Abstract
Two agents from natural sources, citroflavonoids naringin and naringenin, can target enzymes in pathogenic yeasts responsible for hospital infections and crop failure. The aim of this study was to examine the molecular recognition site for naringin and naringenin on the HMGR and TOPOII enzymes of eleven Candida species and one phytopathogen, U. maydis, and evaluate yeast susceptibility to these flavonoids. The HMGR and TOPOII enzymes were analyzed in silico. The alignment of the two enzymes in the twelve pathogenic organisms with the corresponding enzyme of Homo sapiens revealed highly conserved amino acid sequences. Modeling studies of the enzymes indicated highly conserved structures. According to molecular docking simulations, both citroflavonoids recognized the amino acid residues of the active site of the enzymes. Binding energy values were higher for naringin (−10.75 and −9.38 kcal/mol, respectively) than simvastatin on HMGR (−9.9) and curcumin on TOPOII (−8.37). The appraisal of twenty-nine virtual mutations provided evidence of probable mechanisms of resistance (high binding energy) or susceptibility (low energy) to the drugs and emphasized the role of key residues. An in vitro susceptibility evaluation of the twelve yeasts demonstrated that the two flavonoids have similar or better MIC values than those reported for the reference compounds, obtaining the lowest with Candida dubliniensis (2.5 µg/ml) and U. maydis (5 µg/ml). Based on the present findings, naringin and naringenin could possibly be effective for treating diseases caused by pathogenic yeasts of the Candida species and U. maydis, presumably by inhibition of their HMGR and TOPOII enzymes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12088-021-00980-0.
Keywords: Candida spp., U. maydis, HMGR, TOPOII, Docking, Modeling, Mutagenesis
Introduction
Yeast infections have been on the rise in recent years, being the main cause of high mortality rates in immunocompetent patients hospitalized in medical units [1–4]. The most relevant opportunistic pathogenic yeasts in humans are those belonging to the Candida species, which is the etiological agent of candidiasis [5]. A series of resistance mechanisms of Candida spp. against triazoles have been described. The representative drug in the triazoles, fluconazole [5–8], has been tested in combination with other drugs in various studies to analyze the resulting inhibitory activity. Nevertheless, the resistance mechanisms have led to low effectiveness in the combination antifungal treatments [9–11]. On the other hand, U. maydis is a basidiomycete and phytopathogenic fungus responsible for corn smut disease, which can negatively affect farming and the food industry, as seen in other endophytic fungi. Because of the considerable economic impact of this fungus, its physiology and genetics have been subjects of extensive research [12, 13]. The yeasts Candida spp. and U. maydis have both been employed as models of ascomycete and basidiomycete organisms, respectively, in the search for new therapeutic targets [14–16]. Diverse molecular targets have been utilized for the assessment of new drugs on these model organisms. The present effort focuses on the enzymes 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) and DNA topoisomerase type II (TOPOII).
The HMGR enzyme intervenes in the biosynthetic pathway of the membrane component, cholesterol in humans and ergosterol in fungi. It has been a target for the investigation of potential hypocholesterolemic and antifungal agents [14–16] and was recently described as an alternative target for the evaluation of reference compounds (e.g., simvastatin) and new compounds, including those derived from alpha asarone [15].
Another proposed target is the TOPOII enzyme, known to be inhibited by compounds with anticancer activity such as etoposide and curcumin [17, 18]. Derivatives of the curcumin have shown a potent effect on the function of TOPOII. Inhibitors of this enzyme have been evaluated on clinical isolates of C. albicans, demonstrating inhibition of their viability.
Hence, new and previously known inhibitors of the HMGR [15] and TOPOII enzymes represent possible therapies for hypocholesteremia, fungal infections and cancer [17, 18]. However, alternatives are needed to improve their pharmaceutical activity and reduce their side effects. Nowadays, there is increasing recognition of the importance of compounds from natural sources as inhibitors of enzymes and coadjuvants for other drugs. The advantages of such compounds include their wide-ranging pharmacological activity, the ease of obtaining them, and their capacity to decrease adverse effects [19, 20].
Two compounds with interesting potential are naringin and naringenin, present in citrus fruits (e.g., grapefruits and oranges). Among the diverse biological properties of these flavonoids are their anti-hyperlipidemic and anti-carcinogenic activity, as seen in the extract of other plants [21]. Naringin is a glycosylated flavanone, while naringenin, also a flavanone, is an aglycone of naringin formed by its hydrolysis.
The modification of lipid metabolism is one of the multiple pharmacological effects produced by the two flavonoids [22]. Naringin has exhibited hypolipidemic activity in hamsters with diet-induced hypercholesterolemia, and the combination of this compound with pravastatin (inhibitor of the HMGR enzyme) improves the antiobesogenic effects of the latter [23]. Naringenin has shown hypocholesterolemic action by diminishing the activity of HMGR in rats fed a high-cholesterol diet. Additionally, both naringin and naringenin suppress cell growth in cancer cells in vitro and in vivo (in experimental animal models) [24]. Similar compounds have been shown in other work to perform essential functions in plants and microbes [25]. On the other hand, in previous studies, point mutations have been reported in the HMGR and TOPOII enzymes of Pseudomonas mevalonii and Escherichia coli, respectively. So far there are no in silico studies of the generation of point mutants in the HMGR and TOPOII enzymes of opportunistic pathogenic fungi of the genus Candida and U. maydis, this is the first work where such mutations would be addressed, which will help us to know the role of certain amino acids in the molecular recognition of the citroflavonoids naringin and naringenin towards the active site of the HMGR and TOPOII enzymes of Candida spp. and U. maydis and predict a probable mechanism of resistance or susceptibility to the compounds evaluated in this work. The aim of the current study was to examine the molecular recognition site of naringin and naringenin on the HMGR and TOPOII enzymes of eleven Candida species and one phytopathogen, U. maydis, and evaluate the susceptibility of the yeasts to the flavonoid treatment.
Material and Methods
Microorganisms and Compounds
The 12 fungal strains included presently were the following: C. albicans (ATCC 10231), C. auris Val5, C. dubliniensis (CD36), C. glabrata (CBS138), C. guilliermondii (also called Meyerozyma guilliermondii, ATCC 6260), C. haemulonii ENCB-87, C. kefyr ENCB-BMBL (also called C. Kluyveromyces marxianus), C. krusei ATCC 6358, C. lusitaniae (ATCC 38533), C. parapsilosis (ATCC 22019), C. tropicalis (MYA -3404), and U. maydis 521. The flavonoids (naringin and naringenin) and reference compounds (fluconazole, simvastatin and curcumin) were purchased from Sigma-Aldrich. Simvastatin was acquired in the form of a lactone prodrug but was needed in its active form.
Paired and Multiple Sequence Alignments of the HMGR and TOPOII Enzymes of Candida spp., U. maydis and H. sapiens
The alignments were carried out with the amino acid sequences of the HMGR (catalytic fraction) and TOPOII enzymes of Candida spp., U. maydis and H. sapiens from the NCBI database (https://www.ncbi.nlm.nih.gov/) [26]. Subsequently, sequence alignment was performed in pairs to calculate the local alignment (expressed as values of identity and similarity) of each of the HMGR and TOPOII enzymes of the 12 organisms (Candida spp. and U. maydis) with the corresponding enzymes of H. sapiens, using the EMBOSS Water tool (entailing the application of the Smith-Waterman algorithm). Additionally, multiple sequence alignment was done with the HMGR and TOPOII enzymes of the 13 organisms on the Clustal Omega program in order to locate the domains of interest (motif sites) and the amino acid residues participating in the catalytic site of both enzymes.
Homology Modeling of the HMGR and TOPOII Enzymes of Candida spp. and U. maydis
The homology modeling technique was conducted with version 9.24 of the Modeller program [27], designed for comparative modeling of 3D protein structures. The structures utilized were the crystallized HMGR enzyme (PDB: 1DQA) and the TOPOII enzyme of H. sapiens in complex with a 22-base pair DNA duplex (PDB: 3QX3). Both complexes were deposited in the protein data bank (https://www.rcsb.org/). Once downloaded in its PDB format, the scripts necessary to generate the 3D models were executed to construct 24 3D models, consisting of 12 each for HMGR and TOPOII, corresponding to the twelve yeast species. Each of the models was evaluated on three programs: Procheck, ProQ and Verify3D.
Molecular Docking
Molecular docking studies were carried out between the flavonoids (naringin and naringenin) and the enzymes (HMGR and TOPOII of Candida spp. and U. maydis) on the Autodock4 program [28]. The 2D structures of naringin, naringenin and reference compounds (simvastatin and curcumin) were downloaded from the ZINC15 server, then converted into 3D structures with the Open Babel GUI program, which were optimized on the Gaussian 6.0 program with semi-empirical AM1 calculations to obtain the lowest energy conformation. Hydrogen atoms were added to each of the models (12 HMGRs and 12 TOPOIIs) with the MolProbity program. The parameters for simulation were prepared with the Visual Molecular Dynamics program (VMD 1.9.1) and the models were optimized on the nanoscale molecular dynamics (NAMD) program. The docked model with the lowest binding energy (Kcal/mol) was employed in the docking analyses, editing the results in Discovery Studio Visualizer [29].
In Silico Generation and Evaluation of Site-Directed Mutations
Site-directed mutations were performed in silico in the HMGR and TOPOII enzymes that exhibited the best identity and similarity percentages in relation to the H. sapiens enzymes. Point mutations were made and the mutants provided by the Modeller 9.24 program [27].
In Vitro Susceptibility of Candida spp. and U. maydis to Naringin and Naringenin
The minimal inhibitory concentrations (MICs) were ascertained for the flavonoids (naringin and naringenin) and reference compounds (simvastatin and curcumin) on the 12 fungal species (see Sect. 2.1) as described in the M27-A3 document for yeasts from the CLSI guide. In supplementary supporting the methodology is detailed for a better understanding.
Results and Discussion
The Alignment of the Sequences of HMGR and TOPOII of Candida spp. or U. maydis with those of H. sapiens
Suppl. Fig. S1 shows the alignment of the catalytic fraction of the HMGR enzyme of C. dubliniensis with the same fraction of H. sapiens, highlighting the characteristic domains of the catalytic fraction: the enzyme substrate binding site (HMG-CoA) and the dimerization and cofactor binding site (NADPH). Since such domains are highly conserved in the 12 yeast sequences subjected to multiple sequence alignment (Suppl. Fig. S2). In addition to the previously mentioned domains, the amino acid residues participating in catalysis (E97, K231, D307 and H405) were observed in the HMGR of C. dubliniensis and the 12 yeasts currently investigated. Alignment was made between the TOPOII sequences of U. maydis and H. sapiens (Suppl. Fig. S3), as well as multiple alignments with all 12 yeast sequences used presently (Suppl. Fig. S4). In such alignments, the toprim domain (topoisomerase-primase) is marked in green. It is a structurally conserved catalytic domain involved in DNA strand breakage and rejoining. The toprim domain has conserved motifs, one focusing on the conserved glutamate residue (E) and another on the two conserved aspartates (DxD). Two amino acids, lysine (K) and glutamine (N), also play a role in the interaction with the DNA of the enzyme. Given the structural similarity between these enzymes, they could be used as a model for the evaluation of new compounds proposed as TOPOII inhibitors in the search for alternative antifungal and anticancer therapy [17, 18]. The percentages of identity and similarity of each of the HMGR and TOPOII sequences under study with the respective enzymes of H. sapiens is shown in Table 1. The analysis of these two enzymes is presented here in the text, while the results of the other enzymes are included in the supplementary material.
Table 1.
Percentage of identity and similarity of HMGR or TOPOII from the yeasts with the corresponding enzymes of H. sapiens
| Enzyme | Identity (%) | Similarity (%) | Enzyme | Identity (%) | Similarity (%) |
|---|---|---|---|---|---|
| C. albicans HMGR | 60.1 | 76.5 | C. albicans TOPOII | 51.5 | 70.9 |
| C. auris HMGR | 59.9 | 74.6 | C. auris TOPOII | 49.3 | 69.3 |
| C. dubliniensis HMGR | 60.6 | 76 | C. dubliniensis TOPOII | 51.6 | 70.8 |
| C. glabrata HMGR | 60.1 | 74.3 | C. glabrata TOPOII | 49.9 | 69.3 |
| C. guilliermondii HMGR | 59.6 | 74.7 | C. guilliermondii TOPOII | 51.2 | 71.7 |
| C. haemulonii HMGR | 59.7 | 75.1 | C. haemulonii TOPOII | 49.1 | 69.8 |
| C. kefyr HMGR | 59.8 | 75.9 | C. kefyr TOPOII | 50.8 | 69 |
| C. krusei HMGR | 57.9 | 75.4 | C. krusei TOPOII | 48.5 | 67.9 |
| C. lusitaniae HMGR | 58.9 | 74.9 | C. lusitaniae TOPOII | 49.6 | 69.4 |
| C. parapsilosis HMGR | 60.1 | 75.6 | C. parapsilosis TOPOII | 51.6 | 71.9 |
| C. tropicalis HMGR | 60.1 | 76.5 | C. tropicalis TOPOII | 52.5 | 70.8 |
| U. maydis HMGR | 60.7 | 75.9 | U. maydis TOPOII | 55.4 | 71 |
Homology Modeling and Evaluation of the HMGR and TOPOII Yeast Enzymes
Twelve models each of the HMGR and TOPOII enzymes of Candida spp. and U. maydis were generated. The optimal model with the lowest DOPE value was selected. Suppl. Fig. S5 illustrates the overlay of the best twelve models from the HMGR and TOPOII enzymes, indicating a close structural similarity between the yeast and H. sapiens enzymes. The best model selected from the HMGR and TOPOII enzymes of Candida spp. and U. maydis is portrayed in Figs. 1 and 2, respectively. The models of the HMGR and TOPOII of C. dubliniensis are included in the Suppl. Figures S6 and S9, respectively, as are the results of their analysis with the Procheck, ProQ and Verify3D programs.
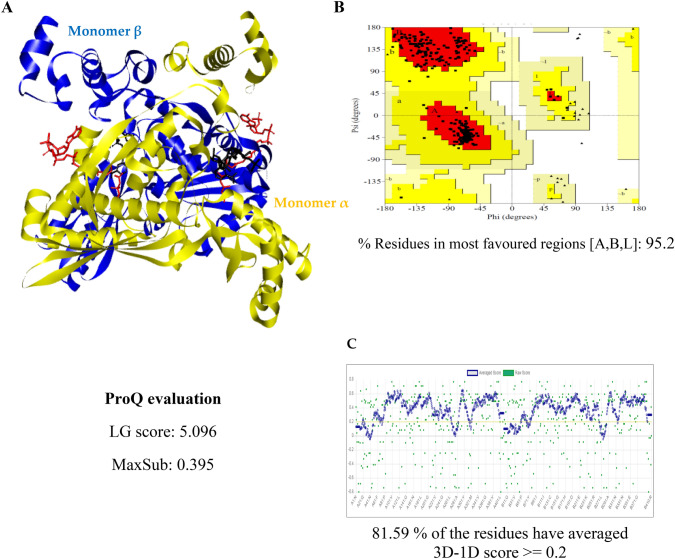
Fig. 1.
Model and assessment of C. dubliniensis HMGR. a The flat ribbon 3D model of C. dubliniensis HMGR is represented in its dimeric form (monomer α in yellow and monomer β in blue). In each monomer, the HMG-CoA (substrate) is depicted in red and the NADPH (cofactor) in black. b Ramachandran plot of C. dubliniensis HMGR. c Portrayal of the verification of the 3D model C. dubliniensis HMGR
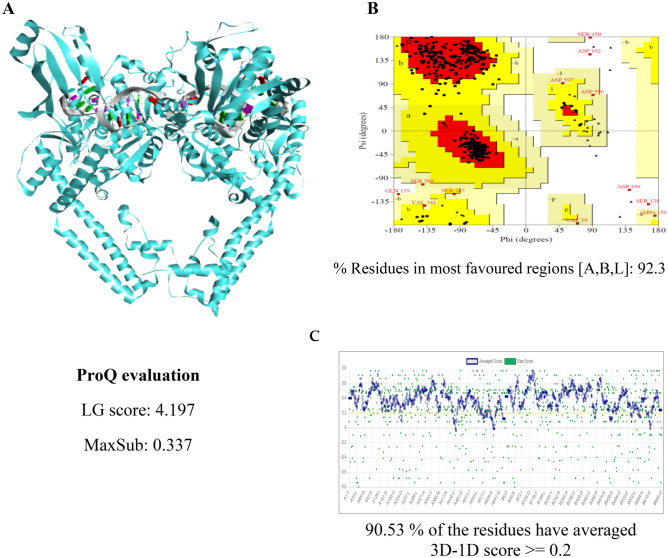
Fig. 2.
Model and assessment of TOPOII U. maydis. a The flat ribbon model of 3D TOPOII U. maydis is represented in complex with DNA. b Ramachandran plot of TOPOII U. maydis. c Portrayal of the verification of 3D TOPOII U. maydis model
The values were above 90% for the Ramachandran plots of the remaining yeast enzymes, evidencing that the models generated are of good quality (Suppl. Figs. S7 and S10). For the HMGR and TOPOII models of Candida spp. and U. maydis, the majority of the values obtained with LG were > 4 (Suppl. Tables 1S and 2S) and with MaxSub > 3. The 3D atomic coordinate profiles were provided by Verify3D, finding that all profiles met the requirement: at least 80% of the amino acids scored ≥ 0.2 in the 3D/1D profile of all the enzymes evaluated (Suppl. Figs. S8 and S11). The models proved to be of good to excellent quality for use in subsequent analyses.
Molecular Docking of the Flavonoids and Reference Compounds with HMGR and TOPOII of Candida spp. and U. maydis
Molecular docking simulations were carried out at the active site of the HMGR and TOPOII enzymes of Candida spp. and U. maydis. The binding modes are illustrated for the flavonoids, simvastatin, and the enzyme substrate (HMG-CoA) at the active site of C. dubliniensis HMGR (Suppl. Fig. S12). The molecular interactions involved in the binding of the flavonoids (naringin and naringenin) to the HMGR enzyme of Candida spp. and U. maydis is included in the supplementary material (Suppl. Fig. S13). The binding energy values of C. dubliniensis HMGR (Suppl. Table S3) with naringin and naringenin (−10.75 and −9.75 kcal/mol, respectively) were better or similar to the value found with simvastatin (−9.9 kcal/mol). Better binding energy values were obtained for naringin and naringenin with the remaining HMGRs of Candida spp. and U. maydis (Suppl. Table S3). The values of the current contribution are better than those reported previously when using alpha-asarone analogs in docking studies with the HMGR from C. glabrata (−4.53 to −6.1 kcal/mol) [15].
An analysis of the interaction of the TOPOII enzyme of the yeasts under study with naringin, naringenin and curcumin demonstrated that the flavonoids bind to the same active site as the reference compound (Suppl. Fig. S14). The 3D and 2D interactions are illustrated for the TOPOII yeast enzymes with the flavonoids and the reference compound (Suppl. Fig. S15). Among the highly conserved residues in such interactions (Suppl. Fig. S15 and Table S4) are Glu26, Asp28, Arg52 and Gly53 (amino acid residues) as well as DC8, DT9, DG10, DA12 and DG13 (DNA). These amino acids have also been reported in H. sapiens TOPOII (Asp479, Arg503 and Gln778) [30, 31]. The binding energy values of U. maydis TOPOII for curcumin, naringin and naringenin were −8.38, −8.38 and −7.85 kcal/mol, respectively (Suppl. Table S4). The current findings suggest that the flavonoids herein evaluated could possibly be used as alternative hypolipidemic agents, anticancer and antifungal treatments due to a remarkable similarity with the mechanism of action of these compounds.
Analysis and Assessment of Virtual Mutants in C. dubliniensis HMGR and U. maydis TOPOII
Each of the mutants was generated (Suppl. Figs. S16 and S19). The subsequent evaluation revealed that over 90% of the residues are displayed in favorable regions for all mutants (Suppl. Figs. S17 and S20). The verification of the model was also performed with ProQ and Verify3D programs (Suppl. Tables S5 and S6), and observing that 80% of the amino acids scored ≥ 0.2 in the 3D/1D profile with the latter (Suppl. Figs. S18 and S21), thus confirming that the models generating mutants are of good quality.
Based on the values found for the interactions, the binding energy values of the mutations in C. dubliniensis HMGR (Suppl. Figs. S22 and S24) and U. maydis were graphed. Studies on H. sapiens HMGR have described similar results, with amino acid residues such as glutamate (E) of the ENVIG and EGCLVAS site favoring diminished activity. Three mutations led to a more positive binding energy value, suggesting a resistance mechanism of simvastatin and naringin in the A102R mutation, of naringenin and naringin in the S103P mutation, and of naringin in the G194Y mutation. These findings correlate with a previous study, in which a mutation change from Serine (S) to Proline (P) in the HMGR gene of Aspergillus fumigatus caused significantly increased resistance to the triazole class of agents [32].
The overlay of the interaction of the two forms of C. dubliniensis HMGR (wild-type and mutant H405E) with naringin is portrayed (Suppl. Figs. S23A), as are the 2D interactions in the wild-type (yellow, Suppl. Figs. S23B) and mutant (fuchsia, Suppl. Figs. S23C) respectively are shown. In the mutant, the modification in amino acids from histidine at position 405 to glutamic acid is depicted. Mutants of this residue in H. sapiens HMGR have exhibited low catalytic activity, thus evidencing their functional role in catalysis.
According to the binding energy values of the nine mutations generated in the U. maydis TOPOII enzyme (Suppl. Figs. S24), most affect the affinity of the compound for the enzyme (causing more negative values), as occurs in C. dubliniensis HMGR mutations. However, a probable resistance mechanism was observed for curcumin with the G53S mutation, as a more positive binding energy was obtained. A similar modification was found for naringenin in the R52G mutation. The binding mode of naringin with the nine mutants resulted in significantly more negative binding energy values. Hence, naringin would likely be a more effective inhibitor than curcumin, because the former significantly affected the form, mode and energy value of binding to the enzyme. A similar phenomenon was described for triflamide derivatives with H. sapiens HMGR [31].
For purposes of clarity, only the result of the interaction between the R52G mutant and naringenin is portrayed in the overlay of the wild-type TOPOII of U. maydis on the mutant (Suppl. Figs. S25A). The 2D structures of the interactions of the respective enzymes are shown as well (Suppl. Figs. S25B and C). As can be appreciated, the hydrophilic interaction between the arginine residue (R52, wild-type) does not exist in the mutant. It was in this interaction that a more positive binding energy value was found, indicating the important role of the corresponding amino acid in the resistance mechanism of naringin to the enzyme. An impact on the type of interaction formed is also evident, finding a hydrophilic interaction in the wild-type amino acid Gly265 and a hydrophobic interaction in the mutant, as well as a hydrophobic interaction in the wild-type Ala267 and a hydrophilic interaction in the mutant. All these changes contribute significantly to the participation of this amino acid in a highly probable resistance mechanism of naringenin.
Antifungal Assessment of Naringin and Naringenin on Candida spp. and U. maydis
The MIC values were determined for each of the compounds applied to the different yeasts (Suppl. Table S7). The MICs of the naringin and naringenin were better than or equal to the reference compound in most cases. They were better than or equal to fluconazole in relation to six yeasts, simvastatin with regard to ten yeasts (2.5–80 µg/ml), and curcumin for twelve yeasts (>160 µg/ml). The best inhibition values of the flavonoids were observed in C. dubliniensis (2.5 µg/ml) and U. maydis (5 µg/ml). On the other hand, the yeast (C. haemulonii, C. lusitaniae and C. tropicalis) were more resistant to naringenin. The results of MICs for naringin and naringenin in some Candida species were the same, while in other species they were different; this is due to the varied susceptibility given their different genetic backgrounds. Previous research has shown an inhibitory effect of naringin on fungi (C. albicans and C. parapsilosis), with MIC values similar to those found presently for such species [33]. Likewise, a similar outcome has been described for naringenin derivatives on C. albicans, Alternaria alternata, Fusarium linii and A. niger [34]. Hence, the flavonoids examined probably constitute a good option for the treatment of infections caused by yeasts. The major contribution of the current effort is to demonstrate the broad spectrum of inhibition produced by naringin and naringenin on the yeasts under study, and to identify the binding mode of these flavonoids with the molecular recognition site.
Conclusions
Two flavonoids, naringin and naringenin, were evaluated for their inhibitory activity on twelve yeasts, including eleven Candida species of medical relevance and U. maydis, of biotechnological interest. Molecular docking simulations displayed the binding of naringin and naringenin to amino acid residues at the active site of the two yeast enzymes herein examined, HMGR and TOPOII. Additionally, the flavonoids showed in vitro inhibition of the yeasts. With most of the yeasts, their MIC values were equal to or better than those of the reference compounds. The flavonoids exhibited their best MIC values in relation to C. dubliniensis and U. maydis. According to the current results, the test compounds probably interact with the catalytic site of the HMGR and TOPOII enzymes. It can be concluded that naringin, naringenin, or their respective derivatives could possibility be administered as alternative antifungal treatments for pathogenic yeasts, either alone or in combination with other drugs. Moreover, the present findings validate the use of the HMGR and TOPOII yeast enzymes as models for research on new compounds proposed as an option in antifungal or anticancer therapy, respectively.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Bruce Allan Larsen for proofreading the manuscript.
Author Contributions
DMAP designed and carried out the study and wrote the manuscript. JOGG and LVT analyzed the data and reviewed the manuscript.
Funding
This work was supported by SIP-IPN Grants [20200204, 20210765]. SNI-CONACyT awarded Grants to DMAP, JOGG and LVT, COFAA-IPN and EDI-IPN provided Grants to LVT.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
We have carefully considered all the comments and suggestions made by yourself and the reviewers, which have helped to greatly improve the manuscript. The point-by-point response to each recommendation is included in a separate file. We hope that the manuscript is now suitable for publication by Indian Journal of Microbiology and look forward to hearing from you.
In honor of my father Leonardo Andrade Capetillo.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dulce Andrade-Pavón, Email: dandradep@ipn.mx, Email: andrade_eclud88@hotmail.com.
Omar Gómez-García, Email: dandradep@ipn.mx, Email: andrade_eclud88@hotmail.com, Email: gogamanj@hotmail.com.
References
- 1.Lockhart SR, Guarner J. Emerging and reemerging fungal infections. Semin Diagn Pathol. 2019;36:177–181. doi: 10.1053/j.semdp.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Suleyman G, Alangaden GJ. Nosocomial fungal infections: epidemiology, infection control, and prevention. Infect Dis Clin North Am. 2016;30:1023–1052. doi: 10.1016/j.idc.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Ostrosky-Zeichner L, Sobel JD. Fungal Infections. Infect Dis Clin North Am. 2016 doi: 10.1016/j.idc.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Garbee DD, Pierce SS, Manning J. Opportunistic fungal infections in critical care units. Crit Care Nurs Clin North Am. 2017;29:67–79. doi: 10.1016/j.cnc.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Köhler JR, Hube B, Puccia R, Casadevall A, Perfect JR. Fungi that Infect Humans Microbiol Spectr. 2017;5:1–29. doi: 10.1128/microbiolspec.FUNK-0014-2016. [DOI] [PubMed] [Google Scholar]
- 6.Jensen RH. Resistance in human pathogenic yeasts and filamentous fungi: prevalence, underlying molecular mechanisms and link to the use of antifungals in humans and the environment. Dan Med J. 2016;63:B5288. [PubMed] [Google Scholar]
- 7.Prasad R, Nair R, Banerjee A. Multidrug transporters of Candida species in clinical azole resistance. Fungal Genet Biol. 2019;132:103252. doi: 10.1016/j.fgb.2019.103252. [DOI] [PubMed] [Google Scholar]
- 8.Al-Baqsami ZF, Ahmad S, Khan Z. Antifungal drug susceptibility, molecular basis of resistance to echinocandins and molecular epidemiology of fluconazole resistance among clinical Candida glabrata isolates in Kuwait. Sci Rep. 2020;10:6238. doi: 10.1038/s41598-020-63240-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campitelli M, Zeineddine N, Samaha G, Maslak S. Combination antifungal therapy: a review of current data. J Clin Med Res. 2017;9:451–456. doi: 10.14740/jocmr2992w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghanem-Zoubi N, Qasum M, Khoury J, Zorbavel D, Arnon M, Geffen Y, Paul M. The association between fluconazole dose and MIC with mortality and persistence in candidemia. Eur J Clin Microbiol Infect Dis. 2019;38:1773–1780. doi: 10.1007/s10096-019-03611-1. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Gao L, Yu P, Kosgey JC, Jia L, Fang Y, Xiong J, Zhang F. In vitro synergy of azole antifungals and methotrexate against Candida albicans. Life Sci. 2019;235:116827. doi: 10.1016/j.lfs.2019.116827. [DOI] [PubMed] [Google Scholar]
- 12.Lanver D, Tollot M, Schweizer G, Lo Presti L, Reissmann S, Ma L-S, Schuster M, Tanaka S, Liang L, Ludwig N, Kahmann R. Ustilago maydis effectors and their impact on virulence. Nat Rev Microbiol. 2017;15:409–421. doi: 10.1038/nrmicro.2017.33. [DOI] [PubMed] [Google Scholar]
- 13.Meena H, Mishra R, Ranganathan S, Sarma VV, Ampasala DR, Kalia VC, Jung-Kul Lee J-K, Siddhardha B. Phomopsis tersa as inhibitor of quorum sensing system and biofilm forming ability of Pseudomonas aeruginosa. Indian J Microbiol. 2020;60(1):70–77. doi: 10.1007/s12088-019-00840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrade-Pavón D, Cuevas-Hernández RI, Trujillo-Ferrara JG, Hernández-Rodríguez C, Antonio Ibarra J, Villa-Tanaca L. Recombinant 3-hydroxy 3-methyl glutaryl-CoA reductase from Candida glabrata (Rec-CgHMGR) obtained by heterologous expression, as a novel therapeutic target model for testing synthetic drugs. Appl Biochem Biotechnol. 2017;182:1478–1490. doi: 10.1007/s12010-017-2412-9. [DOI] [PubMed] [Google Scholar]
- 15.Andrade-Pavón D, Ortiz-Álvarez J, Sánchez-Sandoval E, Tamariz J, Hernández-Rodríguez C, Antonio Ibarra J, Villa-Tanaca L. Inhibition of recombinant enzyme 3-hydroxy-3-methylglutaryl-CoA reductase from Candida glabrata by α-asarone-based synthetic compounds as antifungal agents. J Biotechnol. 2019;292:64–67. doi: 10.1016/j.jbiotec.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Muneeswaran G, Patel SKS, Kondaveeti S, Shanmugam R, Gopinath K, Kumar V, Kim S-Y, Lee JK, Kalia VC, Kim I-W. Biotin and Zn2+ increase xylitol production by Candida tropicalis. Indian J Microbiol. 2021;61:331–337. doi: 10.1007/s12088-021-00960-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delgado JL, Hsieh CM, Chan NL, Hiasa H. Topoisomerases as anticancer targets. Biochem J. 2018;475:373–398. doi: 10.1042/BCJ20160583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mittal A, Kumar N, Chauhan N-S. Curcumin encapsulated PEGylated nanoliposomes: a potential anti-infective therapeutic agent. Indian J Microbiol. 2019;59:336–343. doi: 10.1007/s12088-019-00811-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choksi J, Vora J, Shrivastava N. Bioactive pigments from isolated bacteria and its antibacterial, antioxidant and sun protective application useful for cosmetic products. Indian J Microbiol. 2020;60:379–382. doi: 10.1007/s12088-020-00870-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Zhao B, Liu C-J, Yang E. Optimization of biosynthesis conditions for the production of exopolysaccharides by Lactobacillus plantarum SP8 and the exopolysaccharides antioxidant activity test. Indian J Microbiol. 2020;60:334–345. doi: 10.1007/s12088-020-00865-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma B, Singh I, Bajar S, Gupta S, Gautam H, Kumar P. Biogenic silver nanoparticles: evaluation of their biological and catalytic potential. Indian J Microbiol. 2020;60:468–474. doi: 10.1007/s12088-020-00889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi R, Kulkarni YA, Wairkar S. Pharmacokinetic, pharmacodynamic and formulations aspects of Naringenin: an update. Life Sci. 2018;215:43–56. doi: 10.1016/j.lfs.2018.10.066. [DOI] [PubMed] [Google Scholar]
- 23.Raffoul-Orozco AK, Ávila-González AE, Rodríguez-Razón CM, García-Cobian TA, Pérez-Guerrero EE, García-Iglesias T, Rubio-Arellano ED. Combination effect naringin and pravastatin in lipid profile and glucose in obese rats. Life Sci. 2018;193:87–92. doi: 10.1016/j.lfs.2017.11.044. [DOI] [PubMed] [Google Scholar]
- 24.Yoshinaga A, Kajiya N, Oishi K, Kamada Y, Ikeda A, Chigwechokha PK, Kibe T, Kishida M, Kishida S, Komatsu M, Shiozaki K. NEU3 inhibitory effect of naringin suppresses cancer cell growth by attenuation of EGFR signaling through GM3 ganglioside accumulation. Eur J Pharmacol. 2016;782:21–29. doi: 10.1016/j.ejphar.2016.04.035. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Z, Jin G, Ge Y, Guo Z. Naringenin inhibits migration of breast cancer cells via inflammatory and apoptosis cell signaling pathways. Inflammopharmacology. 2019;27:1021–1036. doi: 10.1007/s10787-018-00556-3. [DOI] [PubMed] [Google Scholar]
- 26.Sharma S, Ciufo S, Starchenko E, Darji D, Chlumsky L, Karsch-Mizrachi I, Schoch CL. The NCBI BioCollections database. Database. 2019 doi: 10.1093/database/baz057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. 2016 doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bitencourt-Ferreira G, Oliveira Pintro V, Filgueira de Azevedo W. Docking with AutoDock4. Methods Mol Biol. 2019;2053:125–148. doi: 10.1007/978-1-4939-9752-7_9. [DOI] [PubMed] [Google Scholar]
- 29.Dassault Systems BIOVIA, Discovery Studio Modeling Environment, Release 2017, San Diego: Dassault Systems, 2016.
- 30.Khanderao Jadhav A, Mohan Karuppayil S. Molecular docking studies on thirteen fluoroquinolines with human topoisomerase II a and b. Silico Pharmacol. 2016;5:4. doi: 10.1007/s40203-017-0024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrade-Pavón D, Gómez-García O, Álvarez-Toledano C. Exploring the binding mode of triflamide derivatives at the active site of Topo I and Topo II enzymes: In silico analysis and precise molecular docking. J Chem Sci. 2020;132:1–20. doi: 10.3390/molecules23030599. [DOI] [Google Scholar]
- 32.Rybak JM, Ge W, Wiederhold NP, Parker JE, Kelly SL, Rogers PD, del Fortwen JR. Mutations in hmg1, challenging the paradigm of clinical triazole resistance in aspergillus fumigatus. MBio. 2019;10:e00437-19. doi: 10.1128/mBio.00437-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutiérrez-Venegas G-M, Meraz-Rodríguez MA, Flores-Sánchez MA, Ortiz-Miranda LF. Effect of flavonoids on antimicrobial activity of microorganisms present in dental plaque. Heliyon. 2019;5:e03013. doi: 10.1016/j.heliyon.2019.e03013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozłowska J, Potaniec B, Żarowska B, Anioł M. Synthesis and biological activity of novel O-alkyl derivatives of naringenin and their oximes. Molecules. 2017;22:1485. doi: 10.3390/molecules22091485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.