Abstract
In neurodegenerative diseases, extracellular vesicles (EVs) transfer pathogenic molecules and are consequently involved in disease progression. We have investigated the proteomic profiles of EVs that were isolated from four different human‐induced pluripotent stem cell‐derived neural cell types (excitatory neurons, astrocytes, microglia‐like cells, and oligodendrocyte‐like cells). Novel cell type‐specific EV protein markers were then identified for the excitatory neurons (ATP1A3, NCAM1), astrocytes (LRP1, ITGA6), microglia‐like cells (ITGAM, LCP1), and oligodendrocyte‐like cells (LAMP2, FTH1), as well as 16 pan‐EV marker candidates, including integrins and annexins. To further demonstrate how cell‐type‐specific EVs may be involved in Alzheimer's disease (AD), we performed protein co‐expression network analysis and conducted cell type assessments for the proteomes of brain‐derived EVs from the control, mild cognitive impairment, and AD cases. A protein module enriched in astrocyte‐specific EV markers was most significantly associated with the AD pathology and cognitive impairment, suggesting an important role in AD progression. The hub protein from this module, integrin‐β1 (ITGB1), was found to be significantly elevated in astrocyte‐specific EVs enriched from the total brain‐derived AD EVs and associated with the brain β‐amyloid and tau load in independent cohorts. Thus, our study provides a featured framework and rich resource for the future analyses of EV functions in neurodegenerative diseases in a cell type‐specific manner.
1. INTRODUCTION
Alzheimer's disease (AD) is a pervasive neurodegenerative brain disease, and alongside other types of dementia, it affects an estimated 44 million people worldwide (Collaborators, G.B.D.D., 2019). Although AD is currently characterized by the presence of amyloid‐β (Aβ) plaques and tau neurofibrillary tangles in the brain (Jack et al., 2018), the molecular and cellular changes during the progression of the disease beyond Aβ and tau depositions have remained elusive.
Recently, extracellular vesicles (EVs) have attracted considerable interest as they play a critical role in intercellular communication during both physiological and pathological conditions (Delpech et al., 2019; Thompson et al., 2016; You & Ikezu, 2019). Mounting evidence has revealed that the cargo contents of EVs that are isolated from brain tissues and biofluids, are dynamically altered by neurodegenerative diseases, including AD, and they reflect a disease‐specific molecular signature. For instance, AD brain‐derived EVs have been shown to contain pathogenic proteins, such as Aβ, hyper‐phosphorylated tau, and α‐synuclein (Muraoka, Deleo, et al., 2020; Sardar Sinha et al., 2018). The combination of annexin A5 (ANXA5), VGF nerve growth factor inducible (VGF), glycoprotein M6A (GPM6A), and actin‐related protein 1A (ACTZ) in AD brain‐derived EVs has enabled them to be distinguished from controls with 88% accuracy (Muraoka, Deleo, et al., 2020). A more recent study using 16‐plex tandem mass tag (TMT)‐based quantitative proteomics of cerebrospinal fluid (CSF)‐derived EVs identified significant elevations in the heat shock protein family A (Hsp70) member 1A (HSPA1A) and aminopeptidase puromycin‐sensitive (NPEPPS) in AD CSF EVs when compared to CSF EVs isolated from patients with mild cognitive impairment (MCI), and this enabled AD to be distinguished from MCIs with a 91% accuracy (Muraoka, Jedrychowski, Yanamandra, et al., 2020). Therefore, EVs may serve as a reliable resource for future developments in disease diagnosis, prognosis, and therapy.
Every cell type in the central nervous system (CNS), including the neurons and glia, are known to shed EVs (Budnik et al., 2016; You & Ikezu, 2019) and they are then secreted into biofluids such as blood, urine, and CSF (Chiasserini et al., 2014; A. G. Thompson et al., 2016). A growing number of studies have reported the immuno‐based capture of cell type‐specific EVs from biofluids, mostly plasma samples, to assess the pathological processes in neurodegenerative disorders. For example, plasma neuron‐derived EVs isolated with anti‐L1 cell adhesion molecule (L1CAM) antibody showed significantly increased levels of Aβ1–42 in AD cohorts 1–10 years before disease onset and could thus possibly be used to predict AD (Fiandaca et al., 2015). In addition, plasma astrocyte‐derived EVs isolated with anti‐GLAST/EAAT1 (an astrocyte specific glutamate aspartate transporter/excitatory amino acid transporter 1) antibodies showed significantly increased levels of complement components in the AD cohorts (Goetzl et al., 2018; Nogueras‐Ortiz et al., 2020). Thus, capturing cell type‐specific EVs from patient‐derived biofluids or biopsies and profiling their contents could be utilized as a novel method for studying the pathophysiology of AD. However, despite previous studies on the immunocapture of brain cell‐derived EVs from the patient plasma or serum, a consensus on cell type‐specific EV markers is lacking as information on cell type‐specific EV proteomic datasets from human samples is currently limited (Norman et al., 2021). To address these concerns, we sought to define human brain cell type‐specific EV protein signatures that can distinguish EVs from distinct subsets of brain cells.
Here we isolated EVs that were secreted from cultured excitatory neurons, astrocytes, microglia‐like cells, and oligodendrocyte‐like cells derived from human induced pluripotent stem cells (hiPSCs). We analyzed the EV samples using label‐free mass spectrometry (MS) and identified 16 common proteins shared by all EV samples, which will be useful to help improve EV isolation from human brain tissues. Moreover, our data revealed very distinct protein profiles across the different brain cell‐derived EVs that reflect the unique EV cargo loading of each cell type, providing comprehensive and unbiased information for cell type‐specific EV biomarker discovery.
To demonstrate the utility of these EV protein signatures, we analyzed 30 human brain‐derived EV samples that were purified from the frozen brain tissues of healthy controls (HC), MCI, and AD patients, using a MS‐based protein quantification approach with isobaric multiplex TMTs (Johnson et al., 2020; Ting et al., 2011). We then used weighted correlation network analysis (WGCNA) to identify significant modules of co‐expressed proteins linked to specific cell types, biological pathways, and disease processes (Langfelder & Horvath, 2008; Miller et al., 2008). The most centrally correlated proteins with these co‐expression modules are enriched in key drivers that may play prominent roles in pathogenesis (Barabási et al., 2011; Johnson et al., 2020). We generated AD brain‐derived EV protein co‐expression networks related to distinct functions and brain cell types using cell type‐specific EV proteins. One of the most strongly altered AD EV protein co‐expression modules was enriched with astrocyte‐derived EVs and plasma membrane molecules, which overlap with the EV profiles from the activated astrocytes, as we have previously reported (You et al., 2020). Our results demonstrate an important role for cell type‐specific EVs in assessing AD progression and provide a framework for future EV studies on neurodegenerative diseases.
2. MATERIALS AND METHODS
2.1. Human induced pluripotent stem cell (hiPSC) cultures
The hiPSC cell line ADRC iPS clone 4 (male; healthy individual; APOE; Apolipoprotein E, ε3/3 allele) was provided by UCI (University of California, Irving) Alzheimer's Disease Research Center (ADRC) Induced Pluripotent Stem Cell Core (http://stemcells.mind.uci.edu/). Pluripotency of the iPSCs was verified using the Human Pluripotent Stem Cell Marker Antibody Panel (anti‐OCT3/4; octamer transcription factor‐3/4, anti‐NANOG; homeobox protein NANOG, anti‐alkaline phosphatase, anti‐SSEA‐4, stage‐specific embryonic antigen‐4, R&D Systems, SC008) according to the manufacturer's instructions. The iPSCs were maintained in feeder‐free conditions coated with human embryonic stem cell‐qualified Matrigel (Corning, 354277) in complete mTeSR1 (StemCell Technologies, 05825) or TeSR‐E8 medium (StemCell Technologies, 05990) in a humidified incubator (5% CO2, 37°C). hiPSCs were fed fresh media daily and passaged every 5 days. All cell lines used were authenticated and tested negative for mycoplasma.
2.2. Differentiation of hiPSCs into brain cell types
Rapid induction of the hiPSCs into human excitatory neurons was performed via a doxycycline‐inducible neurogenin‐2 (NGN2) system as previously described (Guix et al., 2018). hiPSCs were differentiated into hematopoietic progenitor cells (HPCs) and subsequently to microglia as previously described with minor modifications (Mcquade et al., 2018); hiPSCs were differentiated into neural progenitor cells (NPCs) and subsequently to astrocytes as previously described, with some modifications (Tcw et al., 2017); rapid induction of NPCs into oligodendrocytes was performed via a doxycycline‐inducible overexpression of oligodendroglial transcriptional factors (Sox10; SRY‐Box transcription factor 10, and Olig2; oligodendrocyte transcription factor 2) as previously described, with minor modifications (Ehrlich et al., 2017). Methodological details are provided in the Supplementary Materials and Methods.
2.3. Immunocytochemistry (ICC)
hiPSCs and differentiated neural cells were washed three times with 1 × Dulbecco's phosphate‐buffered saline (DPBS) and fixed in 4% paraformaldehyde at 37°C for 15 min. Cells were permeabilized and blocked in a blocking solution (1 × PBS, 5% bovine serum albumin [BSA, Sigma‐Aldrich, A7906], 5% goat [ThermoFisher Scientific, 16210‐064] or donkey serum [Sigma‐Aldrich, D9663], and 1% Triton X‐100 [Sigma‐Aldrich, T9284]) at room temperature (25℃) for 1 h. Cells were then labelled overnight at 4°C and the primary antibodies used for hiPSCs were anti‐OCT3/4, anti‐NANOG, anti‐alkaline phosphatase, and anti‐SSEA‐4 (all 1:100, R&D Systems, SC008). The NPCs were anti‐paired Box 6 (PAX6, 1:1000, R&D, AF8150), anti‐ SRY‐Box transcription factor 2 (SOX2, 1:1000, R&D, AF1828), and anti‐NESTIN (1:1000, R&D, MAB1259); the hiPSC‐neurons were anti‐MAP2 (1: 1000, Millipore, AB5622), anti‐RNA binding Fox‐1 homolog 3 (NeuN, 1:50, Chemicon, MAB377B), and anti‐synaptophysin (SYP, 1:1000, Sigma‐Aldrich, S5768); the hiPSC‐microglia‐like cells were anti‐Triggering Receptor Expressed On Myeloid Cells 2 (TREM2, 1:1000, R&D, AF1828) and anti‐Purinergic Receptor P2Y12 (P2RY12, 1:1000, Sigma, HPA014518); for the hiPSC‐astrocytes they were anti‐glial fibrillar acidic protein (GFAP, 1:700, Agilent, Z033429), anti‐Vimentin (1:200, Thermo Fisher Scientific, 14‐9897‐82), and anti‐APOE (1:500, Sigma‐Aldrich, 178479); the iPSC‐oligodendrocytes were anti‐O4 (1:500, R&D Systems, MAB1326) and anti‐myelin basic protein (MBP, 1:500, R&D Systems, MAB42282). Alexa Fluor® secondary antibodies (Life Technologies) were conjugated to the target species at room temperature (25℃) for 1 h in blocking buffer. DAPI (4′,6‐diamidino‐2‐phenylindole, 1:2500, Thermo Fisher Scientific, D1306) was used to label the nuclei. Images were acquired using a Leica SP8 confocal microscope (Leica).
2.4. Isolation of iNeuron‐, iMGL‐, iAstrocyte‐, and iOligo‐derived extracellular vesicles
iNeurons were cultured in a 75‐cm (Jack et al., 2018) flask at a density of 7.5 × 106 cells/flask with three independent replicates. Media for the iNeurons D17–21 was collected and processed for further iNeuron‐EV isolation. iMGLs were plated in a six‐well plate at a density of 1 × 106 cells/well, and media on the D31–36 were collected for iMGL‐EV isolation with three replicates. iAstrocytes were cultured in a 75‐cm (Jack et al., 2018) flask at a density of 7.5 × 106 cells/flask with three replicates. Conditioned media on D60–D66 were then collected for iAstrocyte‐EV isolation. iOligo cells were plated in a 75‐cm (Jack et al., 2018) flask at a density of 7.5 × 106 cells/flask with three replicates. Conditioned media on D14–D24 were then collected for iOligo‐EV isolation. All fresh media used were ultra‐centrifuged at 100,000g for 18 h at 4°C (Beckman Coulter, Brea, CA) to remove exogenous exosomes and other EVs. Thirty minutes before collecting the conditioned media, 25 mM potassium chloride (KCl, P9333, all from Sigma–Aldrich) was added to stimulate the cells for complete EV release (Fauré et al., 2006; Stebbing et al., 2015). iNeuron‐EVs, iMGL‐EVs, iAstrocyte‐EVs, and iOligo‐EVs were isolated by differential centrifugation, as described in Figure 1b. First, the medium was centrifuged at 300g for 10 min at room temperature (25℃) to remove floating cells. The remaining supernatant was centrifuged at 2000g for 10 min at 4°C to remove cell debris, sequentially 10,000g for 30 min at 4°C to remove large microvesicles and apoptotic bodies, and then ultra‐centrifuged at 100,000g overnight at 4°C in a 41Ti rotor (Optima‐XE SW41, Beckman Coulter) to obtain small EV pellets. The final pellets were resuspended in 1 mL cold PBS and loaded on qEV original size exclusion chromatography columns (qEVoriginal, Izon Science, Christchurch, New Zealand) to harvest fractions (500 μl each) according to the manufacturer's instructions. Each fraction was analyzed using a nanoparticle tracking analysis to determine the pooled EV‐enriched fractions (f8‐11).
FIGURE 1.
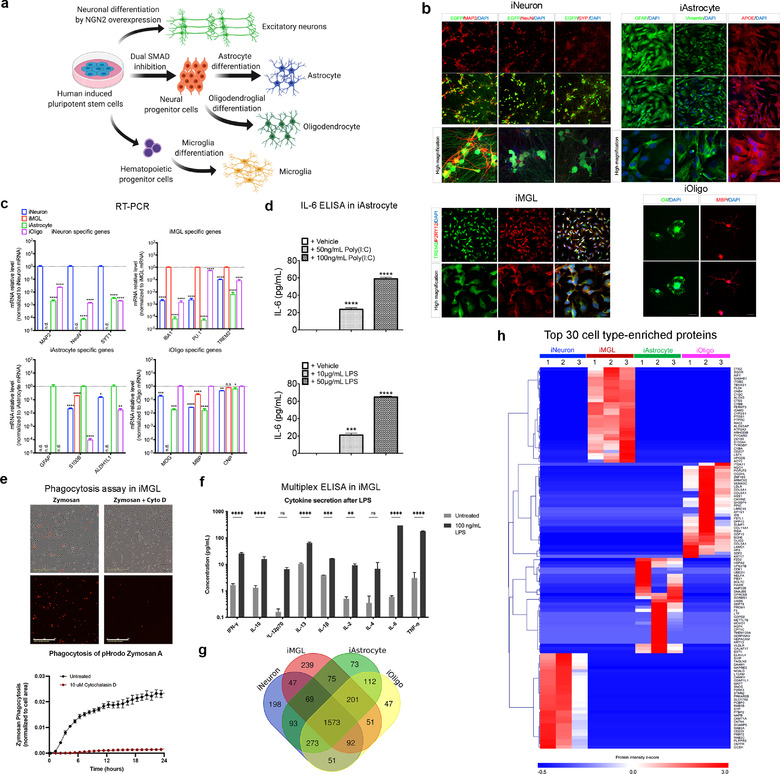
Brain cell type differentiation and characterization. (a) Schematics for the excitatory neuron, astrocyte, microglia, and oligodendrocyte differentiation from hiPSCs. (b) Representative immunofluorescent images of cell type specific markers. iPSC‐induced excitatory neurons (iNeurons) were stained for neuron‐specific markers (MAP2, NeuN, and SYP; synaptophysin). Scale bar, 75 and 20 μm (high magnification). iPSC‐derived microglia‐like cells (iMGLs) were stained for microglia‐specific markers (TREM2 and P2RY12). Scale bar, 30 μm (upper) and 10 μm (lower). iPSC‐derived astrocytes (iAstrocytes) were stained for astrocyte‐specific markers (GFAP, Vimentin, and APOE). Scale bar, 75 and 20 μm (high magnification). iPSC‐derived oligodendrocyte‐like cells (iOligos) were stained for oligodendrocyte‐specific markers (O4 and MBP). Scale bar, 75 μm. (c) qRT‐PCR of cell type‐specific markers for iNeuron (MAP2, NeuN and SYT1), iMGL (IBA1, PU.1 and TREM2), iAstrocyte (GFAP, S100B and ALDH1L1), and iOligo (MOG, MBP and CNP) showing the relative mRNA levels of these genes normalized by the specified cell type. Data are presented as mean ± SEM. n.d., non‐determined; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001, determined by one‐way ANOVA with Dunnett's multiple comparisons. (d) IL‐6 secretion following 24 h of treatment with poly(I:C) and lipopolysaccharide in iAstrocytes (n = 3). Data are presented as the mean ± SEM using one‐way ANOVA with Tukey multiple comparisons. ***p < 0.001 and ****p < 0.0001. (e) Phagocytosis of pHrodo‐tagged Zymosan A over 24 h (representative images at 24 h) in iMGLs determined by fluorescence intensity. Inhibition of actin polymerization with Cytochalasin D (10 μM, 30 min pre‐treatment) attenuated phagocytosis. n = 4 wells (four images per well, imaged hourly on Incucyte S3). Scale bar, 200 μm. (f) Stimulation of iMGLs with 100 ng/ml LPS for 24 h increased pro‐inflammatory cytokine secretion (n = 4). Data are presented as mean ± SEM using two‐way ANOVA with Sidak's multiple comparisons. n.s., no significance, **p < 0.01, ***p < 0.001, and ****p < 0.0001. (g) Venn diagram showing the number of differentially identified proteins in cell lysates from iNeuron, iMGL, iAstrocyte, and iOligo. (h) Heatmap illustrating the top 30 enriched proteins across the four cell types based on their protein intensity as determined from mass spectrometry. Some common cell type‐specific protein markers were identified in iNeuron (e.g., MAPT), iMGL (e.g., AIF1, PU.1), iAstrocyte (e.g., AQP4), and iOligo (e.g., OLIG2). MAP2, microtubule associated protein2; NeuN, RNA binding Fox‐1 homolog 3; SYP, synaptophysin; TREM2, Triggering Receptor Expressed On Myeloid Cells 2; P2RY12, purinergic receptor P2RY12; GFAP, glial fibrillar acidic protein; APOE, apolipoprotein E; MBP, myelin basic protein; SYT1, synaptotagmin 1; IBA1/AIF1, ionized calcium binding adaptor molecule 1/allograft inflammatory factor 1; PU.1, transcription factor PU.1; S100B, S100 calcium‐binding protein B; ALDH1L1, 10‐formyltetrahydrofolate dehydrogenase; MOG, myelin oligodendrocyte glycoprotein; MBP, myelin basic protein; CNP, 2′,3′‐cyclic nucleotide 3′‐phosphodiesterase; SEM, standard error of the mean; ANOVA, analysis of variances; LPS, lipopolysaccharide; MAPT, microtubule associated protein tau; AQP4, aquaporin 4; OLIG2, oligodendrocyte transcription factor 2
2.5. Nanoparticle tracking analysis (NTA) of extracellular vesicles
Both the size distribution and concentration of the EVs were analyzed by NTA using a NanoSight NS300 instrument (Malvern, Worcestershire, UK), as previously described (You et al., 2020). Briefly, EVs were pre‐diluted in PBS to 600 μl to achieve a concentration within the 107–108 range for optimal analysis. Diluted EVs were injected into the sample‐carrier cell, and the particles were automatically tracked and sized using Brownian motion and diffusion coefficients for each sample. The parameters were set at 22.5°C ± 0.5°C for detection level 5 and camera level 13–15. Four recordings of 30 s were acquired for each sample. The mean size (nm) and concentration (particles/mL) of the EVs were both calculated by integrating the data from the four records.
2.6. Transmission electron microscopy (TEM)
The cell type‐specific and human brain‐derived EVs were analyzed using TEM at the Electron Microscopy Facility, Harvard Medical School, as previously described (Muraoka, Jedrychowski, Yanamandra, et al., 2020). A total of 5 μl of an EV sample was adsorbed for 1 min on a carbon‐coated mesh grid (Electron Microscopy Sciences; www.emsdiasum.com) that had been made hydrophilic by a 20‐s exposure to a glow discharge (25 mA). After removing excess liquid, the grid was floated briefly on a drop of water to wash away the phosphate or salt, blotted on a filter paper, and then stained with 0.75% uranyl formate (Electron Microscopy Sciences) for 30 s. After removing the excess uranyl formate, the grids were examined, and random fields were photographed using a JEOL 1200EX TEM with an AMT 2k CCD camera.
2.7. Processing of cell type‐specific cell lysates and EV proteins and label‐free quantitative LC‐MS/MS proteomics
Cell lysates were extracted from hiPSC‐derived cells in RIPA buffer (Thermo Fisher Scientific, 89900) with proteasome inhibitors (Thermo Fisher Scientific, PI78443). EV‐enriched fractions were resuspended in 1% Triton X‐100 with proteasome inhibitors in PBS for protein extraction. The EVs were then sonicated to form a homogeneous protein suspension. Proteins in cell lysates or EVs were quantified using the Pierce BCA assay kit according to the manufacturer's protocol (Thermo Fisher Scientific, 23225). Details of the label‐free quantitative LC‐MS/MS proteomics and bioinformatics analysis are provided in the Supplementary Materials and Methods.
2.8. Brain sample acquisitions
Two cohorts of brain tissue (frontal cortical grey matter) were used in this study. The first cohort for the proteomics study was obtained from the Mount Sinai NIH Brain Bank and Tissue Repository (NBTR; nine AD, eight MCI, and six healthy controls) and the Greater Los Angeles Veteran's Affairs Hospital (two AD and five healthy controls) through the National Institutes of Health (NIH) NeuroBioBank, which were matched for age and sex. The second cohort used in the validation stage was obtained from the University of Maryland (five AD and five healthy controls). All AD, MCI, and control cases were diagnosed neuropathologically. The demographic information for all cases, including age, sex, postmortem interval (PMI), cognitive function, neuropathological criteria, and disease status, is provided in Supplementary Table S8. The Institutional Review Board at the Mount Sinai NBTR, the Greater Los Angeles Veteran's Affairs Hospital, the University of Maryland, and the NIH NeuroBioBank approved the brain acquisitions, and all participants provided informed consent.
2.9. Isolation of EVs from human brain tissues
Unfixed frozen tissue (frontal cortical grey matter, 0.5 g) from deceased AD, MCI, or control cases were processed for EV extraction using our published protocol (Muraoka, Deleo, et al., 2020; Muraoka, Lin, et al., 2020). Methodological details are provided in Supplementary Materials and Methods.
2.10. Western blot
Brain tissues (total with collagenase) and EV fractions were lysed in TENT buffer (50 mM Tris HCl [pH 7.5], 2 mM EDTA, 150 mM NaCl, 1% Triton X‐100) and sonicated for 20 min. Equivalent amounts of brain or EV proteins (5 μg) were determined using the BCA assay in Laemlli sample buffer (Cat# 1610737, BioRad) without a reducing agent, and they were loaded on 4%–20% TGX stain‐free SDS‐PAGE gels (Cat# 4568096, BioRad) or normal SDS‐PAGE gels (Cat# 4561096, Bio‐Rad) and then electrotransferred onto 0.45 μm nitrocellulose membranes (Cat# 1620115, Bio‐Rad). The membranes were then blocked in freshly prepared 8% non‐fat milk (Cell Signaling Technology, 9999S) diluted in PBS and immunoblotted with specific primary antibodies overnight at 4°C. These primary antibodies were anti‐neural cell adhesion molecule 1 (NCAM1, 1:200, Thermo Fisher Scientific, MA5‐11563), anti‐ ATPase Na+/K+ transporting subunit alpha 3 (ATP1A3, 1:500, Thermo Fisher Scientific, MA3915), anti‐L1 cell adhesion molecule (L1CAM, 1:200, eBioscience, 14‐1719‐82), anti‐EAAT1 (1:250, Abcam, ab416), anti‐ low‐density lipoprotein receptor‐related protein 1 (LRP1, 1:250, Thermo Fisher Scientific, 37–7600), anti‐integrin alpha 6 (ITGA6/CD49f, 1:50, Novus Biologicals, NBP1‐85747), anti‐ITGAM/CD11b (1:200, Thermo Fisher Scientific, 14‐0112‐85), anti‐L‐Plastin (LCP1, 1:50, Thermo Fisher Scientific, MA5‐11921), anti‐ferritin heavy chain 1 (FTH1, 1:500, Abcam, ab65080), anti‐myelin oligodendrocyte glycoprotein (MOG, 1:500, Abcam, ab32760), anti‐lysosome associated membrane protein 2 (LAMP2, 1:200, Thermo Fisher Scientific, PA1‐655), anti‐CD81 (1:500, BioLegend, 349502), anti‐CD9 (1:200, Millipore, CBL162), anti‐Cytochrome C (CytoC, 1:500, Cell Signaling Technology, 11940S), anti‐Golgin A2 (GM130, 1:200, BD Biosciences, 610822), anti‐H2A. Z‐variant histones (H2A.Z, 1:200, Cell Signaling Technology, 2718S), anti‐β‐actin (Santa Cruz Biotechnology, 1:500, sc‐47778), anti‐intercellular adhesion molecule 1 (ICAM1, 1:250, Thermo Fisher Scientific, MA5407), anti‐integrin alpha 5 (ITGA5, 1:200, Thermo Fisher Scientific, 10569‐1‐AP), anti‐caveolin 1 (CAV1, 1:250, Abcam, ab2910), and anti‐integrin beta 1 (ITGB1, 1:250; Thermo Fisher Scientific, PA5‐29606). The membrane was further incubated with horseradish peroxidase (HRP)‐labelled secondary antibodies (anti‐rat IgG, HRP‐linked antibody, Novus Biologicals, NB7126; anti‐mouse IgG, HRP‐linked antibody, Cell Signaling Technology, 7076S; anti‐rabbit IgG, HRP‐linked antibody, Cell Signaling Technology, 7074S) for 1 h, and immunoreactivity was detected using enhanced chemiluminescence solutions (Millipore, WBKLS0100). The membrane was visualized using a digital chemiluminescent imager (C300, Azure Biosystems). The band densities were digitally measured using ImageJ software (NIH).
2.11. Isolation of astrocyte‐specific EVs from human brain‐derived EV samples
Antibodies against astrocyte‐specific EV markers LRP1 (Thermo Fisher Scientific, 37–7600) and EAAT1 (Abcam, ab416) were conjugated to Dynabeads M‐270 Epoxy using the Dynabeads Antibody Coupling Kit (Thermo Fisher Scientific, 14311D) according to the manufacturer's instructions. Equal amounts of total EVs (30 μg) isolated from five HC and five AD patients were pretreated with 20 μl human Fc receptor blocking solution (Miltenyi Biotec, 130‐059‐901) and then incubated with antibody‐bead complex (5 μg/mg). As a negative control, immunoglobulin G (IgG)‐conjugated beads were added to the mixed EVs from either the HC or AD samples. After 12 h of incubation at 4°C, immunoprecipitated astrocyte‐specific EVs were eluted using 20 μl IgG elution buffer (Thermo Fisher Scientific, 21028), followed by the addition of 2 μl of 1 M Tris buffer for pH neutralization. The total EVs and astrocyte‐specific EVs were subjected to immunoblotting as previously described.
2.12. Processing of brain‐derived EV proteins and 16‐plex isobaric TMT peptide labelling
Isolated brain‐derived EV fractions were incubated in a final concentration of 20% (w/v) TCA (trichloroacetic acid; Cat# T6399, Sigma‐Aldrich) for 30 min on ice and precipitated by centrifuging at 15,000g for 20 min at 4°C, and then washed twice with ice‐cold acetone (Cat# 179124, Sigma‐Aldrich). After drying, the pellet was resuspended in Laemmli sample buffer with 5 mM dithiothreitol (Cat# 43815, Sigma‐Aldrich), reduced for 20 min at 65°C, and alkylated with 15 mM iodoacetamide (Cat# I1149, Sigma‐Aldrich) in the darkness for 45 min at room temperature in the dark. Subsequently, EV samples were run in a 4%–20% gradient gel (Cat# 4561095, BioRad) until the dye front was 15 mm from the top of the gel. Following GelCode Blue Stain Reagent (Cat# 24590, Thermo Fisher Scientific), the entire protein region of the gel was excised and subjected to in‐gel trypsin digestion (Cat# 03708985, Roche, USA) overnight at 37°C. The digested peptides were extracted with 70% acetonitrile /1% formic acid, and were removed the gel by Ultrafree‐MC Centrifugal Filter (Cat# UFC30L, Millipore). The digested peptides were reconstituted in 25 μl of 200 mM HEPPS (pH 8.0; Cat# E9502, Sigma‐Aldrich) and vortexed for 5 min.
Next, two sets of 16‐plex Tandem mass tag (TMT)‐labelling were performed according to the manufacturer's instructions (Cat# A44520, Thermo Fisher Scientific). In each set, there were fifteen individual samples and one additional sample mixed from 12 individuals which was used for normalization between two sets. Briefly, 4 μl of TMT label reagent (20 ng/μl) was added to the digested peptides in 30 μl of 200 mM HEPPS (pH 8.0) at room temperature for 1 h, and the reaction was quenched with 2 μl of 5% hydroxylamine in water for 15 min. The combined samples were added to 100 μl of 20% formic acid, 2 ml of 1% formic acid, desalted via in‐house StageTip (Cat# 14‐386‐2, Thermo Fisher Scientific), dried by vacuum centrifugation, and resuspended in 20 μl of 5% acetonitrile and 5% formic acid for nano liquid chromatography and tandem mass‐spectrometry (Nano LC‐MS/MS/MS).
2.13. Weighted correlation network analysis (WGCNA) of the brain‐derived EV proteome
The WGCNA algorithm was used to define protein co‐expression networks based on the normalized protein abundance of brain‐derived EV samples using the R package as previously described (Johnson et al., 2020; Langfelder & Horvath, 2008). Methodological details are provided in Supplementary Materials and Methods.
2.14. Gene ontology (GO) enrichment analysis
GO enrichment analysis of the co‐expressed proteins within the corresponding WGCNA modules was performed to gain a better biological understanding of the identified modules using the GO Elite database (Zambon et al., 2012). Z scores were used for overrepresentation of the ontologies within a module and the enrichment p‐value or Bonferroni corrected p‐value was set to ≤0.05 to assess the significance. The set of all proteins identified and considered in the network was used as the background.
2.15. Cell type EV marker enrichment analyses
Cell type enrichment was assessed by cross‐referencing module genes with lists of cell type‐specific EV protein markers provided in Supplementary Table S7. Proteins with a log2 abundance >5 in the specified cell type when compared to the other three cell types were used. Activated human primary astrocyte EV markers were obtained from our previous publication (You et al., 2020) and are described in Supplementary Table S16. The significance of the cell type enrichment was assessed using a one‐tailed Fisher's exact test and corrected for multiple comparisons using the FDR (Benjamini‐Hochberg) method.
2.16. Ingenuity pathway analysis (IPA)
We used the Ingenuity Pathway Analysis (IPA) (version 01–12) (https://www.qiagenbioinformatics.com/products/ingenuity‐pathway‐analysis/) to analyze, integrate, and categorize the protein data within the M7 module according to the software manual guidelines. A standard IPA core analysis was performed, including canonical pathways, upstream regulators, and disease and function annotation.
2.17. Statistical analysis
Graphical data were analyzed using Prism 6 (GraphPad) and are presented as mean ± SEM where indicated. All WGCNA related analyses were performed in R (v.3.6.3). Correlations in WGCNA were performed using the “biweight midcorrelation” function as implemented in the WGCNA R package. Correlations of ITGB1 with AD hallmarks were performed using nonparametric Spearman correlations. Boxplots represent the median, 25th, and 75th percentiles and whiskers represent measurements to the 5th and 95th percentiles. Comparisons between two groups were performed using a two‐tailed t‐test; comparisons among three or more groups were performed using an ordinary one‐way analysis of variances (ANOVA) with a Tukey's or Dunnett's post hoc test, and Kruskal–Wallis nonparametric ANOVA with Dunn's post hoc test for significance. Comparisons between two factors were performed using two‐way ANOVA with Sidak's multiple comparisons. P values in the cell type enrichment analyses were adjusted for multiple comparisons using a false discovery rate (FDR) correction using the Benjamini‐Hochberg method where indicated.
3. RESULTS
3.1. Differentiation and characterization of hiPSC‐derived brain cells
In this study, we attempted to identify cell type‐specific EV protein markers in the brain cells. To ensure uniformity of the genetic background, an identical iPSC line was differentiated into four major brain cell types: astrocytes, microglia, neurons, and oligodendrocytes using published protocols (Ehrlich et al., 2017; Mcquade et al., 2018; Tcw et al., 2017; Zhang et al., 2013) (Figure 1a). The pluripotency of the hiPSC lines was confirmed using immunocytochemical staining of the pluripotent stem cell‐specific markers (Supplementary Figure S1a). We generated induced excitatory neurons (iNeurons) by overexpressing the neuronal transcriptional factor neurogenin‐2 with minor modifications (Zhang et al., 2013) (Supplementary Figure S1c). iMGL cells were differentiated from hiPSC‐derived HPCs as previously described (Abud et al., 2017) (Supplementary Figure S1c). To generate astrocytes and oligodendrocyte‐like cells (iOligos), we differentiated hiPSCs into NPCs (Supplementary Figure S1b), and subsequently induced into astrocytes (iAstrocytes and iOligos, respectively (Ehrlich et al., 2017; Tcw et al., 2017) (Supplementary Figure S1c). These differentiated cells were validated with cell type‐specific markers using both immunocytochemistry (Figure 1b) and quantitative real‐time PCR (Figure 1c). We also performed a functional characterization of the iAstrocytes and iMGLs. The production of interleukin‐6 (IL‐6), a key effector of astrocytes in response to neuroinflammatory factors (Van Wagoner & Benveniste, 1999), was examined in iAstrocytes by ELISA. Treatment with polyinosinic‐polycytidylic acid (polyI:C; 50 or 100 ng/ml) and lipopolysaccharide (LPS; 10 or 50 μg/ml) for 24 h resulted in a dose‐dependent elevation of IL‐6 secretion in iAstrocytes (Figure 1d). The pHrodo‐tagged Zymosan A, a TLR 2/Dectin 1 agonist from Saccharomyces cerevisiae, induced a high level of phagocytosis in the iMGLs, which was attenuated by the actin polymerization inhibitor cytochalasin D (Figure 1e). Treatment of the iMGLs with LPS (100 ng/ml) significantly increased the secretion of multiple cytokines (Figure 1f).
Furthermore, we performed label‐free quantitative mass spectrometry to obtain comprehensive proteomic profiles for each of the four cell types. A total of 3206 proteins were identified with a peptide and protein FDR of 1% (Supplementary Table S1). Among them, 198 proteins were unique to iNeuron, 239 to iMGL, 73 to iAstrocytes, and 47 to iOligo (Figure 1g). Moreover, some common cell‐type‐specific protein markers were abundant in iNeuron (e.g., MAPT), iMGL (e.g., AIF1, PU.1), iAstrocytes (e.g., AQP4), and iOligo (e.g., OLIG2), respectively (Figure 1h). We have provided a list of the most abundant (log2 abundance >5 as compared to the other three cell types) proteins across all four cell types (Supplementary Table S2). Taken together, our results revealed the valid differentiation of hiPSCs into the major brain cell types.
3.2. Isolation and characterization of extracellular vesicles from hiPSC‐derived brain cells
We then used sequential ultracentrifugation and size‐exclusion chromatography to isolate the EVs (three biological replicates per cell line) from the conditioned media of the hiPSC‐derived cells for label‐free quantitative proteomics, as previously described (You et al., 2020) (Figure 2a). The purified EVs showed a cap‐shaped morphology with the transmission electron microscopy (TEM; Figure 2a and Supplementary Figure S2a). Subsequently, the EV populations were characterized using nanoparticle tracking analysis (NTA; Supplementary Figure S2b). We found that most human brain cell‐derived EVs were in the expected diameter range of 50–150 nm, except for the iAstrocyte‐derived EVs, of which a majority had diameters greater than 150 nm (Figure 2b), revealing distinct EV populations from the iAstrocytes. The mean diameter of 222.0 ± 2.6 nm for the iAstrocyte‐EV was also significantly higher than that of the other neural cell types (Figure 2c). To further compare EV protein compositions across the cell types, equal amounts of proteins from the isolated EVs were analyzed by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) followed by label‐free quantitative proteomics analysis (Figure 2d). A combined analysis of the replicates with a peptide and protein FDR of 1% identified 109 proteins unique to iNeuron‐derived EVs (iNeuron‐EV), 197 proteins unique to iMGL‐derived EVs (iMGL‐EV), 378 proteins unique to iAstrocyte‐EVs, and 117 proteins unique to iOligo‐EVs (Figure 2d and Supplementary Table S3). The number of proteins detected in the iNeuron‐EV and iOligo‐EV was consistently lower than the iMGL‐EV and iAstrocyte‐EV, suggesting a less diverse protein content in these EVs. To rule out potential contaminating proteins in the EV samples, we compared the expression of non‐EV protein markers (e.g., nucleus, mitochondria, secretory pathway, and others) as indicated in the minimal information for studies of extracellular vesicles 2018 (MISEV2018) guideline (Théry et al., 2018) between EV samples and their cellular origins. Notably, these non‐EV proteins were highly enriched in cell lysates, while most of them were not found in EV lysates (Figure 2e), thereby demonstrating the good quality of the EV fractions isolated from the hiPSC‐derived brain cells.
FIGURE 2.
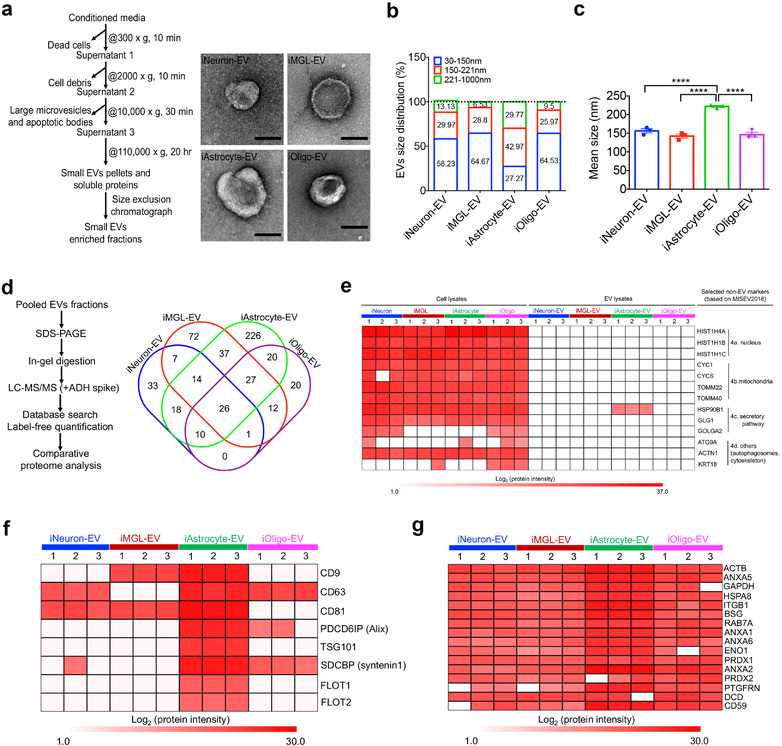
Isolation and characterization of extracellular vesicles from hiPSC‐derived brain cells. (a) Workflow for the EV isolation and transmission electron microscopy (TEM) images of isolated iNeuron‐, iMGL‐, iAstrocyte‐, and iOligo‐EVs. Scale bar: 100 nm. (b) Size distribution of the isolated EVs from four cell types determined using a nanoparticle tracking analysis (NTA, n = 3 for each cell type). (c) Comparison of the EV mean size among the four cell types by NTA (n = 3 for each cell type). iAstrocyte showed the significant increase in diameter of the EV size when compared to the other three cell types. Data are presented as the mean ± SEM. **** p < 0.0001. (d) Workflow for the label‐free proteomics and Venn diagram showing the number of EV proteins differentially identified in iNeuron, iMGL, iAstrocyte, and iOligo. (e) Heatmap illustrating the expression of non‐EV protein markers present in different brain cell‐derived EVs and their cellular origins based on the protein intensity determined using mass spectrometry. Four types of non‐EV protein markers, including proteins from the nucleus, mitochondria, secretory pathway, and others, were selected as indicated in the MISEV2018 guideline (Théry et al., 2018). (f) Heatmap illustrating the enrichment of conventional exosome protein markers present in different brain cell‐derived EVs based on their protein intensity from mass spectrometry. (g) Sixteen newly defined pan‐EV marker candidates from different cell types based on their protein intensity, determined using mass spectrometry. Proteins represented in at least two of the three replicates within each cell type were selected. The white cells indicate no presence of the protein in the specified cell type. hiPSC, human induced pluripotent stem cells; EV, extracellular vesicle; TEM, transmission electron microscopy; iNeuron, hiPSC‐derived excitatory neuron; iMGL, hiPSC‐derived microglia‐like cell; iAstrocyte, hiPSC‐derived astrocyte; iOligo, hiPSC‐derived oligodendrocyte‐like cells; NTA, nanoparticle tracking analysis; SEM, standard error of the mean; MISEV2018, Minimal information for studies of extracellular vesicles 2018
To further identify ubiquitous pan‐EV marker candidates among human neural cell‐derived EVs, we investigated the highly represented proteins found in the EVs across different cell types as was previously described (Hoshino et al., 2020). We first examined the frequency of eight conventional EV markers as indicated in the MISEV2018 guidelines (Théry et al., 2018), including tetraspanins (CD9, CD63, and CD81), programmed cell death 6‐interacting protein (PDCD6IP/ALIX), tumour susceptibility gene 101 protein (TSG101), syntenin1 (SDCBP), flotillin 1 (FLOT1), and 2 (FLOT2), which are involved in EV biogenesis and release (You & Ikezu, 2019). The established EV markers were all found in at least one cell‐type‐specific EV sample (Figure 2f). Only iAstrocyte‐EVs showed enrichment of all these traditional markers, highlighting the need to identify novel pan‐EV markers to detect EVs from all the neural cell types. We searched for proteins that were represented in at least two of the three replicates across all EV samples and identified 16 proteins (Figure 2g). These include β‐actin (ACTB), glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH), enolase 1 (ENO1), heat shock protein (HSPA8), and ras‐related protein 7a (RAB7A), which are all involved in the formation and trafficking of EVs (Van Niel et al., 2018). We also identified integrins (ITGB1) and annexins (ANXA1, 2, 5, 6) that mediate EV binding and fusion to the plasma membrane (Mulcahy et al., 2014). In addition, plasma membrane proteins such as basigin (BSG), peroxiredoxin 1 (PRDX1 and 2), prostaglandin F2 receptor negative regulator (PTGFRN), dermcidin (DCD), and CD59 were also identified in this group. These molecules will be useful for the future development of potential pan‐brain EV markers.
3.3. Comparison of neural cell type specific EV proteomes
To further determine the differences in the protein composition of the EVs isolated from hiPSC‐derived neural cells, we compared the protein abundances across the different cell types using the label‐free intensity‐based absolute quantification (iBAQ) method with all samples normalized by either total ion current or by the total intensity of a spiked yeast alcohol dehydrogenase 1 (ADH1) protein digest (You et al., 2020) (Supplementary Table S4). Principal component analysis (PCA) showed good discrimination among different cell type‐specific EV samples; all biological replicates of the EV proteomes formed independent clusters according to their expression profiles (Figure 3a), revealing the high reproducibility of the EV isolation and label‐free quantitative proteomics. Additionally, Pearson correlation coefficients were higher between biological replicates and much lower between different cell types (Figure 3b). Collectively, our analyses demonstrated excellent biological reproducibility and potential cell‐type‐specific protein signatures in the EVs.
FIGURE 3.
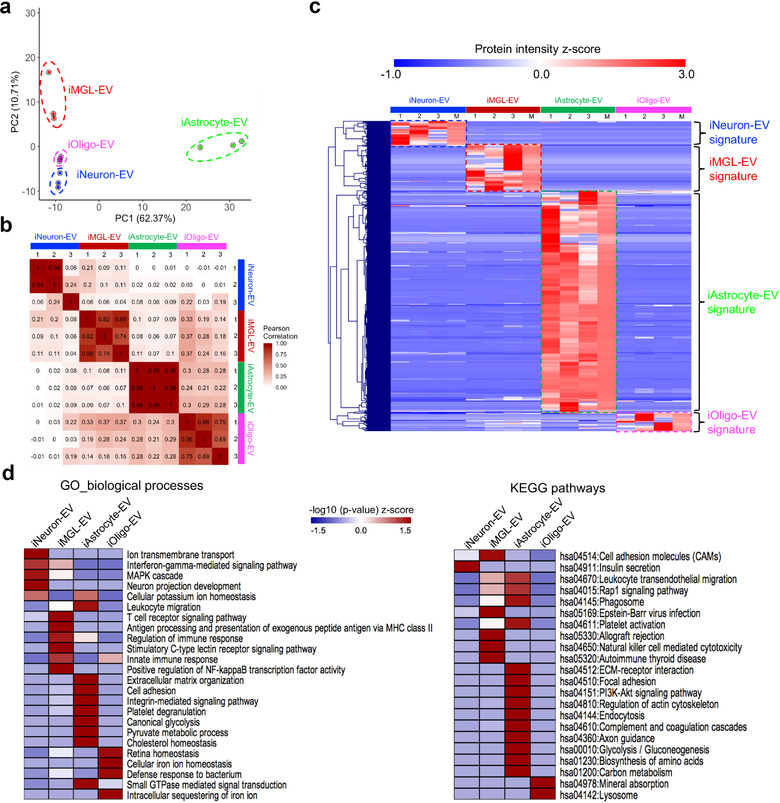
Comparative analysis of the hiPSC‐derived cell‐type‐specific EV proteome. (a) Principal component analysis (PCA). The EV proteome of all cell types and their differentiation states were measured in triplicate and classified into four major cell types based on component 1 (62.37% variability) and component 2 (10.71% variability). (b) Heatmap of the Pearson correlations between each sample, showing high correlation coefficients for the protein intensity determined from the triplicate measurements. (c) Heatmap of the z‐scores for the protein intensities of the cell type‐specific EV proteins after unsupervised hierarchical clustering (n = 3 for each cell type). M represents the median protein abundance of the triplicate measurements. Proteins are divided into four clusters showing the different EV protein signatures across different cell types. (d) Heatmap of the functional enrichments [z‐scored (‐log10 p‐value)] of the EV protein signatures across different cell types for the gene ontology (biological process) terms and KEGG pathways using DAVID Bioinformatics Resources 6.8. PCA, principal component analysis; EV, extracellular vesicle; KEGG, Kyoto Encyclopedia of Genes and Genomes; DAVID, Database for Annotation, Visualization, and Integrated Discovery
We then defined the differentially expressed proteins across different cell types using unsupervised hierarchical clustering of the 495 identified EV proteins after the removal of the non‐EV contaminants, such as keratins and albumin, and identified the cell type‐specific EV protein signatures (Figure 3c). These cell type‐specific proteins were then selected for gene enrichment analysis including Gene Ontology Biological Processes (GO_BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Supplementary Table S5; Figure 3d). We found that the iNeuron‐EVs were enriched in proteins associated with ion transport, interferon‐γ‐mediated signalling, MAPK cascade, neuron projection development, and insulin secretion (Supplementary Figure S3a; Supplementary Table S6). For the iMGL‐EVs, the biological functions and pathways were mostly related to immune processes, including T cell receptor signalling, nuclear factor kappa B (NF‐κB)‐mediated signalling, antigen processing and presentation, immune responses, or autoimmune disease (Supplementary Figure S3b; Supplementary Table S6). For iAstrocyte‐EVs, the enriched proteins were mainly related to the extracellular matrix, including cell adhesion, extracellular matrix (ECM)‐receptor interaction, and integrin‐mediated signaling, and biological metabolism including glycolysis, pyruvate metabolic process, cholesterol homeostasis, biogenesis of amino acids, and carbon metabolism (Supplementary Figure S3c; Supplementary Table S6). Notably, iAstrocyte‐EV proteins are also enriched in the regulation of platelet activation, cytoskeleton, axon guidance, and complement cascades. Finally, enriched biological processes and pathways in iOligo‐EVs involved iron ion homeostasis and lysosomal function (Supplementary Figure S3d; Supplementary Table S6). Together, these comparative analyses may represent distinct cellular functions unique to each cell type captured in the EVs.
3.4. Identification of hiPSC‐derived cell type‐specific EV proteins
Given that EV protein signatures were strongly observed across different hiPSC‐derived neural cell types, we proceeded to identify the specific signature proteins that could be used as cell type‐specific biomarkers for future EV studies in human brains. We performed a quantitative comparison of the EV proteomes and plotted the proteins using protein intensity (normalized iBAQ value) and the fold change in protein abundance within one cell type against the others (Figure 4a). We provided a list of the most abundant (log2 abundance >5 as compared to the other three cell types) cell‐type‐specific EV proteins, which showed a total of 30 proteins in iNeuron‐EV, 70 proteins in iMGL‐EV, 300 proteins in iAstrocyte‐EV, and 14 proteins in iOligo‐EV (Supplementary Table S7). Among the most abundant and enriched EV proteins, there are some well‐known cell type‐specific proteins (Sharma et al., 2015) (Figure 4b), revealing how EV samples can excellently reflect cell origin. For example, proteins of the cytoskeleton (MAP1LC3B2, microtubule‐associated protein 1 light chain 3 beta; TUBB2A, tubulin beta 2A class IIa), synapses (VAMP2, vesicle‐associated membrane protein 2; STX1B, syntaxin 1B), and the energy processes (ATP1A3, ATPase Na+/K+ Transporting Subunit Alpha 3; ATP1B1, ATPase Na+/K+ transporting subunit beta 1), as well as other neuronal specific proteins (RTN1, reticulon‐like protein 1; NCAM1, neural cell adhesion molecule 1) were unique to iNeuron‐EV. We found that microglia‐specific cell surface proteins, including ITGB2, ITGAM/CD11b (integrin alpha‐M), and LCP1 (lymphocyte cytosolic protein 1), were also highly enriched in iMGL‐EV. For iAstrocyte‐EV, metabolic transporters (SLC2A1, solute carrier family 1 member 1; SLC1A5, solute carrier family 1 member 5), lipoprotein pathway including APOE and LRP1 (low‐density lipoprotein receptor‐related protein 1), astrocyte‐specific cell surface antigen integrin alpha 6 (ITGA6/CD49f) (Barbar et al., 2020), as well as intermediate filaments (Vimentin) were abundant. The oligodendrocyte‐specific proteins of myelin sheath such as PLP1, CNP, and MBP were not identified in the iOligo‐EV, likely due to the lack of myelin formation by the iOligos in vitro. However, we observed that the proteins with significant abundance in the iOligo‐EVs included lysosomal proteins (LAMP2; lysosomal associated membrane protein 2, ACP2; acid phosphatase 2, lysosomal), which have been reported to play important roles in oligodendrocytes (Mckenzie et al., 2017; Raddatz et al., 2014), and ferritin heavy chain (FTH1), which was recently identified as a neuroprotective molecule secreted in oligodendrocyte‐derived EVs (Mukherjee et al., 2020). Of note, proteome data from cell lysates also demonstrated the relative enrichment of these cell type‐specific EV marker candidates in their cellular origins (Supplementary Figure S4).
FIGURE 4.
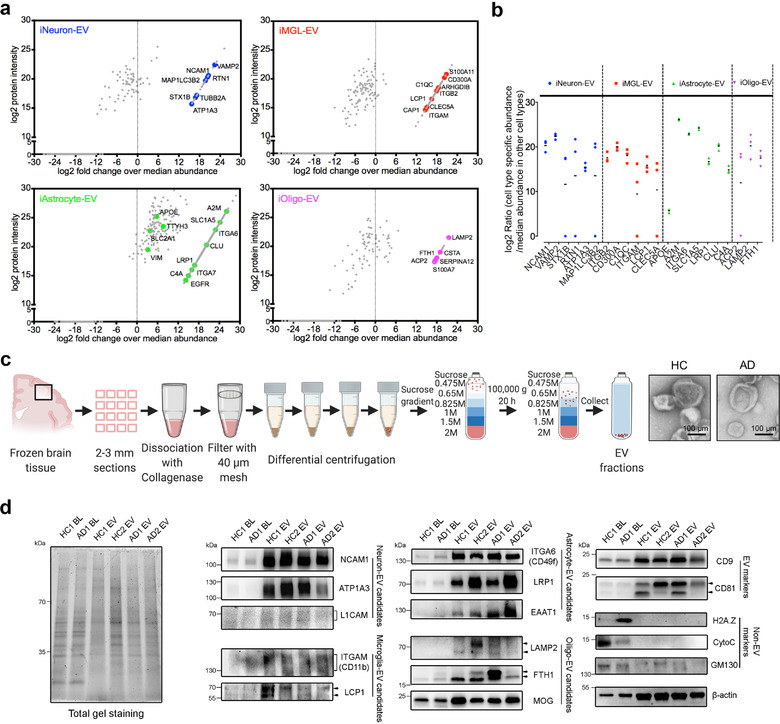
Abundant and enriched proteins were identified in the cell type specific EVs. (a) Scatter plot of the identified proteins in the indicated cell type using log2 protein intensity versus log2 fold change over the protein abundance in the other three cell types. The coloured dots highlight several enriched proteins that are cell type‐specific (Sharma et al., 2015) as potential cell type‐specific EV marker candidates. (b) Log2 fold change of selected marker candidates in individual replicates for the specified cell types when compared to the other cell types. (c) Schematic of the EV isolation method from human frozen brain tissues for immunoblotting and TEM images of isolated brain derived EVs from healthy controls (HC) and AD patients. Scale bar; 100 nm. (d) Western blot analysis of the brain lysates and associated EVs isolated from both HC and AD patients for conventional and newly identified neural cell type specific EV proteins, as well as common EV (CD9, CD81) and non‐EV (H2A.Z, CytoC, GM130) protein markers, and the reference protein β‐actin (ACTB). Equivalent proteins from brain lysates and EVs were loaded as loading controls and indicated in the total gel staining (left panel). The original western blot images are shown in Supplementary Figure S8. BL, brain lysate; EV, extracellular vesicle; HC, healthy control; AD, Alzheimer's disease
The presence of several newly identified cell type‐specific EV protein candidates was then validated, as well as some common cell type‐specific EV markers (e.g., L1CAM, GLAST/EAAT1, and myelin oligodendrocyte glycoprotein) using human brain‐derived EV samples isolated from both healthy controls and AD patients by immunoblotting (Figure 4c). The isolated brain‐derived EVs showed a good cap‐shaped morphology by TEM (Figure 4c) and enriched common EV markers (CD9 and CD81) (Figure 4d). In addition, some non‐EV markers such as the mitochondrial protein cytochrome C (CytoC) and histone protein H2A.Z were negative in these EV samples, while the Golgi apparatus marker GM130 was present, but its levels were very low. An internal control protein, β‐actin (ACTB), was present in both the brain lysates and EV samples. Furthermore, these newly identified cell type‐specific proteins including NCAM1, ATP1A3, LCP1, LRP1, ITGA6, LAMP2, and FTH1, as well as common cell type‐specific proteins including EAAT1 and MOG showed significantly higher enrichment in the EVs over brain tissues; while putative neuronal EV protein L1CAM and microglial EV protein ITGAM were present in smeared bands in the EV samples with the tested antibodies (Figure 4d), likely due to the heterogenous posttranslational modifications. Our results revealed that NCAM1, ATP1A3, LCP1, LRP1, ITGA6, EAAT1, LAMP2, FTH1, and MOG may represent brain cell‐type EV markers, as they were also identified in human CSF‐derived EVs (Muraoka, Jedrychowski, Yanamandra, et al., 2020; Muraoka, Lin, et al., 2020). Collectively, our data provide direct evidence of the cell type‐specific EV proteins in different brain cell types, which could be employed to develop novel markers for human neural cell type‐specific EV isolation from human biofluids and tissues.
3.5. Proteomic analysis of human brain tissue derived EVs
To strengthen our proteomic findings and explore their potential translation toward clinical applications, we then analyzed the EVs derived from post‐mortem human brain tissues from a cohort of 30 age‐and sex‐matched individuals consisting of 11 healthy controls (HC), eight patients with mild cognitive impairment and 11 patients with AD (Figure 5a). The demographic information for all cases, including the Clinical Dementia Rating Scale (CDR), amyloid‐β plaque load, and tau pathology (Braak stage), is provided in Supplementary Table S8. EV enrichment from the frozen brain tissues was performed using discontinuous sucrose gradient ultracentrifugation, following a previously published protocol (Muraoka, Deleo, et al., 2020; Muraoka, Lin, et al., 2020). EV proteomic profiling was identified and quantified using multiple sets of isobaric tandem mass tags with a 16‐plex TMT mass spectrometry approach (A. Thompson et al., 2019). A total of 4286 unique brain‐derived EV proteins were identified in two TMT‐labeling sets with 2645 common proteins, which were used for proteomic analysis (Supplementary Figure S5a; Supplementary Table S9). The PCA showed a marginal separation of the three groups based on protein abundance (Figure 5b).
FIGURE 5.
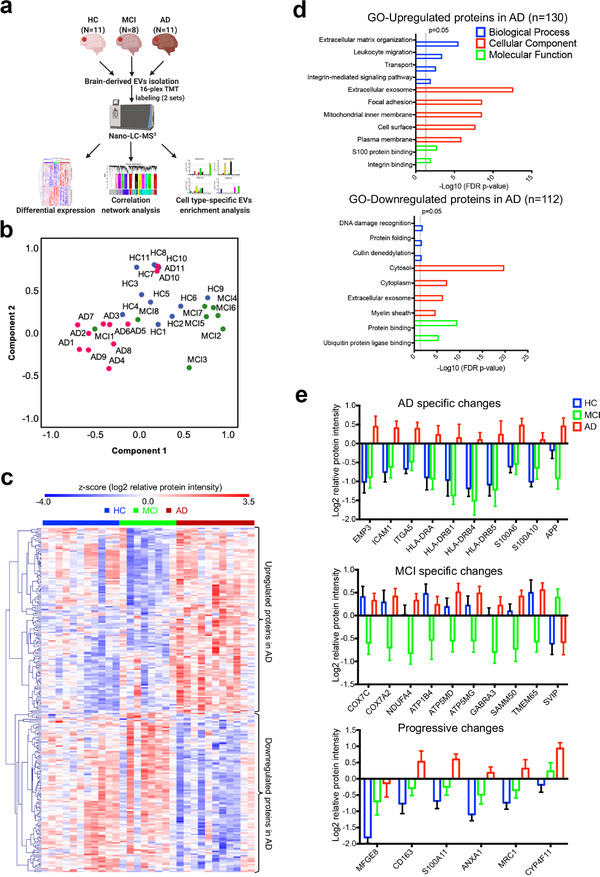
Proteomic analyses of the EVs isolated from the brains of healthy controls, mild cognitive impairment and Alzheimer's disease cases. (a) Graphic illustration of the workflow for brain derived EV proteomes. Brain tissue was sectioned from the postmortem frontal cortex of HC (n = 11), MCI (n = 8), and AD (n = 11) patients for EV isolation. Protein levels were measured and quantified using 16‐plex TMT labelled mass spectrometry and analyzed using differential expression, correlation network analysis, and cell type‐specific EV enrichment analysis. (b) PCA of the total EV samples. HC, blue symbols; MCI, green symbols; AD, red symbols. (c) Heatmap of the z‐scored log2 relative protein intensities within each of the EV sample after unsupervised hierarchical clustering showing the significantly altered proteins in the three comparisons (HC vs. AD, HC vs. MCI, and MCI vs. AD) determined by ANOVA (p < 0.05) followed by Tukey's post hoc test (p < 0.01). Proteins are divided into two clusters showing upregulated and downregulated EV proteins in AD. (d) Gene ontology (GO) analysis of the upregulated and downregulated proteins in AD brain derived EVs by using DAVID Bioinformatics Resources 6.8. The top significant (FDR p‐value < 0.05) GO terms in biological Process, Cellular Component and Molecular Function are listed. (e) List of several significantly differentially expressed proteins (Tukey's post hoc test p < 0.05, fold change over other group > 2) that have AD specific changes, MCI specific changes, and progressive changes across the comparisons of HC, MCI, and AD groups. Data are presented as mean ± SEM. HC, healthy control; MCI, mild cognitive impairment; AD, Alzheimer's disease; EV, extracellular vesicle; TMT, tandem mass tag; PCA, principal component analysis; ANOVA, analysis of variances; GO, gene ontology; DAVID, Database for Annotation, Visualization, and Integrated Discovery; FDR, false discovery rate; SEM, standard error of the mean
Our previous work revealed distinct brain‐derived EV proteomic profiles between AD and the controls (Muraoka, Deleo, et al., 2020). Here, we sought to identify the significantly altered EV proteins among the three groups using one‐way ANOVA (p < 0.05) followed by Tukey's comparison post hoc test (p < 0.01 (Supplementary Figure 5b). In total, 242 significantly altered proteins were identified (Supplementary Table S10). Using unsupervised hierarchical clustering of these significantly differentially expressed proteins (DEPs), we observed that a cluster of 130 DEPs was upregulated, while a distinct cluster of 112 DEPs was downregulated in AD EVs when compared to HC or MCI EVs (Figure 5c). GO biological process term enrichment analysis indicated that these upregulated DEPs in AD were associated with the extracellular matrix, leukocyte migration, transport, and integrin‐mediated signalling pathways, whereas the downregulated DEPs were related to DNA damage recognition, protein folding, and Cullin deneddylation (Figure 5d; Supplementary Table S11). Notably, they were both over‐represented in the GO molecular function terms for the extracellular exosomes, further demonstrating the significantly altered EV proteomic profile in AD brains.
To narrow down the list of significant DEPs, we performed the three comparisons mentioned above, using the criteria of Tukey's post hoc test p < 0.05, and fold change >2. A total of 42 DEPs were identified (Supplementary Figure S6a; Supplementary Table S12) and a heatmap highlighting the differences across the three groups was generated (Supplementary Figure S6b). We identified several disease‐specific DEPs among the three groups (Figure 5e). The AD EV‐specific changes include increases in proteins of the major histocompatibility class II cell surface receptors (HLA‐DRA, HLA‐DRB1, 4, 5), S100 calcium‐binding proteins (S100A6 and S100A10), epithelial membrane protein 3 (EMP3), intercellular adhesion molecule 1 (ICAM1), and integrin alpha 5 (ITGA5), most of which are strongly implicated in AD (Cristóvão & Gomes, 2019; Steele et al., 2017; Verbeek et al., 1994). Notably, amyloid precursor protein (APP) was also specifically increased in AD EVs. In contrast, DEPs involved in oxidative phosphorylation (COX7C, COX7A2, NDUFA4) and ATPase Na+/K+ transporting family members (ATP1B4, ATP5MD, ATP5MG) displayed a specific decrease in MCI EVs, whereas small VCP/p97‐interacting protein (SVIP) showed a unique increase in MCI EVs. Additionally, several DEPs showed a progressive increase across HC, MCI, and AD, including lactadherin (MFGE8), CD163, S100A11, ANXA1, macrophage mannose receptor 1 (MRC1), and cytochrome P450 protein (CYP4F11). Overall, these results suggest that these disease‐specific DEPs may provide a unique molecular signature to distinguish the EVs isolated from HC, MCI, and AD brains.
3.6. Construction of an AD EV protein co‐expression network with cell type specificity
Recent studies have reported the utility of weighted protein co‐expression network analysis (WGCNA) of proteomic datasets as a powerful tool to identify key molecular pathways and potential therapeutic targets for neurodegenerative diseases (Johnson et al., 2020; Seyfried et al., 2017). It enables the classification of protein co‐expression modules that are linked to disease phenotypes, specific cell types, and biological pathways. Here, we applied WGCNA to construct an AD EV protein co‐expression network using 2645 common proteins. We identified 11 modules from the co‐expressed protein groups, ranging in size from the largest module M1 at 720 proteins to the smallest module M11 at 42 proteins (Figure 6a; Supplementary Table S13). GO analysis of the module proteins showed a specific ontology for each module based on biological processes, cellular components, and molecular functions (Figure 6a; Supplementary Table S14). To assess the significance of the modules with AD pathology, we correlated the module eigenprotein (the first principal component of the given module) with neuropathological hallmarks of AD, including Aβ plaque load and Braak stage (neurofibrillary tangle load), and CDR (clinical dementia rating) scores across HC, MCI, and AD samples (Figure 6b; Supplementary Table S13). We identified three modules (M1, M6, and M9) that were negatively correlated with all three AD traits and revealed a significant decrease in AD. These modules were associated with the GO terms “Microtubule” (M1), “Cellular response to nitrogen starvation” (M6), and “Glycerol biosynthesis process” (M9), which is consistent with previous studies showing the loss of microtubule function and metabolic processes in AD (Cai et al., 2012; Feinstein & Wilson, 2005). Moreover, M7, which was associated with the intrinsic component of the plasma membrane, had the strongest positive correlation with all AD traits (CDR: cor = 0.68, p = 4e‐5; plaque load: cor = 0.72, p = 6e‐6; Braak Stage: cor = 0.6, p = 4e‐4). M2, M8, and M10 were related to “Integral component of membrane,” “Mitochondrion,” and “Oxidative phosphorylation,” respectively, and showed positive correlations exclusively with Plaque Load, whereas M4 with the ontology of ‘Cell junction assembly’ had a positive correlation exclusively with Braak Stage (Figure 6b).
FIGURE 6.
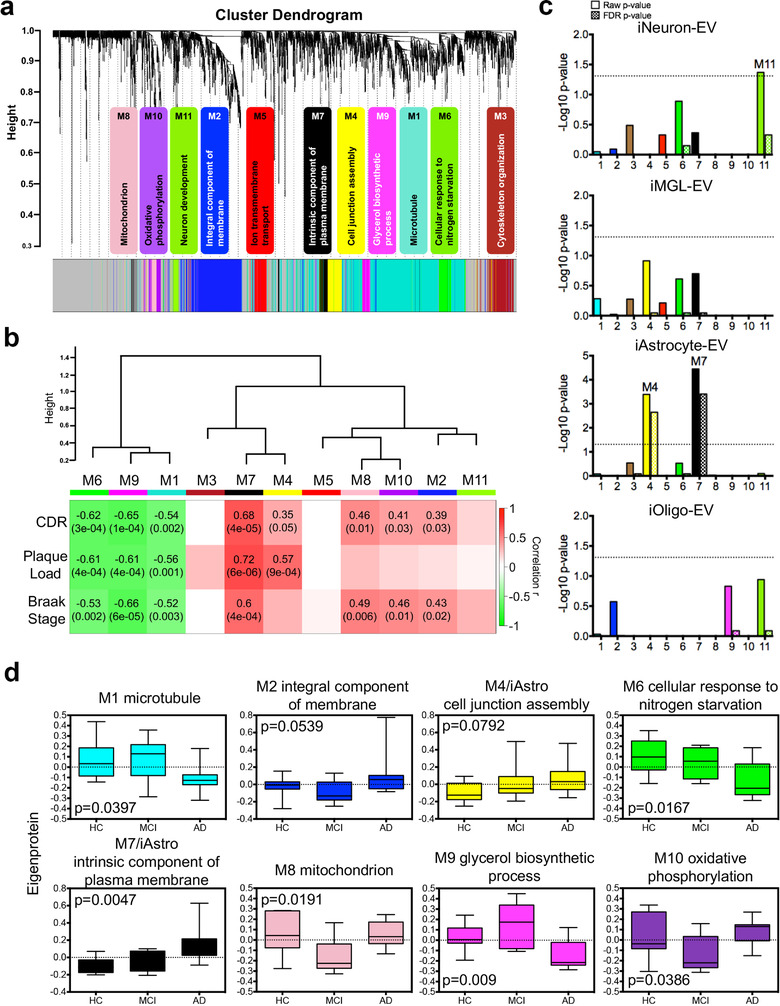
Protein co‐expression network analysis of the brain derived EV proteome identified modules associated with specific gene ontologies and brain cell types. (a) WGCNA cluster dendrogram of the total identified brain‐derived EV proteins (n = 2645) from three groups with distinct protein modules (M1–11) defined by dendrogram branch cutting. The significant gene ontologies associated with these modules in biological processes or cellular components were noted. (b) Brain‐derived EV protein modules were clustered to assess module correlation with neuropathological hallmarks of AD based on protein co‐expression eigenproteins. The Pearson correlation and p‐value between module eigenprotein expression and CDR, plaque load value, and Braak stage. (c) Cell‐type enrichment was assessed by cross‐referencing module proteins (via matching gene symbols) using the one‐tailed Fisher's exact test against the lists of proteins determined as cell type‐specific EV markers derived from neurons, microglia, astrocytes, and oligodendrocytes. Both the raw p‐value and the FDR p‐value (correction for multiple comparisons by the Benjamini‐Hochberg method) are shown; bars extending above the dashed line represent p < 0.05. (d) Module eigenprotein level by case status for each protein module that had significant correlation with at least two traits in b. Cell‐type associated modules are highlighted. Boxplots are displayed for each of the case samples among the three groups (HC, MCI, and AD) and represent the median, 25th and 75th percentiles and whiskers represent the 5th and 95th percentiles, respectively. Significance was measured using one‐way nonparametric ANOVA, Kruskal‐Wallis p‐values. WGCNA, weighted gene co‐expression network analysis; EV, extracellular vesicle; CDR, cognitive dementia rating; FDR, false discovery rate; HC, healthy control; MCI, mild cognitive impairment; AD, Alzheimer's disease; ANOVA, analysis of variances
Given our finding of cell type‐specific EV protein markers, we further evaluated the enrichment of cell type markers in each co‐expression module of the brain‐derived EV proteins (Figure 6c). We observed that M11 was associated with “Neuron development” and had nominally significant enrichment in iNeuron‐EV markers. Notably, significant enrichment of iAstrocyte‐EV markers was found in both M4 and M7, which were significantly positively correlated with AD traits. These results suggest that astrocyte‐derived EVs have the potential to serve as AD biomarkers and might contribute to pathology development, which is consistent with recent findings that reported the neurotoxicity of astrocyte‐derived EVs isolated from AD patients (Goetzl et al., 2018; Nogueras‐Ortiz et al., 2020) and its application as a plasma EV biomarker for AD diagnosis (Elahi et al., 2020). We subsequently refer to those modules with strong cell type enrichment as the “M11/iNeuron” module, the “M4/iAstro” and “M7/iAstro” module. In addition, we also measured the module eigenprotein values by disease status to identify the EV protein networks that may affect changes in MCI to AD (Figure 6d; Supplementary Table S15). The M1 and M2 modules were significantly changed in the AD cases when compared to HC or MCI, revealing a disruption of the microtubules and integral components of the membrane specifically in AD. Intriguingly, the M4, M6, and M7 modules showed the same trends for increases or decreases in the MCI samples as in the AD samples when compared to HC, which implies that the changes in cell adhesion assembly, cellular response to nitrogen starvation, and the intrinsic components of the plasma membrane occur early in the preclinical phase of AD. Notably, astrocyte‐derived EVs, enriched in M4 and M7, might be important mediators of AD progression. The M8, M9, and M10 modules were particularly altered in the MCI samples (Figure 6d), indicating a transient and specific regulation of these protein networks in cognition decline, and their importance in cognitive function. Collectively, we generated the first brain‐derived EV protein co‐expression networks mapped with human neural cell‐type‐specific EV markers that are strongly linked to neuropathological hallmarks and cognitive function in AD.
3.7. M7 astrocyte‐EV module is enriched in markers of activated astrocyte‐derived EVs and associated with AD pathology
Our previous study characterized the proteomic profiles of human primary astrocyte‐derived EVs and defined many significantly upregulated EV proteins from ΙL‐1β induced astrocytes as activated astrocyte‐derived EV markers (You et al., 2020) (Supplementary Table S16). Since the M7 module showed the strongest association with AD traits and astrocyte‐derived EVs, we next asked whether this AD‐related module is related to the activated astrocyte‐derived EV profile. We found that the M7 and M6 modules, showed significant enrichment in the activated astrocyte‐derived EV markers, even after FDR correction (Figure 7a). Furthermore, we also observed that the activated astrocyte‐derived EV markers in the M7 module interacted closely with the top 10 hub proteins, which were ranked by the protein correlation significance with AD traits (Figure 7b). These findings suggest that the protein co‐expression network in M7 may reflect the specific EV profile of the reactive astrocytes and serve as important mediators in AD development and progression. To further explore the potential biological functions and upstream pathways of these EV proteins in M7, we performed Ingenuity Pathway Analysis. Multiple significant canonical pathways associated with endocytosis, integrin signalling, and NF‐κB activation were identified (Figure 7c; Supplementary Table S17). Additionally, the protein co‐expression network, including several core hub proteins such as ITGB1, ITGB5, and SLC7A2, was predicted to be regulated by pro‐inflammatory cytokines, including tumour necrosis factor (TNF)‐α, IL‐6, and IL‐1β, which are upstream regulators involved in the activation of phagocytes, inflammation of organs, and inflammatory responses (Figure 7d).
FIGURE 7.
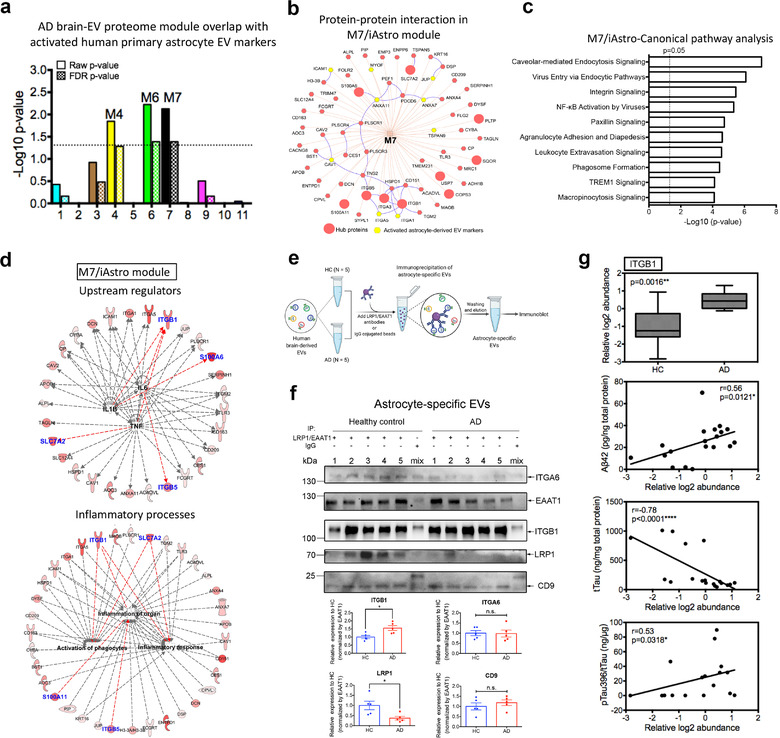
The M7 module is enriched in activated astrocyte derived EV markers and involved in inflammatory processes. (a) The enrichment of proteins, identified as EV markers for the activated astrocytes, was calculated for each module in the AD protein network using a one‐tailed Fisher's exact test. Both the raw p‐value and the FDR p‐value (correction for multiple comparisons by the Benjamini‐Hochberg method) are shown; The dashed line represents a p‐value <0.05, above which enrichment was considered statistically significant. Enrichment was assessed by cross‐referencing module proteins against lists of proteins determined to be differentially expressed in activated astrocyte derived EVs provided in Supplementary Table S16. (b) Protein‐protein interaction network within the M7 module generated by the ToppCluster tool (Kaimal et al., 2010) (https://toppcluster.cchmc.org/). The top 10 hub proteins are ranked by correlation significance with AD traits in the module and are highlighted in the larger red circle. Proteins highlighted in yellow are marker proteins from activated astrocyte derived EVs. (c) Ingenuity canonical pathway analysis of the proteins within the M7 module. The top 10 significantly enriched canonical pathways are listed. A statistically significant p‐value of 0.05 is indicated on the plot as a vertical dashed line, at the x‐value of 1.30. (d) Involvement of the protein co‐expression network within the M7 module in mediating inflammation. These proteins were regulated by potential pro‐inflammatory factors including TNF‐α, IL‐6, and IL‐1β and involved in inflammatory processes including the activation of phagocytes, inflammation of organ and inflammatory responses. The red color indicates the significance of the proteins in the M7 module; Blue indicates the top 10 hub proteins within the module; Red lines indicate leading to activation by a predicted relationship. (e) Schematic of the immunoprecipitation of astrocyte specific EVs from an independent cohort of HC (n = 5) and AD patients (n = 5) for immunoblotting. Combined antibodies against astrocyte‐EV proteins LRP1 and EAAT1 were conjugated to magnetic beads and then added for the immunocapture of astrocyte‐derived EVs. As a negative control, combined mouse and rabbit IgG were used to immunoprecipitate EVs mixed from the HC and AD groups. (f) Western blot analysis of the astrocyte‐specific EVs from the HC and AD patients for the hub protein ITGB1 in the M7/astrocyte‐EV module. The newly identified astrocyte specific EV marker ITGA6, common EV marker CD9, and immunoprecipitated protein EAAT1 and LRP1 were also detected. The original western blot images are shown in Supplementary Figure S9b. Equivalent brain derived EV proteins (30 μg) were isolated from healthy controls and AD patients were used to initiate the immunoprecipitation assay. The ITGB1 level was enriched and significantly elevated in the astrocyte specific EVs from AD when compared to the HC samples (p = 0.032). The LRP1 level was significantly reduced in astrocyte specific EVs from AD when compared to the HC samples (p = 0.032). The western blot signals were calculated using ImageJ and normalized by EAAT1 intensity. Data are presented as mean ± SEM, *p < 0.05, n.s., no significance, and determined using the Mann‐Whitney non‐parametric test. (g) Relative ITGB1 protein level in the human brain‐derived EVs isolated from HC (n = 9) and AD (n = 11) samples using our previously published proteomics dataset (Muraoka, Deleo, et al., 2020) and its correlation with AD pathogenic hallmarks (Aβ42, total tau and pSer396 tau) and evaluated using ELISA kits. The proteomics data and measurements are provided in Supplementary Table S18. Significance in the ITGB1 protein level was measured using a two‐sided Mann‐Whitney t‐test. Correlations were performed using nonparametric Spearman correlation analysis. Box plots represent the median, 25th and 75th percentiles and whiskers indicate the 5th and 95th percentiles, respectively. *p < 0.05, **p < 0.01, ****p < 0.0001. EV, extracellular vesicle; AD, Alzheimer's disease; FDR, false discovery rate; TNF‐α, tumour necrosis factor alpha; IL‐1β, interleukin 1 beta; IL‐6, interleukin 6; HC, healthy control; AD, Alzheimer's disease
To further validate whether EV proteins from the M7 astrocyte module were associated with AD pathology, we focused on the activated astrocyte‐derived EV markers (CAV1, ICAM1, and ITGA5) and the hub protein (ITGB1) within M7 and analyzed their protein levels in brain‐derived EVs from an independent cohort consisting of five HC and five AD patients. No difference was observed in the total brain EV samples from the HC and AD cases with immunoblotting after normalization of the signals with the reference protein β‐actin (Supplementary Figure S7a). In addition, as the M7 module is highly linked to astrocytes, we also enriched astrocyte‐specific EVs from the brain‐derived total EVs by immunoprecipitation using antibodies against astrocyte‐specific EV markers LRP1 and EAAT1 (Figure 7e). The newly identified astrocyte marker ITGA637 and common EV markers CD9, EAAT1, and LRP1 were present in the immunoprecipitated EV samples (Figure 7f). Notably, other cell‐type‐specific EV proteins (ATP1A3, LCP1, and FTH1) were undetectable in the immunoprecipitated EV samples (Supplementary Figure S7b). These data demonstrate the successful immunoprecipitation of astrocyte‐derived EVs from the total brain‐derived EVs using antibodies against EAAT1 and LRP1. We observed that ITGB1 levels were significantly increased in astrocyte‐specific EVs in AD samples when compared to controls after normalization of the intensity with EAAT1, while the levels of ITGA6 and CD9 were unchanged (Figure 7f). Interestingly, LRP1 levels were significantly reduced in the astrocyte‐specific EVs in the AD samples when compared to the controls, likely indicating a reduction in astrocytic expression of LRP1. Since LRP1 mediates extracellular tau uptake (Rauch et al., 2020) and astrocytic LRP1 reportedly promotes brain Aβ clearance (Liu et al., 2017), it may indicate the reduced clearance of tau by astrocytes in the AD brain. Notably, ITGB1 levels were also significantly correlated with AD pathogenic hallmarks, including Aβ42, total tau, and pSer396 tau levels using our previously published human brain‐derived EV proteomic dataset and measurements (Figure 7g; Supplementary Table S18) (Muraoka, Deleo, et al., 2020). In summary, our findings suggest a leading contribution of EV protein co‐expression network within the M7 astrocyte module in AD pathogenesis. Furthermore, the identified M7 EV proteins (e.g., ITGB1), particularly in astrocyte‐specific EVs, could be utilized as potential AD biomarkers.
4. DISCUSSION
In this study, we have reported the proteomic profiles of EVs isolated from four different hiPSC‐derived neural cell types (iNeurons, iAstrocytes, iMGL, and iOligos) using label‐free quantitative mass spectrometry. With these unique proteomic datasets, a total of 16 universal proteins were identified as novel brain pan‐EV marker candidates, including HSPA8, ITGB1, BSG, and ENO1. We also characterized neural cell‐type‐specific EV proteins that could be used as potential markers for brain cell‐type‐specific EV isolation. TMT‐based high‐resolution proteomic profiling of the AD, MCI, and HC brain‐derived EVs was conducted to explore alterations in the cellular composition and the disease‐associated status that occurs during AD progression, by combining the two proteomic and pathological datasets using WGCNA. The EV protein co‐expression network was correlated most strongly to the AD trait that was significantly enriched in astrocyte‐derived EV profiles and intrinsic components of the plasma membrane. Moreover, the M7 module is most correlated with AD traits and enriched in astrocytic‐EV markers as well as reactive astrocyte‐derived EV proteins, which are implicated in neuroinflammation, suggesting a potential causative role for this EV protein network in disease progression.
We used hiPSCs to differentiate into four major neural cell types, including astrocytes, excitatory neurons, microglia, and oligodendrocytes, to collect homogenous cell‐type‐specific EV protein profiling. Intriguingly, we found an increased diameter in iAstorcyte‐EVs with NTA, which was validated by TEM imaging. This is likely due to the higher portion of microvesicles secreted by iAstrocytes when compared to other cell types, as with the current EV isolation methods it is difficult to separate exosomes and microvesicles (Brennan et al., 2020). Microvesicle‐specific EV markers flotillin‐1 and 2 (FLOT1 and FLOT2) were uniquely identified in iAstrocyte‐EVs, supporting a potential difference in the synthesis and secretion of different EV populations from astrocytes. This was further supported by a recent report showing that microvesicles contain proteins distinct from those of exosomes (Haraszti et al., 2016).
Neurodegenerative disorders, including AD, are driven by specific cell types in the brain (Wilson & Nairn, 2018). Our EV proteome data identified not only potential brain pan‐EV markers for general EV purification and detection from major neural cell types, but also specific markers to improve the accuracy and availability of EV isolation from the cell of origin. For instance, proteins involved in synaptic function and neuron development were expressed at higher levels or found uniquely in iNeuron‐EVs, such as SYT1, STX1B, ATP1A3, NCAM1, and RTN1. In contrast, iMGL‐EVs are enriched in proteins associated with antigen presentation processing, immune response, and T cell signalling, including ITGAM, ITGB2, CD300A, and LCP1, whereas iAstrocyte‐EVs were enriched in proteins related to the extracellular matrix, cell metabolism, and integrin signalling (e.g., A2M, SLC2A1, SLC1A5, APOE, LRP1, and ITGA6). Interestingly, FTH1 was uniquely enriched in iOligo‐EVs and may be considered a novel EV marker for oligodendrocytes. A recent study reported that FTH1 is secreted by murine oligodendrocyte‐derived EVs and provides an antioxidant defense against neuroprotection. Importantly, some of these highlighted molecules were also enriched in the cell lysates of each cell type and human brain‐derived EVs, further illustrating their existence and the cell‐type specificity of EVs. Of note, the putative neuron‐derived EV marker L1CAM and astrocyte‐derived EV marker EAAT1/GLAST (Goetzl et al., 2015, 2016), were not found in our label‐free cell‐type‐specific EV proteome data but were present in cell lysates. It is possible that our methods may lack the sensitivity needed to identify low‐abundance proteins (Nguyen et al., 2018). It is also possible that L1CAM is not associated with neuron‐derived EVs, as was recently reported (Norman et al., 2021) and the weak signal shown in our immunoblotting. Thus, different markers need to be explored to develop a high‐affinity immunocapture method. Astrocytes co‐cultured with neurons in vitro may potentially enhance EAAT1/GLAST expression in EVs, as evidence has shown that EAAT1/GLAST expression in astrocytes is significantly increased during neuronal co‐culture (Hasel et al., 2017; Swanson et al., 1997). Additionally, oligodendrocyte‐specific markers such as PLP1, MOG, and MBP were not identified in either the iOligo cell lysate or iOligo‐EV, likely due to the lack of myelin sheaths in the in vitro iOligo culture without neurons (Ehrlich et al., 2017). Further research on isolating EVs from the co‐cultured systems, or 3D minibrains, might help to expand our knowledge of cell type‐specific EV profiles that are similar to those in the human brain.
In our cell type‐specific AD brain‐derived EV protein co‐expression network analysis, three protein modules (M1/M6/M9) were negatively correlated with AD, suggesting that the disruption of biological functions was reflected by these modules, such as microtubules and cellular metabolism. The EV protein network module M7 most strongly correlated with AD was associated with the component of the plasma membrane and enriched in astrocyte‐derived EV proteins. The M7 module proteins are also increased in MCI patients and showed the strongest positive correlation with cognitive function and AD pathology hallmarks, suggesting that the biological changes in the EV proteins within this module might occur early in the disease and may have significant contributions to AD pathology. For example, SLC7A2, a M7 hub protein strongly associated with the Braak stage (p = 8.65e‐6), is involved in the induction of nitric oxide synthesis (iNOS/NOS2), and its inactivation could exacerbate AD pathology in APP/PS1 mice (Cifuentes et al., 2017). It is an astrocyte‐specific L‐arginine transporter in the brain that is induced by astrocyte activation (Stevens et al., 1996), signifying its relevance for cell type‐specificity and disease association. Another M7 hub protein, S100A6, is highly expressed in astrocytes around Aβ plaques in the AD brain, and likely induces Aβ disaggregation and degradation (Tian et al., 2019). Thus, our results highlight that glia‐derived EVs, particularly those from astrocytes, could be utilized as sensitive markers for monitoring AD progression. This is consistent with the findings of recent EV studies and other protein co‐expression analyses of AD (Johnson et al., 2020; Muraoka, Deleo, et al., 2020; Muraoka, Jedrychowski, Yanamandra, et al., 2020; Seyfried et al., 2017). Despite the accumulating evidence that microglia‐derived EVs mediate neurodegeneration (Asai et al., 2015; Ruan et al., 2020), no significant disease‐associated microglia‐specific EV modules were found in this investigation. We assume that this may be due to the presence of the ageing‐associated microglia phenotype among all age‐matched groups, making the difference insignificant, especially as there was a lack of unique proteomic EV datasets isolated from iMGL with the disease‐associated microglia (DAM) phenotype to integrate with the WGCNA (Olah et al., 2018; Srinivasan et al., 2020). Proteomic profiling of the EVs enriched from the DAM‐induced iMGLs is required in future investigation to identify DAM modules in human brain‐derived EV proteomics modules.
This study has also found that the M7 module was enriched in EV proteins associated with activated astrocytes (You et al., 2020). This protein network is regulated by pro‐inflammatory factors and involved in mediating inflammation. Among them, the hub protein ITGB1 was significantly increased in AD brain‐derived astrocyte‐specific EVs in an independent cohort. Integrins, including ITGB1, are implicated in regulating synaptic plasticity and immune responses in neurodegenerative conditions (Wu & Reddy, 2012). A previous study found that soluble Aβ can interact with and activate ITGB1 in astrocytes to promote astrogliosis by activating the nicotinamide adenine dinucleotide phosphate oxidase 2 activation pathway (Wyssenbach et al., 2016). Disruption of ITGB1 expression in astrocytes significantly alters extracellular Aβ levels, thereby contributing to AD pathology (Sullivan et al., 2019). Therefore, our findings suggest that EVs shed by astrocytes, particularly reactive astrocytes, may be deleterious to brain health and contribute to AD pathology. Future studies targeting M7 protein levels, such as ITGB1 or astrocyte‐derived EV secretion, would provide new insights for AD therapies.
There were some limitations with this study. First, is that we only used a single hiPSC line to characterize cell type‐specific EV protein profiles. Future studies utilizing different cell lines with various backgrounds are warranted to better understand the fundamental signatures of the cell‐type‐specific EVs. Second, the characterization of the EV markers related to disease conditions was limited as we profiled EVs from hiPSC‐derived neural cells under normal conditions. For example, microglia are known to induce DAM in disease conditions, including AD (Clayton et al., 2017; Krasemann et al., 2017). A recent single‐cell RNA sequencing study identified a range of human microglia subtypes some which were altered in AD, and this may be reflected in their EV protein composition. Indeed, our recent study showed that EVs isolated from aged APP/PS1 mice were enriched in disease‐specific molecules, such as APP, PS1, and ITGAX (integrin alpha X)/CD11c, a representative marker of DAM (Muraoka, Jedrychowski, Iwahara, et al., 2021). Further studies are required to profile the EVs isolated from hiPSC‐derived neural cells under various conditions mimicking neurodegenerative disorders for a more comprehensive characterization of the cell type‐specific disease‐associated proteins.
In conclusion, our combined proteomics study of the cell type‐specific EVs isolated from hiPSC‐derived neural cells and AD brain‐derived EVs presents the availability and specificity of cell type‐specific EV markers for EV isolation, and provides a new outlook for the EV proteomic network landscape of AD. We also highlight the key role of astrocyte‐derived EVs, particularly reactive astrocytes, in the pathogenesis of AD. Importantly, we identified that the ITGB1 protein in astrocyte‐derived EVs may serve as a potential AD biomarker. These findings improve our understanding of EV biology in neurodegenerative diseases and show promise for the development of novel diagnostic and therapeutic targets specifically for AD.
CONFLICTS OF INTEREST
T.I. has a sponsored research agreement from Abbvie, Inc. for this study. M.B.J. and W.W.P. are co‐inventors of patent WO/2018/160496, related to the differentiation of human pluripotent stem cells into microglia.
AUTHOR CONTRIBUTIONS
Yang You, Satoshi Muraoka, and Tsuneya Ikezu: designed the research; Yang You, Satoshi Muraoka, Mark P. Jedrychowski, Amanda K. McQuade, and Roshanak Aslebagh: carried out the experiments; Yang You, Satoshi Muraoka, and Jianqiao Hu: analyzed data; Tracy Young‐Pearse: provided the protocol and advice on hiPSC‐derived neuron differentiation; Scott A. Shaffer and Steven P. Gygi: provided oversight and advice on mass spectrometry; Amanda K. McQuade, Mathew Blurton‐Jones, and Wayne W. Poon: provided the hiPSC cell lines, iMGLs, and advice on hiPSC‐derived iMGL differentiation; Yang You and Satoshi Muraoka: wrote the manuscript with input from co‐authors; Tsuneya Ikezu: supervised the study and contributed to manuscript preparation and editing. All authors have read and approved the final version of the manuscript.
Supporting information
FigureS1‐S9
Supporting Information
TableS1‐S18
ACKNOWLEDGEMENTS
We would like to thank the staff members of the Laboratory of Molecular NeuroTherapeutics at Boston University and Mayo Clinic Florida for technical assistance, NIH NeuroBioBank for unfixed human frozen brain tissues, and M. Ericsson (Electron Microscopy Facility, Harvard Medical School) for electron microscope imaging services. We also thank BioRender.com for providing resources and templates for the illustrations. This work was funded in part by AbbVie, Inc. (TI), Alzheimer's Association AARF‐9550302678 (SM), Alzheimer's Research UK (TI), Cure Alzheimer's Fund (TI), NIH RF1 AG054199 (TI), NIH R01 AG054672 (TI), NIH R01 AG066429 (TI), NIH R01 AG072719 (TI), NIH R01 AG067763 (TI), and NIH R21 NS104609 (TI).
You, Y. , Muraoka, S. , Jedrychowski, M. P. , Hu, J. , McQuade, A. K. , Young‐Pearse, T. , Aslebagh, R. , Shaffer, S. A. , Gygi, S. P. , Blurton‐Jones, M. , Poon, W. W. , & Ikezu T., (2022). Human neural cell type‐specific extracellular vesicle proteome defines disease‐related molecules associated with activated astrocytes in Alzheimer's disease brain. Journal of Extracellular Vesicles, 11, e12183. 10.1002/jev2.12183
REFERENCES
- Abud, E. M. , Ramirez, R. N. , Martinez, E. S. , Healy, L. M. , Nguyen, C. H. H. , Newman, S. A. , Yeromin, A. V. , Scarfone, V. M. , Marsh, S. E. , Fimbres, C. , Caraway, C. A. , Fote, G. M. , Madany, A. M. , Agrawal, A. , Kayed, R. , Gylys, K. H. , Cahalan, M. D. , Cummings, B. J. , Antel, J. P. , … Blurton‐Jones, M. (2017). iPSC‐derived human microglia‐like cells to study neurological diseases. Neuron, 94, 278–293 e279. 10.1016/j.neuron.2017.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, H. , Ikezu, S. , Tsunoda, S. , Medalla, M. , Luebke, J. , Haydar, T. , Wolozin, B. , Butovsky, O. , Kügler, S. , & Ikezu, T. (2015). Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nature Neuroscience, 18, 1584–1593. 10.1038/nn.4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási, A.‐L. , Gulbahce, N. , & Loscalzo, J. (2011). Network medicine: A network‐based approach to human disease. Nature Reviews Genetics, 12, 56–68. 10.1038/nrg2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbar, L. , Jain, T. , Zimmer, M. , Kruglikov, I. , Sadick, J. S. , Wang, M. , Kalpana, K. , Rose, I. V. L. , Burstein, S. R. , Rusielewicz, T. , Nijsure, M. , Guttenplan, K. A. , Di Domenico, A. , Croft, G. , Zhang, B. , Nobuta, H. , Hébert, J. M. , Liddelow, S. A. , & Fossati, V. (2020). CD49f is a novel marker of functional and reactive human iPSC‐derived astrocytes. Neuron, 107, 436–453 e412. 10.1016/j.neuron.2020.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, K. , Martin, K. , FitzGerald, S. P. , O'Sullivan, J. , Wu, Y. , Blanco, A. , Richardson, C. , & Mc Gee, M. M. (2020). A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Scientific Reports, 10, 1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik, V. , Ruiz‐Cañada, C. , & Wendler, F. (2016). Extracellular vesicles round off communication in the nervous system. Nature Reviews Neuroscience, 17, 160–172. 10.1038/nrn.2015.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, H. , Cong, W.‐N.a , Ji, S. , Rothman, S. , Maudsley, S. , & Martin, B. (2012). Metabolic dysfunction in Alzheimer's disease and related neurodegenerative disorders. Current Alzheimer Research, 9, 5–17. 10.2174/156720512799015064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiasserini, D. , Van Weering, J. R. T. , Piersma, S. R. , Pham, T. V. , Malekzadeh, A. , Teunissen, C. E. , De Wit, H. , & Jiménez, C. R. (2014). Proteomic analysis of cerebrospinal fluid extracellular vesicles: A comprehensive dataset. Journal of Proteomics, 106, 191–204. 10.1016/j.jprot.2014.04.028 [DOI] [PubMed] [Google Scholar]
- Cifuentes, D. , Poittevin, M. , Bonnin, P. , Ngkelo, A. , Kubis, N. , Merkulova‐Rainon, T. , & Lévy, B. I. (2017). Inactivation of nitric oxide synthesis exacerbates the development of alzheimer disease pathology in APPPS1 mice (Amyloid Precursor Protein/Presenilin‐1). Hypertension, 70, 613–623. 10.1161/HYPERTENSIONAHA.117.09742 [DOI] [PubMed] [Google Scholar]
- Clayton, K. A. , Van Enoo, A. A. , & Ikezu, T. (2017). Alzheimer's disease: The role of microglia in brain homeostasis and proteopathy. Frontiers in Neuroscience, 11, 680. 10.3389/fnins.2017.00680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborators, G.B.D.D. (2019). Global, regional, and national burden of Alzheimer's disease and other dementias, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurology, 18, 88–106. 10.1016/S1474-4422(18)30403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristóvão, J. S. , & Gomes, C. M. (2019). S100 proteins in Alzheimer's disease. Frontiers in Neuroscience, 13, 463. 10.3389/fnins.2019.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpech, J. ‐. C. , Herron, S. , Botros, M. B. , & Ikezu, T. (2019). Neuroimmune crosstalk through extracellular vesicles in health and disease. Trends in Neuroscience (Tins), 42, 361–372. 10.1016/j.tins.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich, M. , Mozafari, S. , Glatza, M. , Starost, L. , Velychko, S. , Hallmann, A.‐L. , Cui, Q.‐L. , Schambach, A. , Kim, K. ‐. P. , Bachelin, C. , Marteyn, A. , Hargus, G. , Johnson, R. M. , Antel, J. , Sterneckert, J. , Zaehres, H. , Schöler, H. R. , Baron‐Van Evercooren, A. , & Kuhlmann, T. (2017). Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proceedings of the National Academy of Sciences of the United States of America, 114, E2243–E2252. 10.1073/pnas.1614412114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi, F. M. , Casaletto, K. B. , La Joie, R. , Walters, S. M. , Harvey, D. , Wolf, A. , Edwards, L. , Rivera‐Contreras, W. , Karydas, A. , Cobigo, Y. , Rosen, H. J. , Decarli, C. , Miller, B. L. , Rabinovici, G. D. , & Kramer, J. H. (2020). Plasma biomarkers of astrocytic and neuronal dysfunction in early‐ and late‐onset Alzheimer's disease. Alzheimers Dement, 16, 681–695. 10.1016/j.jalz.2019.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauré, J. , Lachenal, G. , Court, M. , Hirrlinger, J. , Chatellard‐Causse, C. , Blot, B. , Grange, J. , Schoehn, G. , Goldberg, Y. , Boyer, V. , Kirchhoff, F. , Raposo, G. , Garin, J. , & Sadoul, R. (2006). Exosomes are released by cultured cortical neurones. Molecular and Cellular Neuroscience, 31, 642–648. 10.1016/j.mcn.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Feinstein, S. C. , & Wilson, L. (2005). Inability of tau to properly regulate neuronal microtubule dynamics: A loss‐of‐function mechanism by which tau might mediate neuronal cell death. Biochimica Et Biophysica Acta, 1739, 268–279. 10.1016/j.bbadis.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Fiandaca, M. S. , Kapogiannis, D. , Mapstone, M. , Boxer, A. , Eitan, E. , Schwartz, J. B. , Abner, E. L. , Petersen, R. C. , Federoff, H. J. , Miller, B. L. , & Goetzl, E. J. (2015). Identification of preclinical Alzheimer's disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case‐control study. Alzheimers Dement, 11, 600–607 e601. 10.1016/j.jalz.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzl, E. J. , Boxer, A. , Schwartz, J. B. , Abner, E. L. , Petersen, R. C. , Miller, B. L. , & Kapogiannis, D. (2015). Altered lysosomal proteins in neural‐derived plasma exosomes in preclinical Alzheimer disease. Neurology, 85, 40–47. 10.1212/WNL.0000000000001702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzl, E. J. , Mustapic, M. , Kapogiannis, D. , Eitan, E. , Lobach, I. V. , Goetzl, L. , Schwartz, J. B. , & Miller, B. L. (2016). Cargo proteins of plasma astrocyte‐derived exosomes in Alzheimer's disease. Faseb Journal, 30, 3853–3859. 10.1096/fj.201600756R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzl, E. J. , Schwartz, J. B. , Abner, E. L. , Jicha, G. A. , & Kapogiannis, D. (2018). High complement levels in astrocyte‐derived exosomes of Alzheimer disease. Annals of Neurology, 83, 544–552. 10.1002/ana.25172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guix, F. , Corbett, G. , Cha, D. , Mustapic, M. , Liu, W. , Mengel, D. , Chen, Z. , Aikawa, E. , Young‐Pearse, T. , Kapogiannis, D. , Selkoe, D. , & Walsh, D. (2018). Detection of aggregation‐competent tau in neuron‐derived extracellular vesicles. International Journal of Molecular Sciences, 19, 663. 10.3390/ijms19030663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraszti, R. A. , Didiot, M. ‐. C. , Sapp, E. , Leszyk, J. , Shaffer, S. A. , Rockwell, H. E. , Gao, F. , Narain, N. R. , Difiglia, M. , Kiebish, M. A. , Aronin, N. , & Khvorova, A. (2016). High‐resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. Journal of Extracellular Vesicles, 5, 32570. 10.3402/jev.v5.32570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasel, P. , Dando, O. , Jiwaji, Z. , Baxter, P. , Todd, A. C. , Heron, S. , Márkus, N. M. , Mcqueen, J. , Hampton, D. W. , Torvell, M. , Tiwari, S. S. , Mckay, S. , Eraso‐Pichot, A. , Zorzano, A. , Masgrau, R. , Galea, E. , Chandran, S. , Wyllie, D. J. A. , Simpson, T. I. , & Hardingham, G. E. (2017). Neurons and neuronal activity control gene expression in astrocytes to regulate their development and metabolism. Nature Communications, 8, 15132 10.1038/ncomms15132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, A. , Kim, H. S. , Bojmar, L. , Gyan, K. E. , Cioffi, M. , Hernandez, J. , Zambirinis, C. P. , Rodrigues, G. , Molina, H. , Heissel, S. , Mark, M. T. , Steiner, L. , Benito‐Martin, A. , Lucotti, S. , Di Giannatale, A. , Offer, K. , Nakajima, M. , Williams, C. , Nogués, L. , …, Lyden, D. (2020). Extracellular vesicle and particle biomarkers define multiple human cancers. Cell, 182, 1044–1061 e1018. 10.1016/j.cell.2020.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, C. R. , Bennett, D. A. , Blennow, K. , Carrillo, M. C. , Dunn, B. , Haeberlein, S. B. , Holtzman, D. M. , Jagust, W. , Jessen, F. , Karlawish, J. , Liu, E. , Molinuevo, J. L. , Montine, T. , Phelps, C. , Rankin, K. P. , Rowe, C. C. , Scheltens, P. , Siemers, E. , Snyder, H. M. , … Silverberg, N. (2018). NIA‐AA research framework: Toward a biological definition of Alzheimer's disease. Alzheimers Dement, 14, 535–562. 10.1016/j.jalz.2018.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E. C. B. , Dammer, E. B. , Duong, D. M. , Ping, L. , Zhou, M. , Yin, L. , Higginbotham, L. A. , Guajardo, A. , White, B. , Troncoso, J. C. , Thambisetty, M. , Montine, T. J. , Lee, E. B. , Trojanowski, J. Q. , Beach, T. G. , Reiman, E. M. , Haroutunian, V. , Wang, M. , Schadt, E. , … Seyfried, N. T. (2020). Large‐scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nature Medicine, 26, 769–780. 10.1038/s41591-020-0815-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaimal, V. , Bardes, E. E. , Tabar, S. C. , Jegga, A. G. , & Aronow, B. J. (2010). ToppCluster: A multiple gene list feature analyzer for comparative enrichment clustering and network‐based dissection of biological systems. Nucleic Acids Research, 38, W96–W102. 10.1093/nar/gkq418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann, S. , Madore, C. , Cialic, R. , Baufeld, C. , Calcagno, N. , El Fatimy, R. , Beckers, L. , O'loughlin, E. , Xu, Y. , Fanek, Z. , Greco, D. J. , Smith, S. T. , Tweet, G. , Humulock, Z. , Zrzavy, T. , Conde‐Sanroman, P. , Gacias, M. , Weng, Z. , Chen, H. , … Butovsky, O. (2017). The TREM2‐APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity, 47, 566–581 e569. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder, P. , & Horvath, S. (2008). WGCNA: An R package for weighted correlation network analysis. Bmc Bioinformatics, 9, 559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. ‐. C. , Hu, J. , Zhao, N.a , Wang, J. , Wang, N.a , Cirrito, J. R. , Kanekiyo, T. , Holtzman, D. M. , & Bu, G. (2017). Astrocytic LRP1 mediates brain abeta clearance and impacts amyloid deposition. Journal of Neuroscience, 37, 4023–4031. 10.1523/JNEUROSCI.3442-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenzie, A. T. , Moyon, S. , Wang, M. , Katsyv, I. , Song, W. ‐. M. , Zhou, X. , Dammer, E. B. , Duong, D. M. , Aaker, J. , Zhao, Y. , Beckmann, N. , Wang, P. , Zhu, J. , Lah, J. J. , Seyfried, N. T. , Levey, A. I. , Katsel, P. , Haroutunian, V. , Schadt, E. E. , … Zhang, B. (2017). Multiscale network modeling of oligodendrocytes reveals molecular components of myelin dysregulation in Alzheimer's disease. Molecular Neurodegeneration, 12, 82. 10.1186/s13024-017-0219-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcquade, A. , Coburn, M. , Tu, C. H. , Hasselmann, J. , Davtyan, H. , & Blurton‐Jones, M. (2018). Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Molecular Neurodegeneration, 13, 67. 10.1186/s13024-018-0297-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. A. , Oldham, M. C. , & Geschwind, D. H. (2008). A systems level analysis of transcriptional changes in Alzheimer's disease and normal aging. Journal of Neuroscience, 28, 1410–1420. 10.1523/JNEUROSCI.4098-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, C. , Kling, T. , Russo, B. , Miebach, K. , Kess, E. , Schifferer, M. , Pedro, L. D. , Weikert, U. , Fard, M. K. , Kannaiyan, N. , Rossner, M. , Aicher, M. ‐. L. , Goebbels, S. , Nave, K. ‐. A. , Krämer‐Albers, E. ‐. M. , Schneider, A. , & Simons, M. (2020). Oligodendrocytes provide antioxidant defense function for neurons by secreting ferritin heavy chain. Cell Metabolism, 32, 259–272 e210. 10.1016/j.cmet.2020.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy, L. A. , Pink, R. C. , & Carter, D. R. F. (2014). Routes and mechanisms of extracellular vesicle uptake. Journal of Extracellular Vesicles, 3, 24641. 10.3402/jev.v3.24641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka, S. , Jedrychowski, M. P. , Iwahara, N. , Abdullah, M. , Onos, K. D. , Keezer, K. D. , Hu, J. , Ikezu, S. , Howell, G. R. , Gygi, S. P. & Ikezu , T. (2021). Enrichment of neurodegenerative microglia signature in brain‐derived extracellular vesicles isolated from Alzheimer's disease mouse model. Journal of Proteome Research, 20(3), 1733‐1743. 10.1021/acs.jproteome.0c00934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka, S. , Deleo, A. M. , Sethi, M. K. , Yukawa‐Takamatsu, K. , Yang, Z. , Ko, J. , Hogan, J. D. , Ruan, Z. , You, Y. , Wang, Y. , Medalla, M. , Ikezu, S. , Chen, M. , Xia, W. , Gorantla, S. , Gendelman, H. E. , Issadore, D. , Zaia, J. , & Ikezu, T. (2020). Proteomic and biological profiling of extracellular vesicles from Alzheimer's disease human brain tissues. Alzheimers Dement, 16, 896–907. 10.1002/alz.12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka, S. , Jedrychowski, M. P. , Yanamandra, K. , Ikezu, S. , Gygi, S. P. , & Ikezu, T. (2020). Proteomic profiling of extracellular vesicles Derived from cerebrospinal fluid of Alzheimer's Disease patients: A pilot study. Cells, 9, 1959. 10.3390/cells9091959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka, S. , Lin, W. , Chen, M. , Hersh, S. W. , Emili, A. , Xia, W. , & Ikezu, T. (2020). Assessment of separation methods for extracellular vesicles from human and mouse brain tissues and human cerebrospinal fluids. Methods (San Diego, Calif.), 177, 35–49. 10.1016/j.ymeth.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, D. C. , Lewis, H. C. , Joyner, C. , Warren, V. , Xiao, H. , Kissick, H. T. , Wu, R. , Galipeau, J. , & Lee, F. E.‐H. (2018). Extracellular vesicles from bone marrow‐derived mesenchymal stromal cells support ex vivo survival of human antibody secreting cells. Journal of Extracellular Vesicles, 7, 1463778. 10.1080/20013078.2018.1463778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueras‐Ortiz, C. J. , Mahairaki, V. , Delgado‐Peraza, F. , Das, D. , Avgerinos, K. , Eren, E. , Hentschel, M. , Goetzl, E. J. , Mattson, M. P. , & Kapogiannis, D. (2020). Astrocyte‐ and neuron‐derived extracellular vesicles from Alzheimer's Disease patients effect complement‐mediated neurotoxicity. Cells, 9, 1618. 10.3390/cells9071618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman, M. , Ter‐Ovanesyan, D. , Trieu, W. , Lazarovits, R. , Kowal, E. J. K. , Lee, J.u H. , Chen‐Plotkin, A. S. , Regev, A. , Church, G. M. , & Walt, D. R. (2021). L1CAM is not associated with extracellular vesicles in human cerebrospinal fluid or plasma. Nature Methods, 18, 631–634. 10.1038/s41592-021-01174-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah, M. , Patrick, E. , Villani, A. ‐. C. , Xu, J. , White, C. C. , Ryan, K. J. , Piehowski, P. , Kapasi, A. , Nejad, P. , Cimpean, M. , Connor, S. , Yung, C. J. , Frangieh, M. , Mchenry, A. , Elyaman, W. , Petyuk, V. , Schneider, J. A. , Bennett, D. A. , De Jager, P. L. , & Bradshaw, E. M. (2018). A transcriptomic atlas of aged human microglia. Nature Communications, 9, 539. 10.1038/s41467-018-02926-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz, B. B. R. , Hansmann, F. , Spitzbarth, I. , Kalkuhl, A. , Deschl, U. , Baumgärtner, W. , & Ulrich, R. (2014). Transcriptomic meta‐analysis of multiple sclerosis and its experimental models. Plos One, 9, e86643. 10.1371/journal.pone.0086643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch, J. N. , Luna, G. , Guzman, E. , Audouard, M. , Challis, C. , Sibih, Y. E. , Leshuk, C. , Hernandez, I. , Wegmann, S. , Hyman, B. T. , Gradinaru, V. , Kampmann, M. , & Kosik, K. S. (2020). LRP1 is a master regulator of tau uptake and spread. Nature, 580, 381–385. 10.1038/s41586-020-2156-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, Z. , Delpech, J. ‐. C. , Venkatesan Kalavai, S. , Van Enoo, A. A. , Hu, J. , Ikezu, S. , & Ikezu, T. (2020). P2RX7 inhibitor suppresses exosome secretion and disease phenotype in P301S tau transgenic mice. Molecular Neurodegeneration, 15, 47. 10.1186/s13024-020-00396-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar Sinha, M. , Ansell‐Schultz, A. , Civitelli, L. , Hildesjö, C. , Larsson, M. , Lannfelt, L. , Ingelsson, M. , & Hallbeck, M. (2018). Alzheimer's disease pathology propagation by exosomes containing toxic amyloid‐beta oligomers. Acta Neuropathologica, 136, 41–56. 10.1007/s00401-018-1868-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried, N. T. , Dammer, E. B. , Swarup, V. , Nandakumar, D. , Duong, D. M. , Yin, L. , Deng, Q. , Nguyen, T. , Hales, C. M. , Wingo, T. , Glass, J. , Gearing, M. , Thambisetty, M. , Troncoso, J. C. , Geschwind, D. H. , Lah, J. J. , & Levey, A. I. (2017). A multi‐network approach identifies protein‐specific co‐expression in asymptomatic and symptomatic Alzheimer's disease. Cell Systems, 4, 60–72 e64. 10.1016/j.cels.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, K. , Schmitt, S. , Bergner, C. G. , Tyanova, S. , Kannaiyan, N. , Manrique‐Hoyos, N. , Kongi, K. , Cantuti, L. , Hanisch, U. ‐. K. , Philips, M. ‐. A. , Rossner, M. J. , Mann, M. , & Simons, M. (2015). Cell type‐ and brain region‐resolved mouse brain proteome. Nature Neuroscience, 18, 1819–1831. 10.1038/nn.4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, K. , Friedman, B. A. , Etxeberria, A. , Huntley, M. A. , Van Der Brug, M. P. , Foreman, O. , Paw, J. S. , Modrusan, Z. , Beach, T. G. , Serrano, G. E. , & Hansen, D. V. (2020). Alzheimer's patient microglia exhibit enhanced aging and unique transcriptional activation. Cell Reports, 31, 107843. 10.1016/j.celrep.2020.107843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing, M. J. , Cottee, J. M. , & Rana, I. (2015). The role of ion channels in microglial activation and proliferation—A complex interplay between ligand‐gated ion Channels, K(+) Channels, and Intracellular Ca(2.). Frontiers in Immunology, 6, 497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele, N. Z. R. , Carr, J. S. , Bonham, L. W. , Geier, E. G. , Damotte, V. , Miller, Z. A. , Desikan, R. S. , Boehme, K. L. , Mukherjee, S. , Crane, P. K. , Kauwe, J. S. K. , Kramer, J. H. , Miller, B. L. , Coppola, G. , Hollenbach, J. A. , Huang, Y. , & Yokoyama, J. S. (2017). Fine‐mapping of the human leukocyte antigen locus as a risk factor for Alzheimer disease: A case‐control study. Plos Medicine, 14, e1002272. 10.1371/journal.pmed.1002272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, B. R. , Kakuda, D. K. , Yu, K. , Waters, M. , Vo, C. B. , & Raizada, M. K. (1996). Induced nitric oxide synthesis is dependent on induced alternatively spliced CAT‐2 encoding L‐arginine transport in brain astrocytes. Journal of Biological Chemistry, 271, 24017–24022. 10.1074/jbc.271.39.24017 [DOI] [PubMed] [Google Scholar]
- Sullivan, S. E. , Liao, M. , Smith, R. V. , White, C. , Lagomarsino, V. N. , Xu, J. , Taga, M. , Bennett, D. A. , De Jager, P. L. , & Young‐Pearse, T. L. (2019). Candidate‐based screening via gene modulation in human neurons and astrocytes implicates FERMT2 in Abeta and TAU proteostasis. Human Molecular Genetics, 28, 718–735. 10.1093/hmg/ddy376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, R. A. , Liu, J. , Miller, J. W. , Rothstein, J. D. , Farrell, K. A. , Stein, B. , & Longuemare, M. C. (1997). Neuronal regulation of glutamate transporter subtype expression in astrocytes. Journal of Neuroscience, 17, 932–940. 10.1523/JNEUROSCI.17-03-00932.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcw, J. , Wang, M. , Pimenova, A. A. , Bowles, K. R. , Hartley, B. J. , Lacin, E. , Machlovi, S. I. , Abdelaal, R. , Karch, C. M. , Phatnani, H. , Slesinger, P. A. , Zhang, B. , Goate, A. M. , & Brennand, K. J. (2017). An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Reports, 9, 600–614. 10.1016/j.stemcr.2017.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. ‐. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , …, Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7, 1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, A. G. , Gray, E. , Heman‐Ackah, S. M. , Mäger, I. , Talbot, K. , Andaloussi, S. E.l , Wood, M. J. , & Turner, M. R. (2016). Extracellular vesicles in neurodegenerative disease—Pathogenesis to biomarkers. Nature Reviews Neurology, 12, 346–357. 10.1038/nrneurol.2016.68 [DOI] [PubMed] [Google Scholar]
- Thompson, A. , Wölmer, N. , Koncarevic, S. , Selzer, S. , Böhm, G. , Legner, H. , Schmid, P. , Kienle, S. , Penning, P. , Höhle, C. , Berfelde, A. , Martinez‐Pinna, R. , Farztdinov, V. , Jung, S. , Kuhn, K. , & Pike, I. (2019). TMTpro: Design, synthesis, and initial evaluation of a proline‐based isobaric 16‐plex tandem mass tag reagent set. Analytical Chemistry, 91, 15941–15950. 10.1021/acs.analchem.9b04474 [DOI] [PubMed] [Google Scholar]
- Tian, Z. ‐. Y. , Wang, C. ‐. Y. , Wang, T. , Li, Y. ‐. C. , & Wang, Z.‐Y. (2019). Glial S100A6 degrades beta‐amyloid aggregation through targeting competition with zinc ions. Aging and Disease, 10, 756–769. 10.14336/AD.2018.0912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, L. , Rad, R. , Gygi, S. P. , & Haas, W. (2011). MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nature Methods, 8, 937–940. 10.1038/nmeth.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Niel, G. , D'angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology, 19, 213–228. 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- Van Wagoner, N. J. , & Benveniste, E. N. (1999). Interleukin‐6 expression and regulation in astrocytes. Journal of Neuroimmunology, 100(1‐2), 124–139. 10.1016/S0165-5728(99)00187-3 [DOI] [PubMed] [Google Scholar]
- Verbeek, M. M. , Otte‐Höller, I. , Westphal, J. R. , Wesseling, P. , Ruiter, D. J. , & de Waal, R. M. (1994). Accumulation of intercellular adhesion molecule‐1 in senile plaques in brain tissue of patients with Alzheimer's disease. American Journal of Pathology, 144, 104–116 [PMC free article] [PubMed] [Google Scholar]
- Wilson, R. S. , & Nairn, A. C. (2018). Cell‐type‐specific proteomics: A neuroscience perspective. Proteomes, 6, 51. 10.3390/proteomes6040051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , & Reddy, D. S. (2012). Integrins as receptor targets for neurological disorders. Pharmacology & Therapeutics, 134, 68–81. 10.1016/j.pharmthera.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyssenbach, A. , Quintela, T. , Llavero, F. , Zugaza, J. L. , Matute, C. , & Alberdi, E. (2016). Amyloid beta‐induced astrogliosis is mediated by beta1‐integrin via NADPH oxidase 2 in Alzheimer's disease. Aging Cell, 15, 1140–1152. 10.1111/acel.12521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, Y. , Borgmann, K. , Edara, V. V. , Stacy, S. , Ghorpade, A. , & Ikezu, T. (2020). Activated human astrocyte‐derived extracellular vesicles modulate neuronal uptake, differentiation and firing. Journal of Extracellular Vesicles, 9, 1706801. 10.1080/20013078.2019.1706801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, Y. , & Ikezu, T. (2019). Emerging roles of extracellular vesicles in neurodegenerative disorders. Neurobiology of Disease, 130, 104512. 10.1016/j.nbd.2019.104512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon, A. C. , Gaj, S. , Ho, I. , Hanspers, K. , Vranizan, K. , Evelo, C. T. , Conklin, B. R. , Pico, A. R. , & Salomonis, N. (2012). GO‐Elite: A flexible solution for pathway and ontology over‐representation. Bioinformatics, 28, 2209–2210. 10.1093/bioinformatics/bts366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Pak, C. , Han, Y. , Ahlenius, H. , Zhang, Z. , Chanda, S. , Marro, S. , Patzke, C. , Acuna, C. , Covy, J. , Xu, W. , Yang, N. , Danko, T. , Chen, L.u , Wernig, M. , & Südhof, T. C. (2013). Rapid single‐step induction of functional neurons from human pluripotent stem cells. Neuron, 78, 785–798. 10.1016/j.neuron.2013.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FigureS1‐S9
Supporting Information
TableS1‐S18


