Abstract
Aims
Nutritional and lifestyle interventions can contribute to prevent and treat obesity and its complications; however, genetic background may influence the success of a therapy. The aim of this pilot study is to evaluate the effects of the interaction between nutrigenetic variants and nutritional intervention, as well as the changes in clinical parameters and the adherence to Mediterranean diet (MedDiet) and to physical activity, of 18 overweight or obese subjects affected by T2D or dysglycemia included in a nutritional program.
Methods
The subjects’ clinical parameters as well as their PREDIMED score and physical activity levels were recorded and compared at baseline, at 6 months and at the end of the intervention. Rs9939609 in FTO, rs17782313 near MC4R, rs326 in LPL, rs16147 in NPY, rs2943641 near IRS-1 were genotyped.
Results
The subjects carrying the A allele in FTO lost less weight (p = 0.022) and had a lower BMI decrease from baseline to 12 months (p-interaction = 0.047) than TT carriers. In addition, there was a significant PREDIMED score modification over time, according to genotypes for FTO rs9939609 (p = 0.025) and NPY rs16147 (p = 0.039), respectively.
Conclusions
These preliminary findings show a significant interaction between genetic variants and the PREDIMED score, suggesting that individuals carrying the FTO variant may lose less weight than non-carriers through diet/lifestyle intervention.
Keywords: FTO, Rs9939609, Gene–diet interaction, Nutrition, Mediterranean diet, Type 2 diabetes, Obesity, Nutrigenetics
Introduction
Obesity is a complex and multifactorial disease with an increasing global prevalence, which has been taking on epidemic proportions over the last decades [1]. The adverse metabolic effects of overweight and obesity contribute to an increased risk of several other health conditions such as diabetes mellitus type 2 (T2D), hypertension, hyperlipidemia, and coronary heart disease [2, 3]. The pathophysiology of both obesity and obesity-related complications is complex: hereditary factors as well as socioeconomic and sociocultural milieus, have been shown to affect the risk of obesity. Some modifiable environmental factors including a sedentary lifestyle and unhealthy nutrition can cause a significant imbalance between energy intake and expenditure [4]. Moreover, due to differences in obesity susceptibility among individuals exposed to the same obesogenic environment, it has been demonstrated that genetics also plays a role in the development and the complications of this condition. In fact, the Genome-Wide Association Studies (GWAS) have identified several common single nucleotide polymorphisms (SNPs) of small impact, whose combined effects on adiposity levels, body compositions and obesity susceptibility can be captured by a polygenic risk score [5–8]. Public health strategies focus on weight management and the promotion of healthy lifestyles. Weight reduction, based on nutrition optimization and physical activity can have a beneficial effect against the overall risk and contribute to prevent and treat obesity and its complications [9]. Nevertheless, it is known that most weight loss programs are generally unsuccessful. Significantly, obese patients may be classified as normo-responders, hypo-responders, or hyper-responders, depending on their response to diet or surgical treatment [10]. This can indicate that not only physiological and behavioral factors, but also genetic background may influence the success of weight loss and the effectiveness of a shift in lifestyle behavior. In this context, Nutrigenetics is concerned with the effects of genetic variants on adverse or beneficial response to dietary components and nutrient requirements, thus identifying at-risk individuals for targeted, preventive, and early intervention strategies [11–15]. In this scenario, in order to improve the efficacy of interventions so that they became fitter to the nutritional and metabolic needs of each patient, personalized nutrition based on the metabolic profile (via clinical and biochemical assessment), the genetic profile (via nutrigenetic and nutrigenomic assessment), the body composition, and a nutritional status assessment has been suggested [16, 17]. Several studies found gene–diet interactions involving genetic variants mainly located in genes associated with obesity development, energy expenditure, adipogenesis, and eating behavior or appetite control. The most interesting issues in this field could pave the way toward personalized obesity therapy. Although the association between several SNPs and obesity/body weight has been increasingly clarified, the role of genetic risk on the effects of behavioral or environmental exposures such as diet and exercise should be better explored. Such discoveries could provide novel insight into a possible interaction among diet, genetic make-up, and other environmental factors, so as to understand the subsequent metabolic dysregulation and the effects of obesity treatment. In this light, the aim of the present pilot study is to evaluate the effects of the interaction between variants in 5 candidate genes (namely rs9939609 in FTO, rs17782313 near MC4R, rs326 in LPL, rs16147 in NPY, rs2943641 near IRS-1) and nutritional intervention as well as the impact/changes of the PREDIMED score and physical activity levels on the mid-term changes in the anthropometric and clinical parameters of overweight or obese subjects affected by T2D or impaired glucose regulation (IGR) over one-year period/12 months.
Materials and methods
Study design and participants
18 overweight or obese individuals, affected by T2D or IGR, attended by the Diabetes, Nutrition, and Metabolism Unit at “Gabriele d’Annunzio” University Hospital in Chieti, Italy, were recruited. Study protocols were approved by the Ethics Committee of the Province di Chieti and Pescara. In accordance with the Declaration of Helsinki, all participants gave their written informed consent prior to their inclusion in the study. The inclusion criteria were overweight or obese (BMI ≥ 25 and ≥ 30 kg/m2, respectively) subjects (male or female), ≥ 18 years of age, affected by T2D or Impaired Glucose Regulation (IGR) (Impaired Fasting Glucose or Impaired Glucose Tolerance). The exclusion criteria were the presence of Type 1 Diabetes, Eating Behavior Disorders, or other conditions that might have interfered with the development and completion of the project or cause failure to adherence to the protocol.
Interventions and outcome assessment
All participants were included in an educational and nutritional program as described below. At baseline, each subject underwent an individual interview focusing on Medical Nutrition Therapy (MNT) and healthy lifestyle with a physician and a dietitian from the team. Demographic characteristics as well as clinical and anthropometric parameters were collected. In detail, body weight and height, Body Mass Index (BMI), waist and hip circumferences (WC and HC, respectively), HbA1c, and blood pressure were included. Moreover, adherence to the Mediterranean diet (MedDiet) was assessed using a validated 14-item questionnaire (PREDIMED) which generates a range of possible scores: no adherence (score ≤ 5), medium adherence (6 ≤ score ≤ 9), and maximum adherence (score ≥ 10) [18]. In addition, we evaluated the physical activity level using a short version of the International Physical Activity Questionnaire (IPAQ) [19].
Participants were actively involved in face-to-face individual and group-based intervention through a theoretical and practical course which included content on healthy nutrition and lifestyle.
The objectives to be achieved were, according to the MedDiet: (i) an improvement in the composition of the meals, in particular emphasizing non-starchy vegetable, whole foods over highly processed foods to the extent possible, minimizing added sugars and refined grain; (ii) a 7–10% weight decrease compared to the initial body weight achieved through an individualized eating plan; (iii) increasing moderate-intensity physical activity to at least 150 min per week [24]. The program was conducted using a nutritional and educational approach in accordance with the current literature [20–23]. The first face-to-face individual intervention was a clinical check-up and the recording of the individual’s clinical history. Also, during this first encounter, participants were instructed on how to keep a journal on dietary self-monitoring, based on the scientific literature [20–23]. The journal entries were discussed by the participant and the physician at the next meeting. To ensure the active involvement of patients in the change process, educational group sessions intervention was considered essential to improve the lifestyle of these obese and/or T2D subjects. Group-based intervention was organized in sessions performed with small groups of up to 10 people in the presence of a trained facilitator. Therefore, the endpoint of this study became the comparison between the subjects’ anthropometric and clinical parameters as well as PREDIMED and IPAQ scores at baseline, with those at 6 months (T6) and 12 months (T12) and its relationship with genetic variants.
Gene and SNP selection
Nutrigenetic variants from five genetic loci, identified by previous GWAS or replication studies as associated with T2DM, obesity, lipid, and carbohydrate metabolism and dietary intake were selected for this study. In details, two of these variants, namely rs9939609 (T > A) in FTO and rs17782313 (T > C) near MC4R, were associated with BMI and hunger control [25–28]; rs326 (A > G) in LPL was involved in lipid metabolism [29–31]; rs16147 (T > C) in NPY was involved in adiposity and obesity [32–34]; rs2943641 (C > T) near IRS-1 was involved in obesity, insulin resistance, T2DM risk [35].
Genetic analysis
A blood sample from each patient was collected in a sterile tube containing EDTA and stored at + 4 °C before analysis. The genetic analysis was conducted at the Laboratory of Molecular Genetics, School of Medicine and Health Sciences, ‘‘G. d’Annunzio” University of Chieti-Pescara. Genomic DNA was automatically isolated from peripheral blood lymphocytes using MagPurix 12sAutomatedNucleicAcid Purification System (Zinexts Life Science Corp., Taiwan). Nucleic acids were quantified by measuring UV absorption using a spectrophotometer. In detail, DNA samples were amplified by PCR performed in 30 ul reaction volume containing 30 ng of genomic DNA in a Simpli-Amp™ thermal cycler (Applied Biosystems™), using the AmpliTaq Gold DNA Polymerase. Specific primers were designed on the reference gene sequence. PCR conditions were as follows: initial denaturation at 95 °C for 10 min, followed by 35 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 10 min. The amplification products were submitted to direct sequencing procedure using BigDye Term v3.1 CycleSeq Kit (Life Technologies, Monza, Italy) followed by automatic sequencing analysis (SeqStudio™ Genetic Analyzer).
Statistical analysis
Descriptive statistics relied, after Shapiro–Wilk’s test to evaluate the departures from normality distribution for each variable, on median and interquartile range (IQR) for continuous variables and on absolute and relative frequencies (%) for qualitative variables.
Friedman test or chi-square was applied to evaluate a statistically significant variation over time for quantitative and qualitative variables, respectively.
Several linear mixed models tested the effect of different genotypes and time on features of interest; namely weight, BMI, waist-hip ratio, total cholesterol, LDH, HDL and triglyceride levels. Moreover, within linear mixed models, the interaction between genotype and time was also considered. In order to test differences in PREDIMED and IPAQ, the Friedman test was used. Three models of inheritance (i.e., dominant, recessive, and additive) were tested. If the alleles of the gene of interest are A and B in haploid, and B is the ‘risk’ allele, the dominant genetic model assumes that the risk is the same for heterozygotes, carrying 1 copy of the high-risk allele B, as for homozygotes, and the data are dichotomized into “carriers” versus “non-carriers” (i.e., ‘BB + AB’ versus ‘AA’). The recessive genetic model assumes 2 copies of B are required for the risk to be different from the baseline risk (i.e., ‘BB’ versus ‘AB + AA’); the additive model hypothesizes that AA, AB, and BB are associated with the lowest, the intermediate, and the highest risk, respectively (‘AA’ versus ‘AB’ versus ‘BB’) [36–38]. All tests were two-sided and a level of statistical significance was set at p < 0.05. Analyses were performed using the R software environment for statistical computing and graphics (version 4.0.5; http://www.r-project.org/).
Results
The characteristics of 18 participants (n = 18, 10 males and 8 females) at baseline, at T6, and at the end of the nutritional intervention are summarized in Table 1. The median age of the participants was 64.5 (IQR: 57.5–66.8) years. Significant variations over time were observed for weight (p < 0.001), BMI (p = 0.001) and PREDIMED scores (p = 0.009). The median of PREDIMED score, on the scale of 0–14, was 7.5 and 8.5 at baseline and at the end of the study, respectively (p = 0.009).
Table 1.
Baseline characteristics of participants
| Variable | Baseline | T6 | T12 | p-valuea |
|---|---|---|---|---|
| Weight (Kg) | 96.2 (82.6,107.9) | 96.5 (79.5,105.5) | 91.5 (79.6, 103.2) | < 0.001 |
| BMI (kg/m2) | 31.6 (28.6, 38.0) | 31.6 (28.0, 38.0) | 31.1 (27.8, 36.1) | < 0.001 |
| Waist circumference (cm) | 114.0 (101.5, 118.8) | 109.0 (97.0, 116.0) | 109.0 (93.0, 116.0) | 0.264 |
| Hip circumference (cm) | 113.5 (105.2,121.2) | 115.0 (102.0,122.0) | 112.0 (101.5,118.5) | 0.936 |
| WHR | 1.0 (0.9, 1.0) | 1.0 (1.0, 1.1) | 1.0 (0.9, 1.0) | 0.694 |
| Systolic blood pressure (mmHg) | 145.0 (117.5, 150.0) | 132.5 (128.8, 140.0) | 125.0 (120.0, 136.2) | 0.135 |
| Diastolic blood pressure (mmHg) | 80.0 (71.2, 90.0) | 80.0 (73.8, 85.0) | 78.0 (70.0, 81.2) | 0.359 |
| PREDIMED | 7.5 (7.0, 8.0) | 9.0 (9.0, 10.0) | 8.5 (8.0, 9.8) | 0.009 |
| PREDIMED_CLASS, n(%) | 0.761b | |||
| - No adherence | 1 (5.6%) | 0 (0.0%) | 0 (0.0%) | |
| - Adherence | 14 (77.8%) | 10 (58.8%) | 13 (72.2%) | |
| - Max adherence | 3 (16.7%) | 7 (41.2%) | 5 (27.8%) | |
| IPAQ, n(%) | 0.717b | |||
| - low | 9 (50.0%) | 5 (29.4%) | 4 (22.2%) | |
| - moderate | 6 (33.3%) | 8 (47.1%) | 7 (38.9%) | |
| - high | 3 (16.7%) | 4 (23.5%) | 7 (38.9%) | |
| Fasting blood glucose (mg/dl) | 138.0 (115.0, 149.0) | 107.0 (105.0, 108.0) | 110.0 (103.5, 125.0) | 0.106 |
| Hba1c (%) | 6.5 (6.3, 7.4) | 6.2 (6.1, 6.3) | 6.2 (5.4, 6.5) | 0.267 |
| Total cholesterol (mg/dL) | 201.0 (196.0, 222.0) | 208.5 (197.5, 231.8) | 195.5 (164.5, 223.5) | 0.606 |
| HDL (mg/dL) | 40.0 (37.0, 54.0) | 48.5 (41.0, 54.5) | 47.5 (41.8, 52.8) | 0.139 |
| TG (mg/dL) | 139.5 (102.8, 213.8) | 118.5 (94.8, 186.8) | 113.0 (94.8, 145.8) | 0.078 |
| LDL (mg/dL) | 122.4 (104.6, 142.2) | 132.7 (121.9, 155.4) | 124.9 (101.5, 140.2) | 0.952 |
Statistically significant values are given in bold
Data are expressed as median (Q1–Q3). a p-value derived from Friedman Test; b p-value derived from chi-squared test
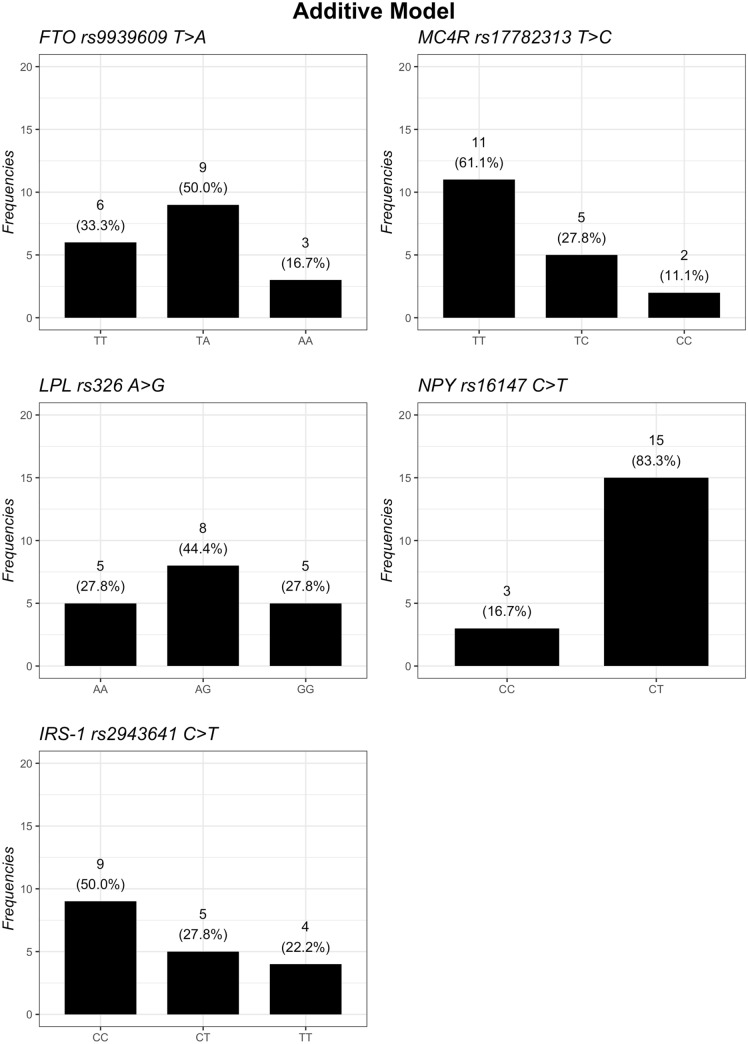
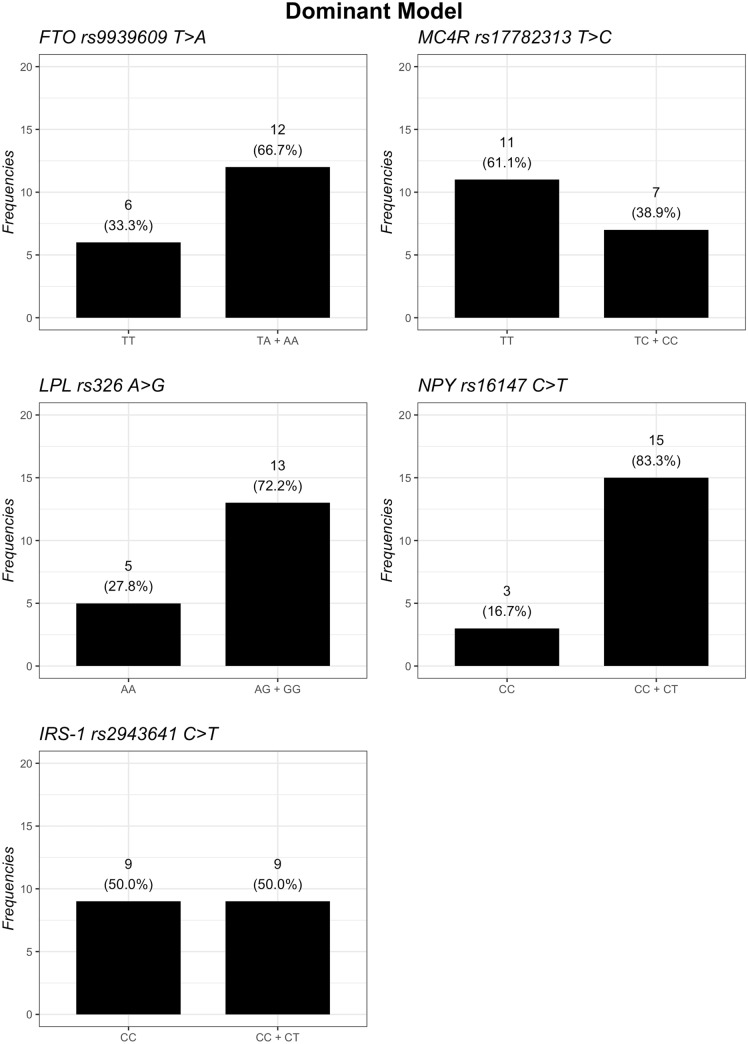
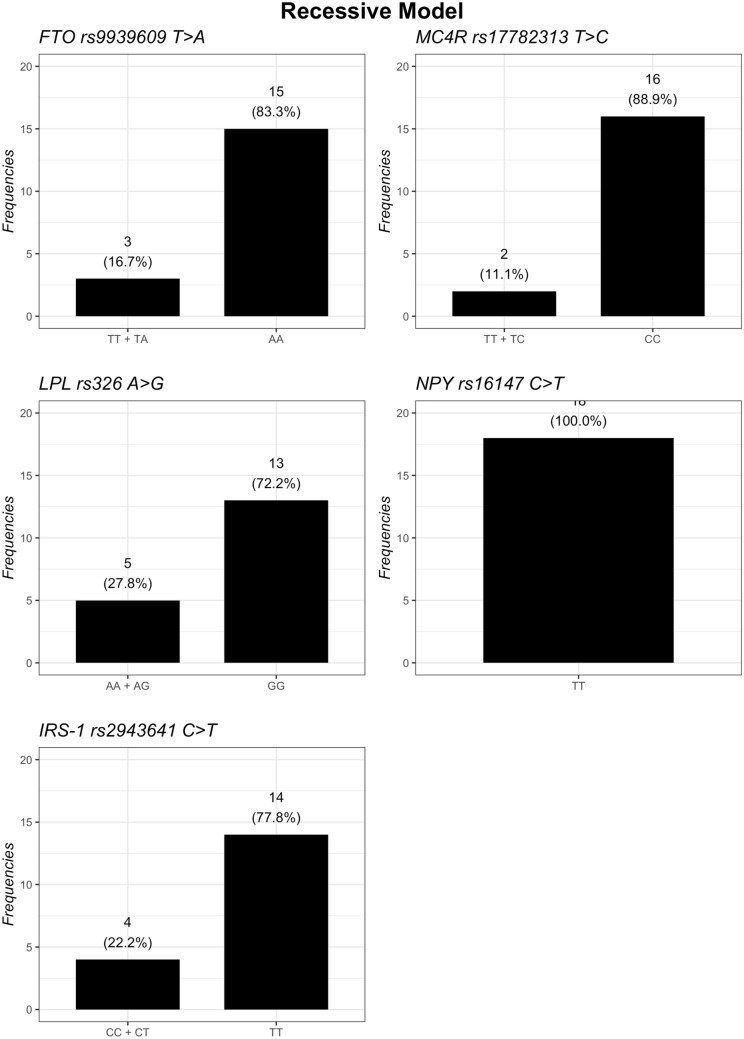
The distribution of the genotypes based on additive, dominant, and recessive models is summarized in Figs. 1, 2 and 3. Regarding the FTO gene, the additive model showed that homozygous subjects for A risk allele had the highest baseline BMI and a statistical tendency to have lowest body weight loss after 12 months of nutritional intervention, compared to TT and TA genotypes (p-interaction = 0.056) (Table 2). However, the dominant model (TT vs. TA.AA) showed that subjects carrying the A risk allele in FTO lost less weight (p-interaction = 0.022) (Table 3) and had a lower decrease BMI from baseline to T12 (p-interaction = 0.047) (Table 4) than TT carriers. However, no difference in the recessive model (TT.TA vs AA) was found.
Fig. 1.
Frequencies of genetic variants according to additive genetic model
Fig. 2.
Frequencies of genetic variants according to dominant genetic model
Fig. 3.
Frequencies of genetic variants according to recessive genetic model
Table 2.
The comparison between the subjects’ BMI at baseline with those at 6 months (T6) and 12 months (T12) and its relationship with genetic variants
| p-value | ||||||
|---|---|---|---|---|---|---|
| FTO rs9939609 T > A | TT | TA | AA | Genotypea | Timeb | Interactionc |
|
Baseline T6 T12 |
33.3 (27.7, 38.0) 36.5 (27.8, 37.2) 31.5 (26.6, 35.8) |
31.3 (28.5, 35.6) 30.6 (28.0, 35.1) 30.4 (27.8, 34.6) |
36.7 (33.1, 37.7) 38.0 (33.5, 38.2) 36.1 (32.1, 37.2) |
0.907 | < 0.001 | 0.056 |
| MC4R rs17782313 T > C | TT | TC | CC | Genotypea | Timeb | Interactionc |
|
Baseline T6 T12 |
29.4 (28.4, 36.7) 29.5 (27.9, 36.2) 28.2 (27.0, 35.3) |
31.9 (31.3, 36.7) 31.6 (30.6, 38.0) 31.9 (30.4, 36.1) |
42.6 (40.3, 44.9) 41.6 (39.4, 43.7) 40.8 (38.0, 43.6) |
0.077 | < 0.001 | 0.175 |
| LPL rs326 A > G | AA | AG | GG | Genotypea | Timeb | Interactionc |
|
Baseline T6 T12 |
28.7 (28.2, 37.9) 32.2 (27.1, 37.6) 27.8 (26.1, 36.0) |
29.6 (28.2, 35.9) 29.5 (27.3, 35.6) 29.1 (27.1, 34.7) |
38.6 (31.9, 41.3) 38.3 (31.6, 40.3) 36.9 (31.9, 38.3) |
0.162 | < 0.001 | 0.966 |
| NPY rs16147 C > T | CC | CT | TT | Genotypea | Timeb | Interactionc |
|
Baseline T6 T12 |
28.7 (28.4, 30.3) 29.8 (28.9, 30.7) 27.8 (27.0, 29.8) |
35.6 (29.0, 38.3) 35.1 (28.4, 38.2) 34.6 (28.1, 36.5) |
– | 0.251 | 0.004 | 0.662 |
| IRS-1 rs2943641 C > T | CC | CT | TT | Genotypea | Timeb | Interactionc |
|
Baseline T6 T12 |
31.3 (28.7, 36.7) 31.1 (27.9, 37.4) 30.4 (26.1, 35.1) |
35.6 (28.2, 37.9) 35.1 (28.0, 36.5) 34.6 (27.9, 36.0) |
34.2 (29.4, 40.8) 34.1 (28.8, 40.2) 34.2 (28.7, 40.4) |
0.653 | < 0.001 | 0.342 |
Data are expressed as median and interquartile range. Probability that the effect of nutritional intervention on BMI is influenced by:
a time, for each variable, the differences have been tested between baseline, T6 and T12 of the three Genotypes
b groups, for each variable, the differences have been tested between Genotypes over time
c Probability that the effects of nutritional intervention is greater in one distinct group (interaction Time *genotype)
Table 3.
The comparison between the subjects’ weight at baseline with those at 6 months (T6) and 12 months (T12) and its relationship with FTO gene according to dominant genetic model
| p-value | |||||
|---|---|---|---|---|---|
| FTO rs9939609 T > A | TT | TA + AA | Genotypea | Timeb | Interactionc |
| Baseline | 96.8 (82.2, 101.9) | 95.0 (82.9, 112.9) | 0.720 | < 0.001 | 0.022 |
| T6 | 96.5 (79.5, 100.5) | 96.2 (80.0, 111.5) | |||
| T12 | 88.8 (81.5, 91.8) | 94.5 (77.4, 110.4) | |||
Statistically significant value is given in bold
Data are expressed as median and interquartile range. Probability that the effect of nutritional intervention on weight is influenced by:
a time, for each variable, the differences have been tested between baseline, T6 and T12 of the two Genotypes
b groups, for each variable, the differences have been tested between Genotypes over times
c Probability that the effects of nutritional intervention is greater in one distinct group (interaction Time *genotype)
Table 4.
The comparison between the subjects’ BMI at baseline with those at 6 months (T6) and 12 months (T12) and its relationship with FTO gene according to dominant genetic model
| p-value | |||||
|---|---|---|---|---|---|
| FTO rs9939609 T > A | TT | TA + AA | Genotypea | Timeb | Interactionc |
| Baseline | 33.3 (27.7, 38.0) | 31.6 (29.2, 37.2) | 0.945 | < 0.001 | 0.047 |
| T6 | 36.5 (27.8, 37.2) | 31.1 (28.8, 38.1) | |||
| T12 | 31.5 (26.6, 35.8) | 31.1 (28.1, 36.7) | |||
Statistically significant value is given in bold
Data are expressed as median and interquartile range. Probability that the effect of nutritional intervention on BMI is influenced by:
a time, for each variable, the differences have been tested between baseline, T6 and T12 of the two Genotypes
b groups, for each variable, the differences have been tested between Genotypes over times
c Probability that the effects of nutritional intervention is greater in one distinct group (Time *genotype)
There was a significant change in the PREDIMED score over time, according to the genotypes for FTO rs9939609 (p = 0.025) and NPY rs16147 (p = 0.039), respectively (data not shown). Regarding other participants’ characteristics relating genetic variants, no differences were detected among basal and post-treatment values of waist circumference, lipid profile, and IPAQ scores assuming a dominant, recessive and additive genetic models of inheritance (data not shown). Nevertheless, subjects with the risk-conferring CC genotype in IRS-1 gene had greater decrease in total cholesterol (TC) than those without this genotype (p-interaction = 0.058) (Table 5) across one-year intervention, although not statistically significant.
Table 5.
The comparison between the subjects’ total cholesterol at baseline with those at 6 months (T6) and 12 months (T12) and its relationship with genetic variants
| p-value | ||||||
|---|---|---|---|---|---|---|
| FTO rs9939609 T > A | TT | TA | AA | Genotypea | Timeb | Interactionc |
|
Baseline T6 T12 |
212.0 (197.0, 224.8) 205.0 (196.0, 212.0) 195.5 (189.2, 202.5) |
199.0 (176.5, 203.2) 199.0 (198.0, 252.0) 176.0 (159.5, 209.8) |
222.0 (213.5, 234.5) 243.0 (233.0, 253.0) 243.0 (233.0, 253.0) |
0.332 | 0.859 | 0.214 |
| MC4R rs17782313 T > C | TT | TC | CC | Genotypea | Timeb | Interactionc |
|
Baseline T6 T12 |
207.5 (199.2, 224.8) 198.0 (195.0, 217.5) 198.0 (168.2, 218.2) |
199.0 (183.0, 201.0) 257.0 (228.0, 260.0) 176.0 (164.0, 200.8) |
204.0 (197.5, 210.5) 228.5 (216.8, 240.2) 221.0 (204.5, 237.5) |
0.614 | 0.669 | 0.283 |
| LPL rs326 A > G | AA | AG | GG | Genotypea | Timeb | Interactionc |
|
Baseline T6 T12 |
199.0 (196.0, 201.0) 225.0 (218.5, 241.0) 158.0 (151.0, 204.0) |
210.0 (195.5, 226.5) 198.0 (195.0, 214.0) 195.5 (189.2, 198.0) |
205.0 (199.0, 217.0) 225.5 (212.2, 238.8) 225.0 (186.0, 254.0) |
0.894 | 0.660 | 0.347 |
| NPY rs16147 C > T | CC | CT | Genotypea | Timeb | Interactionc | |
|
Baseline T6 T12 |
199.0 (197.5, 199.0) 225.0 (225.0, 225.0) 166.0 (155.0, 185.0) |
207.5 (193.2, 223.5) 205.0 (197.0, 237.5) 198.0 (186.0, 225.0) |
0.729 | 0.199 | 0.161 | |
| IRS-1 rs2943641 C > T | CC | CT | TT | Genotypea | Timeb | Interactionc |
|
Baseline T6 T12 |
201.0 (196.0, 225.0) 223.0 (202.0, 241.0) 198.5 (187.5, 223.5) |
199.5 (185.8, 206.0) 203.0 (198.5, 207.5) 154.5 (149.2, 168.0) |
207.5 (193.0, 211.8) 198.0 (165.5, 225.0) 226.0 (188.5, 256.2) |
0.416 | 0.609 | 0.058 |
Data are expressed as median and interquartile range. Probability that the effect of nutritional intervention on total cholesterol is influenced by:
a time, for each variable, the differences have been tested between baseline, T6 and T12 of the three Genotypes
b groups, for each variable, the differences have been tested between Genotypes over times
c Probability that the effects of nutritional intervention is greater in one distinct group (Time *genotype)
Discussion
The main aim of the present pilot study was to assess the interaction between variants in 5 candidate genes (namely rs9939609 in FTO, rs17782313 near MC4R, rs326 in LPL, rs16147 in NPY, rs2943641 near IRS-1) and nutritional intervention on the changes in anthropometric and clinical parameters as well as in the PREDIMED score and the physical activity levels of overweight or obese subjects affected by T2D or IGR over one-year period/twelve months. The additive model showed that homozygous subjects for A risk allele in FTO gene presented the highest baseline BMI and had a statistical tendency to lowest body weight loss after one year of nutritional intervention, compared to TT and TA genotypes. The dominant model showed that subjects carrying the A risk allele in FTO lost less weight and had a lower BMI decrease from baseline to T12 than TT carriers, in spite of the same nutritional intervention. In addition, we observed a significant change in the PREDIMED score over time according to genotypes for FTO rs9939609. To date, the exact function of FTO remains undefined, but it should be noted that polymorphisms located in the FTO gene not only represent genetic risk factors for obesity, but have been linked with BMI, gestational diabetes mellitus (GDM), T2D, and eating behavior [39–41]. In addition, it has been suggested that the FTO variants may influence the expression of other genes such as the homeobox gene Iroquois-class homeobox protein 2 (IRX3) rather than the FTO itself [42]. Some studies showed that the FTO rs9939609 is associated with energy homeostasis and body composition, increasing food intake as well as appetitive behaviors reducing response in hunger and satiety after a meal [43–48]. It has also been reported that diet/lifestyle induced weight loss differs among FTO genotypes, although results are contradictory [49–56]. On the other hand, a systematic review and meta-analysis reported significant differences in weight loss between the AA and TT genotypes after dietary intervention [57]. Hosseini-Esfahani et al. [58] demonstrated that a higher adherence to the MedDiet was associated with lower obesity risk in subjects with more genetic predisposition to obesity, compared to those with lower MedDiet adherence and lower genetic risk score. Our results agree with more recent studies, suggesting, even in a non-significant manner, that homozygous carriers of FTO obesity-predisposing allele lose less weight after having followed a 4 week dietary intervention based on the Mediterranean model than non-carriers [59]. To the best of our knowledge, few and contradictory studies have assessed the effects of LPL rs326, NPY rs16147 and near IRS-1 rs2943641 on metabolic response and weight change after a dietary intervention, thus evidence is scarce and limited. In the present study, no difference among basal and post-treatment values of anthropometric variables (BMI, weight, waist circumference, and IPAQ scores) based on genotype in LPL, NPY and IRS-1 was found. Nevertheless, subjects with the risk-conferring CC genotype in IRS-1 gene had greater decreases in Total Cholesterol than those without this genotype across one-year intervention, although not statistically significant. Our finding is in line with a previous study [60], which showed that HDL cholesterol decreased, and serum triglycerides increased for each copy of the rs2943641 risk allele among T2D subjects. The insulin receptor substrate 1 (IRS1) is considered a major mediator between the insulin receptor and phosphatidylinositol 3-kinase (PI3K) in the insulin signaling pathway and it has been reported that rs2943641, located in inter-genic region 500 kb upstream from the IRS1 may have an effect on IRS1 expression associated PI3K activity [61, 62]. Subjects with CC genotype of the rs2943641 showed a higher improvement in insulin resistance than T allele carriers when adhering to a high-carbohydrate and low-fat diet in a 2 year randomized trial [63]. Thus, although the mechanisms underlying the interaction between the IRS1 gene variant and diet on insulin action remain to be elucidated, the activation of IRS1 associated PI3K activity may be enhanced in rs2943641 CC subjects in response to a low-fat and high-carbohydrate diet. In our study, we showed a significant change of the PREDIMED score from baseline to month 12 according to genotypes for NPY rs16147. NPY is an orexigenic neuropeptide that regulates the hypothalamic control of energy, immune function, and cardiovascular function [64–66]. Variants in the NPY gene have been associated with several human diseases stimulating food intake, decreasing energy expenditure, and increasing energy stored [67]. In this view, lymphoblastoid cell lines showed higher NPY expression in risk allele carriers of rs16147, demonstrating allele-specific effects on NPY gene expression and NPY peptide concentrations [68, 69]. Regarding body weight regulation, it has been showed that the rs16147 variant affected anthropometric, biochemical and inflammatory parameters in response to dietary interventions in overweight or obese subjects [70, 71], and its effect on central obesity and abdominal fat distribution were modified by dietary fat, suggesting that individuals carrying the C allele of the NPY rs16147 SNP might benefit more by taking a high-fat weight loss diet [72]. Recently, Martin et al. [73] demonstrated that the A allele of this NPY variant produces a better metabolic response in terms of insulin resistance and basal insulin secondary to weight loss with two different hypocaloric diets in obese subjects, with improvement being higher with the Mediterranean diet. Further studies are needed to elucidate whether genetic variants may influence the outcomes of the nutritional intervention. To date, it should be emphasized that there are discrepancies in previous weight loss clinical studies due to: (i) the types of interventions, including energy-restricted diets, duration of the intervention, and physical activity interventions; (ii) the participants’ heterogeneity in what concerns age (adults, older adults), ancestry, nutritional status (overweight, obese, severely obese), pathologic situation/status (prediabetic, diabetic); (iii) the dietary assessment method that can produce inaccurate dietary data collection [74]. Therefore, with regard to the analysis of gene–diet interactions, all the aspects mentioned above make comparisons between findings difficult and should be considered in order to achieve comparability across different studies. The strength of our study is that it is based on intensive nutritional intervention. However, several limitations of this pilot study warrant consideration. One limitation is that the sample size of the current study is small for a genetic association study. In addition, although personalized nutrition refers mainly to a person’s genetic background, the composition of gut microbiota and epigenetic markers may modify gene expression and could be involved in the outcome of weight loss interventions. In the era of precision medicine, the integration of nutrigenetic, epigenetic, as well as metagenomic data may offer meaningful opportunities for the design of more personalized dietary treatments to optimize an individual’s response to dietary interventions, as well as to improve the prevention and treatment of metabolic disturbances, such as T2M, GDM, hypertension, and cardiovascular diseases [13, 15]. In summary, our study showed an interaction between the modification of the PREDIMED score over time and FTO as well as NPY. Although it remains unknown whether genetic risk for obesity is associated with timing of food choices [75], genetic variants with central nervous system functions could affect the control of food intake and food-choice behavior. Future clinical intervention studies should explore whether food choices may influence the causal pathway between genetic risk and weight gain. In addition, our preliminary findings support the impact of the interaction between FTO gene and diet/lifestyle intervention in the regulation of body weight. Even though the suboptimal statistical power resulting from the small sample size prevents us from clearly demonstrating the effect of the genetic variants on nutritional intervention, we believe that further studies on the role of FTO, a key gene in food intake and appetite, are highly needed with regard to lifestyle intervention. Therefore, an extension of our preliminary findings in larger, multicenter cohorts is warranted.
If FTO rs9939609 is confirmed to influence the decreasing of BMI and body composition during nutritional treatments, this notion can encourage a shift toward both the development of personalized nutritional advice and monitoring based on joint associations of dietary pattern and genotypes. It should be noted that favorable lifestyle should be universally recommended in obesity prevention, regardless of genetic make-up, thus supporting current public health guidelines.
Conclusions
In summary, our pilot study showed an interaction between the modification of the PREDIMED score over time and FTO as well as NPY. In addition, our findings support the interaction between FTO gene and diet/lifestyle intervention in the regulation of body weight. So, we can say that if FTO rs9939609 is confirmed to influence the decreasing of BMI and body composition during nutritional treatments, this can encourage a shift toward both the development of personalized nutritional advice and monitoring based on joint associations of dietary pattern and genotypes.
Acknowledgements
Marica Franzago was supported by Fondazione Umberto Veronesi
Author contributions
Conceptualization, MF and EV; methodology, MF,FF,EV; software, MDN,MM.; formal analysis, MF, MDN, MM; investigation, MF; data curation, MF, FF, EV; writing—original draft preparation, MF and EV; writing—review and editing, MF, MDN,EV, LS; visualization, MM; supervision, EV; project administration EV; Funding acquisition, EV and LS. All authors have read and agreed to the published version of the manuscript.”
Funding
Open access funding provided by Università degli Studi G. D'Annunzio Chieti Pescara within the CRUI-CARE Agreement. This research received no external funding.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
The study was approved by the Ethics Committee of the University of Chieti-Pescara, Italy.
Human and animal rights
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (Ethics Committee of the University of Chieti-Pescara, Italy) and with the Helsinki Declaration of 1975, as revised in 2008.
Informed consent
Written informed consent was obtained from all participants being included in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hruby A, Hu FB. The epidemiology of obesity: a big picture. Pharmacoeconomics. 2015;33(7):673–689. doi: 10.1007/s40273-014-0243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seravalle G, Grassi G. Obesity and hypertension. Pharmacol Res. 2017;122:1–7. doi: 10.1016/j.phrs.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 3.La Sala L, Pontiroli AE. Prevention of diabetes and cardiovascular disease in obesity. Int J Mol Sci. 2020;21(21):8178. doi: 10.3390/ijms21218178.PMID:33142938;PMCID:PMC7663329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romieu I, Dossus L, Barquera S, et al. On behalf of the IARC working group on Energy Balance and Obesity. Energy balance and obesity: what are the main drivers? Cancer Causes Control. 2017;28(3):247–258. doi: 10.1007/s10552-017-0869-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9(3):1–17. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Sayed Moustafa JS, Froguel P. From obesity genetics to the future of personalized obesity therapy. Nat Rev Endocrinol. 2013;9(7):402–413. doi: 10.1038/nrendo.2013.57.Erratum.In:NatRevEndocrinol.2014,10(1):4. [DOI] [PubMed] [Google Scholar]
- 7.Min J, Chiu DT, Wang Y. Variation in the heritability of body mass index based on diverse twin studies: a systematic review. Obes Rev. 2013;14:871–882. doi: 10.1111/obr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177.PMID:25673413;PMCID:PMC4382211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tchang BG, Saunders KH, Igel LI. Best Practices in the Management of Overweight and Obesity. Med Clin North Am. 2021;105(1):149–174. doi: 10.1016/j.mcna.2020.08.018. [DOI] [PubMed] [Google Scholar]
- 10.Nicoletti CF, Cortes-Oliveira C, Pinhel MA, Nonino CB. Bariatric surgery and precision nutrition. Nutrient. 2017;9(9):974. doi: 10.3390/nu9090974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simopoulos AP. Nutrigenetics/Nutrigenomics. Annu Rev Public Health. 2010;31:53–68. doi: 10.1146/annurev.publhealth.031809.130844. [DOI] [PubMed] [Google Scholar]
- 12.San-Cristobal R, Milagro FI, Martinez JA. Future challenges and present ethical considerations in the use of personalized nutrition based on genetic advice. J Acad Nutr Diet. 2013;113:1447–1454. doi: 10.1016/j.jand.2013.05.028. [DOI] [PubMed] [Google Scholar]
- 13.Peña-Romero AC, Navas-Carrillo D, Marín F, Orenes-Piñero E. The future of nutrition: nutrigenomics and nutrigenetics in obesity and cardiovascular diseases. Crit Rev Food Sci Nutr. 2018;58(17):3030–3041. doi: 10.1080/10408398.2017.1349731. [DOI] [PubMed] [Google Scholar]
- 14.Goni L, Cuervo M, Milagro FI, Martínez JA. Future perspectives of personalized weight loss interventions based on nutrigenetic, epigenetic, and metagenomic data. J Nutr. 2015;146(4):905S–912S. doi: 10.3945/jn.115.218354. [DOI] [PubMed] [Google Scholar]
- 15.Franzago M, Santurbano D, Vitacolonna E, Stuppia L. Genes and diet in the prevention of chronic diseases in future generations. Int J Mol Sci. 2020;21(7):2633. doi: 10.3390/ijms21072633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Renzo L, De Lorenzo A (2016) Up to Date Focus on Prevention in Nutrition. 10.19252/000000029
- 17.Di Renzo L, Gualtieri P, Romano L, Marrone G, Noce A, Pujia A, et al. Role of personalized nutrition in chronic-degenerative diseases. Nutrients. 2019;11(8):1707. doi: 10.3390/nu11081707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-González MA, García-Arellano A, Toledo E, et al. A 14-item Mediterranean diet assessment tool and obesity indexes among high risk subjects: the PREDIMED trial. PLoS ONE. 2012;7(8):e43134. doi: 10.1371/journal.pone.0043134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mannocci A, Di Thiene D, Del Cimmuto A, et al. International physical activity questionnaire: validation and assessment in an Italian sample. Italian J Public Health. 2010;7(4):369–376. [Google Scholar]
- 20.Trento M, Passera P, Borgo E, Tomalino M, Bajardi M, Cavallo F, et al. A 5-year randomized controlled study of learning, problem solving ability, and quality of life modifications in people with type 2 diabetes managed by group care. Diabetes Care. 2004;27(3):670–675. doi: 10.2337/diacare.27.3.670. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Xue H, Huang Y, Huang L, Zhang DA. Systematic review of application and effectiveness of health interventions for obesity and diabetes treatment and self-management. Adv Nutr. 2017;8(3):449–462. doi: 10.3945/an.116.014100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trento M, Passera P, Tomalino M, Bajardi M, Pomero F, Allione A, et al. Group visits improve metabolic control in type 2 diabetes: a 2-year follow-up. Diabetes Care. 2001;24(6):995–1000. doi: 10.2337/diacare.24.6.995. [DOI] [PubMed] [Google Scholar]
- 23.Trento M, Passera P, Bajardi M, Tomalino M, Grassi G, Borgo E, et al. Lifestyle intervention by group care prevents deterioration of type ii diabetes: a 4-year randomized controlled clinical trial. Diabetologia. 2002;45(9):1231–1239. doi: 10.1007/s00125-002-0904-8. [DOI] [PubMed] [Google Scholar]
- 24.Evert AB, Dennison M, Gardner CD et al (2019) Nutrition Therapy for Adults With Diabetes or Prediabetes: A Consensus Report. Diabetes Care 42(5):731–754. 10.2337/dci19-0014 [DOI] [PMC free article] [PubMed]
- 25.Sentinelli F, Incani M, Coccia F, Capoccia D, Cambuli VM, Romeo S, et al. Association of FTO polymorphisms with early age of obesity in obese Italian subjects. Exp Diabetes Res. 2012;2012:872176. doi: 10.1155/2012/872176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng S, Zhu Y, Xu F, Ren X, Li X, Lai M. FTO gene polymorphisms and obesity risk: a meta-analysis. BMC Med. 2011;9:71. doi: 10.1186/1741-7015-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scuteri A, Sanna S, Chen WM et al (2007) Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genetics 3(7):e115. 10.1371/journal.pgen.0030115 [DOI] [PMC free article] [PubMed]
- 28.Qi Q, Kilpeläinen TO, Downer MK, Tanaka T, Smith CE, Sluijs I, et al. FTO genetic variants, dietary intake and body mass index: insights from 177,330 individuals. Hum Mol Genet. 2014;20(23):6961–6972. doi: 10.1093/hmg/ddu411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stutzmann F, Cauchi S, Durand E, Calvacanti-Proença C, Pigeyre M, Hartikainen AL, et al. Common genetic variation near MC4R is associated with eating behaviour patterns in European populations. Int J Obes (Lond) 2009;33:373–378. doi: 10.1038/ijo.2008.279. [DOI] [PubMed] [Google Scholar]
- 30.Tang W, Apostol G, Schreiner PJ, Jacobs DR, Jr, Boerwinkle E, Fornage M. Associations of lipoprotein lipase gene polymorphisms with longitudinal plasma lipid trends in young adults: the coronary artery risk development in young adults (CARDIA) study. Circ Cardiovasc Genet. 2010;3(2):179–186. doi: 10.1161/CIRCGENETICS.109.913426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Zhou D, Zhang Z, et al. Effects of genetic variants on lipid parameters and dyslipidemia in a Chinese population. J Lipid Res. 2011;52(2):354–360. doi: 10.1194/jlr.P007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zain SM, Mohamed Z, Jalaludin MY, Fauzi F, Hamidi A, Zaharan NL. Comprehensive evaluation of the neuropeptide-Y gene variants in the risk of obesity: a case-control study and meta-analysis. Pharmacogenet Genomics. 2015;25:501–510. doi: 10.1097/FPC.0000000000000164. [DOI] [PubMed] [Google Scholar]
- 33.Katus U, Villa I, Ringmets I, Veidebaum T, Harro J. Neuropeptide Y gene variants in obesity, dietary intake, blood pressure, lipid and glucose metabolism: A longitudinal birth cohort study. Peptides. 2021;139:170524. doi: 10.1016/j.peptides.2021.170524. [DOI] [PubMed] [Google Scholar]
- 34.Shah SH, Freedman NJ, Zhang L, et al. Neuropeptide Y gene polymorphisms confer risk of early-onset atherosclerosis. PLoS Genet. 2009;5:e1000318. doi: 10.1371/journal.pgen.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41:1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 36.Attia J, Thakkinstian A, D'Este C. Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol. 2003;56:297–303. doi: 10.1016/S0895-4356(03)00011-8. [DOI] [PubMed] [Google Scholar]
- 37.Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol. 2005;34:1319–1328. doi: 10.1093/ije/dyi169. [DOI] [PubMed] [Google Scholar]
- 38.Salanti G, Southam L, Altshuler D et al (2009) Underlying genetic models of inheritance in established type 2 diabetes associations. Am J Epidemiol 170(5):537–545. 10.1093/aje/kwp145 [DOI] [PMC free article] [PubMed]
- 39.Castellini G, Franzago M, Bagnoli S, Lelli L, Balsamo M, Mancini M, et al. Fat mass and obesity-associated gene (FTO) is associated to eating disorders susceptibility and moderates the expression of psychopathological traits. PLoS ONE. 2017;12(3):e0173560. doi: 10.1371/journal.pone.0173560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franzago M, Fraticelli F, Nicolucci A, et al. Molecular analysis of a genetic variants panel related to nutrients and metabolism: association with susceptibility to gestational diabetes and cardiometabolic risk in affected women. J Diabetes Res. 2017 doi: 10.1155/2017/4612623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fairbrother U, Kidd E, Malagamuwa T, Walley A. Genetics of severe obesity. Curr DiabRep. 2018;18(10):85. doi: 10.1007/s11892-018-1053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gholamalizadeh M, Jarrahi AM, Akbari ME, Rezaei S, Doaei S, Mokhtari Z, Torki A. The possible mechanisms of the effects of IRX3 gene on body weight: an overview. Arch Med Sci Atherosclerotic Dis. 2019;4:e225. doi: 10.5114/amsad.2019.87545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Church C, Moir L, McMurray F, et al. Overexpression of Ftoleads to increased food intake and results in obesity. Nat Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi Q, Kilpelainen TO, Downer MK, et al. FTO genetic variants, dietary intake and body mass index: insights from 177,330 individuals. Hum Mol Genet. 2014;23(6961–72):10. doi: 10.1093/hmg/ddu411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wardle J, Carnell S, Haworth CM, Farooqi IS, O’Rahilly S, Plomin R. Obesity associated genetic variation in FTO is associated with di-minished satiety. J Clin Endocrinol Metab. 2008;93:3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- 46.Qi Q, Downer MK, Kilpelainen TO, Taal HR, Barton SJ, Ntalla I, Standl M, et al. Dietary in-take, FTO genetic variants, and adiposity: a combined analysis of over16,000 children and adolescents. Diabetes. 2015;64:2467–2476. doi: 10.2337/db14-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.den Hoed M, Westerterp-Plantenga MS, Bouwman FG, Mariman EC, Westerterp KR. Postprandial responses in hunger and satiety are as-sociated with the rs9939609 single nucleotide polymorphism in FTO. Am J Clin Nutr. 2009;90:1426–1432. doi: 10.3945/ajcn.2009.28053. [DOI] [PubMed] [Google Scholar]
- 48.Karra E, O’Daly OG, Choudhury AI, et al. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J Clin Invest. 2013;123:3539–3551. doi: 10.1172/JCI44403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grau K, Hansen T, Holst C, Astrup A, et al. Macronutrient-specific effect of FTO rs9939609 in response to a 10-week randomized hypo-energetic diet among obese Europeans. Int J Obes (Lond) 2009;33:1227–1234. doi: 10.1038/ijo.2009.159. [DOI] [PubMed] [Google Scholar]
- 50.Razquin C, Martinez JA, Martinez-Gonzalez MA, Bes-Rastrollo M, Fernández-Crehuet J, Marti A. A 3-year intervention with a Mediterranean diet modified the association between the rs9939609 gene variant in FTO and body weight changes. Int J Obes (Lond) 2010;34(2):266–272. doi: 10.1038/ijo.2009.233. [DOI] [PubMed] [Google Scholar]
- 51.Mitchell JA, Church TS, Rankinen T, Earnest CP, Sui X, Blair SN. FTOgenotype and the weight loss benefits of moderate intensity exercise. Obesity (Silver Spring) 2010;18:641–643. doi: 10.1038/oby.2009.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Luis DA, Aller R, Conde R, Izaola O, Gonzalez Sagrado M, Castrodeza SJ. The rs9939609 gene variant in FTO modified the metabolic response of weight loss after a 3-month intervention with a hypocaloric diet. J Investig Med. 2013;61:22–26. doi: 10.2310/JIM.0b013e318276161d. [DOI] [PubMed] [Google Scholar]
- 53.Verhoef SP, Camps SG, Bouwman FG, Mariman EC, Westerterp KR. Genetic predisposition, dietary restraint and disinhibition in relation toshort and long-term weight loss. Physiol Behav. 2014;128:247–251. doi: 10.1016/j.physbeh.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X, Qi Q, Zhang C, Smith SR, Hu FB, Sacks FM, Bray GA, Qi L (2012) FTO genotype and 2-year change in body composition and fat distribution in response to weight-loss diets: the POUNDS LOST Trial. Diabetes 61(11):3005–3011. 10.2337/db11-1799 [DOI] [PMC free article] [PubMed]
- 55.Ortega-Azorín C, Sorlí JV, Asensio EM, et al. Associations of the FTO rs9939609 and the MC4R rs17782313 polymorphisms with type 2 diabetes are modulated by diet, being higher when adherence to the Mediterranean diet pattern is low. Cardiovasc Diabetol. 2012;11:137. doi: 10.1186/1475-2840-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livingstone KM, Celis-Morales C, Lara J et al (2015) Associations between FTO genotype and total energy and macronutrient intake in adults: a systematic review and meta-analysis. Obes rev 16(8):666–678. 10.1111/obr.12290 [DOI] [PubMed]
- 57.Xiang L, Wu H, Pan A, Patel B, Xiang G, Qi L, Kaplan RC, Hu F, Wylie-Rosett J, Qi Q. FTO genotype and weight loss in diet and lifestyle interventions: a systematic review and meta-analysis. Am J Clin Nutr. 2016;103(4):1162–1170. doi: 10.3945/ajcn.115.123448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hosseini-Esfahani F, Koochakpoor G, Daneshpour MS, Sedaghati-Khayat B, Mirmiran P, Azizi F. Mediterranean dietary pattern adherence modify the association between FTO genetic variations and obesity phenotypes. Nutrients. 2017;9:E1064. doi: 10.3390/nu9101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Renzo L, Cioccoloni G, Falco S, et al. Influence of FTO rs9939609 and Mediterranean diet on body composition and weight loss: a randomized clinical trial. J Transl Med. 2018;16:308. doi: 10.1186/s12967-018-1680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma R, Prudente S, Andreozzi F, Powers C, Mannino G, Bacci S et al (2011) The type 2 diabetes and insulin-resistance locus near IRS1 is a determinant of HDL cholesterol and triglycerides levels among diabetic subjects. Atherosclerosis 216(1):157–160. 10.1016/j.atherosclerosis.2011.01.022 [DOI] [PMC free article] [PubMed]
- 61.Copps KD, White MF. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia. 2012;55(10):2565–2582. doi: 10.1007/s00125-012-2644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boucher J, Kleinridders A, Kahn CR. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014;6(1):a009191. doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qi Q, Bray GA, Smith SR, Hu FB, Sacks FM, Qi L. Insulin receptor substrate 1 gene variation modifies insulin resistance response to weight-loss diets in a 2-year randomized trial: the preventing overweight using novel dietary strategies (POUNDS LOST) trial. Circulation. 2011;124:563–571. doi: 10.1161/CIRCULATIONAHA.111.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dryden S, Pickavance L, Frankish HM, Williams G. Increased neu-ropeptide Y secretion in the hypothalamic paraventricular nucleus ofobese (fa/fa) Zucker rats. Brain Res. 1995;690(185–8):4. doi: 10.1016/0006-8993(95)00628-4. [DOI] [PubMed] [Google Scholar]
- 65.Kuo LE, Kitlinska JB, Tilan JU, Li L, Baker SB, Johnson MD, et al. Neuropeptide Y actsdirectly in the periphery on fat tissue and mediates stress-inducedobesity and metabolic syndrome. Nat Med. 2007;13:803–811. doi: 10.1038/nm1611. [DOI] [PubMed] [Google Scholar]
- 66.Wheway J, Mackay CR, Newton RA, Sainsbury A, Boey D, Herzog H, Mackay F. A fundamental bimodal role for neuropeptide Y1 receptor in the immune system. J Exp Med. 2005;202:1527–1538. doi: 10.1084/jem.20051971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X, Qi Q, Liang J, Hu FB, Sacks FM, Qi L. Neuropeptide ypromoter polymorphism modifies effects of a weight-loss diet on 2-year changes of blood pressure: the preventing overweight using noveldietary strategies trial. Hypertension. 2012;60:1169–1175. doi: 10.1161/HYPERTENSIONAHA.112.197855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou Z, Zhu G, Hariri AR, Enoch MA, Scott D, Sinha R, et al. Genetic variation in humanNPY expression affects stress response and emotion. Nature. 2008;452:997–1001. doi: 10.1038/nature06858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buckland PR, Hoogendoorn B, Guy CA, et al. A high proportion ofpolymorphisms in the promoters of brain expressed genes influencestranscriptional activity. Biochim Biophys Acta. 2004;1690:238–249. doi: 10.1016/j.bbadis.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 70.de Luis D, Izaola O, de la Fuente B, et al. Polymorphism of neuropeptide Y gene rs16147 modifies the response to a hypocaloric diet on cardiovascular risk biomarkers and adipokines. J Hum Nutr Diet. 2017;30:159–165. doi: 10.1111/jhn.12406. [DOI] [PubMed] [Google Scholar]
- 71.de Luis DA, Izaola O, Primo D, Aller R. Polymorphism rs16147 of the neuropeptide Y gene modifies the response of cardiovascular risk biomarkers and adipokines to two hypocaloric diets. Lifestyle Genomics. 2017;10(1–2):63–72. doi: 10.1159/000478528. [DOI] [PubMed] [Google Scholar]
- 72.Lin X, Qi Q, Zheng Y, et al. Neuropeptide Y genotype, central obesity, and abdominal fat distribution: the POUNDS LOST trial. Am J Clin Nutr. 2015;102:514–519. doi: 10.3945/ajcn.115.107276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Primo Martín D, Izaola Jáuregui O, López Gómez JJ et al (2020) Effect of a Mediterranean-pattern diet on the metabolic response secondary to weight loss; role of the single nucleotide polymorphism (rs16147) of neuropeptide Y. Nutr Hosp 37(4):742–749. Spanish. 10.20960/nh.02941 [DOI] [PubMed]
- 74.Archer E, Marlow ML, Lavie CJ. Controversy and Debate: Memory Based Methods Paper 3: Nutrition’s ‘Black Swans’: Our reply. J Clin Epidemiol. 2018;104:130–135. doi: 10.1016/j.jclinepi.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 75.Dashti HS, Hivert M-F, Levy DE, McCurley JL, Saxena R, Thorndike AN. Polygenic risk score for obesity and the quality, quantity, and timing of workplace food purchases: A secondary analysis from the ChooseWell 365 randomized trial. PLoS Med. 2020;17(7):e1003219. doi: 10.1371/journal.pmed.1003219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.