Under appropriate temperature and concentration conditions, charge-neutral homopolymer solutions can segregate into two distinct phases: a polymer-dilute (solvent-rich) and a polymer-dense phase (1, 2). This phenomenon, known as liquid-liquid phase separation (LLPS), is widespread in hetero-polymeric protein solutions as well. The LLPS of folded proteins has been well known for many decades (3, 4). In contrast, the LLPS of newly found classes of proteins that do not fold into unique “native” structures, intrinsically disordered proteins (IDPs) or proteins with intrinsically disordered regions (IDRs), has been reported only in the last decade. Although recently discovered, there is growing interest in LLPS of IDPs and IDRs due to their critical role in a multitude of biological processes including the formation of membraneless organelles (5). In many instances, the presence of only low-complexity domains in the participating proteins is sufficient for the solution system to undergo phase separation. However, inside a cell, phase separation is often multicomponent, involving different IDP or IDR sequences and often RNA as well.
Complexation between different biomolecules near the infinitely dilute limit, far from the solution behavior described above, is another example of multicomponent phenomena that dictate biological function. Complexation is well known to take place between two types of folded proteins or between a disordered and a folded protein, and it has been recently reported that two types of disordered proteins can also form a disordered complex (6). Complexation between two oppositely charged synthetic polymers is well known and a topic of active interest in materials science applications (7, 8) as well. A defining feature of complexation between two types (A and B) of molecules is adherence to a fixed stoichiometry; for example, m number of A and n number of B molecules can form a complex .
Given the preponderance of complexation and LLPS in biology, it is likely that a two-component biopolymer system in solution (three component including the solvent) can undergo both complexation and LLPS (at higher concentration). How do we bridge the two regimes and couple the two different processes? Lin, Chan, and colleagues provide a deeply insightful discussion of this problem by combining mathematical modeling and experiments (9). The richness and the difficulty of the problem can be appreciated by recognizing the sets of all allowed interactions. The LLPS of a two-component (A and B) biopolymer system is typically driven by homotypic (, ) and heterotypic () interactions, in addition to polymer-solvent interactions. When A and B are both IDPs or IDRs, the interactions are typically labile or “stochastic,” primarily due to substantial disorder and conformational fluctuations. However, if A and B form complexes, an additional component (complexed molecules, i.e., ) arises. Complexes are typically expected to lose substantial conformational entropy, limiting the ability to form “stochastic” interactions between two complexes. Consequently, complexes may interact in a more “specific” manner. How do these two types of interactions—specific versus stochastic—dictate LLPS? Do stoichiometric complexes form first and subsequently interact in a specific manner to drive LLPS? Or does the process of LLPS involve both “specific” and “nonspecific or stochastic” interactions?
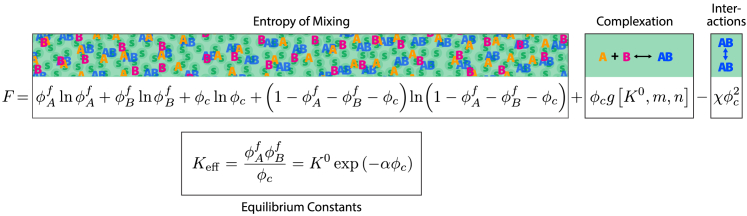
The article (9) by Lin et al. answers these fundamental questions by studying the model system of two proteins, Syn-Gap and PSD-95, that complex with a 3:2 stoichiometry and serve as the simplest model of postsynaptic densities (PSD). Previous studies made four critical observations: (1) three Syn-Gap molecules (say A) form a trimer that subsequently complexes with two PSD-95 molecules (say B), forming the complex ; (2) Syn-Gap and PSD-95 together can undergo LLPS; (3) Syn-Gap and PSD-95 individually cannot phase separate (up to a concentration of 100 micro molar); and (4) mutations that disrupt trimerization of Syn-Gap also prevent LLPS. These observations together suggest the hypothesis that the process of LLPS is primarily driven by specific interaction between complexes (), and not driven so much by stochastic or nonspecific interactions between individual A and B molecules. To test this hypothesis, Lin et al. performed experiments to determine the concentrations of A and B in two coexisting phases, yielding tie lines. The novelty of the experimental data is noteworthy, given the paucity of data in systems in which complexation and LLPS are coupled. Next, they introduced a simple, yet insightful mean-field model capturing the essential physics. The mean-field free energy is given by the free energy F provided in Fig. 1, where are volume fractions of free (noncomplexed) molecules of A and B, and is the volume fraction of complexed species. The first four terms describe the entropy of mixing for four species (free , complex , and solvent). The fifth term accounts for the physics of complexation and is linearly proportional to the number of complexes . Furthermore, this term depends on the equilibrium dissociation constant () of complexation near the infinitely dilute limit, and stoichiometry , as expected. Finally, the sixth term accounts for interaction between complexes mediated by an effective χ parameter and is proportional to the square of the density, similar to the interaction term in Flory Huggins mean-field free energy of a polymer solution. Several numerical prefactors have been omitted for simplicity. The equilibrium constant (near the infinitely dilute limit) gets renormalized to an effective dissociation constant due to interaction among complexes, an essential ingredient of the model. See Fig. 1 for . Note that the stoichiometry-dependent constant terms and χ have been absorbed in the prefactor α (see equation 10 in the paper by Lin et al. for the derivation). Attractive interaction among complexes enhances the binding constant or decreases the dissociation constant . is not a free variable: instead, it is constrained by , and . Phase diagrams are controlled by “total” volume fraction ( and ) of A and B, respectively. Fig. 2 A shows a typical phase diagram (ignoring numerical details) predicted by this model. The two defining features of the phase diagram are (1) its “L” shape and (2) the convergence of tie lines toward the line of stoichiometry (red line). In this model, favorable intercomplex interaction drives the formation of complexes in the concentrated phase causing the tie lines to approach the red line in the high concentration regime. Experimental phase diagrams, however, exhibit “diverging” tie lines, away from the stoichiometric line (see Fig. 2 B). The qualitative disagreement between model and data is sufficient to reject the model of LLPS driven solely by specific interaction among complexes. The failure of this model suggests that additional interactions must be accounted for. Consequently, Lin et al. introduce auxiliary, nonstoichiometric interactions between complexes in the model. These interactions relax the strict requirement of following stoichiometry in the high concentration regime. Consequently, tie lines are not forced to converge: instead, they diverge, consistent with the experimental data. In fact, when additional homotypic interactions and are introduced, the tie lines diverge even further because homotypic interactions compete with heterotypic ones. The model with homotypic and nonstoichiometric heterotypic binding agrees best with data, determined by slope of tie lines.
Figure 1.
The free energy (F) consists of six terms. The first four terms describe the entropy of mixing (among four species , and solvent S). The fifth and sixth terms account for the complexation equilibrium and intercomplex interaction, respectively. For simplicity, complex is denoted as in the picture and subscript c in the equation.
Figure 2.
Theoretical phase diagram (left) created by considering only interaction among complexes, which predicts converging tie lines in contrast to experimentally determined (right) tie lines, which are diverging. The red line in the L-shaped theoretical phase diagram is the line of stoichiometry. The axes indicate total concentration of species A and B.
New questions now emerge: what type of interactions are responsible for nonstoichiometric interactions? How are the complexes partitioned in the dilute and concentrated phases? Is the interaction among complexes sufficient to cause gelation (10)? If so, how does gelation couple with multicomponent LLPS both in bio- and synthetic polymers (8)? The finding also indicates phase diagrams in the space of densities of both (A and B) polymeric species could be insightful for synthetic systems as well. Another interesting question emerges when IDP or IDR (say A) competitively binds to two other proteins (say B and C). If B and C are ingredients of different condensates formed under different conditions, competitive binding may couple formation of different condensates, a feature generally not considered at present. For competitive complexation reactions, fluctuations in the number of complexes can increase (11). Does the process of LLPS help suppress these fluctuations and provide a buffering mechanism similar to LLPS in a one component system (12)?
Finally, the approach taken by Lin et al. is a strong reminder of the long-standing tradition in theoretical physics where valuable insights are gained from a mismatch between a model and the data. Failure of a model is critical in building new models, and physical biology is rife with such examples (13). These approaches should be embraced and not abandoned.
Acknowledgments
We acknowledge support from NIH (R01GM138901) and help from Sarina Bromberg on creating the figures. Author KG declares no competing interests.
Editor: Robert Best
References
- 1.Flory P. Thermodynamics of high polymer solutions. J. Chem. Phys. 1942;10:51. [Google Scholar]
- 2.Huggins M.L. Some properties of solutions of long-chain compounds. J. Phys. Chem. 1942;46:151–158. [Google Scholar]
- 3.Tanaka T., Ishimoto C., Chylack L.T. Phase separation of a protein-water mixture in cold cataract in the young rat lens. Science. 1977;197:1010–1012. doi: 10.1126/science.887936. [DOI] [PubMed] [Google Scholar]
- 4.Broide M., Berland C., Pande J., Ogun O., Benedek G. Binary-liquid phase separation of lens protein solutions. Proc. Natl. Acad. Sci. 1991;88:5660–5664. doi: 10.1073/pnas.88.13.5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeynaems S., Alberti S., Fawzi N., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Bosch L., Tompa P., Fuxreiter M. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borgia A., Borgia M., Bugge K., Kissling V., Heidarsson P., Fernandes C., Sottini A., Soranno A., Buholzer K., Nettels D., Kragelund R., Best, Schuler B. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018;555:61–66. doi: 10.1038/nature25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srivastava S., Tirrell M. Polyelectrolyte complexation. Adv. Chem. Phys. 2016;161:499–543. [Google Scholar]
- 8.Muthukumar M. 50th anniversary perspective: a perspective on polyelectrolyte solutions. Macromolecules. 2017;50:9528–9560. doi: 10.1021/acs.macromol.7b01929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Y.H., Wu H., Jia B., Zhang M., Chan H.S. Assembly of model postsynaptic densities involves interactions auxiliary to stoichiometric binding. Biophysical J. 2022;121:157–171. doi: 10.1016/j.bpj.2021.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harmon T., Holehouse A., Rosen M., Pappu R. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. eLife. 2017:e30294. doi: 10.7554/eLife.30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firman T., Ghosh K. Competition enhances stochasticity in biochemical reactions. J. Chem. Phys. 2013;139:121915. doi: 10.1063/1.4816527. [DOI] [PubMed] [Google Scholar]
- 12.Klosin A., Oltsch F., Harmon T., Honigmann A., Julicher F., Hyman A., Zechner C. Phase separation provides a mechanism to reduce noise in cells. Science. 2020;367:464–468. doi: 10.1126/science.aav6691. [DOI] [PubMed] [Google Scholar]
- 13.Phillips R., Kondev J., Theriot J. Garland Science; 2009. Physical Biology of the Cell. [Google Scholar]