Abstract
Autosomal-dominant Alzheimer’s disease (ADAD) may be associated with atypical amyloid beta deposits in the brain. In vivo amyloid imaging using 11C-Pittsburgh compound B (PiB) tracer has shown differences in binding between brains from ADAD and sporadic Alzheimer’s disease (sAD) patients. To gain further insight into the various pathological characteristics of these genetic variants, we performed large frozen hemisphere autoradiography and brain homogenate binding assays with 3H-PiB, 3H-MK6240-3H-THK5117, and 3H-deprenyl for detection of amyloid fibrils, tau depositions, and activated astrocytes, respectively, in two AβPParc mutation carriers, one PSEN1ΔE9 mutation carrier, and three sAD cases. The results were compared with Abeta 40, Abeta 42, AT8, and GFAP immunostaining, respectively, as well as with Congo red and Bielschowsky. PiB showed a very low binding in AβPParc. A high binding was observed in PSEN1ΔE9 and in sAD tissues but with different binding patterns. Comparable 3H-THK5117 and 3H-deprenyl brain homogenate binding was observed for AβPParc, PSEN1ΔE9, and sAD, respectively. Some differences were observed between 3H-MK6240 and 3H-THK5117 in ADAD. A positive correlation between 3H-deprenyl and 3H-THK5117 binding was observed in AβPParc, while no such correlation was found in PSEN1ΔE9 and sAD. Our study demonstrates differences in the properties of the amyloid plaques between two genetic variants of AD and sAD. Despite the lack of measurable amyloid fibrils by PiB in the AβPParc cases, high regional tau and astrocyte binding was observed. The lack of correlation between 3H-deprenyl and 3H-THK5117 binding in PSEN1ΔE9 and sAD in contrast of the positive correlation observed in the AβPParc cases suggest differences in the pathological cascade between variants of AD that warrant further exploration in vivo.
Subject terms: Diagnostic markers, Neuroscience
Introduction
In a small percentage of patients, Alzheimer’s disease (AD) is characterized by an early onset due to a mutation in one of three identified genes: amyloid-beta precursor protein (AβPP), Presenilin 1 (PSEN1), and Presenilin 2 (PSEN2). Mutation-specific features have been described, both in vitro and in vivo, showing a faster progression of the disease in Autosomal-dominant Alzheimer’s disease (ADAD) variants in comparison to the sporadic form of Alzheimer’s disease (sAD) [1]. However, not all identified mutations will result in the clinical phenotype of AD [2].
In PSEN1DE9 brains, (due to an exon 9 deletion in PSEN1 (PSEN1DE9) identified in the FINN2 family [3]) accumulation of large plaques composed of amyloid beta 42 and amyloid beta 40 without a compact amyloid core so called ‘cotton wool’ plaques have been reported [4]. Significant difference was observed between PSEN1DE9 mutation and sAD with significantly higher Abeta 42/40 ratio in PSEN1DE9 [4]. PSEN1DE9 is distinct from the sAD phenotype and could also result for some cases, in a clinical phenotype that includes spastic paraparesis [3]. The AβPParc mutation (p. E693G) is pathologically characterized by the presence of ring-shaped amyloid plaques without amyloid core [5]. Those plaques are Congo red negative but amyloid beta 42-positive on the ring as observed using amyloid beta 1–42 antibodies [1, 6]. The clinical phenotype in AβPParc is similar to that in patients with sAD but with an earlier onset of the disease (45–57 years) [3]. Previous in vitro studies have shown increased levels of oligomeric and protofibrillar forms of amyloid beta in AβPParc oligomeric preparations [7, 8]. Philipson et al., in 2012 compared both the amyloid plaques structure and the accumulations of N- and C-truncated Aβ in AβPParc, PSEN1DE9 and sAD, and they could observe differences in length accumulation of N- and C-truncated Aβ40 and 42 between the two mutation and sAD in parenchymal plaques as well as in cerebral amyloid angyopathy [9]. The in vivo PET binding of 11C-Pittsburgh compound B (PiB) is low in both symptomatic and nonsymptomatic AβPParc carriers in comparison to patients with sAD while levels of amyloid beta 42, total tau and P-tau in the cerebrospinal fluid and cerebral metabolism as measured by 18F-fluorodeoxyglucose (FDG) PET are comparable with sAD [6, 10, 11].
In the PSEN1DE9 mutation carriers, 11C-PIB PET has demonstrated increased amyloid plaque deposition in comparison to control and with a distinct pattern than sAD with a higher 11C-PIB binding in the putamen in comparison to sAD patients [12]. 18F-FDG hypometabolism and pathological levels of CSF biomarkers were comparable to those observed in sAD patients [12].
In this study, our aim was to further characterize the neuropathological features of two AβPParc and one PSEN1DE9 mutation carrier using autoradiography with PET tracers as well as immunohistochemistry, in order to assess the extent and regional distribution of plaques, tangles, and activated astrocytes. An ante-mortem/postmortem binding comparison was performed for one AβPParc case.
Material and methods
Autopsy material
Large frozen whole left hemisphere sections were obtained from two AβPParc (AβPParc1, provided by the Brain Bank at Karolinska Institutet; AβPParc2, provided by the Uppsala University brain bank), one PSEN1DE9 (provided by the Department of Pathology, University of Helsinki, Helsinki, Finland) and from the right hemisphere for three sAD brains (provided by the Neuropathology of Dementia Laboratory, Indiana University School of Medicine, Indianapolis, IN, USA). Direct comparison between the cases should be taken with caution due to the fact that large frozen hemisphere sections are rare material and the sections were not from the exact same coronal anatomical level. For the two AβPParc, and the three sAD pieces of frontal, temporal, and entorhinal cortices, as well as hippocampus and caudate nucleus were dissected and used for binding assay studies. For the PSEN1DE9, frontal, temporal cortices, and caudate were available. Frozen homogenates of frontal cortex, temporal cortex, hippocampus, and caudate nucleus from two nondemented controls (obtained from the Netherlands Brain Bank) were used for binding assays. The demographic data are presented in Table 1. Clinical description of the patients with AβPParc and PSEN1ΔE9 mutations is available on Supplementary data 1.
Table 1.
Table representing the clinical information.
| Sex | Age of onset (years) | Age at death (years) | Braak stage | APOE | PMI (h) | |
|---|---|---|---|---|---|---|
| AβPParc1 | F | 53 | 66 | VI | 3/3 | >30 |
| AβPParc2 | M | 61 | 64 | VI | 3/3 | 12 |
| PSEN1ΔE9 | F | 51 | 66 | N/A | 3/3 | 5 |
| AD1 | F | 59 | VI | 3/3 | 4 | |
| AD2 | F | 73 | V | 3/3 | 1.5 | |
| AD3 | F | 59 | V | 3/4 | 10 | |
| Control 1 | M | 62 | I | 3/3 | 7 | |
| Control 2 | F | 71 | I | 3/2 | 7 | |
| Control 3 | M | 79 | II | 3/3 | 9 |
Demographic information for patients with autosomal-dominant Alzheimer’s disease, patients with sporadic Alzheimer’s disease and normal controls.
AD Alzheimer’s disease, APOE apolipoprotein E, AβPParc Arctic amyloid-β protein precursor mutation, F female, M male, N/A not applicable, PMI postmortem interval, PSEN1ΔE9 PS1 exon 9 deletion.
Chemicals
3H-THK5117 and unlabeled THK5117 were synthesized by Novandi chemistry AB (Södertälje, Sweden; specific activity (SA) = 75 Ci/mmol). 3H-MK6240 and unlabeled MK6240 were synthesized by Merck & Co; SA: 44 Ci/mmol. 3H-PIB was custom synthesized by Novandi (Södertälje, Sweden; SA = 73 Ci/mmol). 3H-l-deprenyl was custom synthesized by Quotient Bioresearch (Cardiff, UK; SA = 85 Ci/mmol). Unlabeled (R)-(−)-deprenyl was purchased from Tocris Bioscience and BTA-1 was purchased from Sigma-Aldrich.
Autoradiography
Autoradiography from all cases was carried out at room temperature (RT) on frozen coronal sections (100 μm thick using a Cryomacrotome Leica CM3600XP, Leica Biosystems, USA) after allowing them to dry. For 3H-PIB, the sections were preincubated for 15 min with Phosphate Buffer Saline (PBS) + 1% BSA, then incubated for 45 min with 3H-PIB (1 nM) in PBS + 0.1% BSA. Nonspecific (NSP) binding was determined with 1 µM BTA-1. For 3H-THK5117 and 3H-MK6240, the sections were preincubated 15 min with PBS + 0.1% BSA, then incubated for 1 h with 3H-THK5117 (3 nM) or 3H-MK6240 (1 nM) with the same buffer. NSP was determined with 1 µM unlabeled THK5117 or MK6240, respectively. Finally, for 3H-deprenyl autoradiography the sections were incubated with 3H-deprenyl (10 nM) for 1 h in Na–K phosphate buffer. NSP was determined with 1 µM of unlabeled deprenyl.
For all the radioligands, the binding reaction was terminated by washing 3 × 5 min with cold binding buffer (4 °C) followed by one dip in cold distilled water (4 °C). The sections were then dried and apposed on photostimulable phosphor-plates for 4 days for 3H-deprenyl and 3H-THK5117 and 7 days for 3H-PIB and 3H-MK6240. The photostimulable phosphor-plates were then read using a BAS-2500 imager and the results were analyzed using multigauge software to draw the regions of interest manually.
Immunohistochemistry for neuropathological evaluation
Immunohistochemistry for neuropathological evaluation was performed on small paraffin-embedded sections from right brain hemisphere obtained from the same cases mentioned above (AβPParc1, AβPParc2, and PSEN1DE9) (For AβPParc1 6 μm thick sections on no coated slides were used; for AβPParc2 7 μm thick sections on coated slides were used and for PSEN1DE9 7 μm thick sections on superfrost slides were used).
A routine deparaffinization protocol was used. AT8 (Phospho-Tau, Ser202, Thr205) monoclonal antibody from Thermofisher, amyloid beta 1–42 (antiamyloid β42 antibody, clone G2-11 from Merck Millipore), and amyloid beta 1–40 (antiamyloid β40 antibody, clone G2-10 from Merck milipore) antibodies were used as follows: AT8 dilution 1:2500 stained in Roche Ventana immunostainer; amyloid beta 1–42 dilution 1:750 and amyloid beta 1–40 dilution 1:500 stained in Roche Ventana immunostainer but with 10 min in formic acid first after deparaffination.
In vitro brain regional binding studies
Regional binding studies were carried out on fresh frozen tissues from frontal, temporal, and entorhinal cortices, caudate nucleus and hippocampus using 3H-PIB, 3H-deprenyl, and 3H-THK5117. Each compound was incubated with the brain homogenates and filtered after a tracer-specific incubation time, as follow: 3H-PIB (1 nM) was incubated in PBS for 2 h at RT with 0.1 mg/ml of tissue. NSP was determined using 1 µM of BTA-1. 3H-deprenyl (10 nM) was incubated for 1 h at 37 °C with 0.2 mg/ml of tissue using Na–K phosphate buffer. NSP was determined using 1 µM unlabeled deprenyl. 3H-THK5117 (3 nM) was incubated for 2 h at RT with 0.2 mg/ml of tissue using PBS + 0.1% BSA. NSP was determined using 1 µM unlabeled THK5117.
All experiments were terminated by filtering through glass fiber filter paper presoaked with polyethylenimine 0.3%, rinsing three times with cold binding buffer and then counting on a Beckman scintillation counter. Each experiment was performed at least in triplicate and analyzed using graph pad prism software.
AβPParc in vivo imaging
Two years before death, the AβPParc1 patient underwent a structural T1 MPRAGE MRI on a 3T (Siemens Trio) scanner at the Karolinska University Hospital, Huddinge (Sweden), and PET examinations with 11C-deprenyl, 11C-PIB, and 18F-FDG at the Uppsala PET Centre, Uppsala University (Sweden), on an ECAT EXACT HR + (Siemens/CTI) PET/CT scanner. Productions of the tracers and image acquisitions have been previously described [6].
Results
Autoradiography and immunostaining comparison
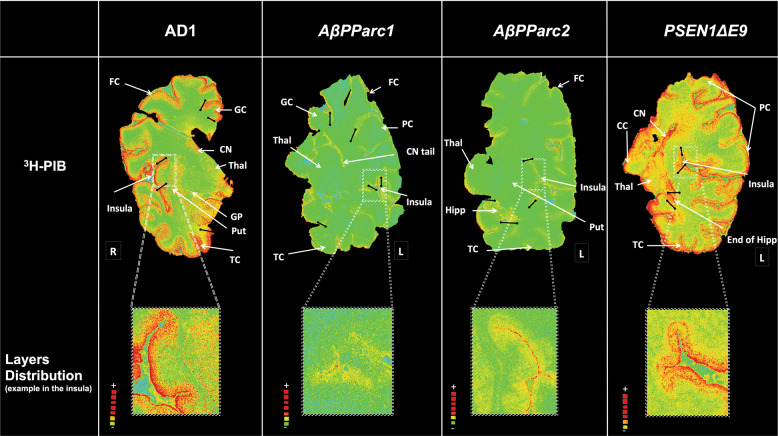
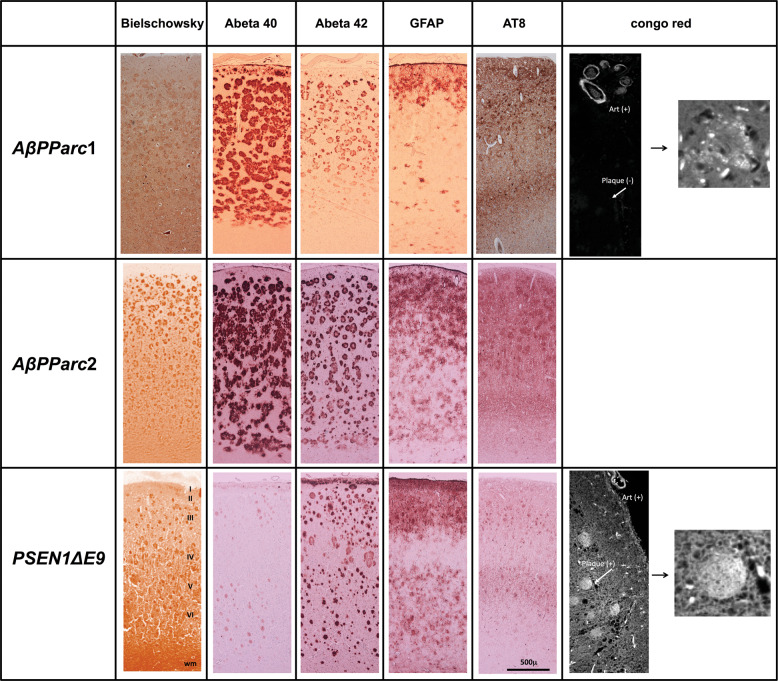
The regional binding of 3H-PIB on large frozen brain sections of the two AβPParc mutation carriers, the PSEN1DE9 mutation carrier and one sAD case is presented in Fig. 1. 3H-PIB binding was very low in the whole brain hemisphere sections of the two AβPParc brains in comparison to the sAD brain. In contrast, the binding in the PSEN1DE9 brain was higher and comparable to that in the sAD brain, but with a more uneven distribution (‘cloudy’ pattern) (see enlargement in Fig. 1). Some 3H-PIB binding could also observed in the white matter of the PSEN1DE9 brain. A comparison of the immunostaining using amyloid β 40, amyloid β 42, and Bielschowsky staining is presented in Fig. 2. The Bielschowsky stain bound to senile plaques and neurofibrillary tangles more intensely in the two AβPParc brains than in the PSEN1DE9 brain. The amyloid β 40 and 42 staining was also intense in all layers for the two AβPParc brains, with more spreading in the upper layers. In the PSEN1DE9 brain, amyloid β 42 staining was more intense than amyloid β 40 staining; amyloid staining was especially intense in layer 1 (corresponding to subpial amyloid accumulation). Amyloid beta 42 antibodies resulted in more compact, filled-in ‘cotton wool’ plaques in the PSEN1DE9 brain than the ring-shaped plaques in the two AβPParc brains (Fig. 2). In the AβPParc brains, the specific ring shape of the plaques was observed predominantly with amyloid β42 staining, which was located on the outer rims of the plaques; amyloid β 40 binding was homogeneously distributed throughout the plaque formations. The prominent difference between the ‘cotton wool’ plaques in the PSEN1DE9 brain and the ring-shaped plaques in the AβPParc brains was clearly illustrated by Congo red (see Fig. 2).
Fig. 1. Representation of total binding autoradiography on large frozen sections with 3H-Pittsburg compound B.
Autoradiography on large frozen sections from one Alzheimer’s disease brain (AD1), two AβPParc brains and one PSEN1DE9 deletion brain. The top panel shows 3H-Pittsburgh compound B (PIB) autoradiography (standard: + = 4700 fmol/mg, − = 50 fmol/mg) and the bottom panel shows enlargements of the insula region to illustrate distribution in the layers. All images were put on the same threshold (47,545) from the raw images (16 bits: 0–65,535 (color scale)) for comparison. CN caudate nucleus, CC corpus callosum, FC frontal cortex, GC gyrus cingulate, GP globus pallidus, Hipp hippocampus, Put putamen, PC parietal cortex, TC temporal cortex, Thal thalamus.
Fig. 2. Immunostaining of AβPParc1, AβPParc2, and PSEN1DE9 brain tissue.
From left to right, Bielschowsky, Abeta 40, Abeta 42, glial fibrillary acidic protein (GFAP), and AT8 stains and Congo red.
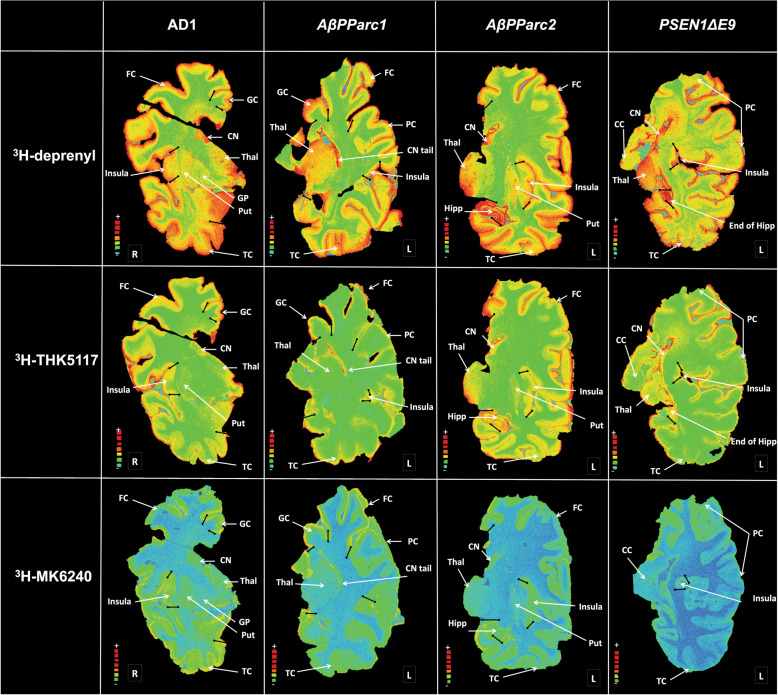
The regional binding pattern of 3H-deprenyl, 3H-THK5117, and 3H-MK6240 are presented in the Fig. 3. 3H-deprenyl autoradiography showed the most intensity of binding compared with the two-tau tracers using the same color scale. An interesting difference between the two genetic variants and sAD was the binding of 3H-deprenyl in the white matter of the PSEN1DE9 as confirmed by intensive immunostaining using the glial fibrillary acidic protein (GFAP) stain (Fig. 2). While comparing the two-tau tracers 3H-THK5117 and 3H-MK6240, lower general binding intensity was observed for 3H-MK6240 due to different SA (44 Ci/mmol for 3H-MK6240 and 85 Ci/mmol for 3H-THK5117) as well as incubation concentration (1 nM for 3H-MK6240 and 4 nM for 3H-THK5117). Regional distribution binding was similar for the two-tau tracers in the sAD. For AβPParc1, 3H-MK6240 binding is higher than the one of 3H-THK5117. For AβPParc2 and PSEN1DE9 differences were observed between the two-tau tracers. Indeed, in PSEN1DE9 lower 3H-MK6240 binding was observed in temporal area in comparison to other cases. 3H-THK5117 binding was more extensive throughout the cortical ribbon in the AβPParc2 brain than in the AβPParc1 brain, again confirmed by GFAP and AT8 immunostaining (Fig. 2). GFAP and AT8 immunostaining also showed differential bilayer distribution in the superficial and deep pyramidal layers of the three ADAD (data not shown). In the PSEN1DE9 brain, AT8 staining was similar to that in the sAD brain but was less intense than that in the two AβPParc brains. In the AβPParc2 brain, GFAP and AT8 immunostaining was more intense throughout all the layers, including the superficial layer, than in the AβPParc1 brain. In both the AβPParc brains, AT8 staining followed the distribution of the amyloid plaques, depicting binding in the neuritic plaques. GFAP staining was more intense in the PSEN1DE9 brain than in the two AβPParc brains, with intense binding in the upper layers (including the molecular layer with subpical positivity) and in the white matter.
Fig. 3. Representation of total binding autoradiography on large frozen sections with 3H-deprenyl, 3H-THK5117, and 3H-MK6240.
Autoradiography on large frozen sections from one Alzheimer’s disease (AD) brain, two AβPParc brains and one PSEN1DE9 deletion brain. The top panel shows 3H-deprenyl autoradiography (Standard: + = 4100 fmol/mg, − = 50 fmol/mg), middle panel shows 3H-THK5117 autoradiography (standard: + = 4500 fmol/mg, − = 30 fmol/mg) and the bottom panel shows 3H-MK6240 autoradiography (standard: + = 6500 fmol/mg, − = 70 fmol/mg). All images from deprenyl and THK5117 tracers were put on the same threshold (50,372) from the raw images (16 bits :0–65,535 (color scale)) for comparison. For MK6240 threshold were putted at 46,517 to allow comparison with other tracers. CN caudate nucleus, CC corpus callosum, FC frontal cortex, GC gyrus cingulate, GP globus pallidus, Hipp hippocampus, Put putamen, PC Parietal cortex, TC temporal cortex, Thal thalamus.
Quantitative assessment of the regional binding distribution of 3H-PIB, 3H-THK5117, and 3H-deprenyl
Regional binding of the three PET tracers was quantitatively assessed using brain homogenates in binding assays; results are shown in Fig. 4. Low 3H-PIB binding was observed in the two AβPParc brains compared with intermediate binding in the PSEN1DE9 brain and high binding in sAD (Fig. 4a). The binding in the caudate was in general similar to the cortical binding. In the AD cases the caudate showed an intermediate binding while both higher and lower cortical binding were observed, probably due to larger intra-cases variability. Higher 3H-THK5117 binding was detected in the frontal and temporal cortices in the sAD brains compared with the AβPParc and PSEN1DE9 brains (Fig. 4b). The highest 3H-THK5117 mean binding was observed in the hippocampus of the AβPParc1 brain, in the entorhinal cortex of the AβPParc2 brain, and in the caudate nucleus of the PSEN1DE9 brain. Binding in the frontal and temporal cortices was similar for the two AβPParc and slightly higher in the PSEN1DE9 brains. The binding in caudate is higher than in the cortex in all mutation brains except in sAD and control.
Fig. 4. Regional-specific binding distribution assays.

Regional-specific binding distribution assays using single concentrations of (a) 3H-Pittsburgh compound B (PIB); (b) 3H-THK5117 and (c) 3H-deprenyl in two AβPParc brains, one PSEN1DE9 brain, three sporadic Alzheimer’s disease (AD) brains and three normal control brains. Frontal cortex, entorhinal cortex, caudate nucleus, temporal cortex, and hippocampus were investigated depending on the availability of the tissue.
3H-deprenyl binding was similar in the frontal cortex and in the caudate nucleus for both sporadic and mutation cases. Greater 3H-deprenyl binding was observed in the hippocampus of the AβPParc1 case and the entorhinal cortex of AβPParc2 compared with sAD cases (Fig. 4c). Caudate binding is higher than the cortex binding in all AD brains while lower than the entorhinal cortex and hippocampus in the two AβPParc.
Relationship between 3H-deprenyl and 3H-THK5117 regional binding
A significant positive correlation between 3H-deprenyl and 3H-THK5117 binding was observed for the two AβPParc in different brain regions, while no such significant correlations were observed for the sAD or PSEN1DE9 brains (Fig. 5). No significant correlations were seen between 3H-PIB and 3H-THK5117 in any group (Supplementary data 2a). Between 3H-PIB and 3H-Deprenyl significant negative correlation could be observe only in sAD (Supplementary data 2b).
Fig. 5. Correlation between 3H-THK5117 and 3H-deprenyl binding.
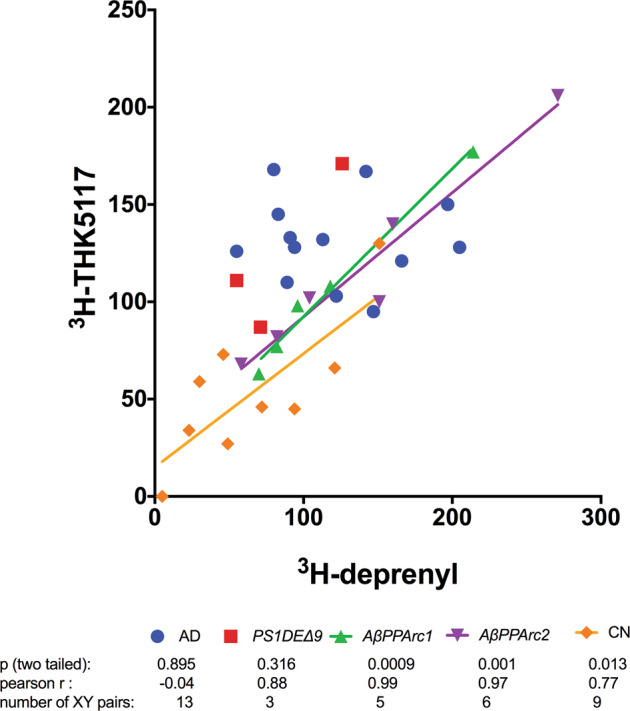
Comparison of deprenyl and THK5117 regional binding distributions in two AβPParc brains, one PSEN1DE9 brain, three Alzheimer’s disease brains, and three normal control brains. Frontal cortex, temporal cortex, entorhinal cortex, caudate nucleus, and hippocampus tissue was used depending on availability. n = number of samples investigated.
AβPParc1 brain multitracer PET scan
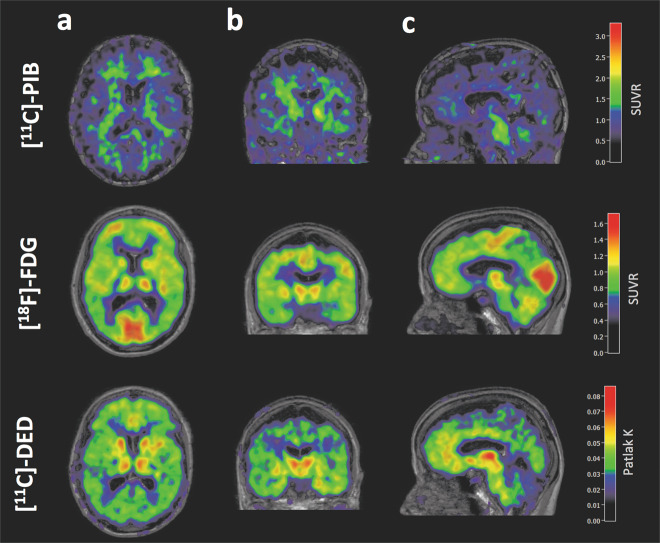
Parametric images of PET acquisitions using 11C-L-deprenyl, 18F-FDG, and 11C-PIB in the AβPParc1 carrier performed 2 years prior to death are shown in Fig. 6. The 11C-PIB PET scan showed very low binding and considered to be amyloid negative according to used cutoff values [13]. The 18F-FDG PET scan showed cortical hypometabolism, predominantly in the temporo-parietal regions, and 11C-deprenyl showed high binding in the cortical regions and basal ganglia (Fig. 6) (see Supplementary data 3 for in vivo–in vitro correlation for AβPParc1).
Fig. 6. Multitracer PET scans for AβPParc1 mutation carrier Multitracer PET scan for an AβPParc mutation carrier (AβPParc1) using 11C-Pittsburgh compound B (PIB), 18F-fludeoxyglucose (FDG), and 11C-deprenyl tracers.
(11C-PIB and 18F-FDG late sum images (40–60 and 30–45 min, respectively) were created and co-registered onto the T1 MRI image. The whole pons was used as a reference for both tracers as it was found to be preserved from pathology in both autosomal-dominant and sporadic Alzheimer’s disease [14]. The 11C-deprenyl dynamic PET images were co-registered onto the T1 MRI image, and a modified reference Patlak model [15] was applied to the 20–60 min image using PMOD software, with the cerebellum as the modified reference region [16] to generate individual 3D parametric Patlak slope images (unit = min−1)). a horizontal section; b coronal section; c sagittal section.
Discussion
The aim of the study was to compare the neuropathological features of the AβPParc and PSEN1DE9 mutations with those of sAD using in vitro binding studies of autopsied brain tissue with PET ligands, in order to be able to understand the relationships between the different hallmarks of AD pathology.
Autoradiographies with 3H-PIB binding confirmed the in vivo data: the lack of 3H-PIB binding in the two AβPParc brains reflects the absence of fibrillar form (Congo positive) amyloid plaques in the two AβPParc brains. However, 3H-PIB autoradiography showed relatively intense binding in the PSEN1DE9 brain, comparable to sAD brains. 3H-PIB binding in the PSEN1DE9 brain was distributed mostly in the upper layers of the cortex, with specific binding also detected in the white matter. The PSEN1DE9 brain has been shown to have rich amyloid angiopathy as observed with thioflavin-S [3] as well as AβPParc [1]. Interestingly, we confirmed these differences in the 3H-PIB binding pattern using amyloid β 42 and 40 antibody immunostaining. A striking difference between AβPParc and PSEN1DE9 was observed with the amyloid β 40 antibody; there was much less staining in the PSEN1DE9 brain than in the two AβPParc brains. This major difference illustrates the different biological compositions of the amyloid plaques in AβPParc and PSEN1DE9 mutations. These differences were confirmed with Congo red staining; there was a total Congo red negative core for the AβPParc brains and some positivity for the PSEN1DE9 brain, but it was still more diffusely and evenly distributed (like cotton wool) in comparison to the sAD brains. In previous studies, Philipson et al. [9] have also demonstrated the differences between the biological composition of the amyloid plaques of AβPParc and PSEN1DE9 mutations. Moreover, Verkkoniemi et al., showed that both noncored, cored as well as diffuse plaques were found in the cerebral cortex of a patient with the FINN2 PSEN1DE9 mutation [14].
The other characteristic pathological hallmarks of AD, such as tau deposits and activated astrocytes, measured by 3H-THK5117/3H-MK6240 and 3H-deprenyl, respectively, showed more similarities across the AβPParc, PSEN1DE9 and less similar with sAD brains. The most striking similarities is that both tau deposits and activated astrocytes binding are the highest in the limbic areas > caudate > cortical areas for the two AβPParc and PSEN1DE9, when in the sAD, these differences are not pronounced.
For both the PSEN1DE9 and the AβPParc mutations, similar laminar distributions throughout the cortical ribbon were observed for 3H-THK5117 and 3H-deprenyl. The similar results have already been described for sAD autoradiography using 3H-THK5117 and 3H-deprenyl [15]. This bilayer pattern was confirmed using GFAP and AT8 immunostaining. Similarly, the activated astrocytes and tau deposits seemed to both describe a bilayer distribution for both mutations. Interestingly, a positive correlation between 3H-deprenyl and 3H-THK5117 binding was only observed in the two AβPParc and the control brains, while there was no significant correlation in either the PSEN1DE9 or the sAD brains. These observations suggest that it might be a closer relationship between activated astrocytes and tau deposits in AβPParc brains than in PSEN1DE9 brains. Since some off-target binding to monoamine oxidase B (MAO-B) has been reported for the first generation tau tracer, THK5117, we also included MK6240, a tau tracer from the second generation, with no MAO-B off-target binding [16]. No regional differences in regional binding pattern were observed between the two-tau tracers in sAD except for a difference in intensity of binding that can be due both to the difference in specific radioactivity between the two tracers as well as probably more NSP binding for THK5117. However, some differences were observed between the two-tau PET tracers since 3H-MK6240 showed lower binding in the PSEN1DE9 and AβPParc2 variants in comparison to AβPParc1 cases and the sAD cases. This observation might suggest some difference in binding properties between these two-tau PET tracers at least in AD mutation cases. It will be interesting to study further the underlying cause of the difference between the bindings of the two-tau PET tracers in the familial form of AD. Recent Cryo-EM studies have demonstrated similar tau fibril folding structure in sAD and inherited AD (V717FAPP) [13] and it would be interesting to have similar studies comparing the tau folds in AβPParc and PSEN1DE9.
It should be born in mind that the different cutting levels in the large frozen brain sections for autoradiography and in the paraffin sections from the contralateral hemisphere used for immunostaining could have affected the comparison between the AβPParc, PSEN1DE9, and sAD brains as the same regions cannot be compared directly.
The results of the binding studies in brain homogenates were in general similar to those of the autoradiography studies on large frozen sections. The homogenization process of the brain most probably let us access to more binding sites than for the autoradiography. Indeed, on the autoradiography the tracer can reach only the accessible binding site, than when the brain is homogenized binding site that where inaccessible might become accessible. During the homogenization process, conformational and structural changes most probably occur in the cotton wool plaques and the 3H-PIB do not have access to the similar binding sites in comparison to amyloid plaques found in sAD brains. In silico computer modeling has suggested that there may be several binding sites on the amyloid fibril [17], as also suggested by in vitro binding with various amyloid ligands [18]. In this study, we observed the highest binding of 3H-PIB in the caudate nucleus of the PSEN1DE9 brain in comparison with AβPParc and, despite the limitation of there being only one brain examined, this observation appears to be in agreement with in vitro results by Ni et al. [19] and in vivo results by Koivunen et al. [12]. The binding pattern for amyloid deposition is confirmed by binding assay as well as autoradiography and immunostaining. Binding of 3H-THK5117 and 3H-deprenyl in the entorhinal cortex of the AβPParc2 brain was much higher than that in the AβPParc1 brain and the sAD brain. When the results from the in vivo and in vitro studies were compared in AβPParc1, we observed a trend toward a negative association between 18F-FDG and 3H-THK5117 binding; no statistical analysis could be performed due to few data points. The time differences between in vivo/in vitro measurements might add some limitations. Increased tau deposits (as observed with 3H-THK5117) seemed to occur in regions with lower 18F-FDG PET data, implying lower cerebral metabolism and neuronal dysfunction.
There were several differences in amyloid plaque, tau deposition and activated astrocyte between the AβPParc and PSEN1DE9 brains as well as in comparison with the sAD brains. A positive association was observed between activated astrocyte and tau loads for the AβPParc brains but no such correlation was observed, respectively, neither the PSEN1DE9 nor the sAD brains. Although both mutations present abnormal plaque shapes (a ring shape for AβPParc and ‘cotton wool’ for PSEN1DE9), these mutations in the APP and PSEN1 genes, respectively, lead to major differences in the amyloid-beta composition and also in the tau deposits and activated astrocytes hallmarks in the development of AD pathology. Finally the use of two-tau tracers showed differences in regional binding both in the PSEN1DE9 but also between the two AβPParc brains suggesting difference in AD variants, which deserves further exploration in vivo.
Supplementary information
Acknowledgements
The authors would like to thank Pr. Hannu Kalimo and Dr. Liisa T Myllykangas from the Department of Pathology, University of Helsinki, Helsinki, Finland, for providing the PSEN1DE9 tissue. The authors would like to thank Pr. Bernardino Guetti and Rose Marie Richardson from the Department of Pathology & Laboratory Medicine, Indiana University School of Medicine, and Indianapolis, IN, USA for provinding the sporadic AD brain tissue. The Netherlands Brain Bank is also thanked for providing the control human brains used in the binding assay.
Funding
This study was financially supported by the Swedish Foundation for Strategic Research (SSF), the Swedish Research Council (project 05817, 2017-02965, 2017-06086), the Stockholm County Council-Karolinska Institutet regional agreement on medical training and clinical research (ALF grant), the Swedish Brain Foundation, the Alzheimer Foundation in Sweden, Karolinska Institutet StratNeuro, the Foundation for Old Servants, Gun and Bertil Stohne’s Foundation, Gunvor och Josef Anérs stiftelsen, the Loo and Hans Osterman’s Foundation, the Tore Nilsson Foundation, the KI Foundation for Geriatric Diseases, and The Swedish Dementia Association, Demensfonden, CIMED Karolinska Institutet-Stockholm County Council.
Author contributions
LL, PGG, and AN designed the study. LL performed and analyzed the autoradiography and binding assay studies. LL, PGG, and NB analysed the autoradiography and draw the region of interest. NB and IN provided analyze and interpretations for immunostaining. LSA provide the images and analyze for in vivo PET. CG, MV, and MI provided the AβPParc1, PSEN1DE9, and AβPParc2 case, respectively. LL, PGG, and AN, drafted the first version of the MS. All authors provided input and feedback during the process of writing the paper. All authors read and approved the final paper.
Data availability
All data generated or analysed during this study are included in this published article.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All experiments on autopsied human brain tissue were carried out in accordance with ethical permission obtained from the regional human ethics committee in Stockholm (permission number 2011/962/31-1; 2006/901-31/3 and 2017/2301-32), Uppsala (2005/103, 2011/044), the medical ethics committee of the VU Medical Center for the Netherlands Brain Bank tissue (permission number 1998-06/5), Indiana University Institutional Review Board, and Helsinki University Hospital DNr 86/2014 and Valvira (National Supervisory Authority for Welfare and Health) Dnr 2855/2012.
Informed consent
Informed consent was obtained for all cases investigated.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41380-020-0817-2) contains supplementary material, which is available to authorized users.
References
- 1.Basun H, Bogdanovic N, Ingelsson M, Almkvist O, Naslund J, Axelman K, et al. Clinical and neuropathological features of the arctic APP gene mutation causing early-onset Alzheimer disease. Arch Neurol. 2008;65:499–505. doi: 10.1001/archneur.65.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bettens K, Sleegers K, Van Broeckhoven C. Genetic insights in Alzheimer’s disease. Lancet Neurol. 2013;12:92–104. doi: 10.1016/S1474-4422(12)70259-4. [DOI] [PubMed] [Google Scholar]
- 3.Crook R, Verkkoniemi A, Perez-Tur J, Mehta N, Baker M, Houlden H, et al. A variant of Alzheimer’s disease with spastic paraparesis and unusual plaques due to deletion of exon 9 of presenilin 1. Nat Med. 1998;4:452–5. doi: 10.1038/nm0498-452. [DOI] [PubMed] [Google Scholar]
- 4.Mann DM, Takeuchi A, Sato S, Cairns NJ, Lantos PL, Rossor MN, et al. Cases of Alzheimer’s disease due to deletion of exon 9 of the presenilin-1 gene show an unusual but characteristic beta-amyloid pathology known as ‘cotton wool’ plaques. Neuropathol Appl Neurobiol. 2001;27:189–96. doi: 10.1046/j.1365-2990.2001.00316.x. [DOI] [PubMed] [Google Scholar]
- 5.Kalimo H, Lalowski M, Bogdanovic N, Philipson O, Bird TD, Nochlin D, et al. The Arctic AbetaPP mutation leads to Alzheimer’s disease pathology with highly variable topographic deposition of differentially truncated Abeta. Acta Neuropathol Commun. 2013;1:60. doi: 10.1186/2051-5960-1-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scholl M, Wall A, Thordardottir S, Ferreira D, Bogdanovic N, Langstrom B, et al. Low PiB PET retention in presence of pathologic CSF biomarkers in Arctic APP mutation carriers. Neurology. 2012;79:229–36. doi: 10.1212/WNL.0b013e31825fdf18. [DOI] [PubMed] [Google Scholar]
- 7.Dahlgren KN, Manelli AM, Stine WB, Jr., Baker LK, Krafft GA, LaDu MJ. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J Biol Chem. 2002;277:32046–53. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 8.Nilsberth C, Westlind-Danielsson A, Eckman CB, Condron MM, Axelman K, Forsell C, et al. The ‘Arctic’ APP mutation (E693G) causes Alzheimer’s disease by enhanced Abeta protofibril formation. Nat Neurosci. 2001;4:887–93. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- 9.Philipson O, Lord A, Lalowski M, Soliymani R, Baumann M, Thyberg J, et al. The Arctic amyloid-beta precursor protein (AbetaPP) mutation results in distinct plaques and accumulation of N- and C-truncated Abeta. Neurobiol Aging. 2012;33:1010.e1–13. doi: 10.1016/j.neurobiolaging.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 10.Thordardottir S, Kinhult Stahlbom A, Almkvist O, Thonberg H, Eriksdotter M, Zetterberg H, et al. The effects of different familial Alzheimer’s disease mutations on APP processing in vivo. Alzheimers Res Ther. 2017;9:9. doi: 10.1186/s13195-017-0234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thordardottir S, Stahlbom AK, Ferreira D, Almkvist O, Westman E, Zetterberg H, et al. Preclinical cerebrospinal fluid and volumetric magnetic resonance imaging biomarkers in Swedish familial Alzheimer’s disease. J Alzheimers Dis. 2015;43:1393–402. doi: 10.3233/JAD-140339. [DOI] [PubMed] [Google Scholar]
- 12.Koivunen J, Verkkoniemi A, Aalto S, Paetau A, Ahonen JP, Viitanen M, et al. PET amyloid ligand [11C]PIB uptake shows predominantly striatal increase in variant Alzheimer’s disease. Brain. 2008;131:1845–53. doi: 10.1093/brain/awn107. [DOI] [PubMed] [Google Scholar]
- 13.Falcon B, Zhang W, Schweighauser M, Murzin AG, Vidal R, Garringer HJ, et al. Tau filaments from multiple cases of sporadic and inherited Alzheimer’s disease adopt a common fold. Acta Neuropathol. 2018;136:699–708. doi: 10.1007/s00401-018-1914-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verkkoniemi A, Kalimo H, Paetau A, Somer M, Iwatsubo T, Hardy J, et al. Variant Alzheimer disease with spastic paraparesis: neuropathological phenotype. J Neuropathol Exp Neurol. 2001;60:483–92. doi: 10.1093/jnen/60.5.483. [DOI] [PubMed] [Google Scholar]
- 15.Lemoine L, Saint-Aubert L, Nennesmo I, Gillberg PG, Nordberg A. Cortical laminar tau deposits and activated astrocytes in Alzheimer’s disease visualised by (3)H-THK5117 and (3)H-deprenyl autoradiography. Sci Rep. 2017;7:45496. doi: 10.1038/srep45496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leuzy A, Chiotis K, Lemoine L, Gillberg PG, Almkvist O, Rodriguez-Vieitez E, et al. Tau PET imaging in neurodegenerative tauopathies-still a challenge. Mol Psychiatry. 2019;24:1112–34. doi: 10.1038/s41380-018-0342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murugan NA, Halldin C, Nordberg A, Langstrom B, Agren H. The culprit is in the cave: the core sites explain the binding profiles of amyloid-specific tracers. J Phys Chem Lett. 2016;7:3313–21. doi: 10.1021/acs.jpclett.6b01586. [DOI] [PubMed] [Google Scholar]
- 18.Ni R, Gillberg PG, Bergfors A, Marutle A, Nordberg A. Amyloid tracers detect multiple binding sites in Alzheimer’s disease brain tissue. Brain. 2013;136:2217–27. doi: 10.1093/brain/awt142. [DOI] [PubMed] [Google Scholar]
- 19.Ni R, Gillberg PG, Bogdanovic N, Viitanen M, Myllykangas L, Nennesmo I, et al. Amyloid tracers binding sites in autosomal dominant and sporadic Alzheimer’s disease. Alzheimers Dement. 2017;13:419–30. doi: 10.1016/j.jalz.2016.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article.