Summary
Directed neuronal differentiation of human pluripotent stem cells (hPSCs), neural progenitors, or fibroblasts using transcription factors has allowed for the rapid and highly reproducible differentiation of mature and functional neurons. Exogenous expression of the transcription factor Neurogenin-2 (NGN2) has been widely used to generate different populations of neurons, which have been used in neurodevelopment studies, disease modeling, drug screening, and neuronal replacement therapies. Could NGN2 be a “one-glove-fits-all” approach for neuronal differentiations? This review summarizes the cellular roles of NGN2 and describes the applications and limitations of using NGN2 for the rapid and directed differentiation of neurons.
Keywords: Neurogenin-2, NGN2, neural differentiation, induced neurons, transcription factor, pluripotent stem cells, neural progenitors
Graphical abstract
Forced expression of the transcription factor NGN2 has been widely used to directly convert hPCSs, neural progenitors, or fibroblasts to functionally mature neurons of the central and peripheral nervous system (PNS). The article by Dottori and colleagues summarizes the cellular roles of NGN2 and describes the applications and limitations of using NGN2 for directed differentiation of neurons.
Introduction
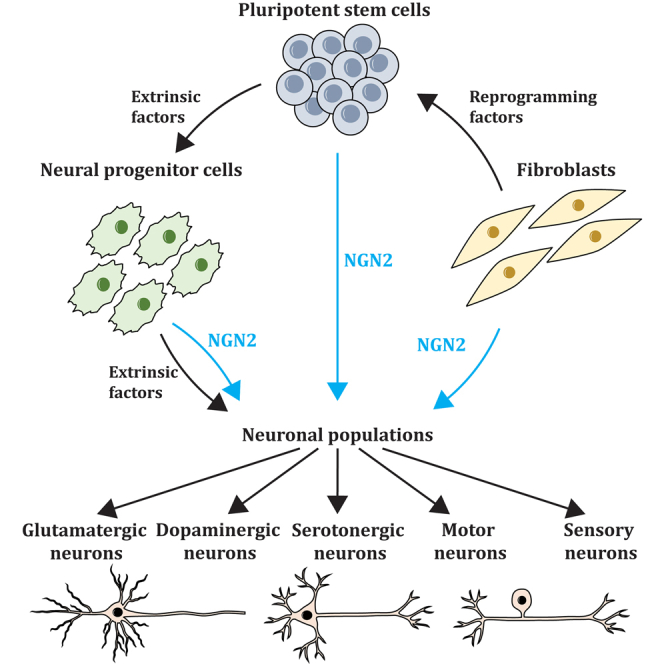
Understanding the complex molecular mechanisms regulating neurogenesis at the cellular level is integral to providing insights into the processes that govern normal development and, conversely, are disrupted in neurological diseases. Much of our understanding of neurodevelopment and neurodegeneration has been from studies in rodents and established cell lines; however, such approaches are limited due to inherent differences between rodent and human genomes and neuronal subtypes. Thus, the ability to compare and translate findings from animal studies to humans is impaired (Hodge et al., 2019). Due to the ethical and technical difficulties associated with gaining access to human brain tissue and the limited availability of post-mortem samples, human pluripotent stem cells (hPSCs) have become an alternative approach to study neurons in vitro. Human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) are widely used to generate different types of neurons in vitro by mimicking the developmental processes that occur in vivo in a controlled manner (Chambers et al., 2009). Two main methods exist to generate neurons from hPSCs. The first approach mimics the stepwise processes of neurogenesis through timely addition of extrinsic factors, usually cocktails of small molecule inhibitors and growth factors, to direct cells through different progenitor states and toward distinct cell lineages (Chambers et al., 2009; Denham and Dottori, 2011; Mica et al., 2013). The second approach utilizes the induced expression of lineage-specifying transcription factors to directly and rapidly differentiate cells to a specific fate, often bypassing the progenitor stage (Blanchard et al., 2015; Chen et al., 2020; Fernandopulle et al., 2018; Zhang et al., 2013).
The use of extrinsic factors only has been widely used in the differentiation of hPSCs into different neuronal classes, including cortical (Denham and Dottori, 2011), dopaminergic (Yan et al., 2005; Zhang et al., 2014), serotonergic (Lu et al., 2016), GABAergic (Lin et al., 2015), motor (Amoroso et al., 2013; Karumbayaram et al., 2009), and peripheral sensory neurons (Abu-Bonsrah et al., 2019; Alshawaf et al., 2018). Recent studies have also utilized combinations of extrinsic factors to generate 3D organoids including cerebral (Lancaster et al., 2013; Paşca et al., 2015; Qian et al., 2016; Velasco et al., 2019), dorsal root ganglion (Mazzara et al., 2020), and motor neuron organoids (Faustino Martins et al., 2020; Kawada et al., 2017). However, extrinsic-based differentiation protocols are limited by low reproducibility and yields, with inductions often being costly and time consuming to achieve mature, functional neurons. In contrast, induced expression of transcription factors can rapidly generate functional neurons from progenitors, stem cells, and fibroblasts at timescales significantly faster than by extrinsic-factor-mediated differentiation (Blanchard et al., 2015; Busskamp et al., 2014; Ho et al., 2016). Forced overexpression of relevant transcription factors is performed using plasmids, viruses, synthetic mRNA, or CRISPR technologies to obtain constitutive or inducible expression. While methods of transfecting and transducing cells can cause genetic modifications to the cell genome, these approaches provide a fast and robust method to generate various functional neurons.
A master regulator of neurogenesis, Neurogenin-2 (NGN2), has been widely used to rapidly differentiate a vast range of functional neurons from hPSCs, neural progenitors, glial cells, or fibroblasts with high reproducibility. NGN2 overexpression alone, in combination with other transcription factors, or in the presence of small molecules has been used to generate populations of glutamatergic neurons (Fernandopulle et al., 2018; Ho et al., 2016; Nehme et al., 2018; Zhang et al., 2013), motor neurons (Fernandopulle et al., 2018; Garone et al., 2019; Goparaju et al., 2017; Hester et al., 2011; Liu et al., 2013; Mazzoni et al., 2013; Son et al., 2011), peripheral sensory neurons (Blanchard et al., 2015; Hulme et al., 2020; Nickolls et al., 2020; Schrenk-Siemens et al., 2015), dopaminergic neurons (Liu et al., 2012; Park et al., 2008; Xue et al., 2019), and serotonergic neurons (Vadodaria et al., 2016). The rapid generation of induced neurons (iNs) has allowed for applications in neurodevelopmental and disease modeling, high-throughput drug screening, and neuron replacement therapies.
Extrinsic-factor-mediated differentiation: Slow and steady
Protocols for the differentiation of hPSCs commonly attempt to recapitulate events in embryonic development. Initially, protocols involved the generation of embryoid bodies (EBs), which are 3D aggregates relying on spontaneous differentiation and cell patterning of the three primary germ layers (Itskovitz-Eldor et al., 2000); however, EB formation is inefficient and has a limited ability to derive specific cell populations (Dang et al., 2002). Modeling embryonic developmental processes with hPSC-derived EBs uncovered relevant signaling pathways that could be manipulated to better direct cellular differentiation using exogenous growth factors, cytokines, and small molecule inhibitors (Schuldiner et al., 2000). In recapitulating the developmental path of neurogenesis, differentiation methods first rely on the differentiation of hPSCs to neural progenitor cells (NPCs) and then subsequent specification of neuronal subtypes. For example, dual SMAD inhibition using SB431542 (a Lefty/Activin/TGF-β signaling inhibitor) and noggin or LDN193189 (BMP pathway inhibitors) drives the generation of a highly enriched NPC population (Chambers et al., 2009; Li et al., 2011). Furthermore, the application of recombinant Sonic-hedgehog (SHH) and the agonists purmorphamine (PUR) and smoothened agonist (SAG) have been demonstrated in ventralization, while the use of BMP and canonical WNT pathway modulators such as dorsomorphin, LDN193189, CHIR99021, and XAV939 are utilized for dorsalization (Imaizumi et al., 2015; Qi et al., 2017). Small molecules have also been used to generate neural crest cells at high efficiency through the timely activation of WNT signaling using the GSK3β inhibitor, CHIR99021 (Denham et al., 2015; Lee et al., 2010). Optimization of the precise concentrations, timing, and combinations of these growth factors and small molecules have led to defined protocols for the generation of specific neuronal subpopulations. In recent years, extrinsic-factor differentiation protocols have been essential in the generation of specific tissue-like organoids including the maturation of cerebral (Lancaster et al., 2013; Salick et al., 2017; Velasco et al., 2019), midbrain (Jo et al., 2016), vestibular-tissue-like (Mattei et al., 2019), dorsal root ganglia (DRG) (Mazzara et al., 2020), and neuromuscular organoids (Faustino Martins et al., 2020; Kawada et al., 2017). Extrinsic-factor-based differentiation protocols are valuable in organoid maturation, as they mimic the developmental cues that occur in the specific tissue development in vivo and enable for complex heterogeneity of progenitors, mature cells, and supporting cell types.
More recently, cell culture protocols have been developed that allow for greater control over cell differentiation, with defined, stepwise culture methods to induce neuronal cell fate. Despite these advances, it is critical to note that none of the examined morphogens or growth factor cocktails exclusively directs differentiation to only one cell type, but rather alters the relative proportions of specific cell types (as reviewed by Cohen and Melton, 2011). A multi-site study of the reproducibility of neuronal differentiation with identical iPSC lines used across five distinct laboratories showed high variability in cell populations via single-cell transcriptomics (Volpato et al., 2018). In addition, hPSC lines are demonstrated to respond differently to the same neuronal differentiation protocols (Wu et al., 2007). Extrinsic-factor-based neuronal differentiations are also limited by multi-step processes that are time consuming and result in functionally variable neurons (Johnson et al., 2007). The inherent limitations of extrinsic-factor-based differentiations have led to a shift toward transcription-factor-driven differentiation methods, owing to their ability to specifically target relevant genes involved in neuronal development for the rapid and reproducible generation of specific neuronal subtypes.
Differentiation by forced expression of NGN2
NGN2: A key regulator of neurogenesis
The neurogenin (NGN) family, comprising NGN1, NGN2, and NGN3, is a class of proneural basic-helix-loop-helix (bHLH) transcription factors that bind to DNA in heterodimeric complexes to activate gene transcription (as reviewed in Bertrand et al., 2002; Ma et al., 1996; Sommer et al., 1996). The NGN family is expressed throughout the developing nervous system in a region- and cellular-context-specific manner. The NGN family have major roles in the commitment of progenitors to neurons by (1) inhibiting glial fate (Nieto et al., 2001; Sun et al., 2001), (2) inducing a cascade of pan-neuronal genes, (3) promoting cell cycle exit (Farah et al., 2000), (4) promoting neuronal migration (Ge et al., 2006; Seo et al., 2007), and (5) promoting the expression of neuron subtype-specific genes. While all NGNs are important throughout development, NGN2 has been heavily utilized for its ability to rapidly induce neuronal differentiation. The role of NGN2 and its use in neuronal differentiation approaches is the primary focus of this review.
NGN2 is transiently expressed in multiple NPC types and is downregulated during the final differentiation stages to promote proneural pathways and subtype specification. NGN2 has been described to regulate differentiation of glutamatergic (Bel-Vialar et al., 2007; Roybon et al., 2009; Scardigli et al., 2003), motor (Lee et al., 2009; Lee and Pfaff, 2003; Mizuguchi et al., 2001; Parras et al., 2002; Scardigli et al., 2001), sensory (Ma et al., 1999; Perez et al., 1999; Sommer et al., 1996; Zirlinger et al., 2002), and dopaminergic neurons (Andersson et al., 2006a, 2006b; Kele et al., 2006) and inhibit the generation of GABAergic neurons (Fode et al., 2000; Jo et al., 2007; Schuurmans et al., 2004), astrocytes (Sun et al., 2019a), and oligodendrocytes (Jiang et al., 2020) through the activation or inhibition of various transcription factors and pathways (Figure 1). Post-translational modifications and the presence of morphogens and cofactors can affect the role NGN2 plays during neuronal differentiation, depending on the cellular context (Hand et al., 2005; Hindley et al., 2012; Lo et al., 2002; Ma et al., 2008; Parras et al., 2002). For example, during the development of motor neurons, NGN2 is phosphorylated on two serine residues that enables the interaction of NGN2 with a combination of transcription factors, which results in the activation of motor neuron gene cascades (Ma et al., 2008). Tyrosine residue 241 of NGN2 is selectively phosphorylated within cortical progenitors during cortex development, and this phosphorylation allows NGN2 to drive the radial migration and dendritic morphology of pyramidal neurons (Hand et al., 2005). The phosphorylation of tyrosine 241 does not alter the proneural activity of NGN2 and instead allows NGN2 to operate in a regional and subtype-specific manner (Hand et al., 2005). Additionally, the role of NGN2 can be altered by the expression of different cofactors. For example, the expression of NGN1 or NGN2 in the presence of low concentrations of BMP2 is sufficient for the formation of sensory neurons during differentiation of neural tube cultures, whereas the same cultures incubated in the presence of high BMP2 concentrations acquire an autonomic neuronal fate (Lo et al., 2002). Thus, the cellular context is critical for the function and specification of neurons by NGN2.
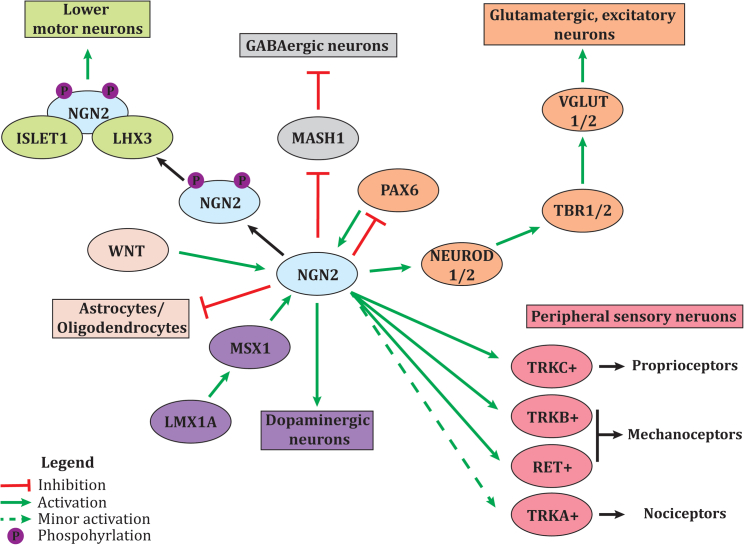
Figure 1.
Summary of the role NGN2 plays in the differentiation of different neuronal subtypes
During the differentiation of glutamatergic cortical neurons, PAX6 activates NGN2, which in turn inhibits PAX6 and results in the activation of the cortical transcription factor cascade (orange pathway). NGN2 is phosphorylated at two serine residue sites and forms a complex with LHX3 and ISLET1, activating genes required for motor neuron differentiation (green pathway). NGN2 plays a role in dopaminergic neuron differentiation but is not a critical regulator (purple pathway). NGN2 plays a role in the waves of sensory neurogenesis, in turn activating the genes required for differentiation of specific peripheral sensory neuron subtypes (pink pathway). NGN2 inhibits MASH1, a transcription factor required for GABAergic neuron generation (gray pathway). WNT signaling activates NGN2, which represses the differentiation of astrocytes and oligodendrocytes by the downregulation of astrocyte genes and OLIG2, respectively (cream pathway).
While there are multiple bHLH transcription factors, NGN2 has received particular attention for its use in in vitro differentiation of fibroblasts, hPSCs, and progenitors into a range of neurons including glutamatergic, dopaminergic, serotonergic, motor, and sensory neurons. Published protocols using the overexpression of NGN2 in the generation of specific neuronal subtypes are summarized in Table 1. However, since the generation of neurons relies on complex interactions of intrinsic and extrinsic factors in a region- and cellular-context-dependent manner, care must be taken when using a differentiation protocol to generate the desired neuronal type. To differentiate hPSCs into a specific type of neuron, it is critical to first understand how the neuron develops in vivo so that the best combination of transcription factors, small molecules, growth factors, and co-cultures can be used to generate the neuronal subtype. It is imperative to profile the generated neurons both transcriptionally and functionally to confirm the neuronal type identity. The section below provides a background to the role of NGN2 during the development of specific neuronal lineages and summarizes current efforts of how NGN2 can be used alone, or in combination with other transcription factors, to generate particular neuronal subtypes.
Table 1.
Current protocols of neuronal differentiation using NGN2 overexpression
| Neuronal subtype | Source/species | Transcription factors used and method of delivery | NGN2 induction duration | Time to functional neurons | Developmental patterning | Growth factors and co-culture | Reference |
|---|---|---|---|---|---|---|---|
| Bipolar neurons | Human ESCs and iPSCs | Ngn2 and Ngn1 via lentiviral gene delivery | 1–4 days | 4 days for single action potentials, 14 days for action potentials trains | NA | Co-cultured with rat astrocytes | (Busskamp et al., 2014) |
| Glutamatergic excitatory neurons | Human ESC and iPSCs | Ngn2 via lentiviral gene delivery | Entire duration | 2 weeks | NA | BDNF | (Zhang et al., 2013) |
| Mouse ESCs | Transient Ngn2 expression and clonal transgenic line | Entire duration | 7 days | NA | NA | (Thoma et al., 2012) | |
| NPCs derived from human iPSCs | Ngn2 or NGN2 via lentiviral gene delivery | 2 days–3 weeks | 2 weeks | SB431542 and LDN193189 | NA | (Ho et al., 2016) | |
| Human iPSCs | NGN2 transgenic line (i3 neurons) | 72 h | 2 weeks | NA | BDNF, NT-3 co-cultured with primary mouse astrocytes (also recommend using astrocyte conditioned media) | (Fernandopulle et al., 2018) | |
| Human ESCs and iPSCs | Ngn2 via lentiviral gene delivery | Entire duration | 2–3 weeks | SB431542, XAV939, and LDN193189 | Co-cultured with mouse primary cortical glial cells | (Nehme et al., 2018) | |
| Human ESCs or ESC-derived anterior and posterior NPCs | NGN2 alone or in addition to one of the following: EMX1, EMX2, OTX1, OTX2, EOMES (TBR2), LHX2, or FOXG1 | 4 days or for single-cell RNA sequencing 14 days | 28 days | SB431542, LDN193189 and CHIR99021 | Co-cultured with mouse glia for long term cultures | (Ang et al., 2020) | |
| Dopaminergic neuron-like | Human fibroblasts | NGN2, ASCL1, SOX2, NURR1, PITX3 via lentiviral gene delivery | Transient | 20 days | SHH, FGF8 | NA | (Liu et al., 2012) |
| Midbrain dopaminergic neurons | Human iPSCs | NGN2, ATOH1 via synthetic mRNA delivery | Multiple mRNA transfections: ATOH1 for 3 days, NGN2 for 1 day | 20–49 days | SHH, FGF8b, and DAPT | BDNF, GDNF, TGFb-3, cAMP, ascorbic acid, and DAPT | (Xue et al., 2019) |
| Dopaminergic neuron | Mouse ESC-derived NPCs | Ngn2, Nurr1 via retroviral gene delivery | 3–10 days | 9–16 + days | bFGF, EGF | NA | (Park et al., 2008) |
| Serotonergic neurons | Human dermal fibroblasts | NGN2, ASCL1, NKX2.2, FEV, GATA2, LMX1B via lentiviral gene delivery | 3–4 weeks | 6 weeks | Dibutyryl cyclic-AMP, noggin, LDN193189, A83-1, CHIR99021, SB431542, forskolin | GDNF, BDNF, dibutyryl cyclic-AMP co-culture with primary rodent astrocytes for electrophysiological analyses | (Vadodaria et al., 2016) |
| Motor neurons | Human ESC and iPSCs | NGN2 NGN1, NGN3, ND1, ND2 via synthetic mRNA delivery | 2–4 transient transfections | 7–10 days | Forskolin, SB431542, dorsomorphin, retinoic acid | BDNF, GDNF, NT-3 | (Goparaju et al., 2017) |
| Human ESC and iPSC-derived NPCs | NGN2, ISL1, LHX3 via adenoviral gene delivery | Transduce day 0 and 4 | 21 days | Retinoic acid, forskolin, SHH | Retinoic acid, SHH | (Hester et al., 2011) | |
| Spinal motor neurons | Mouse ESC-derived EBs | Ngn2, Isl1, Lhx3 via inducible transgenic line | 2 days | 7–11 day | NA | Co-culture with mouse astrocytes 7–10 days | (Mazzoni et al., 2013) |
| Mouse and human fibroblasts | Ngn2, Ascl1, Brn2, Mytl1, Lhx3, Hb9, Isl1 via retroviral transduction | Undefined | 21 + days | NA | GDNF, BDNF, CNTF | (Son et al., 2011) | |
| Human iPSCs | Ngn2, Isl1, Lhx3 inducible line via a piggyBac transposon vector | 5 days | 13 days | DAPT, SU5402 | BDNF, GDNF, L-ascorbic acid | (Garone et al., 2019) | |
| Cranial motor neurons | Mouse ESCs-derived EBs | Ngn2, Isl1, Phox2a via inducible transgenic line | 2 days | 11 days | NA | Co-culture with mouse astrocytes 7–10 days | (Mazzoni et al., 2013) |
| Human iPSCs | Ngn2, Isl1, Phox2a inducible line via a piggyBac transposon vector | 5 days | 13 days | DAPT, SU5402 | BDNF, GDNF, L-ascorbic acid | (Garone et al., 2019) | |
| Lower motor neurons | Human iPSCs | NGN2, ISL1, LHX3 inducible transgenic line | 72 h | 14 + days | NA | Optional co-culture of astrocytes | (Fernandopulle et al., 2018) |
| Cholinergic motor neurons | Human fetal lung fibroblasts or human postnatal and adult skin fibroblasts | NGN2 (+Sox11 for skin fibroblasts) using retroviral gene delivery | 2–35 days | 50 days | Dorsomorphin and forskolin | Co-cultured with C2C12 murine myoblast line | (Liu et al., 2013) |
| Mixed population of sensory neurons | Human and mouse fibroblasts | Ngn2 and BRN3A or Ngn1 and BRN3A via lentiviral transduction | 8 days | 14–22 days | NA | BDNF, GDNF, NGF | (Blanchard et al., 2015) |
| Mixed population of sensory neurons | Human ESC neural crest progenitors | NGN2 via lentiviral transduction | 4 days | 32 days | SB431524, CHIR99021, BMP2, FGF2 | BDNF, GDNF, NT-3, bNGF, Y-27632 | (Hulme et al., 2020) |
| Cold-sensing mechanoreceptors | Human iPSCs or iPSC-derived neural crest cells | NGN2 and BRN3A via an inducible line | 14 days | 20 days | Y-27632, bFGF, EGF, SB431524 | BDNF, GDNF, NT-3, bNGF, Y-27632 | (Nickolls et al., 2020) |
| Mechanoreceptors | Human iPSC-derived neural crest cells | NGN2 and BRN3A via an inducible line | 1 day | 20 days | Y-27632, bFGF, EGF, SB431524 | BDNF, GDNF, NT-3, bNGF Retinoic acid | (Nickolls et al., 2020) |
| Human ESC and iPSC neural crest progenitors | NGN2 via lentiviral transduction | 1 day | 21– 30 days | FGF, EGF | BDNF, GDNF, NT-3, bNGF Retinoic acid | (Schrenk-Siemens et al., 2015) |
The role of NGN2 in the generation of glutamatergic neurons
NGN2 and the cortical cascade
NGN2 is a key regulator of the transcription factor cascade required for corticogenesis. NGN2 expression is directly regulated by PAX6, a transcription factor essential for cortical progenitor differentiation. PAX6 directly binds to the E1 enhancer element of Ngn2 in the ventral spinal cord and dorsal telencephalon of mice and increases expression of the proneural gene (Scardigli et al., 2003). NGN2 also negatively regulates PAX6, decreasing its expression to allow for neuronal differentiation (Bel-Vialar et al., 2007). The decrease in PAX6 and upregulation of NGN2 activates other transcription factors required for the specification and differentiation of cortical neurons, including NEUROD1 and TBR1/2 (Roybon et al., 2009). NGN2 activation in precursor cells also promotes cell cycle exit by reducing the expression of cell cycle regulators including CCND1, CCNE1/2, and CCNA2 (Lacomme et al., 2012). NGN2 also plays a role in the developing cortex by regulating dorsal and ventral specification (Fode et al., 2000). Early born cortical neurons lose expression of dorsal-specific markers with NGN2 loss and express markers specific for the ventral-telencephalon (Fode et al., 2000). Furthermore, NGN2 overexpression induces the ectopic expression of cortical genes in the ventral-telencephalon to promote cortical neuronal identity (Mattar et al., 2008). In Ngn2 alone and dual Ngn1 and Ngn2 mutants, cortical-specific genes Math2, Nscl1, NeuroD1/2, Tbr1/2, Slc17a7 (encodes VGlut1), and Slc17a6 (encodes VGlut2) were downregulated, indicating the role NGN2 plays in activating transcription factors responsible for cortical neuron generation (Schuurmans et al., 2004).
There are conflicting reports on whether NGN2 and NGN1 have redundant roles in cortical development. Schuurmans et al. (2004) showed that in early cortical development, Ngn1 and Ngn2 have redundant roles in the specification of glutamatergic phenotypes in neocortical projection neurons and the specification of deep-layer neurons. Conversely, NGN2 has been shown to be involved in the positive regulation of NGN1 and negative regulation of MASH1 (encoded by Ascl1) in cortical progenitors, with the opposite effect not being true (Fode et al., 2000). Ngn2 has also been described to be the most important gene in the NGN family in cortical neurogenesis, as only Ngn2 mutants presented distinct defects in cerebral cortex development (Fode et al., 2000; Nieto et al., 2001). In addition, genes specific to cortical and glutamatergic phenotypes are not activated in Ngn2 mutants or Ngn2 and Ascl1 double mutants, indicating that NGN2 plays an essential role for cortical neuron development independent of MASH1 (Schuurmans et al., 2004). Together, these studies describe a feedback system of gene regulation governed by NGN2 to promote glutamatergic cortical neuron development through the induction of transcription factors that favor cortical neuron differentiation such as NEUROD1 and TBR1/2 and the negative regulation of genes that favor the progenitor state (Pax6) and GABAergic neurons (Mash1).
Use of NGN2 overexpression in glutamatergic neuronal differentiation
Ectopic expression of NGN2 to generate excitatory glutamatergic neurons was first demonstrated by Thoma et al. (2012). NGN2 overexpression differentiated mouse PSCs into functionally active neurons evidenced by voltage-activated potassium currents, even in the presence of pluripotency-promoting conditions (Thoma et al., 2012). The Ngn2 iNs expressed cortical markers DCX, MATHS3, NEUN, OLIG2, SOX1, TUJ1, MAP2AB, and VGlut1/2 (Thoma et al., 2012). Interestingly, iNs did not express the GABAergic marker GAD1 and had low levels of TH expression, suggestive of a mixed population of glutamatergic and dopaminergic neurons. Overexpression of NGN2 has also resulted in the differentiation of functional glutamatergic neurons that were successfully integrated into the mouse brain (Zhang et al., 2013). iNs were classed as glutamatergic excitatory neurons, as they expressed the layer II and III cortical markers BRN2, CUX1, and FOXG1; however, they lacked the expression of cortical layer VI markers TBR1 and FOG2. iNs expressed AMPA glutamate receptors but lacked expression of NMDA receptors following 3 weeks of differentiation. Moreover, nearly all NGN2 iNs expressed the vesicular glutamate transporter VGlut2, whereas only 20% expressed VGlut1. VGlut1 and VGlut2 are expressed in excitatory neurons that occupy different regions of the brain, with VGlut1 expression localized to the cerebral cortex, hippocampus, and cerebellum and VGlut2 centralized to neurons in the thalamus and brainstem (Balaram et al., 2011; Fremeau et al., 2001; Vigneault et al., 2015). Thus, the NGN2 iNs molecular profile suggests that overexpression of NGN2 alone generates a mixed population of excitatory neurons of the thalamus and brainstem. To bypass issues of random integration and lentiviral efficiency, Fernandopulle et al. (2018) generated inducible, integrated, and isogenic (i3) stem cell lines by inserting NGN2 into a safe-harbor locus (a genomic site that maintains transgene expression without affecting the host's genome). The transgenic lines were able to differentiate into functional glutamatergic neurons following 72 h of NGN2 induction and 14 days of maturation. After co-culture with astrocytes, they exhibited spontaneous excitatory currents, which were inhibited by the glutamate receptor antagonist CNQX (Fernandopulle et al., 2018). Thus, overexpression of NGN2 can rapidly and reproducibly generate excitatory neurons from hPSCs. As NGN2 plays a broad role in the generation of multiple neuronal lineages, additional approaches are often required to generate specific neuronal populations.
To promote further specification in NGN2 iNs, different methods have been tested including the use of NPCs as a starting cell population, adding small-molecule inhibitors to mimic developmental patterning and the overexpression of various cofactors. Ho et al. (2016) developed a protocol to generate glutamatergic-excitatory neurons from hPSC-derived NPCs committed to forebrain neural fate. The NPC-derived NGN2 iNs expressed SYN1, SLC17A6 (encodes VGlut2), and had a high expression of SLC17A7 (encodes VGlut1) (Ho et al., 2016), suggesting that there is a mixed population of excitatory neurons typically found in the cerebral cortex, hippocampus, and cerebellum (Balaram et al., 2011; Fremeau et al., 2001; Vigneault et al., 2015). In addition, Ho et al. (2016) showed that the induction of NGN2 for 48 h was sufficient in driving neuronal differentiation, as it mimicked the transient expression profile seen in neurodevelopment in vivo (Ozen et al., 2007). Nehme et al. (2018) modeled developmental patterning using dual SMAD inhibitors SB431542 and LDN193189 and the WNT inhibitor XAV939 during NGN2 induction coupled with cell sorting for CAM2KA generating a homogeneous population of excitatory, glutamatergic postnatal-like neurons. This study identified that using NGN2 induction alone produced a heterogeneous culture of neurons of varying maturity and that developmental patterning is required to drive differentiation toward a specific lineage. Interestingly, NGN2 iNs derived from hESCs consisted of three different groups of glutamatergic neurons, whereas neurons generated by the co-expression of NGN2 with EMX1 produced a homogeneous population of functional glutamatergic neurons, with a more forebrain phenotype than NGN2 alone (Ang et al., 2020). Collectively, these protocols indicate the importance of profiling derived iNs, starting cell type, duration of NGN2 activation, expression of additional cofactors, and developmental patterning in generating a specific neuronal lineage.
The role of NGN2 in the generation of lower motor neurons
NGN2 and the activation of genes specific for lower motor neurons
Lower motor neurons are a group of cholinergic neurons located in the ventral spinal cord that innervate skeletal muscle and control movement. The region-specific signaling of SHH is critical for the formation of motor neuron progenitors, as it is involved in the activation of a cascade of transcription factors such as NKX6.2, PAX6, OLIG2, NKX6.1, and NGN2 (as reviewed by Briscoe and Ericson, 2001; Cave and Sockanathan, 2018; Jessell, 2000). Together, this combination of transcription factors in turn promotes neurogenesis and the expression of motor neuron specifiers, such as the LIM homeodomain proteins ISLET1 (encoded by ISL1) and LIM homeobox 3 (encoded by LHX3) and prevents the formation of other cell types (Briscoe et al., 2000; Cave and Sockanathan, 2018; Dubreuil et al., 2000; Lee et al., 2012; Pattyn et al., 2003; Pfaff et al., 1996). While NGN2 was initially considered to simply have a proneuronal role in motor neuron development (Novitch et al., 2001), NGN2 is also critical for motor neuron specification (Lee et al., 2009; Lee and Pfaff, 2003; Mizuguchi et al., 2001; Parras et al., 2002; Scardigli et al., 2001). For example, a loss of NGN2 expression disrupts motor neuron development and specification, which cannot be compensated for by other proneural factors and thus must play a specification role (Lee and Pfaff, 2003; Parras et al., 2002; Scardigli et al., 2001). Furthermore, when NGN2 is phosphorylated at two specific serine residues, NGN2 can directly interact with the LIM complex (ISLET1 and LHX3 transcription factors) that coordinates the activation of motor-neuron-specific genes, resulting in the specification of motor neurons (Lee and Pfaff, 2003; Ma et al., 2008). Additionally, NGN2 induces the expression of EBF and ONECUT transcription factors, allowing the LIM complex to bind to previously inaccessible chromatin sites relevant for motor neuron specification (Velasco et al., 2017). Thus, NGN2 in combination with other transcription factors directly acts in the formation and specification of lower motor neurons, making NGN2 a promising transcription factor to use in motor neuron differentiation.
Use of NGN2 overexpression in direct differentiation of lower motor neurons
Protocols to differentiate fibroblasts, PSCs, and PSC-derived progenitors to lower motor neurons have utilized NGN2 by using a mix of proneural and/or lineage-specific transcription factors (Fernandopulle et al., 2018; Garone et al., 2019; Goparaju et al., 2017; Hester et al., 2011; Liu et al., 2013; Mazzoni et al., 2013; Son et al., 2011). Son et al. (2011) utilized a combination of the proneural transcription factors, MASH1, BRN2, MYTL1, and the motor-neuron-specific transcription factors NGN2, LHX3, HB9, and ISLET1 to convert mouse and human fibroblasts into functional spinal motor neurons. Importantly, successful motor neuron differentiation was confirmed by transcriptional profiling and in-depth functional profiling, demonstrating that (1) 90% of neurons fired action potentials, (2) neurons responded to motor-neuron-associated neurotransmitters (GABA, glycine, kainite), (3) neurons formed neuromuscular junctions when co-cultured with C2C12-derived myotubes and primary chick myotubes, and (4) neurons projected axons out from the spinal cord toward musculature when engrafted into chick embryos (Son et al., 2011). Liu et al. (2013) utilized small molecules forskolin and dorsomorphin in combination with NGN2 overexpression to convert human fetal lung fibroblasts into cholinergic motor neurons. The derived neurons expressed cholinergic motor neuron markers HB9 (91.7%), ISLET1/2 (90.2%), choline acetyltransferase (ChAT, 98.5%), and vesicular acetylcholine transporter (VAChT, 98.2%) and were also positive for the glutamatergic marker VGlut1 (Liu et al., 2013). The cholinergic motor neurons were co-cultured with mouse myotubes, formed synaptic-like structures, and caused rhythmic contractions, suggestive of neuromuscular junctions (Liu et al., 2013). However, the addition of SOX11 was required to convert postnatal and adult fibroblasts to neurons efficiently, suggesting that different starting cell types may require distinct combinations of transcription factors to achieve the same neuronal subtype.
Alternatively, motor neurons have been differentiated from either PSC-derived progenitors or directly from PSCs by expressing just the components of the NGN2/LIM complex responsible for the activation of motor-neuron-specific genes (Garone et al., 2019; Hester et al., 2011; Mazzoni et al., 2013; De Santis et al., 2018). For example, Hester et al. (2011) transduced hPSC progenitors at day 0 and day 4 with Ngn2, Isl1, and Lhx3 in the presence of the morphogens SHH and retinoic acid (RA) generating neurons that expressed motor neuron markers, HB9, ChAT, and HOX. These were further able to form neuromuscular junctions when co-cultured with C2C12-derived myotubes (Hester et al., 2011). The combination of NGN2, ISLET1, and LHX3 transcription factors results in the formation of spinal motor neurons; however, replacement of LHX3 with PHOX2A in mouse ESC-derived progenitors changes the neuronal cell fate toward cranial motor neurons (Mazzoni et al., 2013), illustrating how NGN2 can be used in conjunction with a variety of transcription factors to derive different motor neuron subtypes. Notably, Mazzoni et al. (2013) verified the identity and function of the motor neurons by transplanting the neurons in vivo into chick embryos, with cranial motor subtypes successfully integrating with spinal accessory neurons. Overall, the generation of motor neurons using the expression of NGN2 requires the combination of specific transcription factors, which synergistically act together to activate motor neuron gene expression pathways.
The role of NGN2 in the generation of DRG sensory neurons
NGN2 and waves of sensory neurogenesis
Sensory neurons located within the DRG are classed into three general subtypes: nociceptors that detect pain perception and express the neurotrophin receptor tropomyosin kinase A (TRKA), mechanoreceptors that detect touch and hair deflection and express TRKB, and proprioceptors that detect muscle pressure and tension and express TRKC. Sensory neurons arise from neural crest cells that migrate out from the neural tube and undergo multiple specification steps (as reviewed by Lallemend and Ernfors, 2012; Marmigère and Ernfors, 2007; Simões-Costa and Bronner, 2015). Sensory neurogenesis begins in two distinct waves of neural crest cells: the first wave is controlled by transient expression of NGN2 resulting in the differentiation of proprioceptors and mechanoreceptors, and the second is controlled by transient expression of NGN1, resulting in the production of peptidergic and nonpeptidergic nociceptors (Ma et al., 1999; Perez et al., 1999; Sommer et al., 1996; Zirlinger et al., 2002). The expression of NGN2 and NGN1 is essential during sensory neuron formation, since Ngn1/Ngn2-null mice are unable to form DRG sensory neurons (Ma et al., 1999). While NGN2 and NGN1 bias the neural crest cells toward a DRG sensory neuron fate, a combination of other transcription factors including BRN3A, ISLET1, KLF7, FOXS1, RUNX3, SHOX2, and RUNX1 are involved in the final differentiation, specification, and survival of the sensory neuron subgroups (Abdo et al., 2011; Dykes et al., 2011; Kramer et al., 2006; Lei et al., 2005; Montelius et al., 2007; Sun et al., 2008). Overall, NGN2 expression is critical for DRG sensory neurogenesis, with subtype specification controlled by combinations of other transcription factors.
Forced expression of NGN2 drives differentiation of DRG sensory neurons
Since NGN2 expression is critical during sensory neurogenesis, the differentiation of hPSCs into DRG sensory neurons often combines the expression of NGN2 with other sensory-specific transcription factors at the pluripotent or the neural crest stage to mimic DRG sensory neurogenesis. Notably, NGN2 has been used in combination with the pan-sensory transcription factor BRN3A, generating functional DRG sensory neurons from fibroblasts, iPSCs, and iPSC-derived neural crest cells (Blanchard et al., 2015; Nickolls et al., 2020). Expression of NGN2 and BRN3A in fibroblasts produced a mixed population of DRG sensory neurons expressing general sensory neuron and subtype markers TRKA, TRKB, and TRKC, albeit at a low efficiency (Blanchard et al., 2015). Additionally, populations of neurons were selectively responsive to nociceptive stimuli such as heat (capsaicin, 6% of neurons), cooling (menthol, 6%), or itch (mustard oil, 9%), with a small number of cells responsive to multiple conditions (Blanchard et al., 2015). These results indicate the generation of a mixed population of functional DRG sensory neurons; however, profiling the proportion of neurons unresponsive to the tested stimuli for other sensory stimuli, such as mechanical stimulation, could be of interest.
NGN2 has been transiently expressed in hPSC-derived neural crest cells, mimicking developmental cues, to generate functional neurons capable of firing multiple action potentials and expressing the general sensory markers BRN3A and ISLET1 as well as subtype-specific markers such as TRKA, TRKB, and TRKC (Hulme et al., 2020; Nickolls et al., 2020; Schrenk-Siemens et al., 2015). Since these protocols mimic development in which the NGN2 directed wave of neurogenesis drives the differentiation of mechanoreceptors and proprioceptors, Schrenk-Siemens et al. (2015) hypothesized that using NGN2 would bias the neural crest cells into mechanically sensitive neurons. The generated sensory neurons displayed a change in current density in response to mechanical stimulation that could be reduced by using the broad-spectrum mechanosensitive inhibitor, ruthenium red, or completely removed when knocking out the mechanosensitive channel PIEZO2 (Schrenk-Siemens et al., 2015). Additionally, altering the starting cell type and length of NGN2 and BRN3A expression, different DRG sensory subtypes could be generated (Nickolls et al., 2020). Expression of NGN2 and BRN3A in neural crest cells for 14 days generated cold-sensing mechanoreceptors, while expression for only 1 day resulted in a general mechanoreceptor profile (Nickolls et al., 2020). It is apparent that the combination of transcription factors, neurotrophins, length of expression, and the progenitor state during differentiation play a critical role in directing the differentiation of different DRG sensory neuron subtypes.
The role of NGN2 in the generation of dopaminergic neurons
NGN2 is a proneural activator for dopaminergic neuronal fate
Dopaminergic neurons have critical roles in modulating motor control, cognition, behavior, decision making, and in reward pathways. They are characterized by the expression of TH, NURR1, dopa decarboxylase (DDC), and vesicular monoamine transporter 2 (VMAT2) (Björklund and Dunnett, 2007). The development of dopaminergic neurons occurs through a time- and location-specific cascade of morphogens and transcription factors (as reviewed by Abeliovich and Hammond, 2007; Blaess and Ang, 2015; Hegarty et al., 2013). Briefly, dopaminergic precursors are formed by the coordinated actions of SHH, FGF8, and WNTs as well as the transcription factors OTX2, LMX1A, GLI factors, FOXA1/2, MSX1, and NGN2 that initiate expression cascades to establish dopaminergic progenitors. Next, the transcription factors NURR1, PITX3, LMX1B, and ENGRAILED are involved in the maturation of dopaminergic neurons (as reviewed by Abeliovich and Hammond, 2007; Blaess and Ang, 2015).
During dopaminergic neuron development, NGN2 plays a role as a proneural activator rather than as a specifier of dopaminergic neuron fate (Andersson et al., 2006a, 2006b; Kele et al., 2006). Instead, LMX1A has a critical role during dopaminergic neuron generation by activating the expression of dopaminergic-specific gene cascades as well as inducing MSX1 expression, which in turn upregulates NGN2 expression in neural progenitors that then activate proneural pathways (Andersson et al., 2006a). Interestingly, NGN2 is important for dopaminergic neuron development, as a loss of NGN2 causes a decrease in the number of dopaminergic neurons (Andersson et al., 2006b; Kele et al., 2006); however, this loss can be partially compensated for by expression of a different proneural transcription factor, MASH1 (Kele et al., 2006). Additionally, NGN2 overexpression in neurospheres generated from embryonic rat midbrain promoted cell cycle exit and neuronal differentiation (Roybon et al., 2008). NGN2 overexpression increased VMAT2 expression in a NURR1 independent pathway (Roybon et al., 2008). Furthermore, expression of NGN2 in mouse dorsal midbrain cultures increases the expression of pan-neuronal markers but not dopaminergic markers, indicating that NGN2 in midbrain progenitors is not sufficient to drive dopaminergic neuronal fate (Andersson et al., 2006b). NGN2 has also been considered to have an inhibitory role during dopaminergic development as co-expression of NGN2 with NURR1 in rat neuronal precursor cultures represses NURR1-induced dopaminergic markers (Park et al., 2006). Thus, while NGN2 is expressed and has a role in the development of dopaminergic neurons, it is not critical for the specification of such neurons.
The use of NGN2 in dopaminergic neuron differentiation
The use of NGN2 to derive dopaminergic neurons in vitro has been limited to date, although some studies have shown it is possible to generate dopaminergic neurons with high efficiency by combining NGN2 expression with other transcription factors, growth factors, and morphogens to generate dopaminergic neurons (Liu et al., 2012; Park et al., 2008; Xue et al., 2019). Liu et al. (2012) derived dopaminergic neurons from human fibroblasts by using a combination of the dopaminergic-relevant transcription factors NGN2, MASH1, SOX2, NURR1, and PITX3 and the morphogens FGF8 and SHH. While the conversion efficiency from fibroblasts was only 1%–2%, the generated neurons expressed the dopaminergic markers ENGRAILED, TH, DAT, and VMAT2 and were capable of dopamine uptake and production (Liu et al., 2012). Interestingly, the inclusion of NGN2, MASH1, and SOX2 was critical for the formation of neurons (Liu et al., 2012), suggesting that NGN2 and MASH1 were likely key in driving neurogenesis while SOX2, NURR1, and PITX3 were important in the specification of dopaminergic neurons. Alternatively, the combination of NGN2 with another proneural transcription factor, ATOH1, has also been used to generate dopaminergic neurons) alongside multiple morphogens and growth factors such as FGF8, SHH, BDNF, GDNF, TGF-3, cAMP, ascorbic acid, and DAPT (Xue et al., 2019). The generated neurons expressed dopaminergic markers; 59% displayed spontaneous firing activity and released dopamine in response to KCl-induced depolarization (Xue et al., 2019). Interestingly, NGN2 in combination with NURR1 can differentiate mouse neural progenitors into functional dopaminergic neurons; however, the same combination in rat neuron progenitors inhibited the differentiation of dopaminergic neurons (Park et al., 2008). This differential role of NGN2 between two similar species poses an interesting question; is the role of NGN2 species dependent or can differences in the cellular context of the progenitors, such as passage number, transduction efficiency, or plating density (Ko et al., 2005), alter the role of NGN2 in vitro? Overall, the overexpression of NGN2 is not commonly utilized for dopaminergic differentiation, as there are other more critical transcription factors expressed in dopaminergic progenitors required for commitment to a dopaminergic fate. For example, combinations of just ASCL1, NURR1, and LMX1A or LMX1B can efficiently drive the generation of dopaminergic neurons (Ng et al., 2021; Powell et al., 2021). Thus, it is critical to consider the combinations of transcription factors when utilizing dopaminergic neuron protocols.
NGN2 in serotonergic neuronal differentiation
The role NGN2 plays in the generation of serotoninergic neurons is not clearly defined; however, its overexpression in combination with MASH1, NKX2.2, FEV, GATA2, and LMX1B has been shown to generate functional serotonergic neurons (Vadodaria et al., 2016). Human dermal fibroblasts were delivered the combination of genes in conjunction to patterning with small molecule inhibitors dibutyryl cyclic-AMP, noggin, LDN193189, A83-1, CHIR99021, SB431542, and forskolin and were matured for 6 weeks. After 3 weeks of neuronal conversion, 60% of fibroblasts expressed ß-III-TUBULIN (Vadodaria et al., 2016). The derived iNs expressed serotonergic markers including TPH (61% out of the MAP2AB + cells), serotonin (5-HT; 38% out of the MAP2AB + cells), and serotonin transporter, SERT (Vadodaria et al., 2016). Whole transcriptomic analysis of iNs indicated an enriched expression of neuronal and serotonergic genes and low-level expression of other neural subtype markers (Vadodaria et al., 2016). Generated iNs matured for 6 weeks and co-cultured with astrocytes displayed spontaneous firing activity and current evoked action potential firing (Vadodaria et al., 2016). iNs released 5-HT in response to citalopram and tryptophan (Vadodaria et al., 2016). This study highlights that NGN2 may play a role in serotonergic differentiation but is not a key regulator and requires the action of different transcription factors and developmental patterning.
NGN2 inhibits GABAergic neuronal differentiation
NGN2 has been described to play a role in the generation of many neuronal subtypes; interestingly, NGN2 inhibits the differentiation of GABAergic interneurons. Ngn2 and Ngn1;Ngn2 mutants showed upregulation of genes involved in the generation of subcortical GABAergic neurons including Ascl1, Dlx1, Dlx2, Dlx5, Brn4, GAD1/2, and GABA-T1, suggesting that NGN2 inhibits GABAergic neuron differentiation (Fode et al., 2000; Jo et al., 2007; Schuurmans et al., 2004). Roybon et al. (2010) provide evidence in an in vitro neurosphere model that not only NGN2 prevents GABAergic neuron differentiation but also the downstream effectors of the NEUROD1/2 function through the repression of MASH1. These proteins may function downstream to NGN2, maintaining the repression of MASH1 and favoring glutamatergic neuron differentiation (Roybon et al., 2010). Collectively, these studies highlight the crossregulatory mechanisms of transcription factors in cortical neuron development, where NGN2 and its downstream effectors repress Ascl1 and other genes required for GABAergic neuron generation.
Applications of NGN2 iNs
The development of NGN2 differentiation protocols has enabled access to hPSC-derived neurons to a wider range of scientists, without requiring prior expertise in handling hPSC lines. As NGN2 iNs are easy to use, rapid, reproducible, and scalable, experiments that were previously performed in cancer or established cell lines can now be completed in more biologically relevant cell types. This has allowed for use in a wide range of applications, including disease modeling, high-throughput screening, and neuronal replacement therapies.
NGN2 iNs use in modeling neurodegenerative, neurodevelopmental, and neuropsychiatric diseases
The promise of NGN2 iNs for use in modeling of neurodevelopment and disease relies on the ability to generate defined neuronal types (Figure 2A). Excitatory NGN2 iNs have been co-cultured with inhibitory GABAergic interneurons (Yang et al., 2017), useful for modeling cortical development, given the importance of inhibitory neurotransmission in healthy and disease states. Similarly, an NGN2 iN 3D co-culture with astrocytes allowed integration of astrocyte-neuron functional properties such as synapse modulation and homeostatic functions occurring during fetal brain development (Krencik et al., 2017). NGN2 iNs have also been used for electrophysiological assays in a “fully humanized” co-culture system with iPSC-derived astrocytes (Shih et al., 2021). Further, NGN2 iNs have been utilized at a single-cell resolution in culture to study neuronal plasticity and synaptic transmission (Meijer et al., 2019).
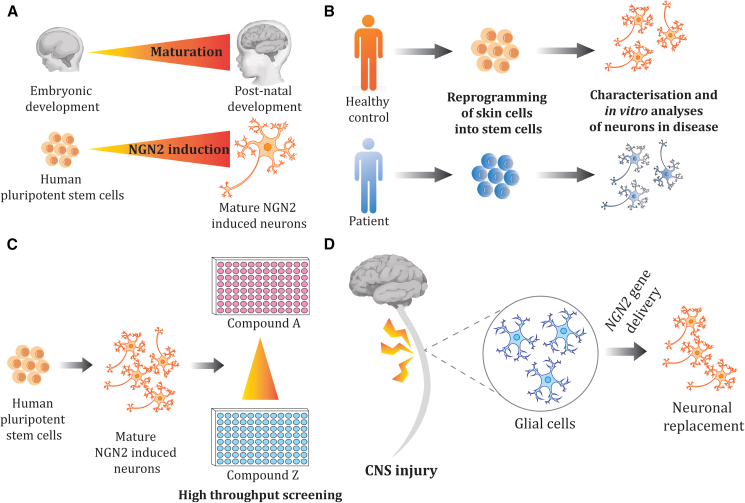
Figure 2.
Summary of the applications of NGN2 iNs
(A and B). NGN2 overexpression in the differentiation of neurons has been applied to development (A) and disease modeling of various neurodegenerative and neuropsychiatric diseases in vitro (B).
(C). Reduced differentiation time and scalability of iNs have allowed NGN2 iNs to be applied for high-throughput compound screening (C).
(D). Following injury to the central nervous system, NGN2 overexpression has been applied to convert glial cells to neurons for neuronal replacement therapies (D).
NGN2 iNs have proven a valuable tool for neurodevelopmental and neurodegenerative studies (Figure 2B). For example, in sporadic Alzheimer's disease, Lin et al. (2018) and Meyer et al. (2019) employed iNs to investigate APOE and other disease-related gene effects in patient iPSC-derived cortical neurons. In neurodevelopmental and neuropsychiatric disorders, NGN2 iNs have been employed to study disease phenotypes and potential therapeutics in autism (Deneault et al., 2018), Down syndrome (Hirata et al., 2020), fragile X syndrome (Graef et al., 2020), and schizophrenia (Xu et al., 2018), signifying the wide applicability of the NGN2 iNs. Nickolls et al. (2020) utilized NGN2 in combination with BRN3A to produce cold- and mechano-sensing iNs, uncovering a novel class of sensory neurons present in humans, and successfully modeled a rare somatosensory disease caused by inactivating mutations in PIEZO2. As such, iNs have demonstrated utility for the study of both neurodevelopment and disease.
The use of NGN2 iNs to improve high-throughput screening
Reduced batch-to-batch and line-to-line variability makes NGN2 overexpression particularly suited for high-throughput microscopy screens and rapidly evolving omics technologies (Figure 2C). Efforts have been made in recent years to optimize NGN2 overexpression to improve scalability and reproducibility—both important necessities for successful screening assays. Inducible cell lines can be generated through integrating a NGN2 expression cassette into a safe harbor locus, variability in NGN2 expression can be minimized, and neurons can be obtained in a rapid and scalable manner (Fernandopulle et al., 2018; Tian et al., 2019; Wang et al., 2017). Wang et al. (2017) used an iPSC line that harbors an inducible NGN2 expression cassette to identify tau-lowering compounds in human neurons as a potential therapeutic strategy for Alzheimer's disease and associated tauopathies. Tian et al. (2019) used iNs generated from i3 stem cell lines to perform CRISPR-interference-based genetic screening of over 2,000 genes, investigating genes involved in neuronal survival. Large batches of cryo-banked iNs performed similarly to freshly prepared iNs in a high-throughput screening neurite outgrowth assay (Sridharan et al., 2019). However, scalability remains challenging for more complex phenotypic assays concerning synaptic activity. In such cases, NGN2 iNs might be required to be kept in culture for longer or co-cultured with glial cells, which would increase assay complexity.
Application of NGN2 overexpression in neuronal replacement therapy
Central nervous system damage or neurodegenerative diseases are linked to the irreversible loss of neurons, leading to permanent functional decline. NGN2 iNs could serve as a valuable source of standardized neurons for application in regenerative medicine (Figure 2D). Classically, cell-replacement approaches involved transplanting neurons derived in vitro into the injured brain. Zhang et al. (2013) demonstrated that NGN2 iNs can functionally integrate into mice brains and NGN2-reprogrammed olfactory ensheathing cells survive and mature after spinal cord transplantation (Sun et al., 2019b). However, classical cell-replacement approaches still face major barriers, related to safety and feasibility (Lindvall, 2012).
Alternatives involving the direct in vivo conversion of non-neuronal cells residing within the affected tissue have emerged as a potential novel therapy. Hu et al. (2019) successfully converted both postnatal and adult astrocytes from cortex, cerebellum, and spinal cord in mice to neurons using NGN2 overexpression, albeit induction efficiency appeared lower in adult astrocytes than that of postnatal astrocytes. Following cortical stab wound injury to both gray and white matter in adult mice, Mattugini et al. (2019) successfully converted astrocytes to neurons by combining NGN2 with NURR1 overexpression. Interestingly, no astrocyte to neuron conversion was observed in the white matter (Mattugini et al., 2019). Similarly, Zhou et al. (2021) overexpressed NGN2 and ISL1 in astrocytes in the mouse spinal cord, which converted astrocytes to motor neurons with high efficiency, with no conversion detected in white matter astrocytes. This indicates possible regional limitations in reprogramming. It is important to note that the effect of NGN2 iNs, from either direct cell replacement or in vivo conversion, on improvements of lost function, memory, cognition, and behavior has yet to be explored. NGN2 overexpression has also been explored as a potential treatment in malignant glioma, a CNS tumor, by forcing glioma cells to undergo terminal differentiation to neuronal cells (Guichet et al., 2013; Su et al., 2014) to stop tumor growth. To make NGN2 iNs clinically relevant, many hurdles linked to the integration of foreign DNA must be overcome, such as varying expression levels and random integrations in coding or regulatory sequences. Recent efforts have been made to overcome those limitations by testing modified messenger RNA-based methods to produce NGN2 iNs (Tolomeo et al., 2021). Overall, NGN2 iNs offer an exciting avenue in neuronal replacement therapy for both CNS injury or tumors; however, there are many hurdles including the ethical implications and the effects of newly converted neurons on function, memory, cognition, and behavior.
Too good to be true? Limitations of using NGN2 iNs
The emerging field of iNs raises several key questions; are NGN2 iNs a biologically relevant model and do the derived iNs represent the homogeneous population of neurons they are intended to be? NGN2 iNs provide a rapid model for neuronal differentiation, as they can bypass the progenitor stage of development; however, there are questions surrounding whether this limits their biological relevance. Notably, the use of NGN2 iNs may not be suitable for the study of diseases in which progenitors underpin disease progression, with extrinsic-factor-based models being more appropriate to recapitulate each stage of neurodevelopment. Rosa et al. (2020) assessed the electrophysiological properties of NGN2 iNs and neurons derived from an EB-based dual SMAD inhibition protocol (EB-derived neurons), comparing these to human and rodent neurodevelopmental studies. Functional readouts for NGN2 iNs were recorded at 2–3 weeks and EB-derived neurons at 3–6 months maturation. EB-derived neurons were more mature on a ranking system based on shooting amplitude of action potentials and firing frequency, with ∼90% of EB-derived neurons and ∼67% of NGN2 iNs reaching a type 5 maturity (Rosa et al., 2020). EB neuron recordings had a significantly lower resting membrane potential and action potential threshold, with input resistance readings resembling adult human neurons, whereas the NGN2 iNs resembled second trimester human brain tissue (Rosa et al., 2020). However, co-culture of murine astrocytes with the NGN2 iNs improved viability and allowed recording to be performed at 6–7 weeks maturation showing similar firing, capacitance, and input resistance to EB-derived neurons (Rosa et al., 2020). These results showed that EB-derived neurons cultured for 6 months have more mature electrophysiological properties compared with NGN2 iNs matured for 2–3 weeks. However, 6–7 week co-cultures with astrocytes exhibited similar functional properties to the 6-month matured EB neurons. The EB neuronal differentiation produced glial cells naturally, which may have allowed these cultures to be viable for longer and promote maturation. NGN2 iNs have been successfully cultured for 10 weeks in the presence of astrocytes (Chen et al., 2020). Nonetheless, this study provides two important lessons: the co-culture of NGN2 iNs with astrocytes should be considered in vitro to promote viability, and more mature cultures can provide insight into the developmental stage they represent.
Owing to the plethora of neuronal cell types that can be derived using NGN2, the ectopic expression of NGN2 for transdifferentiation of neurons raises the question of purity and fidelity of derived iN populations. Lin et al. (2021) used single-cell transcriptomics on NGN2 iNs derived from multiple iPSC lines comparing the cell identity and purity of neuronal cultures. Neuronal cultures constituted a heterogeneous population of peripheral nervous system (PNS) neurons, represented by clusters expressing genes abundantly found in the PNS including PRPH (encodes PERIPHERIN), PHOX2B (encodes paired like homeobox 2B), and POU4F1 (encodes BRN3A) compared with a cluster marked by GPM6A (encodes glycoprotein M6A), which is expressed both in the CNS and spinal cord (Lin et al., 2021). The duration of NGN2 induction also affected neuronal specification, with a shorter duration favoring GPM6A population and longer durations resulting in more abundant populations of PRPH+/POUF41 + neurons. Consistent with these results, NGN2 iN cultures were identified to represent a mixture of PNS and CNS neurons (Chen et al., 2020). NGN2 overexpression alone was shown to result in cultures with 95% MAP2+ cells but contained 10% of cells positive for PERIPHERIN (Chen et al., 2020). A short patterning period with the dual SMAD inhibitors SB431542 and LDN193189, prior to NGN2 transduction, resulted in a homogeneous population of excitatory forebrain neurons, as evidenced by the absence of PERIPHERIN expression and significantly higher expression of forebrain markers, including TBR1 via immunostaining and SIX1 and FOXG3 by RNA-sequencing compared with NGN2 alone (Chen et al., 2020). Whereas, patterning with SB431542 and CHIR99021 resulted in a population of hindbrain and spinal cord V2A interneurons. The interneurons contained a population of VSX2+/SHOX2+ neurons representing excitatory neurons that control motor neurons, and a population of VSX2+/SHOX2-interneurons involved in left-right coordination (Chen et al., 2020). This highlights the importance of developmental patterning in NGN2 iNs differentiation in regionally specifying subtypes of neurons to generate a more homogeneous population. These studies also raise the importance of thorough profiling of iNs to confirm that the cells represent the desired neuronal population, as NGN2 is dynamically and widely expressed throughout the developing nervous system and is involved in differentiation of a diverse range of neurons. Profiling should incorporate single-cell sequencing to define the heterogeneity and maturation of derived iN populations and compare these cells to bona-fide human neurons. Further investigation is required to understand whether the starting cell type (hPSCs versus progenitors versus fibroblasts versus glial cells) affects the single-cell transcriptomic profiles of derived iNs.
Conclusion
The use of NGN2 overexpression has reconfigured the way neuronal differentiations occur, shifting from using extrinsic factors that mimic developmental patterning cues to the rapid, scalable, and reproducible systems currently used. These protocols have allowed for widespread applications including development and disease modeling of neurodegenerative disease and neuropsychiatric disorders, high-throughput compound screening, and neuronal replacement therapies. With the rapidly evolving field, it is becoming clear that NGN2 alone is not a “one-glove-fits-all” approach for neuronal differentiation but requires the combination of other transcription factors and developmental patterning to generate homogeneous neuronal populations.
Acknowledgments
A.J.H., S.M., and M.S.C.G. are each supported by the Australian Government Research Training Program Scholarship. M.S.C.G. is by a joint scholarship from Australian Rotary Health and the Rotary Club of West Wollongong. This review was also supported by funding from Friedreich's Ataxia Research Alliance USA, Friedreich Ataxia Research Association Australia, Australian Research Council Linkage Grant (LP190101139), Medical Research Future Fund's Stem Cell Therapies Mission, (2007421), Illawarra Health and Medical Research Institute, and University of Wollongong.
Conflicts of interest
The authors have no financial interests to declare.
References
- Abdo H., Li L., Lallemend F., Bachy I., Xu X.J., Rice F.L., Ernfors P. Dependence on the transcription factor Shox2 for specification of sensory neurons conveying discriminative touch. Eur. J. Neurosci. 2011;34:1529–1541. doi: 10.1111/j.1460-9568.2011.07883.x. [DOI] [PubMed] [Google Scholar]
- Abeliovich A., Hammond R. Midbrain dopamine neuron differentiation: factors and fates. Dev. Biol. 2007;304:447–454. doi: 10.1016/j.ydbio.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Abu-Bonsrah K.D., Viventi S., Newgreen D.F., Dottori M. Generation of neural crest progenitors from human pluripotent stem cells. Methods Mol. Biol. 2019;1976:37–47. doi: 10.1007/978-1-4939-9412-0_3. [DOI] [PubMed] [Google Scholar]
- Alshawaf A.J., Viventi S., Qiu W., D’Abaco G., Nayagam B., Erlichster M., Chana G., Everall I., Ivanusic J., Skafidas E., Dottori M. Phenotypic and functional characterization of peripheral sensory neurons derived from human embryonic stem cells. Sci. Rep. 2018;8:603. doi: 10.1038/s41598-017-19093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso M.W., Croft G.F., Williams D.J., O'Keeffe S., Carrasco M.A., Davis A.R., Roybon L., Oakley D.H., Maniatis T., Henderson C.E. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 2013;33:574–586. doi: 10.1523/JNEUROSCI.0906-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E., Jensen J.B., Parmar M., Guillemot F., Björklund A. Development of the mesencephalic dopaminergic neuron system is compromised in the absence of neurogenin 2. Development. 2006;133:507–516. doi: 10.1242/dev.02224. [DOI] [PubMed] [Google Scholar]
- Andersson E., Tryggvason U., Deng Q., Friling S., Alekseenko Z., Robert B., Perlmann T., Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Ang C.E., Olmos V.H., Zhou B., Lee Q.Y., Sinha R., Narayanaswamy A., Mall M., Chesnov K., Sudhof T., Wernig M. The dynamic interplay between homeodomain transcription factors and chromatin environment regulates proneural factor outcomes. bioRxiv. 2020 doi: 10.1101/2020.12.02.398677. [DOI] [Google Scholar]
- Balaram P., Hackett T.A., Kaas J.H. VGLUT1 mRNA and protein expression in the visual system of prosimian galagos (Otolemur garnetti) Eye Brain. 2011;3:81. doi: 10.2147/EB.S23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bel-Vialar S., Medevielle F., Pituello F. The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord. Dev. Biol. 2007;305:659–673. doi: 10.1016/j.ydbio.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Bertrand N., Castro D.S., Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Björklund A., Dunnett S.B. Dopamine neuron systems in the brain: an update. Trends Neurosci. 2007;30:194–202. doi: 10.1016/j.tins.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Blaess S., Ang S.L. Genetic control of midbrain dopaminergic neuron development. Wiley Interdiscip. Rev. Dev. Biol. 2015;4:113–134. doi: 10.1002/wdev.169. [DOI] [PubMed] [Google Scholar]
- Blanchard J.W., Eade K.T., Szűcs A., Sardo V.L., Tsunemoto R.K., Williams D., Sanna P.P., Baldwin K.K. Selective conversion of fibroblasts into peripheral sensory neurons. Nat. Neurosci. 2015;18:25–35. doi: 10.1038/nn.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., Ericson J. Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 2001;11:43–49. doi: 10.1016/s0959-4388(00)00172-0. [DOI] [PubMed] [Google Scholar]
- Briscoe J., Pierani A., Jessell T.M., Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Busskamp V., Lewis N.E., Guye P., Ng A.H., Shipman S.L., Byrne S.M., Sanjana N.E., Murn J., Li Y., Li S. Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol. 2014;10:760. doi: 10.15252/msb.20145508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cave C., Sockanathan S. Transcription factor mechanisms guiding motor neuron differentiation and diversification. Curr. Opin. Neurobiol. 2018;53:1–7. doi: 10.1016/j.conb.2018.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Maimaitili M., Habekost M., Gill K.P., Mermet-Joret N., Nabavi S., Febbraro F., Denham M. Rapid generation of regionally specified CNS neurons by sequential patterning and conversion of human induced pluripotent stem cells. Stem Cell Res. 2020;48:101945. doi: 10.1016/j.scr.2020.101945. [DOI] [PubMed] [Google Scholar]
- Cohen D.E., Melton D. Turning straw into gold: directing cell fate for regenerative medicine. Nat. Rev. Genet. 2011;12:243–252. doi: 10.1038/nrg2938. [DOI] [PubMed] [Google Scholar]
- Dang S.M., Kyba M., Perlingeiro R., Daley G.Q., Zandstra P.W. Efficiency of embryoid body formation and hematopoietic development from embryonic stem cells in different culture systems. Biotechnol. Bioeng. 2002;78:442–453. doi: 10.1002/bit.10220. [DOI] [PubMed] [Google Scholar]
- Deneault E., White S.H., Rodrigues D.C., Ross P.J., Faheem M., Zaslavsky K., Wang Z., Alexandrova R., Pellecchia G., Wei W., et al. Complete disruption of autism-susceptibility genes by gene editing predominantly reduces functional connectivity of isogenic human neurons. Stem Cell Rep. 2018;11:1211–1225. doi: 10.1016/j.stemcr.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denham M., Dottori M. In: Neurodegeneration: Methods and Protocols. Manfredi G., Kawamata H., editors. Humana Press; 2011. Neural differentiation of induced pluripotent stem cells; pp. 99–110. [DOI] [PubMed] [Google Scholar]
- Denham M., Hasegawa K., Menheniott T., Rollo B., Zhang D., Hough S., Alshawaf A., Febbraro F., Ighaniyan S., Leung J., et al. Multipotent caudal neural progenitors derived from human pluripotent stem cells that give rise to lineages of the central and peripheral nervous system. Stem Cells. 2015;33:1759–1770. doi: 10.1002/stem.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis R., Garone M.G., Pagani F., de Turris V., Di Angelantonio S., Rosa A. Direct conversion of human pluripotent stem cells into cranial motor neurons using a piggyBac vector. Stem Cell Res. 2018;29:189–196. doi: 10.1016/j.scr.2018.04.012. [DOI] [PubMed] [Google Scholar]
- Dubreuil V., Hirsch M.R., Pattyn A., Brunet J.F., Goridis C. The Phox2b transcription factor coordinately regulates neuronal cell cycle exit and identity. Development. 2000;127:5191–5201. doi: 10.1242/dev.127.23.5191. [DOI] [PubMed] [Google Scholar]
- Dykes I.M., Tempest L., Lee S.I., Turner E.E. Brn3a and Islet1 act epistatically to regulate the gene expression program of sensory differentiation. J. Neurosci. 2011;31:9789–9799. doi: 10.1523/jneurosci.0901-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah M.H., Olson J.M., Sucic H.B., Hume R.I., Tapscott S.J., Turner D.L. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Faustino Martins J.-M., Fischer C., Urzi A., Vidal R., Kunz S., Ruffault P.-L., Kabuss L., Hube I., Gazzerro E., Birchmeier C., et al. Self-organizing 3D human trunk neuromuscular organoids. Cell Stem Cell. 2020;26:172–186.e6. doi: 10.1016/j.stem.2019.12.007. [DOI] [PubMed] [Google Scholar]
- Fernandopulle M.S., Prestil R., Grunseich C., Wang C., Gan L., Ward M.E. Transcription factor-mediated differentiation of human iPSCs into neurons. Curr. Protoc. Cell Biol. 2018;79:e51. doi: 10.1002/cpcb.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C., Ma Q., Casarosa S., Ang S.-L., Anderson D.J., Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. doi: 10.1101/gad.14.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fremeau R.T., Troyer M.D., Pahner I., Nygaard G.O., Tran C.H., Reimer R.J., Bellocchio E.E., Fortin D., Storm-Mathisen J., Edwards R.H. The expression of vesicular glutamate transporters defines two classes of excitatory synapse. Neuron. 2001;31:247–260. doi: 10.1016/S0896-6273(01)00344-0. [DOI] [PubMed] [Google Scholar]
- Garone M.G., de Turris V., Soloperto A., Brighi C., De Santis R., Pagani F., Di Angelantonio S., Rosa A. Conversion of human induced pluripotent stem cells (iPSCs) into functional spinal and cranial motor neurons using PiggyBac vectors. J. Vis. Exp. 2019 doi: 10.3791/59321. [DOI] [PubMed] [Google Scholar]
- Ge W., He F., Kim K.J., Blanchi B., Coskun V., Nguyen L., Wu X., Zhao J., Heng J.I., Martinowich K., et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl. Acad. Sci. U S A. 2006;103:1319–1324. doi: 10.1073/pnas.0510419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goparaju S.K., Kohda K., Ibata K., Soma A., Nakatake Y., Akiyama T., Wakabayashi S., Matsushita M., Sakota M., Kimura H., et al. Rapid differentiation of human pluripotent stem cells into functional neurons by mRNAs encoding transcription factors. Sci. Rep. 2017;7:42367. doi: 10.1038/srep42367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef J.D., Wu H., Ng C., Sun C., Villegas V., Qadir D., Jesseman K., Warren S.T., Jaenisch R., Cacace A., Wallace O. Partial FMRP expression is sufficient to normalize neuronal hyperactivity in Fragile X neurons. Eur. J. Neurosci. 2020;51:2143–2157. doi: 10.1111/ejn.14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichet P.O., Bieche I., Teigell M., Serguera C., Rothhut B., Rigau V., Scamps F., Ripoll C., Vacher S., Taviaux S., et al. Cell death and neuronal differentiation of glioblastoma stem-like cells induced by neurogenic transcription factors. Glia. 2013;61:225–239. doi: 10.1002/glia.22429. [DOI] [PubMed] [Google Scholar]
- Hand R., Bortone D., Mattar P., Nguyen L., Heng J.I., Guerrier S., Boutt E., Peters E., Barnes A.P., Parras C., et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Hegarty S.V., Sullivan A.M., O'Keeffe G.W. Midbrain dopaminergic neurons: a review of the molecular circuitry that regulates their development. Dev. Biol. 2013;379:123–138. doi: 10.1016/j.ydbio.2013.04.014. [DOI] [PubMed] [Google Scholar]
- Hester M.E., Murtha M.J., Song S., Rao M., Miranda C.J., Meyer K., Tian J., Boulting G., Schaffer D.V., Zhu M.X., et al. Rapid and efficient generation of functional motor neurons from human pluripotent stem cells using gene delivered transcription factor codes. Mol. Ther. 2011;19:1905–1912. doi: 10.1038/mt.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindley C., Ali F., McDowell G., Cheng K., Jones A., Guillemot F., Philpott A. Post-translational modification of Ngn2 differentially affects transcription of distinct targets to regulate the balance between progenitor maintenance and differentiation. Development. 2012;139:1718–1723. doi: 10.1242/dev.077552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata K., Nambara T., Kawatani K., Nawa N., Yoshimatsu H., Kusakabe H., Banno K., Nishimura K., Ohtaka M., Nakanishi M., et al. 4-Phenylbutyrate ameliorates apoptotic neural cell death in Down syndrome by reducing protein aggregates. Sci. Rep. 2020;10:14047. doi: 10.1038/s41598-020-70362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.-M., Hartley B.J., Tcw J., Beaumont M., Stafford K., Slesinger P.A., Brennand K.J. Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells. Methods. 2016;101:113–124. doi: 10.1016/j.ymeth.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge R.D., Bakken T.E., Miller J.A., Smith K.A., Barkan E.R., Graybuck L.T., Close J.L., Long B., Johansen N., Penn O., et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Qin S., Huang X., Yuan Y., Tan Z., Gu Y., Cheng X., Wang D., Lian X.-F., He C., Su Z. Region-restrict astrocytes exhibit heterogeneous susceptibility to neuronal reprogramming. Stem Cell Rep. 2019;12:290–304. doi: 10.1016/j.stemcr.2018.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme A.J., McArthur J.R., Maksour S., Miellet S., Ooi L., Adams D.J., Finol-Urdaneta R.K., Dottori M. Molecular and functional characterization of neurogenin-2 induced human sensory neurons. Front. Cell. Neurosci. 2020;14:600895. doi: 10.3389/fncel.2020.600895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi K., Sone T., Ibata K., Fujimori K., Yuzaki M., Akamatsu W., Okano H. Controlling the regional identity of hPSC-derived neurons to uncover neuronal subtype specificity of neurological disease phenotypes. Stem Cell Rep. 2015;5:1010–1022. doi: 10.1016/j.stemcr.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itskovitz-Eldor J., Schuldiner M., Karsenti D., Eden A., Yanuka O., Amit M., Soreq H., Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 2000;6:88–95. doi: 10.1007/BF03401776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell T.M. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Jiang M., Yu D., Xie B., Huang H., Lu W., Qiu M., Dai Z.-M. WNT signaling suppresses oligodendrogenesis via Ngn2-dependent direct inhibition of Olig2 expression. Mol. Brain. 2020;13:155. doi: 10.1186/s13041-020-00696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo A.Y., Park C.-H., Aizawa S., Lee S.-H. Contrasting and brain region-specific roles of neurogenin2 and mash1 in GABAergic neuron differentiation in vitro. Exp. Cell Res. 2007;313:4066–4081. doi: 10.1016/j.yexcr.2007.08.026. [DOI] [PubMed] [Google Scholar]
- Jo J., Xiao Y., Sun A.X., Cukuroglu E., Tran H.-D., Göke J., Tan Z.Y., Saw T.Y., Tan C.-P., Lokman H., et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell. 2016;19:248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.A., Weick J.P., Pearce R.A., Zhang S.C. Functional neural development from human embryonic stem cells: accelerated synaptic activity via astrocyte coculture. J. Neurosci. 2007;27:3069–3077. doi: 10.1523/jneurosci.4562-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karumbayaram S., Novitch B.G., Patterson M., Umbach J.A., Richter L., Lindgren A., Conway A.E., Clark A.T., Goldman S.A., Plath K. Directed differentiation of human-induced pluripotent stem cells generates active motor neurons. Stem Cells. 2009;27:806–811. doi: 10.1002/stem.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada J., Kaneda S., Kirihara T., Maroof A., Levi T., Eggan K., Fujii T., Ikeuchi Y. Generation of a motor nerve organoid with human stem cell-derived neurons. Stem Cell Rep. 2017;9:1441–1449. doi: 10.1016/j.stemcr.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kele J., Simplicio N., Ferri A.L., Mira H., Guillemot F., Arenas E., Ang S.-L. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development. 2006;133:495–505. doi: 10.1242/dev.02223. [DOI] [PubMed] [Google Scholar]
- Ko J.-Y., Lee J.-Y., Park C.-H., Lee S.-H. Effect of cell-density on in-vitro dopaminergic differentiation of mesencephalic precursor cells. Neuroreport. 2005;16 doi: 10.1097/00001756-200504040-00016. [DOI] [PubMed] [Google Scholar]
- Kramer I., Sigrist M., de Nooij J.C., Taniuchi I., Jessell T.M., Arber S. A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron. 2006;49:379–393. doi: 10.1016/j.neuron.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Krencik R., Seo K., van Asperen J.V., Basu N., Cvetkovic C., Barlas S., Chen R., Ludwig C., Wang C., Ward M.E., et al. Systematic three-dimensional coculture rapidly recapitulates interactions between human neurons and astrocytes. Stem Cell Rep. 2017;9:1745–1753. doi: 10.1016/j.stemcr.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme M., Liaubet L., Pituello F., Bel-Vialar S. NEUROG2 drives cell cycle exit of neuronal precursors by specifically repressing a subset of cyclins acting at the G1 and S phases of the cell cycle. Mol. Cell Biol. 2012;32:2596–2607. doi: 10.1128/mcb.06745-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemend F., Ernfors P. Molecular interactions underlying the specification of sensory neurons. Trends Neurosci. 2012;35:373–381. doi: 10.1016/j.tins.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.-A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Chambers S.M., Tomishima M.J., Studer L. Derivation of neural crest cells from human pluripotent stem cells. Nat. Protoc. 2010;5:688–701. doi: 10.1038/nprot.2010.35. [DOI] [PubMed] [Google Scholar]
- Lee S., Cuvillier J.M., Lee B., Shen R., Lee J.W., Lee S.K. Fusion protein Isl1-Lhx3 specifies motor neuron fate by inducing motor neuron genes and concomitantly suppressing the interneuron programs. Proc. Natl. Acad. Sci. U S A. 2012;109:3383–3388. doi: 10.1073/pnas.1114515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee B., Lee J.W., Lee S.K. Retinoid signaling and neurogenin2 function are coupled for the specification of spinal motor neurons through a chromatin modifier CBP. Neuron. 2009;62:641–654. doi: 10.1016/j.neuron.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Pfaff S.L. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Lei L., Laub F., Lush M., Romero M., Zhou J., Luikart B., Klesse L., Ramirez F., Parada L.F. The zinc finger transcription factor Klf7 is required for TrkA gene expression and development of nociceptive sensory neurons. Genes Dev. 2005;19:1354–1364. doi: 10.1101/gad.1227705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sun W., Zhang Y., Wei W., Ambasudhan R., Xia P., Talantova M., Lin T., Kim J., Wang X., et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc. Natl. Acad. Sci. U S A. 2011;108:8299–8304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.-C., He Z., Ebert S., Schörnig M., Santel M., Weigert A., Hevers W., Kasri N.N., Taverna E., Camp J.G., Treutlein B. Ngn2 induces diverse neuronal lineages from human pluripotency. Stem Cell Rep. 2021 doi: 10.1016/j.stemcr.2021.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Yuan J., Sander B., Golas M.M. In vitro differentiation of human neural progenitor cells into striatal GABAergic neurons. Stem Cells Transl. Med. 2015;4:775–788. doi: 10.5966/sctm.2014-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-T., Seo J., Gao F., Feldman H.M., Wen H.-L., Penney J., Cam H.P., Gjoneska E., Raja W.K., Cheng J., et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron. 2018;98:1141–1154.e7. doi: 10.1016/j.neuron.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindvall O. Why is it taking so long to develop clinically competitive stem cell therapies for CNS disorders? Cell Stem Cell. 2012;10:660–662. doi: 10.1016/j.stem.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Liu M.-L., Zang T., Zou Y., Chang J.C., Gibson J.R., Huber K.M., Zhang C.-L. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li F., Stubblefield E.A., Blanchard B., Richards T.L., Larson G.A., He Y., Huang Q., Tan A.-C., Zhang D., et al. Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 2012;22:321–332. doi: 10.1038/cr.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L., Dormand E., Greenwood A., Anderson D.J. Comparison of the generic neuronal differentiation and neuron subtype specification functions of mammalian achaete-scute and atonal homologs in cultured neural progenitor cells. Development. 2002;129:1553–1567. doi: 10.1242/dev.129.7.1553. [DOI] [PubMed] [Google Scholar]
- Lu J., Zhong X., Liu H., Hao L., Huang C.T.-L., Sherafat M.A., Jones J., Ayala M., Li L., Zhang S.-C. Generation of serotonin neurons from human pluripotent stem cells. Nat. Biotech. 2016;34:89–94. doi: 10.1038/nbt.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Fode C., Guillemot F., Anderson D.J. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Kintner C., Anderson D.J. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Ma Y.C., Song M.R., Park J.P., Henry Ho H.Y., Hu L., Kurtev M.V., Zieg J., Ma Q., Pfaff S.L., Greenberg M.E. Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron. 2008;58:65–77. doi: 10.1016/j.neuron.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmigère F., Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat. Rev. Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- Mattar P., Langevin L.M., Markham K., Klenin N., Shivji S., Zinyk D., Schuurmans C. Basic helix-loop-helix transcription factors cooperate to specify a cortical projection neuron identity. Mol. Cell Biol. 2008;28:1456–1469. doi: 10.1128/MCB.01510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei C., Lim R., Drury H., Nasr B., Li Z., Tadros M.A., D'Abaco G.M., Stok K.S., Nayagam B.A., Dottori M. Generation of vestibular tissue-like organoids from human pluripotent stem cells using the Rotary Cell Culture System. Front Cell Dev. Biol. 2019;7:25. doi: 10.3389/fcell.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattugini N., Bocchi R., Scheuss V., Russo G.L., Torper O., Lao C.L., Götz M. Inducing different neuronal subtypes from astrocytes in the injured mouse cerebral cortex. Neuron. 2019;103:1086–1095.e5. doi: 10.1016/j.neuron.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzara P.G., Muggeo S., Luoni M., Massimino L., Zaghi M., Valverde P.T.-T., Brusco S., Marzi M.J., Palma C., Colasante G., et al. Frataxin gene editing rescues Friedreich’s ataxia pathology in dorsal root ganglia organoid-derived sensory neurons. Nat. Commun. 2020;11:4178. doi: 10.1038/s41467-020-17954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni E.O., Mahony S., Closser M., Morrison C.A., Nedelec S., Williams D.J., An D., Gifford D.K., Wichterle H. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 2013;16:1219–1227. doi: 10.1038/nn.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer M., Rehbach K., Brunner J.W., Classen J.A., Lammertse H.C.A., van Linge L.A., Schut D., Krutenko T., Hebisch M., Cornelisse L.N., et al. A single-cell model for synaptic transmission and plasticity in human iPSC-derived neurons. Cell Rep. 2019;27:2199–2211.e6. doi: 10.1016/j.celrep.2019.04.058. [DOI] [PubMed] [Google Scholar]
- Meyer K., Feldman H.M., Lu T., Drake D., Lim E.T., Ling K.-H., Bishop N.A., Pan Y., Seo J., Lin Y.-T., et al. REST and neural gene network dysregulation in iPSC models of Alzheimer’s disease. Cell Rep. 2019;26:1112–1127.e9. doi: 10.1016/j.celrep.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mica Y., Lee G., Chambers Stuart M., Tomishima Mark J., Studer L. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hESCs and patient-specific iPSCs. Cell Rep. 2013;3:1140–1152. doi: 10.1016/j.celrep.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R., Sugimori M., Takebayashi H., Kosako H., Nagao M., Yoshida S., Nabeshima Y., Shimamura K., Nakafuku M. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–771. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- Montelius A., Marmigère F., Baudet C., Aquino J.B., Enerbäck S., Ernfors P. Emergence of the sensory nervous system as defined by Foxs1 expression. Differentiation. 2007;75:404–417. doi: 10.1111/j.1432-0436.2006.00154.x. [DOI] [PubMed] [Google Scholar]
- Nehme R., Zuccaro E., Ghosh S.D., Li C., Sherwood J.L., Pietilainen O., Barrett L.E., Limone F., Worringer K.A., Kommineni S., et al. Combining NGN2 programming with developmental patterning generates human excitatory neurons with NMDAR-mediated synaptic transmission. Cell Rep. 2018;23:2509–2523. doi: 10.1016/j.celrep.2018.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng Y.H., Chanda S., Janas J.A., Yang N., Kokubu Y., Südhof T.C., Wernig M. Efficient generation of dopaminergic induced neuronal cells with midbrain characteristics. Stem Cell Rep. 2021;16:1763–1776. doi: 10.1016/j.stemcr.2021.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickolls A.R., Lee M.M., Espinoza D.F., Szczot M., Lam R.M., Wang Q., Beers J., Zou J., Nguyen M.Q., Solinski H.J., et al. Transcriptional programming of human mechanosensory neuron subtypes from pluripotent stem cells. Cell Rep. 2020;30:932–946.e7. doi: 10.1016/j.celrep.2019.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M., Schuurmans C., Britz O., Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- Novitch B.G., Chen A.I., Jessell T.M. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Ozen I., Galichet C., Watts C., Parras C., Guillemot F., Raineteau O. Proliferating neuronal progenitors in the postnatal hippocampus transiently express the proneural gene Ngn2. Eur. J. Neurosci. 2007;25:2591–2603. doi: 10.1111/j.1460-9568.2007.05541.x. [DOI] [PubMed] [Google Scholar]
- Park C.-H., Kang J.S., Yoon E.-H., Shim J.-W., Suh-Kim H., Lee S.-H. Proneural bHLH neurogenin 2 differentially regulates Nurr1-induced dopamine neuron differentiation in rat and mouse neural precursor cells in vitro. FEBS Lett. 2008;582:537–542. doi: 10.1016/j.febslet.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Park C.H., Kang J.S., Kim J.S., Chung S., Koh J.Y., Yoon E.H., Jo A.Y., Chang M.Y., Koh H.C., Hwang S., et al. Differential actions of the proneural genes encoding Mash1 and neurogenins in Nurr1-induced dopamine neuron differentiation. J. Cell Sci. 2006;119:2310–2320. doi: 10.1242/jcs.02955. [DOI] [PubMed] [Google Scholar]
- Parras C.M., Schuurmans C., Scardigli R., Kim J., Anderson D.J., Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paşca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.-Y., O'Rourke N.A., Nguyen K.D., et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A., Vallstedt A., Dias J.M., Samad O.A., Krumlauf R., Rijli F.M., Brunet J.F., Ericson J. Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes Dev. 2003;17:729–737. doi: 10.1101/gad.255803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez S.E., Rebelo S., Anderson D.J. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development. 1999;126:1715–1728. doi: 10.1242/dev.126.8.1715. [DOI] [PubMed] [Google Scholar]
- Pfaff S.L., Mendelsohn M., Stewart C.L., Edlund T., Jessell T.M. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Powell S.K., O’Shea C., Townsley K., Prytkova I., Dobrindt K., Elahi R., Iskhakova M., Lambert T., Valada A., Liao W., et al. Induction of dopaminergic neurons for neuronal subtype-specific modeling of psychiatric disease risk. Mol. Psychiatry. 2021:1–13. doi: 10.1038/s41380-021-01273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Zhang X.-J., Renier N., Wu Z., Atkin T., Sun Z., Ozair M.Z., Tchieu J., Zimmer B., Fattahi F., et al. Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat. Biotech. 2017;35:154–163. doi: 10.1038/nbt.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C.J.C. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa F., Dhingra A., Uysal B., Mendis G.D.C., Loeffler H., Elsen G., Mueller S., Schwarz N., Castillo-Lizardo M., Cuddy C., et al. In vitro differentiated human stem cell-derived neurons reproduce synaptic synchronicity arising during neurodevelopment. Stem Cell Rep. 2020;15:22–37. doi: 10.1016/j.stemcr.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybon L., Deierborg T., Brundin P., Li J.Y. Involvement of Ngn2, Tbr and NeuroD proteins during postnatal olfactory bulb neurogenesis. Eur. J. Neurosci. 2009;29:232–243. doi: 10.1111/j.1460-9568.2008.06595.x. [DOI] [PubMed] [Google Scholar]
- Roybon L., Hjalt T., Christophersen N.S., Li J.-Y., Brundin P. Effects on differentiation of embryonic ventral midbrain progenitors by Lmx1a, Msx1, Ngn2, and Pitx3. J.Neurosci. 2008;28:3644–3656. doi: 10.1523/jneurosci.0311-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybon L., Mastracci T.L., Ribeiro D., Sussel L., Brundin P., Li J.-Y. GABAergic differentiation induced by Mash1 is compromised by the bHLH proteins Neurogenin2, NeuroD1, and NeuroD2. Cereb. Cortex. 2010;20:1234–1244. doi: 10.1093/cercor/bhp187. [DOI] [PubMed] [Google Scholar]
- Salick M.R., Wells M.F., Eggan K., Kaykas A. Modelling Zika virus infection of the developing human brain in vitro using stem cell derived cerebral organoids. J. Vis. Exp. 2017;56404 doi: 10.3791/56404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli R., Bäumer N., Gruss P., Guillemot F., Le Roux I. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development. 2003;130:3269–3281. doi: 10.1242/dev.00539. [DOI] [PubMed] [Google Scholar]
- Scardigli R., Schuurmans C., Gradwohl G., Guillemot F. Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron. 2001;31:203–217. doi: 10.1016/s0896-6273(01)00358-0. [DOI] [PubMed] [Google Scholar]
- Schrenk-Siemens K., Wende H., Prato V., Song K., Rostock C., Loewer A., Utikal J., Lewin G.R., Lechner S.G., Siemens J. PIEZO2 is required for mechanotransduction in human stem cell–derived touch receptors. Nat. Neurosci. 2015;18:10–16. doi: 10.1038/nn.3894. [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Yanuka O., Itskovitz-Eldor J., Melton D.A., Benvenisty N. Effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2000;97:11307–11312. doi: 10.1073/pnas.97.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurmans C., Armant O., Nieto M., Stenman J.M., Britz O., Klenin N., Brown C., Langevin L.-M., Seibt J., Tang H., et al. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Lim J.-W., Yellajoshyula D., Chang L.-W., Kroll K.L. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih P.-Y., Kreir M., Kumar D., Seibt F., Pestana F., Schmid B., Holst B., Clausen C., Steeg R., Fischer B., et al. Development of a fully human assay combining NGN2-inducible neurons co-cultured with iPSC-derived astrocytes amenable for electrophysiological studies. Stem Cell Res. 2021;54:102386. doi: 10.1016/j.scr.2021.102386. [DOI] [PubMed] [Google Scholar]
- Simões-Costa M., Bronner M.E. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer L., Ma Q., Anderson D.J. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol. Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- Son E.Y., Ichida J.K., Wainger B.J., Toma J.S., Rafuse V.F., Woolf C.J., Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan B., Hubbs C., Llamosas N., Kilinc M., Singhera F.U., Willems E., Piper D.R., Scampavia L., Rumbaugh G., Spicer T.P. A simple procedure for creating scalable phenotypic screening assays in human neurons. Sci. Rep. 2019;9:9000. doi: 10.1038/s41598-019-45265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Zang T., Liu M.L., Wang L.L., Niu W., Zhang C.L. Reprogramming the fate of human glioma cells to impede brain tumor development. Cell Death Dis. 2014;5:e1463. doi: 10.1038/cddis.2014.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Zhu X.J., Huang H., Guo W., Tang T., Xie B., Xu X., Zhang Z., Shen Y., Dai Z.M. WNT signaling represses astrogliogenesis via Ngn2-dependent direct suppression of astrocyte gene expression. Glia. 2019;67:1333–1343. doi: 10.1002/glia.23608. [DOI] [PubMed] [Google Scholar]
- Sun X., Tan Z., Huang X., Cheng X., Yuan Y., Qin S., Wang D., Hu X., Gu Y., Qian W.-J., et al. Direct neuronal reprogramming of olfactory ensheathing cells for CNS repair. Cell Death Dis. 2019;10:646. doi: 10.1038/s41419-019-1887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Dykes I.M., Liang X., Eng S.R., Evans S.M., Turner E.E. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat. Neurosci. 2008;11:1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nadal-Vicens M., Misono S., Lin M.Z., Zubiaga A., Hua X., Fan G., Greenberg M.E. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Thoma E.C., Wischmeyer E., Offen N., Maurus K., Sirén A.-L., Schartl M., Wagner T.U. Ectopic expression of neurogenin 2 alone is sufficient to induce differentiation of embryonic stem cells into mature neurons. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R., Gachechiladze M.A., Ludwig C.H., Laurie M.T., Hong J.Y., Nathaniel D., Prabhu A.V., Fernandopulle M.S., Patel R., Abshari M., et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron. 2019;104:239–255.e2. doi: 10.1016/j.neuron.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolomeo A.M., Laterza C., Grespan E., Michielin F., Canals I., Kokaia Z., Muraca M., Gagliano O., Elvassore N. NGN2 mmRNA-based transcriptional programming in microfluidic guides hiPSCs toward neural fate with multiple identities. Front. Cell. Neurosci. 2021;15 doi: 10.3389/fncel.2021.602888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadodaria K.C., Mertens J., Paquola A., Bardy C., Li X., Jappelli R., Fung L., Marchetto M.C., Hamm M., Gorris M., et al. Generation of functional human serotonergic neurons from fibroblasts. Mol. Psychiatry. 2016;21:49–61. doi: 10.1038/mp.2015.161. [DOI] [PubMed] [Google Scholar]
- Velasco S., Ibrahim M.M., Kakumanu A., Garipler G., Aydin B., Al-Sayegh M.A., Hirsekorn A., Abdul-Rahman F., Satija R., Ohler U., et al. A multi-step transcriptional and chromatin state cascade underlies motor neuron programming from embryonic stem cells. Cell Stem Cell. 2017;20:205. doi: 10.1016/j.stem.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A., et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneault É., Poirel O., Riad M., Prud'homme J., Dumas S., Turecki G., Fasano C., Mechawar N., El Mestikawy S. Distribution of vesicular glutamate transporters in the human brain. Front. Neuroanat. 2015;9 doi: 10.3389/fnana.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpato V., Smith J., Sandor C., Ried J.S., Baud A., Handel A., Newey S.E., Wessely F., Attar M., Whiteley E., et al. Reproducibility of molecular phenotypes after long-term differentiation to human iPSC-derived neurons: a multi-site omics study. Stem Cell Rep. 2018;11:897–911. doi: 10.1016/j.stemcr.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Ward M.E., Chen R., Liu K., Tracy T.E., Chen X., Xie M., Sohn P.D., Ludwig C., Meyer-Franke A., et al. Scalable production of iPSC-derived human neurons to identify tau-lowering compounds by high-content screening. Stem Cell Rep. 2017;9:1221–1233. doi: 10.1016/j.stemcr.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Xu J., Pang Z.P., Ge W., Kim K.J., Blanchi B., Chen C., Südhof T.C., Sun Y.E. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc. Natl. Acad. Sci. U S A. 2007;104:13821–13826. doi: 10.1073/pnas.0706199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Hartley B.J., Kurup P., Phillips A., Topol A., Xu M., Ononenyi C., Foscue E., Ho S.M., Baguley T.D., et al. Inhibition of STEP61 ameliorates deficits in mouse and hiPSC-based schizophrenia models. Mol. Psychiatry. 2018;23:271–281. doi: 10.1038/mp.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Zhan X., Sun S., Karuppagounder S.S., Xia S., Dawson V.L., Dawson T.M., Laterra J., Zhang J., Ying M. Synthetic mRNAs drive highly efficient iPS cell differentiation to dopaminergic neurons. Stem Cell Transl. Med. 2019;8:112–123. doi: 10.1002/sctm.18-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Yang D., Zarnowska E.D., Du Z., Werbel B., Valliere C., Pearce R.A., Thomson J.A., Zhang S.-C. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells. 2005;23:781–790. doi: 10.1634/stemcells.2004-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Chanda S., Marro S., Ng Y.-H., Janas J.A., Haag D., Ang C.E., Tang Y., Flores Q., Mall M., et al. Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods. 2017;14:621. doi: 10.1038/nmeth.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Xia N., Pera R.A.R.J.J. Directed dopaminergic neuron differentiation from human pluripotent stem cells. J. Vis. Exp. 2014 doi: 10.3791/51737.e51737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J., et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Tao X., Sui M., Cui M., Liu D., Wang B., Wang T., Zheng Y., Luo J., Mu Y., et al. Reprogramming astrocytes to motor neurons by activation of endogenous Ngn2 and Isl1. Stem Cell Rep. 2021;16:1777–1791. doi: 10.1016/j.stemcr.2021.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirlinger M., Lo L., McMahon J., McMahon A.P., Anderson D.J. Transient expression of the bHLH factor neurogenin-2 marks a subpopulation of neural crest cells biased for a sensory but not a neuronal fate. Proc. Natl. Acad. Sci. U S A. 2002;99:8084–8089. doi: 10.1073/pnas.122231199. [DOI] [PMC free article] [PubMed] [Google Scholar]