Summary
Adult skeletal muscle stem cells (MuSCs) are important for muscle regeneration and constitute a potential source of cell therapy. However, upon isolation, MuSCs rapidly exit quiescence and lose transplantation potency. Maintenance of the quiescent state in vitro preserves MuSC transplantation efficiency and provides an opportunity to study the biology of quiescence. Here we show that Tubastatin A (TubA), an Hdac6 inhibitor, prevents primary cilium resorption, maintains quiescence, and enhances MuSC survival ex vivo. Phenotypic characterization and transcriptomic analysis of TubA-treated cells revealed that TubA maintains most of the biological features and molecular signatures of quiescence. Furthermore, TubA-treated MuSCs showed improved engraftment ability upon transplantation. TubA also induced a return to quiescence and improved engraftment of cycling MuSCs, revealing a potentially expanded application for MuSC therapeutics. Altogether, these studies demonstrate the ability of TubA to maintain MuSC quiescence ex vivo and to enhance the therapeutic potential of MuSCs and their progeny.
Keywords: muscle stem cells, Tubastatin A, quiescence, transplantation, HDAC, primary cilium
Highlights
-
•
TubA prevents primary cilium resorption and maintains MuSC quiescence ex vivo
-
•
TubA treatment maintains the transcriptional characteristics of quiescent MuSCs
-
•
Treatment with TubA sustains MuSC engraftment potential for transplantation studies
-
•
TubA induces a return to quiescence of cycling MuSC progeny
In this article, Arjona et al. show that TubA, an HDAC6 inhibitor, can inhibit primary cilium resorption and preserve MuSC quiescence and stem cell potency ex vivo. Indeed, treatment with TubA maintains MuSC engraftment potential and induces a return to quiescence in cycling MuSCs, revealing a potentially valuable approach to enhancing the therapeutic potential of MuSCs.
Introduction
Skeletal muscle is characterized by a remarkable regenerative capacity mediated by tissue-specific muscle stem cells (MuSCs) (Dhawan and Rando, 2005; Schmidt et al., 2019). MuSCs reside under the muscle fiber basal lamina and are distinguished by the expression of the transcription factor Pax7 (Mauro, 1961; Seale et al., 2000). Under physiological conditions, MuSCs persist in a state of quiescence, but they become activated in response to different types of stress or injury (Cheung and Rando, 2013). Following activation, MuSCs enter the cell cycle and rapidly proliferate to produce myogenic progenitor cells that either differentiate into mature myofibers or self-renew and return to a quiescent state, preserving the MuSC pool (Motohashi and Asakura, 2014; Olguin and Olwin, 2004).
Quiescence is a reversible state characterized by cell-cycle arrest in which cells retain the ability to respond to different signals from their environment and re-enter the cell cycle (Cheung and Rando, 2013; van Velthoven and Rando, 2019). Quiescent cells exhibit enhanced resistance to several stresses, increased autophagic activity, small cell size, low protein synthesis, low total RNA content, and low turnover (Gray et al., 2004; Valcourt et al., 2012; van Velthoven and Rando, 2019). Quiescent MuSCs are distinguished by the expression of different myogenic regulatory factors, low transcriptional activity, and low expression of proliferation markers (Fukada et al., 2007; Olguin and Olwin, 2004; van Velthoven and Rando, 2019).
As soon as MuSCs are isolated and grown in conventional culture conditions ex vivo, they quickly activate out of quiescence and undergo cell division (Montarras, 2005). Important changes in intracellular trafficking and signaling, metabolic pathways, and gene expression are observed during MuSC exit out of quiescence, resulting in profound molecular alterations. Therefore, the ability to maintain stem cells in a quiescent state in vitro would significantly enhance the ability to study the biology of quiescence and the mechanisms that govern this state (Quarta et al., 2016). Simultaneously, a better characterization of MuSC quiescence would allow the development of novel approaches to improve homeostasis and repair of damaged or diseased muscle.
Another important potential application of maintaining cultured MuSCs in a quiescent state includes the ability to perform ex vivo gene editing for stem cell-based gene therapy. Additionally, a promising therapeutic strategy to treat many muscle diseases and traumatic injuries is the transplantation of MuSCs (Rinaldi and Perlingeiro, 2014). After transplantation, MuSCs are able to divide and either contribute to new myonuclei in growing myofibers or repopulate the stem cell compartment (Judson and Rossi, 2020). However, ex vivo expansion of MuSCs not only results in a loss of stemness but also in a considerable loss of their regenerative potential and reduction of engraftment ability (Montarras, 2005). Therefore, the ability to improve the engraftment potential of MuSCs that have been expanded in vitro is required to significantly enhance their therapeutic efficiency (Quarta et al., 2016).
Many types of quiescent cells exhibit a primary cilium, which is a microtubule-based organelle, anchored at the cellular membrane (Tucker et al., 1979a). A major function of the primary cilium is to integrate and transduce extracellular signals and to coordinate intracellular signaling pathways. One of the most important developmental pathways regulated by this organelle is the Hedgehog (Hh) signaling pathway (Anvarian et al., 2019; Huangfu et al., 2003). Primary cilia assembly and disassembly are intimately linked to cell cycle progression, with the primary cilia beginning to disassemble as cells enter the cell cycle (Kim and Tsiokas, 2011; Quarmby and Parker, 2005; Tucker et al., 1979b). The histone deacetylase 6 (Hdac6) plays a crucial role in promoting ciliary resorption by inducing the deacetylation of axonemal microtubules (de Diego et al., 2014; Pugacheva et al., 2007).
The primary cilium is present at the surface of MuSCs during quiescence but disassembles upon MuSC activation out of quiescence and entry into the cell cycle (Jaafar Marican et al., 2016). Ablation of primary cilia in adult MuSCs causes impaired muscle regeneration, lower engraftment capacity, and increased expression of cell-cycle-related genes (Palla et al., 2020). Furthermore, it has been described that repression of Hh signaling by the primary cilium is important in maintaining MuSCs in a quiescent state (Betania Cruz-Migoni et al., 2019; Palla et al., 2020). Activation of Hh signaling triggers MuSC activation and proliferation (Betania Cruz-Migoni et al., 2019; Palla et al., 2020).
Because Hdac6 appears to be an important mediator of ciliary resorption, the regulation of either Hdac6 expression or activity has been previously used to modulate ciliary dynamics and signaling, which in turn modulate cellular proliferation and, in the setting of cancer, tumor growth (Gradilone et al., 2013; Pugacheva et al., 2007; Rao et al., 2014). Tubastatin A (TubA) has been recently developed as a potent and specific small-molecule inhibitor of Hdac6 (Butler et al., 2010). Indeed, Hdac6 inhibition by TubA has been shown to increase primary cilium formation in different cell lines, and to decrease cellular proliferation in vitro and to reduce tumor growth in vivo (Gradilone et al., 2013; Pham et al., 2019; Rao et al., 2014; Tao et al., 2018; Woan et al., 2015). These findings show that TubA can be used as a powerful and effective tool to control both primary cilium dynamics and cell cycle progression.
Here we show that TubA prevents MuSC activation and progression into the cell cycle and preserves the typical phenotypic and transcriptomic characteristics of quiescent cells. Our results show that TubA, through its Hdac6 inhibitory function, impedes primary cilium resorption in quiescent MuSCs ex vivo, and that TubA improves MuSC engraftment potential in transplantation studies. Furthermore, TubA is able to induce a return to quiescence and improve the transplantation efficiency of cycling MuSCs, revealing a potentially valuable approach to enhancing the therapeutic potential of MuSCs.
Results
TubA prevents MuSC entry into the cell cycle
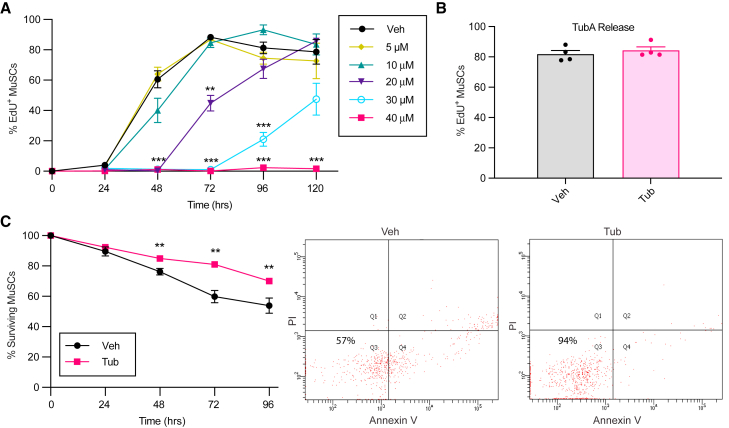
It has been described that the primary cilium is present in quiescent MuSCs but disassembles as soon as MuSCs exit out of quiescence and that the presence of the primary cilium induces the repression of Hh signaling and maintenance of the quiescence state (Betania Cruz-Migoni et al., 2019; Jaafar Marican et al., 2016; Palla et al., 2020). Furthermore, TubA has been shown to increase primary cilium formation and to downregulate Hh signaling (Gradilone et al., 2013). Based on these studies, we wondered whether TubA could maintain MuSCs in a quiescent state ex vivo by inhibiting primary cilium resorption and thus repressing Hh signaling. To investigate this, we first examined the effects of TubA on MuSC activation ability. Freshly isolated MuSCs (MuSCsFI) are characterized by reduced cell size, low RNA content, low cellular metabolism, and decreased protein synthesis, exhibiting typical characteristics of quiescent cells (van Velthoven and Rando, 2019). We cultured MuSCsFI in the presence of different doses of TubA for up to 120 h. Entry into the cell cycle was measured by EdU (5-ethynyl-2′-deoxyuridine) incorporation. At the highest dose tested (40 μM), TubA prevented almost 100% of the cells from exiting quiescence, and this effect was sustained up to 120 h (Figure 1A).
Figure 1.
TubA prevents MuSC entry into the cell cycle and enhances MuSC survival
(A) MuSCsFI were treated with different doses of TubA while being cultured continuously in EdU to assess S-phase progression. Cells were fixed at different time points, and the percentage of EdU+ MuSCs was quantified (n = 8 mice in Veh and 40 μM TubA; n = 4 mice in other conditions).
(B) MuSCs were treated with vehicle (Veh) or with 40 μM of TubA for 120 h (Tub). Cells were then cultured for 40 h in absence of TubA but the presence of EdU, after which the number of MuSCs+ for EdU was quantified (n = 4 mice).
(C) MuSCsFI were cultured for different periods of times in the absence or presence of 40 μM of TubA and then stained with Annexin V and propidium iodide (PI) to determine viability by flow cytometry. Shown are the quantification of the fraction of surviving cells at the different time points (left) and representative FACS plots (right) at 72 h with indicated survival percentages (n = 8 mice). Error bars represent ±s.e.m. ∗∗p < 0.01, ∗∗∗p < 0.001; no asterisk: not significant; two-tailed paired t test.
In order to test if this effect was reversible, we treated MuSCsFI with 40 μM of TubA for 120 h and then washed TubA out and maintained the cells for 40 h in the presence of EdU (Figure 1B). We found that release from TubA allowed the cells to activate and incorporate EdU similarly to vehicle-treated MuSCs (MuSCsVeh) exiting from quiescence. These functional studies show that TubA is able to maintain isolated MuSCs in a state of reversible quiescence for extended periods of time.
TubA prevents FAP entry into the cell cycle
In order to determine whether the effects of TubA were specific to MuSCs, we tested the effects of TubA on fibroadipogenic progenitors (FAPs). Freshly isolated FAPs were cultured in the presence of increasing doses of TubA for up to 48 h (Figure S1A). We found that 20 μM of TubA was enough to block EdU incorporation in almost 100% of FAPs. Similar to MuSCs, FAPs were able to activate and incorporate EdU after being released from TubA (Figure S1B). Altogether, these results demonstrate the ability of TubA to prevent entry into the cell cycle of multiple cell types.
TubA improves MuSC survival
Adult stem cells maintained in a quiescent state in vivo exhibit a remarkable ability to withstand environmental stress (Cheung and Rando, 2013; Der Vartanian et al., 2019; Scaramozza et al., 2019). However, in conventional ex vivo culture conditions, the absence of signals derived from the niche leads to considerable MuSC death (Liu et al., 2018; White et al., 2018). We therefore wondered whether TubA, by sustaining quiescence, would improve cell viability in isolated MuSCs over time. To test this, we cultured MuSCsFI in the presence of 40 μM TubA for up to 96 h and analyzed the cultures for cell death by Annexin V staining. TubA significantly reduced the percentage of MuSCs succumbing to apoptotic cell death over time (Figure 1C). These findings demonstrate the ability of TubA to reduce apoptotic cell death in MuSCs cultured ex vivo.
TubA prevents MuSC activation
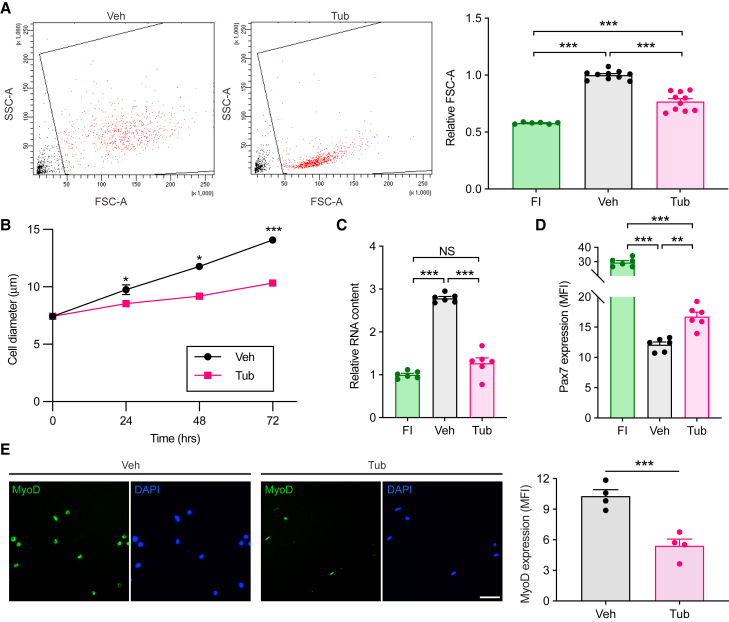
Because TubA was able to prevent cell cycle entry of MuSCsFI, we sought to further characterize the phenotypic features of TubA-treated MuSCs (MuSCsTub). We studied the behavior of MuSCsVeh during the process of activation compared with MuSCsTub maintained in culture for the same amount of time. Cell growth is one of the first processes that occurs upon MuSC activation and was measured by using fluorescence-activated cell sorting (FACS) and analyzing forward scatter area (FSC-A) (Brett et al., 2020). We found that, after 72 h in culture, MuSCsTub were much smaller than MuSCsVeh (Figure 2A). We confirmed this result by assessing cell size using a Coulter counter. Whereas MuSCsVeh increased in size over time, MuSCsTub remained small and exhibited negligible growth up to 72 h in culture (Figure 2B). Another feature of MuSC activation is an increase in RNA content (Dell’Orso et al., 2019; Tang and Rando, 2014). We found that, whereas total RNA content increased by over two-fold in MuSCsVeh over 72 h in culture, no increase was observed in MuSCsTub (Figure 2C). The MuSC-specific transcription factor Pax7 is highly expressed in quiescence but downregulated as MuSCs activate out of quiescence and enter the cell cycle (Seale et al., 2000) (Figure 2D). We found that TubA maintains Pax7 expression at significant higher levels compared with MuSCsVeh after 72 h in culture (Figure 2D). However, Pax7 expression levels were reduced by 50% in MuSCsTub compared with those of MuSCsFI (Figure 2D). We then analyzed MyoD, a master regulator of myogenic differentiation known to start being expressed at the protein level soon after MuSCs activate out of quiescence (de Morrée et al., 2017; Hausburg et al., 2015). Compared with MuSCsVeh, MuSCsTub exhibited significantly less MyoD protein expression up to 72 h in culture (Figure 2E). These data indicate that MuSCsTub exhibit typical phenotypic attributes of quiescent MuSCs. Taken together, these experiments show that TubA prevents MuSC activation and maintains the typical features of quiescence in MuSCs grown ex vivo for up to 72 h.
Figure 2.
TubA-treated cells exhibit typical phenotypic attributes of quiescent MuSCs
MuSCsFI were cultured in the presence (Tub) or absence (Veh) of 40 μM of TubA for 72 h.
(A) Cells were analyzed for cellular size by flow cytometry based on forward scatter area (FSC-A). Representative FACS plots (left) and a graph with relative FSC-A values (right) are shown. Data were normalized to the mean level in MuSCsVeh (n = 10 mice in Veh and Tub; n = 6 mice in FI).
(B) Cell size was measured with a Coulter counter at 0, 24, 48, or 72 h (n = 12 mice at 0 h, n = 10 mice at 24 and 48 h, and n = 4 mice at 72 h).
(C) Cells were assayed for RNA content based on Pyronin Y intensity in flow cytometry. Data were normalized to the mean level in MuSCsFI (n = 6 mice).
(D) FACS-isolated MuSCs were cultured for 72 h and then analyzed for Pax7 expression by immunofluorescence. Shown is the quantification of Pax7 mean fluorescence intensity (MFI) in MuSCsFI, MuSCsVeh, and MuSCsTub (n = 6 mice).
(E) MyoD immunofluorescence of either MuSCsVeh or MuSCsTub fixed after 72 h. Representative images are shown on the left. Quantification of MyoD mean fluorescence intensity (MFI) is shown on the right (n = 4 mice). Scale bar in e, 50 μm. Error bars represent ±s.e.m. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; two-tailed paired t test in (A)–(E).
TubA decreases MuSC primary cilium resorption
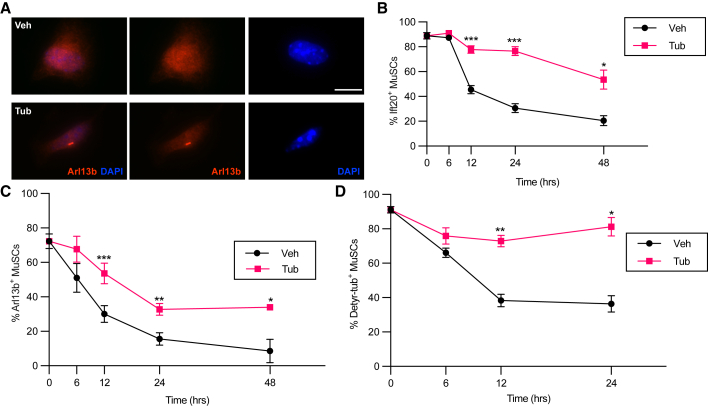
Previous findings suggest an important role of the primary cilium in MuSCs by regulating their exit from quiescence and controlling MuSC proliferation and differentiation (Betania Cruz-Migoni et al., 2019; Palla et al., 2020). Hdac6 has been proposed to facilitate primary cilium resorption (de Diego et al., 2014; Pugacheva et al., 2007), and its inhibition by TubA has been shown to restore the formation of primary cilia and to decrease cell proliferation in different cell lines (Gradilone et al., 2013; Rao et al., 2014). Consequently, we hypothesized that treatment of MuSCs with TubA ex vivo would hinder the resorption of the primary cilium associated with MuSC activation. To test this, we treated MuSCs with TubA and examined the cells for the presence of the primary cilium by immunofluorescence. To visualize this organelle, we used antibodies directed against intraflagellar transport protein 20 (Ift20) and ADP-ribosylation factor-like 13b (Arl13b), both of which are involved in cilia assembly and maintenance (Caspary et al., 2007; Follit et al., 2006), and an antibody directed against detyrosinated tubulin, a posttranslational modification involved in the anterograde intraflagellar transport (Follit et al., 2006; Sirajuddin et al., 2014). Ift20 is localized to both the basal body and the primary cilium, whereas Arl13b, a small guanosine triphosphatase, is localized to the ciliary membrane (Caspary et al., 2007; Follit et al., 2006). Detyrosinated tubulin is enriched on the outer doublets B tubules of the axonemal microtubules (Johnson, 1998). We found that TubA treatment was able to maintain a higher proportion of ciliated MuSCs (exhibiting Ift20, Arl13b, and detyrosinated tubulin staining) compared with MuSCsVeh after being cultured ex vivo (Figures 3A–3D). Thus, these experiments indicate that TubA inhibits the resorption of the primary cilium in conventional ex vivo MuSC culture conditions.
Figure 3.
TubA maintains a higher proportion of ciliated MuSCs
(A–C) MuSCsFI were cultured in the presence or absence of 40 μM of TubA and fixed and stained for primary cilium markers at 0, 6, 12, 24, and 48 h. (A) Representative images of MuSCsVeh and MuSCsTub cultured for 48 h and stained for Arl13b and DAPI. (B) MuSCs were assayed for Ift20 expression based on immunocytochemistry (n = 8 mice at 0, 12, and 24 h; n = 4 mice at 6 and 48 h). (C) MuSCs were stained for Arl13b by immunocytochemistry (n = 8 mice at 0, 12 and 24 h; n = 4 mice at 6 and 48 h).
(D) MuSCsFI were cultured in the presence or absence of 40 μM of TubA and fixed and stained for detyrosinated tubulin (Detyr-tub) at 0, 6, 12, and 24 h (n = 4 mice). Scale bar in (A), 10 μm. Error bars represent ±s.e.m. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; no asterisk: not significant; two-tailed paired t test in (B)–(D).
TubA-treated MuSCs exhibit a quiescent transcriptome signature
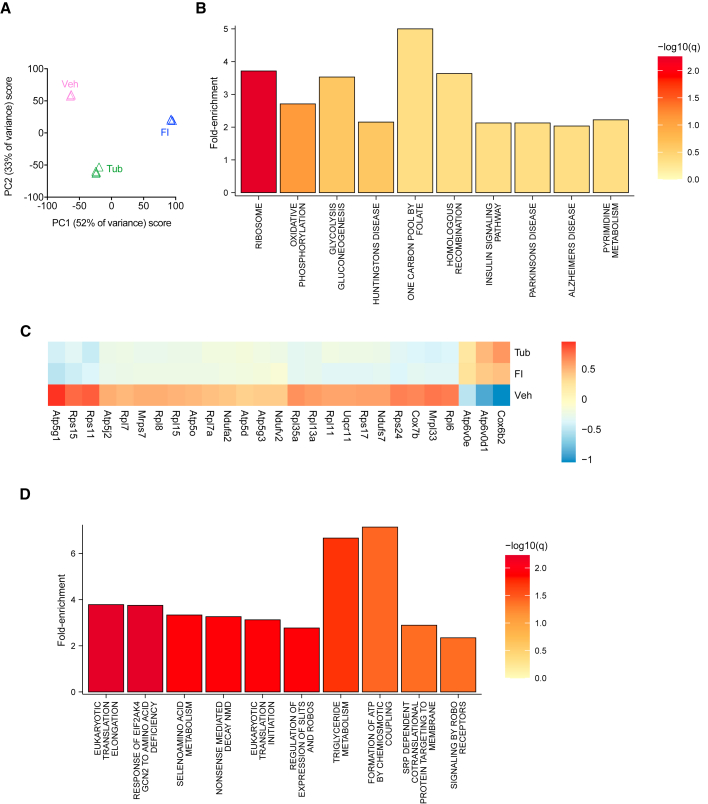
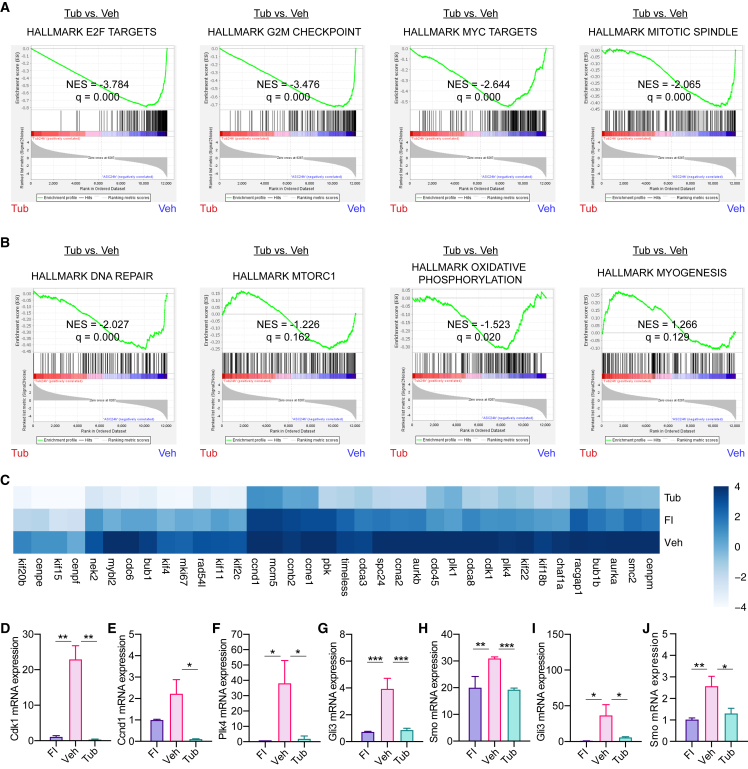
To examine the state of quiescence preserved by TubA at the transcriptome level, we performed RNA-seq on MuSCsVeh or MuSCsTub after being cultured for 24 h, as well as on freshly isolated cells for comparison. Principal component analysis (PCA) of all expressed genes revealed a higher correlation of the transcriptome of MuSCsTub with the transcriptome of MuSCsFI than that of MuSCsVeh (Figure 4A). We found that 2,572 genes were similarly expressed between MuSCsFI and MuSCsTub (Benjamini-Hochberg-corrected p value >0.05, fold-change <2). Specifically, of these 2,572 similarly expressed genes, 1,266 genes were significantly different in MuSCsVeh compared with MuSCsFI (FDR 1%, fold-change ≥4). To examine potential pathways involved in the maintenance of quiescence by TubA, we performed over-representation analysis using the KEGG gene set collection (Kanehisa, 2000). The top hits of this analysis were ribosome- and oxidative phosphorylation-related genes (Figures 4B and 4C). To further characterize these TubA-regulated genes, we used the REACTOME gene set collection (Jassal et al., 2019). We found an enrichment in transcripts involved in translation and ATP synthesis (Figure 4D), which correlated with the findings from the KEGG gene set collection. Furthermore, metabolism-related gene sets such as triglyceride metabolism, selenoamino acid metabolism, and metabolism of water-soluble vitamins and cofactors were also found to be enriched using the REACTOME gene set collection (Figure 4D).
Figure 4.
Similarities between the transcriptomes of MuSCsTub and MuSCsFI
(A) PCA of RNA-seq profiles of MuSCsFI, and of MuSCsVeh, or MuSCsTub cultured for 24 h. Each profile represents the MuSCs of an individual mouse (n = 3 mice for MuSCsFI, n = 3 mice for MuSCsVeh, and n = 4 mice for MuSCsTub).
(B) Summary plot for the over-representation analysis using the KEGG database. Over-represented signaling pathways of the similarly expressed genes between MuSCsFI and MuSCsTub are shown.
(C) Heatmap of similarly expressed genes between the MuSCsFI and the MuSCsTub transcriptomes but differentially expressed in the MuSCsVeh.
(D) Summary plot for the over-representation analysis using the REACTOME gene set collection. Over-represented signaling pathways of the similarly expressed genes between MuSCsFI and MuSCsTub are shown.
To investigate the molecular mechanisms underlying the effects of TubA on MuSCs in more detail, we examined the transcriptional differences between MuSCsTub and MuSCsVeh in the RNA-seq data. We performed gene set enrichment analysis (GSEA) using the Hallmark gene set collection (Subramanian et al., 2005). Compared with MuSCsVeh, MuSCsTub exhibited different expression levels of cell-cycle-promoting signaling gene sets including E2F targets, G2/M checkpoint, Myc targets, and mitotic spindle gene sets (Figure 5A). Furthermore, GSEA revealed that DNA repair, mTORC1 signaling, metabolism-related (cholesterol homeostasis, oxidative phosphorylation, and fatty acid metabolism), and myogenesis factors were among the differentially expressed gene sets (Figure 5B). Most of the cell-cycle-promoting signaling genes consisted of centrosome, microtubule, and primary cilium associated proteins involved in cell cycle regulation (Figure 5C). We performed RT-qPCR to validate the changes for select genes implicated in cell cycle progression and centriole duplication and found that the expression levels of some of these genes, such as Cdk1, Ccnd1, and Plk4, were lower in MuSCsTub compared with MuSCsVeh, confirming the results obtained in the RNA-seq analysis (Figures 5D–5F).
Figure 5.
Transcriptional differences between MuSCsTub and MuSCsVeh
(A and B) GSEA results for the Hallmark gene sets in comparisons of RNA-seq profiles for MuSCsTub versus MuSCsVeh after being cultured for 24 h. (A) GSEA enrichment plots for the E2F targets gene set, the G2/M checkpoint gene set, the Myc targets gene set, and the mitotic spindle gene set are shown. (B) GSEA enrichment plots for the DNA repair gene set, the mTORC gene set, the oxidative phosphorylation gene set, and the myogenesis gene set.
(C) Heatmap of differentially expressed genes between the MuSCsTub and the MuSCsVeh transcriptomes.
(D–F) RT-qPCR analyses were done in MuSCsFI, and in MuSCsVeh and MuSCsTub after being cultured for 24 h. Ct values were normalized first to the mean of Gapdh and then to the mean of MuSCsFI level in each experiment. RT-qPCR analysis of Cdk1 in (D), Ccnd1 in (E), and Plk4 in (F) (n = 3 mice).
(G and H) mRNA expression levels of Hh signaling genes such as Gli3 (G) and Smo (H) in MuSCsFI and in MuSCsVeh and MuSCsTub after being cultured for 24 h. Data obtained from our RNA-seq analysis (n = 3 mice for MuSCsFI, n = 3 mice for MuSCsVeh, and n = 4 mice for MuSCsTub).
(I and J) RT-qPCR analysis of either Gli3 (I) or Smo (J) in MuSCs. Ct values were normalized first to the mean of Gapdh and then to the mean of MuSCsFI level in each experiment (n = 6 mice). NES, normalized enrichment score in (A) and (B). Error bars represent ±s.e.m. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; no asterisk: not significant; one-tailed t test in (D)–(J).
Because activation of ciliary-mediated Hh signaling has been associated with MuSC exit from quiescence and entry into the cell cycle (Betania Cruz-Migoni et al., 2019; Palla et al., 2020), we then analyzed the expression of Hh signaling genes in our RNA-seq data. We found an upregulation of Smo and Gli3 expression in MuSCsVeh (Figures 5G and 5H), as has been previously shown (Betania Cruz-Migoni et al., 2019; Palla et al., 2020). However, the expression of these specific genes in MuSCsTub was similar to the expression levels in quiescent MuSCsFI (Figures 5G and 5H). These results were then confirmed by RT-qPCR analysis (Figures 5I and 5J). Ptch1 mRNA was nearly undetectable in both RNA-seq and RT-qPCR assays. These findings, together with the low expression levels of cell-cycle-related genes in response to TubA treatment, suggest that the ability of TubA in holding MuSCs in a quiescent state ex vivo could be due to the maintenance of the primary cilium and the subsequent repression of Hh signaling preventing MuSC entry into the cell cycle.
TubA improves MuSC engraftment ability
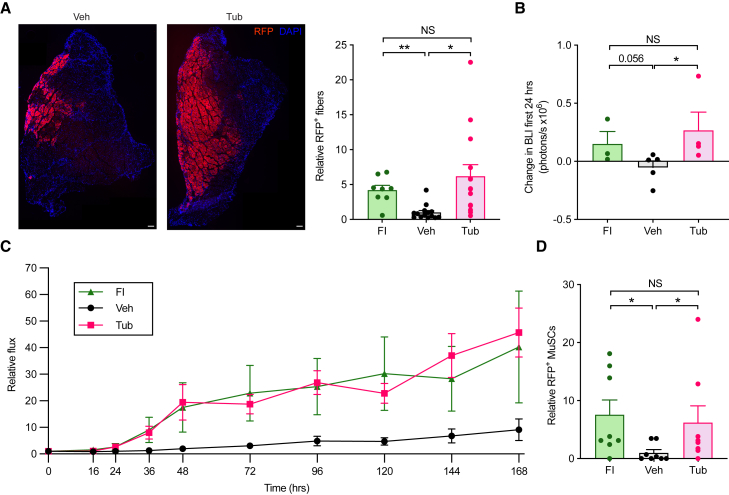
Because of their important role in muscle repair and regeneration (Lepper et al., 2011; Relaix and Zammit, 2012; Sambasivan et al., 2011), MuSCs have been considered as potential sources of cell therapy for muscle injury and for the treatment of muscular dystrophies (Collins et al., 2005; Sacco et al., 2008; Webster et al., 2016). After being transplanted into damaged or diseased muscles, MuSCs are able to expand and fuse with host myofibers (Collins et al., 2005; Sacco et al., 2008; Webster et al., 2016). However, MuSCs rapidly lose engraftment and regenerative potential as they activate out of quiescence when expanded in culture prior to transplantation (Gilbert et al., 2010; Ikemoto et al., 2007; Montarras, 2005; Quarta and Rando, 2015). Because MuSCs maintain a quiescent phenotype when cultured in the presence of TubA, we sought to determine whether TubA treatment would improve the engraftment potential of MuSCs in transplantation experiments. In order to test this, we used, as donors, Pax7CreER; R26RRFP mice, which express RFP specifically in adult MuSCs after tamoxifen administration. Freshly isolated RFP+ MuSCs were cultured in the presence of TubA for 72 h and transplanted into injured tibialis anterior (TA) muscles of NSG mice. Ten days later, the TAs were collected and analyzed. We found that TubA significantly increased the total number of RFP+ muscle fibers relative to control (Figure 6A), suggesting that TubA enhances the regenerative potential of MuSCs maintained in vitro. Indeed, no differences in the number of RFP+ fibers were observed between transplanted MuSCsFI and MuSCsTub (Figure 6A).
Figure 6.
TubA treatment enhances the regenerative potential of MuSCs maintained in vitro
(A) FACS-isolated MuSCs from Pax7CreER; R26RRFP donor mice were cultured in the presence or absence of 40 μM of TubA for 72 h and then transplanted into injured tibialis anterior (TA) muscles of NSG mice. We injected MuSCsTub in one leg of a given mouse and MuSCsVeh in the contralateral leg of that same mouse. MuSCsFI were isolated from Pax7CreER; R26RRFP mice and transplanted into injured TA muscles of recipient mice as a control. Ten days after transplantation, TA muscles were collected, sectioned, and assayed for RFP+ myofibers by immunohistochemistry. DAPI was used to stain nuclei. Representative images of TA muscles transplanted with either MuSCsVeh or MuSCsTub are shown on the left. Quantification of the number of RFP+ fibers in TA muscles transplanted with either MuSCsFI, MuSCsVeh, or MuSCsTub is shown on the right. Data were normalized to the mean level in MuSCsVeh (n = 14 mice in Veh and Tub; n = 8 mice in FI).
(B and C) FACS-isolated MuSCs that express a luciferase reporter were cultured in the presence or absence of 40 μM of TubA for 72 h and then transplanted into injured TA muscles of NSG mice. MuSCsTub were injected in one leg of a given mouse and MuSCsVeh in the contralateral leg of that same mouse. MuSCsFI were transplanted into injured TA muscles of recipient mice as a control. (B) Bioluminescence analysis was done at 0 and 24 h after transplantation. The bioluminescence signal obtained at 0 h (baseline bioluminescence signal) was then subtracted from that obtained at 24 h, and the results from this calculation are shown (n = 5 mice in Veh; n = 4 mice in Tub; n = 3 mice in FI). (C) Transplanted mice were assayed for bioluminescence for 168 h. Data were normalized to the mean level at 0 h (n = 5 mice in Veh; n = 4 mice in Tub; n = 3 mice in FI).
(D) FACS-isolated MuSCs from Pax7CreER; R26RRFP donor mice were cultured in the presence or absence of 40 μM of TubA for 72 h and then transplanted into injured TA muscles of NSG mice. We injected MuSCsTub in one leg of a given mouse and MuSCsVeh in the contralateral leg of that same mouse. MuSCsFI were isolated from Pax7CreER; R26RRFP mice and transplanted into injured TA muscles of recipient mice as a control. Four weeks after transplantation, TA muscles were collected, sectioned, and assayed for RFP+ MuSCs by immunohistochemistry. The total number of RFP+ MuSCs in TA muscles transplanted with either MuSCsFI, MuSCsVeh, or MuSCsTub was quantified. Data were normalized to the mean level at MuSCsVeh (n = 8 mice). Scale bar in (A), 100 μm. Error bars represent ± SEM. ∗p < 0.05, ∗∗p < 0.01; NS: not significant; two-tailed paired t test in (A)–(D).
We then tested the effect of TubA treatment on MuSC survival immediately after transplantation. MuSCs that expressed a luciferase reporter were treated with TubA for 72 h and then transplanted into recipient NSG mice. Bioluminescence (BLI) analysis 24 h after transplantation showed that BLI signal from MuSCsTub was significantly higher than that of MuSCsVeh but comparable to that of MuSCsFI (Figure 6B), suggesting similar survival rates between 72-h-treated MuSCsTub and quiescent MuSCs. Next, we investigated the in vivo proliferation rate of MuSC populations after transplantation. Non-invasive imaging for seven days showed comparable expansions of MuSCsTub and MuSCsFI but a much lower rate of expansion of MuSCsVeh (Figure 6C). To test whether, following TubA treatment and subsequent withdrawal, the progeny of the treated cells behave as the progeny of MuSCsFI do, we compared the in vitro proliferation rate as well as the differentiation potential of MuSCsTub following their release from TubA treatment with those of MuSCsFI cultured ex vivo. We found that the in vitro proliferation and differentiation characteristics of progeny of MuSCsTub and MuSCsFI were indistinguishable (Figures S2A and S2B).
To determine to which extent TubA treatment preserves MuSC self-renewal capacity, RFP+ MuSCs isolated from Pax7CreER; R26RRFP mice were cultured in the presence of TubA for 72 h and then transplanted into previously injured TA muscles of NSG mice. Four weeks after transplantation, the TA muscles were collected and the numbers of RFP+ MuSCs were quantified. The number of self-renewed MuSCs obtained from the transplantation of MuSCsTub was significantly higher than that from transplantation of MuSCsVeh but comparable to that following MuSCFI transplantation (Figure 6D), indicating that maintenance of a quiescent state by TubA treatment preserves MuSC potency and allows efficient self-renewal ability in vivo.
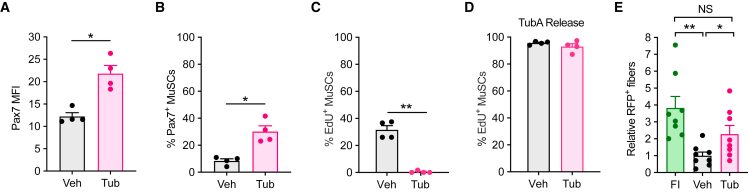
TubA induces re-quiescence in activated MuSCs
Given the rarity of MuSCs following isolation from muscle samples, the expansion of these cells in vitro is required to obtain enough cells for cellular therapy (Charville et al., 2015; Judson and Rossi, 2020). However, as noted, in vitro expansion of MuSCs results in a marked reduction in transplantation efficacy compared with freshly isolated cells (Montarras, 2005). To explore if TubA could induce a return to quiescence of MuSCs that had already activated and begun to proliferate, we allowed MuSCs to expand in culture for 72 h and then treated them with TubA for 48 h. We found that TubA induced Pax7 expression and decreased the number of cycling MuSCs as measured by EdU incorporation (Figures 7A–7C). To confirm that the non-cycling cells were indeed in a state of quiescence, we washed out TubA and maintained the cells in culture for an additional 48 h in the presence of EdU (Figure 7D). We found that the cells then entered into the cell cycle similarly to control cells. These experiments show that TubA addition to cycling MuSCs induces their return to a quiescent state.
Figure 7.
TubA induces re-quiescence in cycling MuSCs
(A–C) MuSCsFI were grown for 72 h and then cultured in the presence or absence of 40 μM of TubA for 48 h. EdU was added to the cells 15 min before fixation. MuSCs were stained for Pax7 by immunocytochemistry. Pax7 mean fluorescence intensity (MFI) (A), the number of Pax7+ cells (B), and the percentage of EdU+ cells (C) were quantified (n = 4 mice).
(D) MuSCsFI were expanded in culture for 72 h and then cultured in the presence or absence of 40 μM of TubA for 48 h. Cells were then cultured for 48 h in the absence of TubA but the presence of EdU, after which the number of EdU+ MuSCs was quantified (n = 4 mice).
(E) FACS-isolated MuSCs from Pax7CreER; R26RRFP donor mice were grown for 72 h and then treated with or without 40 μM of TubA for 48 h. These cells were then transplanted into injured TA muscles of NSG mice. We injected MuSCsTub in one leg of a given mouse and MuSCsVeh in the contralateral leg of that same mouse. MuSCsFI were isolated from Pax7CreER; R26RRFP mice and transplanted into injured TA muscles of recipient mice as a control. Ten days after transplantation, TA muscles were collected, sectioned, and assayed for RFP+ myofibers by immunohistochemistry. DAPI was used to stain nuclei. The total number of RFP+ fibers in TA muscles transplanted with either MuSCsFI, MuSCsVeh, or MuSCsTub was quantified. Data were normalized to the mean level at MuSCsVeh (n = 8 mice). Error bars represent ± s.e.m. ∗p < 0.05, ∗∗p < 0.01; no asterisk: not significant; two-tailed paired t test.
We therefore wondered whether TubA, by inducing a return to quiescence, would improve the transplantation ability of ex vivo expanded MuSCs. To investigate this, freshly isolated, RFP+ MuSCs were expanded in culture for 72 h and then treated with TubA for 48 h. These cells were then transplanted into previously injured TA muscles of NSG mice. Ten days after transplantation, the TA muscles were collected and the numbers of RFP+ fibers were quantified. TubA treatment significantly increased the number of RFP+ fibers relative to vehicle treatment (Figure 7E). Indeed, MuSCsTub yielded similar numbers of RFP+ fibers as did MuSCsFI (Figure 7E). These findings suggest that TubA could be used for cellular therapy by improving the transplantation efficacy of in vitro expanded MuSCs.
Discussion
Here we provide evidence that TubA is able to prevent MuSCs from activation and progression into S-phase. Further characterization of MuSCsTub revealed features typical of quiescent cells, such as small cell size, low RNA content, low expression levels of myogenic differentiation factors, and enhanced resilience. Transcriptomic analysis of MuSCsTub also revealed patterns typical of quiescent cells, further demonstrating the ability of TubA to maintain MuSCs in a quiescent state ex vivo and to enhance their regenerative potential in transplantation experiments. Additionally, TubA treatment was able to induce re-quiescence and improve the transplantation efficacy of activated MuSCs.
Our experiments show that TubA impedes primary cilium resorption in quiescent MuSCs and maintains low expression levels of centrosome, microtubule, and primary cilium-related genes, including Cdk1/Cyclin B, AurkA, and NEK2, involved in cell cycle regulation. By coordinating centriole duplication and mitotic spindle apparatus formation, these cell-cycle-related proteins control the balance between ciliary assembly and disassembly. Cdk1/Cyclin B inhibits Plk4-induced centriole duplication, AurkA starts the disassembly of the primary cilium by activating Hdac6, and NEK2 facilitates ciliary disassembly by activating kinesins (Hong et al., 2015; Pugacheva et al., 2007; Zitouni et al., 2016). Low expression levels of each of these genes in response to TubA treatment ensure both the maintenance of the primary cilium in MuSCs cultured ex vivo and the persistence of the ciliary repression on mitogenic signaling. Additionally, given the strong association of the primary cilium with Hh signaling and the importance of Hh signaling in MuSC exit from quiescence (Betania Cruz-Migoni et al., 2019; Palla et al., 2020), the preservation of quiescent expression levels of Hh signaling genes by TubA conditions could be contributing to the preservation of MuSC quiescence. Indeed, our results reveal that TubA promotes primary cilium formation and decreases cell proliferation in MuSCs. These findings are in accordance with recent studies showing decreased cellular proliferation after TubA treatment in various cellular types, such as melanoma cells, cholangiocarcinoma cells, and esophageal cancer cell lines (Gradilone et al., 2013; Tao et al., 2018; Woan et al., 2015). The effects of TubA treatment on the primary cilium could be associated with the maintenance of MuSC quiescence; however, this association has not yet been causally demonstrated. Further experiments need to be done in order to test this causal relationship.
MuSCs constitute an important cellular source for muscle therapy in the context of muscle injury or in the treatment of muscular dystrophies (Webster et al., 2016). However, as soon as MuSCs are isolated and cultured in conventional conditions ex vivo, they rapidly exit quiescence, activate, and proliferate, losing their stem cell potency (Ikemoto et al., 2007; Montarras, 2005; Quarta and Rando, 2015). The ability of TubA to induce a return to quiescence of cycling MuSCs, enhancing MuSC potential for cellular therapy after their expansion in vitro, demonstrates the importance of TubA applicability, constituting a potential valuable and effective tool in the context of muscle cell therapeutics.
Experimental procedures
Detailed methods can be found in the supplemental experimental procedures.
Animals
Animals were housed in pathogen-free rooms and maintained with a 12-h light-dark cycle in the Veterinary Medical Unit at the Veterans Affairs Palo Alto Health Care System. Animal protocols and care were approved by the Institutional Animal Care and Use Committee. Two- to three-month-old C57BL/6J mice (strain 000664) and R26RRFP mice on the C57BL/6J background (strain 007914) were purchased from The Jackson Laboratory. Two- to three-month-old FVB-Tg(CAG-luc,-GFP)L2G85Chco/J (strain 008450) male mice and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (strain 005557) male mice were obtained from Jackson Laboratory. Pax7CreER mice are on the C57BL/6 x 129/SvJ background (Brack et al., 2007). Pax7CreER; Rosa26RRFP were generated by mating Pax7CreER and R26RRFP mice. Two- to four-month-old Pax7CreER; Rosa26RRFP male mice were used for transplantation experiments.
Cellular purification by FACS
MuSC isolation protocol was performed as previously described (Liu et al., 2015). Hindlimb and triceps muscles were finely chopped with scissors and digested in Collagenase II and Dispase (Invitrogen). A 20G needle was used to dissociate MuSCs from myofibers, and the resulting solution was filtered using a 40-μm cell strainer and labeled with specific antibodies. MuSCs were then purified by surface antigen-based isolation (CD31- CD45- Sca1- VCAM+). In transplantation experiments, RFP-based isolation was used to purify MuSCs. Aria II and Aria III machines (BD Biosciences) were used to obtain pure MuSC populations. In order to check the purity of the MuSC population collected, an aliquot of cells was stained with Pax7 antibodies (1:50, DSHB AB_528428). FAPs were also purified by surface antigen-based isolation (CD31- CD45- Sca1+ VCAM-).
MuSC culture and treatment with Tubastatin A
Purified MuSCs were immediately plated on glass chamber slides coated with poly-D-lysine (0.1 mg/mL, EMD Millipore) and extracellular matrix (ECM, 25 μg/mL, Sigma) for immunofluorescence assays, or on plastic tissue-culture plates coated with ECM for all other experiments. Ex vivo culture of MuSCs was performed in wash media (Ham's F-10 media containing 10% horse serum, 100 U/mL penicillin, and 100 μg/mL streptomycin). Tubastatin A (Cayman Chemical) was dissolved in DMSO and, unless otherwise indicated, used at a concentration of 40 μM. An equal volume of DMSO was added to control cells.
Data and code availability
The data that support the findings of this study are available from the corresponding author upon request. RNA-seq data have been deposited in the NCBI Gene Expression Omnibus.The accession number for the RNA-seq data reported in this paper is GEO: GSE178070.
Author contributions
M.A. and T.A.R. designed experiments. M.A., A.G., C.R-M., J.O.B., P.B., and H.I. conducted and analyzed experiments. M.A. and T.A.R. wrote the manuscript.
Conflict of interests
The authors declare no conflict of interest.
Acknowledgments
We thank all the people in the Rando Lab for intellectual support.
This research was funded by grants from the Stanford University School of Medicine Medical Scientist Training Program (T32 GM007365) to J.O.B. and from the NIH (P01 AG036695 and R01 AR062185) and the Department of Veterans Affairs (Merit Review) to T.A.R.
Published: December 30, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.11.012.
Supplemental information
References
- Anvarian Z., Mykytyn K., Mukhopadhyay S., Pedersen L.B., Christensen S.T. Cellular signalling by primary cilia in development, organ function and disease. Nat. Rev. Nephrol. 2019;15:199–219. doi: 10.1038/s41581-019-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betania Cruz-Migoni S., Imran K.M., Wahid A., Rahman O., Briscoe J., Borycki A.-G. A switch in cilia-mediated Hedgehog signaling controls muscle stem cell quiescence and cell cycle progression. bioRxiv. 2019 doi: 10.1101/2019.12.21.884601. [DOI] [Google Scholar]
- Brack A.S., Conboy M.J., Roy S., Lee M., Kuo C.J., Keller C., Rando T.A. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- Brett J.O., Arjona M., Ikeda M., Quarta M., de Morrée A., Egner I.M., Perandini L.A., Ishak H.D., Goshayeshi A., Benjamin D.I., et al. Exercise rejuvenates quiescent skeletal muscle stem cells in old mice through restoration of Cyclin D1. Nat. Metab. 2020;2:307–317. doi: 10.1038/s42255-020-0190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler K.V., Kalin J., Brochier C., Vistoli G., Langley B., Kozikowski A.P. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J. Am. Chem. Soc. 2010;132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T., Larkins C.E., Anderson K.V. The graded response to sonic Hedgehog depends on cilia architecture. Dev. Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Charville G.W., Cheung T.H., Yoo B., Santos P.J., Lee G.K., Shrager J.B., Rando T.A. Ex vivo expansion and in vivo self-renewal of human muscle stem cells. Stem Cell Reports. 2015;5:621–632. doi: 10.1016/j.stemcr.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung T.H., Rando T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C.A., Olsen I., Zammit P.S., Heslop L., Petrie A., Partridge T.A., Morgan J.E. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Dell’Orso S., Juan A.H., Ko K.-D., Naz F., Perovanovic J., Gutierrez-Cruz G., Feng X., Sartorelli V. Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. Development. 2019;146:dev174177. doi: 10.1242/dev.174177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan J., Rando T.A. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 2005;15:666–673. doi: 10.1016/j.tcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- de Diego A.S., Alonso Guerrero A., Martínez-A C., van Wely K.H.M. Dido3-dependent HDAC6 targeting controls cilium size. Nat. Commun. 2014;5:3500. doi: 10.1038/ncomms4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., Tuft R.A., Fogarty K.E., Pazour G.J. The intraflagellar transport protein IFT20 is associated with the golgi complex and is required for cilia assembly. Mol. Biol. Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada S., Uezumi A., Ikemoto M., Masuda S., Segawa M., Tanimura N., Yamamoto H., Miyagoe-Suzuki Y., Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Gilbert P.M., Havenstrite K.L., Magnusson K.E.G., Sacco A., Leonardi N.A., Kraft P., Nguyen N.K., Thrun S., Lutolf M.P., Blau H.M. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradilone S.A., Radtke B.N., Bogert P.S., Huang B.Q., Gajdos G.B., LaRusso N.F. HDAC6 inhibition restores ciliary expression and decreases tumor growth. Cancer Res. 2013;73:2259–2270. doi: 10.1158/0008-5472.CAN-12-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J.V., Petsko G.A., Johnston G.C., Ringe D., Singer R.A., Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausburg M.A., Doles J.D., Clement S.L., Cadwallader A.B., Hall M.N., Blackshear P.J., Lykke-Andersen J., Olwin B.B. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. Elife. 2015;4:e03390. doi: 10.7554/eLife.03390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Kim J., Kim J. Myosin heavy chain 10 (MYH10) is required for centriole migration during the biogenesis of primary cilia. Biochem. Biophys. Res. Commun. 2015;461:180–185. doi: 10.1016/j.bbrc.2015.04.028. [DOI] [PubMed] [Google Scholar]
- Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., Anderson K.V. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Ikemoto M., Fukada S., Uezumi A., Masuda S., Miyoshi H., Yamamoto H., Wada M.R., Masubuchi N., Miyagoe-Suzuki Y., Takeda S. Autologous transplantation of SM/C-2.6+ satellite cells transduced with micro-dystrophin CS1 cDNA by lentiviral vector into mdx mice. Mol. Ther. 2007;15:2178–2185. doi: 10.1038/sj.mt.6300295. [DOI] [PubMed] [Google Scholar]
- Jaafar Marican N.H., Cruz-Migoni S.B., Borycki A.-G. Asymmetric distribution of primary cilia allocates satellite cells for self-renewal. Stem Cell Rep. 2016;6:798–805. doi: 10.1016/j.stemcr.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2019;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.A. The axonemal microtubules of the Chlamydomonas flagellum differ in tubulin isoform content. J. Cell Sci. 1998;111:313–320. doi: 10.1242/jcs.111.3.313. [DOI] [PubMed] [Google Scholar]
- Judson R.N., Rossi F.M.V. Towards stem cell therapies for skeletal muscle repair. NPJ Regen. Med. 2020;5:10. doi: 10.1038/s41536-020-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Tsiokas L. Cilia and cell cycle re-entry. Cell Cycle. 2011;10:2683–2690. doi: 10.4161/cc.10.16.17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C., Partridge T.A., Fan C.-M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Cheung T.H., Charville G.W., Rando T.A. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc. 2015;10:1612–1624. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Charville G.W., Cheung T.H., Yoo B., Santos P.J., Schroeder M., Rando T.A. Impaired notch signaling leads to a decrease in p53 activity and mitotic catastrophe in aged muscle stem cells. Cell Stem Cell. 2018;23:544–556.e4. doi: 10.1016/j.stem.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- de Morrée A., van Velthoven C.T.J., Gan Q., Salvi J.S., Klein J.D.D., Akimenko I., Quarta M., Biressi S., Rando T.A. Staufen1 inhibits MyoD translation to actively maintain muscle stem cell quiescence. Proc. Natl. Acad. Sci. U S A. 2017;114:E8996–E9005. doi: 10.1073/pnas.1708725114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi N., Asakura A. Muscle satellite cell heterogeneity and self-renewal. Front. Cell Dev. Biol. 2014;2:1. doi: 10.3389/fcell.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olguin H.C., Olwin B.B. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev. Biol. 2004;275:375–388. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palla A.R., Hilgendorf K.I., Yang A.V., Kerr J.P., Hinken A.C., Demeter J., Kraft P., Mooney N.A., Yucel N., Jackson P.K., et al. Ciliation of muscle stem cells is critical to maintain regenerative capacity and is lost during aging. bioRxiv. 2020 doi: 10.1101/2020.03.20.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T.Q., Robinson K., Xu L., Skapek S.X., Chen E.Y. HDAC6 promotes self-renewal and migration/invasion of rhabdomyosarcoma. bioRxiv. 2019 doi: 10.1101/823864. [DOI] [Google Scholar]
- Pugacheva E.N., Jablonski S.A., Hartman T.R., Henske E.P., Golemis E.A. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby L.M., Parker J.D.K. Cilia and the cell cycle? J. Cell Biol. 2005;169:707–710. doi: 10.1083/jcb.200503053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarta M., Rando T.A. Mimicking the niche: cytokines expand muscle stem cells. Cell Res. 2015;25:761–762. doi: 10.1038/cr.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarta M., Brett J.O., DiMarco R., De Morree A., Boutet S.C., Chacon R., Gibbons M.C., Garcia V.A., Su J., Shrager J.B., et al. An artificial niche preserves the quiescence of muscle stem cells and enhances their therapeutic efficacy. Nat. Biotechnol. 2016;34:752–759. doi: 10.1038/nbt.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y., Hao R., Wang B., Yao T.-P. A mec17-myosin II effector axis coordinates microtubule acetylation and actin dynamics to control primary cilium biogenesis. PLoS One. 2014;9:e114087. doi: 10.1371/journal.pone.0114087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F., Zammit P.S. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 2012;139:2845–2856. doi: 10.1242/dev.069088. [DOI] [PubMed] [Google Scholar]
- Rinaldi F., Perlingeiro R.C.R. Stem cells for skeletal muscle regeneration: therapeutic potential and roadblocks. Transl. Res. 2014;163:409–417. doi: 10.1016/j.trsl.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A., Doyonnas R., Kraft P., Vitorovic S., Blau H.M. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., Guenou H., Malissen B., Tajbakhsh S., Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- Scaramozza A., Park D., Kollu S., Beerman I., Sun X., Rossi D.J., Lin C.P., Scadden D.T., Crist C., Brack A.S. Lineage tracing reveals a subset of reserve muscle stem cells capable of clonal expansion under stress. Cell Stem Cell. 2019;24:944–957.e5. doi: 10.1016/j.stem.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Schüler S.C., Hüttner S.S., von Eyss B., von Maltzahn J. Adult stem cells at work: regenerating skeletal muscle. Cell. Mol. Life Sci. 2019;76:2559–2570. doi: 10.1007/s00018-019-03093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M., Rice L.M., Vale R.D. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A.H., Rando T.A. Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J. 2014;33:2782–2797. doi: 10.15252/embj.201488278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H., Chen Y., Sun Z., Chen H., Chen M. Silence of HDAC6 suppressed esophageal squamous cell carcinoma proliferation and migration by disrupting chaperone function of HSP90. J. Cell. Biochem. 2018;119:6623–6632. doi: 10.1002/jcb.26841. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tucker R.W., Pardee A.B., Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- Tucker R.W., Scher C.D., Stiles C.D. Centriole deciliation associated with the early response of 3T3 cells to growth factors but not to SV40. Cell. 1979;18:1065–1072. doi: 10.1016/0092-8674(79)90219-8. [DOI] [PubMed] [Google Scholar]
- Valcourt J.R., Lemons J.M.S., Haley E.M., Kojima M., Demuren O.O., Coller H.A. Staying alive: metabolic adaptations to quiescence. Cell Cycle. 2012;11:1680–1696. doi: 10.4161/cc.19879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der Vartanian A., Quétin M., Michineau S., Auradé F., Hayashi S., Dubois C., Rocancourt D., Drayton-Libotte B., Szegedi A., Buckingham M., et al. PAX3 confers functional heterogeneity in skeletal muscle stem cell responses to environmental stress. Cell Stem Cell. 2019;24:958–973.e9. doi: 10.1016/j.stem.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velthoven C.T.J., Rando T.A. Stem cell quiescence: dynamism, restraint, and cellular idling. Cell Stem Cell. 2019;24:213–225. doi: 10.1016/j.stem.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M.T., Manor U., Lippincott-Schwartz J., Fan C.-M. Intravital imaging reveals ghost fibers as architectural units guiding myogenic progenitors during regeneration. Cell Stem Cell. 2016;18:243–252. doi: 10.1016/j.stem.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.P., Billin A.N., Campbell M.E., Russell A.J., Huffman K.M., Kraus W.E. The AMPK/p27Kip1 Axis regulates autophagy/apoptosis decisions in aged skeletal muscle stem cells. Stem Cell Reports. 2018;11:425–439. doi: 10.1016/j.stemcr.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woan K.V., Lienlaf M., Perez-Villaroel P., Lee C., Cheng F., Knox T., Woods D.M., Barrios K., Powers J., Sahakian E., et al. Targeting histone deacetylase 6 mediates a dual anti-melanoma effect: enhanced antitumor immunity and impaired cell proliferation. Mol. Oncol. 2015;9:1447–1457. doi: 10.1016/j.molonc.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitouni S., Francia M.E., Leal F., Montenegro Gouveia S., Nabais C., Duarte P., Gilberto S., Brito D., Moyer T., Kandels-Lewis S., et al. CDK1 prevents unscheduled PLK4-STIL complex assembly in centriole biogenesis. Curr. Biol. 2016;26:1127–1137. doi: 10.1016/j.cub.2016.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. RNA-seq data have been deposited in the NCBI Gene Expression Omnibus.The accession number for the RNA-seq data reported in this paper is GEO: GSE178070.