Abstract
Wastewater surveillance has emerged as a useful tool in the public health response to the COVID-19 pandemic. While wastewater surveillance has been applied at various scales to monitor population-level COVID-19 dynamics, there is a need for quantitative metrics to interpret wastewater data in the context of public health trends. 24-hour composite wastewater samples were collected from March 2020 through May 2021 from a Massachusetts wastewater treatment plant and SARS-CoV-2 RNA concentrations were measured using RT-qPCR. The relationship between wastewater copy numbers of SARS-CoV-2 gene fragments and COVID-19 clinical cases and deaths varies over time. We demonstrate the utility of three new metrics to monitor changes in COVID-19 epidemiology: (1) the ratio between wastewater copy numbers of SARS-CoV-2 gene fragments and clinical cases (WC ratio), (2) the time lag between wastewater and clinical reporting, and (3) a transfer function between the wastewater and clinical case curves. The WC ratio increases after key events, providing insight into the balance between disease spread and public health response. Time lag and transfer function analysis showed that wastewater data preceded clinically reported cases in the first wave of the pandemic but did not serve as a leading indicator in the second wave, likely due to increased testing capacity, which allows for more timely case detection and reporting. These three metrics could help further integrate wastewater surveillance into the public health response to the COVID-19 pandemic and future pandemics.
Keywords: Wastewater surveillance, Wastewater-based epidemiology, SARS-CoV-2, Temporal dynamics
Graphical abstract
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), continues to affect all aspects of global life since its emergence in late 2019. Efforts to contain its spread have relied on public health measures like social distancing, stay-at-home orders, mandatory wearing of face coverings in public, and community-based SARS-CoV-2 testing (Nussbaumer-Streit et al., 2020). In the face of this public health emergency, multiple methods have been designed to assess the impact of SARS-CoV-2 on critical health infrastructure, implement rapid and robust individual testing, and predict potential surges to inform hospital preparedness and health policy makers.
Clinical surveillance data is the gold standard for evaluating the state of the pandemic. However, clinical data can be limited by clinical testing capacity and availability, human behavior, and the presence of asymptomatic infections. In contrast, wastewater surveillance, also known as wastewater-based epidemiology (WBE), is a potentially powerful strategy for near real-time monitoring of viral burden in the population, as it captures viral shedding from infected individuals, irrespective of clinical presentation. Such surveillance has been shown to detect SARS-CoV-2 in wastewater before widespread clinical reporting (Bar-Or et al., 2020; Kocamemi et al., 2020; Medema et al., 2020; Randazzo et al., 2020; Wurtzer et al., 2020). It has been previously demonstrated that viral copy numbers in wastewater were higher than expected from confirmed clinical cases (Peccia et al., 2020a; Randazzo et al., 2020; Wu et al., 2020b; F. 2020a) and that they preceded clinical reporting of new cases and hospital admissions (Peccia et al., 2020a; Randazzo et al., 2020; F. Wu et al., 2020a), suggesting that wastewater surveillance could be used as an early warning system. Wastewater surveillance has also been used in combination with clinical data to infer average population-level viral shedding dynamics (Schmitz et al., 2021; F. Wu et al., 2020a). Wastewater surveillance has been mostly implemented at municipal wastewater treatment plants, but as colleges and universities reopened, it has also been applied in dormitories as an early warning system to prevent large-scale outbreaks of COVID-19 (Betancourt et al., 2020; Gibas et al., 2021; Harris-Lovett et al., 2021; P. Liu et al., 2020).
To interpret wastewater surveillance results in terms of population health, it is critical to understand the relationship between wastewater SARS-CoV-2 concentrations and infections in the population. Early work focused on evaluating the correlation between wastewater SARS-CoV-2 concentrations and clinically reported cases and characterizing the lead time of wastewater surveillance data. Various groups used correlation methods to estimate this lead time, with reports of wastewater leading clinical cases by 0–16 days (Medema et al., 2020; Nemudryi et al., 2020; Peccia et al., 2020b; Randazzo et al., 2020; F. Wu et al., 2020a; Wurtzer et al., 2020). While the correlation between the wastewater and clinical curves is relatively easy to compute, it does not take into account autocorrelation in the datasets, so further work has employed more sophisticated statistical models to estimate the time lag (Galani et al., 2021; Peccia et al., 2020a). Some groups have used wastewater data to infer the number of infected cases in the catchment (Ahmed et al., 2020; Chavarria-Miró et al., 2021; Galani et al., 2021; Gerrity et al., 2021; Krivoňáková et al., 2021), while others have used wastewater data to model average population shedding rates (Cavany et al., 2021; Wu et al., 2022).
These modeling studies shed important light on the relationship between wastewater viral concentrations and clinical cases. However, municipalities often rely on qualitative trends when using wastewater SARS-CoV-2 concentrations to inform decisions (Cambridge Public Schools, 2021; Centers for Disease Control, 2021; Ohio Coronavirus Wastewater Monitoring Network, 2021), whereas wastewater data can be a richer source of public health information when integrated with other datasets. There remains a need for quantitative metrics to help municipalities interpret wastewater data in the context of clinical trends and public health interventions and evaluate their pandemic response. Such metrics would help municipalities more effectively incorporate wastewater data into the public health decision making toolkit, beyond observing the qualitative trends in wastewater SARS-CoV-2 copy numbers. Here, we developed three new metrics and applied them to a 14-month-long time series of SARS-CoV-2 wastewater SARS-CoV-2 copy numbers in Massachusetts. These three metrics – (1) the ratio between wastewater SARS-CoV-2 copy numbers and clinical cases (WC ratio), (2) the time lag between wastewater and clinical reporting, and (3) a transfer function between the wastewater and clinical case curves – aptly describe dynamic changes in the relationship between wastewater data and clinical data as the pandemic evolves and management strategies adapt. Our results show that wastewater data preceded clinically reported cases in the first wave of the pandemic but did not serve as a leading indicator in the second wave, likely due to increased testing capacity, which allows for more timely case detection and reporting. Thus, these metrics can be useful tools for officials to evaluate the public health response over time.
2. Materials and methods
2.1. Sample collection and viral inactivation
700 24-hour composite samples of raw sewage were collected from the Deer Island Wastewater Treatment Plant (comprised of northern and southern influents) in Massachusetts from March 4, 2020 to May 13, 2021. The Deer Island Wastewater Treatment Plant serves approximately 2.3 million individuals in the greater Boston area, comprising primarily Norfolk, Suffolk, and Middlesex counties. The average flow rate is 178 million gallons per day (MGD) for the northern influent and 93 MGD for the southern influent. Historical samples from March 4 to March 17, 2020 were stored at 4 °C at the WWTP and delivered to the lab. Starting on March 18, the sewage samples were transported to the lab on the same day of sample collection with ice. If the samples were collected on weekends or holidays, they were stored at 4 °C and transported to the lab the next business day with ice. Upon receipt, samples were brought to 60 °C and pasteurized for 1 h to inactivate the virus due to biosafety regulations at MIT. Several studies have found that heat treatment at 60 °C did not negatively impact quantification of SARS-CoV-2 (Y. Liu et al., 2020; Pastorino et al., 2020) or PMMoV (Shirasaki et al., 2020).
2.2. Lab analysis
Samples were analyzed using previously described methods (Duvallet et al., 2021; F. Wu et al., 2020a). Briefly, samples were filtered to remove large particulate matter using a 0.2uM vacuum-driven filter (EMD-Millipore SCGP00525 or Corning 430,320, depending on sample turbidity). We used Amicon Ultra-15 centrifugal ultrafiltration units (Millipore UFC903096) to concentrate 15 ml of wastewater approximately 100x. Viral particles in this concentrate were immediately lysed by adding AVL Buffer containing carrier RNA (Qiagen 19,073) to the Amicon unit before transfer and >10 min incubation in a 96-well 2 mL block. 100% ethanol was added to the lysate, and samples were applied to RNeasy Mini columns or RNeasy 96 cassettes (Qiagen 74,106 or 74,181). RNA samples were subjected to one-step RT-qPCR (ThermoFisher 4,444,436) analysis in triplicate for N1, N2, and PMMoV amplicons on CFX96 and/or CFX-Connect instruments based on the following protocol: 50 °C 10 min for reverse transcription, 95 °C 20 s for RT inactivation and initial denaturation, and 48 cycles of denature (95 °C 1 s) and anneal/extend (55 °C 30 s). Cts were called from raw fluorescence data using the Cy0 algorithm from the qpcR package (v1.4–1) in R (Guescini et al., 2008), and manually inspected for agreement with the raw traces in the native BioRad Maestro software. Previous work with murine hepatitis virus (MHV) spike-ins have shown the recovery efficiency of our process to be 31.42 ± 2.59% (Wu et al., 2021).
An extraction blank was processed in parallel to samples to detect contamination of extraction reagents. If the extraction blank had detection of SARS-CoV-2 by the N1 or N2 primers with Ct < 40, or PMMV with Ct < 35, the data were discarded, and the samples in the batch were re-extracted starting from filtered wastewater.
Each RT-qPCR plate has a specified number of controls. Two no-template controls were assayed to detect contamination of the RT-qPCR reagents. If either of the no-template controls had detection of SARS-CoV-2 by the N1 or N2 primers with Ct < 40, or PMMV with Ct < 35, the data were discarded, and RNA samples were re-assayed with a fresh mix of RT-qPCR reagent. Four replicates of synthetic SARS-CoV-2 RNA were assayed as positive controls. If the average Ct values from these controls was not within range (+/- 1 Ct from expected, lot-specific Ct-value), the data were discarded, and RNA samples were re-assayed with a fresh mix of RT-qPCR reagent. Matrix inhibition was assessed by manually reviewing the raw qPCR curves.
In addition to these laboratory controls, a thorough data review process was implemented for quality control. Firstly, we used PMMoV as a proxy measure for per-extraction-batch recovery and flagged any plate with unusually low PMMoV values for further review and potential repeat processing. Furthermore, each sample would be manually reviewed if they met any of the following criteria: -PMMoV above 99th or below 1st percentile of all previously processed sample values; -SARS-CoV-2 concentrations changing more than 5-fold since prior sample at the same location; -suspected inhibition based on manual inspection of raw fluorescence curves; -discordant virus concentrations obtained from N1 and N2 primers; -pigmentation present in extracted RNA. During manual review, individual RT-qPCR replicates and timelines of SARS-CoV-2 and PMMoV concentrations were inspected. A small fraction of samples manually inspected in this manner were flagged for rerun. If sufficient filtered wastewater remained in the initially-processed tube, a second 15 mL aliquot was processed. An aliquot from one of the back-up tubes was always processed. If the rerun results were significantly different from the initial results, the initial results were discarded. If the rerun results recapitulated the initial results, the average of all results was reported and used in this work.
2.3. Data processing
A standard curve was generated using serial dilutions of Twist Bioscience synthetic SARS-CoV-2 RNA control 2 (MN908947.3) and used to convert Ct values into copies per well. We used pepper mild mottle virus (PMMoV) as a fecal indicator and quantified it relative to SARS-CoV-2 in each sample using the standard curve for N1, as synthetic RNA for PMMoV was not available and our PMMoV normalization method is dependent on ratios of PMMoV values rather than absolute values. Ct values above 40 were considered as non-detect. The copies per well were multiplied by a dilution factor accounting for the volume changes described above (RNA extraction, concentration, etc.) and then divided by the original sewage volume (15 ml) to convert to a sewage concentration (copies per liter). Concentrations of N1 and N2 replicates were averaged first within each primer set and then across primers to get the final SARS-CoV-2 concentration; replicates of the PMMoV amplicon were averaged. Samples were required to have at least two quantified replicates between N1 or N2, and at least one detected PMMoV replicate to be considered a detection. For median normalization using PMMoV, SARS-CoV-2 concentrations were divided by the PMMoV levels and multiplied by a reference PMMoV value derived as the median of our dataset. For example, samples that are more diluted will have lower PMMoV concentration and will be normalized up, while samples that are less diluted will have higher PMMoV concentration and will be normalized down. Normalization using PMMoV has previously been shown to account for variability in wastewater flow (Wu et al., 2020b), and allows us to correct for dilution and laboratory variation simultaneously. Data on the concentration of SARS-CoV-2 viral RNA in Massachusetts wastewater are publicly available at https://www.mwra.com/biobot/biobotdata.htm.
2.4. Clinical case analysis
Clinical case data from March 1, 2020 to May 16, 2021 from Norfolk, Suffolk, and Middlesex Counties was downloaded from Mass.gov (C.D. Massachusetts Department of Public Health, 2020a; C.D. 2020b, 2021a). Clinical cases are counted based on positive PCR tests. Clinical cases from each county were summed to represent the cases in the catchment of the wastewater treatment plant. The difference between adjacent days was taken to calculate the new cases per day. Positive test rates by age and state level death data were downloaded from Mass.gov (Massachusetts Department of Public Health, 2021a; C.D. 2021b).
2.5. Approximate Bayesian computation for estimating delay distribution
Approximate Bayesian computation (ABC) was used to find the delay distribution between when an infected individual's viral shedding appears in wastewater and when they are counted as a clinical case before and after August 15, 2020. ABC was chosen because it is extremely flexible and relatively easy to implement, allowing the inference to be carried out for complex models without need for evaluating the likelihood function, which could be computationally intractable (Csilléry et al., 2010; Fearnhead and Prangle, 2011; Toni et al., 2009). Instead, ABC uses the computational efficiency of modern simulation techniques by comparing the observed and simulated data (Csilléry et al., 2010; Fearnhead and Prangle, 2011; Toni et al., 2009). First, wastewater viral copy numbers were normalized by their sum for each portion of the data. The delay δ was assumed to be normally distributed with mean μ and standard deviation σ. Prior distributions for μ and σ were chosen as follows: μ ∼ Norm(0,10) and σ ∼ Exp(0.1). 10,000 values of μ and σ were sampled from the priors. For each iteration, a delay for each clinically reported case was sampled. Simulated wastewater data was generated by adding the delay for each case to the date they were actually reported. The number of delayed cases per day was normalized by the sum. The average per day sum of squared errors (SSE) between the simulated wastewater data and the actual wastewater data was calculated. The values of μ and σ were accepted if the average SSE was less than some cutoff value ε. ε was tuned so approximately 10–20% of the iterations were accepted.
2.6. Convolution to estimate the transfer function
A similar approach as we previously reported was used to find the transfer function that describes the relationship between the shape of the wastewater data and clinical data (F. Wu et al., 2020a). This concept was borrowed from the field of signal processing, where a transfer function represents the mathematical relationship between the numerical input to a dynamic system and the resulting output Pollock (2011). Because the shapes rather than the magnitudes of the curves are of greatest relevance, the wastewater data was divided by the median ratio between wastewater and clinical data. Clinically reported cases C(t) were modeled as the convolution between the scaled wastewater data W(t) and the unknown transfer function T(t) before and after August 15, 2020: log10(C(t)) = log10([T * W](t)). It was hypothesized that the transfer function could be fit by a beta distribution with parameters α, β, and scaling factor c because beta distributions harbor a rich variety of shapes with only two parameters. The score function was defined as the sum of squared errors (SSE) between log10(clinically reported cases) and log10([T * W](t)). The l-BFGS method (Liu and Nocedal, 1989) in the scipy.optimize.minimize function was used to find parameters α, β, c of the beta distribution that minimized the SSE. l-BFGS was chosen because of its computational speed and memory requirements (Liu and Nocedal, 1989). For initial parameter guesses, a combination of α = [2, 20, 50, 100, 200], β = [2, 20, 50, 100, 200], and c = [0.01, 0.1] was used, which gives a wide variety of starting shapes for the transfer function.
2.7. Markov chain Monte Carlo (MCMC) simulation to quantify uncertainty in transfer function
Under the assumption of normally distributed errors, minimizing the SSE is equivalent to maximizing the likelihood Berkson (1956). MCMC simulation was used to investigate the uncertainty landscape around the maximum likelihood estimation of the parameters for the inferred transfer function. MCMC methods have previously been used to quantify uncertainty in various fields (Bardsley and Fox, 2012; Hassan et al., 2009; Maybank et al., 2020). The Metropolis-Hastings algorithm, a ubiquitous and versatile MCMC algorithm Hastings (1970), was employed. Briefly, the algorithm started at the maximum likelihood estimate for each parameter α, β, and c. The transition function was defined as a normal distribution centered around the previous parameters, with standard deviation (1, 1, 0.001) for α, β, and c, respectively. At each iteration, a new set of parameters was selected using the transition function and the log likelihood was computed. The new parameters were accepted based on the Metropolis-Hastings algorithm acceptance rules. New parameters were always accepted if the log likelihood was higher. If the log likelihood was lower for the new parameters, they were accepted with probability exp(-delta(SSE)). 100 random accepted parameter sets were selected and plotted in Fig. 4B and 4D to illustrate the uncertainty around the maximum likelihood estimate of the transfer function. MCMC simulation was done with python 3.6.5, numpy 1.14.3, pandas 0.23.0, and scipy 1.1.0.
Fig. 4.
Transfer function between wastewater and clinical cases becomes more peaked in the second wave of the pandemic. We modeled clinically reported new cases as the convolution between wastewater viral copy numbers and an unknown transfer function. (A, C) Our model finds parameters of a beta function that minimizes the sum of squared error (SSE) between the model prediction (orange) and the observed (blue) clinical new cases. (B, D) The maximum likelihood estimate of the transfer function (black) with 100 accepted Markov Chain Monte Carlo (MCMC) parameter sets in blue. Before 8/15, the transfer function has a broad peak and long tail. After 8/15, the transfer function becomes more sharply peaked.
3. Results
3.1. Wastewater viral copy numbers mirror disease incidence and can depict the impact of social activities more sensitively than clinical cases
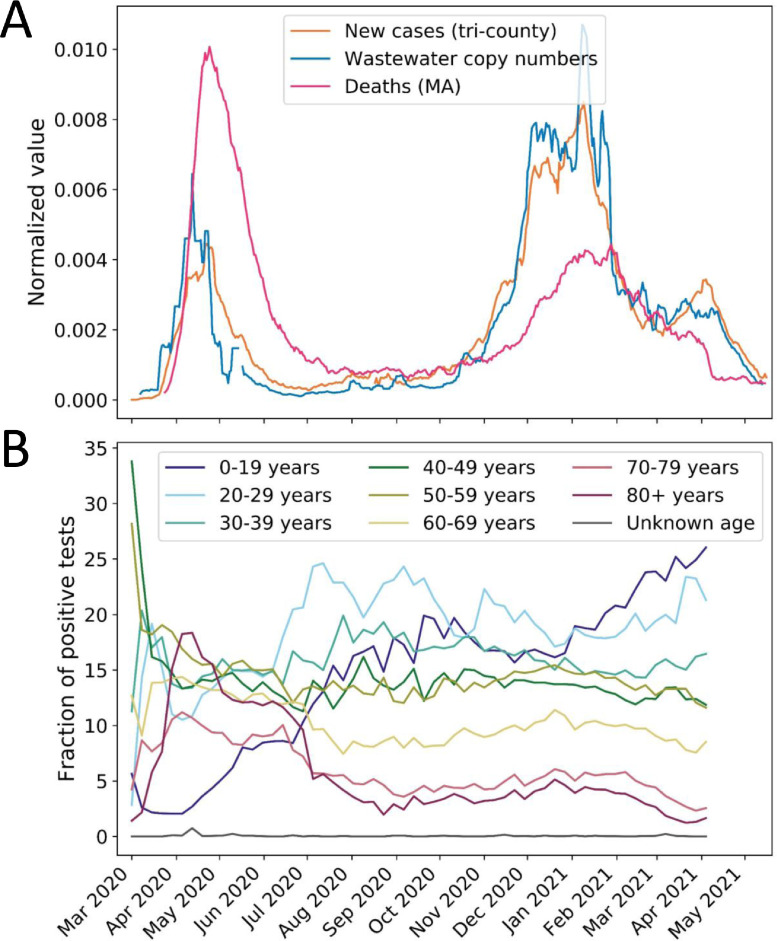
Analysis of our 14-month-long time series for wastewater copy numbers of SARS-CoV-2 viral gene fragments spanning March 4, 2020 to May 13, 2021 showed two distinct waves of SARS-CoV-2 in the Boston Area. Similar trends appeared in wastewater viral copy numbers and clinical cases: exponential rise from March to mid-April, a decline through July, a slow increase over the summer, followed by a sharper increase in the fall and second peak in the winter (Fig. 1 ), indicating that wastewater viral copy numbers generally mirrored trends in disease incidence. Previous reports have shown that SARS-CoV-2 concentrations in wastewater may be affected by flow rate and other physicochemical properties, such as total suspended solids, to varying degrees across different wastewater treatment plants (Amoah et al., 2022; Paul et al., 2021). In both the northern and southern influents of Deer Island Treatment Plant, SARS-CoV-2 concentrations did not have high correlation with flow or physicochemical properties (Table S2, Table S3). The robustness of the SARS-CoV-2 measurements to variation in flow and total solids is likely due to the normalization with PMMoV which corrects for dilution of human fecal material.
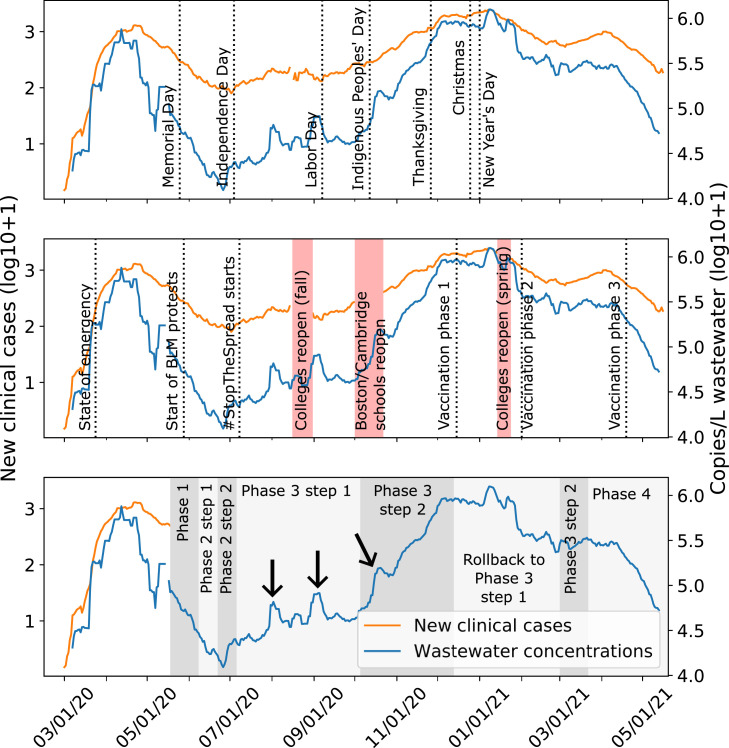
Fig. 1.
SARS-CoV-2 RNA copy numbers in Massachusetts wastewater and new clinical cases. Seven-day averages of wastewater viral copy numbers (blue) and new clinical cases reported for the three counties in the catchment (orange). We marked major holidays (top), major social events (middle), and state reopening phases (bottom) in the three panels, respectively (Baker, 2021a, p. 2, C.D. 2021b, C.D. 2020a, C.D. 2020b, 2020c, 2020d, 2020e, 2020f, n.d.). Black arrows indicate peaks in wastewater SARS-CoV-2 RNA copy numbers that were not reflected in clinical case counts.
Wastewater and clinical data were compared with dates of known policy changes and social gatherings in the Boston Area and key trends were noted. For example, wastewater viral copy numbers and clinical cases continued to decline overall after Memorial Day (May 25, 2020), despite the potential for large gatherings to celebrate the holiday (Fig. 1). Similarly, the social justice protests during late May and June did not immediately spark an increase in clinical cases or wastewater viral copy numbers (Fig. 1). The start of Phase 2 Step 2 (Table S1) marked the start of a steady increase in wastewater viral copy numbers and clinical cases during the summer (Fig. 1). There was also a steeper increase in wastewater viral copy numbers and clinical cases after the Indigenous Peoples’ Day holiday (October 12, 2020). This increase continued through reopening Phase 3 Step 2 and peaked in late November to January around the time of Thanksgiving, Christmas, and New Year holidays, perhaps due to increased indoor gatherings (Fig. 1).
However, trends in wastewater data differed from clinical data after some key events, suggesting a decoupling of wastewater and clinical trends which we hypothesize provides insight into dynamics of COVID-19 in the community. There was a short peak in wastewater viral copy numbers at the start of August, which was only slightly reflected in the clinical data (Fig. 1). Similarly, after colleges and universities welcomed students back in late August/early September, there was another peak in wastewater viral copy numbers, but not in clinical cases (Fig. 1). After the start of Phase 3 Step 2 reopening, wastewater viral copy numbers increased steeply, while clinical cases had a shallower slope (Fig. 1). These differences could be due to the inherent difference between wastewater and clinical data. For example, wastewater measurements of SARS-CoV-2 may vary based on viral shedding differences between asymptomatic, symptomatic, mild, or severe cases, whereas clinical cases are either reported or not. However, we hypothesized that the observation of distinct trends in wastewater copy numbers over clinical data may suggest that wastewater viral copy numbers could indicate the impact of social activities more sensitively than clinical cases. This observation prompted us to derive a metric for detecting discordance between wastewater and clinical trends.
3.2. Ratio between wastewater viral titer and clinical cases (WC ratio) may indicate changes in testing capacity or demographics of clinical cases
Wastewater surveillance captures all individuals who are shedding the virus regardless of disease manifestation, their access to testing, or representation in clinical case data. As such, wastewater trends are commonly used for benchmarking against clinical cases. However, there is a lack of a quantitative measure for comparing wastewater and clinical trends. Here, we propose using the ratio of wastewater viral copy numbers to clinical cases (WC ratio) as a metric for detecting differences between wastewater and clinical trends.
Changes in the WC ratio can serve as an indicator of potential under- or over-estimation of disease incidence. Under-counting of clinical cases could occur when clinical tests are limited, when people are not seeking testing or cannot access convenient testing locations, or when the proportion of asymptomatic infections is high. Over-estimation of disease incidence could occur when rapid expansion of testing infrastructure allows many people who were infected in the prior weeks to get tested, so their results show up as new cases even though they were actually infected weeks prior. This situation could occur especially because throat and nasal swab PCR tests for SARS-CoV-2 can remain positive for up to 20 days after symptom onset (Wölfel et al., 2020).
Because viral copy numbers will vary based on the number of people in the catchment, the magnitude and range of the WC ratio must be established for each catchment considered. When this ratio is high, it implies that the existing testing capacity has not kept pace with exponentially rising new cases, which nevertheless are detected in wastewater surveillance. A high WC ratio can occur in situations where wastewater viral copy numbers are increasing but clinical cases are not rising equivalently, or conversely when wastewater viral copy numbers are stable but clinical testing is decreasing. Conversely, a low WC ratio indicates that clinical tests are capturing the majority of infections reflected in wastewater viral copy numbers. When this ratio is stable and low, it implies that the existing testing capacity is sufficient to assess the extent of new infections. Importantly, changes in the WC ratio relative to a stable baseline may provide early indications of changing epidemic dynamics. Positive changes may highlight new bursts of infections before they are captured in clinical data or identify periods where clinical tests are not capturing the full extent of new infections, while negative changes may highlight periods where clinical testing is over-estimating disease incidence when counting previously infected cases as new cases. Notably, this ratio has changed by approximately two orders of magnitude over the course of the pandemic, demonstrating its utility as a metric to assess the public health response.
At the beginning of the pandemic (March 2020), the ratio between wastewater viral copy numbers (viral genome copies (GC) per L) and reported clinical cases was very high (>10^3 for the Boston Area), indicating that cases were likely undercounted due to extremely limited testing (Fig. 2 A). In fact, during March 2020, the seven-day average of new molecular tests administered per day in the state did not exceed 5000 tests per day (Fig. 2B). As testing ramped up throughout April and May, the WC ratio dipped by approximately two orders of magnitude (Fig. 2A). In this phase, delayed clinical test results may have been “catching up” to the more instantaneous wastewater viral copy numbers, which infected individuals had contributed to many days prior to getting their test results. Thus, these individuals were no longer contributing to wastewater viral copy numbers (numerator of the WC ratio) but were now being counted as reported cases (denominator of the WC ratio), leading to a much lower ratio.
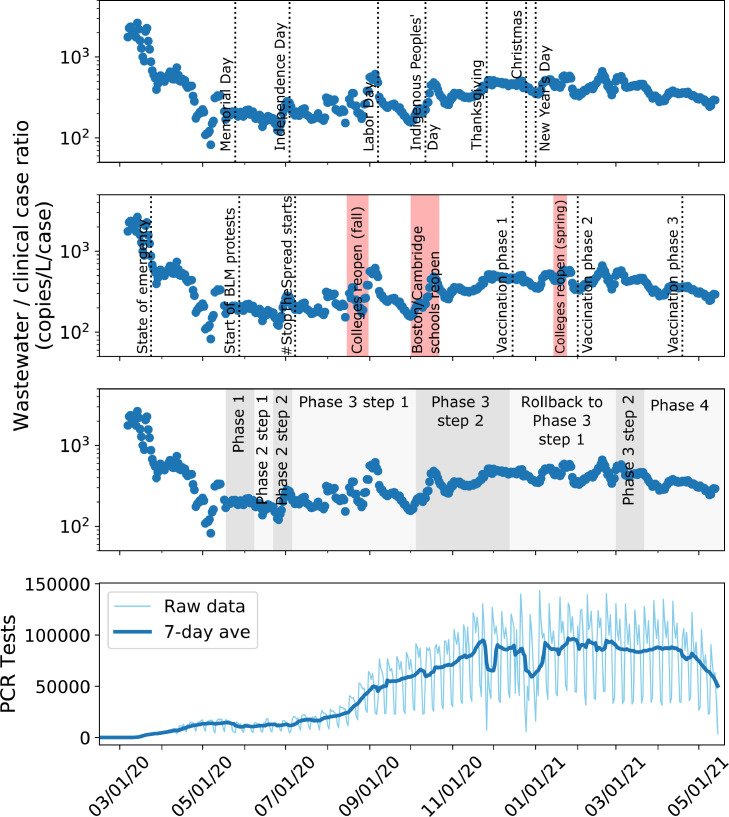
Fig. 2.
Ratio between wastewater viral copy numbers and clinically reported new cases changes with testing availability. (A) Ratio between seven-day averages of wastewater viral concentration (genome copies/L) and clinically reported new cases changes over the course of the pandemic, with some spikes after key holidays, important events, and reopening phases. (B) PCR tests conducted each day in Massachusetts throughout the pandemic (Massachusetts Department of Public Health, 2021a).
As the public health response in Massachusetts began to ramp up over the summer and individual clinical testing became more available, the ratio between wastewater and clinical cases remained fairly consistent between July 2020 and November 2020 (Fig. 2A). During this time, Massachusetts’ clinical testing capacity became fully established, and percent positivity remained stable below 2% (C.D. Massachusetts Department of Public Health, 2021b). Such periods could be used to determine baseline WC ratios that indicate sufficient public health capacity for testing. Interestingly, even during the pronounced peak of the second wave (November 2020 - March 2021), the WC ratio did not spike, indicating that testing capacity was sufficient to capture the scope of exponentially rising new infections.
However, there were a few increases in the ratio even with fully established testing capacity, notably in early September and early October (Fig. 2A). These increases in the WC ratio could be related to community events, such as reopening of businesses and universities, or to changes in testing availability. These increases could also be due to a combination of factors including shifts in population demographics. For example, college students returning for classes in late August/early September may have been more likely to be asymptomatic due to their younger age, and thus less likely to be reflected in clinical case counts (Leidman et al., 2021; Leidner et al., 2021). Similarly, after Phase 3 Step 2 reopening allowed bars and entertainment venues to open (Baker, 2021a), young adults may have had higher degrees of social contacts. In these instances, wastewater surveillance may have detected a silent and short-lived peak in community transmission that was missed by clinical surveillance. Importantly, these short increases became more apparent when analyzing the ratio between the two datasets.
3.3. Wastewater lead time is affected by increased clinical testing
While the WC ratio could identify whether trends in clinical cases are concordant with trends in wastewater copy numbers, it does not provide information on the timeliness of clinical reporting, which would be an important measure of the public health response. To address this gap, we introduce two methods to characterize this time lag between wastewater and clinical trends: (1) a model of the distribution of time lags for each new case, and (2) a transfer function to describe the relationship between the wastewater and clinical curves.
In our first metric to assess the time-varying relationship between wastewater data and clinical cases (Fig. 3 A), approximate Bayesian computation (ABC) was used to model the distribution of the time lag between when an infected person's viral shedding is detectable in wastewater and when they receive a clinical test (Methods). In this model, it is assumed that the viral shedding detected in wastewater occurs on a single day. The assumption that wastewater reflects viral shedding early in infection is supported by animal models, meta-analysis of clinical data, and wastewater surveillance in dormitories (Bao et al., 2020; Hoffmann and Alsing, 2021; Schmitz et al., 2021). The date of clinical cases in this analysis corresponds to the date of specimen collection. Negative time lags indicate that wastewater signal precedes clinical testing and vice versa. To compare the relationship between wastewater data and clinical data during the first and second waves of the pandemic, time series was split on August 15, the approximate midpoint between the end of the first wave and the start of the second wave.
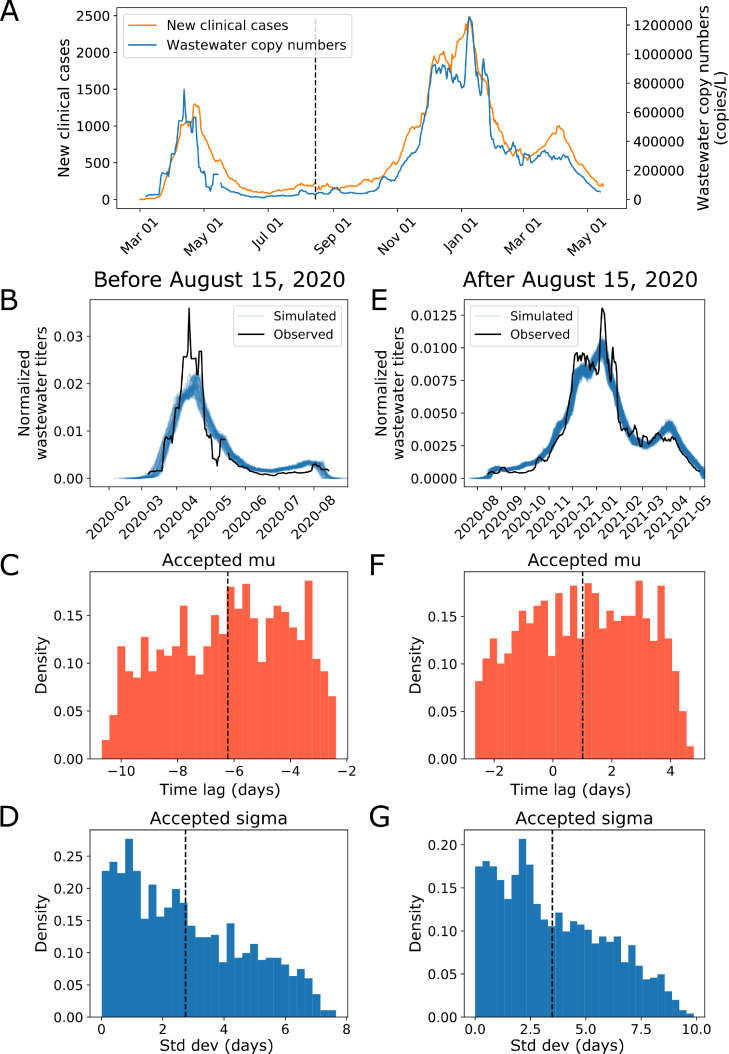
Fig. 3.
Modeling reveals that time delay between clinical case reporting and wastewater data changes over the course of the pandemic. We use approximate Bayesian computation to determine the distribution of the time lag between when a case shows up in the wastewater surveillance data and when they are clinically reported. (A) Seven-day averages of wastewater data and new clinical case data from Mass.gov shown on a linear scale (Massachusetts Department of Public Health, 2021a; C.D. 2020a; C.D. 2020b). (B, C, D) Modeling results on data before August 15, 2020. (B) Simulated vs observed wastewater viral copy numbers for data before 8/15. (C) Accepted values for the mean time lag (−6.2 days, 95% CI: −10.1, −2.7) and (D) standard deviation (2.7 days, 95% CI: 0.1, 6.7) of the time lag for data before 8/15. We used 10,000 iterations with a distance threshold of 1.3e-5 and 11.1% of parameter sets were accepted. (E, F, G) Modeling results on data from August 15, 2020 and after. (E) Simulated vs observed wastewater viral copy numbers for data after 8/15. (F) Accepted values for the mean time lag (1.0 days, 95% CI: −2.4, 4.2) and (G) standard deviation (3.5 days, 95% CI: 0.2, 8.4) of the time lag for data after 8/15. We used 10,000 iterations with a distance threshold of 9.5e-7 and 15.3% of parameter sets were accepted. Negative time lags indicate that wastewater signal precedes clinical case reporting and vice versa.
Before August 15, 2020, the wastewater signal preceded clinical cases by approximately 6.2 days (95% CI: −10.1, −2.7) with a standard deviation of 2.7 days (95% CI: 0.1, 6.7) (Fig. 3B-D). This finding is consistent with our previous report that wastewater data preceded clinical data by 4–10 days (F. Wu et al., 2020a). After August 15, 2020, the wastewater signal was more in phase with clinical cases (mean time lag 1.0 days, 95% CI: −2.4, 4.2; standard deviation 3.5 days, 95% CI: 0.2, 8.4) (Fig. 3E-G). We repeated the modeling while varying the date of the split and found similar results (Figure S1–4).
To confirm these modeling results, we next investigated the transfer function T(t) that transforms wastewater viral copy numbers to clinical cases. This concept was borrowed from the field of signal processing, where a transfer function represents the mathematical relationship between the numerical input to a dynamic system and the resulting output Pollock (2011). While the previous ABC analysis modeled the delay for each individual case to find the distribution of delays, the transfer function analysis focuses on relating the shapes of the wastewater signal and clinical case signal. The shape of T(t) can provide insight on the time delay between the two signals and can reflect factors such as how long it takes someone to request testing and the probability that someone gets a positive test result over the course of their infection (Fig. 4 A and C).
The shape of the transfer function changed between the first and second waves, reflecting changing relationships between infections and clinical testing. Using data before August 15, the inferred transfer function had a broad peak and long tail, with the peak at approximately 3 days and an average of 10 days, which is within the confidence interval of the ABC model (Fig. 4B). The broad shape implies that the process of infected individuals getting counted as cases has a broad distribution, with some individuals getting reported very quickly but others taking up to weeks. In this situation, wastewater viral copy numbers could be an early indicator of disease dynamics before clinical test results come back positive. As the pandemic progressed, the inferred transfer function became more sharply peaked around a 1-day time lag, which is consistent with the results of the ABC model. This sharp distribution indicates that wastewater and reported cases track each other closely (Fig. 4D). In this case, wastewater viral copy numbers have less utility as an early warning system because increased clinical testing capacity effectively captures new infections in a timely manner.
Taken together, these results suggest that the relationship and time lag between wastewater viral copy numbers and positive clinical tests changed over the course of the pandemic. Wastewater was more effective as a leading indicator in the first wave of the pandemic, and this early warning effect diminished drastically in the second wave, perhaps as clinical testing availability increased. Thus, parameters such as the time lag between wastewater signal and clinical signal and the transfer function describing their relationship could also be used to evaluate clinical test availability or capacity.
3.4. Relationship between wastewater data, new cases, and deaths is different in the first and second waves
Given that wastewater SARS-CoV-2 copy numbers correlated with new clinical cases, we next investigated whether we could use wastewater copy numbers to predict COVID-19 deaths. The relationships between wastewater copy numbers, new cases, and deaths were different in the first and second waves, suggesting that wastewater may have a direct, mechanistic relationship with new cases, but an indirect relationship with deaths, indicating that wastewater data must be used in conjunction with other public health datasets for making policy decisions.
In the first surge (Mar-May 2020), both new cases and deaths peaked shortly after wastewater peaked (Fig. 5 A). In this situation, wastewater copy numbers could be a predictive indicator of COVID-19 deaths. However, in the second surge (Nov 2020-Mar 2021), wastewater viral copy numbers and new cases increased to levels higher than in the first surge, while deaths remained lower than the first wave (Fig. 5A). In this situation, wastewater copy numbers were an indicator of new cases but not deaths. This difference could be due to a combination of several factors. First, the first wave strongly affected people in the 80+ year bracket, while the second wave has seen an increase in positivity in the 0–29 year bracket (Wikle et al., 2020). Older adults with comorbidities have higher mortality rates from COVID-19 (Centers for Disease Control and Prevention, n.d.). Second, medical professionals have gained experience in treating and managing the disease since the first wave, thus leading to reduced deaths in the second wave (Q. Liu et al., 2020). Third, changes in human practices, such as improved hygiene and social distancing, could reduce the viral inoculum, resulting in less severe disease (Spinelli et al., 2021). Fourth, the increase in testing means a higher proportion of cases are being diagnosed, possibly including those who are asymptomatic or only mildly ill, so the proportion of deaths to new cases would be lower. This explanation is consistent with the decrease in WC ratio (Fig. 2) seen in the second wave. We explored the possibility that variants of concern with increased transmissibility like B.1.1.7 (Frampton et al., 2021; Graham et al., 2021), could have influenced the ratio of deaths to new cases. However, the proportion of B.1.1.7 in the last week of January 2021, during the peak of the second wave, was estimated to make up only an average of ∼2.1% COVID-19 cases in the U.S (Washington et al., 2021). Furthermore, B.1.1.7 did not make up a large proportion of wastewater viral copy numbers in the Boston Area until Feb-March 2021, towards the end of the second wave (Lee et al., 2021).
Fig. 5.
Wastewater is an early warning of new cases but not deaths, perhaps due to a changing demographic of the pandemic. (A) Seven-day averages of wastewater viral copy numbers, new clinical cases in the three counties served by the WWTP (Massachusetts Department of Public Health, 2021a; C.D. 2020a; C.D. 2020b), and new reported deaths in the state of Massachusetts (C.D. Massachusetts Department of Public Health, 2021b). All three datasets are normalized by their sums for comparison. (B) Fraction of positive tests in Massachusetts by age bracket (Massachusetts Department of Public Health, 2021c). Positivity peaked among 80+ year-olds in the first wave of the pandemic, whereas the second wave saw an increase in positivity in 0–29 year-olds.
Notably, wastewater viral copy numbers reflect the number of new cases regardless of the demographics or symptoms of cases. With the changing demographics of the disease, changes in clinical practice, and increased testing capacity, the power of wastewater to predict new cases and COVID-related deaths also changes. Therefore, wastewater should not be used alone to predict public health outcomes, but rather should be used in combination with other data sources to understand the pandemic and inform decision making.
4. Discussion
Many groups have demonstrated that wastewater SARS-CoV-2 copy numbers reflect trends in new COVID-19 cases, and wastewater viral copy numbers have been observed to precede trends from clinical surveillance (Bar-Or et al., 2020; Kocamemi et al., 2020; Medema et al., 2020; Randazzo et al., 2020; Wurtzer et al., 2020). Here, we developed three new metrics to integrate wastewater and clinical data to quantitatively understand their relationship and the public health response. We introduced the ratio of wastewater viral copy numbers to clinical cases (WC ratio) as a simple metric which may reflect testing capacity. We also applied two independent types of modeling to quantify the lead time between wastewater data and clinical data. These models show that wastewater's lead time changes over the course of the pandemic as public health responses adapt. Finally, we showed that a third public health data stream, COVID-19 deaths, also has changing relationships with cases and wastewater over the pandemic, suggesting that wastewater data cannot be used alone and should be integrated with public health data to make policy decisions. Together, this work demonstrates the utility of combining wastewater surveillance with multiple public health data streams to provide a more nuanced view of the changing public health response to the COVID-19 pandemic.
The ratio between wastewater viral copy numbers and clinical cases (WC ratio) is a useful indicator of the public health response and testing coverage and could be used to gauge the intensity of public health interventions in the face of changing disease incidence. In addition, short-lived trends in increased transmission may be more easily detected when analyzing the WC ratio compared to analyzing wastewater and clinical data independently. For example, the WC ratio showed a small spike in early September and early October. These short-lived spikes could be due to community events, such as university reopening and economic reopening phases. These spikes could also be due to changing demographics of disease. As younger people are more likely to have mild or asymptomatic infections, they are less likely to be captured by clinical testing infrastructure, even when tests are readily available. Therefore, the WC ratio should be considered when interpreting wastewater surveillance data because it may help detect asymptomatic disease transmission among the population.
We also introduced two models that showed that the delay between wastewater data and clinical reporting shrank from an average of 6.2 days to no significant difference between the wastewater and clinical trend after August 15, 2020. In addition, the inferred transfer function between wastewater viral copy numbers and reported cases shifted from a broad to a sharp peak, suggesting quicker access to clinical testing. These results suggest that wastewater surveillance can be useful as an early warning indicator of disease incidence when clinical testing is limited and also as a method to understand ramp up and scale down of community based testing capacity in different phases of the pandemic (Olesen et al., 2021). Importantly, modeling the time lag and transfer function between the wastewater viral copy numbers and clinical cases provides a more quantitative method to understand their relationship and assess the pandemic response.
There could be many factors contributing to the decreasing delay between trends in wastewater and trends in clinical cases. Changing criteria to qualify for clinical testing, individual behavior in requesting tests, availability of convenient testing locations, and lab turnaround time can all affect the time lag between when a patient is infected and when their positive result is reported. In the beginning of the first wave, clinical testing was largely limited to those who met a restrictive combination of symptoms and exposure, gradually expanding to those who had exposure history (Becker, 2020; Brown et al., 2020). However, the initiation of the Massachusetts #StopTheSpread program on July 8, 2020 and its expansion on August 7, 2020 made walkup, on-demand testing available to the public, aiding in the identification of asymptomatic and pre-symptomatic cases (Murphy, 2020; Office of Governor Charlie Baker and Lt. Governor Karyn Polito et al., 2020). Widespread testing by local colleges further expanded this segment of identified cases through fall and winter (Broad Institute, 2020). Reported case counts thus depended heavily on public health resources and policy. Wastewater surveillance is not subject to these social and logistical limitations and can therefore serve as a more instantaneous and unbiased readout of new cases during the pandemic. We and others have shown that wastewater likely detects a short period of high viral shedding early in infection (Hoffmann and Alsing, 2021; F. Wu et al., 2020a), whereas patients can test positive during PCR testing of respiratory samples for longer periods of time (Wölfel et al., 2020; Zheng et al., 2020), suggesting that wastewater could be more specific to newly infected patients. However, wastewater surveillance does not necessarily provide a readout of hospitalizations or deaths because these numbers also depend on who is infected and their access to healthcare, which cannot be distinguished via wastewater monitoring (Olesen et al., 2021). Therefore, wastewater should be used in conjunction with additional clinical data streams when making public health decisions related to hospitalizations and mortality.
This study has several limitations. First, the interpretation of the WC ratio relies on the assumption that the viral shedding rate did not drastically change over the course of the pandemic. While some variants of SARS-CoV-2 have been reported to have higher shedding rates or longer shedding duration (Frampton et al., 2021; Kissler et al., 2021), the B.1.1.7 variant did not make up a large proportion of wastewater viral copy numbers in the Boston Area until March 2021, well past the periods in the summer where we described the notable peaks in the WC ratio (Lee et al., 2021). There have been mixed reports of the difference in shedding rate between symptomatic and asymptomatic people (Han et al., 2020; Van Vinh Chau et al., 2020; Zhou et al., 2020), but in any case, it is unlikely that the ratio of asymptomatic to symptomatic cases would swiftly change in a catchment of 2.25 million people. Second, there are some fluctuations in the WC ratio throughout summer 2020 and spring 2021 that we were not able to tie back to community events that potentially increased social contacts and disease transmission. These fluctuations could arise from the noise in both datasets and from community behaviors that were not considered. Private gatherings would be hard to monitor, but perhaps more detailed analysis of mobility data could enhance our interpretations of these fluctuations. Third, in practice, the WC ratio is particularly useful when considering deviations from a stable baseline value. However, assessing such deviations in real time could be difficult, especially in the beginning stages of a pandemic before a baseline is reached. Additionally, it could be difficult to distinguish a shifting baseline from true long-term trends. It is also difficult to use clinical data to verify the silent community spread detected by the WC ratio in the early summer, particularly if those affected were asymptomatic and did not seek hospitalization. In any case, increases in the WC ratio could prompt officials to increase their public health messaging to quell these silent peaks and to remain on alert for further trends.
Application of these metrics to other municipalities requires several considerations. Differences in sewer system residence time, wastewater sample matrix, and sample processing methods could change the relationship between measured wastewater SARS-CoV-2 genome copies and clinical new cases. For example, longer sewer residence times may stretch out the wastewater curve, as the positive signals in wastewater would take longer to reach the sampling point at the WWTP. Viral degradation in wastewater during transport would also affect the measured wastewater concentrations, particularly if there were hot spots of infection in the sewershed. Quantifying SARS-CoV-2 genome copies from sludge rather than primary influent could also lead to a delayed and extended wastewater signal, as it takes some time for solids to settle into sludge and the persistence of SARS-CoV-2 in sludge is longer than in the aqueous fraction (Balboa et al., 2021). Either of these situations may lead to a smaller time lag between wastewater and clinical data. The persistence of SARS-CoV-2 viral particles is affected by environmental factors in different water matrices, such as temperature, UV, exposure, organic matter, disinfectants, and adversarial microorganisms (Amoah et al., 2022; Paul et al., 2021), which affects the baseline magnitude of the WC ratio. Finally, laboratory methods for sample processing, such as pasteurization and choice of quantification method, could affect the measured wastewater SARS-CoV-2 concentrations based on viral degradation or different limits of detection. Thus, while these metrics could still be informative for decision makers, it will be important for municipalities to establish baseline values for the metrics in their specific catchments before using them to make public health decisions.
There are many extensions to this work that will make wastewater surveillance data more integrated and useful for the public health response. For example, wastewater viral copy numbers and clinically reported cases are inherently linked to the number of true infections in the population. More mechanistic modeling is necessary to infer the underlying trend of true infections from the observable wastewater and clinical data (Fernandez-Cassi et al., 2021). Similarly, wastewater viral copy numbers are linked to true infections by a transfer function that describes population-level shedding, while clinical cases are linked to true infections by a transfer function that describes population-level testing availability and turnaround time (Olesen et al., 2021). Inferring these two transfer functions will allow us to understand these parameters separately. Combining wastewater data with clinical and demographic data may also allow us to infer these transfer functions per demographic group, giving us finer resolution understanding of viral shedding parameters and access to testing. As the pandemic has progressed and public health measures have changed, the utility of wastewater surveillance has also changed. In this time-varying context, integrative models can make wastewater data more flexible, useful, and predictive.
5. Conclusions
We introduced three new metrics to assess the time-varying relationship between wastewater data and clinical data. Applying these metrics to a 14-month time series of wastewater surveillance data in Massachusetts, we conclude:
-
•
The relationship between wastewater viral copy numbers and clinically reported cases changed over the course of the COVID-19 pandemic.
-
•
Wastewater surveillance data served as a leading indicator in the first wave (∼6 days) but not the second wave, likely due to substantially increased testing capacity.
-
•
Evaluating the relationships between wastewater and various public health data streams using these new metrics can provide a real-time evaluation of public health responses.
-
•
More integrative models can help increase the utility and application of wastewater surveillance for managing the ongoing COVID-19 pandemic as well as future pandemics.
Declaration of Competing Interest
MM and NG are cofounders of Biobot Analytics. EJA, TBE, PRC, and WPH are advisors to Biobot Analytics. CD, NE, MI, and KAM are employees at Biobot Analytics, and all these authors hold shares in the company.
Acknowledgments
We thank the Deer Island wastewater treatment facility team for providing the samples for analysis, Penny Chisholm (MIT) and Allison Coe (MIT) for access to equipment and other supplies, Mathilde Poyet (MIT) and Shandrina Burns (MIT) for logistical support, and Shijie Zhao (MIT), Ethan Evans (MIT), Yanjia Jason Zhang (MIT), Scott Olesen, Stephen Estes-Smargiassi (MWRA), and Stefan Wuertz (NTU) for helpful discussion. We express our deep gratitude to all healthcare professionals and first-line responders who have been caring for patients with COVID-19.
This work was supported by the Center for Microbiome Informatics and Therapeutics and Intra-CREATE Thematic Grant (Cities) grant NRF2019-THE001–0003a to JT and EJA; National Institute on Drug Abuse of the National Institutes of Health award numbers K23DA044874 to PRC; and R44DA051106 to MM and PRC, Hans and Mavis Psychosocial Foundation funding, and e-ink corporation funding to PRC; funding from the Morris-Singer Foundation and NIH award R01AI106786 to WPH; funds from the Massachusetts Consortium on Pathogen Readiness and China Evergrande Group to TBE, PRC, MM, and EJA. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding institutions.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2022.118070.
Appendix. Supplementary materials
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoah I.D., Abunama T., Awolusi O.O., Pillay L., Pillay K., Kumari S., Bux F. Effect of selected wastewater characteristics on estimation of SARS-CoV-2 viral load in wastewater. Environ. Res. 2022;203 doi: 10.1016/j.envres.2021.111877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, C.D., 2021a. Order advancing all communities to Phase III, Step 2 of the Commonwealth's reopening plan. Commonwealth of Massachusetts Office of the Governor.
- Baker, C.D., 2021b. Order advancing all communities to Phase IV, Step 1 of the Commonwealth's reopening plan and transitioning to a travel advisory policy. Commonwealth of Massachusetts Office of the Governor.
- Baker, C.D., 2020a. Order implementing a phased reopening of workplaces and imposing workplace safety measures to address COVID-19. Commonwealth of Massachusetts Office of the Governor.
- Baker C.D. Commonwealth of Massachusetts Office of the Governor; 2020. Order Authorizing the Re-Opening of Phase II enterprises. [Google Scholar]
- Baker C.D. Commonwealth of Massachusetts Office of the Governor; 2020. Order Further Advancing the Re-Opening of Phase II enterprises. [Google Scholar]
- Baker C.D. Commonwealth of Massachusetts Office of the Governor; 2020. Order Authorizing the Re-Opening of Phase III Enterprises. [Google Scholar]
- Baker, C.D., 2020e. Order further advancing Phase III re-openings in municipalities with reduced incidence of COVID-19 infection. Commonwealth of Massachusetts Office of the Governor.
- Baker, C.D., 2020f. Order returning all municipalities to Phase III, Step 1 COVID-19 safety rules. Commonwealth of Massachusetts Office of the Governor.
- Baker, C.D., n.d. Executive Order No. 591: Declaration of a State of Emergency to Respond to COVID-19. Commonwealth of Massachusetts Office of the Governor.
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. The fate of SARS-COV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., Qu Y., Li F., Lv Q., Wang W., Xue J., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao Linna, Liu P., Zhao Li, Ye F., Wang H., Zhou W., Zhu N., Zhen W., Yu H., Zhang X., Guo L., Chen L., Wang C., Wang Y., Wang X., Xiao Y., Sun Q., Liu, Zhu F., Ma C., Yan L., Yang M., Han J., Xu W., Tan W., Peng X., Jin Q., Wu G., Qin C. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Bardsley J.M., Fox C. An MCMC method for uncertainty quantification in nonnegativity constrained inverse problems. Inverse Probl. Sci. Eng. 2012;20:477–498. doi: 10.1080/17415977.2011.637208. [DOI] [Google Scholar]
- Bar-Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shirazi R., Kramarsky-Winter E., Nir O., Abu-Ali H., Ronen Z., Rinott E., Lewis Y.E., Friedler E., Paitan Y., Bitkover E., Berchenko Y., Kushmaro A. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. medRxiv. 2020 doi: 10.1101/2020.04.26.20073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D. WBUR News; 2020. State: Massachusetts Has Tested More Than 200 People For Coronavirus. [Google Scholar]
- Berkson J. Proc. Third Berkeley Symp. Math. Stat. Probab. Vol. 1 Contrib. Theory Stat. 3.1. 1956. Estimation by least squares and by maximum likelihood; pp. 1–11. [Google Scholar]
- Betancourt W.W., Schmitz B.W., Innes G.K., Pogreba Brown K.M., Prasek S.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. Wastewater-based epidemiology for averting COVID-19 outbreaks on The University of Arizona campus. medRxiv. 2020 doi: 10.1101/2020.11.13.20231340. [DOI] [Google Scholar]
- Broad Institute, 2020. Broad Institute provides COVID-19 Screening For students, faculty, and Staff At More Than 100 Colleges and Universities [WWW Document]. URL https://www.broadinstitute.org/news/broad-institute-provides-covid-19-screening-students-faculty-and-staff-more-100-colleges-and.
- Brown, C.M., Madoff, L., Smole, S., Department of Public Health, 2020. Testing of Persons with Suspect COVID-19.
- Cambridge Public Schools, 2021. COVID-19 Data Dashboard [WWW Document]. URL https://www.cpsd.us/covid19data (accessed 10.15.21).
- Cavany S., Bivins A., Wu Z., North D., Bibby K., Perkins T.A. Inferring SARS-CoV-2 RNA shedding into wastewater relative to time of infection (preprint) Epidemiology. 2021 doi: 10.1101/2021.06.03.21258238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control National Wastewater Surveillance System [WWW Document] Cent. Dis. Control Prev. 2021 URL https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/wastewater-surveillance.html (accessed 10.15.21) [Google Scholar]
- Centers for Disease Control and Prevention, n.d. Older adults at greater risk of requiring hospitalization or dying if diagnosed with COVID-19 [WWW Document]. URL https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/older-adults.html.
- Chavarria-Miró G., Anfruns-Estrada E., Martínez-Velázquez A., Vázquez-Portero M., Guix S., Paraira M., Galofré B., Sánchez G., Pintó R.M., Bosch A. Time evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Wastewater during the first pandemic wave of COVID-19 in the Metropolitan Area of Barcelona, Spain. Appl. Environ. Microbiol. 2021;87 doi: 10.1128/AEM.02750-20. e02750-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csilléry K., Blum M.G.B., Gaggiotti O.E., François O. Approximate Bayesian Computation (ABC) in practice. Trends Ecol. Evol. 2010;25:410–418. doi: 10.1016/j.tree.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Duvallet C., Wu F., McElroy K.A., Imakaev M., Endo N., Xiao A., Zhang J., Floyd-O'Sullivan R., Powell M.M., Mendola S., Wilson S.T., Cruz F., Melman T., Sathyanarayana C.L., Olesen S.W., Erickson T.B., Ghaeli N., Chai P., Alm E., Matus M. Nationwide trends in COVID-19 cases and SARS-CoV-2 wastewater concentrations in the United States (preprint) Epidemiology. 2021 doi: 10.1101/2021.09.08.21263283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead P., Prangle D. Constructing summary statistics for approximate bayesian computation: semi-automatic ABC. ArXiv10041112 Stat. 2011 [Google Scholar]
- Fernandez-Cassi X., Scheidegger A., Bänziger C., Cariti F., Corzon A.T., Ganesanandamoorthy P., Lemaitre J.C., Ort C., Julian T.R., Kohn T. Wastewater monitoring outperforms case numbers as a tool to track COVID-19 incidence dynamics when test positivity rates are high. medRxiv. 2021 doi: 10.1101/2021.03.25.21254344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton D., Rampling T., Cross A., Bailey H., Heaney J., Byott M., Scott R., Sconza R., Price J., Margaritis M., Bergstrom M., Spyer M.J., Miralhes P.B., Grant P., Kirk S., Valerio C., Mangera Z., Prabhahar T., Moreno-Cuesta J., Arulkumaran N., Singer M., Shin G.Y., Sanchez E., Paraskevopoulou S.M., Pillay D., McKendry R.A., Mirfenderesky M., Houlihan C., Nastouli E. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani A., Aalizadeh R., Kostakis M., Markou A., Alygizakis N., Lytras T., Adamopoulos P.G., Peccia J., Thompson D.C., Kontou A., Karagiannidis A., Lianidou E.S., Avgeris M., Paraskevis D., Tsiodras S., Scorilas A., Vasiliou V., Dimopoulos M.-.A., Thomaidis N.S. SARS-CoV-2 wastewater surveillance data can predict hospitalizations and ICU admissions. Sci. Total Environ. 2021;804 doi: 10.1016/j.scitotenv.2021.150151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. X. 2021;10 doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Brazell L.R., Hinton K., Lontai J., Stark N., Young I., Quach C., Russ M., Kauer J., Nicolosi B., Akella S., Tang W., Chen D., Schlueter J., Munir M. Implementing Building-Level SARS-CoV-2 Wastewater Surveillance on a University Campus. medRxiv. 2021 doi: 10.1101/2020.12.31.20248843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, M.S., Sudre, C.H., May, A., Antonelli, M., Murray, B., Varsavsky, T., Kläser, K., Canas, L.s, Molteni, E., Modat, M., Drew, D.A., Nguyen, L.H., Polidori, L., Selvachandran, S., Hu, C., Capdevila, J., COVID-19 Genomics UK (COV-UK) Consortium, Hammers, A., Chan, A.T., Wolf, J., Spector, T.D., Steves, C.J., Ourselin, S., 2021. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study 6, E335–E345. https://doi.org/ 10.1016/S2468-2667(21)00055-4. [DOI] [PMC free article] [PubMed]
- Guescini M., Sisti D., Rocchi M.B.L., Stocchi L., Stocchi V. A new real-time PCR method to overcome significant quantitative inaccuracy due to slight amplification inhibition. BMC Bioinform. 2008;30:326. doi: 10.1186/1471-2105-9-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.S., Seong M.-.W., Kim N., Shin S., Cho S.I., Park H., Kim T.S., Park S.S., Choi E.H. Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19, Seoul, South Korea. Emerg. Infect. Dis. 2020;26:2497–2499. doi: 10.3201/eid2610.202449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Lovett S., Nelson K., Beamer P., Bischel H.N., Bivins A., Bruder A., Butler C., Camenish T.D., De Long S.K., Karthikeyan S., Larsen D.A., Meierdiercks K., Mouser P., Pagsuyoin S., Prasek S., Radniecki T.S., Ram J.L., Roper D.K., Safford H., Sherchan S.P., Shuster W., Stalder T., Wheeler R.T., Korfmacher K.S. Wastewater surveillance for SARS-CoV-2 on college campuses: initial efforts, lessons learned and research needs. medRxiv. 2021 doi: 10.1101/2021.02.01.21250952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.E., Bekhit H.M., Chapman J.B. Using Markov chain Monte Carlo to quantify parameter uncertainty and its effect on predictions of a groundwater flow model. Environ. Model. Softw. 2009;24:749–763. doi: 10.1016/j.envsoft.2008.11.002. [DOI] [Google Scholar]
- Hastings W.K. Monte Carlo sampling methods using markov chains and their applications. Biometrika. 1970;57:97–109. doi: 10.2307/2334940. [DOI] [Google Scholar]
- Hoffmann T., Alsing J. Faecal shedding models for SARS-CoV-2 RNA amongst hospitalised patients and implications for wastewater-based epidemiology. medRxiv. 2021 doi: 10.1101/2021.03.16.21253603. [DOI] [Google Scholar]
- Kissler S.M., Fauver J.R., Mack C., Tai C.G., Breban M.I., Watkins A.E., Samant R.M., Anderson D.J., Ho D.D., Grubaugh N.D., Grad Y.H. Densely sampled viral trajectories suggest longer duration of acute infection with B.1.1.7 variant relative to non-B.1.1.7 SARS-CoV-2. medRxiv. 2021 [Google Scholar]
- Kocamemi B.A., Kurt H., Hacioglu S., Yarali C., Saatci A.M., Pakdemirli B. First Data-Set on SARS-CoV-2 Detection for Istanbul Wastewaters in Turkey. medRxiv. 2020 doi: 10.1101/2020.05.03.20089417. [DOI] [Google Scholar]
- Krivoňáková N., Šoltýsová A., Tamáš M., Takáč Z., Krahulec J., Ficek A., Gál M., Gall M., Fehér M., Krivjanská A., Horáková I., Belišová N., Bímová P., Škulcová A.B., Mackuľak T. Mathematical modeling based on RT-qPCR analysis of SARS-CoV-2 in wastewater as a tool for epidemiology. Sci. Rep. 2021;11:19456. doi: 10.1038/s41598-021-98653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., McElroy K.A., Armas F., Imakaev M., Gu X., Duvallet C., Chandra F., Chen H., Leifels M., Mendola S., Floyd-O'Sullivan R., Powell M.M., Wilson S.T., Wu F., Xiao A., Moniz K., Ghaeli N., Matus M., Thompson J., Alm E.J. Quantitative detection of SARS-CoV-2 B1.1.7 variant in wastewater by allele-specific RT-qPCR. medRxiv. 2021 doi: 10.1101/2021.03.28.21254404. [DOI] [Google Scholar]
- Leidman, E., Duca, L.M., Omura, J.D., Proia, K., Stephens, J.W., Sauber-Schatz, E.K., 2021. COVID-19 Trends Among Persons Aged 0-24 Years – United States, March 1-December 12, 2020 (No. 70(3)), Morbidity and Mortality Weekly Report. [DOI] [PMC free article] [PubMed]
- Leidner, A.J., Barry, V., Bowen, V.B., Silver, R., Trieste, M., Kang, G.J., Ritchey, M.D., Fletcher, K., Barrios, L., Pevzner, E., 2021. Opening of Large Institutions of Higher Education and County-Level COVID-19 Incidence – United States, july 6-September 17, 2020 (No. 70(1)), Morbidity and Mortality Weekly Report. [DOI] [PMC free article] [PubMed]
- Liu D.C., Nocedal J. On the limited memory BFGS method for large scale optimization. Math. Program. 1989;45:503–528. doi: 10.1007/BF01589116. [DOI] [Google Scholar]
- Liu P., Ibaraki M., VanTassell J., Geith K., Cavallo M., Kann R., Moe C. A novel COVID-19 early warning tool: Moore swab method for wastewater surveillance at an institutional level. medRxiv. 2020 doi: 10.1101/2020.12.01.20238006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Luo D., Haase J.E., Guo Q., Wang X.Q., Liu S., Xia L., Liu Z., Yang J., Yang B.X. The experiences of health-care providers during the COVID-19 crisis in China: a qualitative study. Lancet Glob Health. 2020;8:E790–E798. doi: 10.1016/S2214-109X(20)30204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Cao, Z., Chen, M., Zhong, Y., Luo, Y., Shi, G., Xiang, H., Luo, J., Zhou, H., 2020. Effect of Heat inactivation on Real-Time Reverse Transcription PCR of the SARS-COV-2 Detection. https://doi.org/10.1101/2020.05.19.20101469.
- Massachusetts Department of Public Health, 2021a. Archive of COVID-19 Cases in Massachusetts- COVID-19 Raw Data - May 16, 2021.
- Massachusetts Department of Public Health, 2021b. COVID-19 Interactive Data Dashboard [WWW Document]. Mass.gov. URL https://www.mass.gov/info-details/covid-19-response-reporting.
- Massachusetts Department of Public Health, 2021c. Archive of COVID-19 Cases in Massachusetts- COVID-19 Raw Data - April 13, 2021.
- Massachusetts Department of Public Health, 2020a. Archive of COVID-19 Cases in Massachusetts- COVID-19 Raw Data - May 7, 2020.
- Massachusetts Department of Public Health, 2020b. Archive of COVID-19 Cases in Massachusetts- COVID-19 Raw Data - August 11, 2020.
- Maybank, P., Peltzer, P., Naumann, U., Bojak, I., 2020. MCMC for Bayesian uncertainty quantification from time-series data. ArXiv200514281 Stat 12143. https://doi.org/ 10.1007/978-3-030-50436-6.
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Murphy M. WBUR News; 2020. “Stop The Spread” Initiative Will Increase Testing Capacity In Eight Mass. Towns. [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaumer-Streit B., Mayr V., Dobrescu A.I., Chapman A., Persad E., Klerings I., Wagner G., Siebert U., Christof C., Zachariah C., Gartlehner G. Quarantine alone or in combination with other public health measures to control COVID-19: a rapid review. Cochrane Database Syst Rev. 2020;4 doi: 10.1002/14651858.CD013574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Office of Governor Charlie Baker and Lt. Governor Karyn Polito, Governor's Press Office, Massachusetts Executive Office of Health and Human Services, Massachusetts Executive Office of Housing and Economic Development, Massachusetts Executive Office of Public Safety and Security, 2020. Baker-Polito Administration Announces New Initiatives to Stop Spread of COVID-19.
- Ohio Coronavirus Wastewater Monitoring Network, 2021. COVID-19 Dashboard [WWW Document]. URL https://public.tableau.com/views/COVIDWastewater/Dashboard2?:embed=y&:showVizHome=no&:host_url=https%3A%2F%2Fpublic.tableau.com%2F&:embed_code_version=3&:tabs=no&:toolbar=yes&:display_count=yes&:language=en&:loadOrderID=0 (accessed 10.15.21).
- Olesen S.W., Imakaev M., Duvallet C. Applications of wastewater-based epidemiology as a leading indicator for COVID-19. arXiv. 2021 doi: 10.1016/j.watres.2021.117433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino B., Touret F., Gilles M., de Lamballerie X., Charrel R.N. Heat inactivation of different types of SARS-CoV-2 samples: what protocols for biosafety, molecular detection and serological diagnostics? Viruses. 2020;12:E735. doi: 10.3390/v12070735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Kolar P., Hall S.G. A review of the impact of environmental factors on the fate and transport of coronaviruses in aqueous environments. Npj Clean Water. 2021;4:1–13. doi: 10.1038/s41545-020-00096-w. [DOI] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Omer S.B. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics (preprint) Epidemiology. 2020 doi: 10.1101/2020.05.19.20105999. [DOI] [Google Scholar]
- Pollock D.S. School of Business, University of Leicester; 2011. Transfer Functions. Presented at the Discussion Papers in Economics 11/15, Division of Economics. [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B.W., Innes G.K., Prasek S.M., Betancourt W.W., Stark E.R., Foster A.R., Abraham A.G., Gerba C.P., Pepper I.L. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. medRxiv. 2021 doi: 10.1101/2021.04.16.21255638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki N., Matsushita T., Matsui Y., Koriki S. Suitability of pepper mild mottle virus as a human enteric virus surrogate for assessing the efficacy of thermal or free-chlorine disinfection processes by using infectivity assays and enhanced viability PCR. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116409. [DOI] [PubMed] [Google Scholar]
- Spinelli M.A., Glidden D.V., Gennatas E.D., Bielecki M., Beyrer C., Rutherford G., Chambers H., Goosby E., Gandhi M. Importance of non-pharmaceutical interventions in lowering the viral inoculum to reduce susceptibility to infection by SARS-CoV-2 and potentially disease severity. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(20)30982-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toni T., Welch D., Strelkowa N., Ipsen A., Stumpf M.P.H. Approximate Bayesian computation scheme for parameter inference and model selection in dynamical systems. J. R. Soc. Interface. 2009;6:187–202. doi: 10.1098/rsif.2008.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vinh Chau N., Lam V.T., Dung Nguyen Thanh, Yen L.M., Minh N.N.Q., Hung L.M., Ngoc N.M., Dung Nguyen Tri, Man D.N.H., Nguyet L.A., Nhat L.T.H., Nhu L.N.T., Ny N.T.H., Hong N.T.T., Kestelyn E., Dung N.T.P., Xuan T.C., Hien T.T., Phong N.T., Tu T.N.H., Geskus R.B., Thanh T.T., Truong N.T., Binh N.T., Thuong T.C., Thwaites G., Van Tan L., Oxford University Clinical Research Unit COVID-19 Research Group The natural history and transmission potential of asymptomatic severe acute respiratory syndrome coronavirus 2 infection. Clin. Infect. Dis. 2020;71:2679–2687. doi: 10.1093/cid/ciaa711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington N.L., Gangavarapu K., Zeller M., Bolze A., Cirulli E.T., Schiabor Barrett K.M., Larsen B.B., Anderson C., White S., Cassens T., Jacobs S., Levan G., Nguyen J., Ramirez J.M., III, Rivera-Garcia C., Sandoval E., Wang X., Wong D., Spencer E., Robles-Sikisaka R., Kurzban E., Hughes L.D., Deng X., Wang C., Servellita V., Valentine H., De Hoff P., Seaver P., Sathe S., Gietzen K., Sickler B., Antico J., Hoon K., Liu J., Harding A., Bakhtar O., Basler T., Austin B., Isaksson M., Febbo P.G., Becker D., Laurent M., McDonald E., Yeo G.W., Knight R., Laurent L.C., de Feo E., Worobey M., Chiu C., Suchard M.A., Lu J.T., Lee W., Andersen K.G. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B1.1.7 in the United States. medRxiv. 2021 doi: 10.1101/2021.02.06.21251159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikle N., Tran T.N.-A., Gentilesco B., Leighow S.M., Albert J., Strong E.R., Břinda K., Inam H., Yang F., Hossain S., Chan P., Hanage W.P., Messick M., Pritchard J.R., Hanks E.M., Boni M.F. SARS-CoV-2 epidemic after social and economic reopening in three US states reveals shifts in age structure and clinical characteristics. medRxiv. 2020 doi: 10.1101/2020.11.17.20232918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggermos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Nagler J., Rhode S.F., Santillana M., Tucker J.A., Wuertz S., Zhao S., Thompson J., Alm E.J. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Nagler J., Rhode S.F., Santillana M., Tucker J.A., Wuertz S., Zhao S., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. medRxiv. 2020 doi: 10.1101/2020.06.15.20117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., McElroy K.A., Rhode S.F., Matus M., Wuertz S., Thompson J., Alm E.J. Wastewater surveillance of SARS-CoV-2 across 40 U.S. states from February to June 2020. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T., Chai P., Thompson J., Alm E. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5:e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv. 2020 doi: 10.1101/2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., Chen W., Wang Q., Zhang D., Liu Y., Gong R., Ma Z., Lu S., Xiao Y., Gu Y., Zhang J., Yao H., Xu K., Lu X., Wei G., Zhou J., Fang Q., Cai H., Qiu Y., Sheng J., Chen Y., Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Li F., Chen F., Liu H., Zheng J., Lei C., Wu X. Viral dynamics in asymptomatic patients with COVID-19. Int. J. Infect. Dis. 2020;96:288–290. doi: 10.1016/j.ijid.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.