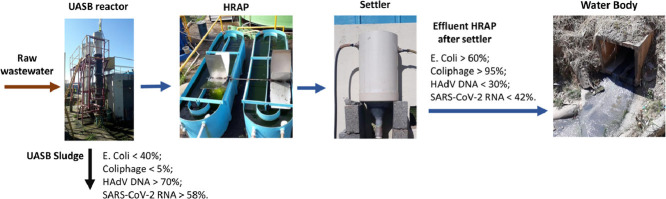
Abstract
Investigating waterborne viruses is of great importance to minimizing risks to public health. Viruses tend to adsorb to sludge particles from wastewater processes by electrostatic and hydrophobic interactions between virus, aquatic matrix, and particle surface. Sludge is often re-used in agriculture; therefore, its evaluation is also of great interest to public health. In the present study, a pilot scale system treating real domestic wastewater from a large city in Brazil was used to evaluate the removal, the overall reduction, and liquid-solid partitioning of human adenovirus (HAdV), the novel coronavirus (SARS-CoV-2) and fecal indicators (F-specific coliphages and E. coli). The system consists of a high-rate algal pond (HRAP) post-treating the effluent of an upflow anaerobic sludge blanket (UASB) reactor. Samples were collected from the influent and effluent of each unit, as well as from the sludge of the UASB and from the microalgae biomass in the HRAP. Pathogens and indicators were quantified by quantitative polymerase chain reaction (qPCR) (for HAdV), qPCR with reverse transcription (RTqPCR) (for SARS-CoV-2), the double agar plaque assay (for coliphages), and the most probable number (MPN) method (for E. coli). The removal and overall reduction of HAdV and SARS-CoV-2 was greater than 1-log10. Almost 60% of remaining SARS-CoV-2 RNA and more than 70% of remaining HAdV DNA left the system in the sludge, demonstrating that both viruses may have affinity for solids. Coliphages showed a much lower affinity to solids, with only 3.7% leaving the system in the sludge. The system performed well in terms of the removal of organic matter and ammoniacal nitrogen, however tertiary treatment would be necessary to provide further pathogen reduction, if the effluent is to be reused in agriculture. To our knowledge, this is the first study that evaluated the reduction and partitioning of SARS-CoV-2 and HAdV through the complete cycle of a wastewater treatment system consisting of a UASB reactor followed by HRAPs.
Keywords: SARS-CoV-2, Adenovirus, Sludge, Virus removal, Sewage, UASB, High-rate algal ponds
Graphical abstract
1. Introduction
The study of waterborne viruses is of great importance to minimizing risks to public health, especially in developing countries, where sanitation infrastructure is scarce and wastewater is commonly discharged without any previous treatment (Pandey et al., 2021). For instance, Brazil treats 49% of the generated wastewater (SNIS, 2019), India only 37%, and the situation in South-East Asian countries is even worse, as only 10% of wastewater is treated (Pandey et al., 2021).
Human adenoviruses (HAdV), a non-enveloped and double-stranded DNA virus, have been suggested as preferred candidates as indicators for viral pathogens because, compared to other viruses, HAdV are frequently present in contaminated waters and their concentration in wastewater is high (Allard, 2017). Also, they do not fluctuate seasonally (Verbyla et al., 2016) and may survive long periods in aquatic environments, not to mention that they are highly resistant to different processes such as UV radiation (Allard, 2017).
New viral pathogens are constantly emerging or being discovered in the environment. One example is the novel coronavirus (SARS-CoV-2) that caused the COVID-19 pandemic. It is an enveloped and single-stranded RNA virus, which viral particles have been detected in stool and urine samples of infected people that are subsequently discharged into the wastewater (Saawarn and Hait, 2021). The detection of SARS-CoV-2 has been reported in wastewater samples (Ahmed et al., 2020a; Medema et al., 2020; Mota et al., 2021) and in sludge samples (Peccia et al., 2020; Serra-Compte et al., 2021) from wastewater treatment plants.
The detection of HAdV in wastewater and sludge has also been widely studied (Allard, 2017; Fong et al., 2010; Yin et al., 2018). Nevertheless, there is still a lack of knowledge and literature data about the removal of viruses (including SARS-CoV-2 and HAdV) in wastewater treatment processes, especially in developing countries (Verbyla et al., 2017). This may be attributed to the high costs and expertise required to perform the necessary quantitative molecular analyses.
Upflow anaerobic sludge blanket (UASB) reactors are one of the most widely used wastewater treatment technologies in Latin America and the Caribbean, mainly due to their low operating costs (Noyola et al., 2012). High-rate algal ponds (HRAP) have been recently used as post treatment for UASB reactors. Organic matter, solids, nutrients, pathogen indicators and other pollutants have been reported for this combined system (Espinosa et al., 2021; Vassalle et al., 2020b, 2020a) but there are still very limited data available on the removal of pathogens.
An important factor which is often widely ignored in the literature is the viruses’ affinity toward solids in the wastewater and their removal in the solid phase. Viruses tend to adsorb to solids due to electrostatic interactions influenced by the surface charge of the virus, and hydrophobic interactions influenced by the hydrophobic proteins of the viral capsid (Verbyla and Mihelcic, 2015). If a virus is highly resistant to a treatment process but has a high affinity toward solids and is discharged in the sludge, there may be an illusion of high reduction based on the observed differences in the concentration of the virus at the influent and liquid effluent points, but there could still be a need to ensure sufficient reduction of viruses in the sludge before it can be safely reused or discharged. In our recently study (Espinosa et al., 2021), we proposed a mass balance model, which helps estimating a mass balance of the microorganisms entering and leaving the system. The model is based on the microbial influent and effluent concentrations in the liquid phase, and the microbial fraction in the solid phase. This model can be used by other WWTP since the concentrations of microorganisms in the liquid phase are commonly reported. As the fraction of pathogens in the sludge is not usually reported, it can be predicted using the total solid yields with respect to the TS produced or COD treated in the system. A complete explanation of the model is reported in (Espinosa et al., 2021).
The aim of this study was to fill important knowledge gaps regarding the removal and liquid-solid partitioning of HAdV, SARS-CoV-2, F-specific coliphages, and E. coli in a pilot-scale UASB reactor followed by parallel HRAPs treating real domestic wastewater in Belo Horizonte, Brazil.
2. Materials and methods
2.1. Experimental design
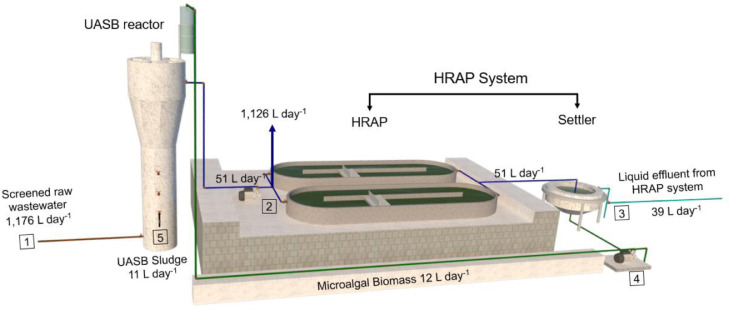
Experimental data were gathered in a demonstration scale system located at the Research and Training Center for Sanitation (CePTS) in Belo Horizonte, Brazil. The system consists of a UASB reactor followed by twin high-rate algal ponds (Vassalle et al., 2020a) fed with real wastewater from a full-scale wastewater treatment plant (WWTP) in the city of Belo Horizonte, as shown in Fig. 1 . The system was operating normally during the pandemic.
Fig. 1.
Diagram of the pilot-scale upflow anaerobic sludge blanket (UASB) reactor followed by twin high-rate algal ponds (HRAPs) with return of algal biomass, showing the measured flow rates and sample collection points (1 – 5).
The UASB reactor worked with a volume of 343 L (height 4.0 m; diameter 0.3 m) and was operated at a flow rate of 50 L.h−1 and a hydraulic retention time (HRT) of 7 h. The HRAPs worked in parallel with a volume of 205 L each (height 0.5 m; length 1.7 m; width 0.24 m) and were operated at a flow rate of 25.5 L.day−1 (each) and an HRT of 8 days. The biomass settler worked with a volume of 30 L and was operated at an HRT of 14 hours. For the anaerobic co-digestion, 12 L of harvested microalgae biomass were pumped to a plexiglass column located 4 m above the UASB reactor and recirculated to the reactor at flow rate of 0.5 L.h−1. The system has been operating since July 2018 on a continuous basis.
A total of 13 samples were collected from each sample collection point during a total period of 6 weeks (i.e., approximately two samples collected each week), between July and August 2020, during the “first wave” of the COVID-19 outbreak in the city of Belo Horizonte. This sampling period of six weeks was chosen because of the high concentrations of SARS-CoV-2 in the wastewater due to the outbreak, which permitted its detection in both untreated and treated wastewater, allowing for the calculation of removal rates. The samples were composite samples, with subsamples collected every 20 minutes for 1 h, at the influent (point 1, Fig. 1) and at the effluent of the UASB reactor (point 2, Fig. 1), from the UASB sludge (point 5, Fig. 1), from the HRAP liquid effluent (point 3, Fig. 1), and from the algal biomass at the bottom of the settler (point 4, Fig. 1). These sample collection points were chosen to perform a mass balance of microorganisms entering and leaving each reactor, in the liquid and sludge/biomass phases. All samples were stored at 4 °C and the microbial analyses were conducted within 24 h of sample collection at the Laboratory of Microbiology of the Sanitary and Environmental Engineering Department at Federal University of Minas Gerais.
2.2. Physical-chemical analysis
Liquid phase samples from the raw wastewater and the effluents of the UASB reactor and the HRAPs were collected every sample day at 10:00 am ± 2 h. Physical-chemical parameters analyzed were pH, temperature, dissolved oxygen (DO), chemical oxygen demand (COD), total and volatile suspended solids (TSS and VSS), total nitrogen (TN) and ammonium nitrogen (NH4 +-N). Temperature, pH and DO were determined in-situ using a portable Hach® sensor - (HQ30D). COD was measured through a Hach® kit COD at high range. TSS and VSS were determined according to Standard Methods (APHA-AWWA-WEF, 2017). TN and NH4 +-N were analyzed by ionic chromatography (Metrohm® - 940 professional IC Vario). These chemical parameters were used to determine the loadings of microorganisms present in the solid phase system and to evaluate the treatment efficiency.
For microalgae biomass characterization, samples were taken once a week from the settler. Total and volatile solids (TS and VS) and total Kjeldahl nitrogen (TKN) were analyzed according to standard procedures (APHA-AWWA-WEF, 2017). Total COD was analyzed using Hach® kit COD at high range. For protein content, a conversion factor of 5.95 was used based on the results of TKN (López et al., 2010). Microalgae production was calculated using the methodology presented by Vassalle et al. (2020a).
2.3. Viral and bacterial indicators analysis
F-specific coliphages were used as viral indicators. Their quantification was performed using the double agar plaque assay based on the protocols described in 9224B and 9224C of Standard Methods (APHA SMWW, 2017). Results were measured as PFU per 100 mL. For sludge samples, the coliphage quantification was based on Guzmán et al. (2007) and the results were quantified as PFU per g of dry matter. A complete description of the coliphage method is in Espinosa et al. (2021). E. coli was used as bacterial indicator. It was quantified using the Colilert and Quanti-Tray 2000 most probable number (MPN) method (IDEXX, Maine, EUA) and results were measured as MPN per 100 mL.
2.4. Viral nucleic acids concentration
Viral DNA/RNA concentration was performed using the adsorption-extraction method (Ahmed et al., 2020b; Symonds et al., 2014), which is a modified method of the adsorption-elution method described by Katayama et al. (2002). The final elution volume of each sample was 100 µL. A description of the method is in the Supplementary Material.
2.5. Molecular quantification
2.5.1. Human adenovirus
The concentration of human adenovirus (HAdV) from wastewater samples was determined by quantitative polymerase chain reaction (qPCR) using an assay published by Jothikumar et al. (2005), adapted for SYBR Green chemistry. Sequences of primers were as published by Jothikumar et al. (2005): (Forward 5′-GGACGCCTCGGAGTACCTGAG-3′ and Reverse 5′- ACIGTGGGGTTTCTGAACTTGTT-3′). Reaction conditions were adapted from Jothikumar et al. (2005), Verbyla et al. (2016), and the PowerUp SYBR Green Master Mix User Guide, resulting in a pre-cycling stage of 50 °C for 2 min and 95 °C for 2 min, followed by 40 cycles of: denaturation at 95 °C for 15 s, annealing at 55 °C for 30 s, and extension at 72 °C for 15 s. At the end of the 40 cycles, a melting curve analysis was performed.
Reactions were executed using a 7500 Real-Time PCR System (Applied Biosystems) with a final volume of 20 µL containing: 7.66 µL of ultra-pure water (RNase free), 0.17 µL of each primer (final concentration of 0.25 mM), 10 µL of PowerUp SYBR Green Master Mix and 2 µL of the template DNA. Each sample was analyzed in triplicate. Ultra-pure water was used as the negative control (no template) for each assay. An artificial template using gBlocks™ (detailed in the Supplementary material) was used as positive control for each plate and to generate the standard curve.
2.5.2. Standard curve
Six serial dilutions (1:10) from the gBlocks™ working stock were performed to generate the standard curve. All points were analyzed in triplicate. The standard curve Cq values and respective melt curves are presented in the Supplementary material. The final standard curve had an R2 value that was >0.99 and an efficiency of 91.1%.
2.5.3. SARS-CoV-2
The concentration of SARS-CoV-2 RNA was determined by a reverse transcription quantitative polymerase chain reaction (RT-qPCR) with Taqman chemistry using primers and probes published by CDC (2020) (Forward 5′- GACCCCAAAATCAGCGAAAT-3′, Reverse 5′- TCTGGTTACTGCCAGTTGAATCTG-3′ and probe 5′- ACCCCGCATTACGTTTGGTGGACC-3′). The analysis of the concentrations was done using the N1 (nucleocapsid) region as it shown to be more sensitive than the N2 assay for raw wastewater collected from the same treatment plant (Calábria et al., 2020). The cycling conditions were adapted from the CDC (2020) protocol and the recommendations provided by the Master mix manufacturer (Biorad) and was carried out as follows: 50 °C for 10 min (for reverse transcription), 95 °C for 2 min (for enzyme activation/inactivation), followed by 45 cycles of 95 °C for 3 s (denaturation) and 55 °C for 30 s (annealing and extension).
All analyses were performed using a 7500 Real-Time PCR System (Applied Biosystems). For the reaction, a MasterMix iTaq Universal Probes One Step Kit (Biorad) was used. This mix features a combination of iScript RNase H + reverse transcriptase and iTaq hot-start DNA polymerase to complete the real time reaction in a single step. All reactions were carried out with a final volume of 20 µL containing: 3 µL of ultra-pure water (RNase free), 1.5 µL of the primer/probe mixture (0.50 mM final concentration for each primer; 0.125 mM final concentration for the probe), 0.5 µL of iScript reverse transcriptase (Biorad), 10 µL of MasterMix iTaq Universal Probes One Step (Biorad), and 5 µL of the RNA template. Each sample was analyzed in triplicate. Ultra-pure water was used as the negative (no template) control for each assay. Plasmid containing the full sequence of the SARS-COV-2 nucleocapsid (N) gene (IDT, USA) was used to generate the standard curve and as positive control for each plate. Six serial dilutions from the plasmid working stock were done to generate the standard curve and the final curve had an R2 value that was >0.99 and a q-PCR efficiency of 90.9%. The standard curve points with their Cq values are in the Supplementary material.
Samples that did not amplify any or just one replicate were marked as “not detected” (ND). For quantitative calculations, the LOD was defined as the number of copies corresponding with 95% probability of amplification (Bustin et al., 2009). The calculation for the LOD was carried out following the exponential model established by Verbyla et al. (2016). Values obtained for the LOD in this study are presented in the Supplementary material for both viruses (SARS-CoV-2 and HAdV). In order to calculate removal, reduction, and partitioning values, the concentrations of all ND samples were substituted with the LOD for the respective assay.
2.6. Analysis of removal, reduction, and partitioning of the microrganisms
The removal and reduction of the four microbial constituents analyzed in this study were calculated as previously explained in Espinosa et al (2021), using the log10 removal or the log10 reduction of the geometric mean concentrations or loadings (see Eq. S1 and Eq. S2 in the Supplementary material). Here, removal is defined as the removal of the microorganism from the liquid fraction of influent and effluent streams, whereas reduction is defined as the overall difference in the loadings coming into the reactor vs. leaving the reactor (including both liquid and sludge/biomass streams). Solids partitioning was calculated using the fraction of geometric mean loadings discharged in sludge or biomass, where the loadings were calculated from the concentrations multiplied by the respective volumetric flow rates (for liquid samples) or dry mass loading rates (for sludge/biomass samples) (see Eq. S3 in the Supplementary material).
2.7. Statistical analysis
The ANOVA and post-hoc Tukey test were performed in Minitab® 19 to determine whether log removals and log reductions were significantly different from each other (p < 0.05) for the different microorganisms throughout the treatment stages. Furthermore, the presence or absence of significant differences (p < 0.05) between the influent loadings of the four microorganisms were analyzed.
3. Results and discussion
3.1. Effluent quality and system monitoring
The results of the physical-chemical analyses of raw wastewater, UASB effluent and HRAP effluent are presented in Table 1 . Physical-chemical parameters of the sludge and biomass are shown in the Supplementary material. An overall average efficiency of 71% was observed in COD removal. This result agrees with the average values referenced in the literature, which ranging from 64 to 75% for UASB followed by HRAP (Espinosa et al., 2021; Vassalle et al., 2020a; Villar-Torres et al., 2018).
Table 1.
Physical-chemical characterization of raw wastewater, UASB effluent and final effluent from the treatment system (total number of samples = 13).
| Raw wastewater | UASB effluent | HRAP effluent | ||||
|---|---|---|---|---|---|---|
| Mean (SD) | Min/Max | Mean (SD) | Min/Max | Mean (SD) | Min/Max | |
| TSS (mg.L−1) | 162.8 (47.5) | 110.0/247.9 | 34.4 (9.3) | 19.0/54.0 | 74.5 (44.0) | 26.4/194.0 |
| VSS (mg.L−1) | 134.5 (47.6) | 82.7/223.8 | 29.3 (7.0) | 15.5/39.0 | 56.6 (34.1) | 19.3/148.0 |
| TS (mg.L−1) | 697.5 (691.6) | 267.5/2750.0 | 306.6 (117.2) | 195.0/430 | 475.5 (117.2) | 250.0/675.0 |
| VS (mg.L−1) | 542.5 (710.0) | 175.0/2740.0 | 191.5 (92.5) | 56.0/390 | 313.6 (131.4) | 85.5/480.5 |
| COD (mg.L−1) | 402.8 (28.1) | 355.5/460 | 135.3 (33.2) | 94.0/415.0 | 116.8 (31.2) | 185.0/80.0 |
| NH4+-N (mg.L−1) | 30.1 (5.8) | 20.4/42.3 | 38.5 (6.5) | 24.3/45.5 | 16.1 (3.8) | 13.1/20.8 |
| TN (mg.L−1) |
46.5 (11.1) | 38.2/52.9 | 50.1 (8.9) | 42.3/58.8 | 28.6 (6.9) | 25.1/34.8 |
| pH | 7.7 (0.1) | 7.6/7.8 | 7.8 (0.2) | 7.4/8.0 | 8.5 (0.4) | 8.0/9.0 |
| DO (mg.L−1) | 0.8 (0.3) | 0.4/1.1 | 0.2 (0.1) | 0.2/1.1 | 13.8 (1.6) | 12.0/16.0 |
| Temp. ( °C) | 23.3(1.8) | 21.0/27.2 | 21.8 (1.8) | 17.9/23.0 | 21.7 (2.0) | 17.8/25.0 |
Note: UASB – Upflow anaerobic sludge blanket; HRAP – High rate algal pond; DO – Dissolved Oxygen; COD – Chemical Oxygen Demand; TSS – Total Suspended Solids; VSS – Volatile Suspended Solids; N-NH4+ - Ammonium nitrogen; TN – Total Nitrogen.
Regarding the removal of TSS and VSS in the system, the average concentrations in the UASB effluent were 34.4 mg.L−1 and 29.3 mg.L−1, respectively. These values are below those reported in the literature, 50–160 mg TSS.L−1 and 30 mgVSS.L−1 (Chernicharo, 2007). After the post-treatment of the effluent by HRAP, there is an increase in TSS and VSS concentrations to 74.5 mg.L−1 and 56.6 mg.L−1, respectively . The increase in both TSS and VSS is due to microalgae biomass production in the HRAPs during effluent treatment, which has been reported previously (Vassalle et al., 2020a; Villar-Navarro et al., 2018). The ammonium N concentration increased slightly in the anaerobic reactor (from 30 to 38 mg N-NH4 +.L−1), a likely result of the hydrolysis of proteins and urea (Tchobanoglous et al., 2003). In the HRAP, the average N-NH4 + removal was 57%. Previous studies have shown similar results (64%) in HRAPs operated at an HRT of 6 days treating anaerobic effluent (Gonçalves et al., 2020). Total Nitrogen (TN) removal in the HRAP was on average 44%. The same removal efficiency was observed in another UASB+HRAP system treating domestic wastewater with HRT of 6 days in the HRAP (Gonçalves et al., 2020). It is important to highlight that the overall concentration of pollutants in the treated effluent met the Brazilian regulatory requirements, which are 180 mg COD.L−1, 150 mg TSS.L−1 and 20 mg N-NH4 +.L−1 (CONAMA, 2011).
Microalgae production was calculated from the average TSS concentration into the HRAP (155 mg.L−1), which resulted in an average value of 9.2 g TSS.m−2.day−1 over the year. This value is similar to that reported for this system over 1 year of operation (Vassalle et al, 2020a). In terms of biomass characterization, the average composition was as follow: 1.75 g TS.L−1, 1.19 g VS.L−1, 3.15 g COD.L−1, 206 mg TKN.L−1 and 1226 mg.L−1 of proteins. The main microalgae species found in harvested biomass was Scenedesmus sp., which has been reported to be the most common species in microalgae-based wastewater treatment systems (Mohsenpour et al., 2021). Physical-chemical parameters of UASB sludge and microalgae biomass are in the Supplementary material.
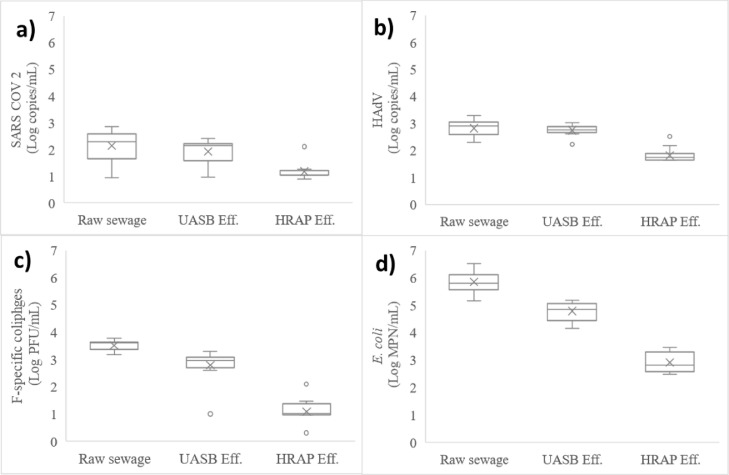
3.2. Concentrations and removals of the microrganisms
Measured concentrations of microbial constituents and the resulting log10 removal values (using Eq. S1) are shown in Tables 2 and 3 , respectively. The geometric mean concentrations of SARS-CoV-2 RNA in the influent (raw wastewater) and liquid effluent of the UASB reactor were 2.09 and 1.92 log(copies)/mL, respectively, which results in a calculated removal of 0.18-log10 units. The geometric mean concentrations of HAdV DNA were slightly higher than SARS-CoV-2 RNA, with 2.81 and 2.74 log(copies)/mL in the influent and effluent, respectively, and a log10 removal of less than 0.1 log10 units. Geometric mean influent and liquid effluent concentration of F-specific coliphages were 3.51 and 2.79 log (PFU)/mL, respectively, with a removal of 0.73 log10 units. ANOVA + Tukey test showed significant difference (p = 0.000) between influent concentration of coliphages and the two human viruses. No significant difference (p = 0.193) was observed between the influent concentrations of HAdV and SARS-CoV-2.
Table 2.
Microbial concentrations throughout the different stages of the pilot-scale wastewater treatment system.
| Stage | Statistics1 | SARS-CoV-2 RNA | HAdV DNA | F-spec. coliphages | E. coli |
|---|---|---|---|---|---|
| Log10 copies/mL | Log10 copies/mL | Log10 PFU/mL | Log10 MPN/mL | ||
| Raw wastewater | N | 13 | 13 | 13 | 13 |
| Mean | 2.08 | 2.81 | 3.51 | 5.85 | |
| Max/Min | 2.85/0.93 | 3.28/2.30 | 3.78/3.17 | 6.51/5.17 | |
| UASB effluent | N | 13 | 13 | 13 | 13 |
| Mean | 1.92 | 2.74 | 2.79 | 4.79 | |
| Max/Min | 2.42/0.96 | 3.02/2.23 | 3.30/1.00 | 5.17/4.16 | |
| HRAP effluent | N | 13 | 13 | 13 | 13 |
| Mean | 1.18 | 1.82 | 1.09 | 2.91 | |
| Max/Min | 2.11/0.90 | 2.51/1.65 | 2.10/0.30 | 3.46/2.47 |
Means reported are the arithmetic means of the log10-transformed concentrations (which are equivalent to the geometric means of the non-transformed concentrations). N = number of measurements.
Table 3.
Log10 removals of the microorganisms throughout the different stages of the pilot-scale wastewater treatment system.
| SARS-CoV-2 RNA | HAdV DNA | F-spec. coliphages | E. coli | |
|---|---|---|---|---|
| UASB | 0.16 | 0.07 | 0.73 | 1.07 |
| HRAP | 0.74 | 0.92 | 1.70 | 1.87 |
| Overall system | 0.90 | 1.00 | 2.43 | 2.94 |
Influent concentrations in the raw wastewater for all three viruses are consistent with values reported in the literature. For instance, SARS-CoV-2 concentrations reported previously have ranged from <1 to 5.1 log(copies)/mL (Ahmed et al., 2020a; Medema et al., 2020; Prado et al., 2021), HAdV concentrations generally range from 1.7 to 5.8 log(copies)/mL (García-Aljaro et al., 2018; Kaliakatsos et al., 2019; Prado et al., 2011; Sheludchenko et al., 2016) and coliphage concentrations range from 2.8 to 4.0 log(PFU)/mL (Dias et al., 2018; Jebri et al., 2016). The efficiency of virus removal from wastewater depends on the types of treatment technologies used, the wastewater characteristics (e.g., temperature, pH, organic matter content, etc.) and design/operational factors such as hydraulic retention times (Ali et al., 2021). For instance, El-Senousy and Abou-Elela (2017) obtained a removal of HAdV in a UASB reactor between 0 to 1 log10 units, with a mean of 0.5 log10 units and Fong et al. (2010) obtained a mean removal of HAdV after primary sedimentation of only 0.01 log10 units. Symonds et al. (2014) reported negligible removal for some enteric viruses in a UASB reactor in Bolivia. In a recent review, the viral removal in a UASB reactor treating wastewater was reported between 0 to 0.7 log10 units (Oakley et al., 2017). In the UASB reactor analyzed in this study the viral removal was <1 log10 units, which is expected in these reactors. Recently, Kumar et al. (2021) reported a removal of >1.3 log10 units for SARS-CoV-2 in a UASB reactor, which is higher than the removal obtained in this study.
On the other hand, E. coli had a removal above 1 log10 units, which is similar to values reported in the literature for UASB reactors (Dias et al., 2014; Oakley et al., 2017). The geometric mean influent and effluent concentrations for E. coli were 5.85 and 4.79-log MPN/mL, respectively. Statistical results showed that SARS-CoV-2 RNA and HAdV DNA removals were significantly lower than E. coli (p < 0.05). Coliphages removals were significantly greater than HAdV (p < 0.05), but they did not show a significant difference with SARS-CoV-2 RNA and E. coli removals (p > 0.05). It is expected for this to happen because bacteria are more sensitive to treatment processes compared with viruses and other pathogens (Rodriguez-Manzano et al., 2012; WHO, 2001). Another important factor affecting the results is that the measurement of HAdV DNA and SARS-CoV-2 RNA using (RT)qPCR does not assess viability, whereas the double agar layer plaque assay used to quantify F-specific coliphages only measures coliphages that are viable. It is possible for (RT)qPCR to detect intact segments of genome associated with virus particles that have lost viability. However, once viral RNA is released from a damaged capsid, its persistence in wastewater is likely very limited; one study reported that free viral RNA released into wastewater was no longer detectable after only a few minutes (Limsawat and Ohgaki, 1997). Extracellular DNA (free-DNA and adsorbed-DNA), on the contrary, have more persistence in the environment since it can be adsorbed onto solid particles and organic matter which gives protection against degradation by nuclease (Gutiérrez-cacciabue et al., 2016; Yuan et al., 2019). This may lead to an overestimation of the HAdV DNA in our study. It is important to note that even though culture-based methods were not used to quantify the human viruses in this study, the results presented here advance knowledge as they show the dynamics of the removal of these viruses and indicators in a treatment system.
For the HRAPs, only 2 of the 13 HRAP effluent samples amplified for SARS-CoV-2 RNA and for HAdV DNA. Therefore, the limit of detection (LOD) of 28 and 45 copies/reaction, respectively, was used for the non-detect samples, to calculate a conservative estimate for removal and reduction (i.e., the estimated values are likely less than the actual values). This assumption also likely led to partitioning values that overestimated the fraction present in liquid effluent and underestimated the fraction present in biomass. The geometric means of the effluent concentrations were 1.18 log(copies)/mL, 1.82 log(copies)/mL, 1.09 log(PFU)/mL, and 2.91 log(MPN)/mL, for SARS-CoV-2 RNA, HAdV DNA, F-specific coliphages, and E. coli, respectively. SARS-CoV-2 RNA and HAdV DNA removals were both <1 log10 unit. F-specific coliphages and E. coli had removals of 1.70 and 1.87-log units, respectively. Statistical results showed that SARS-CoV-2 RNA and HAdV DNA removals were significantly lower than E. coli and coliphages removal (p < 0.05).
The removal of microorganisms was higher in the HRAP stage than in the UASB reactor, which is expected due to sunlight exposure and longer hydraulic retention time in the ponds (Symonds et al., 2014; Verbyla and Mihelcic, 2015). The observed removal of 1.70-log10 units for F-specific coliphages in HRAPs with an HRT of 8 days was within the range reported (1–3-log10 units) for HRAPs operating under acidic conditions by (Delanka-Pedige et al., 2020b). The observed removals of HAdV DNA (0.92-log10 unit) and SARS-CoV-2 RNA (0.74-log10 unit) in the current study were more consistent with the removal rates (1-log10 unit) of enteric viruses and bacteriophages reported by Verbyla and Mihelcic (2015) for conventional stabilization ponds (not high rate) with HRTs of 15 to 20 days. However, our observed removals of HAdV DNA and SARS-CoV-2 RNA were slightly lower than those reported by Delanka-Pedige et al (2020a) for enterovirus RNA (1.05-log10 units) and Norovirus GI RNA (1.49-log10 units) in an acidic HRAP with an HRT of 4–5 days. E. coli removal in HRAPs has been reported in the literature between 1.76 to 2.19-log units (Buchanan et al., 2018; Fallowfield et al., 2018; Young et al., 2016) and virus removal between zero and 1.7-log units (Verbyla et al., 2017). Hence, removals observed in the current study are within the range of values previously reported in the literature.
When considering the overall system, UASB+HRAP (influent = point 1, effluent = point 3, Fig. 1), the removal of SARS-CoV-2 RNA was 0.9 log10 units and for HAdV DNA it was 1 log10 units. It is important to note that UASB + HRAP systems are generally not designed to optimize the removal of pathogens. The main design objective is usually to reduce organic matter and nutrients to comply with discharge regulations, which was the case for this system. The viral and bacterial indicators (coliphages and E. coli) had an overall removal of 2.43- and 2.94-log10 units, respectively. There is still a lack of studies in the literature about SARS-CoV-2 removal in different wastewater treatment processes (Saawarn and Hait, 2021). Only a few studies of the removal of SARS-CoV-2 in wastewater treatment systems have mostly focused on conventional activated sludge system, in some cases followed by tertiary treatment processes (Randazzo et al., 2020; Serra-Compte et al., 2021; Sherchan et al., 2020), or for systems using a moving bed biofilm reactor and sequencing batch reactor technologies (Arora et al., 2020; Balboa et al., 2021), with a maximum removal value of 1.97-log. Due to the limited amount of data from the literature on this type of UASB-HRAP system, we compared our results with similar anaerobic reactors followed by similar algal-based treatment technologies. For example, in an anaerobic digester followed by a HRAP, coliphages had a 1-log10 overall removal (Davies-Colley et al., 2005). In a septic tank followed by HRAP the removal was found to be 1.8-log10 (Young et al., 2016). Symonds et al. (2014) reported a 0.8-log10 removal of culturable enteric viruses in a UASB reactor followed by polishing (maturation) ponds. Our findings regarding virus removal are generally within the range of previously published literature, except for coliphages, where our observed removal was higher than what has been reported in similar systems with anaerobic reactors followed by ponds or lagoons. One possible hypothesis is that the combination of a high-rate anaerobic reactor with granular sludge followed by a high-rate algal system may somehow enhance the decay rate of viable coliphages. However, more studies with direct comparison of anaerobic reactor + algal pond systems (e.g., septic tank followed by conventional stabilization pond) and high-rate anaerobic reactor + high-rate algal pond systems (e.g., UASB+HRAP) to be able to test this hypothesis.
On the other hand, for E. coli removal, Santiago et al. (2013) reported a removal of 2-log10 units in a UASB reactor followed by HRAP and Young et al. (2016) reported the same 2-log10 removal in septic tank followed by HRAP. Our finding resulted a little higher than these removals. In an algal-based system (with acidic conditions), the removal of total coliforms was 7-log10 and for pathogenic E. coli was 4-log10 (Delanka-Pedige et al., 2019), These high removals are attributed to the system's low pH (=4). E. coli and also other fecal indicator bacteria have been demonstrated to be less resistant to treatment than viruses (Momba et al., 2019). This state was confirmed with our finding, where E. coli removal was significantly higher than SARS-CoV-2 RNA, HAdV DNA and coliphages removal (p < 0.05). Fig. 2 shows boxplots of the concentrations of the microorganisms in the liquid phase of the overall system.
Fig. 2.
Boxplots of the concentrations in raw wastewater, UASB effluent and HRAP effluent, a) SARS-CoV-2 RNA (log copies/mL), b) HAdV DNA (log copies/mL), c) F-specific coliphages (log PFU/mL) and d) E. coli (log MPN/mL). The lower and upper bars denote minimum and maximum values, respectively. The lower and upper boxes represent the 25th and 75th percentiles, respectively. Mean values are represented by an “x”. The line inside the box denotes the median value.
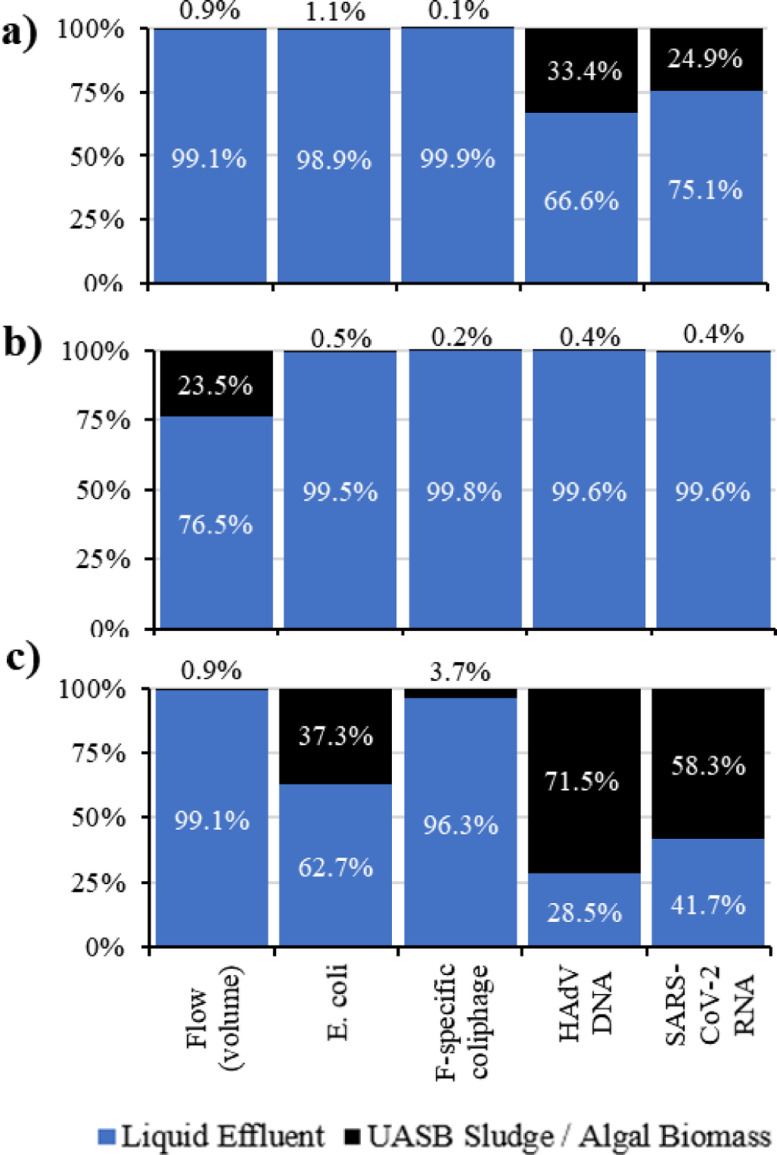
3.3. Overall reductions and liquid-solid partitioning of the microorganisms
Results of influent and effluent loadings and overall reduction including flow rate and the total loading inactivation of each microorganism throughout the UASB reactor, HRAPs and complete system, are presented in Table 4, Table 5, Table 6 , respectively. A diagram with the liquid – solid partitioning in the system is presented in Fig. 3 . No significant correlation (p<0.05) was found between water parameters (OD, COD and SST) and influent, effluent, and sludge loadings (results not shown).
Table 4.
Mass balance of the four microorganisms for the UASB reactor.
| Flow (L/d) | SARS-CoV-2 RNA loading copies/d | HAdV DNA loading copies/d | F-specific coliphages loading PFU/d | E. coli loading MPN/d | |
|---|---|---|---|---|---|
| Raw wastewater | 1176 | 1.40•108 | 7.67•108 | 3.83•109 | 8.37•1011 |
| HRAP biomass (returned to UASB) | 12 | 2.31•103 | 8.95•103 | 9.68•102 | 1.38•105 |
| TOTAL IN | 1188 | 1.40•108 | 7.67•108 | 3.83•109 | 8.37E+11 |
| UASB liquid effluent | 1177 | 9.78•107 | 6.48•108 | 7.19•108 | 7.19•1010 |
| UASB sludge | 11 | 2.55•107 | 2.55•108 | 6.31•105 | 5.90•108 |
| TOTAL OUT | 1188 | 1.34•108 | 1.03•109 | 1.06•109 | 7.27•1010 |
| Log10 Reduction Value | 0.02 | -0.13 | 0.56 | 1.06 |
Table 5.
Mass balance of the four microorganisms for the HRAPs.
| Flow rate (L/d) | SARS-CoV-2 RNA loading copies/d | HAdV DNA loading copies/d | F-specific coliphages loading PFU/d | E. coli loading MPN/d | |
|---|---|---|---|---|---|
| UASB effluent (sent to pilot-scale HRAPs) | 51 | 4.24•106 | 2.81•107 | 3.12•107 | 3.11•109 |
| TOTAL IN | 51 | 4.24•106 | 2.81•107 | 3.12•107 | 3.11•109 |
| Clarified liquid HRAP effluent | 39 | 5.85•105 | 2.56•106 | 4.77•105 | 3.18•107 |
| HRAP microalgae biomass | 12 | 2.31•103 | 8.95•103 | 9.68•102 | 1.38•105 |
| TOTAL OUT | 51 | 5.87•105 | 2.57•106 | 4.78•105 | 3.20•107 |
| Log10 Reduction Value | 0.86 | 1.04 | 1.81 | 1.99 |
Table 6.
Mass balance of the complete system (UASB+HRAP).
| Flow rate (L/d) | SARS-CoV-2 RNA loading copies/d | HAdV DNA loading copies/d | F-specific coliphages loading PFU/d | E. coli loading MPN/d | |
|---|---|---|---|---|---|
| Raw wastewater (partial) | 39.4 | 4.68•106 | 2.57•107 | 1.28•108 | 2.80•1010 |
| TOTAL IN | 39.4 | 4.68•106 | 2.57•107 | 1.28•108 | 2.80•1010 |
| Clarified liquid HRAP effluent | 39.0 | 5.85•105 | 2.56•106 | 4.77•105 | 3.18•107 |
| UASB sludge (partial) | 0.4 | 8.49•105 | 8.46•106 | 2.10•104 | 1.96•107 |
| TOTAL OUT | 39.4 | 1.66•106 | 1.24•107 | 8.14•105 | 5.52•107 |
| Log10 Reduction Value | 0.45 | 0.32 | 2.20 | 2.71 |
Fig. 3.
Liquid-solid partitioning of the microrganisms throughout the different stages, a) UASB reactor, b) HRAP settler and c) UASB reactor + HRAP.
When analyzing the UASB reactor alone (considering influent as the raw wastewater and the microalgae biomass recirculated from the HRAPs, point 1 + point 4, Fig. 1), viruses and E. coli loadings into the reactor came mostly from the raw wastewater, with the loadings from the microalgae biomass contributing only negligible amounts. The majority of coliphage and E. coli effluent loadings also left the UASB reactor in the liquid phase, whereas approximately one-third of SARS-CoV-2 RNA and HAdV DNA left the UASB reactor in the sludge.
Using Eq. S2, the overall reduction in the UASB reactor was less than 1-log10 unit for all the viruses. The reduction of E. coli in the UASB reactor was significantly greater than the reduction of viruses. F-specific coliphages reduction was significantly greater than the reduction of HAdV DNA, demonstrating that coliphages may not be the best indicators for enteric virus nucleic acids.
A mass balance for the HRAPs showed that more than 99% of all microorganisms left the HRAPs in the liquid phase (Fig. 3b). The overall reductions (Eq. S2) in this stage were higher than they were in the UASB reactor. SARS-CoV-2 RNA, HAdV DNA, F-specific coliphages, and E. coli had log10 reductions of 0.86, 1.04, 1.81 and 1.99, respectively. Reduction of the viral and bacterial indicators was significantly higher than the reduction of SARS-CoV-2 RNA and HAdV DNA in the HRAP.
A mass balance for the complete system (UASB+HRAP) showed that the influent loadings of the different viruses were not significantly different from each other, but the influent E. coli loading was higher than influent loadings for viruses (p < 0.05). The percentage of SARS-CoV-2 RNA and HAdV DNA leaving the reactor in the UASB sludge was 58.3% and 71.5%, respectively, indicating that these viruses may have had affinity to solids in the reactor (Fig. 3c). In contrast, F-specific coliphages showed a lower affinity to solids, with only 3.7% of them leaving the system in the sludge. Regarding E. coli, 37.3% were removed in the sludge and 62.7% were removed in the liquid phase. However, the larger percentage of E. coli leaving in the sludge compared to coliphages may have been due to the higher removal of E. coli observed in the liquid fraction, compared to F-specific coliphages. As mentioned before, viruses can adsorb to particles by electrostatic and hydrophobic interactions. These interactions not only depend on the particle surface and the water composition (Arraj et al., 2005; Verbyla and Mihelcic, 2015), but also on the characteristics of the virus (Arraj et al., 2005). Even if viruses are of the same family, they can present different behaviors (Yin et al., 2018). For instance, the percentage of six types of Echoviruses in the solid phase varied from 67 to 99.5% (Gerba et al., 1980), HAdV in primary and secondary sludge was 75.8 and 67.8%, respectively (Yin et al., 2018), and coliphages ranged from 1 to 99% (Arraj et al., 2005). Our results agree with these observations. However, more studies are needed on the liquid-solid partitioning of different viruses to better understand the viral distribution and removal mechanisms in wastewater treatment plants.
Ali et al. (2021) obtained ∼1-log removal of SARS-CoV-2 RNA in a primary sedimentation tank, indicating that most of SARS-CoV-2 RNA were attached to settled solids. Balboa et al. (2021) also suggested that SARS-CoV-2 RNA is mainly adsorbed to the settled solids by the lipid bilayer surrounding the SARS-CoV-2 protein capsid. Other studies have also detected the SARS-CoV-2 in primary, secondary and anaerobically digested sludge samples (Bhattarai et al., 2021; Peccia et al., 2020), indicating that this enveloped virus may have a higher affinity to solids compared to other waterborne viruses. Our findings support these suggestions, indicating that almost 60% of SARS-CoV-2 RNA leaving the reactor were found in the sludge, despite the fact that the sludge removed accounted for <1% of the volumetric flow leaving the reactor. The removal of SARS-CoV-2 and liquid-solid partitioning in the sludge of UASB reactors has not been reported previously. These results were not strongly affected by our assumptions about non-detect values. When using the half the LOD instead of the LOD for non-detect samples, the percentage of SARS-CoV-2 leaving in the sludge varied only slightly, from 58.3% to 66%. For HAdV the difference was only 1%.
Ye et al. (2016) hypothesized that some enveloped viruses may have a higher affinity for solids compared to non-enveloped viruses and microbial indicators. Our findings do not support this statement since DNA from HAdV (a non-enveloped virus) also showed evidence of having a high affinity to solids, with more than 70% of them leaving the UASB reactor in the sludge, which only accounted for <1% of the volumetric flow leaving the reactor. This finding was also reported by Verbyla (2015), who showed that adenovirus was volumetrically concentrated in the sludge from two UASB reactors in Brazil, one pilot-scale, and one full-scale (the pilot-scale UASB was not the same UASB reactor from the present study). On the other hand, the effluent loadings of the microbial indicators (coliphages and E. coli) support the results presented by Ye et al. (2016).
As summarized by Yin et al. (2018), the sorption of viruses to wastewater solids can be highly variable. For that reason, it is important not only to analyze the removal in the liquid phase but also analyze in the solid phase for each microorganism in wastewater treatment systems. This is especially important when biosolids produced from sludge or biomass extracted from wastewater treatment unit processes is reused for beneficial purposes such as soil amendment (Kumar et al., 2017). The fraction of viruses and E. coli leaving in the microalgae biomass in our study was negligible (≤0.5%). This contrasts with previous findings from the literature, such as Young et al. (2016), who reported that microalgae from HRAPs could influence pathogen removal by increasing adsorption to microalgae biomass. There are tools that can be used to assess virus-solid affinity/adsorption from a mechanistic or genetic perspective, in a controlled laboratory setting. This was not the aim of our study, but previous studies have already reported the affinity of viruses to solids using these approaches (Moore et al., 1975; Ye et al., 2016; Yin et al., 2018).
A low reduction (<0.5-log units) of SARS-CoV-2 RNA and HAdV DNA was observed for the overall system. At the same time, viral and bacterial indicators had significantly higher reductions with values of 2.20 and 2.74-log units, respectively. This echoes previous reports that the concentrations of coliphages quantified by plaque assays may not correlate with the concentrations of enteric viruses detected via (RT)qPCR (Sheludchenko et al., 2016). SARS-CoV-2 RNA and HAdV DNA had very similar behavior throughout the system, but no significant correlation was found between both viruses.
In order to find suitable indicators of microbial risk for wastewater reuse and resource recovery activities, further studies are needed to better understand the factors that influence reduction and liquid - solid partitioning of different viruses for different wastewater treatment technologies.
4. Conclusions
A pilot-scale UASB reactor followed by twin HRAPs treating real domestic wastewater showed high removal of organic matter and nutrients (71% COD and 57% N-NH4 +), but low removal and reduction of viruses, demonstrating the potential need for additional tertiary treatment or disinfection processes if resources are to be safely recovered from systems like these. Furthermore, the results of this study showed that E. coli and F-specific coliphages are both inadequate indicators for the liquid-solid partitioning of enteric viruses. Almost 60% of remaining SARS-CoV-2 RNA and more than 70% of HAdV DNA left the system in the sludge, compared to <5% of coliphages, demonstrating that the human viruses may have a higher affinity for solids than the coliphages. This study demonstrates the importance of analyzing concentrations of pathogens and indicators not only in the liquid phase of wastewater treatment processes, but also in the solid phase. A mass balance approach can be used to compare overall reduction and liquid-solids partitioning for different pathogens and indicators. More studies are needed about liquid-solid partitioning in different treatment systems to better understand differences in the affinity of different viruses and viral surrogates to wastewater solids.
There is a limitation of these results since SARS-CoV-2 RNA and HAdV DNA were quantify using molecular methods (RT-qPCR), while for coliphages and E. coli was used culture methods. Thus, these differences with the detection methods must be considered when analyzing the obtained results. Even with this limitation, the results presented here advance knowledge when it shows the dynamics of removing these viruses and indicators in this treatment system.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the National Science Foundation (USA) under Grant No. 1827251 and the following Brazilian organizations: National Research Council – CNPq; to the National Institute of Science and Technology on Sustainable Sewage Treatment Plants - INCT ETEs Sustentáveis; Coordination for the Improvement of Higher Education Personnel – CAPES; Foundation for Research of the State of Minas Gerais – FAPEMIG (Brazil); the National Health Foundation – FUNASA (Brazil), and the National Agency for Water and Sanitation (ANA). Lucas Vassalle would like to acknowledge CNPq scholarship 204026/2018–0. M.F. Espinosa is co-advised by C.R. Mota Filho and M.E. Verbyla.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2022.118069.
Appendix. Supplementary materials
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.v., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J., Simpson S., Smith W.J.M., Symonds E.M., Thomas K.v., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali H.A., Yaniv K., Bar-Zeev E., Chaudhury S., Shaga M., Lakkakula S., Ronen Z., Kushmaro A., Nir O. Tracking SARS-CoV-2 RNA through the wastewater treatment process. Environ. Sci. Technol. Water. 2021;1:1161–1167. doi: 10.1101/2020.10.14.20212837. [DOI] [PubMed] [Google Scholar]
- Allard A. In: Global Water Pathogen Project. Rose J.B., Jiménez-Cisneros B., editors. UNE, Michigan State University; E. Lansing, MI: 2017. Adenoviruses.http://www.waterpathogens.org/book/adenoviruses (J.S Meschke, and R. G. (eds) P. 3V.(Ed.) (Ed.) [Google Scholar]
- APHA SMWW 9224 Detection of coliphages. 2017 doi: 10.2105/SMWW.2882.195. https://doi.org/ [DOI] [Google Scholar]
- APHA-AWWA-WEF . APHA American Public Health Association, American Water Works Association, Water Environment Federation; 2017. Standard methods for the examination of water and wastewater. [Google Scholar]
- Arora S., Nag A., Sethi J., Rajvanshi J., Saxena S., Shrivastava S.K., Gupta A.B. Sewage surveillance for the presence of SARS-CoV-2 genome as a useful wastewater based epidemiology (WBE) tracking tool in India. Water Sci. Technol. 2020;82:2823–2836. doi: 10.2166/wst.2020.540. [DOI] [PubMed] [Google Scholar]
- Arraj A., Bohatier J., Laveran H., Traore O. Comparison of bacteriophage and enteric virus removal in pilot scale activated sludge plants. J. Appl. Microbiol. 2005;98:516–524. doi: 10.1111/j.1365-2672.2004.02485.x. [DOI] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. The fate of SARS-COV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai B., Sahulka S.Q., Podder A., Hong S., Li H., Gilcrease E., Beams A., Steed R., Goel R. Prevalence of SARS-CoV-2 genes in water reclamation facilities: from influent to anaerobic digester. Sci. Total Environ. 2021;796 doi: 10.1016/j.scitotenv.2021.148905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan N.A., Young P., Cromar N.J., Fallowfield H.J. Performance of a high rate algal pond treating septic tank effluent from a community wastewater management scheme in rural South Australia. Algal. Res. 2018;35:325–332. doi: 10.1016/j.algal.2018.08.036. [DOI] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Calábria J., Mota C.R., Chernicharo C., Leal C., Leroy D., Machado E., Fernandes L., Espinosa M.F., Leão T. Detecção e Quantificação Do Novo Coronavírus Em Amostras de Esgoto Nas Cidades de Belo Horizonte e Contagem - Monitoramento COVID Esgotos. INCT ETEs Sustentáveis; Belo Horizonte: 2020. Quantificação do material genético do novo coronavírus: sensibilidade dos ensaios moleculares e correlação das cargas virais com o número de casos de COVID-19. [Google Scholar]
- CDC . Center for Desease Control and Prevention; 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel.https://www.fda.gov/media/134922/download [WWW Document]URL. [Google Scholar]
- Chernicharo C.A., de L. Reatores Anaeróbios. 2nd ed. DESA-UFMG; Belo Horizonte: 2007. Princípios do tratamento biológico de Águas residuárias. [Google Scholar]
- CONAMA, (Conselho Nacional de Meio Ambiente) Resolução CONAMA N° 430. 2011 http://www.mma.gov.br/port/conama/legiabre.cfm?codlegi=646 [WWW Document]URL. [Google Scholar]
- Davies-Colley R.J., Craggs R.J., Park J., Sukias J.P.S., Nagels J.W., Stott R. Virus removal in a pilot-scale “advanced” pond system as indicated by somatic and F-RNA bacteriophages. Water Sci. Technol. 2005;51:107–110. doi: 10.2166/wst.2005.0440. [DOI] [PubMed] [Google Scholar]
- Delanka-Pedige H.M.K., Cheng X., Munasinghe-Arachchige S.P., Abeysiriwardana-Arachchige I.S.A., Xu J., Nirmalakhandan N., Zhang Y. Metagenomic insights into virus removal performance of an algal-based wastewater treatment system utilizing Galdieria sulphuraria. Algal. Res. 2020;47 doi: 10.1016/j.algal.2020.101865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanka-Pedige H.M.K., Munasinghe-Arachchige S.P., Cornelius J., Henkanatte-Gedera S.M., Tchinda D., Zhang Y., Nirmalakhandan N. Pathogen reduction in an algal-based wastewater treatment system employing Galdieria sulphuraria. Algal. Res. 2019;39 doi: 10.1016/j.algal.2019.101423. [DOI] [Google Scholar]
- Delanka-Pedige H.M.K., Munasinghe-Arachchige S.P., Zhang Y., Nirmalakhandan N. Bacteria and virus reduction in secondary treatment: potential for minimizing post disinfectant demand. Water Res. 2020;177 doi: 10.1016/j.watres.2020.115802. [DOI] [PubMed] [Google Scholar]
- Dias D.F.C., Possmoser-Nascimento T.E., Rodrigues V.a.J., von Sperling M. Overall performance evaluation of shallow maturation ponds in series treating UASB reactor effluent: ten years of intensive monitoring of a system in Brazil. Ecol. Eng. 2014;71:206–214. doi: 10.1016/j.ecoleng.2014.07.044. [DOI] [Google Scholar]
- Dias E., Ebdon J., Taylor H. The application of bacteriophages as novel indicators of viral pathogens in wastewater treatment systems. Water Res. 2018;129:172–179. doi: 10.1016/j.watres.2017.11.022. [DOI] [PubMed] [Google Scholar]
- El-Senousy W.M., Abou-Elela S.I. Assessment and evaluation of an integrated hybrid anaerobic–aerobic sewage treatment system for the removal of enteric viruses. Food Environ. Virol. 2017;9:287–303. doi: 10.1007/s12560-017-9286-4. [DOI] [PubMed] [Google Scholar]
- Espinosa M.F., Verbyla M.E., Vassalle L., Rosa-Machado A.T., Zhao F., Gaunin A., Mota C.R. Reduction and partitioning of viral and bacterial indicators in a UASB reactor followed by high rate algal ponds treating domestic sewage. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.144309. [DOI] [PubMed] [Google Scholar]
- Fallowfield H.J., Young P., Taylor M.J., Buchanan N., Cromar N., Keegan A., Monis P. Independent validation and regulatory agency approval for high rate algal ponds to treat wastewater from rural communities. Environ. Sci. 2018;4:195–205. doi: 10.1039/c7ew00228a. [DOI] [Google Scholar]
- Fong T.T., Phanikumar M.S., Xagoraraki I., Rose J.B. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl. Environ. Microbiol. 2010;76:715–723. doi: 10.1128/AEM.01316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Aljaro C., Blanch A.R., Campos C., Jofre J., Lucena F. Pathogens, faecal indicators and human-specific microbial source-tracking markers in sewage. J. Appl. Microbiol. 2018;126:701–717. doi: 10.1111/jam.14112. [DOI] [PubMed] [Google Scholar]
- Gerba C.P., Goyal S.M., Hurst C.J., Labelle R.L. Type and strain dependence of enterovirus adsorption to activated sludge, soils and estuarine sediments. Water Res. 1980;14:1197–1198. doi: 10.1016/0043-1354(80)90176-1. [DOI] [Google Scholar]
- Gonçalves R.F., Assis T.I., Maciel G.B., Borges R.M., Cassini S.T.A. Co-digestion of municipal wastewater and microalgae biomass in an upflow anaerobic sludge blanket reactor. Algal. Res. 2020;52 doi: 10.1016/j.algal.2020.102117. [DOI] [Google Scholar]
- Gutiérrez-cacciabue D., Cid A.G., Rajal V.B. How long can culturable bacteria and total DNA persist in environmental waters ? The role of sunlight and solid particles. Sci. Total Environ. 2016;539:494–502. doi: 10.1016/j.scitotenv.2015.07.138. The. [DOI] [PubMed] [Google Scholar]
- Guzmán C., Jofre J., Blanch A.R., Lucena F. Development of a feasible method to extract somatic coliphages from sludge, soil and treated biowaste. J. Virol. Methods. 2007;144:41–48. doi: 10.1016/j.jviromet.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Jebri S., Hmaied F., Yahya M., Ammar A.ben, Hamdi M. Total coliphages removal by activated sludge process and their morphological diversity by transmission electron microscopy. Water Sci. Technol. 2016;74:318–323. doi: 10.2166/wst.2016.178. [DOI] [PubMed] [Google Scholar]
- Jothikumar N., Cromeans T.L., Hill V.R., Lu X., Sobsey M.D., Erdman D.D. Quantitative real-time pcr assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 2005;71:3131–3136. doi: 10.1128/AEM.71.6.3131-3136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliakatsos A., Kalogerakis N., Manios T., Venieri D. Efficiency of two constructed wetland systems for wastewater treatment: removal of bacterial indicators and enteric viruses. J. Chem. Technol. Biotechnol. 2019;94:2123–2130. doi: 10.1002/jctb.6001. [DOI] [Google Scholar]
- Katayama H., Shimasaki A., Ohgaki S. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 2002;68:1033–1039. doi: 10.1128/AEM.68.3.1033-1039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Kuroda K., Patel A.K., Patel N., Bhattacharya P., Joshi M., Joshi C.G. Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with Upflow Anaerobic Sludge Blanket (UASB) system evaluated through two sample concentration techniques. Sci. Total Environ. 2021;754 doi: 10.1016/j.scitotenv.2020.142329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Chopra A.K., Kumar A. A review on sewage sludge (biosolids) a resource for sustainable agriculture. Arch. Agricul. Environ. Sci. 2017;2:340–347. doi: 10.26832/24566632.2017.020417. [DOI] [Google Scholar]
- Limsawat S., Ohgaki S. Fate of liberated viral RNA in wastewater determined by PCR. Appl. Environ. Microbiol. 1997;63:2932–2933. doi: 10.1128/aem.63.7.2932-2933.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López C.V.G., del Carmen Cerón García M., Fernández F.G.A., Bustos C.S., Chisti Y., Sevilla J.M.F. Protein measurements of microalgal and cyanobacterial biomass. Bioresour. Technol. 2010;101:7587–7591. doi: 10.1016/j.biortech.2010.04.077. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mohsenpour S.F., Hennige S., Willoughby N., Adeloye A., Gutierrez T. Integrating micro-algae into wastewater treatment: a review. Sci. Total Environ. 2021;752 doi: 10.1016/j.scitotenv.2020.142168. [DOI] [PubMed] [Google Scholar]
- Momba M., Edbon J., Kamika I., Verbyla M. In: Global Water Pathogen Project. Rose J.B., Jiménez-Cisneros B., Farnleitner A., Blanch A., editors. UNESCO, Michigan State University; E. Lansing, MI: 2019. Using indicators to assess microbial treatment and disinfection efficacy. Part 2 Indicators and Microbial Source Tracking Markers. [Google Scholar]
- Moore B.E., Sagik B.P., JF M. Viral association with suspended solids. Water Res. 1975;9:197–203. doi: 10.1016/0043-1354(75)90009-3. [DOI] [Google Scholar]
- Mota C.R., Bressani-Ribeiro T., Araújo J.C., Leal C.D., Leroy-Freitas D., Machado E.C., Espinosa M.F., Fernandes L., Leão T.L., Chamhum-Silva L., Azevedo L., Morandi T., Freitas G.T.O., Costa M.S., Carvalho B.O., Reis M.T.P., Melo M.C., Ayrimoraes S.R., Chernicharo C.A.L. Assessing spatial distribution of COVID-19 prevalence in Brazil using decentralised sewage monitoring. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyola A., Padilla-Rivera A., Morgan-Sagastume J.M., Güereca L.P., Hernández-Padilla F. Typology of municipal wastewater treatment technologies in Latin America. Clean - Soil, Air, Water. 2012;40:926–932. doi: 10.1002/clen.201100707. [DOI] [Google Scholar]
- Oakley S., von Sperling M., Verbyla M. In: Global Water Pathogen Project. Mihelcic J.R., Verbyla M.E., Rose J.B., Jiménez-Cisneros B., editors. UNESCO, Michigan State University; E. Lansing, MI: 2017. Anaerobic Sludge Blanket Reactors. Part 4 Management Of Risk from Excreta and Wastewater. [Google Scholar]
- Pandey D., Verma S., Verma P., Mahanty B., Dutta K., Daverey A., Arunachalam K. SARS-CoV-2 in wastewater: challenges for developing countries. Int. J. Hyg. Environ. Health. 2021;231 doi: 10.1016/j.ijheh.2020.113634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Resende P.C., Motta F.C., Eppinghaus A.L.F., Vale Chagas do, Braz V.H., de Andrade R.M.S., da SR J., Maranhão A.G., Miagostovich M.P. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021;191 doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Silva D.M., Guilayn W.C., Rose T.L., Gaspar A.M.C., Miagostovich M.P. Quantification and molecular characterization of enteric viruses detected in effluents from two hospital wastewater treatment plants. Water Res. 2011;45:1287–1297. doi: 10.1016/j.watres.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Manzano J., Alonso J.L., Ferrús M.A., Moreno Y., Amorós I., Calgua B., Hundesa A., Guerrero-Latorre L., Carratala A., Rusiñol M., Girones R. Standard and new faecal indicators and pathogens in sewage treatment plants, microbiological parameters for improving the control of reclaimed water. Water Sci. Technol. 2012;66:2517–2523. doi: 10.2166/wst.2012.233. [DOI] [PubMed] [Google Scholar]
- Saawarn B., Hait S. Occurrence, fate and removal of SARS-CoV-2 in wastewater: Current knowledge and future perspectives. J. Environ. Chem. Eng. 2021;9 doi: 10.1016/j.jece.2020.104870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago A.F., Calijuri M.L., Assemany P.P., Calijuri M.D.C., Reis A.J.D.dos. Algal biomass production and wastewater treatment in high rate algal ponds receiving disinfected effluent. Environ. Technol. (United Kingdom) 2013;34:1877–1885. doi: 10.1080/09593330.2013.812670. [DOI] [PubMed] [Google Scholar]
- Serra-Compte A., González S., Arnaldos M., Berlendis S., Courtois S., Loret J.F., Schlosser O., Yáñez A.M., Soria-Soria E., Fittipaldi M., Saucedo G., Pinar-Méndez A., Paraira M., Galofré B., Lema J.M., Balboa S., Mauricio-Iglesias M., Bosch A., Pintó R.M., Bertrand I., Gantzer C., Montero C., Litrico X. Elimination of SARS-CoV-2 along wastewater and sludge treatment processes. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheludchenko M., Padovan A., Katouli M., Stratton H. Removal of fecal indicators, pathogenic bacteria, adenovirus, Cryptosporidium and Giardia (oo)cysts in waste stabilization ponds in Northern and Eastern Australia. Int. J. Environ. Res. Public Health. 2016;13 doi: 10.3390/ijerph13010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SNIS, 2019. 25° Diagnóstico dos Serviços de Água e Esgotos.
- Symonds E.M., Verbyla M.E., Lukasik J.O., Kafle R.C., Breitbart M., Mihelcic J.R. A case study of enteric virus removal and insights into the associated risk of water reuse for two wastewater treatment pond systems in Bolivia. Water Res. 2014;65:257–270. doi: 10.1016/j.watres.2014.07.032. [DOI] [PubMed] [Google Scholar]
- Tchobanoglous G., Burton F.L., Stensel H.D. McGraw-Hill; New York: 2003. Wastewater Engineering: Treatment and Reuse. [Google Scholar]
- Vassalle L., Díez-Montero R., Machado A.T.R., Moreira C., Ferrer I., Mota C.R., Passos F. Upflow anaerobic sludge blanket in microalgae-based sewage treatment: co-digestion for improving biogas production. Bioresour. Technol. 2020;300 doi: 10.1016/j.biortech.2019.122677. [DOI] [PubMed] [Google Scholar]
- Vassalle L., García-Galán M.J., Aquino S.F., Afonso R.J.de C.F., Ferrer I., Passos F., R Mota C. Can high rate algal ponds be used as post-treatment of UASB reactors to remove micropollutants? Chemosphere. 2020;248 doi: 10.1016/j.chemosphere.2020.125969. [DOI] [PubMed] [Google Scholar]
- Verbyla M., von Sperling M., Maiga Y. J.B. Rose and B. Jiménez-Cisneros, (eds) Global Water Pathogens (Ed.), J.R. Mihelcic and M.E. Verbyla (Eds), Part 4 Management of Risk from Excreta and Wastewater. UNESCO, Michigan State University; Lansing, MI, USA: 2017. Waste stabilization ponds. [Google Scholar]
- Verbyla, M.E., 2015. Pathogen removal in natural wastewater treatment and resource recovery systems: solutions for small cities in an urbanizing world. https://doi.org/10.1017/CBO9781107415324.004.
- Verbyla M.E., Mihelcic J.R. A review of virus removal in wastewater treatment pond systems. Water Res. 2015;71:107–124. doi: 10.1016/j.watres.2014.12.031. [DOI] [PubMed] [Google Scholar]
- Verbyla M.E., Symonds E.M., Kafle R.C., Cairns M.R., Iriarte M., Mercado Guzmán A., Coronado O., Breitbart M., Ledo C., Mihelcic J.R. Managing microbial risks from indirect wastewater reuse for irrigation in urbanizing watersheds. Environ. Sci. Technol. 2016;50:6803–6813. doi: 10.1021/acs.est.5b05398. [DOI] [PubMed] [Google Scholar]
- Villar-Torres M., Montero F.E., Raga J.A., Repullés-Albelda A. Come rain or come shine: Environmental effects on the infective stages of Sparicotyle chrysophrii, a key pathogen in Mediterranean aquaculture. Paras. Vect. 2018;11:1–19. doi: 10.1186/s13071-018-3139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . In: Water Quality: Guidelines, Standards and Health. Fewtrell Lorna, Bartram Jamie., editors. IWA Publishing; London, UK: 2001. Indicators of microbial water quality; pp. 289–316. [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yin Z., Voice T.C., Tarabara V.v., Xagoraraki I. Sorption of human adenovirus to wastewater solids. J. Environ. Eng. (United States) 2018;144:2–7. doi: 10.1061/(ASCE)EE.1943-7870.0001463. [DOI] [Google Scholar]
- Young P., Buchanan N., Fallowfield H.J. Inactivation of indicator organisms in wastewater treated by a high rate algal pond system. J. Appl. Microbiol. 2016;121:577–586. doi: 10.1111/jam.13180. [DOI] [PubMed] [Google Scholar]
- Yuan Q.bin, Huang Y.M., Wu W.bin, Zuo P., Hu N., Zhou Y.Z., Alvarez P.J.J. Redistribution of intracellular and extracellular free & adsorbed antibiotic resistance genes through a wastewater treatment plant by an enhanced extracellular DNA extraction method with magnetic beads. Environ. Int. 2019;131 doi: 10.1016/j.envint.2019.104986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.