Abstract
Enlargement or aneurysm of the aorta predisposes to dissection, an important cause of sudden death. We trained a deep learning model to evaluate the dimensions of the ascending and descending thoracic aorta in 4.6 million cardiac magnetic resonance images from the UK Biobank. We then conducted genome-wide association studies in 39,688 individuals, identifying 82 loci associated with ascending and 47 with descending thoracic aortic diameter, of which 14 loci overlapped. Transcriptome-wide analyses, rare-variant burden tests, and human aortic single nucleus RNA sequencing prioritized genes including SVIL, which was strongly associated with descending aortic diameter. A polygenic score for ascending aortic diameter was associated with thoracic aortic aneurysm in 385,621 UK Biobank participants (HR = 1.43 per s.d.; CI 1.32-1.54; P = 3.3 × 10−20). Our results illustrate the potential for rapidly defining quantitative traits with deep learning, an approach that can be broadly applied to biomedical images.
Aortic aneurysm, a pathologic enlargement of the aorta, is common, having a prevalence of approximately ~1% in industrialized nations1. Over time, the enlarged aorta progressively expands; this process can lead to aortic dissection and rupture, which are the most catastrophic complications of aortic aneurysm and important causes of sudden cardiac death. Currently, the most effective preventive therapy is surgical or endovascular repair of the aorta, morbid procedures that are only performed when aneurysms are detected prior to aortic dissection. However, timely detection is uncommon because thoracic aortic aneurysm is typically asymptomatic until the time of dissection or rupture. Unlike abdominal aortic aneurysm, which has clinical screening guidelines, population screening for thoracic aortic aneurysm is not routinely performed2,3.
Consequently, the epidemiological and genetic contributions to aortic aneurysm have long been of interest to investigators. Clinical studies have suggested the close association of aneurysms of the descending thoracic aorta with atherosclerosis and lifestyle-associated risk factors, while those of the ascending aorta occur in younger patients, sometimes associated with pathogenic genetic predisposition4–6. Mutations in several genes have been associated with ascending aortic aneurysms, but the small number of implicated genes is mostly limited to highly penetrant Mendelian loci identified in family studies7–9. Thus, there is an urgent need to identify the genetic basis for variation in aortic size in order to enable the development of new therapeutic targets for medical intervention and to identify at-risk individuals with aortic aneurysms.
Results
We hypothesized that the size of the thoracic aorta is a complex trait, with contributions from common genetic variants. Because the ascending and descending thoracic aorta have not only separate biological origins10,11, but also distinct clinical risk factors underlying aneurysm formation12, we chose to quantify these aortic regions independently. All analyses were conducted in the UK Biobank unless otherwise stated.
Semantic segmentation of aorta with deep learning.
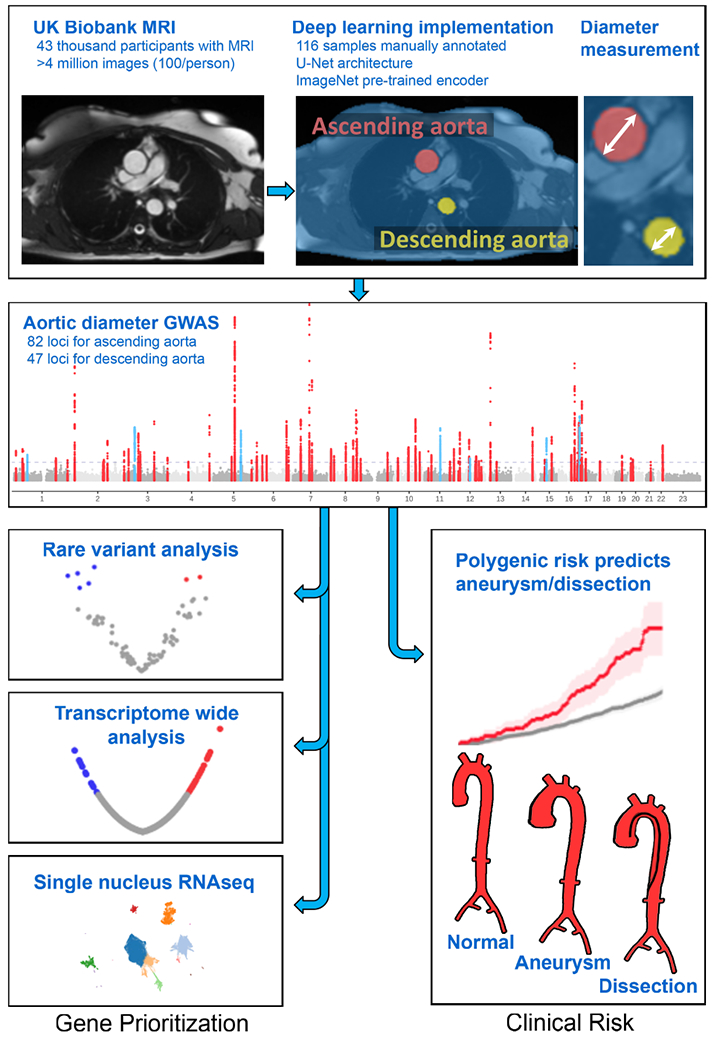
First, 116 cross-sectional cardiovascular magnetic resonance imaging (MRI) still-frame images at the level of the right pulmonary artery from the UK Biobank were manually annotated by a cardiologist (J.P.P.). This annotation is known as semantic segmentation—the task of identifying and labeling all pixels that comprise an object in an image.
We then used those annotations to train a deep learning model to perform the same semantic segmentation task. We chose a U-Net architecture13,14, because it has (i) an encoder that permits the model to recognize the image content (such as the presence of the aorta), and (ii) skip-connections from some of the earliest layers to some of the deepest layers, enabling the fine-grained localization of that semantic label within the input image. This allows the model to precisely identify the boundaries of the aorta, permitting accurate measurements. As a form of transfer learning, this model’s encoder had been pre-trained on ImageNet, which is a natural-image classification dataset. Therefore, instead of starting with random weights, the model was initialized with weights that are helpful for processing images, reducing the amount of manual annotation and model training necessary to achieve informative results13,15.
During training, 92 images were used for training and 24 as a validation set. The model achieved 96.5% pixel categorization accuracy for the ascending aorta and 94.1% for the descending aorta in the validation set. These were typical accuracies based on 10-fold cross validation (ascending aorta accuracy mean 95.2%, range 90.9%-97.2%; descending aorta accuracy mean 92.2%, range 88.9%-95.9%). We also evaluated inter-rater reliability between annotators, compared models trained by different annotators, and assessed the dependence of model performance on the number of training examples (see Supplementary Note, with a visualization of model output in Supplementary Fig. 1).
Having trained a deep learning model to recognize the pixels of ascending and descending aorta using manually annotated images in the UK Biobank, we then applied the model to all dedicated aortic MRI data available in the UK Biobank (Table 1). The model was applied to 4,374,900 images from 43,243 participants who participated in the first UK Biobank imaging visit (Fig. 1). The deep learning model produced pixel labels with the same dimensions as the input MRI image (generally 240px by 196px).
Table 1 |.
Baseline characteristics of UK Biobank GWAS participants
| Women | Men | |||
|---|---|---|---|---|
| Mean (or n) | s.d. (or %) | Mean (or n) | s.d. (or %) | |
| n | 20,909 | 19,842 | ||
| Age at time of MRI | 64.0 | 7.6 | 65.3 | 7.8 |
| BMI (kg/m2) | 25.9 | 4.6 | 27.0 | 3.9 |
| Height (cm) | 163 | 6.2 | 176 | 6.6 |
| Weight (kg) | 68.5 | 12.7 | 83.6 | 13.3 |
| Systolic blood pressure (mmHg) | 132 | 18 | 139 | 17 |
| Diastolic blood pressure (mmHg) | 79.4 | 9.7 | 83.6 | 9.6 |
| American standard drinks/week | 4.9 | 5.5 | 6.1 | 7.1 |
| Smoking status | ||||
| Current | 1,055 | 5% | 1,470 | 7% |
| Never | 13,413 | 64% | 11,216 | 57% |
| Prefer not to answer | 37 | 0% | 35 | 0% |
| Previous | 6,400 | 31% | 7,118 | 36% |
| Unknown | 4 | 0% | 3 | 0% |
| Pack years of smoking | 3.6 | 9.1 | 5.9 | 13.0 |
| Ascending aorta diameter (cm) | 3.04 | 0.31 | 3.32 | 0.34 |
| Descending aorta diameter (cm) | 2.29 | 0.18 | 2.55 | 0.21 |
Demographic information is shown for UK Biobank participants with genetic and cardiac MRI data that passed quality control as detailed in the sample flow diagram in Extended Data Figure 2. For count data, values shown are n (%). For continuous data, values shown are mean (s.d.).
Figure 1 |.
Study overview
Diameter measurement and quality control.
We applied classical computer vision algorithms to post-process the deep learning output in order to measure the aortic diameter16. We considered the elliptical minor axis at its maximum size throughout the cardiac cycle to be the aortic diameter. We computed the diameter of both the ascending and descending thoracic aorta and treated these as our primary phenotypes for subsequent analyses.
Quality control was then performed to exclude measurements from images in which the aorta was deemed to be incorrectly recognized according to one or more heuristics (see Online Methods). In total, 42,518 UK Biobank participants had at least one measurement that passed quality control (40,363 with ascending aortic diameter and 41,415 with descending aortic diameter). 39,260 participants’ measurements passed quality control for both ascending and descending aorta. We identified a subset of 2,976 individuals who had undergone imaging at two different times, and used those data to confirm that our modeling approach yielded reproducible measurements (detailed in the Supplementary Note).
Characteristics of the thoracic aortic diameter.
The median diameter of the ascending aorta in women was higher with advancing age (Extended Data Fig. 1), from 2.9 cm in women under the age of 55 to 3.1 cm in women over the age of 75. In men, the diameter ranged from 3.2 cm under the age of 55 to 3.4 cm over the age of 75. These values are similar to those reported previously using MRI to measure ascending aortic diameter in other cohorts17. For the descending aorta, the median diameter in women ranged from 2.2 cm in women under the age of 55 to 2.3 cm in women over the age of 75. In men, the diameter ranged from 2.4 cm under the age of 55 to 2.6 cm over the age of 75. A standard reference table of aortic diameters by age and sex was computed and is available in Supplementary Table 1. The ascending and descending aortic diameters were modestly positively correlated with one another (r2 = 0.18 after adjusting for sex, detailed in the Supplementary Note and shown in Supplementary Fig. 2). The ascending aortic diameter had greater variance than that of the descending aorta (Supplementary Note), consistent with prior observations18.
Correlation between aortic diameter and other traits.
We characterized the relationship between the aortic diameter and other anthropometric measurements in the UK Biobank (Supplementary Table 2 and Supplementary Fig. 3a, left). The diameter of ascending aorta was strongly positively correlated with traits such as weight, height, and blood pressure, as well as traits that correspond with larger body size such as greater forced expiratory volume in one second, hand grip strength, and food and alcohol consumption, consistent with previous reports19. The diameter of ascending aorta was strongly inversely correlated with heart rate and biomarkers including cholesterol, testosterone, and sex hormone binding globulin. We observed similar associations for the descending aortic diameter (Supplementary Fig. 3a, right).
We also analyzed the association between aortic size and PheCode-based disease labels20. The size of the ascending aorta was associated with cardiovascular diseases such as hypertension, aortic aneurysm, valvular disorders, and cardiac arrhythmias, as well as other traits including varicose veins, obesity, and osteoarthritis, several of which correspond to previous clinical observations21. Descending thoracic aortic size was associated with obesity, hypertension, and varicose veins. Notably, coronary artery disease was inversely associated with descending aortic diameter (P = 1.7 × 10−6), but not associated with ascending diameter (P = 0.6). In addition, the descending aortic size was directly associated with cholelithiasis and headache, and inversely associated with type 1 diabetes, as has previously been observed22,23 (Supplementary Table 3 and Supplementary Fig. 3b, left). While the ascending and descending aortic diameters shared similar correlations with most continuous traits, their relationships with PheCode-based disease phenotypes were more independent (Supplementary Fig. 3b, right).
GWAS of thoracic aortic diameter.
We next sought to understand the common genetic basis for variation in the size of the ascending and descending thoracic aorta in the UK Biobank. We excluded participants from genetic analysis if they had an aortic diameter greater than 5 cm, a known history of aortic disease, or genetic data that did not pass sample-level quality control (Extended Data Fig. 2). 38,694 participants had data that passed quality control and contributed to genetic analyses of the ascending aortic diameter, and 39,688 participants contributed to analyses of the descending aortic diameter (Table 1; participant characteristics stratified by smoking status are displayed in Supplementary Table 4).
We confirmed that both traits were highly heritable: the single nucleotide polymorphism (SNP) heritability of the size of the ascending aorta was 63% (95% CI 60%-67%), while that of the descending aorta was 50% (95% CI 47%-53%).
We then conducted genome-wide association studies (GWAS) of these two traits, testing 16.7 million genotyped and imputed SNPs with MAF > 0.001. We identified 82 independent loci associated with the diameter of the ascending aorta at a commonly used genome-wide significance threshold (P < 5 × 10−8) (Table 2 and Fig. 2a,b). Of these, 75 loci were novel. In the descending aorta, we identified 47 genome-wide significant loci, of which 43 were novel and one was located on the X chromosome (Table 3). In total, we identified 115 loci, of which 14 were associated at genome-wide significance with both traits (Fig. 2c). Test statistic inflation was observed in QQ plots (Extended Data Fig. 3) and the low linkage disequilibrium (LD) score regression (ldsc) intercepts indicated that this inflation was consistent with polygenicity rather than confounding (Supplementary Table 5)24. As a sensitivity analysis, the GWAS was also repeated in a European-only subset of the UK Biobank (Supplementary Note and Supplementary Tables 6 and 7).
Table 2 |.
GWAS loci for the ascending thoracic aorta
| SNP | CHR | BP | Effect allele | Other allele | EAF | INFO | BETA | P | Nearest gene | Prior |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2871651 | 1 | 9434969 | C | T | 0.58 | 0.99 | −0.042 | 5.80E-12 | SPSB1 | |
| rs67631072 | 1 | 38461821 | C | T | 0.45 | 0.99 | 0.041 | 1.40E-12 | SF3A3 | |
| rs3768274 | 1 | 41951383 | C | T | 0.50 | 0.98 | −0.040 | 6.70E-12 | EDN2 | |
| rs11207420 | 1 | 59646524 | G | A | 0.73 | 1.00 | −0.042 | 1.30E-10 | FGGY | Vasan et al. 2009 |
| rs72727759 | 1 | 185663021 | T | C | 0.74 | 0.99 | −0.041 | 2.10E-09 | HMCN1 | |
| rs35534155 | 1 | 237207943 | A | ATT | 0.19 | 0.90 | −0.047 | 9.70E-09 | RYR2 | |
| rs6707048 | 2 | 19720468 | T | C | 0.32 | 1.00 | 0.084 | 3.50E-41 | OSR1 | |
| rs138963986 | 2 | 145752940 | G | A | 0.93 | 0.96 | 0.068 | 4.00E-08 | ZEB2 | |
| rs12992231 | 2 | 148799710 | C | A | 0.64 | 1.00 | −0.039 | 8.10E-09 | MBD5 | |
| rs16849225 | 2 | 164906820 | C | T | 0.77 | 1.00 | −0.053 | 1.90E-15 | FIGN | |
| rs12052878 | 2 | 238227594 | G | A | 0.69 | 1.00 | −0.045 | 1.10E-11 | COL6A3 | |
| rs11712199 | 3 | 14858226 | G | A | 0.91 | 0.99 | 0.069 | 1.40E-12 | FGD5 | |
| rs9847006 | 3 | 41755359 | T | C | 0.83 | 1.00 | −0.075 | 3.80E-20 | ULK4 | Guo et al. 2016 |
| rs545996255 | 3 | 58100423 | G | GT | 0.70 | 0.97 | 0.057 | 5.10E-18 | FLNB | |
| rs2306272 | 3 | 66434643 | T | C | 0.71 | 1.00 | −0.043 | 1.50E-11 | LRIG1 | |
| rs55914222 | 3 | 128202943 | G | C | 0.97 | 0.99 | 0.179 | 3.90E-22 | GATA2 | |
| rs1108450 | 3 | 186995297 | T | G | 0.83 | 0.99 | −0.050 | 1.50E-09 | MASP1 | |
| rs16998073 | 4 | 81184341 | A | T | 0.71 | 1.00 | −0.036 | 3.50E-08 | FGF5 | |
| rs67846163 | 4 | 174656889 | A | G | 0.77 | 0.99 | −0.072 | 2.30E-24 | HAND2 | |
| rs73766539 | 5 | 81722919 | C | T | 0.79 | 1.00 | 0.048 | 6.80E-10 | ATP6AP1L | |
| rs72787618 | 5 | 95591331 | A | G | 0.63 | 0.99 | 0.099 | 3.20E-58 | PCSK1 | |
| rs17470137 | 5 | 122531347 | G | A | 0.73 | 1.00 | −0.058 | 5.80E-19 | PRDM6 | Vasan et al. 2009 |
| rs76888257 | 5 | 169809901 | C | T | 0.90 | 1.00 | 0.062 | 1.30E-08 | KCNMB1 | |
| rs496236 | 6 | 11641601 | A | G | 0.46 | 1.00 | 0.034 | 7.20E-10 | ADTRP | |
| rs1630736 | 6 | 12295987 | C | T | 0.54 | 0.99 | −0.046 | 8.30E-15 | EDN1 | |
| rs12199346 | 6 | 36641546 | C | A | 0.76 | 1.00 | −0.046 | 2.00E-10 | CDKN1A | |
| rs6459130 | 6 | 56055564 | G | T | 0.44 | 1.00 | −0.033 | 3.30E-10 | COL21A1 | |
| rs1570350 | 6 | 143592386 | A | G | 0.56 | 0.99 | −0.059 | 2.90E-22 | AIG1 | |
| rs13203975 | 6 | 152333104 | G | A | 0.89 | 0.99 | 0.070 | 3.30E-13 | ESR1 | |
| rs79215950 | 7 | 35277067 | G | A | 0.62 | 1.00 | 0.065 | 7.80E-23 | TBX20 | |
| rs6974735 | 7 | 73428222 | A | G | 0.55 | 1.00 | −0.111 | 7.90E-77 | ELN | |
| rs1583081 | 7 | 85034227 | G | T | 0.58 | 1.00 | −0.075 | 2.40E-36 | SEMA3D | |
| rs483916 | 8 | 9793601 | A | C | 0.48 | 0.99 | 0.044 | 1.30E-12 | MSRA | |
| rs11785562 | 8 | 23391493 | G | A | 0.80 | 0.97 | −0.043 | 3.70E-10 | SLC25A37 | |
| rs9721183 | 8 | 75781818 | C | T | 0.63 | 0.95 | 0.048 | 1.40E-14 | PI15 | |
| rs16876090 | 8 | 108363596 | G | A | 0.91 | 0.99 | −0.080 | 1.40E-15 | ANGPT1 | |
| rs562291939 | 8 | 120709336 | T | C | 1.00 | 0.80 | 0.744 | 5.10E-26 | ENPP2 | |
| rs10111085 | 8 | 122646152 | G | T | 0.71 | 0.99 | 0.048 | 2.00E-12 | HAS2 | |
| rs34557926 | 8 | 124607159 | C | T | 0.63 | 0.99 | −0.060 | 2.90E-22 | FBXO32 | |
| rs112342612 | 8 | 141047976 | AAC | A | 0.40 | 0.95 | −0.035 | 3.30E-09 | TRAPPC9 | |
| rs4978966 | 9 | 113662374 | C | T | 0.79 | 1.00 | 0.049 | 2.50E-11 | LPAR1 | |
| rs1757223 | 10 | 18514999 | G | A | 0.24 | 0.99 | 0.042 | 2.00E-09 | CACNB2 | |
| rs16916931 | 10 | 63813744 | A | T | 0.69 | 0.98 | 0.045 | 1.20E-12 | ARID5B | |
| rs7090111 | 10 | 65077994 | C | G | 0.58 | 1.00 | 0.044 | 3.10E-13 | JMJD1C | |
| rs71482305 | 10 | 96119130 | C | T | 0.84 | 1.00 | 0.079 | 6.30E-23 | NOC3L | |
| rs1340837 | 10 | 97542035 | A | G | 0.59 | 1.00 | 0.031 | 4.90E-09 | ENTPD1 | |
| rs11196083 | 10 | 114500004 | G | T | 0.77 | 1.00 | −0.049 | 1.60E-11 | VTI1A | |
| rs77889556 | 11 | 17498057 | G | A | 0.83 | 0.91 | −0.056 | 8.40E-12 | ABCC8 | |
| rs3741025 | 11 | 30851976 | C | T | 0.43 | 0.99 | 0.041 | 1.70E-10 | DCDC1 | |
| rs111412755 | 11 | 69819139 | C | T | 0.91 | 0.98 | −0.093 | 7.80E-20 | ANO1 | Wild et al. 2017 |
| rs12286728 | 11 | 113022450 | G | C | 0.90 | 1.00 | 0.056 | 3.10E-08 | NCAM1 | |
| rs747249 | 11 | 130271647 | A | G | 0.36 | 0.99 | −0.044 | 1.30E-12 | ADAMTS8 | |
| rs61907983 | 12 | 15448631 | C | T | 0.91 | 0.97 | 0.062 | 2.60E-08 | RERG | |
| rs2307024 | 12 | 22005003 | T | G | 0.59 | 0.99 | 0.054 | 2.30E-18 | ABCC9 | |
| rs56298756 | 12 | 62777565 | G | T | 0.89 | 1.00 | −0.082 | 8.40E-16 | USP15 | |
| rs10400419 | 12 | 66389968 | T | C | 0.45 | 0.95 | 0.036 | 2.50E-09 | LLPH | Vasan et al. 2009, Wild et al. 2017 |
| rs7302816 | 12 | 89950320 | A | C | 0.80 | 0.98 | −0.043 | 2.50E-08 | GALNT4 | |
| rs2363080 | 12 | 94140463 | C | G | 0.56 | 0.99 | 0.037 | 4.30E-10 | CRADD | |
| rs11112482 | 12 | 105738183 | C | G | 0.77 | 0.99 | −0.039 | 2.10E-08 | C12orf75 | |
| rs61937394 | 12 | 116756670 | T | G | 0.81 | 0.91 | 0.042 | 1.60E-08 | MED13L | |
| rs7994761 | 13 | 22871446 | A | G | 0.78 | 0.99 | 0.109 | 1.30E-52 | FGF9 | |
| rs2687941 | 13 | 50760363 | T | C | 0.55 | 0.99 | −0.032 | 3.70E-08 | DLEU1 | |
| rs4905134 | 14 | 94459845 | A | G | 0.50 | 0.99 | 0.055 | 5.40E-20 | ASB2 | |
| rs3803359 | 15 | 40662748 | G | A | 0.83 | 1.00 | −0.044 | 7.50E-09 | DISP2 | |
| rs2118181 | 15 | 48915884 | T | C | 0.90 | 0.99 | −0.082 | 2.30E-16 | FBN1 | LeMaire et al. 2011, Guo et al. 2016, van ‘t Hof et al. 2016 |
| rs1441358 | 15 | 71612514 | T | G | 0.66 | 1.00 | 0.053 | 8.10E-17 | THSD4 | |
| rs369339295 | 16 | 56322945 | A | AAG | 0.68 | 0.97 | 0.042 | 1.50E-10 | GNAO1 | |
| rs62053262 | 16 | 69969299 | C | G | 0.95 | 0.99 | 0.187 | 4.00E-42 | WWP2 | |
| rs546590249 | 16 | 71104575 | A | C | 0.99 | 0.38 | 0.275 | 2.70E-08 | HYDIN | |
| rs7500448 | 16 | 83045790 | A | G | 0.75 | 0.98 | −0.045 | 2.90E-11 | CDH13 | |
| rs16965180 | 16 | 88989862 | A | G | 0.65 | 0.99 | 0.063 | 1.20E-21 | CBFA2T3 | |
| 17 | 2088848 | CCAGA | C | 0.68 | 1.00 | −0.063 | 6.80E-24 | SMG6 | Vasan et al. 2009, Wild et al. 2017 | |
| rs78180894 | 17 | 7483662 | G | C | 0.93 | 0.94 | −0.072 | 6.60E-09 | CD68 | |
| rs7215383 | 17 | 12182246 | A | G | 0.25 | 0.99 | 0.078 | 4.90E-29 | MAP2K4 | |
| rs6505216 | 17 | 29206421 | G | T | 0.77 | 0.92 | 0.053 | 2.00E-11 | ATAD5 | |
| rs76954792 | 17 | 30033514 | C | T | 0.77 | 0.98 | 0.044 | 3.90E-09 | COPRS | |
| rs264203 | 18 | 10882121 | A | C | 0.38 | 0.99 | −0.035 | 2.50E-08 | PIEZO2 | |
| rs7257694 | 19 | 30314666 | C | T | 0.60 | 0.99 | −0.039 | 3.00E-10 | CCNE1 | |
| rs3063286 | 20 | 10488552 | T | TTA | 0.47 | 0.94 | 0.034 | 2.20E-09 | SLX4IP | |
| rs6075516 | 20 | 19455985 | G | A | 0.75 | 0.97 | 0.040 | 6.30E-09 | SLC24A3 | |
| rs28451064 | 21 | 35593827 | G | A | 0.87 | 0.96 | 0.051 | 4.20E-08 | KCNE2 | |
| rs4402860 | 22 | 40554445 | A | T | 0.80 | 1.00 | 0.057 | 7.30E-14 | TNRC6B |
The lead SNPs from the GWAS for the diameter of the ascending thoracic aorta. SNP, the rsID of the variant, where available; for variants that are not in dbSNP, this column is left blank. BP, genomic position, keyed to GRCh37. EAF, effect allele frequency. INFO, imputation INFO score. BETA, effect size per effect allele on the inverse normal transformed trait. P, the BOLT-LMM association P value. Prior, known from prior publications addressing common genetic variation linked to aortic size, aortic aneurysm, or aortic dissection25–29.
Figure 2 |. Genome-wide association study results for ascending and descending thoracic aorta diameter.
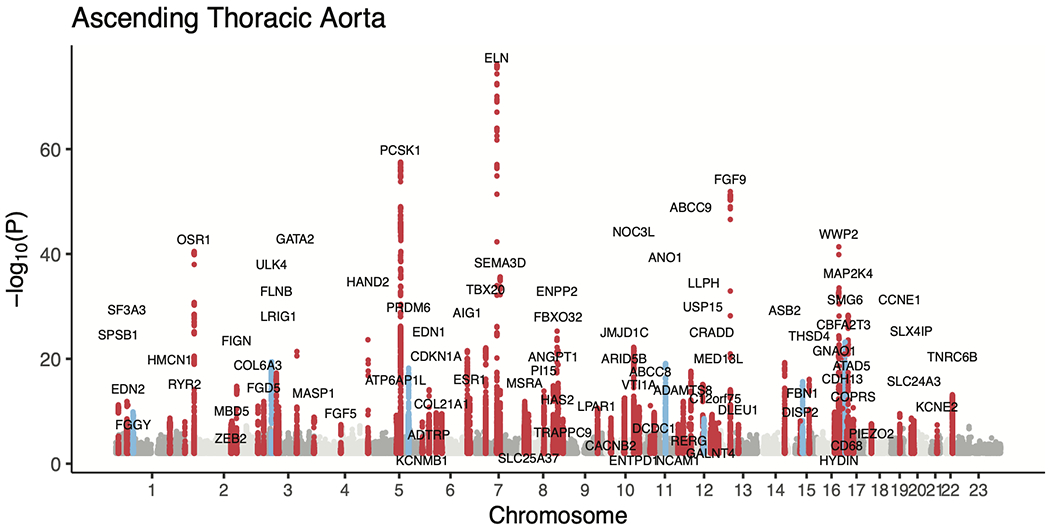
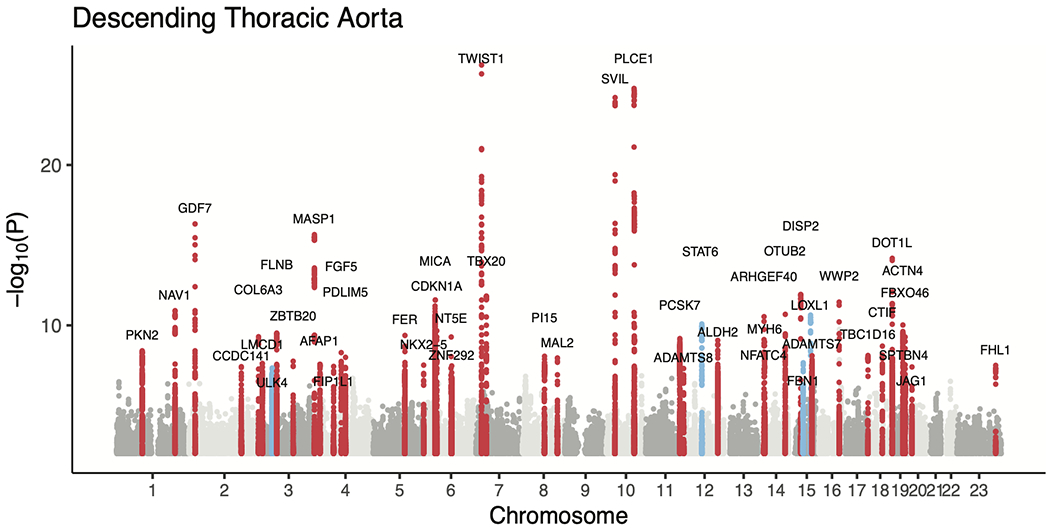
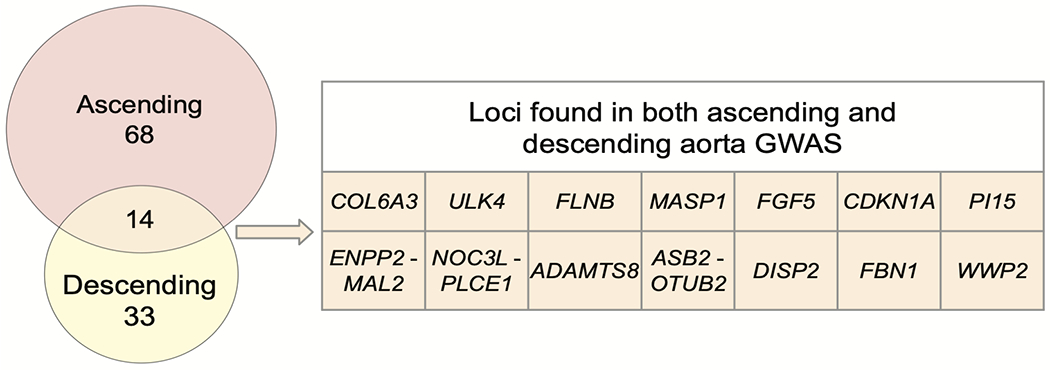
a,b, Loci with P < 5 × 10−8 are shown in red (if not previously reported) or blue (if previously reported in common variant association studies for aortic size or disease status (aneurysm or dissection)). The X chromosome is represented as ‘23’. c, Venn diagram showing the number of loci uniquely associated at P < 5 × 10−8 with either the ascending or descending thoracic aorta. Those in orange are associated with both and are enumerated in the table to the right. Loci whose lead SNP’s nearest gene differs between ascending and descending are demarcated as “Ascending/Descending”.
Table 3 |.
GWAS loci for the descending thoracic aorta
| SNP | CHR | BP | Effect allele | Other allele | EAF | INFO | BETA | P | Nearest gene | Prior |
|---|---|---|---|---|---|---|---|---|---|---|
| rs35584696 | 1 | 89145392 | C | CT | 0.44 | 1.00 | −0.033 | 3.80E-09 | PKN2 | |
| rs527725 | 1 | 201752429 | A | C | 0.60 | 0.97 | 0.036 | 1.20E-11 | NAV1 | |
| rs7255 | 2 | 20878820 | T | C | 0.45 | 1.00 | 0.045 | 4.80E-17 | GDF7 | |
| rs202119031 | 2 | 179744659 | CAG | C | 0.87 | 1.00 | 0.045 | 3.80E-08 | CCDC141 | |
| rs7580831 | 2 | 238219499 | C | A | 0.68 | 1.00 | −0.037 | 5.20E-10 | COL6A3 | |
| rs11707002 | 3 | 8580237 | C | G | 0.55 | 0.99 | 0.031 | 2.40E-08 | LMCD1 | |
| rs5848609 | 3 | 41802815 | G | GTTA | 0.84 | 0.99 | −0.041 | 4.50E-08 | ULK4 | Guo et al. 2016 |
| rs56004178 | 3 | 58101471 | G | A | 0.70 | 0.99 | 0.038 | 3.10E-10 | FLNB | |
| rs2055981 | 3 | 114203969 | T | C | 0.36 | 0.99 | −0.032 | 1.70E-08 | ZBTB20 | |
| rs698099 | 3 | 186987941 | G | A | 0.17 | 1.00 | 0.060 | 2.20E-16 | MASP1 | |
| rs6855532 | 4 | 7908237 | C | T | 0.57 | 1.00 | 0.030 | 2.70E-08 | AFAP1 | |
| rs60991988 | 4 | 54801228 | T | G | 0.89 | 0.99 | −0.047 | 3.70E-08 | FIP1L1 | |
| rs3733336 | 4 | 81207963 | A | G | 0.64 | 0.90 | −0.034 | 5.10E-09 | FGF5 | |
| rs6853490 | 4 | 95544718 | A | G | 0.58 | 0.98 | 0.031 | 1.00E-08 | PDLIM5 | |
| rs9285863 | 5 | 108071655 | T | C | 0.66 | 0.99 | −0.036 | 4.20E-10 | FER | |
| rs35564079 | 5 | 172670611 | C | CT | 0.71 | 0.97 | −0.035 | 3.00E-08 | NKX2-5 | |
| rs2853975 | 6 | 31382717 | A | T | 0.71 | 0.99 | −0.042 | 2.60E-12 | MICA | |
| rs733590 | 6 | 36645203 | T | C | 0.65 | 1.00 | −0.035 | 2.20E-10 | CDKN1A | |
| rs4707174 | 6 | 85987918 | A | C | 0.70 | 0.98 | −0.036 | 5.30E-10 | NT5E | |
| 6 | 87836772 | ACACACACACC | A | 0.65 | 0.77 | 0.035 | 3.40E-08 | ZNF292 | ||
| rs2107595 | 7 | 19049388 | G | A | 0.84 | 0.99 | 0.079 | 5.80E-27 | TWIST1 | |
| rs343044 | 7 | 35508859 | A | G | 0.20 | 0.99 | −0.047 | 1.50E-12 | TBX20 | |
| rs36086322 | 8 | 75735030 | C | T | 0.93 | 1.00 | 0.059 | 8.40E-09 | PI15 | |
| rs574214679 | 8 | 120244723 | A | G | 1.00 | 0.71 | 0.413 | 1.10E-08 | MAL2 | |
| rs10740811 | 10 | 30167754 | G | A | 0.41 | 1.00 | 0.057 | 6.40E-25 | SVIL | |
| rs2901761 | 10 | 95895127 | G | A | 0.59 | 1.00 | 0.058 | 1.70E-25 | PLCE1 | |
| 11 | 117085914 | CTTA | C | 0.94 | 1.00 | −0.068 | 6.60E-10 | PCSK7 | ||
| rs10894192 | 11 | 130266117 | T | A | 0.42 | 0.98 | −0.030 | 4.90E-08 | ADAMTS8 | |
| rs4759275 | 12 | 57525756 | G | A | 0.58 | 1.00 | 0.035 | 8.10E-11 | STAT6 | Guo et al. 2016 |
| rs10744777 | 12 | 112233018 | T | C | 0.66 | 1.00 | −0.035 | 8.80E-10 | ALDH2 | |
| rs12889267 | 14 | 21542766 | A | G | 0.83 | 1.00 | 0.048 | 2.90E-11 | ARHGEF40 | |
| rs422068 | 14 | 23864804 | T | C | 0.64 | 1.00 | 0.036 | 1.10E-09 | MYH6 | |
| rs12590407 | 14 | 24835115 | G | A | 0.29 | 1.00 | 0.034 | 1.40E-08 | NFATC4 | |
| rs12890024 | 14 | 94469801 | A | G | 0.62 | 0.98 | 0.038 | 2.10E-11 | OTUB2 | |
| rs12913300 | 15 | 40655444 | C | T | 0.83 | 1.00 | −0.052 | 1.20E-12 | DISP2 | |
| rs17352842 | 15 | 48694211 | C | T | 0.81 | 1.00 | −0.037 | 2.20E-08 | FBN1 | LeMaire et al. 2011, Guo et al. 2016, van ‘t Hof et al. 2016 |
| rs1048661 | 15 | 74219546 | G | T | 0.66 | 0.99 | −0.038 | 2.30E-11 | LOXL1 | Vasan et al. 2009 |
| rs116901435 | 15 | 79059695 | C | T | 0.58 | 0.98 | −0.032 | 7.90E-09 | ADAMTS7 | |
| rs62053262 | 16 | 69969299 | C | G | 0.95 | 0.99 | 0.087 | 3.50E-12 | WWP2 | |
| rs894871 | 17 | 77910932 | A | G | 0.68 | 0.98 | −0.032 | 7.50E-09 | TBC1D16 | |
| rs8094206 | 18 | 46317137 | G | A | 0.89 | 0.98 | 0.052 | 2.00E-09 | CTIF | |
| rs55678414 | 19 | 2177625 | T | G | 0.94 | 1.00 | 0.088 | 6.70E-15 | DOT1L | |
| rs2303040 | 19 | 39138608 | T | C | 0.51 | 0.99 | −0.037 | 9.50E-11 | ACTN4 | |
| rs1673096 | 19 | 41042755 | A | G | 0.52 | 0.99 | 0.031 | 3.20E-08 | SPTBN4 | |
| rs11668847 | 19 | 46210365 | T | G | 0.48 | 0.98 | 0.033 | 5.30E-10 | FBXO46 | |
| rs76496822 | 20 | 10687240 | G | T | 0.96 | 0.99 | −0.072 | 4.00E-08 | JAG1 | |
| rs76530933 | 23 | 135204774 | G | T | 0.73 | 0.94 | −0.030 | 3.10E-08 | FHL1 |
The lead SNPs from the GWAS for the diameter of the descending thoracic aorta. SNP, the rsID of the variant, where available; for variants that are not in dbSNP, this column is left blank. BP, genomic position, keyed to GRCh37. EAF, effect allele frequency. INFO, imputation INFO score. BETA, effect size per effect allele on the inverse normal transformed trait. P, the BOLT-LMM association P value. Prior, known from prior publications addressing common genetic variation linked to aortic size, aortic aneurysm, or aortic dissection25–29.
Previous analyses of thoracic aortic phenotypes including aortic root diameter, ascending aortic dissection, or thoracic aortic aneurysm have identified only 16 genome-wide significant loci; of these, nine achieved genome-wide significance in our study, including all three loci that have been associated with thoracic aortic dissection (near FBN1, ULK4, and the STAT6-LRP1 locus; Supplementary Table 8)25–29.
We sought to replicate our UK Biobank GWAS findings in 3,287 participants from the Framingham Heart Study (FHS) who had genotyping data and cross-sectional imaging of the ascending and descending thoracic aorta by computed tomography30,31. Since the FHS sample size was an order of magnitude smaller than our discovery population in the UK Biobank, we focused on directional agreement. Of the 82 lead SNPs in the ascending aorta, 72 were identified in the FHS dataset. 60 of these 72 SNPs were directionally consistent in both datasets (two-tailed binomial P = 8.1 × 10−9; Extended Data Fig. 4a). 41 of the 46 autosomal lead SNPs from the descending aorta were identified in FHS, and 36/41 were directionally consistent (two-tailed binomial P = 7.8 × 10−7; Extended Data Fig. 4b and Supplementary Table 9). Thus, despite comprising a significantly smaller sample, as well as using a different imaging modality and measurement technique, the FHS results were aligned with our findings from the UK Biobank.
Genetic correlation with other phenotypes.
We used genetic correlation to gain insight into the relationship between aortic diameter and other cardiovascular and anthropometric phenotypes in the UK Biobank. The ascending and descending aortic phenotypes had a genetic correlation with one another of 0.48 (95% CI 0.45-0.52) as estimated by BOLT-REML32,33. We used LD score regression to assess genetic correlation between the aortic traits and up to 281 additional quantitative phenotypes from the UK Biobank that were precomputed by the Neale Lab (https://ukbb-rg.hail.is/)34. As expected, we observed positive genetic correlations between aortic size and anthropometric measures such as height and weight, as well as related phenotypes such as blood pressure (Supplementary Table 10, Extended Data Fig. 5 and Supplementary Fig. 4).
Given the observed genetic correlation with blood pressure (ldsc rg 0.30 for ascending aortic diameter and 0.17 for descending aortic diameter), we also surveyed the overlap between the aortic loci and genome-wide significant blood pressure loci. Ten of the 82 lead SNPs for ascending aortic diameter were within 500 kb of a lead SNP from a recent GWAS for blood pressure, as were six of the 47 descending aortic lead SNPs (Supplementary Table 11)35. Of the nine adrenoceptor genes, which encode the molecular targets of alpha- and beta-blocking medicines, none were within 500 kb of a lead SNP in our study.
Transcriptome-wide association study.
To gain more insight into the GWAS loci themselves, we took three approaches to prioritize genes at each locus and to link those genes to relevant cell types. First, we conducted a transcriptome-wide association study (TWAS), linking predicted gene expression in aorta (based on GTEx v7) with aortic size (Fig. 3a and Supplementary Tables 12 and 13)36,37. We identified 53 transcripts that were significantly associated with the diameter of the ascending aorta and 15 with the descending aorta at P < 5 × 10−8.
Figure 3 |. Gene-level association tests.
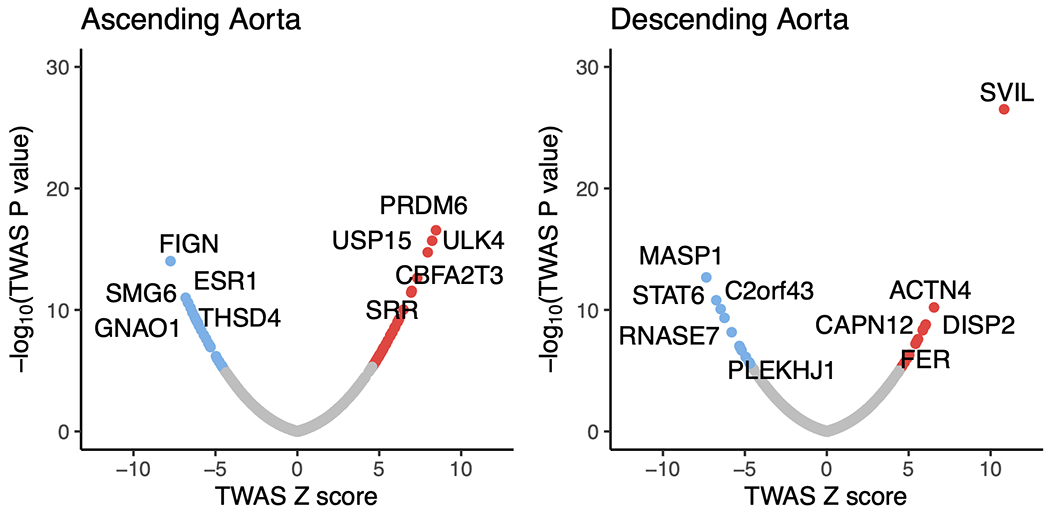
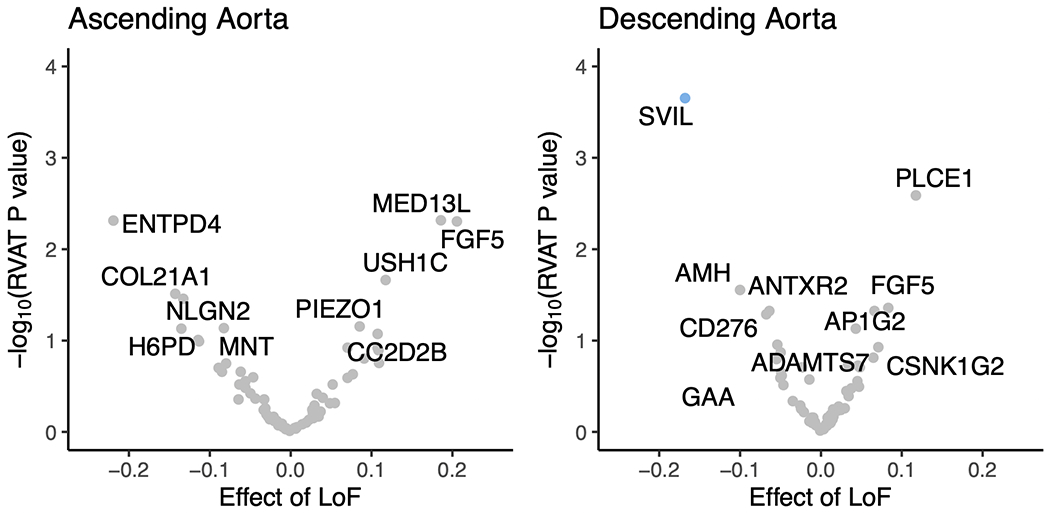
In the top row, protein-coding genes associated with the size of the ascending (left) and descending (right) thoracic aorta based on an integrated gene expression prediction are shown. The x-axis represents the magnitude of the TWAS Z score, while the y-axis represents the −log10 of the TWAS P value. Genes achieving Bonferroni significance are colored red (positive correlation) or blue (negative correlation). The top five positively and negatively correlated genes are labeled. In the bottom row, rare variant collapsing burden test results are depicted for the genes within a 500-kb window around GWAS loci (67 for ascending and 55 for descending). Loss-of-function carrier status in each gene was tested for association with the size of the ascending (left) and descending (right) thoracic aorta. The x-axis represents the effect size of LoF in each gene on aortic size, while the y-axis represents the −log10 of the association P value in a logistic model. SVIL, which achieved P < 0.05/55 in the descending aorta, is colored blue. The top five positively and negatively correlated genes are labeled.
Among the strongest TWAS associations in the ascending aorta were ULK4, a gene previously linked with aortic dissection, and THSD4, whose protein product binds to fibrillin (FBN1) and modulates microfibril assembly38. Also notable was USP15, whose protein product is a deubiquitinating enzyme that acts on the TGF-β receptor and enhances TGF-β signaling39,40; the TWAS results suggest that higher USP15 expression is linked with a greater ascending aortic diameter. In the descending aorta, the strongest TWAS association was with the gene SVIL, of which increased transcription was associated with greater aortic diameter (Fig. 3a).
Rare variant association test.
Second, we conducted a rare variant association test in 12,336 UK Biobank participants with both aortic imaging and exome sequencing data. No gene achieved Bonferroni significance in an exome-wide analysis. Restricting the analysis to genes within a 500-kb window around GWAS loci (67 genes for ascending aorta and 55 genes for descending aorta; Supplementary Table 14), we found that loss of function variants in SVIL were most strongly associated with a smaller mean descending aortic diameter (14 carriers; loss-of-function effect size −0.17 cm, 95% CI −0.08 to −0.26 cm, P = 2.2 × 10−4; Fig. 3b).
Single nucleus RNA sequencing.
Third, we undertook direct analysis of tissue and cell-specific expression patterns to localize and identify relevant cell types. We used tissue-specific LD score regression to test for enrichment of the aortic diameter GWAS results in 53 GTEx v6 tissue types37,41. For the ascending aortic loci, enrichment was significant in aortic and coronary artery tissues (P = 8.8 × 10−5 and P = 1.1 × 10−4, respectively). Enrichment of aortic and coronary artery tissues was also observed for the descending aortic loci (P = 3.1 × 10−4 and P = 1.8 × 10−3; Supplementary Tables 15 and 16). These data are consistent with the expectation that the aorta itself is the most relevant tissue linked with our findings.
Therefore, we incorporated an analysis of single-nucleus RNA sequencing (snRNA-seq) using paired samples from the ascending and descending aorta from three individuals to identify potentially relevant cell types for the genes at aortic GWAS loci (Supplementary Note). We sequenced the transcriptomes of 54,092 nuclei and identified 12 primary cell clusters (Fig. 4a). Through comparison of unique transcriptional profiles in each cluster to canonical cell markers, we identified populations comprising vascular smooth muscle cells, fibroblasts, three distinct types of endothelial cells, as well as macrophages and lymphocytes (Fig. 4b). We then examined the cell type-specific expression of the genes prioritized by the TWAS (Fig. 4c,d).
Figure 4 |. Single nucleus RNA sequencing analyses in human aorta.
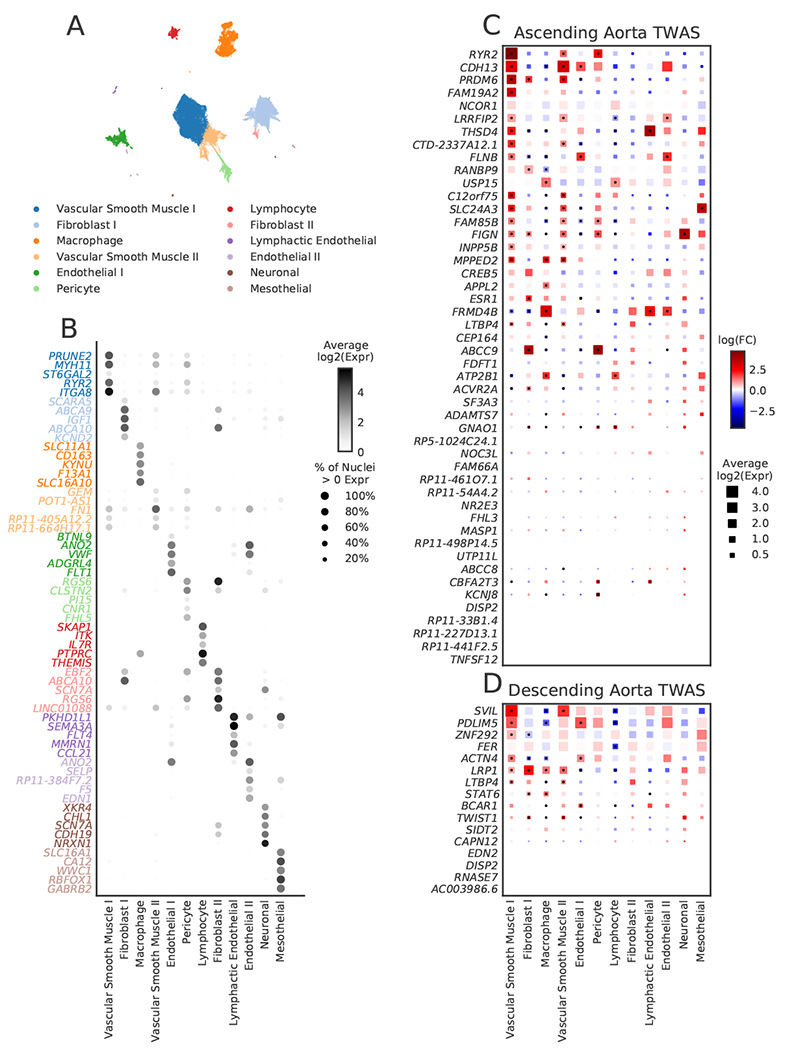
Single nucleus RNA-seq was performed on paired ascending and descending thoracic aortic tissue from three humans. a, Uniform manifold approximation and projection (UMAP) revealed 12 main clusters. Each dot represents an individual nucleus, colored and labeled by putative cell type as identified from Leiden clustering. b, The top five most selectively expressed genes for each cluster were identified as those with the largest fold-change difference in expression comparing the given cluster with all other clusters, only considering genes expressed in at least 30% of nuclei and with a Benjamini-Hochberg corrected P < 0.01. The shade of the dot represents the average log2 expression for a gene across all nuclei in a given cluster and the size of the dot represents the percentage of nuclei in the cluster with non-zero expression. The cell type labels were created by comparing selectively expressed genes in each cluster of nuclei with the literature. c,d, Cell-type specificity of genes with expression data supported by the TWAS in the ascending (c) and descending (d) aorta. The size of each square represents the average log2(Expr) for a gene across all nuclei in a given cluster. The color represents the log fold-change comparing the expression of the given gene in each cluster to all other clusters based on a formal differential expression model. A dot represents significant up- or down-regulation in the given cluster based on a Benjamini-Hochberg correction for multiple testing at FDR < 0.01. Expr, normalized nucleus-level expression calculated as the number of counts of a gene divided by the total number of counts in the nucleus and multiplied by 10,000; FC, fold-change.
Locus prioritization.
The gene SVIL was notable for being in proximity to one of the strongest GWAS signals for the descending aorta. In the TWAS, a predicted increase in SVIL expression corresponded to a larger descending aortic diameter (Fig. 3a), while loss of function variants in SVIL were associated with a smaller descending aortic diameter in the rare variant analysis (Fig. 3b). snRNA-seq revealed that SVIL is most strongly expressed in vascular smooth muscle cells within the aorta (Fig. 4c,d), consistent with a role in aortic size determination. SVIL encodes the protein supervillin, an F-actin and myosin II binding protein that localizes to and coordinates the action of cell surface extensions called ‘invadosomes’. These promote matrix degradation through the localized release of extracellular matrix-lytic enzymes such as disintegrin-and-metalloprotease domain-containing proteins and matrix metalloproteinases42,43.
In the ascending aorta, a lead SNP (rs1441358) was found within an intron of THSD4, which encodes the protein thrombospondin type 1 domain containing 4, a protein that promotes the organized assembly of fibrillin-1 microfibrils38. In the TWAS, a decrease in predicted THSD4 expression was linked to an increase in aortic diameter. The gene was excluded from our rare variant association test because too few UK Biobank participants carried a loss-of-function variant. A recent familial study of thoracic aortic aneurysm and dissection linked loss-of-function variants in THSD4 to ascending aortic aneurysm44, consistent with the expected direction of effect. Our snRNA-seq data suggest that THSD4 is primarily expressed in aortic vascular smooth muscle cells (and a separate cell cluster with lymphatic character), consistent with a role in aortic size (Fig. 4c).
Our genetic and single-nucleus transcriptomic analyses also highlight WWP2, which is linked to the size of both ascending and descending aorta. The lead SNP (rs62053262) is an expression quantitative trait locus (eQTL) in the aorta for WWP237; the rs62053262 G allele corresponds to reduced expression of WWP2 in aorta and smaller aortic size. The protein product of WWP2, NEDD4-like E3 ubiquitin-protein ligase, acts as an E3 ubiquitin ligase for PTEN45 and has previously been shown to regulate cardiac fibrosis through modulation of SMAD signaling46. Examining single-nucleus expression data, we show that WWP2 expression is enriched in aortic vascular smooth muscle cells (Extended Data Fig. 6).
In other cardiovascular phenotypes, GWAS loci have been enriched for Mendelian genes47,48, so we asked whether the loci identified in our study were in closer proximity to more genes implicated in Mendelian aortopathies than expected by chance. We did not find an enrichment of previously described Mendelian thoracic aortic aneurysm and dissection genes49 (23 genes; 2 overlapping with ascending loci, P = 0.14; 1 overlapping with descending loci, P = 0.32 by one-tailed permutation tests). However, our analysis has independently identified loci containing relevant genes such as FBN1, well described as the causal gene in Marfan syndrome50, and loci near genes such as PI15, known to cause arterial dysfunction in rats51, as well as the ABCC9-KCNJ8 locus, linked to Cantú syndrome—a rare recessive cause of aortic aneurysm in humans52. Other loci suggest the involvement of novel genes within networks previously implicated in aortic disease; for instance, the protein product of ASB2 is part of the E3 ligase that targets both filamin B (encoded by FLNB, the nearest gene to a lead SNP on chromosome 3) and the known aortic disease protein filamin A (FLNA) for degradation53. Moreover, TGF-β signaling, heavily implicated in clinical aortic disease, is also represented in our GWAS gene set as indicated by MAGMA analysis (Extended Data Fig. 7 and Supplementary Tables 17 and 18)54.
Polygenic score associated with clinical aortic disease.
Finally, we probed the clinical relevance of the GWAS loci by asking whether a polygenic score for ascending aortic size produced from these loci was associated with thoracic aortic disease risk. We analyzed the remaining UK Biobank participants who had not undergone MRI and who did not have a diagnosis of aortic disease at enrollment. A polygenic score was built from the 89 autosomal, independently significant SNPs from the ascending aorta GWAS (including the lead SNPs as well as other SNPs with P < 5 × 10−8 having r2 < 0.001 with other significant SNPs within the derivation sample; Supplementary Table 19). In 385,621 UK Biobank participants over a median of 11.2 years of follow-up time after enrollment, this polygenic score was strongly associated with the 685 incident cases of thoracic aortic aneurysm or dissection (HR = 1.43 per standard deviation; CI 1.32-1.54; P = 3.3 × 10−20). Participants in the top 10% of the polygenic score had a 2.1-fold hazard ratio compared to the remaining 90% of the cohort (CI 1.8-2.6; P = 7.3 × 10−15; Fig. 5). A descending aortic diameter polygenic score produced from the 46 autosomal lead SNPs had a weaker association with thoracic aortic aneurysm or dissection (HR = 1.15 per standard deviation; CI 1.07-1.24; P = 2.9 × 10−4).
Figure 5 |. Cumulative incidence of thoracic aortic aneurysm or dissection stratified by polygenic score.
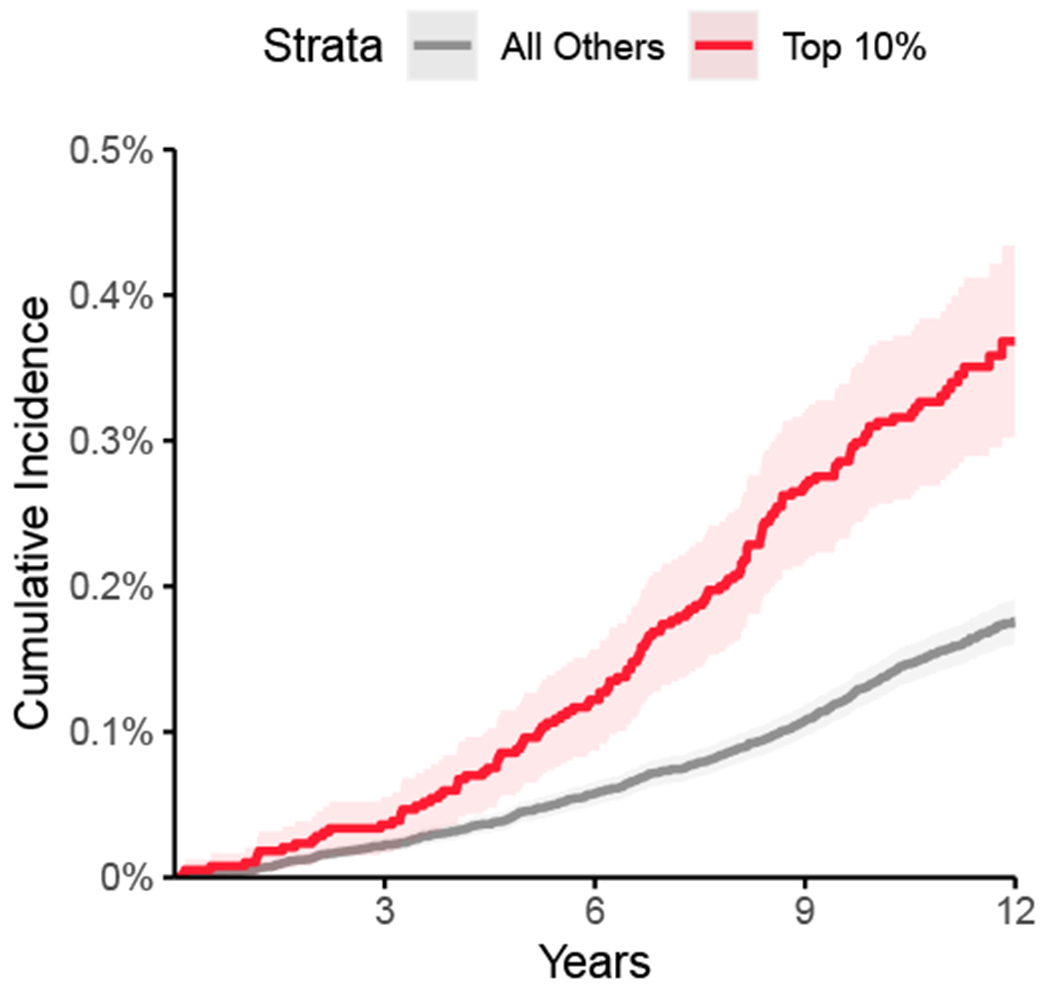
The cumulative incidence (1 minus the Kaplan-Meier survival estimate) of a diagnosis of aortic aneurysm or dissection (y-axis) is plotted against the number of years since UK Biobank enrollment (x-axis). Individuals in the top tenth percentile of the polygenic score for ascending aorta size are shown in red; the remaining 90% are shown in gray. The 95% confidence intervals (from the cumulative hazard standard error) are represented with lighter colors.
Limitations.
Our study is subject to several limitations. The study population largely consisted of European-ancestry UK Biobank participants, limiting generalizability to other populations. The aortic measurements were derived from a deep learning model that was trained on cardiologist-annotated segmentation data, but the vast majority of images were not manually reviewed; nevertheless, genetic results derived from manually annotated FHS imaging data were generally concordant with our findings. Our experiments suggest that increasing the number of training examples would modestly improve the deep learning model, which may enhance our ability to discover genetic associations. The need for additional manually annotated training examples is likely to be particularly important for more complex structures in future work. The human aorta tissue samples for the single-nucleus RNA expression experiments arose from paired samples in three individuals, so there is likely to be considerable variation in expression that is not captured in our analysis. Additional questions of interest, such as the presence of gene-environment interactions, remain for future work. Because only approximately 10% of the UK Biobank population had exome sequencing data available, we were unable to explore the relationship between loss- and gain-of-function variants in genes such as SVIL and disease diagnoses outside of the imaging cohort; this will be interesting to explore when additional sequencing data become available. Finally, because thoracic aortic aneurysm is not routinely assessed in screening tests, the effect estimate of the ascending aortic polygenic score is likely to be biased due to ascertainment in UK Biobank participants; future analyses in external datasets will be required to confirm the observation linking the polygenic score to aortic aneurysm or dissection.
Discussion
In summary, we used deep learning to assess the size of the ascending and descending thoracic aorta using magnetic resonance imaging data in a large population-based biobank. We identified 75 novel loci in the ascending aorta and 43 in the descending aorta, explored their relationships to other traits, and assessed their association with aortic aneurysm or dissection. These findings permit several conclusions. First, these results demonstrate that deep learning is a powerful tool for deriving quantitative phenotypes from raw signal data at a population level. In particular, by using transfer learning from a deep learning model trained on a large but unrelated set of images compiled for a different task, we were able to develop a useful model while manually annotating only a small number of images. Second, these results highlight the value of studying quantitative traits, such as aortic size, in order to gain greater understanding of disease processes underlying aneurysm and dissection. Third, the modest genetic correlation and limited locus overlap of the ascending and descending thoracic aorta highlight their distinct biology. Fourth, we prioritize several potential gene targets based on integration of GWAS, TWAS, and rare variant analyses, and identify their likely cell type of relevance with snRNA-seq. Fifth, a polygenic score for ascending aortic size is an independent risk factor for aneurysmal enlargement of aorta. Future work is warranted to determine whether a model incorporating a polygenic score and clinical risk factors might identify high-risk, asymptomatic individuals who would benefit from thoracic imaging to screen for ascending aortic aneurysm.
Methods
Study design.
All analyses were conducted in the UK Biobank unless otherwise stated. The UK Biobank is a richly phenotyped, prospective, population-based cohort that recruited 500,000 individuals aged 40-69 in the UK via mailer from 2006-201055. In total, we analyzed 487,283 participants with genetic data who had not withdrawn consent as of October 2018. Access was provided under application #7089. Analysis was approved by the Partners HealthCare institutional review board (protocol 2019P003144). GWAS replication was performed in an imaging substudy of the community-based Framingham Heart Study (FHS) Offspring and Third-Generation cohorts; participants were ascertained based on sex-specific age cutoffs (≥ 35 years for men and ≥ 40 years for women), and weight < 350 pounds as described previously and approved by the institutional review boards of the Boston University Medical Center and the Massachusetts General Hospital30. Ascending and descending human aortas were obtained from 5 human patients through a rapid autopsy protocol (DFHCC IRB #13-416) within 4 hours of cardiac death.
Our design was as follows: we manually annotated pixels belonging to the aortic blood pool in 116 cardiac MRIs from the UK Biobank. We then developed a deep learning model, trained on our manual annotations, to perform the same task at scale. The model was then applied to the remainder of the imaging data from the UK Biobank, permitting us to estimate the aortic diameter for every participant with imaging. Genetic discovery of loci related to the diameter of the ascending and descending thoracic aorta, treated as quantitative traits, was performed in this same UK Biobank cohort. A replication GWAS, based on previously performed aortic diameter measurements using computed tomography, was performed in FHS. With the genetic results from the UK Biobank, we performed a transcriptome-wide association study (TWAS) by incorporating publicly available gene expression data in order to prioritize genes at each genomic locus. We also performed a rare-variant association test in just over ~12,000 UK Biobank participants with both imaging and exome sequencing data. A single nucleus RNA sequencing study was then performed (using nuclei from aortas obtained from five human patients through a rapid autopsy protocol) to identify the aortic cell types that were most relevant to the genes highlighted by our bioinformatic analyses. A polygenic score produced from SNPs associated with aortic diameter in the UK Biobank GWAS was used to predict incident aortic disease in the remaining UK Biobank participants who had not undergone cardiac imaging.
Statistical analyses were conducted with R version 3.6 (R Foundation for Statistical Computing, Vienna, Austria).
Cardiac magnetic resonance imaging.
The UK Biobank is conducting an imaging substudy on 100,000 participants which is currently underway56,57. Cardiac magnetic resonance imaging was performed with 1.5 Tesla scanners (MAGNETOM Aera, Siemens Healthcare), using electrocardiographic gating for cardiac synchronization57. A balanced steady-state free precession cine, consisting of a series of exactly 100 images throughout the cardiac cycle, was acquired for each participant at the level of the right pulmonary artery57.
Deep learning for segmentation of the aorta.
Segmentation maps were traced for the ascending and descending thoracic aorta manually by a cardiologist (J.P.P.). To produce the final model used in this manuscript, 116 samples were chosen, manually segmented, and were then used to train a deep learning model with fastai v1.0.5913. The model consisted of a U-Net-derived architecture, where the encoder was a resnet34 model pre-trained on ImageNet13–15,58,59. 80% of the samples were used to train the model, and 20% were used for validation. Development versions prior to this final model are detailed in the following section. Variations on this modeling approach, and inter-rater evaluations, are described in subsequent sections.
During training, all images were resized to be 160 pixels in width by 132 pixels in height for the first half of training (‘small image training’), and then 240 pixels in width by 196 pixels in height, which is the native size of these images, for the second half (‘large image training’), detailed below. The Adam optimizer was used, and the model was trained with a minibatch size of 4 (when training with small images) or 2 (when training with large images)60. Rather than using extensive hyperparameter tuning with a grid search, the model was instead trained using a cyclic learning rate training policy, which alternately decreases and increases the learning rate during training61.
The maximum learning rate (the step size during gradient descent) was chosen with the learning rate finder from the FastAI library13. During small image training, the maximum learning rate was set at 0.002, with 20% of the iterations permitted to have an increasing learning rate during each epoch across 20 epochs. This was performed while keeping all ImageNet-pretrained layers fixed, so that only the final layer was fine-tuned. Then all layers were unfrozen and the model was trained for an additional 15 epochs with the same maximum learning rate. For large image training, the same model was then updated using full-dimension images, and the maximum learning rate was set to 0.0002, with 30% of the iterations permitted to have an increasing learning rate over 8 epochs. Then, all layers were unfrozen and the model was trained for an additional 15 epochs with a maximum learning rate of 0.0002. Additional details about hyperparameter selection are provided in the subsequent sections.
Throughout training, augmentations (random perturbations of the images) were applied as a regularization technique. These augmentations included affine rotation, zooming, and modification of the brightness and contrast. Because medical imaging data are not symmetric across the midline of the human body, we did not permit mirroring transformations. Using the software default settings for splitting samples into training and validation sets, 92 images were used to train the model, and 24 were held out for validation. Segmentation accuracy was assessed separately for the ascending and descending aorta.
This model was then used to infer segmentation of the ascending and descending aorta on all available “CINE_segmented_Ao_dist” images in the UK Biobank. During inference, adaptive pooling was used to permit arbitrary image sizes62, which allows for the production of output that matches the input size, preserving the number of millimeters per pixel as reported in the metadata.
Extraction of aortic diameter from deep learning output.
Having identified which pixels represented aorta, we were able to determine the aorta’s cross-sectional dimensions. The aorta was treated as an ellipse: major and minor axes were computed using classical image moment algorithms63. Separately for the ascending and the descending thoracic aorta, the length of the minor elliptical axis (in centimeters) was ascertained at the point in the cardiac cycle when the aorta was at its maximum size (closely corresponding with end-systole). The minor axis was chosen for analysis because imperfection in the orientation of the plane of image acquisition may falsely elongate the apparent major axis of the ascending and descending aorta; in contrast, the dimension of the minor axis is not affected by such perturbations. The length of the minor axis, in pixels, was converted to an absolute length in centimeters by using the metadata accompanying each image; in the UK Biobank, the reported pixel width and height is 1.58 millimeters for nearly all “CINE_segmented_Ao_dist” images. The length of the minor axis (i.e., the diameter) of the ascending and descending aorta were treated as our primary phenotypes for subsequent analyses.
Characteristics of the thoracic aortic diameter.
The correlation between ascending and descending aortic diameter was assessed with ordinary least squares regression. Because of the strong dependence of aortic diameter on sex, we configured the model to treat sex as a fixed effect, and predicted the ascending aortic diameter from that of the descending aorta. To remove the contribution of sex from the estimate of model fit (r2), we also predicted ascending aortic diameter from sex alone, and then performed an F test (using 1 degree of freedom for the descending aortic diameter) to compare the two nested models.
We also assessed whether the dispersion of the diameters of ascending and descending aorta differed. This analysis was stratified by sex. First, we asked whether the variance was equal between ascending and descending diameter using the F-test in R (implemented as var.test). Because the means of the two diameters were also different, we then tested whether the coefficient of variation, a dimensionless value computed by dividing the standard deviation by the mean, was equivalent between ascending and descending aorta. Significance testing to compare the coefficients of variation was performed using the function asymptotic_test from the cvequality package, the test statistic of which is asymptotically χ2 distributed64,65.
Aortic disease codes.
International Classification of Diseases version 10 (ICD-10) codes and Office of Population Censuses and Surveys Classification of Interventions and Procedures version 4 (OPCS-4) codes used to define aortic procedures and thoracic aortic aneurysm, dissection, or rupture are detailed in Supplementary Table 20. These definitions were used for GWAS participant exclusion and polygenic score assessment.
Correlation between phenotypes and aortic measurements.
We conducted phenome-wide association studies (PheWAS) to assess the relationship between the observed aortic traits and (i) other continuous traits measured in the UK Biobank, and (ii) other disease phenotypes based on ICD-10 and OPCS-4 codes.
All participants with aortic measurements were used in the continuous trait PheWAS. The number of participants modeled for each trait varied based on availability in the UK Biobank. 669 traits had sufficient data for analysis using a linear model accounting for the MRI serial number, sex, PC1-5, age at enrollment, the cubic natural spline of age at the time of MRI, and the genotyping array.
The same covariates were used in a logistic regression model testing the relationship between the aortic traits and 1,333 PheCode-defined diseases derived from hospital billing codes.
Genotyping, imputation, and genetic quality control.
As detailed previously, UK Biobank samples were genotyped on either the UK BiLEVE or UK Biobank Axiom arrays, then centrally imputed into the Haplotype Reference Consortium panel and the UK10K+1000 Genomes panel66. Variant positions were identified using the GRCh37 human genome reference. Genotyped variants with genotyping call rate < 0.95 and imputed variants with INFO score < 0.3 or minor allele frequency ≤ 0.001 in the analyzed samples were excluded. After variant-level quality control, 16,080,416 imputed autosomal variants and 566,283 imputed variants on the X chromosome remained for analysis.
Participants without imputed genetic data, or with a genotyping call rate < 0.98, mismatch between self-reported sex and sex chromosome count, sex chromosome aneuploidy, excessive third-degree relatives, or outliers for heterozygosity as defined centrally by the UK Biobank were excluded66.
We excluded participants with a measured aortic diameter greater than 5 cm, a history of aortic aneurysm or dissection, or a history of aortic surgical procedures. We assessed whether we could also exclude individuals with rare variants likely to lead to Mendelian aortopathy from the GWAS; however, in the subset of ~12,000 participants in the imaging substudy who had exome sequencing data, none had Marfan-related FBN1 variants identified in ClinVar.
The aortic diameters were found to be non-normally distributed (with non-zero skewness and kurtosis). Therefore, for the heritability analysis and genome-wide association study, they were first inverse-normal transformed67.
Heritability and genetic correlation of aortic traits.
BOLT-REML v2.3.4 was used to assess the SNP heritability of the minor axis length of the ascending and descending thoracic aorta and their genetic correlation with one another using the directly genotyped variants in the UK Biobank32.
Genome-wide association study of aortic traits.
We analyzed the inverse-normal transformed values of the diameter of the ascending and descending thoracic aorta at the frame within the cardiac cycle when they were at their largest. Genome-wide association studies for the diameter of the ascending and descending thoracic aorta were conducted using BOLT-LMM version 2.3.4 to account for cryptic population structure and sample relatedness32,33. These traits were adjusted for age at enrollment, age and age2 at the time of MRI, age at enrollment, the first 10 principal components of ancestry, sex, the genotyping array, and the MRI scanner’s unique identifier. We used the full autosomal panel of 714,512 directly genotyped SNPs that passed quality control to construct the genetic relationship matrix (GRM). GWAS covariates included age at enrollment, age and age2 at the time of MRI, the first five principal components of ancestry, sex, the genotyping array, and the MRI scanner’s unique identifier. Associations on the X chromosome were also analyzed, using all autosomal SNPs and X chromosomal SNPs to construct the GRM (n = 732,151 SNPs), with the same covariate adjustments and significance threshold as in the autosomal analysis. In this analysis mode, BOLT treats individuals with one X chromosome as having an allelic dosage of 0/2 and those with two X chromosomes as having an allelic dosage of 0/1/2. Variants with association P < 5 × 10−8, a commonly used threshold, were considered to be genome-wide significant.
In order to identify independently significantly associated variants, linkage disequilibrium (LD) clumping was performed with PLINK-1.968 in the same participants used to conduct the GWAS. We used a wide 5-Mb window (--clump-kb 5000) and a stringent LD threshold (--r2 0.001) in order to identify independently significant SNPs despite long LD blocks (particularly on chromosome 16 near WWP2). Using the independently significant SNPs, distinct genomic loci were defined by starting with the SNP with the strongest P value, excluding other SNPs within 500 kb, and iterating until no SNPs remained. The independently significant SNPs that defined each genomic locus are termed the lead SNPs. Lead SNPs were tested for deviation from Hardy-Weinberg equilibrium at a threshold of P < 1 × 10−6 (ref. 68).
Assessment for test statistic inflation.
Quantile-quantile plots of SNP association test statistics were produced. Linkage disequilibrium (LD) score regression analysis was performed with ldsc version 1.0.024. For both the ascending and descending aorta GWAS, the genomic control factor (lambda GC) was partitioned into polygenic and inflation components using the ldsc software’s defaults.
Genetic correlation with other quantitative traits.
Genetic correlation across traits was assessed using ldsc34 in 281 continuous traits from the UK Biobank whose ldsc-formatted summary statistics were made available by the Neale Lab (https://ukbb-rg.hail.is/). Of the 281 tested traits, genetic correlation with 257 traits was computable in the ascending aorta and with 256 traits in the descending aorta.
Tissue-specific LD score regression.
To address which tissues were most tightly linked to the ascending and descending aorta GWAS results, we applied tissue-specific LD score regression against 53 GTEx v6 tissue types that were preprocessed by the ldsc authors37,41. The ldsc authors identified genes that were specifically expressed in each tissue, retaining the top 10% of genes most specifically expressed from each of the 53 tissues. We then conducted stratified LD score regression with these specifically enriched gene sets (ldsc-SEG) to determine the contribution of the tissue-specific expression to the heritability of the size of the aorta. The returned P value represents the probability of seeing such a large coefficient if the null hypothesis (that the tissue is not enriched) were true, i.e., it tests whether the tissue-specific contribution is distinguishable from zero. Significance was determined using a false discovery rate (FDR) of 5%.
Mendelian aortopathy gene set enrichment.
We considered the 23 thoracic aortic aneurysm and dissection-related genes from Category A, B, or C from Renard et al. to be Mendelian aortopathy genes49. SNPsnap was used to generate 10,000 sets of SNPs that match the lead SNPs from the GWAS based on minor allele frequency, number of SNPs in linkage disequilibrium, distance to the nearest gene, and gene density at the locus69. A lead SNP was considered to be near a Mendelian locus if it was within 500 kb upstream or downstream of any gene on the panel. Significance was assessed by permutation testing across the 10,000 SNP sets to determine the neutral expectation for the number of overlapping genes in loci with characteristics similar to ours, yielding a one-tailed permutation P value.
Transcriptome-wide association study.
For ascending and descending thoracic aorta separately, we performed a TWAS to identify genes whose imputed cis-regulated gene expression corrrelates with aortic size36,70–72. We used FUSION with eQTL data from GTEx v7. Precomputed transcript expression reference weights for the aorta (n = 6,462 genes) were obtained from the FUSION authors’ website (http://gusevlab.org/projects/fusion/)36,37. FUSION was then run with its default settings.
MAGMA gene set analysis.
Using MAGMA 1.07b, we were able to test 7,706 gene sets from MSigDB for enrichment in the ascending and descending aortic GWAS results54,73. We used gene locations for GRCh37 and European reference data that were preprocessed by MAGMA’s authors (https://ctg.cncr.nl/software/magma). We used the composite “GO_PANTHER_INGENUITY_KEGG_REACTOME_BIOCARTA” gene sets from MSigDB provided by the MAGENTA authors74,75.
Exome sequencing in UK Biobank.
We conducted an exome sequencing analysis in the first 50,000 exomes released by the UK Biobank. Samples from the UK Biobank were chosen for exome sequencing based on enrichment for MRI data and linked health records76. Exome sequencing was performed by Regeneron and reprocessed centrally by the UK Biobank following the Functional Equivalent pipeline77. Exomes were captured with the IDT xGen Exome Research Panel v1.0, and sequencing was performed with 75-bp paired-end reads on the Illumina NovaSeq 6000 platform using S2 flowcells. Alignment to GRCh38 was performed centrally with BWA-mem. Variant calling was performed centrally with GATK 3.078. Variants were hard-filtered if the inbreeding coefficient was < −0.03, or if none of the following were true: read depth was greater than or equal to 10; genotype quality was greater than or equal to 20; or allele balance was greater than or equal to 0.2. In total, 49,997 exomes were available. Variants were annotated with the Ensembl Variant Effect Predictor version 95 using the--pick-allele flag79. LOFTEE was used to identify high-confidence loss of function variants: stop-gain, splice-site disrupting, and frameshift variants80.
Rare variant association test.
We conducted a collapsing burden test to assess the impact of loss-of-function variants in up to 12,336 participants who had aortic measurements and exome sequencing data available. For quantitative traits (minor axis length of the ascending and descending thoracic aorta), with heritability of approximately 0.6, we estimated that 13 loss-of-function variant carriers would be sufficient to achieve a power of 0.8 at an alpha of 0.05. Variants with MAF ≥ 0.001 were excluded. Using the LOFTEE “high-confidence” loss-of-function variants, for each of the 3,285 protein-encoding genes with at least 13 carriers of one or more loss-of-function variants in the UK Biobank, we tested whether loss-of-function carrier status was associated with aortic minor axis length using linear regression. The aortic diameter was the dependent variable and the presence or absence of a loss of function variant was the independent variable of interest; the model was adjusted for weight (kg), height (cm), the MRI serial number, age at enrollment, the cubic natural spline of age at the time of MRI, sex, genotyping array, and the first five principal components of ancestry. We performed an additional analysis that subset the gene list to those within a 500-kb window of one of the independently associated SNPs from the GWAS.
Association of aortic polygenic scores with incident disease.
Within a strictly defined European subset of the UK Biobank, we computed a polygenic score from the 89 autosomal, independently significant SNPs from the ascending aorta GWAS (Supplementary Table 20) and another from the 46 autosomal, independently significant SNPs from the descending aorta GWAS (Table 3), excluding participants whose data was used for the GWAS (Supplementary Table 21).
The strict European ancestry was defined using individuals who self-identified in the UK Biobank as British, Irish, or of other European ancestry as previously described81. The R package aberrant was applied to the first three pairs of principal components with the parameter lambda set to 40; only inliers were considered “European” for this analysis82.
We analyzed the relationship between the ascending aorta polygenic score and incident thoracic aortic aneurysm or dissection in 385,621 individuals (685 events) using a Cox proportional hazards model that was also adjusted for clinical risk factors. There is limited data regarding clinical risk factors for thoracic aortic aneurysm outside of associated syndromes and family history, so we chose putatively relevant covariates based in part on inference from evidence in the abdominal aortic aneurysm literature83. These covariates included sex, prevalent diagnoses of type 2 diabetes or hypertension, tobacco smoking history (the number of pack years of tobacco smoking), body mass (the cubic natural spline of BMI), and age (the cubic natural spline of age at enrollment). We also adjusted for other covariates including the cubic natural spline of height, the number of standard alcoholic drinks consumed per week, the genotyping array, and the first five principal components of ancestry. This analysis was performed separately for the ascending and descending aorta polygenic scores.
Extended Data
Extended Data Fig. 1. Aortic size by age and sex.
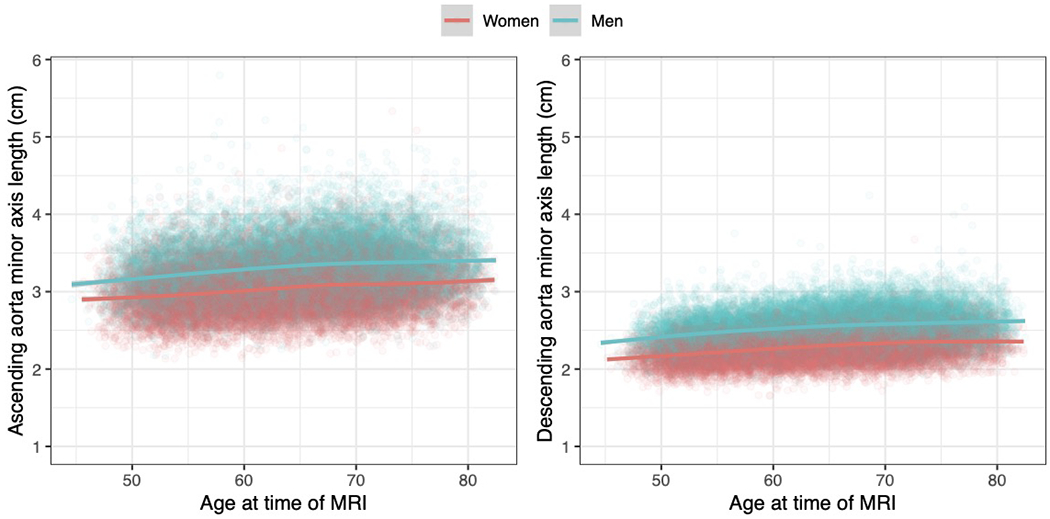
The length of the minor elliptical axis of aorta at its maximum size during the cardiac cycle (i.e., the diameter) is shown for the ascending aorta (left) and the descending aorta (right). The x-axis represents the participant’s age at the time of cardiac MRI, and the y-axis represents the size of aorta. Each point represents one person’s measurements; men are plotted in turquoise and women in red. Sex-specific locally weighted scatterplot smoothing (LOESS) curves are overplotted. Each point represents one of the 42,518 participants who passed imaging quality control for at least one of the ascending or descending aorta measurements: 40,363 had accepted measurements for ascending aorta, and 41,415 had accepted measurements for descending aorta.
Extended Data Fig. 2. GWAS sample flow diagram.
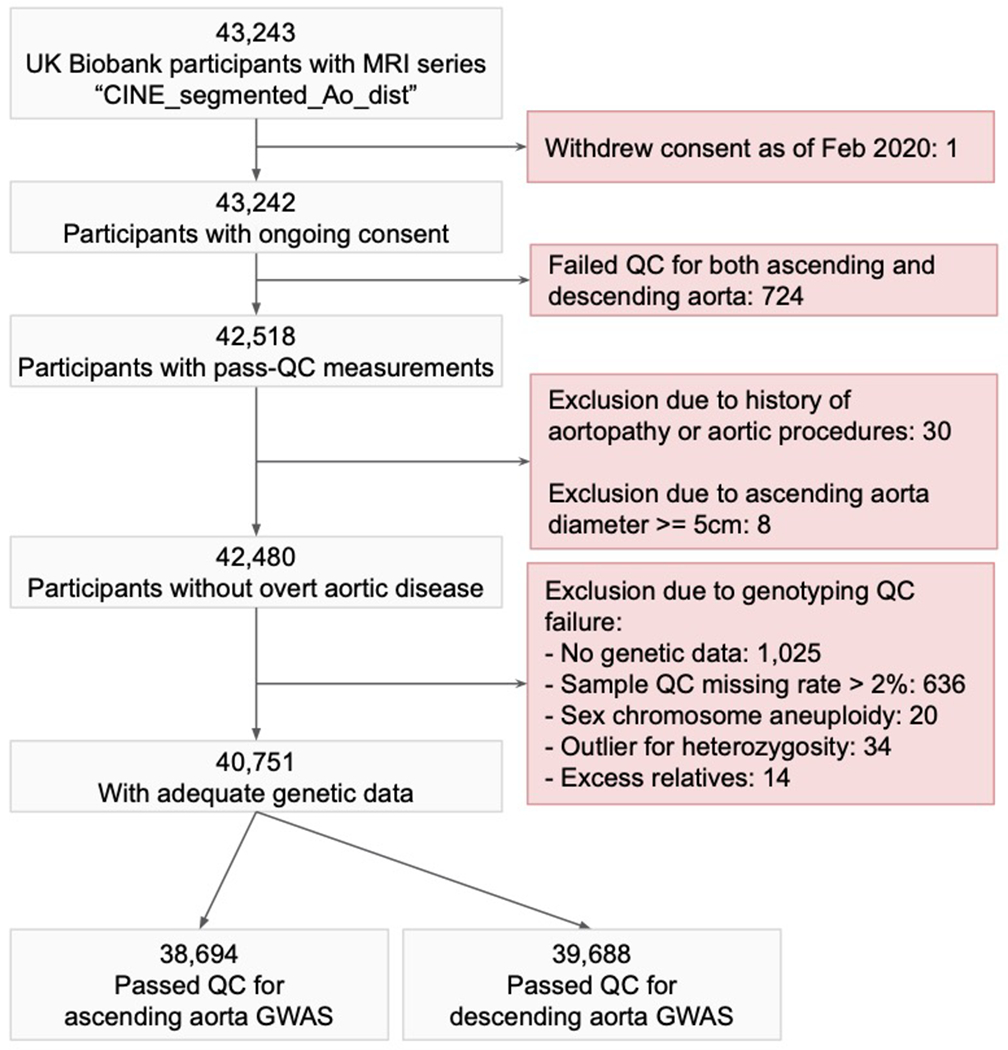
The GWAS sample flow diagram depicts the sample filtering process that led to the specific samples being chosen for the ascending and descending aortic diameter GWAS.
Extended Data Fig. 3. GWAS QQ plots.
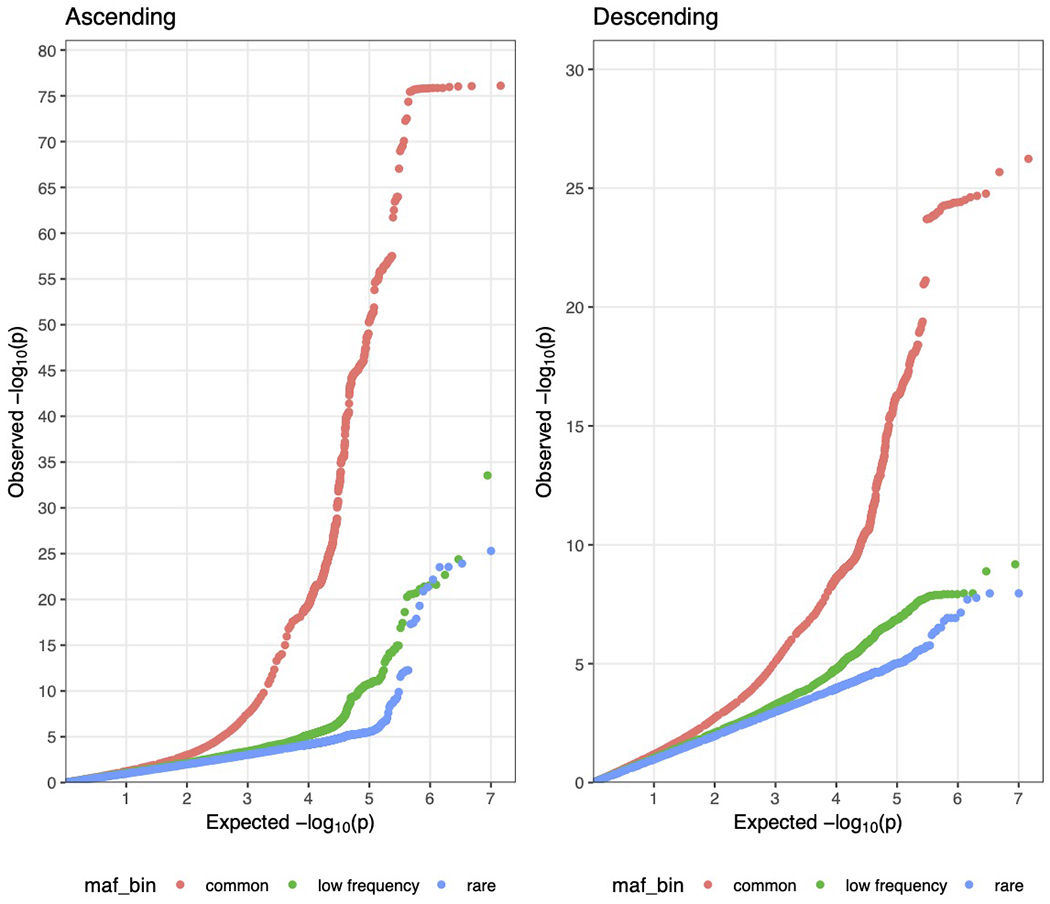
Quantile-quantile plots showing the theoretical distribution of P values under a uniform distribution (x-axis) versus the observed distribution within the sample (y-axis) are displayed for the ascending and descending aorta GWAS summary statistics. The plots are stratified by minor allele frequency (‘maf_bin’): ‘common’ denotes SNPs with MAF > 0.05, low frequency with 0.005 < MAF ≤ 0.05, and rare with 0.001 < MAF ≤ 0.005. Variants with MAF < 0.001 were excluded from the analysis.
Extended Data Fig. 4. GWAS replication in the Framingham Heart Study.
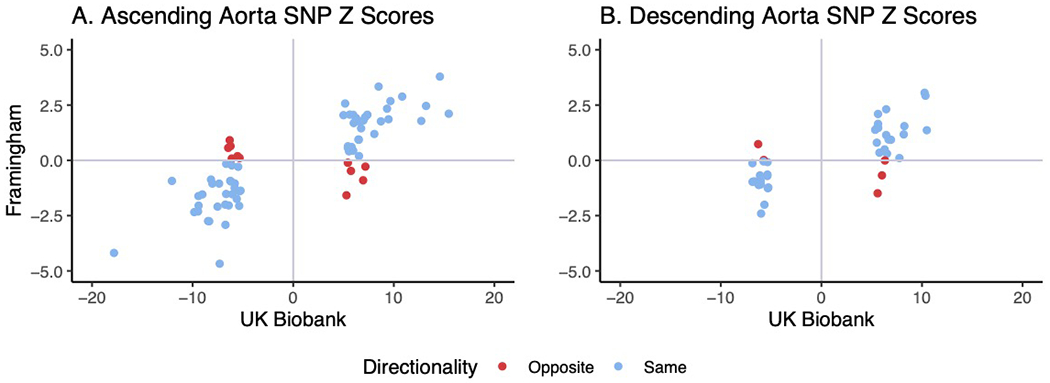
a,b, For lead SNPs from the main UK Biobank GWAS that could be identified in a GWAS from FHS, each SNP is plotted based on the UK Biobank Z score (x-axis) and the FHS Z score (y-axis). 72 SNPs for ascending aortic diameter (a) and 41 SNPs for descending aortic diameter (b) could be identified in FHS and are plotted here. SNPs where the direction of effect is in agreement between FHS and UK Biobank are plotted in blue, while those with opposite direction of effect are marked in red.
Extended Data Fig. 5. Genetic correlation with continuous traits.
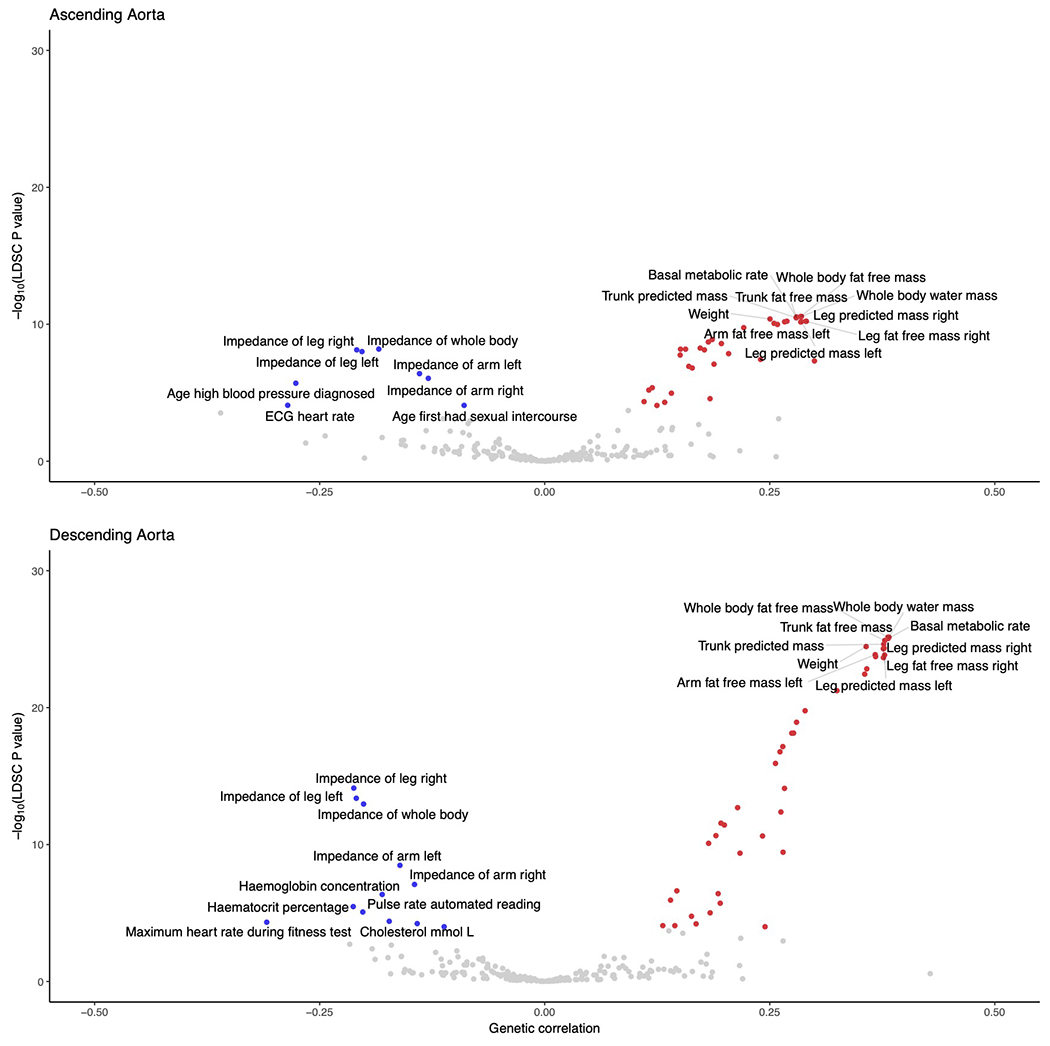
The genetic correlation between continuous traits and the ascending (top) and descending (bottom) thoracic aorta in the UK Biobank are represented in volcano plots. Of the 281 tested traits, genetic correlation with 257 traits was computable in the ascending aorta and with 256 traits in the descending aorta. The x-axis represents the magnitude of genetic correlation, while the y-axis represents the −log10 of the genetic correlation P value, based on ldsc. Traits achieving Bonferroni significance are colored red (for positive genetic correlation) or blue (for negative genetic correlation). The top 10 positively and negatively associated traits are labeled. The underlying data are available in Supplementary Table 10.
Extended Data Fig. 6. Cell type-specific gene expression at the WWP2 locus.
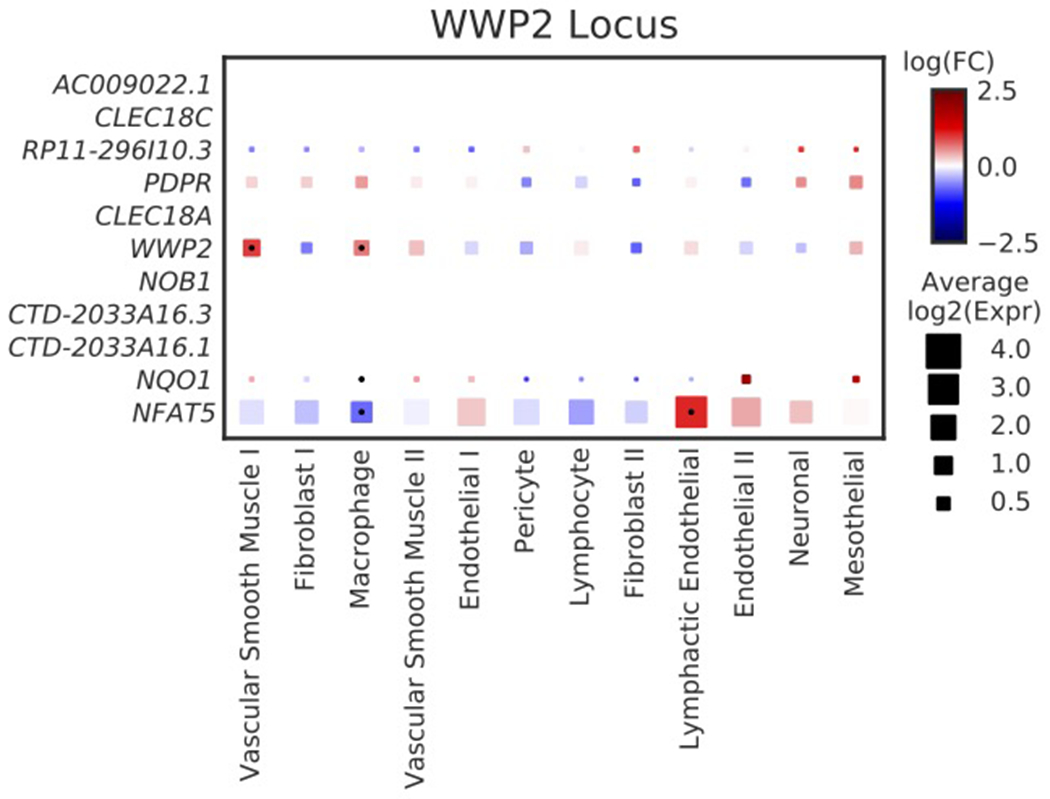
Cell-type specificity of genes with expression data within 500 kb of the lead SNP near WWP2. As with Figure 4, the size of each square represents the average log2(Expr) for a gene across all nuclei in a given cluster. The color represents the log fold-change comparing the expression of the given gene in each cluster to all other clusters based on a formal differential expression model. A dot represents significant up- or down-regulation in the given cluster based on a Benjamini-Hochberg correction for multiple testing at FDR < 0.01. Expr, normalized nucleus-level expression calculated as the number of counts of a gene divided by the total number of counts in the nucleus and multiplied by 10,000; FC, fold-change.
Extended Data Fig. 7. MAGMA gene set associations.
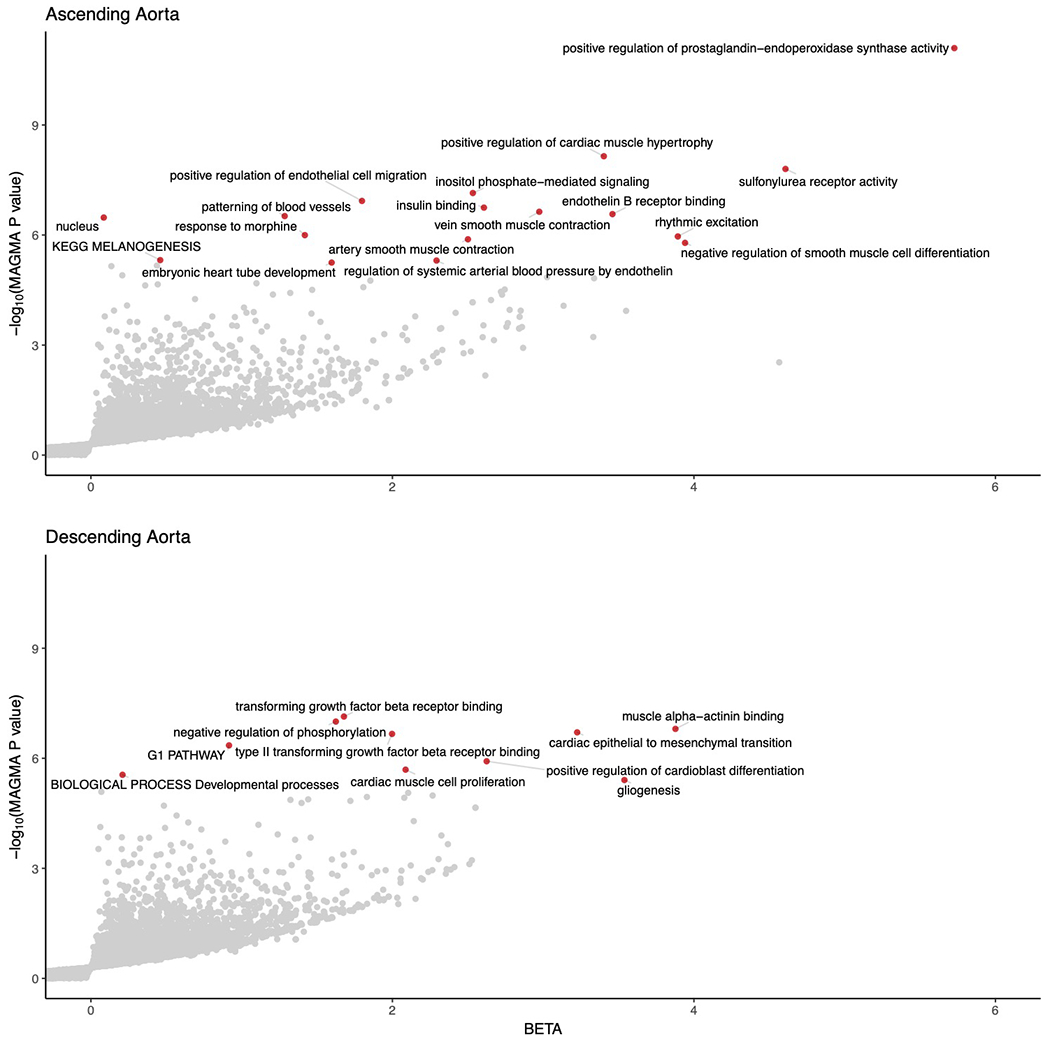
Gene sets enriched in MAGMA analysis of the GWAS of the ascending (top) and descending (bottom) thoracic aorta are represented in volcano plots. The x-axis represents the magnitude of estimated effect of a pathway-based gene set on the aortic trait, while the y-axis represents the −log10 of the MAGMA association P value. Pathways achieving Bonferroni significance are colored red and labeled. The underlying data are available in Supplementary Tables 17 and 18.
Supplementary Material
Acknowledgements
This work was supported by the Fondation Leducq #14CVD01 (P.T.E.); by grants from the National Institutes of Health #1RO1HL092577 (P.T.E.), #R01HL128914 (P.T.E.), #K24HL105780 (P.T.E.), #R01HL134893 (J.E.H.), #R01HL140224 (J.E.H.), #5K01HL140187 (N.R.T.), #T32HL007208 (S.K.), #R01HL128914 (E.J.B.), #2R01HL092577 (E.J.B.), #1R01HL141434 (E.J.B.), #2U54HL120163 (E.J.B.), #1R01HL139731 (S.A.L.), #T32HL007208 (E.L.C.); by a grant from the American Heart Association Strategically Focused Research Networks (P.T.E.); by the American Heart Association #18SFRN34110082 (E.J.B.), #18SFRN34110082 (A.H.), #18SFRN34110082 (L.W.), #18SFRN34250007 (S.A.L.); by a John S LaDue Memorial Fellowship (J.P.P.); by a Career Award for Medical Scientists from the Burroughs Wellcome Fund (A.G.B.); and by the Fredman Fellowship for Aortic Disease (M.E.L.) and the Toomey Fund for Aortic Dissection Research (M.E.L.). The Precision Cardiology Laboratory is a joint effort between the Broad Institute and Bayer AG. The rapid autopsy effort was funded by the Susan Eid Tumor Heterogeneity Initiative.
Competing Interests Statement
J.P.P. and A.G.B. have served as consultants for Maze Therapeutics. A.-D.A. and C.M.S. are employees of Bayer US LLC (a subsidiary of Bayer AG), and may own stock in Bayer AG. D.J. is supported by grants from Genentech, Eisai, EMD Serono, Takeda, Amgen, Celgene, Placon Therapeutics, Syros, Petra Pharma, InventisBio, Infinity Pharmaceuticals, and Novartis. D.J. has also received personal fees from Genentech, Eisai, EMD Serono, Ipsen, Syros, Relay Therapeutics, MapKure, Vibliome, Petra Pharma, and Novartis. A.P.P. is employed as a Venture Partner at GV; he is also supported by a grant from Bayer AG to the Broad Institute focused on machine learning for clinical trial design. J.E.H. is supported by a grant from Bayer AG focused on machine-learning and cardiovascular disease and a research grant from Gilead Sciences. J.E.H. has received research supplies from EcoNugenics. P.B. is supported by grants from Bayer AG and IBM applying machine learning in cardiovascular disease. P.T.E. is supported by a grant from Bayer AG to the Broad Institute focused on the genetics and therapeutics of cardiovascular diseases. P.T.E. has also served on advisory boards or consulted for Bayer AG, Quest Diagnostics, MyoKardia and Novartis. S.A.L. receives sponsored research support from Bristol Myers Squibb / Pfizer, Bayer AG, Boehringer Ingelheim, and Fitbit, and has consulted for Bristol Myers Squibb / Pfizer and Bayer AG, and participates in a research collaboration with IBM. The Broad Institute has filed for a patent on an invention from P.T.E., M.E.L., and J.P.P. related to a genetic risk predictor for aortic disease.
Footnotes
Code availability
The code used to identify connected components is available as a Go library at https://github.com/carbocation/genomisc/tree/master/overlay and a README is provided in that folder to demonstrate library usage.
Data availability
UK Biobank data is made available to researchers from universities and other research institutions with genuine research inquiries, following IRB and UK Biobank approval. Full GWAS summary statistics for ascending and descending thoracic aortic measurements are available at the Broad Institute Cardiovascular Disease Knowledge Portal (http://www.broadcvdi.org). Single nucleus RNA sequencing data are publicly available at the Broad Institute’s Single Cell Portal (https://singlecell.broadinstitute.org/single_cell) and at the National Center for Biotechnology Information’s Gene Expression Omnibus Database (accession # GSE165824). The dbGAP accession number for aortic phenotypes used in FHS replication is phs000007.v30.p11. All other data are contained within the article and its supplementary information, or are available upon reasonable request to the corresponding author.
References
- 1.Benjamin EJ et al. Heart disease and stroke statistics—2019 update: a report from the American Heart Association. Circulation 139, e56–e528 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Isselbacher EM Thoracic and abdominal aortic aneurysms. Circulation 111, 816–828 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Owens DK et al. Screening for abdominal aortic aneurysm: US Preventive Services Task Force Recommendation Statement. JAMA 322, 2211–2218 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Fann JI Descending thoracic and thoracoabdominal aortic aneurysms. Coron. Artery Dis 13, 93–102 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Guo D-C, Papke CL, He R & Milewicz DM Pathogenesis of thoracic and abdominal aortic aneurysms. Ann. N. Y. Acad. Sci 1085, 339–352 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Vapnik JS et al. Characteristics and outcomes of ascending versus descending thoracic aortic aneurysms. Am. J. Cardiol 117, 1683–1690 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Jondeau G & Boileau C Familial thoracic aortic aneurysms. Curr. Opin. Cardiol 29, 492–498 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Pinard A, Jones GT & Milewicz DM Genetics of thoracic and abdominal aortic diseases. Circ. Res 124, 588–606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verstraeten A, Luyckx I & Loeys B Aetiology and management of hereditary aortopathy. Nat. Rev. Cardiol 14, 197–208 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Lindsay ME & Dietz HC Lessons on the pathogenesis of aneurysm from heritable conditions. Nature 473, 308–316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majesky MW Developmental basis of vascular smooth muscle diversity. Arterioscler. Thromb. Vasc. Biol 27, 1248–1258 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Hagan PG et al. The International Registry of Acute Aortic Dissection (IRAD): new insights into an old disease. JAMA 283, 897–903 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Howard J & Gugger S Fastai: a layered API for deep learning. Information 11, 108 (2020). [Google Scholar]
- 14.Ronneberger O, Fischer P & Brox T U-Net: convolutional networks for biomedical image segmentation. ArXiv150504597 Cs (2015). [Google Scholar]
- 15.Deng J et al. ImageNet: a large-scale hierarchical image database. in 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 (2009). doi: 10.1109/CVPR.2009.5206848. [DOI] [Google Scholar]
- 16.Rosenfeld A & Pfaltz JL Sequential operations in digital picture processing. JACM 13, 471–494 (1966). [Google Scholar]
- 17.Turkbey EB et al. Determinants and normal values of ascending aortic diameter by age, gender and race/ethnicity in the Multi-Ethnic Study of Atherosclerosis (MESA). J. Magn. Reson. Imaging 39, 360–368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan S et al. Prevalence of an increased ascending and descending thoracic aorta diameter diagnosed by multislice cardiac computed tomography in men versus women and in persons aged 23 to 50 years, 51 to 65 years, 66 to 80 years, and 81 to 88 years. Am. J. Cardiol 100, 1598–1599 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Campens L et al. Reference values for echocardiographic assessment of the diameter of the aortic root and ascending aorta spanning all age categories. Am. J. Cardiol 114, 914–920 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Wu P et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med. Inform 7, e14325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradley TJ, Bowdin SC, Morel CFJ & Pyeritz RE The expanding clinical spectrum of extracardiovascular and cardiovascular manifestations of heritable thoracic aortic aneurysm and dissection. Can. J. Cardiol 32, 86–99 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Avdic T et al. Reduced long-term risk of aortic aneurysm and aortic dissection among individuals with type 2 diabetes mellitus: a nationwide observational study. J. Am. Heart Assoc 7, e007618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prakash SK, Pedroza C, Khalil YA & Milewicz DM Diabetes and reduced risk for thoracic aortic aneurysms and dissections: a nationwide case-control study. J. Am. Heart Assoc 1, e000323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo D et al. Genetic variants in LRP1 and ULK4 are associated with acute aortic dissections. Am. J. Hum. Genet 99, 762–769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van ‘t Hof FNG et al. Shared genetic risk factors of intracranial, abdominal, and thoracic aneurysms. J. Am. Heart Assoc 5, e002603 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeMaire SA et al. Genome-wide association study identifies a susceptibility locus for thoracic aortic aneurysms and aortic dissections spanning FBN1 at 15q21.1. Nat. Genet 43, 996–1000 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasan RS et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA 302, 168–178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wild PS et al. Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J. Clin. Invest 127, 1798–1812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers IS et al. Distribution, determinants, and normal reference values of thoracic and abdominal aortic diameters by computed tomography (from the Framingham Heart Study). Am. J. Cardiol 111, 1510–1516 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qazi S et al. Increased aortic diameters on multidetector computed tomographic scan are independent predictors of incident adverse cardiovascular events: the Framingham Heart Study. Circ. Cardiovasc. Imaging 10, e006776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loh P-R et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet 47, 284–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh P-R, Kichaev G, Gazal S, Schoech AP & Price AL Mixed model association for biobank-scale data sets. Nat. Genet 50, 906–908 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wain LV et al. Novel blood pressure locus and gene discovery using genome-wide association study and expression data sets from blood and the kidney. Hypertension (2017) doi: 10.1161/HYPERTENSIONAHA.117.09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gusev A et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet 48, 245–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lonsdale J et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsutsui K et al. ADAMTSL-6 is a novel extracellular matrix protein that binds to fibrillin-1 and promotes fibrillin-1 fibril formation. J. Biol. Chem 285, 4870–4882 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou C-K et al. The regulations of deubiquitinase USP15 and its pathophysiological mechanisms in diseases. Int. J. Mol. Sci 18, 483 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eichhorn PJA et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat. Med 18, 429–435 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Finucane HK et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhuwania R et al. Supervillin couples myosin-dependent contractility to podosomes and enables their turnover. J. Cell Sci 125, 2300–2314 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linder S, Wiesner C & Himmel M Degrading devices: invadosomes in proteolytic cell invasion. Annu. Rev. Cell Dev. Biol 27, 185–211 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Elbitar S et al. Pathogenic variants in THSD4, encoding the ADAMTS-like 6 protein, predispose to inherited thoracic aortic aneurysm. Genet. Med 23, 111–122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maddika S et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol 13, 728–733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H et al. WWP2 regulates pathological cardiac fibrosis by modulating SMAD2 signaling. Nat. Commun 10, 3616 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pirruccello JP et al. Analysis of cardiac magnetic resonance imaging in 36,000 individuals yields genetic insights into dilated cardiomyopathy. Nat. Commun 11, 2254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teslovich TM et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renard M et al. Clinical validity of genes for heritable thoracic aortic aneurysm and dissection. J. Am. Coll. Cardiol 72, 605–615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dietz HC et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature 352, 337–339 (1991). [DOI] [PubMed] [Google Scholar]
- 51.Falak S et al. Protease inhibitor 15, a candidate gene for abdominal aortic internal elastic lamina ruptures in the rat. Physiol. Genomics 46, 418–428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parrott A et al. Cantu syndrome: A longitudinal review of vascular findings in three individuals. Am. J. Med. Genet. A. 182, 1243–1248 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Heuzé ML et al. ASB2 targets filamins A and B to proteasomal degradation. Blood 112, 5130–5140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 55.Sudlow C et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petersen SE et al. Imaging in population science: cardiovascular magnetic resonance in 100,000 participants of UK Biobank - rationale, challenges and approaches. J. Cardiovasc. Magn. Reson 15, 46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petersen SE et al. UK Biobank’s cardiovascular magnetic resonance protocol. J. Cardiovasc. Magn. Reson 18, 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He K, Zhang X, Ren S & Sun J Deep residual learning for image recognition. ArXiv151203385 Cs (2015). [Google Scholar]
- 59.Krizhevsky A, Sutskever I & Hinton GE ImageNet classification with deep convolutional neural networks. Communications of the ACM 60, 84–90 (2017). [Google Scholar]
- 60.Kingma DP & Ba J Adam: a method for stochastic optimization. ArXiv14126980 Cs (2017). [Google Scholar]
- 61.Smith LN Cyclical learning rates for training neural networks. ArXiv150601186 Cs (2015). [Google Scholar]
- 62.He K, Zhang X, Ren S & Sun J Spatial pyramid pooling in deep convolutional networks for visual recognition. ArXiv14064729 Cs 8691, 346–361 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Horn B Robot vision. (MIT Press; ; McGraw-Hill, 1986). [Google Scholar]
- 64.Feltz CJ & Miller GE An asympotic test for the equality of coefficients of variation from k populations. Stat. Med 15, 647–658 (1996). [DOI] [PubMed] [Google Scholar]
- 65.Marwick B & Krishnamoorthy K benmarwick/cvequality. (2019). [Google Scholar]
- 66.Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang J et al. FTO genotype is associated with phenotypic variability of body mass index. Nature 490, 267–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pers TH, Timshel P & Hirschhorn JN SNPsnap: a Web-based tool for identification and annotation of matched SNPs. Bioinformatics 31, 418–420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gamazon ER et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet 47, 1091–1098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gusev A et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet 50, 538–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu Z et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet 48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 73.de Leeuw CA, Neale BM, Heskes T & Posthuma D The statistical properties of gene-set analysis. Nat. Rev. Genet 17, 353–364 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Liberzon A et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Segrè AV et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 6, e1001058 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Hout CV et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature 586, 749–756 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Regier AA et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun 9, 4038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van der Auwera GA et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinforma 43, 11.10.1-33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McLaren W et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karczewski KJ et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haas ME et al. Genetic association of albuminuria with cardiometabolic disease and blood pressure. Am. J. Hum. Genet 103, 461–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bellenguez C et al. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics 28, 134–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kent KC et al. Analysis of risk factors for abdominal aortic aneurysm in a cohort of more than 3 million individuals. J. Vasc. Surg 52, 539–548 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UK Biobank data is made available to researchers from universities and other research institutions with genuine research inquiries, following IRB and UK Biobank approval. Full GWAS summary statistics for ascending and descending thoracic aortic measurements are available at the Broad Institute Cardiovascular Disease Knowledge Portal (http://www.broadcvdi.org). Single nucleus RNA sequencing data are publicly available at the Broad Institute’s Single Cell Portal (https://singlecell.broadinstitute.org/single_cell) and at the National Center for Biotechnology Information’s Gene Expression Omnibus Database (accession # GSE165824). The dbGAP accession number for aortic phenotypes used in FHS replication is phs000007.v30.p11. All other data are contained within the article and its supplementary information, or are available upon reasonable request to the corresponding author.


