Abstract
Aromatic amines in nature are typically installed with Glu or Gln as the nitrogen donor. Here we report a pathway that features glycyl-tRNA as the nitrogen donor. During the biosynthesis of pyrroloiminoquinone-type natural products such as ammosamide, peptide-aminoacyl tRNA ligases (PEARLs) append amino acids to the C-terminus of a ribosomally synthesized peptide. First, adds Trp in a Trp-tRNA dependent reaction, and the flavoprotein AmmC1 then carries out three hydroxylations of the indole ring of Trp. After oxidation to the corresponding ortho-hydroxy para-quinone, attaches Gly to the indole ring in a Gly-tRNA dependent fashion. Subsequent decarboxylation and hydrolysis results in an amino-substituted indole. Similar transformations are catalyzed by orthologous enzymes from Bacillus halodurans. This pathway features three previously unknown biochemical processes using a ribosomally synthesized peptide as scaffold for non-ribosomal peptide extension and chemical modification to generate an amino acid derived natural product.
Graphical Abstract
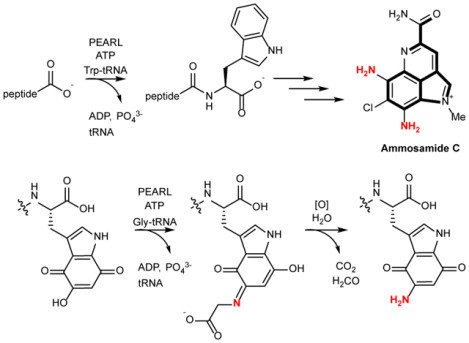
Pyrroloiminoquinones and related natural products are produced by a wide variety of organisms and have diverse bioactivities1–5. The ammosamides are a subset of pyrroloiminoquinones that are produced in various environments (Fig. 1a)6–8. This compound family displays antiproliferative activities against cancer cells with some members binding to myosin or inhibiting quinone reductase 26, 9–11. The core structures of the ammosamides and lymphostin, another pyrroloiminoquinone, are derived from Trp7, 12, 13. Recent studies demonstrated that these compounds are biosynthesized via an unexpected posttranslational modification process in which a peptide aminoacyl-tRNA ligase (PEARL) adds Trp from tryptophanyl-tRNA to the C-terminus of a ribosomally produced scaffold peptide. This Trp is then converted to a pyrroloiminoquinone by an as-of-yet unknown pathway14.
Fig. 1. Structures of pyrroloiminoquinone-derived natural products and activity of PEARL enzymes.
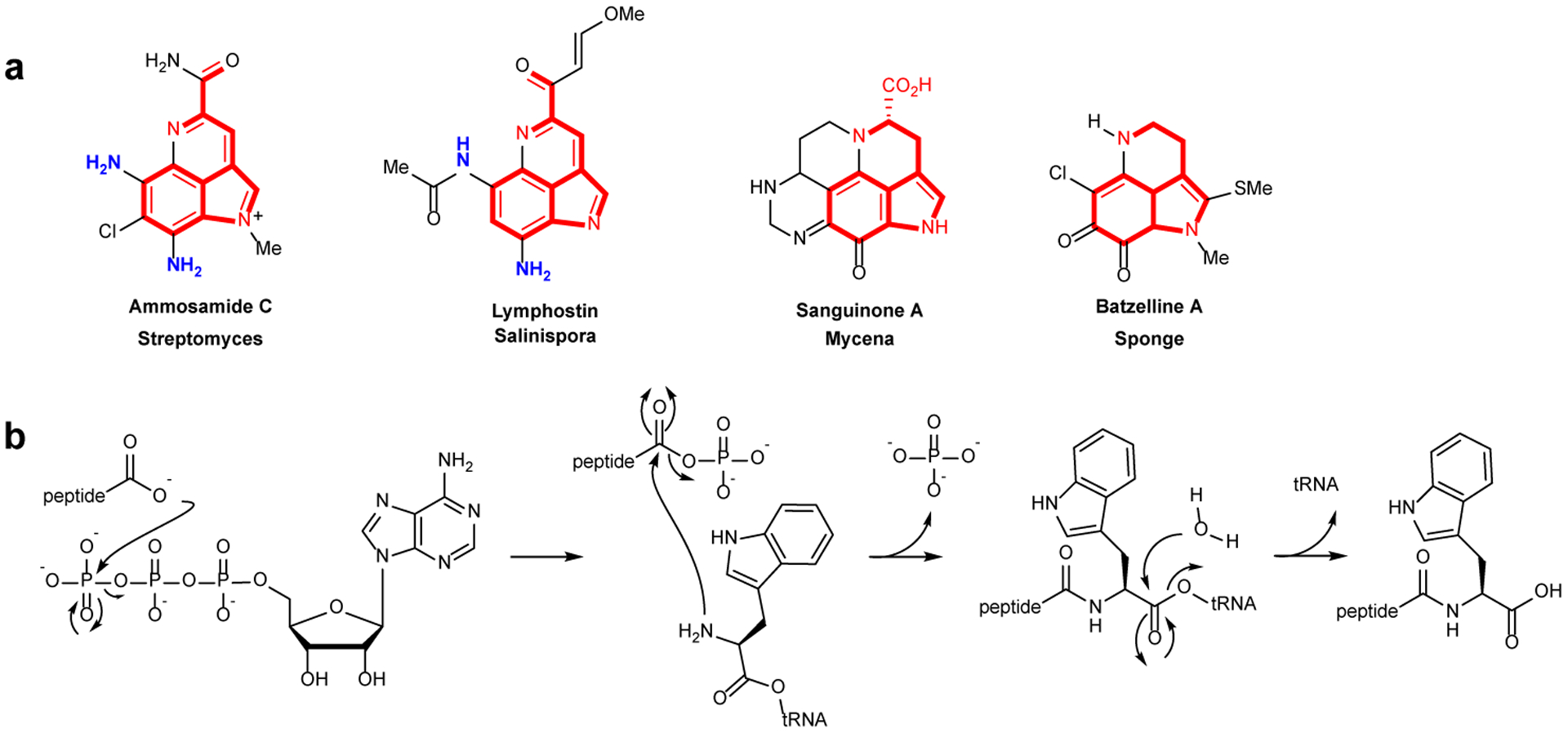
a, Representative examples of pyrroloiminoquinone-type natural products (also called pyrroloquinolines). The part of the structures shown in red highlight the atoms that likely derive from Trp. For family members for which the biosynthetic gene clusters are known, the current work shows that the amino groups depicted in blue likely originate from glycyl-tRNA and are installed by PEARLs. b, PEARL mechanism of non-ribosomal peptide extension during ammosamide biosynthesis.
PEARLs display low sequence similarity to the N-terminal domain of LanB dehydratases involved in lanthipeptide biosynthesis (e.g. 11.2% identity/21.0% similarity between the PEARL TglB and the dehydratase NisB). The N-terminal domain of LanB uses glutamyl-tRNA to glutamylate the side chain hydroxyl groups of Ser and Thr residues in a substrate peptide followed by elimination of the Glu by the C-terminal domain to generate dehydroalanine and dehydrobutyrine, respectively15–19. PEARL enzymes have homology with the LanB glutamyl transferase domain but lack the elimination domain. They also use aminoacyl-tRNA as substrate but catalyze very different chemistry that first involves ATP-dependent phosphorylation of the C-terminus of a scaffold peptide, and then amide bond formation with the amino group of the aminoacyl-tRNA (Fig. 1b)20. This process is involved in the biosynthesis of a range of natural products by phylogenetically diverse bacteria, including the ammosamides and 3-thiaglutamate7, 12, 14.
The ammosamide biosynthetic gene cluster (BGC) in Streptomyces sp. CNR-698 encodes four PEARL enzymes7 (Fig. 2a), even though all carbon atoms found in ammosamide derive from Trp, and hence one PEARL that adds Trp appears sufficient. We demonstrate here a pathway by which PEARLs are utilized to introduce nitrogen atoms onto the indole of Trp, providing a general framework for pyrroloiminoquinone biosynthesis.
Fig. 2 |. Posttranslational modifications during ammosamide biosynthesis.

a, The amm and bha BGCs and the sequences of the precursor peptides. RRE encoding sequences are indicated as white bars in the gene. b, MALDI-TOF mass spectra of AmmA*-Trp (avg. mass calculated m/z 6941.1; observed 6941.8) and AmmA*-Trp co-expressed with AmmC1 (avg. mass calculated m/z 6989.1; observed 6989.6). The two experiments were conducted side-by-side. c, MALDI-TOF mass spectrum of AmmA*-Trp co-expressed with AmmC1 (avg mass calculated 6989.1; observed 6989.6) and AmmA*-Trp with AmmC1 and AmmB3 (avg. mass calculated m/z 7000.1; observed 7000.6). The co-expressions were conducted three times in independent experiments with similar results.
Our studies focused on the ammosamide BGC as well as a gene cluster in Bacillus halodurans C-125 that contains seven PEARL enzymes (Fig. 2a). We demonstrate that both BGCs encode a PEARL that adds Trp to a scaffold peptide. Next, a tetratricopeptide repeat (TPR) domain-containing flavoprotein oxidizes the indole of the tryptophan three times to generate a trihydroxyindole. This electron-rich structure is oxidized to the corresponding hydroxyquinone and converted to an aminoquinone via PEARL-catalyzed addition of Gly-tRNA, providing an unanticipated pathway to aromatic amines. These studies provide an unexpected use of PEARL enzymes for the amination of indoles.
Results and discussion
Analysis of the amm biosynthetic gene cluster
The amm BGC (Fig. 2a, Supplementary Table 1) contains a gene for a precursor peptide (ammA), four genes encoding PEARL enzymes (ammB1–4), two genes encoding tetratricopeptide repeat (TPR) domain containing enzymes (amm7/amm22), a glycine oxidase (amm17), several proteases (amm12/19), a chlorinase (amm3), and several proteins whose functions are not clear (Fig. 2a, Supplementary Table 1)7. A previous study showed that the precursor peptide is first trimmed back to a shorter peptide (termed AmmA*) that has homology with many other peptides that are encoded near PEARL enzymes in various BGCs (Supplementary Fig. 1 and 2). Removal of the seven C-terminal residues from AmmA reveals a C-terminal motif (APLALA) that is conserved in its homologs (Supplementary Fig. 2). AmmB2 was shown to add a Trp from Trp-tRNA to the C-terminus of AmmA*14. The resulting peptide AmmA*-Trp was not a substrate for any of the four PEARL enzymes (Supplementary Fig. 3), which have low pair-wise sequence identities (Supplementary Table 2). Hence, the pathway by which the appended Trp is converted to a pyrroloiminoquinone remained unresolved.
We hypothesized that another posttranslational modification was needed before the next PEARL could act on the peptide. A strict order of PTMs has been reported for other RiPP biosynthetic pathways (e.g.21–24). Therefore, we interrogated the activity of the other proteins encoded in the amm biosynthetic gene cluster by co-expressing His6-AmmA*-Trp with other enzymes in Escherichia coli. Upon purification of the peptides by immobilized metal affinity chromatography (IMAC), we found that co-expression with Amm7 resulted in a mass shift of +48 Da (peptide A) as observed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Fig. 2b). Smaller peaks of +16 and +32 Da were also observed suggesting possibly three hydroxylations. Subsequent treatment with trypsin and high-resolution mass spectrometry (HRMS) analysis revealed that the mass additions occurred on the C-terminal 8-mer fragment (Supplementary Fig. 4). Co-expression of AmmA*-Trp with Amm7 and each of the remaining PEARL enzymes (AmmB1, AmmB3, and AmmB4) individually resulted only in a change in mass of peptide A for AmmB3. A new product was observed that increased in mass by 11 Da (Fig. 2c, Supplementary Fig. 5). This mass increase was puzzling as it did not correspond to the anticipated addition of a proteinogenic amino acid. Poor yield did not allow characterization of the product, but the nature of the chemical transformations catalyzed by Amm7 and AmmB3 were revealed by study of a related BGC in B. halodurans (Fig. 2a) as described in the next sections. The orthologous enzymes from this BGC gave higher product yields in E. coli as well as intermediates that provided key additional insights.
Analysis of the bha biosynthetic gene cluster
The PEARL-containing bha gene cluster in B. halodurans exhibits several similarities to the ammosamide BGC (Fig. 2a, Supplementary Table 3). It contains genes for a precursor peptide (bhaA) with sequence similarity to AmmA*, seven instead of four PEARLs (bhaB1–7), four TPR-domain containing enzymes with sequence homology to AmmC1 (bhaC1–4 ; Supplementary Fig. 6), a glycine oxidase (bhaG), TldD/E-like proteases (bhaDE)25, 26, and several other proteins of unknown function that are not in the amm cluster (Fig. 2a, Supplementary Tables 1 & 3). Six of the seven PEARLs contain a typical RiPP recognition element (RRE)27 that has been shown crystallographically to engage the substrate peptide for LanB enzymes15. The TldD/E-like proteins BhaD and BhaE are anticipated to proteolytically cleave the mature natural product from the scaffold peptide25, 26. Similar BGCs were found in a variety of organisms from several different bacterial phyla (for precursor peptides and accession numbers, see Supplementary Fig. 2a) suggesting a widespread occurrence. Attempts to activate the bha cluster in B. halodurans were unsuccessful (see Supplemental Information), prompting a combined biochemical and synthetic biology study of the enzymes involved.
BhaB1 and BhaB7 are aminoacyl-tRNA dependent amide bond forming enzymes
The BhaA peptide and its homologs (Fig. 2a and Supplementary Fig. 2) are highly acidic peptides (17 out of 39 residues are Asp or Glu). BhaA shares high sequence identity with the precursor peptide AmmA in the ammosamide BGC, but does not contain the seven amino acids at the C-terminus of AmmA (Supplementary Fig. 2a). Compared to its homologs and the truncated AmmA* peptide, BhaA lacks the C-terminal Ala of the APLALA motif (Fig. 2a, Supplementary Fig. 2). To determine which PEARL acts first in the B. halodurans pathway, each PEARL was co-expressed in E. coli with His6-BhaA. Upon purification of the peptide by IMAC, MALDI-TOF MS analysis demonstrated that only co-expression with BhaB1 changed the peptide mass, resulting in an increase of 71 Da (Fig. 3a,b). This increase is consistent with the condensation of an Ala residue. High resolution (HR) tandem mass spectrometry (MS/MS) analysis of the peptide compared to a synthetic standard confirmed that an Ala residue had been appended to the C-terminus of the peptide (Supplementary Fig. 7). The appendage of Ala adds to the known amino acids that are used by PEARLs, which to date included Cys and Trp14. In addition, the activity of BhaB1 completes the APLALA motif that is present in BhaA homologs including AmmA* and compensates for the lack of an Ala at the C-terminus of BhaA.
Fig. 3. Conversion of Trp to aminoquinone.
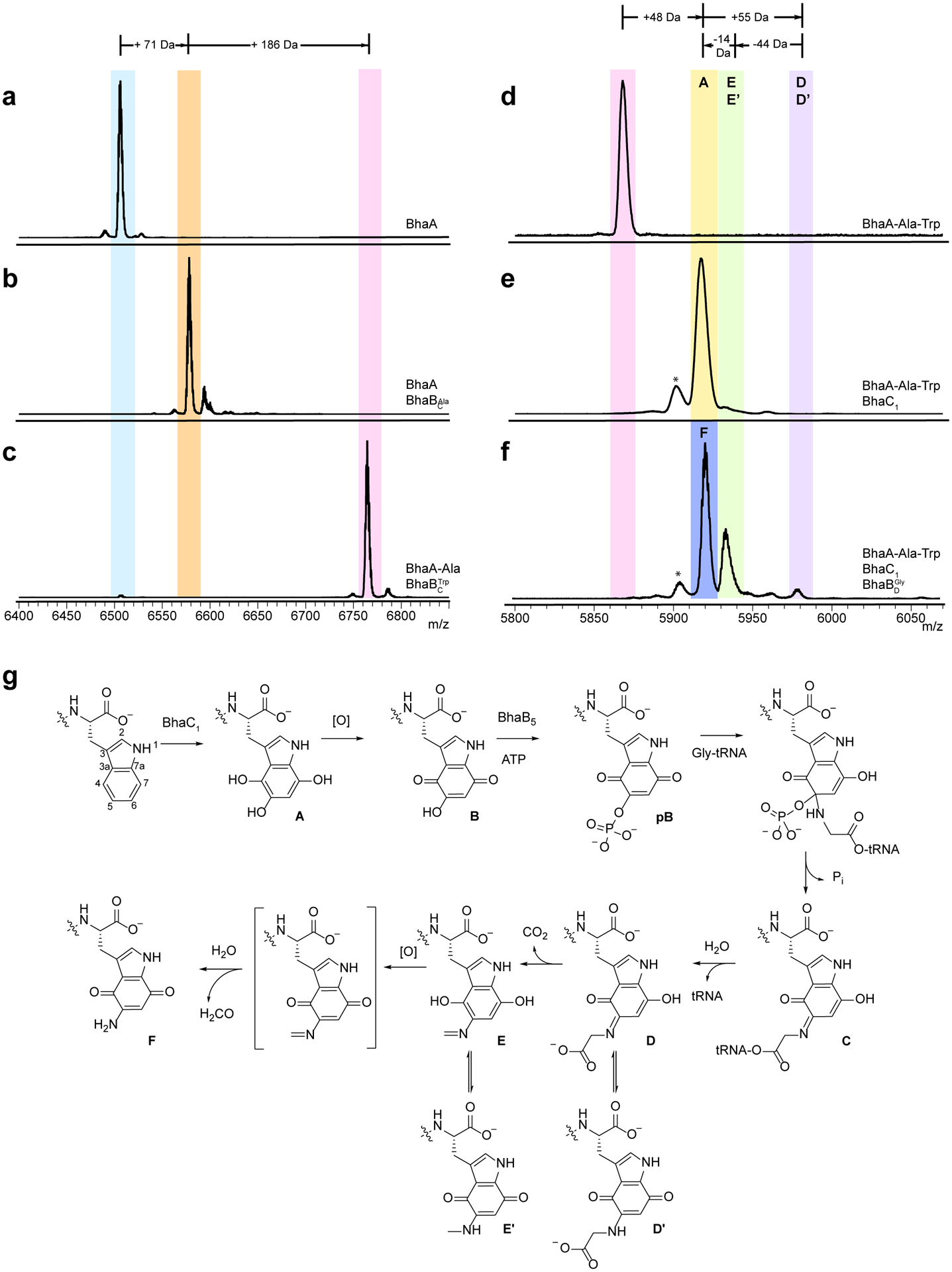
a-f, MALDI-TOF mass spectra of a, BhaA (calculated m/z 6502.1 Da; observed 6502.3 Da), b, BhaA co-expressed with BhaB1 (renamed herein ; calculated m/z 6573.1; observed 6573.2 Da), c, BhaA-Ala peptide co-expressed with BhaB7 (renamed herein ); calculated m/z 6759.2; observed 6760.5, d, BhaA-Ala-Trp (calculated m/z 5869.7; observed 5870.4), e, product from co-expression of BhaA-Ala-Trp with BhaC1 (calculated m/z 5917.7; observed 5917.4), and f, products of co-expression of BhaA-Ala-Trp with BhaC1 and BhaB5 (renamed herein); calculated m/z 5972.7; observed 5972.5. g, proposed mechanism of converting Trp to an aminoquinone. The co-expressions were repeated three times in independent experiments with similar results. The Bha-Ala-Trp peptide in panel c was expressed from a different plasmid (pRSFDuet-1) than the peptide in panel d (pACYCDuet) accounting for their different masses. See coexpression section in Methods. The peaks labeled with * are deamination artifacts (M-17) from analysis by MALDI-TOF MS in positive reflector mode.
We next investigated all remaining BhaB enzymes by co-expression in E. coli with the product of BhaB1, BhaA-Ala, and discovered that BhaB7 increases the peptide mass by 186 Da, consistent with the condensation of a Trp residue (Fig. 3b,c). MS/MS analysis of the peptide compared to a synthetic standard corroborated the appendage of Trp to the C-terminus of BhaA-Ala (Supplementary Fig. 8). Thus, unlike LanB dehydratases that to date have all been shown to transfer glutamate from glutamyl-tRNA to the side chain of Ser and Thr residues, the activities of PEARL enzymes are more diverse. We propose nomenclature that helps to rapidly and concisely indicate the individual activities of these enzymes. We suggest that the term is used for canonical dehydratases that contain both glutamylation and elimination domains and that the term is used for the glutamylation component of “split LanB” enzymes that have the glutamylation and elimination activities in two separate polypeptides (e.g. thiopeptides and goadsporin)21, 28. We suggest the term for PEARL enzymes that conjugate amino acids (Aaa) to the C-terminus of scaffold peptides. Thus, AmmB2 would be , BhaB1 would be , and BhaB7 would be . We will use this terminology in the remainder of this study.
The transformation catalyzed by is akin to the tryptophan condensation by in the ammosamide biosynthetic pathway14, thus providing an example of two PEARL enzymes that catalyze the same conjugation reaction. Sequence similarity analysis reveals that and are quite distinct from one another with only 20% sequence identity (Supplementary Table 2, Supplementary Fig. 9a). This difference in part may reflect the phylogeny of the producing organisms (Actinobacteria and Firmicutes) as is also observed for proteins15, 29, and in part reflect different evolutionary paths given a number of insertions and deletions. Further studies will be required to characterize the molecular basis for amino acid specificity of PEARLs such that their activities may be predicted by bioinformatics.
In vitro incubation of purified His6- with BhaA, Ala, ATP, tRNAAla, and alanyl-tRNA synthetase (AlaRS) from E. coli resulted in the production of BhaA-Ala, just as was observed by co-expression. Omitting reagents from the assay demonstrated that the reaction is ATP-, tRNA-, and AlaRS-dependent (Supplementary Fig. 10). Similarly, in vitro reaction of purified with BhaA-Ala, Trp, ATP, tRNATrp, and tryptophanyl-tRNA synthetase (TrpRS) from E. coli resulted in the production of BhaA-Ala-Trp (Supplementary Fig. 11).
BhaC1 hydroxylates the indole of Trp
Similar to the observations with the amm BGC, no further modification was detected in E. coli upon co-expression of the and modified peptide (BhaA-Ala-Trp) with the remaining PEARLs in the BGC (Supplementary Fig. 12). The AmmC1 homologs BhaC1, -C2, -C3, -C4, are all TPR-domain containing proteins, but analyses using various bioinformatics tools (Pfam, BLAST, HHpred, Phyre2 modeling) do not provide any additional hints regarding function or cofactor use. TPR-domains contain a series of consensus residues that generate alpha-solenoid structures that usually mediate protein-protein interactions30–34. Upon co-expression with His6-BhaA-Ala-Trp, of the four BhaC proteins, only BhaC1 led to a change in the mass of the peptide, increasing it by 48 Da (Fig. 3d,e), consistent with the observations with AmmC1 (Fig. 2b). To evaluate the site of putative oxidation, the peptide product was treated with LysC endoproteinase and the resulting C-terminal 8-mer peptide was subjected to MS/MS analysis, demonstrating that the +48 Da modification was to the C-terminal Trp residue (Supplementary Fig. 13). The enzymatic product readily oxidized resulting in a 2 Da mass loss and alteration of the spectral properties of the peptide, which turned pink in color (peptide B, Supplementary Fig. 14 and 15). NMR and MS data on peptide B after protease treatment are consistent with conversion of the indole of Trp to a 5-hydroxyquinone structure (Fig. 3g; Supplementary Fig. 16 and 17) and also demonstrates the high reactivity of B (see Supplementary Information).
We next expressed His6-BhaC1in E. coli and purified the enzyme by IMAC. Mass spectrometric analysis of the purified protein revealed the presence of flavin mononucleotide (FMN) (Supplementary Fig. 18), consistent with the yellow color of the enzyme. BhaC1 was not predicted bioinformatically to be a flavoprotein and no obvious flavin binding signatures could be detected in its sequence. In vitro His6-BhaC1 also oxidized BhaA-Ala-Trp three times (Supplementary Fig. 19), and intermediates with one (+16 Da) and two hydroxylations (+32 Da) were also observed. These intermediates could not be separated, preventing investigation of the order of the modifications, but the observation of +16 and +32 Da products further supports the assignment of the +48 Da product resulting from three hydroxylations. The oxidation catalyzed by BhaC1 (and AmmC1 as discussed above) is an unusual example of post-translational trihydroxylation of tryptophan by a single enzyme35–40.
BhaB5 adds glycine from Gly-tRNA to the indole of Trp
After observing the hydroxylation of the Trp, we revisited the PEARLs within the cluster to assess whether any of the remaining five BhaB enzymes would act on this modified peptide as was observed with the AmmC1-oxidized AmmA*-Trp and AmmB3 (Fig. 2c). Upon co-expression of BhaA-Ala-Trp, BhaC1 and either BhaB2, -B3, -B4, -B5, or -B6, the only change in the mass of the BhaA peptide was observed with BhaB5 (Fig. 3f). MALDI-TOF MS analysis of the products after IMAC purification showed three products (ions associated with compounds D, E, and F, Fig. 3f,g). Notably, compound E is the equivalent product that was observed upon co-expressing AmmA*-Trp with AmmC1 and AmmB3 (+11 Da from peptide A, Fig. 2c). A minor product (corresponding to peak D) corresponded to an increased mass of +55 Da from the trihydroxylated Trp-containing peptide (or +57 Da from the quinone-containing pink peptide B). Based on the presumption that PEARLs append amino acids to scaffold peptides, the mass shift was consistent with the condensation of Gly to the C-terminus of the peptide. The product peptide was digested with endoproteinase LysC and subjected to HR-MS, which provided data consistent with the addition of Gly (Supplementary Fig. 20). MS/MS analysis corroborated the appendage of a +57 Da moiety to the C-terminal Trp, but the expected y ion corresponding to a glycine fragment by amide bond fragmentation was not detected (Supplementary Fig. 21). The amounts of product D were very small, but scale-up allowed isolation of sufficient material to record an HMBC spectrum, which surprisingly revealed that the appended Gly was attached to C5 of the oxidized indole of the modified Trp residue (Fig. 3g) as indicated by several diagnostic cross peaks (Supplementary Fig. 22).
This transformation at first glance appears very different from the chemistry catalyzed by other PEARLs, which predicted the Gly to be attached to the C-terminal carboxylate. However, as noted above, in vitro the initial trihydroxylated Trp is readily oxidized to the corresponding ortho-hydroxy-para-quinone (B, Fig. 3g). By the concept of vinylogy41, the hydroxyl group at C5 in this intermediate is expected to have similar reactivity as a carboxylic acid. Thus, based on the mechanism previously determined for the PEARL TglB20, we propose that BhaB5 phosphorylates the hydroxyl group at position 5 in the ortho-hydroxy quinone B to generate intermediate pB (Fig. 3g) followed by attack of the amino group of glycyl-tRNAGly onto C5 to form a tetrahedral intermediate and elimination of phosphate to yield intermediate C. Subsequent hydrolysis of the tRNA, as previously demonstrated for TglB14, would give intermediate D. The MS and NMR data (Supplementary Fig. 20–22) are fully consistent with this assignment and are suggestive of the formation of tautomer D’ (Fig. 3g). It is possible that the on-pathway intermediate is D and that a small amount of peptide dissociates from the enzyme and tautomerizes to D’, thereby allowing detection and isolation of the intermediate by preventing decarboxylation. Because BhaB5 does not add an amino acid to the C-terminus of a peptide, we propose that enzymes that catalyze similar chemistry will be termed , with BhaB5 named .
Closer analysis of the other two products formed in the reaction of by MALDI-TOF MS (peaks E/E’ and F, Fig. 3f) provides additional support that the addition of Gly to C5 on the indole of Trp is on pathway to pyrroloiminoquinone formation. At first glance, the mass spectrum suggests that the reaction did not go to completion and that most of the oxidized BhaA-Ala-Trp (B) was unreacted. However, treatment with endoproteinase AspN to shorten the peptide and subsequent high-resolution electrospray ionization (ESI) MS analysis demonstrated that the mass of the peptide had decreased by 1 Da compared to peptide B (Supplementary Fig. 23). The most likely explanation is conversion of the ortho-hydroxy-para-quinone to an ortho-amino-para-quinone (F, Fig. 3g). In vitro experiments discussed below support this assignment.
The third product of the reaction (peak E/E’, Fig. 3g) is the equivalent product observed originally in the AmmB3 reaction (Fig. 2c). The peptide displays an ion with a mass that is increased by 11 Da from the hydroxyquinone and decreased by 44 Da from the Gly adduct D/D’ consistent with decarboxylation of intermediate D to compound E or its tautomer E’. We propose that product F is formed upon oxidation of the electron rich indole of peptide E, which would facilitate hydrolysis of the imine to the amine (Fig. 3g). This oxidation is not seen to the same extent when AmmA*-Trp was co-expressed with AmmC1 and AmmB3 () in E. coli (Fig. 2c), nor is it observed to the same extent in vitro with the Bha enzymes (see below). Thus, we propose that this step is normally catalyzed by one of the redox enzymes in the amm and bha BGCs (Supplementary Tables 1 and 2) and that the oxidation observed in Fig. 3f is due to serendipitous oxidation in E. coli. Indeed, the extent of oxidation also varies in different E. coli expression strains (e.g. Supplementary Fig. 25). The oxidation and hydrolysis of intermediate E/E’ prevented NMR characterization as the intermediate was converted to F during the extended time required (Supplementary Fig. 24). Intermediate F proved very reactive like quinone B (Supplementary Figs. 16 and 17), and decomposed non-enzymatically into multiple products that show fragment ions in the MS-MS that are consistent with cyclization and dimerization products but that we were unable to fully assign.
To provide further support for the mechanism in Fig. 3g and the structural assignments, the BhaB5 reaction was reconstituted in vitro confirming the requirement of glycyl-tRNA and ATP (Supplementary Fig. 26). Use of [15N,13C2, 2H2]-Gly resulted in mass shifts for peptides D/D’, E/E’, and F consistent with their assignments (Supplementary Figs. 27–30).
One aspect of the proposed model requires further discussion. The products of the AmmC1 and BhaC1 enzymes formed in E. coli are colorless and contain Trp with a trihydroxyindole. Only upon purification and exposure to air in the absence of reducing agents do these peptides oxidize and take on a pink color. The question thus arises how AmmB3 and BhaB5 can add Gly in E. coli as observed (Fig. 2c and 3f). We propose that the electron rich trihydroxyindole is oxidized to the quinone in cells but that the reducing environment readily reverses this oxidation. But when AmmB3 or BhaB5 are co-expressed, they convert the small amount of hydroxyquinone-containing peptides to the Gly adduct. We suspect that the oxidation of A to B in the physiological pathway may be carried out by one of several redox enzymes in the BGCs (Supplementary Tables 1 and 3). This hypothesis also suggests that the originally formed product A is not a substrate for AmmB3 or BhaB5. This prediction was confirmed for BhaB5, which did not react with A under anaerobic conditions (Supplementary Fig. 31). Because AmmB3 does not accumulate intermediate D/D’ observed with BhaB5, we could not confirm that the attachment site of Gly is also C5. An alternative possibility is C7 since the hydroxyquinone B could exist in a tautomeric form where phosphorylation could occur at O7. Such putative alternative regiochemistry could explain why the imine product appears to accumulate more in E. coli for AmmA than BhaA (Figures 2c and 3f).
An alternative route to aromatic amines
The current study, and the occurrence of similar BGCs in various bacteria, reveals an unexpected route by which nature accesses aromatic amines in a range of different phyla. Previously characterized pathways to these common structural motifs use the canonical amino group donors Glu and Gln during amino transfer events (Fig. 4). In these pathways, the amino group is introduced prior to aromatization. In an unusual case in roseoflavin biosynthesis42–45, aromatic amine biosynthesis takes place on a substrate that is already aromatic, but the nitrogen donor is still Glu (Fig. 4). An alternative route reported in this work introduces amino groups starting from a phenolic precursor and using glycine from glycyl-tRNA as the amino group donor.
Fig. 4 |. Known pathways to aromatic amines.
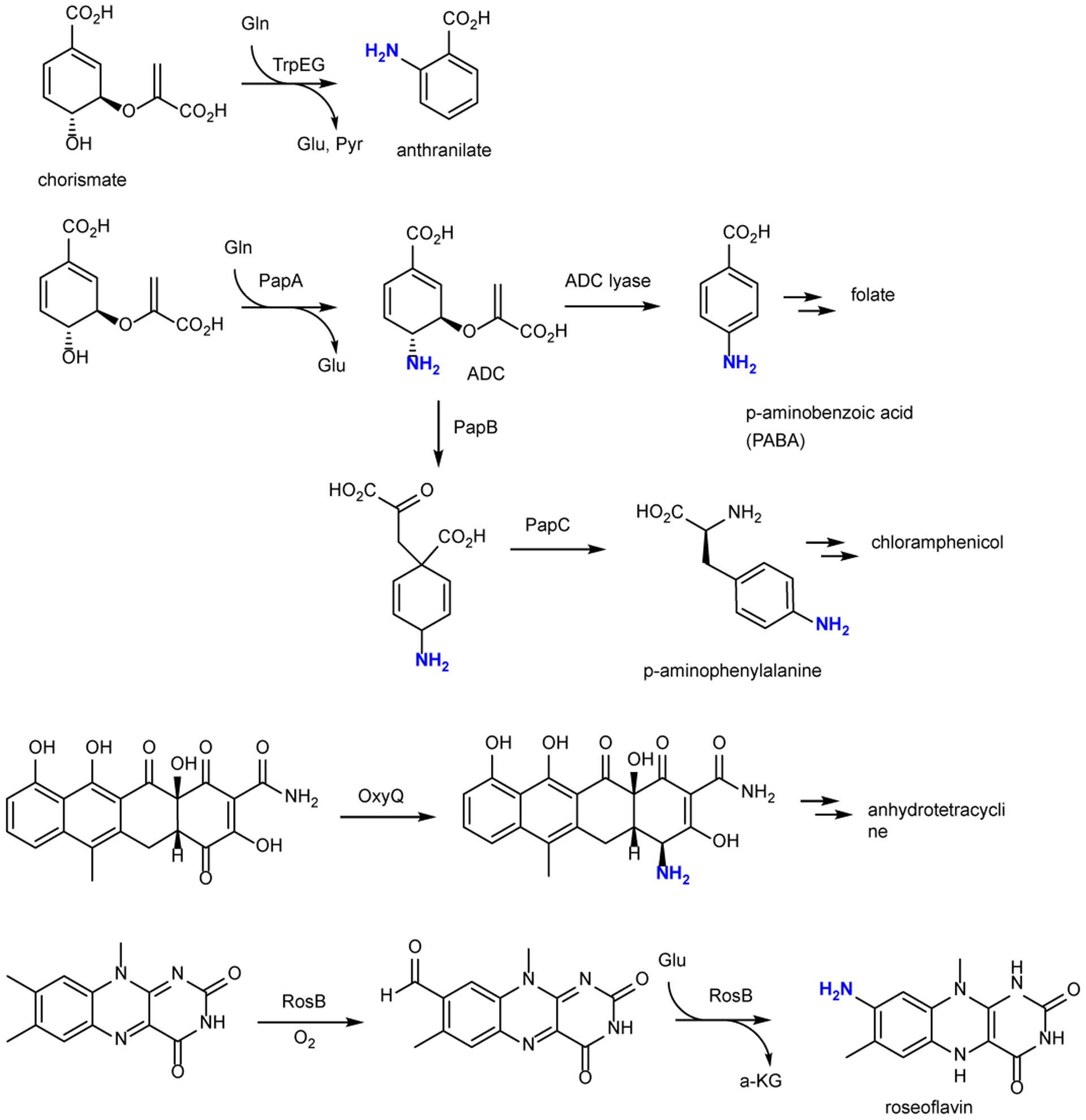
Previously identified pathways to aromatic amines that typically involve Glu or Glu as the nitrogen donors.
Our findings allow a proposed pathway to the pyrroloiminoquinone scaffold of the ammosamides. The current study demonstrates the conversion of Trp to an aminoquinone by AmmC1 and AmmB3. Two more amino groups would need to be introduced to form the skeleton of the ammosamides (Fig. 1a), which can be achieved by two more cycles of addition of glycyl-tRNA by the two remaining AmmB proteins (AmmB1 and AmmB4). Supplementary Fig. 32 presents a pathway to ammosamide C using a combination of the proteins encoded in the amm BGC. Thus, the current study provides insights into how a Trp appended at the C-terminus of a ribosomal peptide can be transformed into a pyrroloiminoquinone using post-translational modifications.
This work also demonstrates an interesting difference between the and enzymes and their BhaB orthologs compared to the and dehydratases. The latter enzymes have been demonstrated to be selective for the tRNAGlu sequence of their producing organisms in both lanthipeptide and thiopeptide biosynthesis. For instance, glutamyl-tRNA dependent dehydratases involved in microbisporicin and thiomuracin biosynthesis showed poor activity with glutamyl-tRNA from E. coli and were fully active only with tRNA from Actinobacteria21, 46. In contrast, and from Streptomyces sp. CNR-698 and and from Bacillus halodurans C-125 were active in E. coli and therefore do not appear to require tRNA sequences from their producing organism (see Supplementary Fig. 33 for a comparison of tRNA sequences). Although more examples will need to be investigated to determine if this conclusion is general, from an evolutionary perspective a relaxed specificity with respect to the tRNA sequence would be beneficial for evolving new activities and selectivities, which may explain the diversity of aminoacyl-tRNAs utilized by LanBC/D enzymes.
Methods
For protein expression, purification, and in vitro assays, see the Supplemental Information.
In vitro transcription of tRNAs
Primers for E. coli tRNAAla, tRNATrp, and tRNAGly were designed according to a previously described method47. The tRNA dsDNA templates were generated from two overlapping synthetic deoxyoligonucleotides (Supplementary Table 4). To prepare the dsDNA template for in vitro transcription, 5′ overhangs were filled in using the following conditions: NEB Buffer 2 (1×), primers (4 μM each), dNTP (100 μM each), and DNA polymerase I large (Klenow) fragment (1 U μg–1 DNA) in a final volume of 50 μL. The reaction was incubated at 25 °C for 15 min, quenched with EDTA (10 mM) at 75 °C for 25 min, and dsDNA templates were precipitated with cold EtOH overnight. In vitro transcription was performed using a previously described method48. The transcribed tRNA was then purified by acidic phenol extraction using a previously described method49.
Co-expression experiments and peptide purification
For plasmid preparation, see the Supplementary Information. E. coli BL21 (DE3) cells (New England Biolabs) expressing N-terminal His6-tagged BhaA variants (BhaA, BhaA-Ala, BhaA-Ala-Trp) were grown with the following antibiotic markers. Cells containing His6-BhaA-pET28b (Fig. 3a) or His6-BhaA-Ala-pET28b were grown with 50 μg/mL kanamycin. Cells containing His6-BhaA-Ala-Trp-pETDuet-1 were grown with 100 μg/mL ampicillin. For co-expression systems of bha, the BhaB1 and BhaB7-modified peptides were obtained by using pRSFDuet-1 with the peptide (His6-BhaA or His6-BhaA-Ala) encoded in multiple cloning site 1 (MCS1) and the PEARL enzyme encoded in MCS2 (Fig. 3b–c). These celles were grown with 50 μg/mL kanamycin. For the co-expression of BhaA-Ala-Trp with BhaC1 we observed that the enzyme did not fully process the substrate when the peptide was expressed from a high copy number plasmid like pETDuet-1 (~40 copies) or pRSFDuet-1 with the enzyme inserted in (MCS2) of the same vector. Therefore, new constructs were redesigned with the peptide BhaA-Ala-Trp expressed from pACYCDuet, which has a lower copy number (10–12 copies), and BhaC1 from pET28b (copy number ~40). This change results in a peptide that has a slightly different N-terminal sequence compared to BhaA-Ala-Trp formed by BhaB1 and BhaB7 expressed from pRSFDuet-1 (pRSFDuet-1, GSSHHHHHHSSGLVPRGSH- Fig. 3c vs pACYCDuet, GSSHHHHHHS- Fig. 3d). Most importantly, this switch resulted in full oxidation of BhaA-Ala-Trp by BhaC1. Cells containing His6-BhaA-Ala-Trp-pACYCDuet-1 were grown with 25 μg/mL chloramphenicol (Fig. 3d–f).
All cells were grown in terrific broth (TB) media (for 1 L total: 20 g bacto tryptone, 24 g yeast extract, 2.3 g KH2PO4, 12.5 g K2HPO4, and 0.4% glycerol). Cultures were grown with shaking at 37 °C to an OD600 = 0.5–0.7, upon which 0.4 mM IPTG was added to induce peptide expression. Cells were harvested after 18 h and resuspended in lysis buffer (50 mM HEPES, 100 mM NaCl, pH 7.5), then lysed by sonication.
Peptides were purified by IMAC. The lysate was clarified by centrifugation at 50,000 relative centrifugal force (rcf) and applied to 3 mL of Ni-NTA-resin gravity flow column (Clonetec). The immobilized peptide was washed with 5 column volumes (CV) of wash buffer (50 mM HEPES, 100 mM NaCl, pH 7.5, 50 mM imidazole), and eluted with 100% elution buffer (50 mM HEPES, 100 mM NaCl, pH 7.5, 500 mM imidazole). The elution fraction was concentrated using a 3 kDa MWCO Amicon spin filter and washed with 10–20 CV of deionized water to remove imidazole, and the peptide solution was lyophilized.
E. coli strains BW25113 ΔtldDE:kan (DE3) or BL21 (DE3) (New England Biolabs) were transformed with the plasmid systems for peptide expression, expressing N-terminal His6-tagged BhaA and variants (BhaA, BhaA-Ala, BhaA-Ala-Trp, BhaA-Ala-Trp*, BhaA-Ala-Trp*Gly). The cultures were grown with 50 μg/mL kanamycin for E. coli BL21 expressing BhaA and Bha-Ala and BhaA-Ala-Trp in pET28b or pRSFDuet-1, 25 μg/mL kanamycin and 12.5 μg/mL chloramphenicol for E. coli BW25113 ΔtldDE:kan (DE3) expressing BhaA-Ala-Trp, 16.6 μg/mL kanamycin, 8.3 μg/mL chloramphenicol, and 33.3 μg/mL carbenicillin for E. coli BW25113 ΔtldDE:kan (DE3) expressing BhaA-Ala-Trp* and BhaA-Ala-Trp*Gly in pACYCDuet-1, 12.5 μg/mL chloramphenicol for E. coli BL21(DE3) expressing BhaA-Ala-Trp, and 12.5 μg/mL chloramphenicol and 50 μg/mL carbenicillin for E. coli BL21(DE3) expressing BhaA-Ala-Trp* and BhaA-Ala-Trp*Gly in pACYCDuet-1 in TB media (for 1 L total: 20 g bacto tryptone, 24 g yeast extract, 2.3 g KH2PO4, 12.5 g K2HPO4, and 0.4% glycerol). Cultures were grown with shaking at 37 °C to an OD600 = 0.5–0.7, upon which 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce peptide and protein expression. Cells were harvested after 18 h and resuspended in lysis buffer, then lysed by sonication. Peptides were purified by IMAC as described above.
For expression of amm peptides, a similar procedure was followed except E. coli BL21 (DE3) cells (New England Biolabs) expressing N-terminal His6-tagged AmmA variants (AmmA* or AmmA*-Trp) from His6-AmmA-pACYCDuet-1 were grown with 33 μg/mL chloramphenicol in LB media (for 1 L total: 10 g bacto tryptone, 5 g yeast extract and 10 g sodium chloride). Modified peptides were obtained by co-expression using pET28a-ammC1 and pACYCDuet-1 plasmid containing ammA* in MCS1 and ammB3 in MCS2. E. coli BL21 (DE3) cells expressing N-terminal His6-tagged modified AmmA* variants were grown with 17 μg/mL chloramphenicol and 25 μg/mL kanamycin in LB media. Cultures were grown with shaking at 37 °C to an OD600 = 0.5–0.7, upon which 0.4 mM IPTG was added to induce peptide and protein expression. Cells were harvested after 18 h and resuspended in lysis buffer, then lysed by sonication. Peptides were purified by IMAC as described above.
Data availability
The raw data associated with the spectra in Figures 2 and 3, and Supplementary Figures 3–5, 7, 8, and 10–15, 17–21, and 23–31 were deposited at Mendeley (van der Donk, Wilfred (2021), “PEARL 2021”, Mendeley Data, V1, doi: 10.17632/mk3ttnbt5t.1).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R37 GM058822 to W.v.d.D., T32 GM070421 to P.N.D, F32 GM105297 to R.S., F32 GM129944 to C.P.T., and R01 GM085770 to B.S.M.). We thank Dr. Michael A. Funk and Kwo-Kwang Abraham Wang for initial attempts to activate the bha cluster and Deborah A. Berthold for help in purifying GlyQS.
Footnotes
Competing financial interests
The authors declare no competing interests.
References
- 1.Lin S et al. Another look at pyrroloiminoquinone alkaloids-perspectives on their therapeutic potential from known structures and semisynthetic analogues. Marine Drugs 15, 98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters S & Spiteller P Sanguinones A and B, blue pyrroloquinoline alkaloids from the fruiting bodies of the mushroom Mycena sanguinolenta. J. Nat. Prod 70, 1274–1277 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Legentil L, Benel L, Bertrand V, Lesur B & Delfourne E Synthesis and antitumor characterization of pyrazolic analogues of the marine pyrroloquinoline alkaloids: wakayin and tsitsikammamines. J. Med. Chem 49, 2979–2988 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Hu JF, Fan H, Xiong J & Wu SB Discorhabdins and pyrroloiminoquinone-related alkaloids. Chem. Rev 111, 5465–5491 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Chen QB, Xin XL, Yang Y, Lee SS & Aisa HA Highly conjugated norditerpenoid and pyrroloquinoline alkaloids with potent PTP1B iinhibitory activity from Nigella glandulifera. J. Nat. Prod 77, 807–812 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Hughes CC, MacMillan JB, Gaudencio SP, Jensen PR & Fenical W The ammosamides: structures of cell cycle modulators from a marine-derived Streptomyces species. Angew. Chem. Int. Ed 48, 725–727 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan PA & Moore BS Biosynthetic pathway connects cryptic ribosomally synthesized posttranslationally modified peptide genes with pyrroloquinoline alkaloids. Cell Chem. Biol 23, 1504–1514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reimer D & Hughes CC Thiol-based probe for electrophilic natural products reveals that most of the ammosamides are artifacts. J. Nat. Prod 80, 126–133 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Reddy PV, Banerjee B & Cushman M Efficient total synthesis of ammosamide B. Org. Lett 12, 3112–3114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes CC, MacMillan JB, Gaudencio SP, Fenical W & La Clair JJ Ammosamides A and B target myosin. Angew. Chem. Int. Ed. Engl 48, 728–732 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo J et al. Discovery of ammosesters by mining the Streptomyces uncialis DCA2648 genome revealing new insight into ammosamide biosynthesis. J. Ind. Microbiol. Biotechnol, 10.1093/jimb/kuab1027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyanaga A et al. Discovery and assembly-line biosynthesis of the lymphostin pyrroloquinoline alkaloid family of mTOR inhibitors in Salinispora bacteria. J. Am. Chem. Soc 133, 13311–13313 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colosimo DA & MacMillan JB Ammosamides unveil novel biosynthetic machinery. Cell Chem. Biol 23, 1444–1446 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Ting CP et al. Use of a scaffold peptide in the biosynthesis of amino acid-derived natural products. Science 365, 280–284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortega MA et al. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517, 509–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garg N, Salazar-Ocampo LM & van der Donk WA In vitro activity of the nisin dehydratase NisB. Proc. Natl. Acad. Sci. U. S. A 110, 7258–7263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Repka LM, Chekan JR, Nair SK & van der Donk WA Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem. Rev 117, 5457–5520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Repka LM, Hetrick KJ, Chee SH & van der Donk WA Characterization of leader peptide binding during catalysis by the nisin dehydratase NisB. J. Am. Chem. Soc 140, 4200–4203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bothwell IR et al. Characterization of glutamyl-tRNA-dependent dehydratases using nonreactive substrate mimics. Proc. Natl. Acad. Sci. U. S. A 116, 17245–17250 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z & van der Donk WA Nonribosomal peptide extension by a peptide amino-acyl tRNA ligase. J. Am. Chem. Soc 141, 19625–19633 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson GA, Zhang Z, Tietz JI, Mitchell DA & van der Donk WA In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin. J. Am. Chem. Soc 137, 16012–16015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bewley KD et al. Capture of micrococcin biosynthetic intermediates reveals C-terminal processing as an obligatory step for in vivo maturation. Proc. Natl. Acad. Sci. USA 113, 12450–12455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sikandar A, Franz L, Melse O, Antes I & Koehnke J Thiazoline-specific amidohydrolase PurAH Is the gatekeeper of bottromycin biosynthesis. J. Am. Chem. Soc 141, 9748–9752 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Severinov K & Nair SK Microcin C: biosynthesis and mechanisms of bacterial resistance. Future Microbiol 7, 281–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allali N, Afif H, Couturier M & Van Melderen L The highly conserved TldD and TldE proteins of Escherichia coli are involved in microcin B17 processing and in CcdA degradation. J. Bacteriol 184, 3224–3231 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghilarov D et al. The origins of specificity in the microcin-processing protease TldD/E. Structure 25, 1549–1561.e1545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burkhart BJ, Hudson GA, Dunbar KL & Mitchell DA A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol 11, 564–570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozaki T et al. Insights into the biosynthesis of dehydroalanines in goadsporin. ChemBioChem 17, 218–223 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q, Yu Y, Velásquez JE & van der Donk WA Evolution of lanthipeptide synthetases. Proc. Natl. Acad. Sci. U. S. A 109, 18361–18366 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M et al. The non-canonical tetratricopeptide repeat (TPR) domain of fluorescent (FLU) mediates complex formation with glutamyl-tRNA reductase. J. Biol. Chem 290, 17559–17565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Riba A & Itzhaki LS The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr. Opin. Struct. Biol 54, 43–49 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Pallen MJ, Francis MS & Futterer K Tetratricopeptide-like repeats in type-III-secretion chaperones and regulators. FEMS Microbiol. Lett 223, 53–60 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Das AK, Cohen PW & Barford D The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J 17, 1192–1199 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamb JR, Tugendreich S & Hieter P Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci 20, 257–259 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Fitzpatrick PF Structural insights into the regulation of aromatic amino acid hydroxylation. Curr. Opin. Struct. Biol 35, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fitzpatrick PF Mechanism of aromatic amino acid hydroxylation. Biochemistry 42, 14083–14091 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perdivara I, Deterding LJ, Przybylski M & Tomer KB Mass spectrometric identification of oxidative modifications of tryptophan residues in proteins: chemical artifact or post-translational modification? J. Am. Soc. Mass Spectrom 21, 1114–1117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basran J et al. The mechanism of formation of N-formylkynurenine by heme dioxygenases. J. Am. Chem. Soc 133, 16251–16257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirose Y et al. Involvement of common intermediate 3-hydroxy-L-kynurenine in chromophore biosynthesis of quinomycin family antibiotics. J. Antibiot 64, 117–122 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Todorovski T, Fedorova M, Hennig L & Hoffmann R Synthesis of peptides containing 5-hydroxytryptophan, oxindolylalanine, N-formylkynurenine and kynurenine. J. Pept. Sci 17, 256–262 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Fuson RC The principle of vinylogy. Chem. Rev 16, 1–27 (1935). [Google Scholar]
- 42.Jhulki I, Chanani PK, Abdelwahed SH & Begley TP A remarkable oxidative cascade that replaces the riboflavin C8 methyl with an amino group during roseoflavin biosynthesis. J. Am. Chem. Soc 138, 8324–8327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarz J, Konjik V, Jankowitsch F, Sandhoff R & Mack M Identification of the key enzyme of roseoflavin biosynthesis. Angew. Chem. Int. Ed 55, 6103–6106 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Konjik V et al. The crystal structure of RosB: insights into the reaction mechanism of the first member of a family of flavodoxin-like enzymes. Angew. Chem. Int. Ed 56, 1146–1151 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Kapoor I & Nair SK Structure-guided analyses of a key enzyme involved in the biosynthesis of an antivitamin. Biochemistry 57, 5282–5288 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Ortega MA et al. Structure and tRNA specificity of MibB, a lantibiotic dehydratase from Actinobacteria involved in NAI-107 biosynthesis. Cell Chem. Biol 23, 370–380 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherlin LD et al. Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. RNA 7, 1671–1678 (2001). [PMC free article] [PubMed] [Google Scholar]
- 48.Rio DC, Ares MJ, Hannon GJ & Nilsen TW RNA: a laboratory manual (Cold Spring Harbor Laboratory Press, 2011). [Google Scholar]
- 49.Walker SE & Fredrick K Preparation and evaluation of acylated tRNAs. Methods 44, 81–86 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data associated with the spectra in Figures 2 and 3, and Supplementary Figures 3–5, 7, 8, and 10–15, 17–21, and 23–31 were deposited at Mendeley (van der Donk, Wilfred (2021), “PEARL 2021”, Mendeley Data, V1, doi: 10.17632/mk3ttnbt5t.1).


