai laser set at 540–600 nm). The first modification was to convert it from a vacuum device to a positive-pressure device by placing a cap above the membrane (Fig. S1 a). The second modification was to drill out the bottom support bracket under the loading post to allow the microscope objective to fit inside the device. The magnitude of the membrane stretch varied linearly with change in pressure, controlled by a pressure regulator programmed using a LabVIEW interface (Fig. S1, b and c). To calculate the substrate deformation, a previously developed technique was used (29). Briefly, the technique tracked the displacement of beads pre-embedded in the membrane during stretching. Subsequently, strain in the substrate was analyzed using the finite element method. Thus, the equibiaxial stretch mode on the membrane and on the cells was validated (Fig. S1, d and e).
Live chondrocyte stretching and imaging
Cells were incubated on the type I collagen-treated membranes for 24 h to allow complete adhesion prior to the stretching experiments. Nuclear DNA was stained with Hoechst 34580 (Life Technologies) before loading the substrate on the stretching device. The cells remained in the culture medium during imaging. Two-photon microscopy (Olympus, 740-nm wavelength, 250-mW power for imaging the nucleus) with a 40× water objective was used to capture three images 3 min apart (Fig. S1 b) prior to stretching. The membrane, and in turn the chondrocytes, were then stretched. Next, three more images were captured after finding the chondrocytes in the new location. For each image captured, the transmitted light was also used to visualize the overall cell shape. All the captured images were 2D.
Spatial strain quantification inside the nucleus
Chondrocyte nucleus images captured before and after stretching were used to quantify 2D spatial strain inside the nucleus with a technique called deformation microscopy (30, 31). Briefly, the image texture of the intranuclear space was utilized to achieve a high-resolution intranuclear displacement map. Several 2D strain measurements were derived, such as local shear strain (angular deformation: Exy), hydrostatic strain (related to volume change: (Exx + Eyy)/2), and the second deviatoric strain (related to shape change: (Eyy − Exx)/2). In this manuscript, second deviatoric strain is reported as “deviatoric strain” for the sake of simplicity. Hydrostatic strain was further categorized into tensile and compressive strain based on the positive and negative values of the strain. A Hill's function-based thresholding of the nuclear image was performed (32) to automatically distinguish the heterochromatin (high chromatin density) and euchromatin (low chromatin density) domains. In summary, the pixel grayscale intensity from the raw image of a nucleus is plotted after sorting by values that can range from 0 to 255. After fitting a sigmoidal curve to this plot, an inflexion point was calculated to find the cutoff intensity that distinguishes the heterochromatin from euchromatin.
Image-based modeling and simulation of stretch-induced strain in chondrocytes
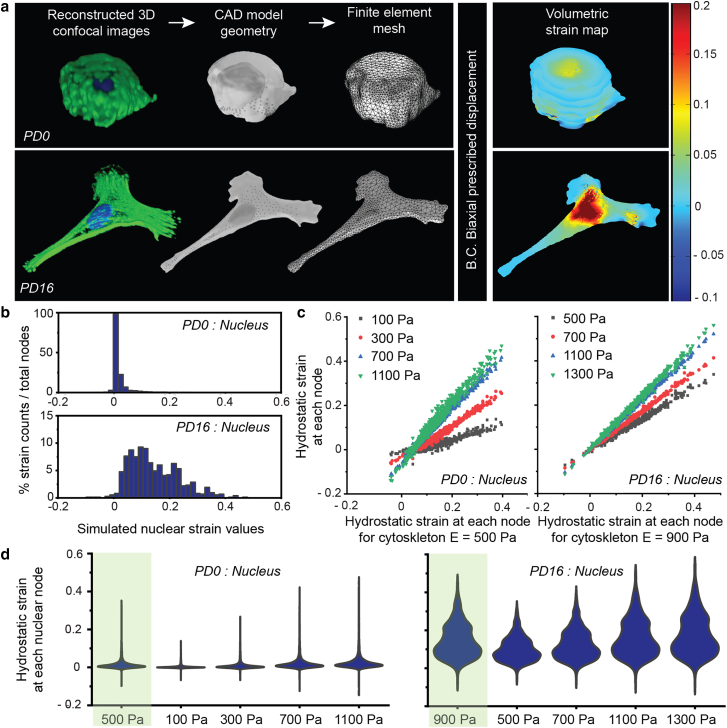
A computational model was developed to quantify the influence of biophysical changes in cell geometry and mechanical properties on the deformation of nucleus at PD0 and PD16 chondrocytes during the chondrocyte equibiaxial stretching experiment. The supplemental note explains the details of the image-based modeling and strain analysis. Briefly, PD0 and PD16 chondrocytes were cultured, fixed, and stained for DNA, F-actin, and vinculin to image the nucleus, cytoplasm, and focal adhesion complex respectively (Fig. 4 a). Three representative cells from each group were chosen at random for subsequent analysis. For each representative cell, the intracellular and intranuclear spaces were segmented to create three separate domains of cytoplasm, heterochromatin, and euchromatin. Based on the confocal image stack, a 3D geometry was created, meshed, and imported into a finite element method software (COMSOL Multiphysics 5.3, Burlington, MA). Further, appropriate governing equations and boundary conditions were used to solve for the strain in the cytoplasm, heterochromatin, and euchromatin.
Figure 4.
The chondrocyte shape change from PD0 to PD16 was a major factor determining spatial patterns of deformation within the cell and nucleus. (a) Confocal image stack of the cell and nucleus, along with the focal adhesion complex, was used to create a 3D model. PD16 cells were flattened with higher area and higher 3D aspect ratio, compared with PD0 cells, which were round with lower area and lower 3D aspect ratio. Appropriate governing equations and boundary conditions were used to analyze intranuclear and intracellular strain using the finite element method. As the cell geometry altered from PD0 to PD16 (i.e., increased 3D aspect ratio and cell area), the tensile strain experienced by the nucleus almost doubled under the same boundary conditions, demonstrating the significant effect of the geometry change. (b) Histogram of the hydrostatic strain burden at the nuclear domain nodes. PD0 chondrocyte nuclei experienced lower value and lower range of strain, whereas PD16 chondrocyte nuclei experienced higher value and higher range of strain. (c) Cell morphology, not mechanical property, was the key determinant of intranuclear strain. Irrespective of the population doubling status and geometry, the distribution of strain at computational nodes did not alter significantly with the variation of cytoskeleton elastic modulus. (d) Violin plots show the complete distribution of hydrostatic strain data at every node of the model, including the average (widest part) for each elastic modulus value. The first violin plot (green background) in each subpanel corresponds to the baseline value of the cytoskeletal elastic modulus used in our model. Also see Figs. S2–S4.
Effect of mechanical property on cell and nuclear strain: parametric study
A parametric study with the in silico cell model was used to investigate the effect of the cell mechanical properties on the intranuclear strain. For the parametric study, the elastic moduli of the cytoskeleton, euchromatin, and heterochromatin domains were varied for the PD0 and PD16 chondrocytes, while Poisson's ratio was held constant. Subsequently, Poisson's ratio was varied independently while other mechanical parameters were held constant. The values chosen for each parametric study are reported in Table S2. The values assigned for each mechanical property are physiologically relevant and represent the extreme ends of the property value. The hydrostatic strain solution at each material node with the baseline mechanical properties (cytoplasm, Young's modulus E = 500 Pa in PD0, and 900 Pa in PD16; euchromatin, E = 1000 Pa in PD0 and PD16; heterochromatin, E = 4000 Pa in PD0 and PD16) was compared against each new solution with a change in the parameter to assess how much the hydrostatic strain solutions differed when the parameters were varied.
Statistical analysis
For cell and nuclear geometry analysis between the population doublings independent of substrate type, t-test was performed with significance at p < 0.01. For strain comparison between population doublings independent of its location at heterochromatin or euchromatin, t-test was performed with significance at p < 0.05. For gene expression analysis study, t-test was performed with significance at p < 0.01. For the analysis of the parametric studies pertaining to the computational model, slope m and coefficient of determination R2 was calculated using linear regression.
Results
Cell and nuclear geometry and morphology
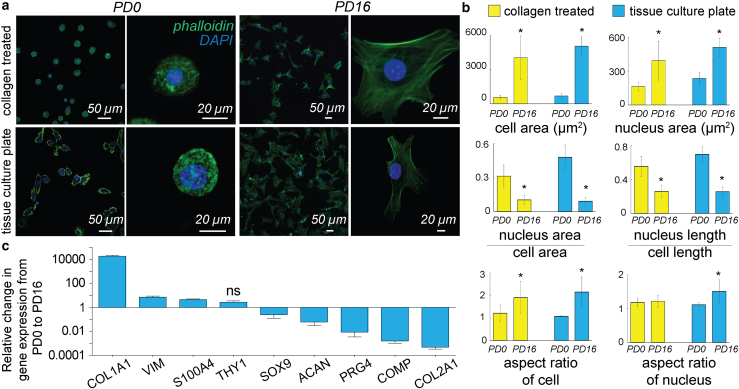
Early-passage (PD0) chondrocytes displayed a round morphology, no visible F-actin stress-fiber formation, and location of the nucleus was centered in the cell on either substrate: type I collagen-treated membranes and tissue culture plastic (TCP) (Fig. 1 a). Late-passage (PD16) cells, in contrast, showed a larger cell area, F-actin stress-fiber formation, and the nucleus was often located off-center. Further quantification of the geometrical measures revealed that, for all substrates, the cell area increased by almost five times and the nuclear area increased by almost two times in PD16 cells compared with PD0 cells (Fig. 1 b). Additionally, the cellular and nuclear morphology of chondrocytes cultured on the type I collagen-treated membranes showed similar phenotypes as TCP, which validated our approach of chondrocyte culture for the mechanical stretching experiment. Along with the cell geometry changes, the nuclear area compared with cell area, and the nuclear linear dimension compared with cell linear dimension, decreased significantly in PD16 compared with PD0, an indicator of deviation from the chondrocyte phenotype to fibroblast-like chondrocyte phenotype, as observed in native cartilage (33). In PD0, the cell aspect ratio was maintained close to 1, signifying a round cell shape. In PD16, the cell aspect ratio deviated significantly from 1, suggesting irregularly shaped cells. The nuclear aspect ratio showed a small deviation from the value 1 in PD16 compared with PD0 chondrocytes on type I collagen-treated membranes, although the effect was not statistically significant. Irrespective of the population doubling number, both the cell and nuclear area increased with the increasing stiffness of the substrate (stiffness of TCP > stiffness of type I collagen-treated membrane), which was expected because a higher stiffness promotes larger cell and nuclear area through higher spreading of cells (34).
Figure 1.
Dedifferentiation of chondrocytes in a monolayer culture was associated with a change in cell and nucleus phenotype as well as changes in extracellular matrix-related gene expression. (a) PD0 (corresponding to passage 0) and PD16 (corresponding to passage 4) chondrocytes were stained for actin (phalloidin-GFP) and DNA (DAPI) to visualize cells and nuclei respectively using confocal microscopy at two different magnifications. Culture conditions included type I collagen-treated membranes and TCP (tissue culture plate). Irrespective of culture condition, the PD0 chondrocytes were smaller, rounder, and displayed fewer F-actin stress fibers than the PD16 chondrocytes. (b) Several biophysical features were used to quantify the cell morphological changes between the PD0 and PD16. From PD0 to PD16, the cell and nuclear area increased, and the nuclear dimensions increased compared with the cell dimensions. Cells lost their round shape and approached a more irregular shape, while nuclei maintained their round shape compared with cells. ∗p < 0.01, number of cells analyzed >60 from three biological replicates. (c) Chondrocyte dedifferentiation from PD0 to PD16 was associated with an increase in the fibroblast-specific extracellular matrix markers, and decrease in chondrocyte-specific extracellular matrix markers. Except THY1, all other gene expression changes were significant with p < 0.01, ns = not significant, number of samples = 4. Error bars represent SD about mean.
Passage-specific gene expression of extracellular matrix proteins
Dedifferentiation of chondrocytes was associated with changes in expression of extracellular matrix proteins and the loss of the hyaline chondrocyte phenotype. The gene expression of chondrocyte-specific proteins (SOX9, ACAN, PRG4, COMP, COL2A1) decreased with dedifferentiation, while the fibroblast marker-specific gene expression (COL1A1, VIM, S100A4, THY1) increased (Fig. 1 c). Our gene expression data from young bovine articular cartilage chondrocyte agree with previous reports of chondrocytes extracted from adult human knee articular cartilage (35), adult pig knee articular cartilage (36), and young bovine meniscus (37).
Complex spatial intranuclear strain pattern
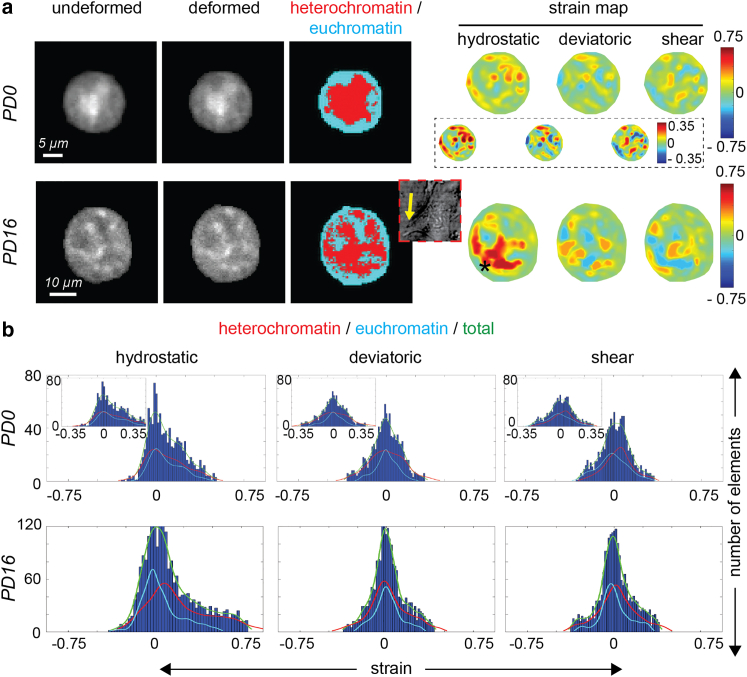
With the significant changes in cell morphology and gene expression between the PD0 and PD16 chondrocyte, we hypothesized that flatter cellular geometry caused by the 2D expansion process might influence strain transfer to the nucleus, contributing to mechanosensitive signaling that drives the dedifferentiation process. High-resolution intranuclear spatial strain maps revealed an inhomogeneous deformation distribution inside PD0 nuclei, while PD16 nuclei displayed a larger strain concentration at specific nuclear locations (Fig. 2 a), although the strain distribution was still inhomogeneous. Shear and deviatoric strain values were comparable in PD0 and PD16. The hydrostatic strain values in the PD0 nuclei were lower overall (up to 0.35) compared with PD16 nuclei (up to 0.75), signifying that the PD16 nuclei displayed a magnified deformation for the same applied stretch on the cell surface. In PD0 chondrocytes, heterochromatin and euchromatin did not show any preferential strain localization. Alternatively, in PD16 chondrocytes, high strain was associated with heterochromatin, which are regions of high-density chromatin. Because heterochromatin is stiffer than euchromatin, such results were surprising. We think such anomalous results in the chondrocyte nuclei in the differentiated state is a consequence of highly heterogeneous stress distribution caused by local stress fibers. Such association of high-strain regions in PD16 nuclei was also predicted from the model, as explained later (Fig. 4). The histograms further showed, in detail, that the high-chromatin-density areas (i.e., the heterochromatin) take more tensile strain burden compared with euchromatin in PD16 cells compared with PD0 cells (Fig. 2 b).
Figure 2.
Passage-dependent and complex spatial strain patterns emerged inside the chondrocyte nucleus upon mechanical stretching of chondrocytes, shown in detail for a representative nucleus from each group. (a) Using a modified FlexCell device, a controlled equibiaxial stretch was applied to a type I collagen-treated membrane with attached chondrocytes (Fig. S1). Nuclei were imaged before stretching (undeformed) and at the stretched condition (deformed), and intranuclear strains were quantified using deformation microscopy, a technique developed previously (30). In PD0 cells, which were round, magnitude of all the strain measures—hydrostatic (volume change related), deviatoric (shape change related), and shear—were comparable with respect to each other. The high-strain pockets were uniformly distributed in the nucleus irrespective of the chromatin density. Inset images shows rescaled strain map in PD0 nucleus. In PD16 cells, which were irregularly shaped, hydrostatic strain was significantly increased with respect to PD0 cells, suggesting strain amplification, while deviatoric and shear strains are comparable in both PD0 and PD16 cell nuclei. High-strain regions (starred) were associated with the heterochromatin (high-chromatin-density regions) in the vicinity of cell protrusions known to be enriched with F-actin stress fibers. Cytoplasm image using the transmitted light is shown in the inset (not in the same scale as the nucleus images), with yellow arrow to show the cell protrusion adjacent to the high-hydrostatic-strain region. (b) Hydrostatic strain in the nucleus was mostly tensile, while deviatoric and shear strains followed mostly a normal distribution commensurate with the tensile cell stretching modality. Plots show histograms of the intranuclear elements for different strain measures for the representative nuclei in (a). In the PD0 nucleus, the heterochromatin (high-density chromatin) and euchromatin (low-density chromatin) strain distribution mostly overlapped, while in the PD16 nucleus the heterochromatin showed a higher tensile strain in most elements. Inset shows rescaled strain distribution for PD0 nucleus.
Abnormal nuclear strain under tension in dedifferentiated cells
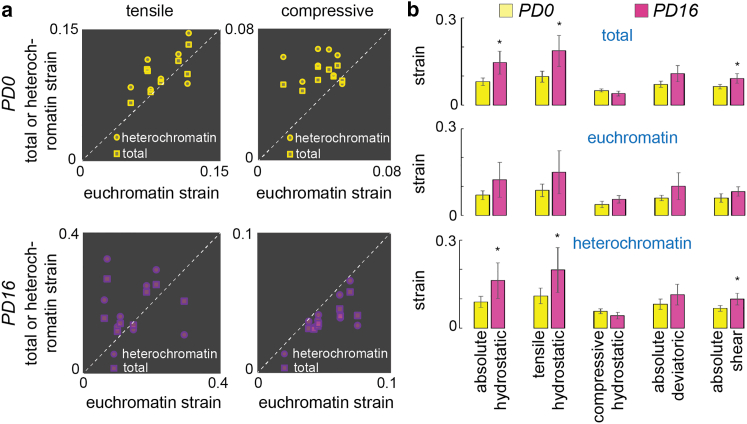
In dedifferentiated chondrocytes, the nucleus not only experienced a higher strain but also experienced an abnormal strain distribution. In PD0 chondrocytes, the heterochromatin showed similar strain (not statistically different) both in the tensile and the compressive modes (Fig. 3). In PD16 chondrocytes, where the stiffer cell most likely imparted higher tension on the nuclear periphery, the heterochromatin experienced a significantly higher strain in tension and a lower strain in compression. Quantitative analysis (Fig. 3 b) further illustrated such strain differences. Overall, in the dedifferentiated state, the heterochromatin experiences higher strain compared with early-passage chondrocytes. Regulation of mechanical homeostasis in heterochromatin is critical for maintaining genome integrity (23, 38). Our results point to an abnormal heterochromatin strain and, independently, abnormal gene expression (Fig. 1 c) in dedifferentiated cells. The present results do not point to any causation between these two events, but future studies will be necessary to elucidate possible relationships between these two observations.
Figure 3.
Dedifferentiation of chondrocytes was associated with abnormal intranuclear strain, most prominently visible in hydrostatic strain measure. (a) Each data point represents the average spatial strain for one specific nucleus. For PD0 chondrocytes, heterochromatin strain and total strain in the nucleus were slightly higher than the euchromatin strain both in the tensile and compressive modes. For PD16 chondrocytes, total strain and heterochromatin strain were lower compared with euchromatin strain in compressive mode, thus signifying a dense, incompressible heterochromatin. In tensile mode, the heterochromatin experienced a higher strain compared with euchromatin. (b) Averaging the strain over multiple nuclei showed that the hydrostatic tensile strain was significantly higher in heterochromatin and in the nucleus as a whole compared with euchromatin, thus signifying an abnormal strain burden in the PD16 nuclei compared with the PD0 nuclei. ∗p < 0.05, number of nuclei analyzed > 6 from three biological replicates. Error bars represent SD about mean.
Computational analysis of cell stretching
To understand why the PD16 chondrocyte nuclei experienced magnified strain, we performed the computational analysis of cell stretching and visualized the intranuclear strain due to the applied cell stretch. The computational analysis and parametric study simulating the chondrocyte biaxial stretch of the chondrocyte reveal that the intracellular and intranuclear strains are primarily influenced by the cell shape and geometry, while the mechanical properties of the cytoskeleton have a smaller effect on the nuclear strain. A lower 3D aspect ratio (maximum linear dimension parallel to the culture surface/maximum cell height) of the cell promotes a lower strain in the nucleus, as is evident in the PD0 chondrocytes, which were round (Fig. 4 a, Fig. S4). On the contrary, a larger 3D aspect ratio promoted a higher strain in the nucleus of PD16 chondrocytes, which were flat (Fig. 4 a, Fig. S4). Volumetric hydrostatic strain maps showed key representative deformation features inside the PD0 and PD16 chondrocyte models (Fig. 4 a, Fig. S4). The spatial hydrostatic strain distribution showed that, on average, larger hydrostatic tensile strains were observed in the PD16 cell nucleus in comparison with the PD0 cell nucleus (Fig. 4 b). The higher strains in the nucleus were primarily associated with the cell protrusions, which were in the primary direction of tension propagation inside the cell (Fig. 4 a). These computational observations matched with the experimental observations, where some areas of the nucleus were associated with larger strains, likely due to the vicinity of the cell protrusions, which were rich in F-actin stress fibers (Fig. 3 a). Overall larger hydrostatic tensile strains were observed in the PD16 cell nucleus in comparison with the PD0 cell nucleus from the computational studies (Fig. 4, a and b), which agreed with the experimental measurement (Fig. 2).
The parametric study showed changing the mechanical properties had a less significant effect on the simulated strain of the system compared with the effect of cell shape and geometry. For example, as the elastic modulus of the cytoskeleton varied in comparison with the baseline initial values (E = 500 Pa for PD0 and E = 900 Pa for PD16), the strain pattern at each individual node in the nucleus remained similar as characterized by the R2 ∼ 0.98 in most cases (Table S3). The range of strain values changed with varying elastic modulus as characterized by deviation of slope m from 1, but, overall, for the same elastic modulus (E = 700 Pa and E = 1100 Pa), the range of strain and maximum strain were higher in PD16 chondrocyte nuclei compared with PD0 chondrocyte nuclei (Fig. 4 c). The change in the heterochromatin and euchromatin elastic modulus had minor effect on the intranuclear strain distribution and values (Fig. S2, Table S3). The varied Poisson's ratio of cytoskeleton, euchromatin, and heterochromatin also did not have a significant effect on the hydrostatic strain solutions at each nuclear node (Fig. S3, Table S4). However, the strain values in the nucleus for the PD0 and PD16 geometries varied by about a factor of two, as shown by the volumetric hydrostatic strain values at each nodal point, and the distribution of the values of the hydrostatic strain (Fig. 4, c and d ). Especially the violin plots (Fig. 4 d) elucidate that the distribution of the hydrostatic strain does not change significantly with the variation of elastic modulus. Therefore, changing the shape of the cell had the primary impact on the resulting nuclear strain range and magnitude compared with the material properties of the cell.
Passage-specific gene expression of nuclear envelope proteins
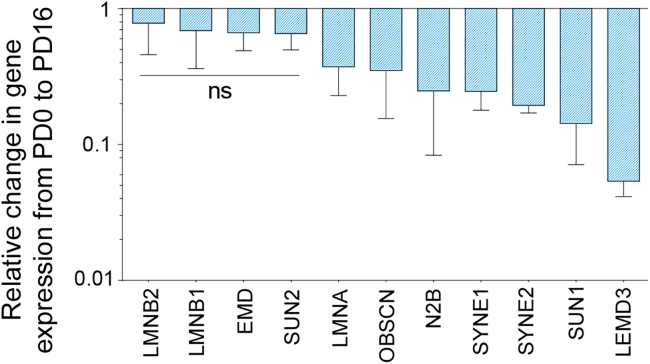
The abnormal strain in heterochromatin can be associated with the loss in genomic integrity and, therefore, abnormal gene expression level (Fig. 1 c). Interestingly, we found that dedifferentiation of chondrocytes was associated with a lower expression of several structural proteins in the nucleus (Fig. 5), in addition to gene expression changes in extracellular matrix-specific proteins (Fig. 1 c). Expression of some key nuclear envelope-related proteins, such as Lamin B1, Lamin B2, Emerin, and SUN2, remained constant in the dedifferentiated state. Expression of some other proteins that provide stability to the nuclear envelope, such as Lamin A/C, Nesprin 1, Nesprin 2, SUN1, and Lemd3, decreased significantly. The highest decrease was associated with Lemd3 protein, which has a direct role in controlling the transforming growth factor beta (TGF-β) and the bone morphogenetic protein (BMP) pathways, both critical factors in chondrogenic expression. SUN1 is one of the two SUN proteins of the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex, which protrudes through the nuclear membrane, and its decreased expression severs the connection between the cytoskeleton and nucleoskeleton (1). Nesprin 1 and Nesprin 2 specifically bind to actin, and Lamin A/C creates a mesh-like structure under the inner nuclear membrane, providing mechanical integrity to the cell nuclear envelope (1). The results of the present study do not provide any further mechanistic insight into whether such lower gene expression of most of the nuclear envelope proteins has any functional significance. However, these results might be suggestive of the lack of genomic integrity caused by abnormal heterochromatin strain, which needs to be investigated in futures studies.
Figure 5.
Dedifferentiation of chondrocyte in monolayer culture caused decrease in nuclear envelope-related gene expression. Most nuclear envelope-specific marker (e.g., Lamin A/C, Nesprin 1 and 2, and SUN1) gene expression decreased in the dedifferentiated state. Significant (p < 0.01) gene expression changes were observed in all cases except for the genes labeled with ns; ns = not significant, number of samples = 4. Error bars represent SD about mean.
Discussion
In this study, we investigated the mechanical behavior of early-passage chondrocytes and dedifferentiated late-passage chondrocytes, to elucidate the passage-dependent gradual deviation from the chondrocyte phenotype. We found that the nuclear mechanics played a role in this elusive chondrocyte behavior. A proposed mechanobiological model summarizes possible contributing factors to the dedifferentiation process. We hypothesize that the fate of chondrocytes is in a positive feedback loop where a chondrocyte is driven toward a fibroblast-like phenotype in two typical scenarios: after implantation in vivo in the ACI environment, and before implantation during 2D cellular expansion in vitro. In the proposed model, the mechanical stretch of the nucleus is a trigger, which could be induced by repeated biomechanical stretching of the scaffold in vivo, or during cell spreading and 2D cell shape maintenance in vitro. The flattened cell shape in the dedifferentiated chondrocytes causes the stretch, resulting in amplified intranuclear strain and abnormal chromatin strain pattern that are opposite to those of early-passage chondrocytes. Such abnormal strain could shift the gene expression pattern to a more fibroblast-specific lineage through the lost genomic integrity, as shown in previous studies (23), which further increases the stretch on the nucleus, thus completing the positive feedback loop. A previous study revealed that chondrocytes show better maintenance of phenotype and less dedifferentiation as measured by gene expression profile and immunofluorescence of cytoskeletal actin filaments, which is in line with the finding of our present study (39).
Several studies thoroughly investigated what molecular changes happen with passage-driven dedifferentiation, and it was found that chondrogenic gene expression decreases and fibroblast-specific expression increases with passaging (40, 41). A time-lapse observation study was performed using chondrocyte-specific reporter Col11α2 to investigate whether the dedifferentiation was triggered by the chondrocyte division and proliferation or by outgrowth of fibroblasts prevailing from the start of culture, but neither of these two mechanisms seemed to be the reason behind the dedifferentiation (42). Attempts have previously been made to understand the dedifferentiation mechanism and subsequently redifferentiate the dedifferentiated cells to chondrocyte phenotype. Three-dimensional encapsulation of late-passage cells showed redifferentiation potential in several hydrogel systems, such as agarose (43), polyethylene glycol and chondroitin sulfate (44), alginate (45, 46), dense collagen (17), and high-density pellet culture (47, 48). There are several proposed mechanisms of how 3D culture promotes redifferentiation, such as the involvement of cytokines (49, 50) and altered expression of microRNAs (51). Our findings from this study suggest a biophysical mechanism in line with the existing reports. Three-dimensional culture decreases the stress-fiber formation inside the cell and also imparts a round shape to the cell and nucleus (52). Therefore, in 3D, tension on the nucleus drastically reduces, changing the loading mode on the nucleus to compressive, potentially halting the dedifferentiation positive feedback loop proposed. Thus, the present study explains the redifferentiation mechanism in 3D. However, the 3D hydrogel-based therapy is not widely accepted for the matrix-induced autologous chondrocyte implantation (MACI) procedure due to a lack of understanding of the passaging limit of chondrocytes in the 2D environment and the inability of chondrocytes to substantially proliferate in the 3D environment (53).
Redifferentiation in 2D culture could be a promising approach as cells can be expanded to a large enough quantity, and their retrieval is straightforward. Several different approaches have been attempted to achieve this goal. Molecular biological interventions include the AIMP1 downregulation using small interfering RNA (siRNA) to restore the TGF-β pathway (54), inhibition of BMP-2 using melanoma inhibitory activity protein (55), and upregulation of the Sonic Hedgehog (Shh) pathway (56). Another approach is to induce a stressed condition to the cells, such as hypothermia (57), hypoxia (58), and applying intermittent hydrostatic pressure (59), but the underlying mechanism of stress-induced redifferentiation has not been completely explored. Importantly, disrupting the contractile function of the cytoskeleton, which in turn would change the deformation of the nucleus, is also a promising technique to prevent dedifferentiation (60, 61). Finally, epigenetic modifications of the dedifferentiated cells is a new approach on the horizon, where the chromatin architecture is directly targeted for redifferentiation (62). The outcome of our study, which suggests a flattened cell and amplified intranuclear strain pushes the chondrocyte phenotype toward a fibroblast-like phenotype, could provide biophysical design strategies to maintain chondrocyte phenotype in 2D culture. Additionally, the extent 2D cultured chondrocytes are able to redifferentiate when introduced into 3D culture after the expansion process is also known, suggesting a possible “mechanical memory” of the cells. This study lays the framework to further investigate how the mechanics gradually affect the chondrocyte phenotype, possibly through a permanent change through intranuclear chromatin architecture, beyond which the chondrocyte memory is completely lost and therefore redifferentiation becomes impossible.
Visualization of intranuclear strain in chondrocytes in the context of dedifferentiation and a weakened nucleus brings new insight into how the heterochromatin and euchromatin distributes the strain in a mechanically challenging condition. This finding can be of potential broad interest to nuclear mechanobiology research, where research efforts have been made specifically for chondrocytes (63) and mesenchymal stem cells with chondrogenic potential (3, 4, 64, 65) over recent years to understand the etiology of cartilage degeneration and regeneration.
Both the modeling and experimental results suggested that the chondrocyte 3D aspect ratio significantly affected the resulting intranuclear strain. In the computational model, as the chondrocyte's 3D aspect ratio increased from PD0 to PD16, the tensile strain experienced by the nucleus increased under the same boundary conditions. The trend was consistent across a sample size of three cell models for both PD0 and PD16. This trend confirmed the experimental data that indicated an overall increase in hydrostatic tensile strain found in the nucleus of the PD16 chondrocytes in comparison with the PD0 chondrocytes. Although major trends were confirmed by the computational model, the exact values of strain reported by the model would not be the same as the experimental results. These inconsistencies were expected due to the simplicity of the isotropic linear elastic model in the computational framework. The parametric study demonstrated that a change in the mechanical properties in the computational model did not have a large effect on the solution of stress and strain at each material node. Therefore, the cell and nuclear geometry changed from PD0 and PD16 was a major factor determining the abnormal increase in tensile strain in the nucleus of the PD16 cell. For example, even if the cytoskeleton stiffness increased in both the PD0 to PD16 cell, the predominant factor that caused an increased nuclear strain was the change in cell geometry toward a flattened shape as the chondrocyte dedifferentiated. It should be noted that the computational model is quite simplified only to understand the effect of cell geometry compared with the cell's mechanical property; therefore, the model has several limitations. For example, contractility of the cells was not considered in the model. If considered, the nucleus in PD16 cells should experience higher strain than what is estimated through our modeling work, because the PD16 cells have a more extensive and fibrous actin filament structure.
Conclusion
In conclusion, we found that the flattened cell shape in expanded chondrocytes leads to an induced abnormal intranuclear strain, specifically in the heterochromatin, which is known to be a key component in maintaining genomic integrity. We propose that this abnormal nuclear stain could be a major factor driving chondrocyte dedifferentiation to a fibroblast-like phenotype, and potentially in mechanically suboptimal tissue graft after implantation. Abnormal intranuclear strain is the key element of the proposed mechanobiological circuit that works in a positive feedback loop to deviate the chondrocyte from its original phenotype. The findings of this study have the potential to advance the ACI procedure by a rational, biophysics-driven chondrocyte engineering design strategy. However, it should be noted that the results of this study may not necessarily apply generally to a cell's differentiation process that involves changes in cell morphology, which may be tissue or cell-type specific. Further studies on differentiation relating to specific cell types are required for achieving a global biophysical model of how morphology changes during (de)differentiation affect the nuclear mechanics.
Author contributions
S.G., B.S., and C.P.N. conceptualized the study. J.H., B.S., J.E.B., B.S.M.M., A.K.S., S.E.S., and S.G. performed the experiments. S.G. analyzed the data with contributions from S.K. and A.K.S. A.K.S. performed the simulation and the computational study. S.G. wrote the manuscript with contributions from all other authors. C.P.N. provided the funding and resources.
Acknowledgments
The authors would like to acknowledge funding from NIH R01 AR063712, NIH R21 AR064178, NIH R21 AR066230, and NSF CAREER1349735. S.E.S. was supported by T32 GM-065103.
Editor: Guy Genin.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.11.018.
Supporting material
References
- 1.Isermann P., Lammerding J. Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 2014;23:R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tajik A., Zhang Y., et al. Wang N. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 2016;15:1287–1296. doi: 10.1038/nmat4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Driscoll T.P., Cosgrove B.D., et al. Mauck R.L. Cytoskeletal to nuclear strain transfer regulates YAP signaling in mesenchymal stem cells. Biophysical J. 2015;108:2783–2793. doi: 10.1016/j.bpj.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heo S.J., Thorpe S.D., et al. Mauck R.L. Biophysical regulation of chromatin architecture instills a mechanical memory in mesenchymal stem cells. Scientific Rep. 2015;5:16895. doi: 10.1038/srep16895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szczesny S.E., Mauck R.L. The nuclear option : evidence implicating the cell nucleus in mechanotransduction. J. Biomechanical Engineering-Transactions ASME. 2017;139:0210061. doi: 10.1115/1.4035350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingal P.C.D.P., Bradshaw A.M., et al. Discher D.E. Fractal heterogeneity in minimal matrix models of scars modulates stiff-niche stem-cell responses via nuclear exit of a mechanorepressor. Nat. Mater. 2015;14:951–960. doi: 10.1038/nmat4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martel-Pelletier J., Barr A.J., et al. Pelletier J. Osteoarthritis. Nat. Rev. Dis. Primers. 2016;2:1–18. doi: 10.1038/nrdp.2016.72. [DOI] [PubMed] [Google Scholar]
- 8.Makris E.A., Gomoll A.H., et al. Athanasiou K.A. Repair and tissue engineering techniques for articular cartilage. Nat. Rev. Rheumatol. 2015;11:21–34. doi: 10.1038/nrrheum.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brittberg M., Lindhal A., et al. Peterson L. Treatment of deep cartilage defects in the knee with autologous chondrocyte transplantation. New Engl. J. Med. 1994;331:889–895. doi: 10.1056/NEJM199410063311401. [DOI] [PubMed] [Google Scholar]
- 10.Brittberg M. Cell carriers as the next generation of cell therapy for cartilage repair: a review of the matrix-induced autologous chondrocyte implantation procedure. Am. J. Sports Med. 2010;38:1259–1271. doi: 10.1177/0363546509346395. [DOI] [PubMed] [Google Scholar]
- 11.Harris J.D., Siston R.A., et al. Flanigan D.C. Failures, re-operations, and complications after autologous chondrocyte implantation: a systematic review. Osteoarthritis and Cartilage. 2011;19:779–791. doi: 10.1016/j.joca.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Akagi M., Nishimura S., et al. Hamanishi C. Cyclic tensile stretch load and oxidized low density lipoprotein synergistically induce lectin-like oxidized ldl receptor-1 in cultured bovine chondrocytes, resulting in decreased cell viability and proteoglycan synthesis. J. Orthopaedic Res. 2006;24:1782–1790. doi: 10.1002/jor.20211. [DOI] [PubMed] [Google Scholar]
- 13.Millward-Sadler S., Wright M.O., Davies L.W., Nuki G., Salter D.M. Mechanotransduction via integrins and interleukin-4 results in altered aggrecan and matrix metalloproteinase 3 gene expression in normal, but not osteoarthritic, human articular chondrocytes. Arthritis Rheum. 2000;43:2091–2099. doi: 10.1002/1529-0131(200009)43:9<2091::AID-ANR21>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 14.Holmvall K., Camper L., et al. Lundgren-Akerlund E. Chondrocyte and chondrosarcoma cell integrins with affinity for collagen type 2 and their response to mechanical stress. Exp. Cell Res. 1995;221:496–503. doi: 10.1006/excr.1995.1401. [DOI] [PubMed] [Google Scholar]
- 15.Sliogeryte K., Botto L., et al. Knight M.M. Chondrocyte dedifferentiation increases cell stiffness by strengthening membrane-actin adhesion. Osteoarthritis and Cartilage. 2016;24:912–920. doi: 10.1016/j.joca.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Caron M.M.J., Emans P.J., et al. Welting T.J.M. Redifferentiation of dedifferentiated human articular chondrocytes : comparison of 2D and 3D cultures. Osteoarthritis and Cartilage. 2012;20:1170–1178. doi: 10.1016/j.joca.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Rosenzweig D.H., Chicatun F., et al. Quinn T.M. Cartilaginous constructs using primary chondrocytes from continuous expansion culture seeded in dense collagen gels. Acta Biomater. 2013;9:9360–9369. doi: 10.1016/j.actbio.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Ofek G., Natoli R.M., Athanasiou K.A. In situ mechanical properties of the chondrocyte cytoplasm and nucleus. J. Biomech. 2009;42:873–877. doi: 10.1016/j.jbiomech.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guilak F., Tedrow J.R., Burgkart R. Viscoelastic properties of the cell nucleus. Biochem. Biophysical Res. Commun. 2000;269:781–786. doi: 10.1006/bbrc.2000.2360. [DOI] [PubMed] [Google Scholar]
- 20.Guilak F. Compression-induced changes in the shape and volume of the chondrocyte nucleus. J. Biomech. 1995;28:1529–1541. doi: 10.1016/0021-9290(95)00100-x. [DOI] [PubMed] [Google Scholar]
- 21.McCreery K.P., Xu X., et al. Neu C.P. Nuclear stiffness decreases with disruption of the extracellular matrix in living tissues. Small. 2021;17:2006699. doi: 10.1002/smll.202006699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds N., McEvoy E., et al. McGarry P. Image-derived modeling of nucleus strain amplification associated with chromatin heterogeneity. Biophysical J. 2021;120:1323–1332. doi: 10.1016/j.bpj.2021.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nava M.M., Miroshnikova Y.A., et al. Wickstrom S.A. Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell. 2020;181:800–817. doi: 10.1016/j.cell.2020.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das R.H.J., Jahr H., et al. Weinans H. In vitro expansion affects the response of chondrocytes to mechanical stimulation. Osteoarthritis and Cartilage. 2008;16:385–391. doi: 10.1016/j.joca.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q., Hu X., et al. Ao Y. Effects of mechanical stress on chondrocyte phenotype and chondrocyte extracellular matrix expression. Scientific Rep. 2016;6:1–8. doi: 10.1038/srep37268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda K., Ohno S., et al. Tanne K. The effects of high magnitude cyclic tensile load on cartilage matrix metabolism in cultured chondrocytes. Eur. J. Cell Biol. 2000;79:601–609. doi: 10.1078/0171-9335-00089. [DOI] [PubMed] [Google Scholar]
- 27.Neu C.P., Reddi A.H., et al. Di Cesare P.E. Increased friction coefficient and superficial zone protein expression in patients with advanced osteoarthritis. Arthritis Rheum. 2010;62:2680–2687. doi: 10.1002/art.27577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neu C.P., Khalafi A., et al. Reddi A.H. Mechanotransduction of bovine articular cartilage superficial zone protein by transforming growth factor beta signaling. Arthritis Rheum. 2007;56:3706–3714. doi: 10.1002/art.23024. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh S., Dutton J.C., Han B. Measurement of spatiotemporal intracellular deformation of cells adhered to collagen matrix during freezing of biomaterials. J. Biomechanical Eng. 2014;136:210251–210258. doi: 10.1115/1.4026180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh S., Seelbinder B., et al. Neu C.P. Deformation microscopy for dynamic intracellular and intranuclear mapping of mechanics with high spatiotemporal resolution. Cell Rep. 2019;27:1607–1620. doi: 10.1016/j.celrep.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh S., Cimino J.G., et al. Goergen C.J. In vivo multiscale and spatially-dependent biomechanics reveals differential strain transfer hierarchy in skeletal muscle. ACS Biomater. Sci. Eng. 2017;3:2798–2805. doi: 10.1021/acsbiomaterials.6b00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh S., Cuevas V.C., et al. Neu C.P. Image based elastography of heterochromatin and euchromatin domains in the deforming cell nucleus. Small. 2021;17:2006109. doi: 10.1002/smll.202006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rim Y.A., Ju J.H. The role of fibrosis in osteoarthritis progression. Life. 2021;11:3. doi: 10.3390/life11010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuvelier D., Thert M., et al. Mahadevan L. The universal dynamics of cell spreading. Curr. Biol. 2007;17:694–699. doi: 10.1016/j.cub.2007.02.058. [DOI] [PubMed] [Google Scholar]
- 35.Lin Z., Fitzgerald J.B., et al. Zheng M.H. Gene expression profiles of human chondrocytes during passaged monolayer cultivation. J. Orthopaedic Res. 2008;26:1230–1237. doi: 10.1002/jor.20523. [DOI] [PubMed] [Google Scholar]
- 36.Darling E.M., Pritchett P.E., et al. Guilak F. Mechanical properties and gene expression of chondrocytes on micropatterned substrates following dedifferentiation in monolayer. Cell Mol. Bioeng. 2009;2:395–404. doi: 10.1007/s12195-009-0077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gunja N.J., Athanasiou K.A. Passage and reversal effects on gene expression of bovine meniscal fibrochondrocytes. Arthritis Res. Ther. 2007;9:1–12. doi: 10.1186/ar2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allshire R.C., Madhani H.D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2018;19:229–244. doi: 10.1038/nrm.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuh E., Kramer J., et al. Rotter N. Effect of matrix elasticity on the maintenance of the chondrogenic phenotype. Tissue Eng. A. 2010;16:1281–1290. doi: 10.1089/ten.TEA.2009.0614. [DOI] [PubMed] [Google Scholar]
- 40.Schnabel M., Marlovits S., et al. Schlegel J. Dedifferentiation-associated changes in morphology and gene expression in primary human articular chondrocytes in cell culture. Osteoarthritis and Cartilage. 2002;10:62–70. doi: 10.1053/joca.2001.0482. [DOI] [PubMed] [Google Scholar]
- 41.Ma B., Leijten J.C.H., et al. Karperien M. Gene expression profiling of dedifferentiated human articular chondrocytes in monolayer culture. Osteoarthritis and Cartilage. 2013;21:599–603. doi: 10.1016/j.joca.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 42.Minegishi Y., Hosokawa K., Tsumaki N. Time-lapse observation of the dedifferentiation process in mouse chondrocytes using chondrocyte-specific reporters. Osteoarthritis and Cartilage. 2013;21:1968–1975. doi: 10.1016/j.joca.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Benya P.D., Shaffer J.D. Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell. 1982;30:215–224. doi: 10.1016/0092-8674(82)90027-7. [DOI] [PubMed] [Google Scholar]
- 44.Smeriglio P., Lai J.H., et al. Bhutani N. 3D hydrogel scaffolds for articular chondrocyte culture and cartilage generation. J. Visualized Experiments. 2015;10:1–6. doi: 10.3791/53085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooke M.E., Pearson M.J., et al. Grover L.M. Geometric confinement is required for recovery and maintenance of chondrocyte phenotype in alginate. APL Bioeng. 2017;1:1–13. doi: 10.1063/1.5006752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh-Bonassera N.D., Wu I., et al. Sah R.L. Expansion and redifferentiation of chondrocytes from osteoarthritic cartilage : cells for human cartilage tissue engineering. Tissue Eng. Part A. 2009;15:3513–3523. doi: 10.1089/ten.tea.2008.0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tallheden T., Karlsson C., et al. Anders L.M. Gene expression during redifferentiation of human articular chondrocytes. Osteoarthritis and Cartilage. 2004;12:525–535. doi: 10.1016/j.joca.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Schulze-Tanzil G., De Souza P., et al. Shakibaei M. Redifferentiation of dedifferentiated human chondrocytes in high-density cultures. Cell Tissue Res. 2002;308:371–379. doi: 10.1007/s00441-002-0562-7. [DOI] [PubMed] [Google Scholar]
- 49.Schulze-Tanzil G. Activation and dedifferentiation of chondrocytes : implications in cartilage injury and repair. Ann. Anat. 2009;191:325–338. doi: 10.1016/j.aanat.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 50.Duan L., Ma B., et al. Wang D. Cytokine networking of chondrocyte dedifferentiation in vitro and its implications for cell-based cartilage therapy. Am. J. Translational Res. 2015;7:194–208. [PMC free article] [PubMed] [Google Scholar]
- 51.Hong E., Reddi A.H. Dedifferentiation and redifferentiation of articular chondrocytes from surface and middle zones : changes in microRNAs-221/-222, -140 and -143/145 expression. Tissue Eng. Part A. 2013;19:1015–1022. doi: 10.1089/ten.TEA.2012.0055. [DOI] [PubMed] [Google Scholar]
- 52.Lele T.P., Dickinson R.B., Gundersen G.G. Mechanical principles of nuclear shaping and positioning. J. Cell Biol. 2018;217:3330–3342. doi: 10.1083/jcb.201804052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang B.J., Hu J.C., Athanasiou K.A. Effects of passage number and post-expansion aggregate culture on tissue engineered, self-assembled neocartilage. Acta Biomater. 2016;43:150–159. doi: 10.1016/j.actbio.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahn J., Kumar H., et al. Lee S.H. AIMP1 downregulation restores chondrogenic characteristics of dedifferentiated/degenerated chondrocytes by enhancing TGF-β signal. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.17. e2099–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Payr S., Tichy B., et al. Albrecht C. Redifferentiation of aged human articular chondrocytes by combining bone morphogenetic protein-2 and melanoma inhibitory activity protein in 3D-culture. PLoS One. 2017;12:1–14. doi: 10.1371/journal.pone.0179729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin L., Shen Q., et al. Yu C. Sonic hedgehog improves redifferentiation of dedifferentiated chondrocytes for articular cartilage repair. PLoS One. 2014;9:1–6. doi: 10.1371/journal.pone.0088550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bomhard A., Von Faust J., Rotter N. Impact of expansion and redifferentiation under hypothermia on chondrogenic capacity of cultured human septal chondrocytes. J. Tissue Eng. 2017;8:1–13. doi: 10.1177/2041731417732655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Babur B.K., Ghanavi P., et al. Doran M.R. The interplay between chondrocyte redifferentiation pellet size and oxygen concentration. PLoS One. 2013;8:1–12. doi: 10.1371/journal.pone.0058865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heyland J., Wiegandt K., et al. Portner R. Redifferentiation of chondrocytes and cartilage formation under intermittent hydrostatic pressure. Biotechnol. Lett. 2006;28:1641–1648. doi: 10.1007/s10529-006-9144-1. [DOI] [PubMed] [Google Scholar]
- 60.Rottmar M., Mhanna R., et al. Maniura-Weber K. Interference with the contractile machinery of the fibroblastic chondrocyte cytoskeleton induces re-expression of the cartilage phenotype through involvement of PI3K, PKC and MAPKs. Exp. Cell Res. 2014;320:175–187. doi: 10.1016/j.yexcr.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 61.Furumatsu T., Matsumoto-Ogawa E., et al. Ozaki T. ROCK inhibition enhances aggrecan deposition and suppresses matrix metalloproteinase-3 production in human articular chondrocytes. Connect. Tissue Res. 2014;55:89–95. doi: 10.3109/03008207.2013.852544. [DOI] [PubMed] [Google Scholar]
- 62.Duan L., Liang Y., et al. Wang D. Epigenetic regulation in chondrocyte phenotype maintenance for cell-based cartilage repair. Am. J. Translational Res. 2015;7:2127–2140. [PMC free article] [PubMed] [Google Scholar]
- 63.Henderson J.T., Shannon G., et al. Neu C.P. Direct measurement of intranuclear strain distributions and RNA synthesis in single cells embedded within native tissue. Biophysical J. 2013;105:2252–2261. doi: 10.1016/j.bpj.2013.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heo S.J., Han W.M., et al. Mauck R.L. Mechanically induced chromatin condensation requires cellular contractility in mesenchymal stem cells. Biophysical J. 2016;111:864–874. doi: 10.1016/j.bpj.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heo S.J., Driscoll T.P., et al. Mauck R.L. Differentiation alters stem cell nuclear architecture, mechanics, and mechano- sensitivity. eLife. 2016;5:1–21. doi: 10.7554/eLife.18207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.