Abstract
Sigma 1 receptor (Sigmar1) is a widely expressed, multitasking molecular chaperone protein that plays functional roles in several cellular processes. Mutations in the Sigmar1 gene are associated with several distal neuropathies with strong manifestation in skeletal muscle dysfunction with phenotypes like muscle wasting and atrophy. However, the physiological function of Sigmar1 in skeletal muscle remains unknown. Herein, the physiological role of Sigmar1 in skeletal muscle structure and function in gastrocnemius, quadriceps, soleus, extensor digitorum longus, and tibialis anterior muscles was determined. Quantification of myofiber cross-sectional area showed altered myofiber size distribution and changes in myofiber type in the skeletal muscle of the Sigmar1−/− mice. Interestingly, ultrastructural analysis by transmission electron microscopy showed the presence of abnormal mitochondria, and immunostaining showed derangements in dystrophin localization in skeletal muscles from Sigmar1−/− mice. In addition, myopathy in Sigmar1−/− mice was associated with an increased number of central nuclei, increased collagen deposition, and fibrosis. Functional studies also showed reduced endurance and exercise capacity in the Sigmar1−/− mice without any changes in voluntary locomotion, markers for muscle denervation, and muscle atrophy. Overall, this study shows, for the first time, a potential physiological function of Sigmar1 in maintaining healthy skeletal muscle structure and function.
Sigma 1 receptor (Sigmar1) is a multifunctional chaperone protein ubiquitously expressed in multiple organs.1, 2, 3, 4, 5, 6 Extensive studies have demonstrated several molecular functions for Sigmar1 in different cell types, including regulation of ion channel function, calcium signaling, mitochondrial function, autophagy, and lipid transport.7, 8, 9, 10 Sigmar1 was discovered over 40 years ago.11,12 Since then, several recessive mutations in Sigmar1 have been associated with amyotrophic lateral sclerosis (ALS), distal hereditary motor neuropathy (dHMN), frontotemporal lobe degeneration, and Silver-like syndrome.
Genetic studies of Sigmar1 have demonstrated an association between Sigmar1 mutations and ALS pathology. The clinical hallmarks of ALS pathology include progressive muscle wasting, speech and swallowing difficulties, fasciculation, altered reflexes, spasticity, and death due to respiratory complications.13 Juvenile cases of ALS have been associated with a missense mutation (c.304G>C, p.E102Q)14 and a frameshift mutation (c.283dupC, p.L95 fs)15 in Sigmar1. Progressive development of skeletal muscle pathology was observed in patients with E102Q mutations, including weakness of the hand and forearm muscles (at the age of 9 to 10 years), leading to paralysis of the forearm extensors and triceps. These patients had no respiratory or bulbar muscle weakness and demonstrated normal sphincteric, sensory, and cerebellar functions.14 Similarly, a patient with the L95 fs mutation developed progressive muscle weakness, with significant atrophy of distal muscles with development of pes cavus and wasting of the calf muscles and intrinsic muscles of the hands.15 Interestingly, examination of vastus lateralis muscle biopsy revealed severe type II fiber predominance, with scattered angular esterase-positive fibers, and showed intense staining with NADH tetrazolium reductase.15 Patients with these mutations show normal brain and spinal cord magnetic resonance imaging.14,15
These clinical skeletal muscle phenotypes, all of which were observed in Sigmar1 mutation-bearing patients, have also been observed in patients with dHMN. In fact, several of the truncations/deletions or point mutations that occur in Sigmar1 have also been reported in association with the development of dHMN.16, 17, 18, 19, 20, 21 dHMN neuropathies comprise a heterogeneous group of diseases accompanied by the common features of slowly progressive, symmetrical, and distal-predominant neurogenic weakness and amyotrophy. Patients with dHMN with the Sigmar1 mutations manifest identical clinical features: progressive muscle wasting/weakness in the lower and upper limbs without sensory loss16, 17, 18, 19, 20, 21 accompanied by normal brain and spine as determined by magnetic resonance imaging.17
Studies have also shown an association between Sigmar1 mutations in the 3′-untranslated region and the frontotemporal lobar degeneration–motor neuron disease–pathologic ubiquitinated inclusion bodies. Although Sigmar1 normally localizes to cytoplasmic membranes in healthy individuals, c.672∗51G>T carriers show intense Sigmar1 immunoreactivity in the nucleus dentate granule and cornu ammonis 1 (CA1) pyramidal cells. However, the details of the clinical features in these patients remain unknown. Patients bearing a homozygous missense variant (c.194T>A, p.Leu65Gln) of Sigmar1 have been associated with autosomal recessive Silver-like syndrome.18 In a patient with this Sigmar1 mutation, the clinical features include bilateral foot drop and frequent falls (at approximately 3 years of age), with the development of progressive muscle weakness and atrophy in the lower limbs. By the age of 17 years, this patient had developed clawed hands with no fixed contractures, bilateral finger and foot drop, knee bobbing, marked muscle atrophy from the midforearms and knees down, and weakness of wrist extension. However, the patient had a normal intellect, no sensory symptoms, and no sphincter problems, with normal brain and spinal cord as indicated by magnetic resonance imaging.
Despite the evidence in these reports, proof of a direct association between mutations in Sigmar1 and human diseases remains elusive, as this association has only been identified in small, isolated families with limited genetic and functional studies. Functional studies performed to determine the possible molecular mechanism showed that ALS-associated Sigmar1 mutations (p.E102Q and p.L95 fs)14,15 were uniformly unstable and nonfunctional when expressed in Neuro2a cells, suggesting that Sigmar1's loss of function plays a role in ALS.14,15 Moreover, expression of the Sigmar1E102Q in Drosophila lacking a Sigmar1 homolog alters locomotor activity and eye development.22 Functional studies using two of dHMN-associated mutations (p.E138Q and p.E150K) in several neuronal cell lines (two human neuroblastoma cell lines, SH-SY5Y and SK-N-BE, and the murine motor neuron-like NSC-34 line) suggests that the pathogenicity of the mutations may involve the alterations in endoplasmic reticulum–mitochondria tethering, calcium homeostasis, and autophagy. The presence of the c.672∗26C>T, c.672∗47G>A, and c.672∗51G>T mutations within the 3′-untranslated region of SIGMAR1 affects transcript stability, resulting in increased Sigmar1 transcript in human neuroblastoma SK-N-MC and HEK293 cells.23 Although studies using Sigmar1 global knockout mice (Sigmar1−/−) have provided a molecular tool to aid in understanding the physiological function of Sigmar1,8,24 these mice do not show any pathologic phenotype associated with the human diseases observed in patients with Sigmar1 mutations. The neuronal dysfunctions reported in Sigmar1−/− mice were locomotor defects,25 nerve denervation,8 loss of motor neurons,8 age-dependent motor dysfunction,15 and development of depressive-like behavior.24,26
The most common clinical feature observed in patients with Sigmar1 mutations is muscle weakness caused possibly by upper or lower motor neuron injury, resulting in denervation, or as the result of myofiber injury, resulting in a primary myopathy process. However, the physiological function of Sigmar1 in the skeletal muscle has not been studied and remains elusive. This study evaluated the physiological role of Sigmar1 in skeletal muscle physiology and function using Sigmar1−/− mice. Extensive evaluations were performed to determine the functional consequences of Sigmar1 ablation in skeletal muscle morphology, histology, ultrastructure, and function in different muscle fibers using Sigmar1−/− mice. This study demonstrated the altered myofiber cross-sectional area (CSA), fibrotic remodeling, presence of abnormal mitochondria, and reduced endurance and exercise capacity in Sigmar1−/− mice compared with wild-type (Wt) littermate controls.
Materials and Methods
Animals
The 9- to 10-month–old global Sigmar1 knockout (Sigmar1−/−) mice were used, as previously reported,8,9 along with their littermate controls, Wt mice on the C57BL/6 background strain. The mice were accommodated in a well-controlled environment in cages following a 12-hour light-dark cycle and provided with water and a regular chow diet ad libitum. Equal numbers of both male and female mice, consisting of Sigmar1−/− and Wt mice, were used for the experiments. The animal handling procedures were in accordance with the Guide for the Care and Use of Laboratory Animals,27 with the protocols being approved by the Animal Care and Use Committee of Louisiana State University Health Sciences Center-Shreveport. The animals were cared for according to the NIH Guide for the Care and Use of the Laboratory Animals.
Muscle Isolation and Morphometry
Mice from both the groups (Wt and Sigmar1−/−) were subjected to isoflurane-mediated anesthesia. Five different muscles, gastrocnemius (Gastro), quadriceps (Quad), soleus (Sol), tibialis anterior (TA), and extensor digitorum longus (EDL), were isolated from both limbs of each group of mice and were processed according to the experimental requirements. For morphometric analysis, the isolated muscles were washed with 1× phosphate-buffered saline (PBS) to remove any fur and any fat or bloodstains. The tissues were soaked using clean paper towels to remove any extra PBS, and the wet tissue weights were taken using an OHAUS electronic balance (OHAUS Corporation, Parsippany, NJ).
Protein Isolation and Western Blot Analysis
Total proteins were prepared from all five different muscles isolated from both Wt and Sigmar1−/− mice. The isolated muscles were lysed with Cell Lytic M (Sigma-Aldrich, St. Louis, MO) lysis buffer supplemented with Complete Protease Inhibitor Cocktail (Roche, Basel, Switzerland), as described previously.28,29 All the muscle sections were homogenized twice using bead homogenizer followed by sonication. The muscle homogenates were then centrifuged at 12,000 × g for 15 minutes to sediment the insoluble cell debris. The protein concentration of the muscle homogenate was measured using the Bradford protocol/reagent (Bio-Rad, Hercules, CA) relative to a bovine serum albumin standard curve (Bio-Rad). Protein samples were then separated on SDS-PAGE using precast 5% to 12% Criterion gels (Bio-Rad) and transferred to polyvinylidene difluoride membranes (Bio-Rad). Membranes were blocked for 1 hour in 5% nonfat dried milk and exposed to primary antibodies overnight. The following primary antibodies were used for immunoblotting: Sigmar1 (1:1000; 61994; Cell Signalling, Danvers, MA), glyceraldehyde-3-phosphate dehydrogenase (1:10,000; MAB374; EMD Millipore, Burlington, MA), β-actin (1:1000; sc-47778; Santa Cruz Biotechnology, Dallas, TX), muscle RING-finger protein-1 (MuRF1; 1:200; sc-398608; Santa Cruz Biotechnology), and desmin (1:5000; V2022; Biomeda, Foster City, CA). Subsequently, membranes were washed, incubated with alkaline phosphatase–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), developed with ECF reagent (Amersham, Amersham, UK), and imaged using a Chemidoc Touch Imaging System (Bio-Rad). Densitometric analysis of the protein bands on the scanned image was performed using ImageJ software version 1.53c (NIH, Bethesda, MD: https://imagej.nih.gov/ij).
RNA Isolation and Quantitative Real-Time PCR Analysis
Total RNA was isolated from all five different skeletal muscles from Wt and Sigmar1−/− mice using TRI reagent (RN-190; Molecular Research Center, Cincinnati, OH) following manufacturer's protocol, as previously described.9,28 The concentration and purity of the isolated RNA were measured using NanoDrop (DeNovix, Wilmington, DE; DS-11 FX+) followed by first-strand cDNA synthesis. Quantitative real-time PCR was done by the CFX-96 instrument (Bio-Rad) to measure Sigmar1 and acetylcholine receptor α (AchRα) abundance in all five skeletal muscles using TaqMan probes (Applied Biosystems, Foster City, CA) and SYBR Green mastermixes (Applied Biosystems), respectively. All data were normalized to β-actin content and expressed as fold change. The primers used for AchRα and β-actin are as follows: AchRα, 5′-CCACAGACTCAGGGGAGAAG-3′ (forward) and 5′-AACGGTGTGTGTTGATG-3′ (reverse); and β-actin, 5′-CTGTCGAGTCGCGTCCACC-3′ (forward) and 5′-TCGTCATCCATGGCGAACTGG-3′ (reverse).
WGA Staining
Wheat germ agglutinin (WGA) staining was used to measure the CSA of the myofibers of different muscles, as described previously.9,29 Briefly, all five different muscles isolated from Wt and Sigmar1−/− mice were fixed in 10% buffered formalin and embedded in paraffin. These paraffin-embedded blocks were cut into 5 μm serial sections, deparaffinized, and hydrated, followed by antigen retrieval by boiling at 100°C in 10 mmol/L sodium citrate buffer (pH 6.0). Subsequently, the sections were blocked for 1 hour with blocking solution (1% bovine serum albumin, 0.1% cold water fish skin gelatin, and 1% Tween 20 in PBS) at room temperature. This was followed by incubation with Alexa Fluor 488 wheat germ agglutinin (5 μg/mL; Invitrogen) for 1 hour at room temperature. The nuclei were counterstained with DAPI (Invitrogen) for 5 minutes. The stained slides were washed with 1× PBS and then mounted using Vectashield Hardset antifade mounting media for fluorescence (Vector Laboratories). The stained sections were then subsequently observed using Nikon A1R high-resolution confocal microscope (Nikon Instruments Inc., Melville, NY) and imaged with Nikon NIS elements software version 4.13.04 with a 20× objective lens. All image acquisition was performed in an investigator-blinded manner. Calculations were performed using 3000 to 4000 myocytes from 4 to 28 images for each muscle from each mice group; these were selected in a blinded manner and used to calculate the average CSA of the muscle fiber using ImageJ software. Central nuclei were quantified by counting the number of DAPI-positive stains inside the myocyte using ImageJ software version 1.53c.
Immunostaining
The serial cut sections (5 μm thick) were used from paraffin-embedded blocks of all five skeletal muscles from Wt and Sigmar1−/− mice to detect the presence of Sigmar1 and evaluate dystrophin expression in muscle sections, as described previously.28,29 Briefly, the muscle sections were deparaffinized, hydrated, and subjected to antigen retrieval by boiling at 100°C in 10 mmol/L sodium citrate buffer (pH 6.0) for 30 minutes. The sections were treated with an enhancer for 30 minutes following 1× PBS wash. The enhanced tissue sections were then washed and blocked with a blocking buffer (1% bovine serum albumin, 0.1% cold water fish skin gelatin, and 1% Tween 20 in PBS) for 1 hour at room temperature. The blocked sections were then incubated with primary antibodies overnight at 4°C in a humidified chamber followed by Alexa Fluor conjugated dyes (1:100) for 1 hour 30 minutes at room temperature in a humidified chamber. Subsequently, the sections were incubated with either second primary antibodies (1 hour 30 minutes) or WGA (5 μg/mL; Invitrogen; 1 hour) at room temperature. Following second antibody incubation, the sections were incubated with Alexa Fluor conjugated secondary antibody (1:100) for 1 hour 30 minutes. All the stained sections were exposed to DAPI (Invitrogen) for 5 minutes at room temperature and mounted with Vectashield Hardset antifade mounting media for fluorescence. The stained sections were then observed using a Nikon A1R high-resolution confocal microscope and imaged with Nikon NIS elements software version 4.13.04 with a 20× objective lens. All image acquisition was performed in an investigator-blinded manner. The primary antibodies used were Sigmar1 N-terminal antibody (1:100; OAAB01426; Aviva, San Diego, CA), Sigmar1 C-terminal antibody (1:100; 61994; Cell Signalling), dystrophin (1:100; D8168; Sigma-Aldrich), α-sarcomeric actinin (skeletal muscle marker; 1:100; A7811; Sigma-Aldrich, San Diego, CA), and OXPHOS (mitochondrial marker; 1:100; ab110413; Abcam, Cambridge, UK). The secondary antibodies used were Alexa Fluor 488 (A11034) and Alexa Fluor 568 (A11031) from Invitrogen.
Muscle Preparation and Myosin Isotype Staining
Myosin isotype staining was done to evaluate the myofiber types in skeletal muscles from Wt and Sigmar1−/− mice, as described previously.30 Briefly, Gastro and Sol isolated from Wt and Sigmar1−/− mice were embedded in O.C.T. compound (Tissue-Tek, Torrance, CA). The embedded blocks were frozen using liquid nitrogen and stored at −80°C. The frozen blocks were cut in serial sections of 20 μm using a cryostat (Leica, Wetzlar, Germany; CM3050 S) maintained at −20°C. To stain for myosin isotypes, the muscle sections were air dried for 10 minutes, followed by blocking with a blocking buffer (10% goat serum in 1× PBS) for 1 hour at room temperature. Subsequently, the blocked sections were incubated with primary antibodies BA-F8 (Myosin Type I; 1:50; DSHB, Iowa City, IA), SC-71 (Myosin Type IIA; 1:600; DSHB), and BF-F3 (Myosin Type IIB; 1:200; DSHB) for 2 hours at room temperature and 6H1 (Myosin Type IIX; 1:50; DSHB) for overnight at 4°C in a humidified chamber. Subsequently, these myosin types I, IIA, IIB, and IIX stained sections were incubated with Alexa Fluor 647, 568, and 555 conjugated goat anti-mouse IgG2b (1:100), IgG (1:500), and IgM (1:700 for Myosin IIB and 1:100 for Myosin IIX) secondary antibodies for 1 hour 30 minutes at room temperature. Subsequently, the stained sections were incubated with Alexa Fluor 488 conjugated WGA (5 μg/mL; Invitrogen) for 1 hour at room temperature. The stained sections were then mounted with Vectashield Hardset antifade mounting media for fluorescence. The stained sections were observed using a Leica TCS SP5 spectral confocal microscope using a 10× objective and imaged using Leica LAS (AF 2.6.3) software. All image acquisition was performed in an investigator-blinded manner.
The myofiber type content was calculated 800 to 900 myocytes for soleus and 3000 to 4000 myocytes for gastrocnemius from each mice group. The images were selected in a blinded manner and quantified using ImageJ software version 1.53c.
Transmission Electron Microscopy
Both Wt and Sigmar1−/− mice were anesthetized with isoflurane, and all five muscles (Gastro, Quad, Sol, TA, and EDL) were collected and cut into small cubes (1 mm3). The cut sections were fixed overnight in 3% glutaraldehyde in 0.1 mol/L sodium cacodylate buffer followed by post-fixation with 1% OsO4 and counterstained with uranyl acetate and lead salts. The sections were embedded using low-viscosity epoxy resins. The thin cut sections were imaged in an investigator-blinded manner using a JEOL JEM-1400 transmission electron microscope (JEOL, Peabody, MA) with an Advanced Microscopy Techniques digital camera (Woburn, MA).
Mitochondrial DNA Analysis
Mitochondrial DNA was measured to determine changes in mitochondrial DNA copy number in all five skeletal muscles from Wt and Sigmar1−/− mice, as described previously.31 Briefly, the total DNA was isolated from all five skeletal muscles using the Quick-gDNA MiniPrep kit (11 to 317B; Zymo Research, Irvine, CA), according to the manufacturer's protocol. The mitochondrial DNA was measured using PCR for cytochrome b (mitochondrial DNA marker) and β-actin (genomic DNA marker). The primers used for cytochrome b and β-actin are as follows: cytochrome b, 5′-CCACTTCATCTTACCATTTATTATCGC-3′ (forward) and 5′-TTTTATCTGCATCTGAGTTTAA-3′ (reverse); and β-actin, 5′-CTGCCTGACGGCCAGG-3′ (forward) and 5′-CTATGGCCTCAGGAGTTTTGTC-3′ (reverse).
Histologic Analysis
Paraffin-embedded blocks for all five different muscles collected from Wt and Sigmar1−/− mice were cut into serial sections (5 μm thick). The sections were deparaffinized, hydrated, and stained with picro-sirius red and Masson trichrome, as previously described,28,29,31 and hematoxylin and eosin (H&E staining kit; H3502; Vector Laboratories) following the manufacturer's protocol. The stained sections were imaged in an investigator-blinded manner using an Olympus BX40 microscope (Tokyo, Japan) in bright-field mode with a 20× objective. The collagen deposition and fibrosis in the muscle sections were quantified using NIH ImageJ software, as described previously.9 Briefly, the quantification for collagen deposition was done by quantifying the red-stained area and nonmyocyte area from each section using color-based thresholding. Fibrosis quantification was done by quantifying blue-stained area and nonmyocyte area from each section using color-based thresholding. The amount of collagen deposition and total fibrosis area was calculated as a percentage of the red-stained area and blue-stained area, respectively, with respect to the total area for each section. H&E staining was used to show the presence of central nuclei in the histologic sections of different muscles from Wt and Sigmar1−/− mice. A total of 4 to 12 high-magnification ×20 images were used for quantification per muscle in each group for each mouse.
Muscle Exercise Tolerance and Endurance Capacity
To assess skeletal muscles' endurance capacity and exercise tolerance, mice were subjected to a grip strength test and treadmill exercise, respectively. Endurance of the skeletal muscle was assessed by measuring forelimb grip strength, as described previously,32,33 using an equal number of mice (n = 9) from both Wt and Sigmar1−/− groups. Briefly, before the tests, mice were acclimated to the experiment room for at least 10 minutes. They were then placed on the mesh grid attached to the grip strength meter (1027SM; Columbus Instruments, Columbus, OH) for the force measurement, and were allowed to hold the grid using their forelimbs. The mice were placed parallel to the grid, held by the tails, and slowly pulled away from the grid. The gauge recorded the tension generated by the mice at the release of the grid. The mice were allowed to rest for 1 minute and were subjected to another trial. Each mouse was subjected to five trials. The data were recorded and analyzed using the grip strength meter software provided by the manufacturer.
An equal number of mice (n = 9) from both Wt and Sigmar1−/− groups were subjected to graded maximal exercise testing, as described previously.34 Briefly, the mice were acclimated to the treadmill (OxyletPro, Panlab; Harvard Apparatus, Holliston, MA), which included three training sessions with 60 hours of recovery time followed by rest for 1 week. During the acclimation period, mice were placed on the motionless treadmill for 3 minutes, followed by activation of the shock grid (1.5 mA). Thereafter, the treadmill was engaged to a walking speed of 6 m/minute for 5 minutes and progressively increased up to 12 m/minute for a total duration of 12 minutes of exercise. After 1 week of rest from acclimation training, mice were placed on the treadmill at 0-degree incline, and the shock grid was activated. The treadmill speeds were then increased until exhaustion, as previously described34: speed, duration, and grade: 0 m/minute, 3 minutes, 0 degrees; 6 m/minute, 2 minutes, 0 degrees; 9 m/minute, 2 minutes, 5 degrees; 12 m/minute, 2 minutes, 10 degrees; 15 m/minute, 2 minutes, 15 degrees; 18, 21, 23, and 24 m/minute, 1 minute, 15 degrees; and +1 m/minute, each 1 minute thereafter, 15 degrees. Exhaustion criteria were defined as mice spending >5 seconds on the shock grid or getting >10 shocks. The data were analyzed for exercise tolerance using Metabolism version 3.0 software (Harvard Apparatus).
Voluntary Locomotion
Total distance was measured to assess the voluntary locomotion of Wt and Sigmar1−/− mice using an open field Plexiglass chamber, as previously described.35 Briefly, the mice were allowed to acclimate in the testing room for 30 minutes before the test. The mice were then individually placed in a testing chamber (length × width × height: 45 × 45 × 30 cm) and were allowed to freely move for 30 minutes. The movement of the mice was recorded as a video using a camcorder. The recorded video was then analyzed for total locomotor activity using TopScan Lite Software 2.0 (CleverSys Behavior Recognition, Reston, VA).
Statistical Analysis and Reproducibility
All in vivo studies were performed with the investigators blinded to the mouse groups. A numerical ear tagging system was used for unbiased data collection. For all imaging studies, paraffin blocks, slides, and acquired microscopic images were labeled alphanumerically. Following the completion of the study, individual mouse identifier and image identifier numbers were cross-referenced with treatment to permit analysis. All statistical analyses were conducted in GraphPad Prism software version 8.4.1 (La Jolla, CA). Data are presented in graphs showing median and interquartile ranges. A two-tailed, unpaired t-test (for two groups) was used, followed by the Tukey multiple-comparisons post hoc test. P < 0.05 (95% CI) were considered significant.
Results
Sigmar1 Expression and Localization in Myofibers
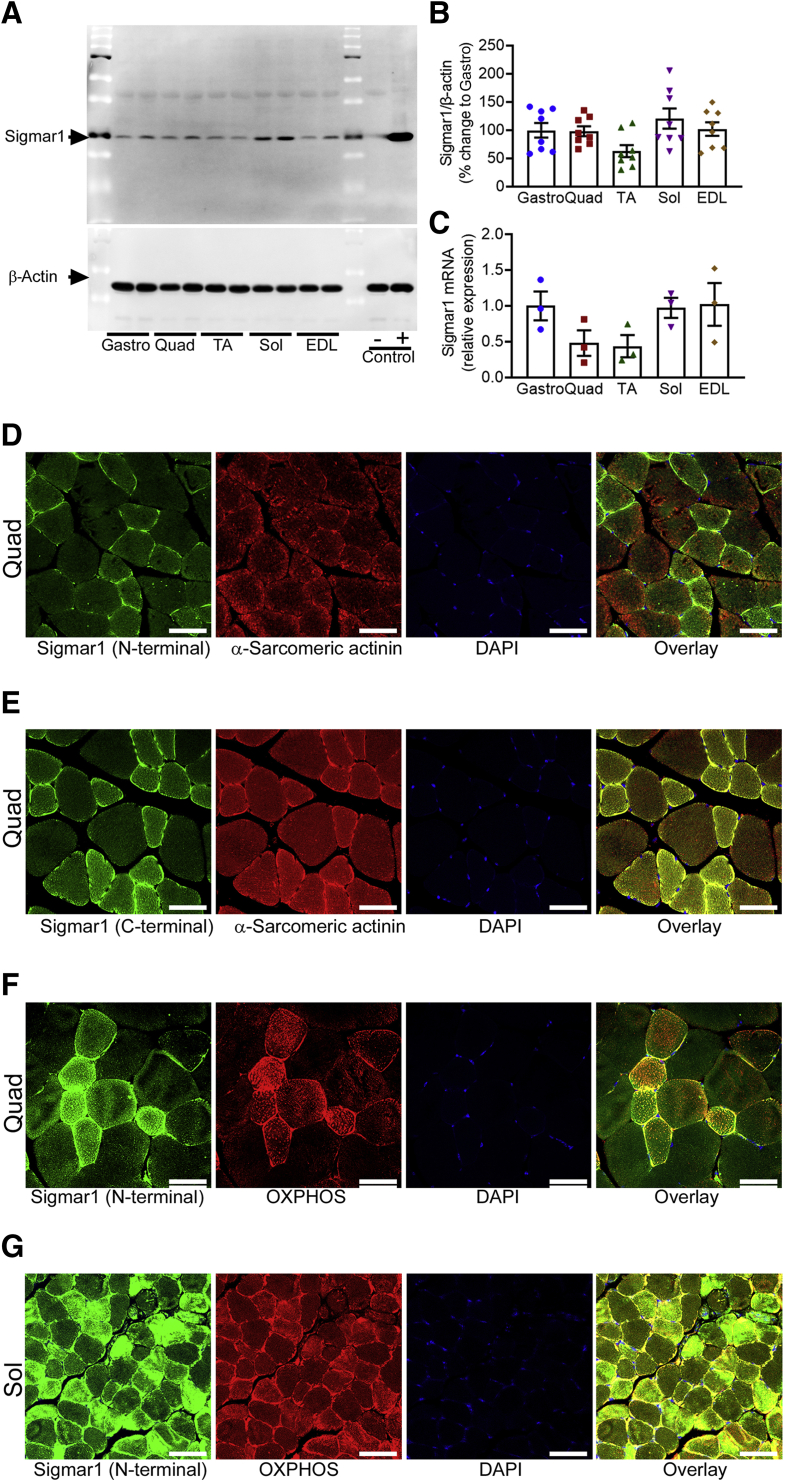
Sigmar1 is a molecular protein widely expressed in mammalian cell systems. In this study, Sigmar1 expression level was first confirmed in five different skeletal muscles, including Gastro, Quad, TA, Sol, and EDL isolated from Wt mice. Sigmar1 protein levels quantified by Western blot analysis (Figure 1, A and B) and mRNA expression levels (Figure 1C) determined using real-time quantitative PCR showed differential expression of Sigmar1 in all five muscles. Next, immunofluorescence was used to visualize Sigmar1 localization and expression pattern in myofibers from quad muscle sections with anti-Sigmar1 antibody targeting the N-terminal (Figure 1D) and C-terminal (Figure 1E) of Sigmar1. Sarcomere-associated anti–α-sarcomeric actinin co-immunostaining was used as a marker for myofibers. Anti-Sigmar1 staining revealed differential expression of Sigmar1 across the myofibers, and some fibers showed a comparatively intense anti-Sigmar1 fluorescence staining (in green). Skeletal muscle fibers consist of slow-twitch, oxidative myofibers (type I) and fast-twitch, glycolytic myofibers (type II).36,37 To confirm the expression of Sigmar1 in mitochondria-rich myofibers, Quad and Sol muscles were stained with N-terminal targeted anti-Sigmar1 antibody and counterstained with an anti-OXPHOS (Figure 1, F and G) antibody. Interestingly, anti-Sigmar1 immunostaining appeared at higher fluorescence intensity (in green) in mitochondria-rich (in red) myofibers (Figure 1, F and G). These data confirmed the expression of Sigmar1 across different types of skeletal muscles and indicated the higher expression of Sigmar1 in mitochondria-rich myofibers.
Figure 1.
Expression and localization of Sigmar1 in the skeletal muscle. A: Representative Western blot analysis images demonstrating Sigmar1 protein level in the gastrocnemius (Gastro), quadriceps (Quad), tibialis anterior (TA), soleus (Sol), and extensor digitorum longus (EDL) muscles isolated from 9- to 10-month–old wild-type (Wt) mice. Cell lysates of Quad muscle isolated from Sigmar1 global knockout (Sigmar1−/−) and Sigmar1-overexpressing transgenic mice were used as a negative and positive control for Sigmar1 protein, respectively. β-Actin was used to verify equal protein loading across the lanes. B and C: Bar graphs represent Sigmar1 protein and mRNA expression levels in the Gastro, Quad, TA, Sol, and EDL muscles extracted from 9- to 10-month–old Wt mice. Dots in the bar graphs represent individual values quantified for muscle. Data are expressed as fold change of Gastro muscle. D and E: Representative immunofluorescence staining for Sigmar1 using antibodies directed against N-terminal and C-terminal of Sigmar1 (green), and α-sarcomeric actinin (red) counterstaining showed expression of Sigmar1 in skeletal muscle fibers in Quad muscle of Wt mouse. F and G: Representative immunofluorescence staining for Sigmar1 (green) with OXPHOS (red) counterstaining showed Sigmar1 fluorescent intensity relatively higher in mitochondria-rich myofibers in Quad and Sol muscles of Wt mouse, respectively. D and G: Quad muscles (D) and Sol muscles (G) isolated from 9- to 10-month–old Wt mice were used for immunostaining. Images are representative of two biological replicates (10 to 12 microscopic fields). DAPI staining was used to counterstain the nucleus. Data are expressed as means ± SEM (B and C). n = 4 mice per skeletal muscle type (A); n = 8 mice per muscle for Western blot analyses (B and C); n = 3 mice per muscle for mRNA (B and C). Scale bars = 50 μm (D–G).
Myofibers from Sigmar1−/− Mice Exhibit Altered Myofiber Areas in Skeletal Muscles
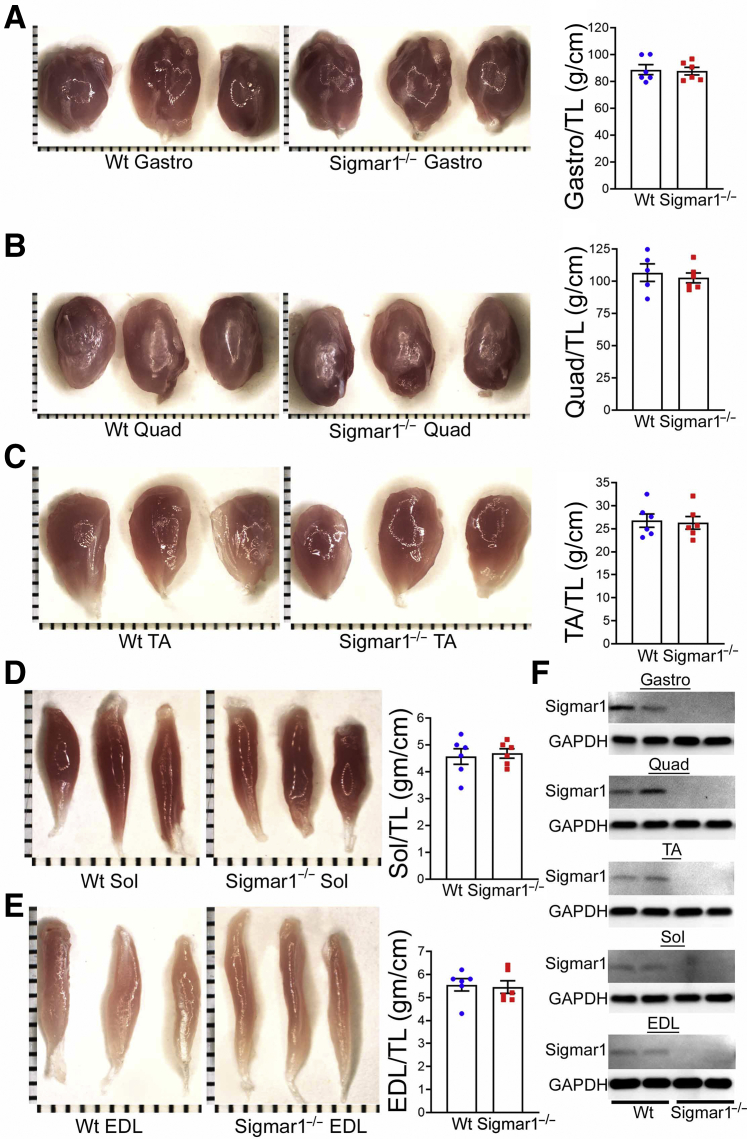
Mammalian skeletal muscle is composed of remarkably heterogeneous myofibers in terms of major contractile motor units, metabolic enzyme activity, and mitochondrial abundance to support a wide variety of physical demands by generating force and movement needed to maintain posture, respiration, locomotion, and intense activities. To investigate whether Sigmar1 plays physiologically essential functions in maintaining normal muscle morphology, histology, or functions in adulthood, genetic Sigmar1 global knockout (Sigmar1−/−) mice were used with respective age-matched littermate Wt mice. Gross and microscopic morphometry of Gastro, Quad, TA, Sol, and EDL muscles, isolated from Wt and Sigmar1−/− mice, was assessed (Figure 2, A–E). Morphologically, the isolated muscles showed no visible difference and had similar weights. Measurements of muscle weight/tibia length ratio by gravimetric studies in freshly isolated skeletal muscles from Wt and Sigmar1−/− mice showed similar muscle mass (Figure 2, A–E). The ablation of Sigmar1 protein in all skeletal muscles isolated from Sigmar1−/− mice was confirmed by Western blot analysis (Figure 2F).
Figure 2.
Morphometric analyses of skeletal muscles in wild-type (Wt) and Sigmar1−/− mice. A–E: Left panels: Gross morphologic microscopic images of gastrocnemius (Gastro), quadriceps (Quad), tibialis anterior (TA), soleus (Sol), and extensor digitorum longus (EDL) muscles isolated from 9- to 10-month–old Wt and Sigmar1−/− mice. Right panels: Bar graphs representing muscle weight/tibia length (TL) ratio of Gastro, Quad, TA, Sol, and EDL muscles. Dots in the bar graphs represent individual values quantified for each Wt and Sigmar1−/− mouse. P values were determined by unpaired t-test. F: Representative Western blot analysis images of Sigmar1 protein in muscle tissue lysates isolated from Wt and Sigmar1−/− mice showing the absence of Sigmar1 in the muscles isolated from Sigmar1−/− mice. Data are expressed as means ± SEM (A–E). n = 6 mice for each muscle per genotype isolated from both of the limbs (A–E); n = 6 mice for each muscle per genotype (F). Scale bars (shown by each small bar across x and y axes) = 1 mm (A–E). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
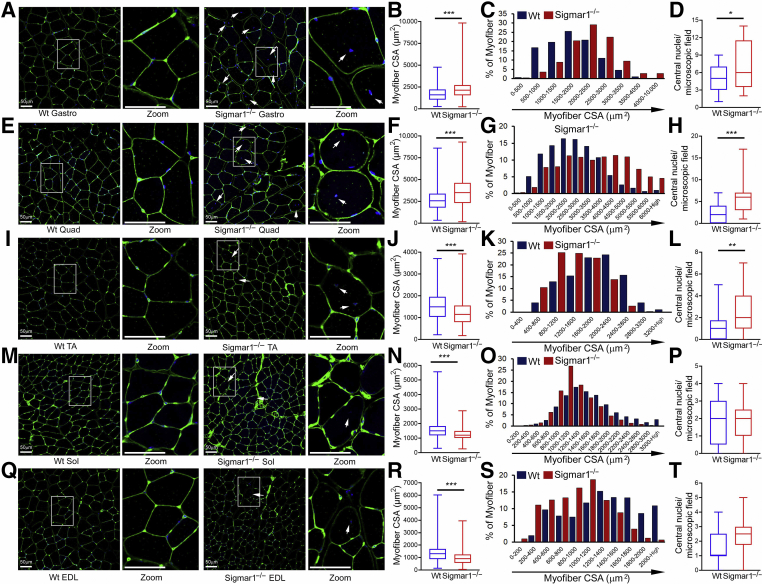
Myofiber CSAs were measured using WGA staining of the histologic sections of skeletal muscles isolated from Wt and Sigmar1−/− mice (Figure 3). Interestingly, the quantified myofibers’ CSA in Sigmar1−/− mice showed differential changes, depending on the anatomically distinct muscle types compared with Wt mice. Gastro (Figure 3, A–C) and Quad (Figure 3, E–G) myofibers in Sigmar1−/− mice showed a significant increase in myofiber CSA with a clear shift toward larger size, as shown by their respective size distribution in muscle sections. However, TA (Figure 3, I–K), Sol (Figure 3, M–O), and EDL (Figure 3, Q–S) myofibers showed a significant decrease in mean myofiber CSA with a shift toward smaller size, as shown by their respective size distribution in muscle sections from Sigmar1−/− mice compared with Wt mice. Central nuclei in skeletal muscles are associated with different myopathies, developmental abnormalities, or muscle regeneration.38, 39, 40, 41 Sigmar1−/− mice showed increased myofiber central nuclei in Gastro (Figure 3D), Quad (Figure 3H), and TA (Figure 3L) muscles compared with the Wt mice. However, the central nuclei number did not change significantly across groups in the Sol (Figure 3P) and EDL (Figure 3T) (although it showed an increasing pattern in EDL). Overall, the Sigmar1−/− mice showed hypertrophied myofibers in the Gastro and Quad muscle sections and an increased number of myofiber central nuclei in Gastro, Quad, and TA muscle sections compared with Wt mice muscles.
Figure 3.
Altered myofiber size, presence of central nuclei, and fiber size distribution in Sigmar1−/− muscles compared with wild-type (Wt) muscles. Representative immunofluorescence images of wheat germ agglutinin (WGA) stained (green) gastrocnemius (Gastro; A), quadriceps (Quad; E), tibialis anterior (TA; I), soleus (Sol; M), and extensor digitorum longus (EDL; Q) muscle cross-sections from Wt and Sigmar1−/− mice. WGA was used to delineate myofiber boundaries, and DAPI was used to counterstain nuclei. Rectangle (white boxed areas) enclosed areas are presented to the right as digitally magnified areas to respective myocyte types, demonstrating the increased presence of central nuclei (white arrows) in Sigmar1−/− mice myocytes. Box plots representing average myocyte cross-sectional areas (CSAs; μm2) for Gastro (B), Quad (F), TA (J), Sol (N), and EDL (R) muscle cross-sections from Wt and Sigmar1−/− mice. Bar graphs represent myofiber cross-sectional area distribution frequency in Gastro (C), Quad (G), TA (K), Sol (O), and EDL (S) muscles of Wt and Sigmar1−/− mice. Cross-sectional areas were measured in 3000 to 4000 myocytes per muscle type in each mouse. Box plots representing the number of central nuclei counts per microscopic field in Gastro (D), Quad (H), TA (L), Sol (P), and EDL (T) muscle cross-sections from Wt and Sigmar1−/− mice. Central nuclei were counted in 4 to 28 high-magnification (depending on the muscle size) microscopic fields (×20) per genotype for each mouse. Boxes represent interquartile ranges, lines represent medians, and whiskers represent ranges. P values were determined by unpaired t-test. n = 3 mice per genotype at the age of 9 to 10 months. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bars = 50 μm (A, E, I, M, and Q).
Changes in Myofiber Type of Sigmar1−/− Mice Muscles
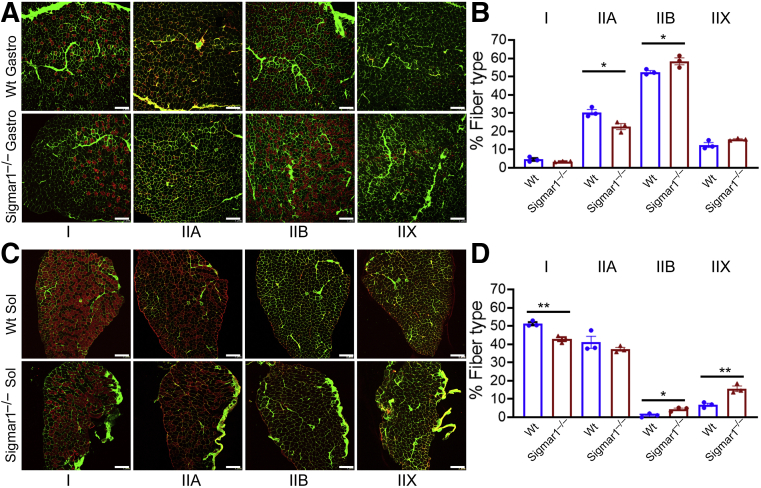
Under different pathologic conditions, myofiber types switch from slow-to-fast or fast-to-slow fiber types.42,43 As Sigmar1 showed higher expression in mitochondria-rich myofibers, histologic sections of the Gastro (type II-rich muscle fiber) and Sol (type I-rich muscle fiber) muscles isolated from Wt and Sigmar1−/− mice were immunostained with antibodies for myosin types I, IIA, IIB, and IIX (red) and WGA (green) (Figure 4). Quantification of the myofibers showed a significant reduction in type IIA myofibers and a substantial increase in type IIB myofibers per microscopic field in Gastro muscles of Sigmar1−/− mice (Figure 4, A and B). Similarly, Sol muscle showed a significant reduction in type I myofibers and a substantial increase in type IIB and IIX myofibers per microscopic field in Sigmar1−/− mice compared with Wt mice (Figure 4, C and D). Overall, these data suggest a significantly reduced number of mitochondria-rich fibers (I and IIA) and an increased number of mitochondria-deficient fibers in the Sigmar1−/− mice compared with Wt mice.
Figure 4.
Altered myofiber isotypes in Sigmar1−/− mice muscles. A and C: Representative immunofluorescence images for anti-myosin types (types I, IIA, IIB, and IIX; left to right in red) and wheat germ agglutinin (in green) stained gastrocnemius (Gastro) and soleus (Sol) muscle sections from wild-type (Wt) and Sigmar1−/− mice. B and D: Bar graphs represent the percentage of the myofibers positive for myosin types I, IIA, IIB, and IIX in Gastro and Sol muscle sections isolated from 9- to 10-month–old Wt and Sigmar1−/− mice. The absolute number of myofibers positive for each myosin type was quantified and expressed as a percentage of total myofibers per microscopic field. The percentage numbers of myosin types I, IIA, IIB, and IIX positive myofibers were quantified from 3000 to 4000 myocytes for Gastro and from 800 to 900 myocytes for Sol muscles for each group. Dots in the bar graphs represent individual values for each Wt and Sigmar1−/− mouse. P values were determined using a two-tailed unpaired t-test. Data are expressed as means ± SEM (B and D). n = 3 mice per genotype. ∗P < 0.05, ∗∗P < 0.01. Scale bars = 200 μm (A and C).
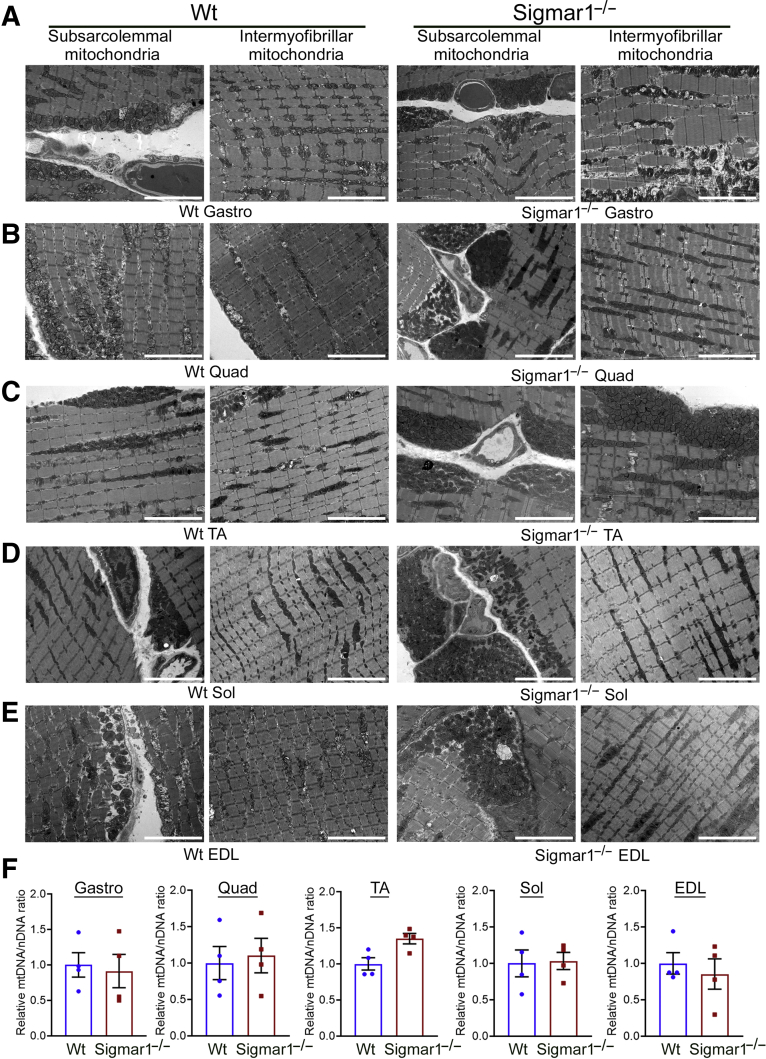
Sigmar1−/− Mice Skeletal Muscles Exhibit Abnormal Mitochondria and Aberrant Dystrophin Localization
Mutations in the SIGMAR1 gene are associated with several neuropathies with severe pathologic manifestations in skeletal muscles.14,20,23,44, 45, 46, 47, 48 Moreover, ultrastructural examination by transmission electron microscopy also showed altered mitochondrial organization in motor neurons8 and cardiac muscles.9 Therefore, the ultrastructure of the Gastro, Quad, TA, Sol, and EDL muscles, isolated from Wt and Sigmar1−/− mice, were examined (Figure 5, A–E). Transmission electron microscopy examination of skeletal muscles from WT mice showed normal ultrastructure, sarcomere organization, and normal mitochondria organization in the subsarcolemmal and intermyofibrillar spaces in all five muscles. Interestingly, Sigmar1−/− mice muscles showed abnormal, amorphously shaped mitochondria accumulation in the subsarcolemmal and intermyofibrillar spaces (Figure 5, A–E). Mitochondrial DNA content, measured by determining mitochondrial DNA/nuclear DNA ratio, showed no significant changes in the five skeletal muscles from Wt and Sigmar1−/− mice (Figure 5F).
Figure 5.
Transmission electron microscopy (TEM) showing increased abnormal mitochondria in Sigmar1−/− mice skeletal muscles. A–E: Representative TEM images of gastrocnemius (Gastro; A), quadriceps (Quad; B), tibialis anterior (TA; C), soleus (Sol; D), and extensor digitorum longus (EDL; E) muscle sections from wild-type (Wt) and Sigmar1−/− mice. All muscles examined were isolated from age-matched 9- to 10-month–old littermate Wt and Sigmar1−/− mice. F: Bar graphs represent relative mitochondrial DNA (mtDNA)/nuclear DNA (nDNA) ratio measured by comparing mtDNA content to total genomic DNA (nDNA) in Gastro, Quad, TA, Sol, and EDL muscles from 9- to 10-month–old Wt and Sigmar1−/− mice. Dots in the bar graphs represent individual values quantified for each muscle. Data are expressed as fold change with respect to Wt mice. Data are expressed as means ± SEM (F). n = 3 mice per genotype (A–E); n = 4 mice per muscle per group (F). Scale bars = 5 μm (A–E).
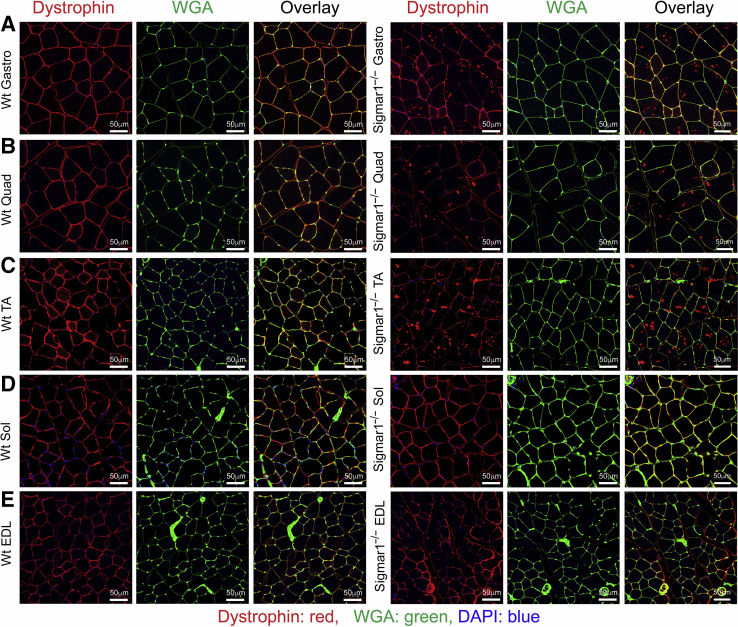
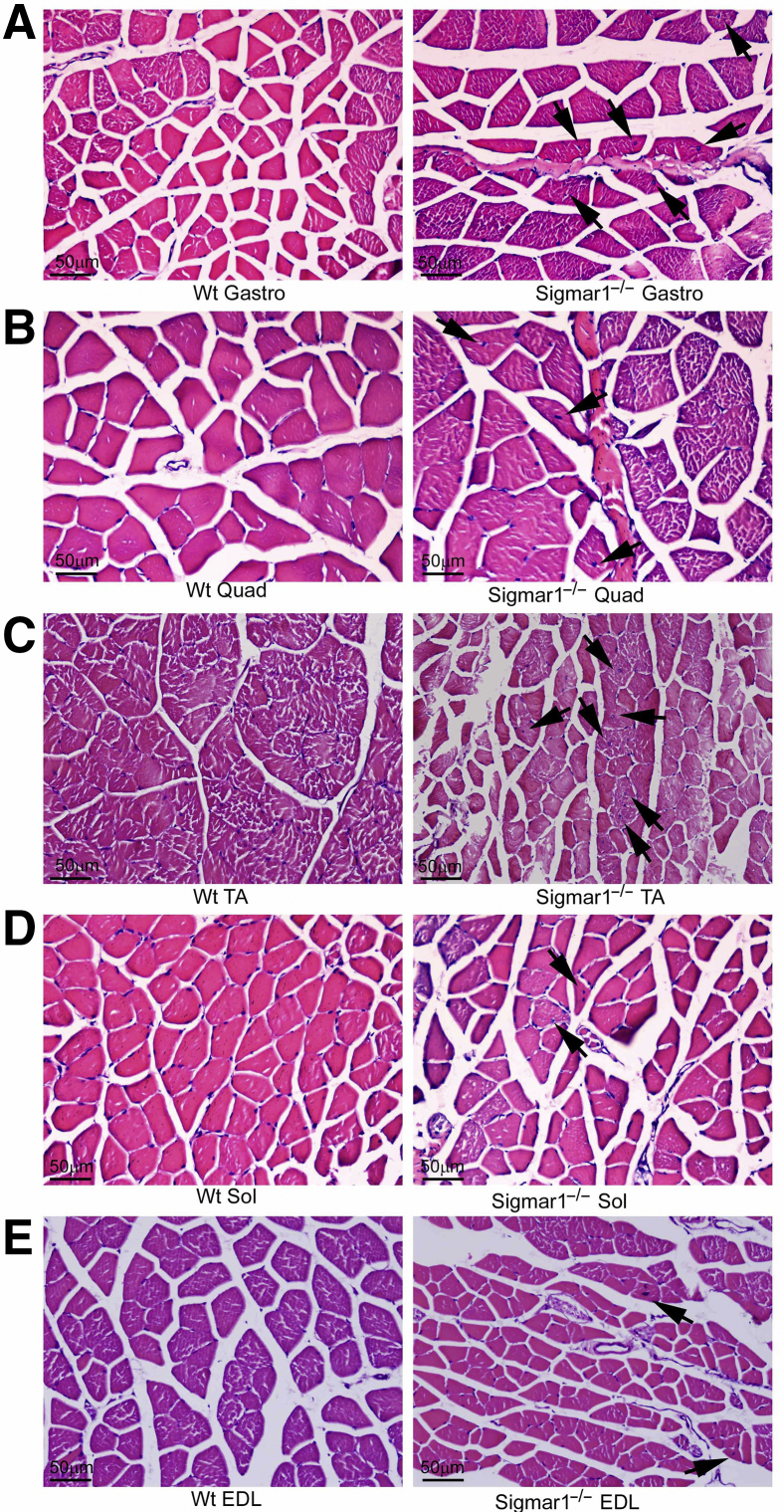
In addition, the sarcolemmal integrity of these muscles was examined using dystrophin immunostaining. In healthy muscle cells, dystrophin is localized between the sarcolemma and the outermost layer of myofilaments in the myofibers. Immunostaining of dystrophin (red) and counterstaining with WGA (green) showed normal sarcolemmal organization in Gastro, Quad, TA, Sol, and EDL muscles isolated from Wt mice (Figure 6). Interestingly, Gastro, Quad, and TA muscles of Sigmar1−/− mice showed the dislocation of dystrophin from sarcolemma and aberrant anti-dystrophin stained aggregates in skeletal muscles (Figure 6, A–C). Sol and EDL muscles showed normal dystrophin localization in both Sigmar1−/− and Wt mice (Figure 6, D and E). Overall, skeletal muscles from Sigmar1−/− mice showed abnormal mitochondria accumulation, and aberrant dystrophin aggregates in the muscles.
Figure 6.
Derangement of dystrophin localization in myofibers from Sigmar1−/− mice. Representative immunofluorescence images of wheat germ agglutinin (WGA; green) with dystrophin (red) counterstained gastrocnemius (Gastro; A), quadriceps (Quad; B), tibialis anterior (TA; C), soleus (Sol; D), and extensor digitorum longus (EDL; E) muscle sections from wild-type (Wt) and Sigmar1−/− mice. WGA staining was used to delineate myofiber boundaries. Anti-dystrophin staining (red) was conducted to delineate dystrophin localization in skeletal muscles. Dystrophin (red) localization appeared in sarcolemma as evident in colocalization with WGA (green) staining on myocyte membrane in Wt mice skeletal muscles. In contrast, aberrant dystrophin aggregates (in red) were detected intracellularly in Gastro, Quad, and TA muscles of Sigmar1−/− mice. Representative images are from 3 to 10 high-magnification microscopic fields (×20). n = 3 mice per genotype at the age of 9 to 10 months (A–E). Scale bars = 50 μm (A–E).
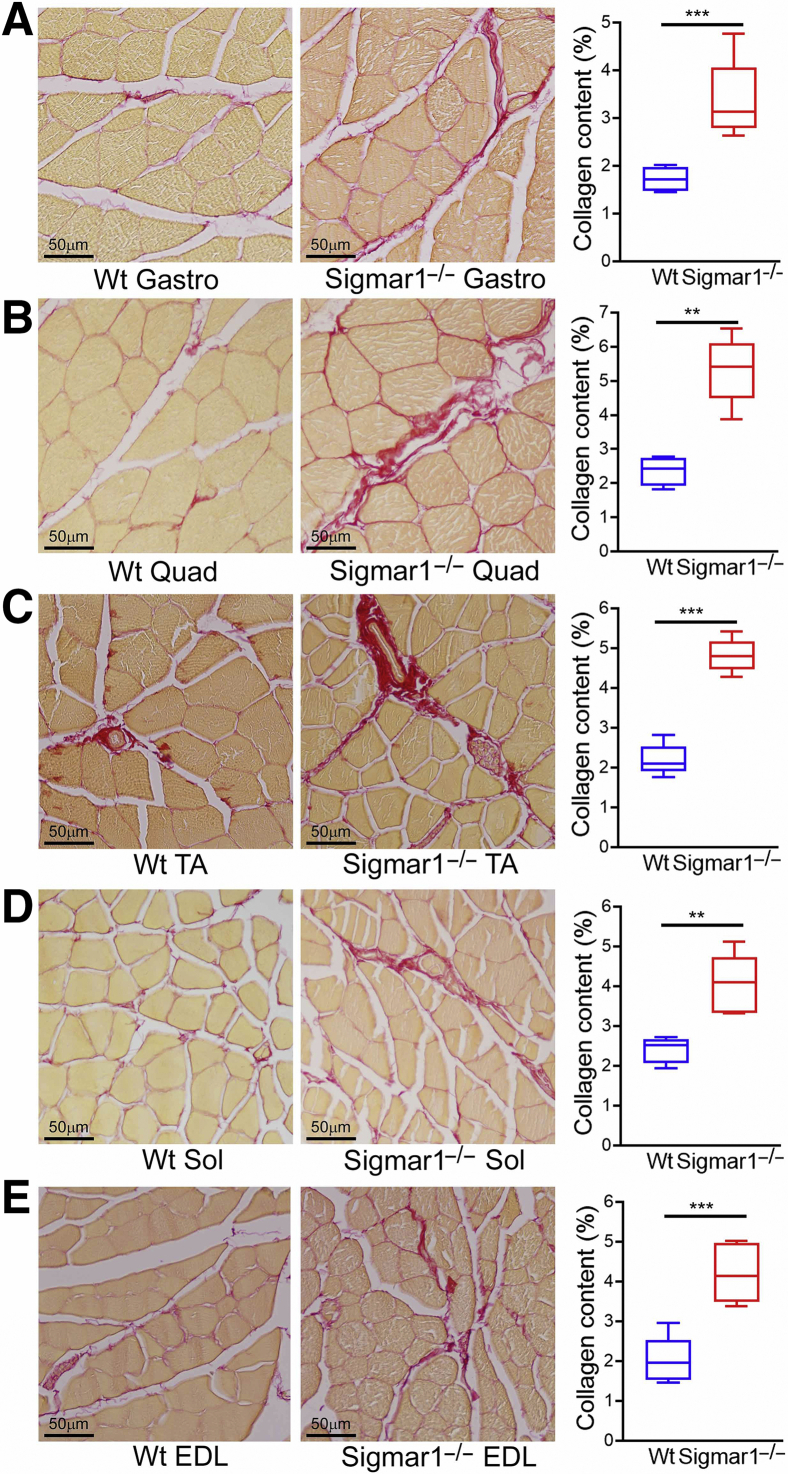
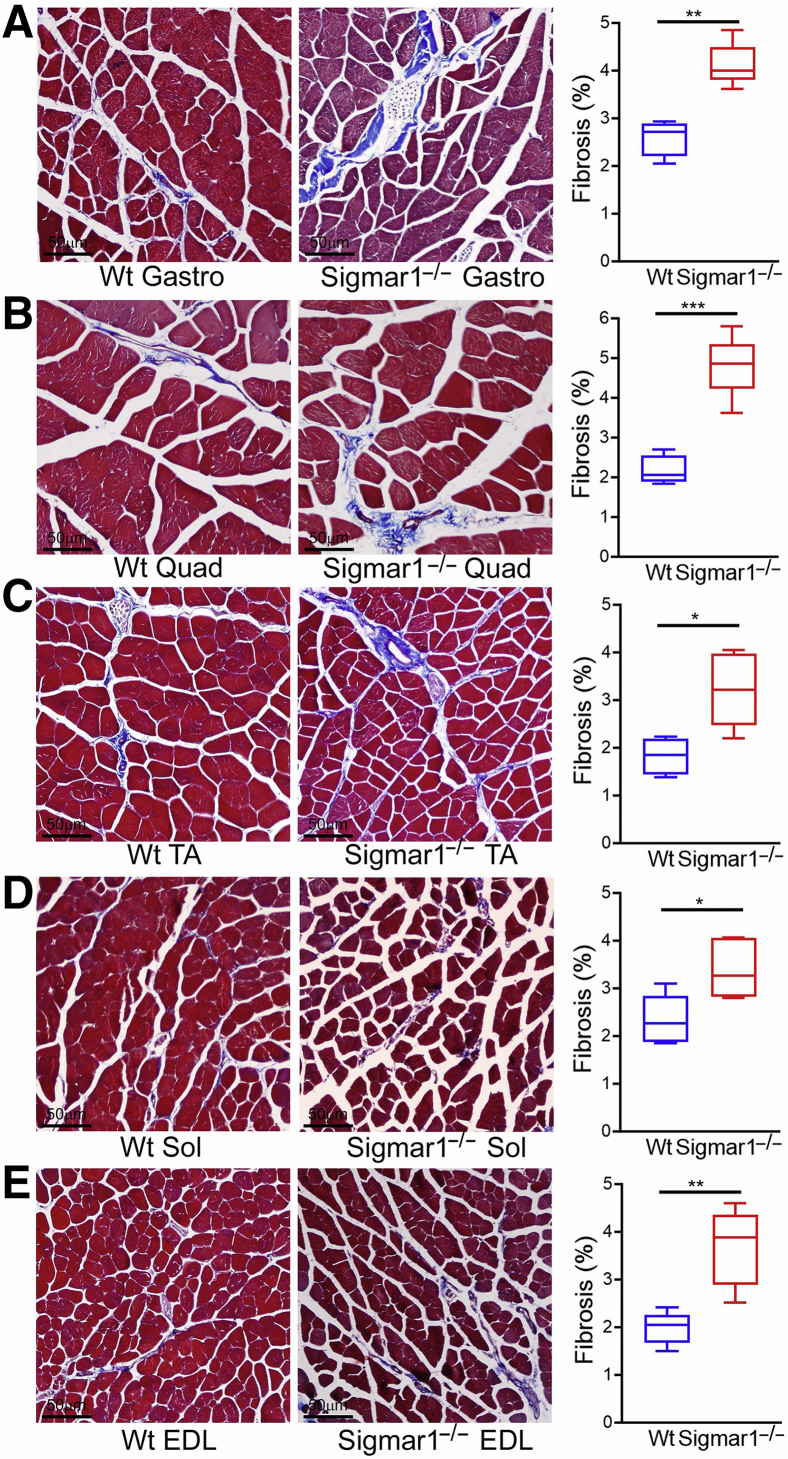
Increased Collagen Deposition and Fibrosis in Sigmar1−/− Mice Skeletal Muscles
Skeletal muscles from Sigmar1−/− mice showed pathologic features with myocyte hypertrophy, and muscle fiber type switching. Fibrotic remodeling in the histologic sections of skeletal muscles (Gastro, Quad, TA, Sol, and EDL muscles) isolated from Wt and Sigmar1−/− mice was evaluated using picro-sirius red (Figure 7) staining and Masson trichrome staining (Figure 8). All five skeletal muscles from Sigmar1−/− mice exhibited significantly increased collagen deposition in the interstitial and perivascular region compared with those in Wt mice (Figure 7). Similarly, Masson trichrome staining showed increased fibrosis areas in all five skeletal muscles of Sigmar1−/− mice compared with those in Wt mice (Figure 8). Moreover, H&E staining (Figure 9), and WGA staining (Figure 3) of Sigmar1−/− mice muscles indicated increased central nuclei (black arrows) in myofibers as compared with those in Wt mice. Moreover, H&E staining also indicated an increased interstitial eosinophilic collagen deposition (bright pink fibers) in Sigmar1−/− mice muscle sections compared to that in Wt muscle sections.
Figure 7.
Increased collagen deposition in the skeletal muscles of Sigmar1−/− mice. A–E: Left panels: Representative images of picro-sirius red (PSR) stained histologic cross-sections of gastrocnemius (Gastro; A), quadriceps (Quad; B), tibialis anterior (TA; C), soleus (Sol; D), and extensor digitorum longus (EDL; E) muscle sections from wild-type (Wt) and Sigmar1−/− mice. Representative images are from 12 high-magnification microscopic fields (×20) per mice muscle sections for Gastro, Quad, and TA muscles, and 5 high-magnification microscopic fields (×20) for Sol and EDL muscles per mice. A–E: Right panels: Box plots represent percentage collagen content measured in respect to PSR stained whole muscle cross-section areas in Wt and Sigmar1−/− mice skeletal muscles. Quantifications of percentage collagen areas were performed on 12 high-magnification microscopic fields (×20) per mice muscle sections for Gastro, Quad, and TA muscles, and on 5 high-magnification microscopic fields (×20) for Sol and EDL muscles per mice. P values indicate statistical significance between Wt and Sigmar1−/− mice. Boxes represent interquartile ranges, lines represent medians, and whiskers represent ranges. P values were determined by unpaired t-test. n = 4 to 5 individual mice at the age of 9 to 10 months per genotype per muscle (A–E). ∗∗P < 0.01, ∗∗∗P < 0.001. Scale bars = 50 μm (A–E).
Figure 8.
Increased fibrosis in the skeletal muscles of Sigmar1−/− mice compared with wild-type (Wt) mice. A–E: Masson trichrome staining was conducted to assess interstitial fibrosis in muscle sections from Wt and Sigmar1−/− mice. A–E: Left panels: Representative micrographs of trichrome-stained histologic cross-sections of gastrocnemius (Gastro; A), quadriceps (Quad; B), tibialis anterior (TA; C), soleus (Sol; D), and extensor digitorum longus (EDL; E) muscle sections from Wt and Sigmar1−/− mice. Images represent 8 to 12 high-magnification microscopic fields (×20) per mice muscle sections for Gastro, Quad, and TA muscles, and from 2 to 6 high-magnification microscopic fields (×20) for Sol and EDL muscles per mice. A–E: Right panels: Box plots represent quantifications of percentage fibrosis area corresponding to total muscle section areas in trichrome-stained whole muscle areas in Wt and Sigmar1−/− mice skeletal muscles. Fibrosis areas (percentages) were quantified on 8 to 12 high-magnification microscopic fields (×20) per mice muscle sections for Gastro, Quad, and TA muscles, and on 2 to 6 high-magnification microscopic fields (×20) for Sol and EDL muscles per mice. P values indicate statistical significance between Wt and Sigmar1−/− mice. Boxes represent interquartile ranges, lines represent medians, and whiskers represent ranges. P values were determined by unpaired t-test. n = 5 individual mice at the age of 9 to 10 months per genotype per muscle (A–E). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bars = 50 μm (A–E).
Figure 9.
Histologic hematoxylin and eosin (H&E) staining in wild-type (Wt) and Sigmar1−/− mice skeletal muscle sections. Representative micrographs of H&E-stained cross-sections of gastrocnemius (Gastro; A), quadriceps (Quad; B), tibialis anterior (TA; C), soleus (Sol; D), and extensor digitorum longus (EDL; E) muscle sections from Wt and Sigmar1−/− mice. Sigmar1−/− mice muscles exhibit central nuclei (black arrows) in myofibers compared with peripheral nuclei in Wt mice muscle sections. Representative images are from 8 to 12 high-magnification microscopic fields (×20) per mice muscle sections for Gastro, Quad, and TA muscles, and from 2 to 6 high-magnification microscopic fields (×20) for Sol and EDL muscles per mice. n = 5 individual mice at the age of 9 to 10 months per genotype per muscle (A–E). Scale bars = 50 μm (A–E).
Sigmar1−/− Mice Exhibit Reduced Endurance and Exercise Capacity
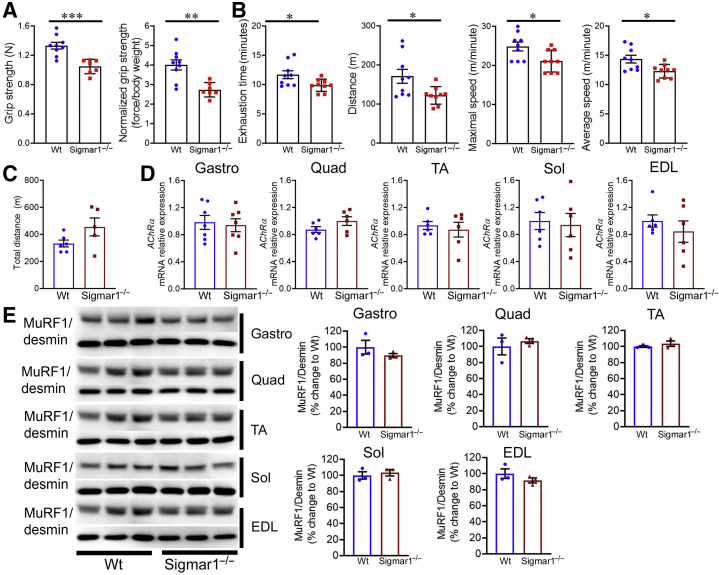
As the myofibers of Sigmar1−/− mice exhibited an array of histopathologic phenotypes, a functional assessment of the skeletal muscle was performed using grip strength measurement and graded maximal exercise testing following acclimatization. Grip strength measurements showed lower values for both absolute values (Figure 10A) and normalized values (Figure 10A) in the Sigmar1−/− mice compared with their littermate control Wt mice, indicating reduced physical endurance in Sigmar1−/− mice. Similarly, forced treadmill running showed that Sigmar1−/− mice had a compromised ability to run on a treadmill, indicated by exhaustion time, maximum distance run, maximal speed, and average speed, compared with littermate Wt mice (Figure 10B). Voluntary locomotion, assessed by measuring the total distance, showed no significant difference between Wt and Sigmar1−/− mice (Figure 10C). Furthermore, the expression of AChRα (subunit of nicotinic acetylcholine receptor), which is rapidly overexpressed after muscle denervation or when muscle electrical activity is absent, was assessed. No significant difference in AChRα expression was seen across all the five muscles (Figure 10D). Western blot analysis of muscle atrophy marker MuRF1 did not show any significant changes across all the five muscle fibers in these mice (Figure 10E). Overall, Sigmar1−/− mice showed reduced muscle strength, corroborating with attenuation in endurance and tolerance to exercise without changes in voluntary locomotion and any sign of muscle denervation and atrophy.
Figure 10.
Sigmar1−/− mice showed reduced grip strength and exercise capacity compared with wild-type (Wt) mice. A: Bar graphs represent summary data showing absolute forelimb grip strength (N) and forelimb grip strength normalized to body weight (force/body weight) in Wt and Sigmar1−/− mice. B: Parameters derived from exercise tolerance tests showing time to exhaustion (minutes), maximum running distance (m), maximum speed attained (m/minute), and average speed (m/minute) in Wt and Sigmar1−/− mice. C: Bar graphs represent summary data showing voluntary locomotion as total distance covered during a 30-minute trial in an open field chamber. Dots in the bar graphs represent individual values for each Wt and Sigmar1−/− mice. D: Bar graphs representing acetylcholine receptor α (AchRα) mRNA expression levels in gastrocnemius (Gastro), quadriceps (Quad), tibialis anterior (TA), soleus (Sol), and extensor digitorum longus (EDL) muscles isolated from 9- to 10-month–old Wt and Sigmar1−/− mice. E: Left panel: Representative Western blot analysis images showing MuRF1 protein level in Gastro, Quad, TA, Sol, and EDL muscles isolated from Wt and Sigmar1−/− mice. Desmin was used to verify equal protein loading across the lanes. Right panels: Bar graphs showing the quantification for protein levels of MuRF1 in Gastro, Quad, TA, Sol, and EDL muscles isolated from Wt and Sigmar1−/− mice. Dots in the bar graphs represent individual values quantified for each Wt and Sigmar1−/− mice. Data are expressed as percentage change to Wt. P values were determined by unpaired t-test. Data are expressed as means ± SEM (A–E). n = 7 to 9 mice per genotype at the age of 9 to 10 months (B); n = 5 to 6 mice per genotype at the age of 9 to 10 months (C); n = 6 mice per genotype at the age of 9 to 10 months (D); n = 3 mice per genotype at the age of 9 to 10 months (E). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
Discussion
This study aimed to extensively characterize the skeletal muscle morphology, histology, ultrastructure, and function in Sigmar1−/− mice to demonstrate the physiological function of Sigmar1 in the skeletal muscles (Gastro, Quad, TA, Sol, and EDL). Several intriguing findings emerged from the study and are as follows: i) Sigmar1 was differentially expressed in skeletal muscles with higher expression in mitochondria-rich muscle fibers; ii) quantification of myofiber CSA showed altered muscle mass and a slow-to-fast fiber-type switch in the skeletal muscle fibers from the Sigmar1−/− mice; iii) ultrastructural analysis by transmission electron microscopy showed the presence of abnormal mitochondria in Sigmar1−/− mice; iv) immunostaining showed a derangement of dystrophin localization in muscles (Gastro, Quad, and TA) of Sigmar1−/− mice; v) Sigmar1−/− mice had an increased number of central nuclei (WGA and H&E staining), increased collagen deposition (picro-sirius red staining), and fibrosis (Masson trichrome staining); and finally vi) functional studies showed that Sigmar1−/− mice had reduced endurance and exercise capacity compared with Wt mice with no changes in voluntary locomotion, markers for muscle denervation, and muscle atrophy. Overall, these studies demonstrated, for the first time, that Sigmar1 has a potential physiological function in the skeletal muscle to maintain healthy muscle structure and function.
Extensive studies of Sigmar1 biology to date have demonstrated an array of beneficial cellular functions in multiple organs, including the brain, heart, liver, and bladder. The most frequently reported neurologic dysfunctions in Sigmar1−/− mice are locomotor defects,25 significant nerve denervation,8 loss of motor neurons,8 age-dependent motor phenotype,15 and a depressive-like behavior.24,26 In addition, the lack of Sigmar1 in the liver show increased oxidative and metabolic stress with increased anaerobic metabolism.49,50 Extensive studies of the cardiac muscles from Sigmar1−/− mice show mitochondrial dysfunction, abnormal mitochondrial architecture, adverse cardiac pathologic remodeling, and development of cardiac contractile dysfunction.9 In addition, Sigmar1 activation by treatment with ligands is responsible for cardioprotective effects that reduce cardiac hypertrophy in an animal model of cardiac injury.1,51, 52, 53, 54, 55 Although there are similarities among the major intracellular components of both cardiac and skeletal muscle cells (ie, mitochondria, sarcoplasmic reticulum, and the myofibrils), the skeletal muscle differs substantially in its structure, regulation, and function. This study explored, for the first time, the effects of Sigmar1 ablation in muscle histochemical and ultrastructural alteration using the Sigmar1−/− mice.
Skeletal muscles consist of a heterogeneous mixture of type I (oxidative) and type II (glycolytic) myofibers. The proportions of these myofibers in skeletal muscle vary depending on the nature, location, and function of skeletal muscle. For instance, the fast-twitch muscles, like Gastro and Quad, consist of a large proportion of type II myofibers, whereas the slow-twitch muscle, like Sol, is composed primarily of type I myofibers. Type I or oxidative myofibers are mitochondria-rich myofibers relying mostly on β-oxidation for energy production, whereas type IIB or glycolytic myofibers have a higher dependency on glycolytic pathways for energy production.36,37 Therefore, the fast-twitch fibers (type II) have a low aerobic potential and are easily fatigued. Type IIA/X fibers have hybrid characteristics between type I and type IIB fibers with intermediate numbers of mitochondria and oxidative potential. Increased Sigmar1 expression was observed in mitochondria-rich myofibers. Gastro muscles of Sigmar1−/− mice showed a significant reduction in type IIA myofibers and a substantial increase in type IIB myofibers. Similarly, Sol muscle of Sigmar1−/− mice showed a significant reduction in type I myofibers and a substantial increase in type IIB and IIX myofibers compared with that in Wt mice. Interestingly, the muscle ultrastructure of Sigmar1−/− mice also showed an increase in abnormal mitochondria, suggesting the inability of efficient energy production in these muscles. In fact, mitochondria isolated from the Sigmar1−/− hearts have decreased mitochondrial respiration and function.9 Therefore, the decreased exercise capacity in Sigmar1−/− mice may be attributed to the decreased level of mitochondrial-rich fibers as well as increased levels of dysfunctional mitochondria.
Current literature indicates that all patients with Sigmar1 mutations develop manifestations of skeletal muscle dysfunction. Among these Sigmar1 mutations, a patient with the L95 fs developed muscle weakness with significant atrophy and wasting of the calf muscles and intrinsic muscles of the hands.15 This patient showed severe type II fiber predominance with scattered angular esterase-positive fibers in a vastus lateralis muscle biopsy, a type 1 myofiber-rich quadriceps muscle in healthy humans.56 Myofibers switching from type I to type II or from type II to type I have also been reported under pathologic conditions.42,43 Interestingly, the p.L95 fs Sigmar1 mutant was unstable and nonfunctional when expressed in Neuro2a cells, suggesting that the loss of Sigmar1 plays a role in the development of pathology. Therefore, changes in myofiber types in muscles from the Sigmar1−/− mice suggest that Sigmar1 may have a role in maintaining myofiber function. The molecular mechanisms of changes in myofiber type in the muscles of Sigmar1–/– mice remain unknown and require further studies.
The most intriguing phenotype observed in the skeletal muscles (Gastro, Quad, and TA) from Sigmar1−/− mice was abnormal mitochondria accumulation and derangement in dystrophin localization. Interestingly, the skeletal muscle pathology observed in Sigmar1−/− skeletal muscles is similar to that observed in tubular aggregate myopathy,56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66 and muscular dystrophy associated with mutations in dystrophin.67, 68, 69, 70, 71, 72, 73 Tubular aggregate myopathy is a rare myopathy characterized by a progressive decline in skeletal muscle function with increased weakness, cramps, and pain.57 Structural abnormalities of mitochondria58, 59, 60, 61 and defects in mitochondrial function60,62,63 have been reported in myopathies with tubular aggregates.58, 59, 60, 61, 62, 63, 64, 65, 66 Mitochondrial dysfunction including aberrant mitochondrial morphology, reduced cristae number, and large mitochondrial vacuoles are well-known pathological features reported prior to the onset of muscle damage in patients with Duchenne and Becker muscular dystrophy67, 68, 69, 70 with mutation(s) in the dystrophin gene, resulting in the expression of an unstable truncated dystrophin protein.71, 72, 73 However, Sigmar1’s direct role and mechanisms associated with mitochondrial morphology, dynamics, and function remained unknown.
In conclusion, this study reported, for the first time, the physiological function of Sigmar1 in skeletal muscles using Sigmar1−/− mice and showed interesting phenotypes, including increased central nuclei number, derangement in dystrophin localization, changes in myofiber isotype composition, and reduced endurance and exercise tolerance without changes in voluntary locomotion, markers for muscle denervation, and muscle atrophy. A significant limitation of this study is the use of global knockout mice, where a possible contribution of cellular signaling due to Sigmar1 ablation in other organs could affect skeletal muscle pathology. For instance, the possible contribution of cardiac contractile dysfunction reported in these Sigmar1−/− mice9 to reduce endurance and exercise tolerance remains unknown. Therefore, future studies using indicate that skeletal muscle fiber-specific knockout mice are required to determine the direct contribution of Sigmar1 in mediating skeletal muscle function under pathophysiological conditions. However, findings from this study explain the physiological function of Sigmar1 in the skeletal muscle, and indicate that skeletal muscle pathology observed in patients with Sigmar1 mutations could be the result of direct effects of dysfunctional Sigmar1 in myofibers.
Footnotes
Supported by NIH grants R00 HL122354, R01 HL145753, HL145753-01S1, HL145753-03S1R00, HL122354. R01 HL145753, HL145753-01S1, HL145753-03S1R00, HL122354, R01 HL145753, HL145753-01S1, HL145753-03S1R00 HL122354, R01 HL145753, HL145753-01S1, and HL145753-03S1 (M.S.B.); Louisiana State University Health Center at Shreveport Center for Cardiovascular Diseases and Sciences (LSUHSC-S CCDS) Finish Line Award; COVID-19 Research Award; Louisiana Addiction Research Center (LARC) Research Award (M.S.B.); NIH grants P20GM121307 and R01 HL149264 (C.G.K.); NIHR01 HL098435, HL133497, and HL141155 (A.W.O.); LSUHSC-S Malcolm Feist Cardiovascular and American Heart Association (AHA) Postdoctoral Fellowship20POST35210789 (C.S.A.); AHA Postdoctoral Fellowship (S.A.); and LSUHSC-S Malcolm Feist Predoctoral Fellowship (R.A.).
R.A. and C.S.A. contributed equally to this work.
Disclosures: None declared.
Author Contributions
R.A., C.S.A., and M.S.B. conceptualized the study; R.A., C.S.A., and M.S.B. designed the experiments; R.A., C.S.A., S.A., N.S.R., M.M., and S.N. performed all experiments and participated in analyses; B.H. and J.K. performed electron microscopic experiments; M.A.N.B. performed statistical analysis; C.G.K. and A.W.O. provided analytic tools and reagents; R.A., C.S.A., and M.S.B. wrote the manuscript; and all of the authors read, edited, and approved the article.
References
- 1.Bhuiyan M.S., Fukunaga K. Stimulation of sigma-1 receptor signaling by dehydroepiandrosterone ameliorates pressure overload-induced hypertrophy and dysfunctions in ovariectomized rats. Expert Opin Ther Targets. 2009;13:1253–1265. doi: 10.1517/14728220903264064. [DOI] [PubMed] [Google Scholar]
- 2.Bhuiyan S., Fukunaga K. Stimulation of Sigma-1 receptor by dehydroepiandrosterone ameliorates hypertension-induced kidney hypertrophy in ovariectomized rats. Exp Biol Med. 2010;235:356–364. doi: 10.1258/ebm.2009.009177. [DOI] [PubMed] [Google Scholar]
- 3.Bhuiyan M.S., Tagashira H., Fukunaga K. Dehydroepiandrosterone-mediated stimulation of sigma-1 receptor activates Akt-eNOS signaling in the thoracic aorta of ovariectomized rats with abdominal aortic banding. Cardiovasc Ther. 2011;29:219–230. doi: 10.1111/j.1755-5922.2010.00196.x. [DOI] [PubMed] [Google Scholar]
- 4.Klouz A., Sapena R., Liu J., Maurice T., Tillement J.P., Papadopoulos V., Morin D. Evidence for sigma-1-like receptors in isolated rat liver mitochondrial membranes. Br J Pharmacol. 2002;135:1607–1615. doi: 10.1038/sj.bjp.0704626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kekuda R., Prasad P.D., Fei Y.J., Leibach F.H., Ganapathy V. Cloning and functional expression of the human type 1 sigma receptor (hSigmaR1) Biochem Biophys Res Commun. 1996;229:553–558. doi: 10.1006/bbrc.1996.1842. [DOI] [PubMed] [Google Scholar]
- 6.Mei J., Pasternak G.W. Molecular cloning and pharmacological characterization of the rat sigma1 receptor. Biochem Pharmacol. 2001;62:349–355. doi: 10.1016/s0006-2952(01)00666-9. [DOI] [PubMed] [Google Scholar]
- 7.Soriani O., Rapetti-Mauss R. In: Sigma Receptors: Their Role in Disease and as Therapeutic Targets. Smith S.B., Su T.-P., editors. Springer International Publishing; Cham: 2017. Sigma 1 receptor and ion channel dynamics in cancer; pp. 63–77. [Google Scholar]
- 8.Bernard-Marissal N., Medard J.J., Azzedine H., Chrast R. Dysfunction in endoplasmic reticulum-mitochondria crosstalk underlies SIGMAR1 loss of function mediated motor neuron degeneration. Brain. 2015;138:875–890. doi: 10.1093/brain/awv008. [DOI] [PubMed] [Google Scholar]
- 9.Abdullah C.S., Alam S., Aishwarya R., Miriyala S., Panchatcharam M., Bhuiyan M.A.N., Peretik J.M., Orr A.W., James J., Osinska H., Robbins J., Lorenz J.N., Bhuiyan M.S. Cardiac dysfunction in the sigma 1 receptor knockout mouse associated with impaired mitochondrial dynamics and bioenergetics. J Am Heart Assoc. 2018;7:e009775. doi: 10.1161/JAHA.118.009775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christ M.G., Huesmann H., Nagel H., Kern A., Behl C. Sigma-1 receptor activation induces autophagy and increases proteostasis capacity in vitro and in vivo. Cells. 2019;8:211. doi: 10.3390/cells8030211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin W.R., Eades C.G., Thompson J.A., Huppler R.E., Gilbert P.E. The effects of morphine- and nalorphine- like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther. 1976;197:517–532. [PubMed] [Google Scholar]
- 12.Su T.P. Evidence for sigma opioid receptor: binding of [3H]SKF-10047 to etorphine-inaccessible sites in guinea-pig brain. J Pharmacol Exp Ther. 1982;223:284–290. [PubMed] [Google Scholar]
- 13.Loeffler J.P., Picchiarelli G., Dupuis L., Gonzalez De Aguilar J.L. The role of skeletal muscle in amyotrophic lateral sclerosis. Brain Pathol. 2016;26:227–236. doi: 10.1111/bpa.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Saif A., Al-Mohanna F., Bohlega S. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann Neurol. 2011;70:913–919. doi: 10.1002/ana.22534. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe S., Ilieva H., Tamada H., Nomura H., Komine O., Endo F., Jin S., Mancias P., Kiyama H., Yamanaka K. Mitochondria-associated membrane collapse is a common pathomechanism in SIGMAR1- and SOD1-linked ALS. EMBO Mol Med. 2016;8:1421–1437. doi: 10.15252/emmm.201606403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Hu Z., Liu L., Xie Y., Zhan Y., Zi X., Wang J., Wu L., Xia K., Tang B., Zhang R. A SIGMAR1 splice-site mutation causes distal hereditary motor neuropathy. Neurology. 2015;84:2430–2437. doi: 10.1212/WNL.0000000000001680. [DOI] [PubMed] [Google Scholar]
- 17.Gregianin E., Pallafacchina G., Zanin S., Crippa V., Rusmini P., Poletti A., Fang M., Li Z., Diano L., Petrucci A., Lispi L., Cavallaro T., Fabrizi G.M., Muglia M., Boaretto F., Vettori A., Rizzuto R., Mostacciuolo M.L., Vazza G. Loss-of-function mutations in the SIGMAR1 gene cause distal hereditary motor neuropathy by impairing ER-mitochondria tethering and Ca2+ signalling. Hum Mol Genet. 2016;25:3741–3753. doi: 10.1093/hmg/ddw220. [DOI] [PubMed] [Google Scholar]
- 18.Horga A., Tomaselli P.J., Gonzalez M.A., Laura M., Muntoni F., Manzur A.Y., Hanna M.G., Blake J.C., Houlden H., Zuchner S., Reilly M.M. SIGMAR1 mutation associated with autosomal recessive Silver-like syndrome. Neurology. 2016;87:1607–1612. doi: 10.1212/WNL.0000000000003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J.J.Y., van Karnebeek C.D.M., Drogemoller B., Shyr C., Tarailo-Graovac M., Eydoux P., Ross C.J., Wasserman W.W., Bjornson B., Wu J.K. Further validation of the SIGMAR1 c.151+1G>T mutation as cause of distal hereditary motor neuropathy. Child Neurol Open. 2016;3 doi: 10.1177/2329048X16669912. 2329048X16669912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almendra L., Laranjeira F., Fernandez-Marmiesse A., Negrao L. SIGMAR1 gene mutation causing distal hereditary motor neuropathy in a Portuguese family. Acta Myol. 2018;37:2–4. [PMC free article] [PubMed] [Google Scholar]
- 21.Nandhagopal R., Meftah D., Al-Kalbani S., Scott P. Recessive distal motor neuropathy with pyramidal signs in an Omani kindred: underlying novel mutation in the SIGMAR1 gene. Eur J Neurol. 2018;25:395–403. doi: 10.1111/ene.13519. [DOI] [PubMed] [Google Scholar]
- 22.Couly S., Khalil B., Viguier V., Roussel J., Maurice T., Lievens J.C. Sigma-1 receptor is a key genetic modulator in amyotrophic lateral sclerosis. Hum Mol Genet. 2020;29:529–540. doi: 10.1093/hmg/ddz267. [DOI] [PubMed] [Google Scholar]
- 23.Luty A.A., Kwok J.B., Dobson-Stone C., Loy C.T., Coupland K.G., Karlstrom H., Sobow T., Tchorzewska J., Maruszak A., Barcikowska M., Panegyres P.K., Zekanowski C., Brooks W.S., Williams K.L., Blair I.P., Mather K.A., Sachdev P.S., Halliday G.M., Schofield P.R. Sigma nonopioid intracellular receptor 1 mutations cause frontotemporal lobar degeneration-motor neuron disease. Ann Neurol. 2010;68:639–649. doi: 10.1002/ana.22274. [DOI] [PubMed] [Google Scholar]
- 24.Langa F., Codony X., Tovar V., Lavado A., Gimenez E., Cozar P., Cantero M., Dordal A., Hernandez E., Perez R., Monroy X., Zamanillo D., Guitart X., Montoliu L. Generation and phenotypic analysis of sigma receptor type I (sigma 1) knockout mice. Eur J Neurosci. 2003;18:2188–2196. doi: 10.1046/j.1460-9568.2003.02950.x. [DOI] [PubMed] [Google Scholar]
- 25.Mavlyutov T.A., Epstein M.L., Andersen K.A., Ziskind-Conhaim L., Ruoho A.E. The sigma-1 receptor is enriched in postsynaptic sites of C-terminals in mouse motoneurons: an anatomical and behavioral study. Neuroscience. 2010;167:247–255. doi: 10.1016/j.neuroscience.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabino V., Cottone P., Parylak S.L., Steardo L., Zorrilla E.P. Sigma-1 receptor knockout mice display a depressive-like phenotype. Behav Brain Res. 2009;198:472–476. doi: 10.1016/j.bbr.2008.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Committee for the Update of the Guide for the Care and Use of Laboratory Animals . Eighth Edition. National Academies Press; Washington, DC: 2011. National Research Council: Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 28.Bhuiyan M.S., Pattison J.S., Osinska H., James J., Gulick J., McLendon P.M., Hill J.A., Sadoshima J., Robbins J. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest. 2013;123:5284–5297. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdullah C.S., Aishwarya R., Alam S., Morshed M., Remex N.S., Nitu S., Kolluru G.K., Traylor J., Miriyala S., Panchatcharam M., Hartman B., King J., Bhuiyan M.A.N., Chandran S., Woolard M.D., Yu X., Goeders N.E., Dominic P., Arnold C.L., Stokes K., Kevil C.G., Orr A.W., Bhuiyan M.S. Methamphetamine induces cardiomyopathy by Sigmar1 inhibition-dependent impairment of mitochondrial dynamics and function. Commun Biol. 2020;3:682. doi: 10.1038/s42003-020-01408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloemberg D., Quadrilatero J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS One. 2012;7:e35273. doi: 10.1371/journal.pone.0035273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alam S., Abdullah C.S., Aishwarya R., Miriyala S., Panchatcharam M., Peretik J.M., Orr A.W., James J., Robbins J., Bhuiyan M.S. Aberrant mitochondrial fission is maladaptive in desmin mutation-induced cardiac proteotoxicity. J Am Heart Assoc. 2018;7:e009289. doi: 10.1161/JAHA.118.009289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castro B., Kuang S. Evaluation of muscle performance in mice by treadmill exhaustion test and whole-limb grip strength assay. Bio Protoc. 2017;7:e2237. doi: 10.21769/BioProtoc.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeshita H., Yamamoto K., Nozato S., Inagaki T., Tsuchimochi H., Shirai M., Yamamoto R., Imaizumi Y., Hongyo K., Yokoyama S., Takeda M., Oguro R., Takami Y., Itoh N., Takeya Y., Sugimoto K., Fukada S.I., Rakugi H. Modified forelimb grip strength test detects aging-associated physiological decline in skeletal muscle function in male mice. Sci Rep. 2017;7:42323. doi: 10.1038/srep42323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrosino J.M., Heiss V.J., Maurya S.K., Kalyanasundaram A., Periasamy M., LaFountain R.A., Wilson J.M., Simonetti O.P., Ziouzenkova O. Graded maximal exercise testing to assess mouse cardio-metabolic phenotypes. PLoS One. 2016;11:e0148010. doi: 10.1371/journal.pone.0148010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klenerova V., Krejci I., Sida P., Hlinak Z., Hynie S. Oxytocin and carbetocin effects on spontaneous behavior of male rats: modulation by oxytocin receptor antagonists. Neuro Endocrinol Lett. 2009;30:335–342. [PubMed] [Google Scholar]
- 36.Scott W., Stevens J., Binder-Macleod S.A. Human skeletal muscle fiber type classifications. Phys Ther. 2001;81:1810–1816. [PubMed] [Google Scholar]
- 37.Guy P.S., Snow D.H. Skeletal muscle fibre composition in the dog and its relationship to athletic ability. Res Vet Sci. 1981;31:244–248. [PubMed] [Google Scholar]
- 38.Mazzotti A.L., Coletti D. The need for a consensus on the locution “central nuclei” in striated muscle myopathies. Front Physiol. 2016;7:577. doi: 10.3389/fphys.2016.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutierrez J.M., Ownby C.L., Odell G.V. Skeletal muscle regeneration after myonecrosis induced by crude venom and a myotoxin from the snake Bothrops asper (Fer-de-Lance) Toxicon. 1984;22:719–731. doi: 10.1016/0041-0101(84)90155-7. [DOI] [PubMed] [Google Scholar]
- 40.Folker E.S., Baylies M.K. Nuclear positioning in muscle development and disease. Front Physiol. 2013;4:363. doi: 10.3389/fphys.2013.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cadot B., Gache V., Gomes E.R. Moving and positioning the nucleus in skeletal muscle - one step at a time. Nucleus. 2015;6:373–381. doi: 10.1080/19491034.2015.1090073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyer J.G., Prasad V., Song T., Lee D., Fu X., Grimes K.M., Sargent M.A., Sadayappan S., Molkentin J.D. ERK1/2 signaling induces skeletal muscle slow fiber-type switching and reduces muscular dystrophy disease severity. JCI Insight. 2019;5:e127356. doi: 10.1172/jci.insight.127356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jennifer G., Masatoshi S. 2018. Skeletal Muscle Fiber Types in Neuromuscular Diseases, London, UK: IntechOpen Limited. [Google Scholar]
- 44.Izumi Y., Morino H., Miyamoto R., Matsuda Y., Ohsawa R., Kurashige T., Shimatani Y., Kaji R., Kawakami H. Compound heterozygote mutations in the SIGMAR1 gene in an oldest-old patient with amyotrophic lateral sclerosis. Geriatr Gerontol Int. 2018;18:1519–1520. doi: 10.1111/ggi.13506. [DOI] [PubMed] [Google Scholar]
- 45.Christodoulou K., Zamba E., Tsingis M., Mubaidin A., Horani K., Abu-Sheik S., El-Khateeb M., Kyriacou K., Kyriakides T., Al-Qudah A.K., Middleton L. A novel form of distal hereditary motor neuronopathy maps to chromosome 9p21.1-p12. Ann Neurol. 2000;48:877–884. [PubMed] [Google Scholar]
- 46.Belzil V.V., Daoud H., Camu W., Strong M.J., Dion P.A., Rouleau G.A. Genetic analysis of SIGMAR1 as a cause of familial ALS with dementia. Eur J Hum Genet. 2013;21:237–239. doi: 10.1038/ejhg.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim H.J., Kwon M.J., Choi W.J., Oh K.W., Oh S.I., Ki C.S., Kim S.H. Mutations in UBQLN2 and SIGMAR1 genes are rare in Korean patients with amyotrophic lateral sclerosis. Neurobiol Aging. 2014;35:1957.e7–1957.e8. doi: 10.1016/j.neurobiolaging.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Ullah M.I., Ahmad A., Raza S.I., Amar A., Ali A., Bhatti A., John P., Mohyuddin A., Ahmad W., Hassan M.J. In silico analysis of SIGMAR1 variant (rs4879809) segregating in a consanguineous Pakistani family showing amyotrophic lateral sclerosis without frontotemporal lobar dementia. Neurogenetics. 2015;16:299–306. doi: 10.1007/s10048-015-0453-1. [DOI] [PubMed] [Google Scholar]
- 49.Couly S., Goguadze N., Yasui Y., Kimura Y., Wang S.M., Sharikadze N., Wu H.E., Su T.P. Knocking out sigma-1 receptors reveals diverse health problems. Cell Mol Neurobiol. 2020 doi: 10.1007/s10571-020-00983-3. [Epub ahead of print] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pal A., Fontanilla D., Gopalakrishnan A., Chae Y.K., Markley J.L., Ruoho A.E. The sigma-1 receptor protects against cellular oxidative stress and activates antioxidant response elements. Eur J Pharmacol. 2012;682:12–20. doi: 10.1016/j.ejphar.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhuiyan M.S., Fukunaga K. Targeting sigma-1 receptor signaling by endogenous ligands for cardioprotection. Expert Opin Ther Targets. 2011;15:145–155. doi: 10.1517/14728222.2011.546350. [DOI] [PubMed] [Google Scholar]
- 52.Bhuiyan M.S., Tagashira H., Shioda N., Fukunaga K. Targeting sigma-1 receptor with fluvoxamine ameliorates pressure-overload-induced hypertrophy and dysfunctions. Expert Opin Ther Targets. 2010;14:1009–1022. doi: 10.1517/14728222.2010.509348. [DOI] [PubMed] [Google Scholar]
- 53.Tagashira H., Bhuiyan M.S., Shioda N., Fukunaga K. Fluvoxamine rescues mitochondrial Ca2+ transport and ATP production through sigma(1)-receptor in hypertrophic cardiomyocytes. Life Sci. 2014;95:89–100. doi: 10.1016/j.lfs.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 54.Tagashira H., Bhuiyan S., Shioda N., Hasegawa H., Kanai H., Fukunaga K. Sigma1-receptor stimulation with fluvoxamine ameliorates transverse aortic constriction-induced myocardial hypertrophy and dysfunction in mice. Am J Physiol Heart Circ Physiol. 2010;299:H1535–H1545. doi: 10.1152/ajpheart.00198.2010. [DOI] [PubMed] [Google Scholar]
- 55.Tagashira H., Shinoda Y., Shioda N., Fukunaga K. Methyl pyruvate rescues mitochondrial damage caused by SIGMAR1 mutation related to amyotrophic lateral sclerosis. Biochim Biophys Acta. 2014;1840:3320–3334. doi: 10.1016/j.bbagen.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 56.Staron R.S., Hagerman F.C., Hikida R.S., Murray T.F., Hostler D.P., Crill M.T., Ragg K.E., Toma K. Fiber type composition of the vastus lateralis muscle of young men and women. J Histochem Cytochem. 2000;48:623–629. doi: 10.1177/002215540004800506. [DOI] [PubMed] [Google Scholar]
- 57.Walter M.C., Rossius M., Zitzelsberger M., Vorgerd M., Muller-Felber W., Ertl-Wagner B., Zhang Y., Brinkmeier H., Senderek J., Schoser B. 50 Years to diagnosis: autosomal dominant tubular aggregate myopathy caused by a novel STIM1 mutation. Neuromuscul Disord. 2015;25:577–584. doi: 10.1016/j.nmd.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 58.Martin J.J., Ceuterick C., Van Goethem G. On a dominantly inherited myopathy with tubular aggregates. Neuromuscul Disord. 1997;7:512–520. doi: 10.1016/s0960-8966(97)00119-3. [DOI] [PubMed] [Google Scholar]
- 59.Muller H.D., Vielhaber S., Brunn A., Schroder J.M. Dominantly inherited myopathy with novel tubular aggregates containing 1-21 tubulofilamentous structures. Acta Neuropathol. 2001;102:27–35. doi: 10.1007/s004010000342. [DOI] [PubMed] [Google Scholar]
- 60.Pierobon-Bormioli S., Armani M., Ringel S.P., Angelini C., Vergani L., Betto R., Salviati G. Familial neuromuscular disease with tubular aggregates. Muscle Nerve. 1985;8:291–298. doi: 10.1002/mus.880080405. [DOI] [PubMed] [Google Scholar]
- 61.Rosenberg N.L., Neville H.E., Ringel S.P. Tubular aggregates: their association with neuromuscular diseases, including the syndrome of myalgias/cramps. Arch Neurol. 1985;42:973–976. doi: 10.1001/archneur.1985.04060090055014. [DOI] [PubMed] [Google Scholar]
- 62.Bendahan D., Pouget J., Pellissier J.F., Figarella-Branger D., Cozzone P.J. Magnetic resonance spectroscopy and histological study of tubular aggregates in a familial myopathy. J Neurol Sci. 1996;139:149–155. [PubMed] [Google Scholar]
- 63.Vielhaber S., Schroder R., Winkler K., Weis S., Sailer M., Feistner H., Heinze H.J., Schroder J.M., Kunz W.S. Defective mitochondrial oxidative phosphorylation in myopathies with tubular aggregates originating from sarcoplasmic reticulum. J Neuropathol Exp Neurol. 2001;60:1032–1040. doi: 10.1093/jnen/60.11.1032. [DOI] [PubMed] [Google Scholar]
- 64.Dobkin B.H., Verity M.A. Familial neuromuscular disease with type 1 fiber hypoplasia, tubular aggregates, cardiomyopathy, and myasthenic features. Neurology. 1978;28:1135–1140. doi: 10.1212/wnl.28.11.1135. [DOI] [PubMed] [Google Scholar]
- 65.Rohkamm R., Boxler K., Ricker K., Jerusalem F. A dominantly inherited myopathy with excessive tubular aggregates. Neurology. 1983;33:331–336. doi: 10.1212/wnl.33.3.331. [DOI] [PubMed] [Google Scholar]
- 66.Sallinger M., Tiffner A., Schmidt T., Bonhenry D., Waldherr L., Frischauf I., Lunz V., Derler I., Schober R., Schindl R. Luminal STIM1 mutants that cause tubular aggregate myopathy promote autophagic processes. Int J Mol Sci. 2020;21:4410. doi: 10.3390/ijms21124410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pellegrini C., Zulian A., Gualandi F., Manzati E., Merlini L., Michelini M.E., Benassi L., Marmiroli S., Ferlini A., Sabatelli P., Bernardi P., Maraldi N.M. Melanocytes–a novel tool to study mitochondrial dysfunction in Duchenne muscular dystrophy. J Cell Physiol. 2013;228:1323–1331. doi: 10.1002/jcp.24290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pant M., Sopariwala D.H., Bal N.C., Lowe J., Delfin D.A., Rafael-Fortney J., Periasamy M. Metabolic dysfunction and altered mitochondrial dynamics in the utrophin-dystrophin deficient mouse model of duchenne muscular dystrophy. PLoS One. 2015 doi: 10.1371/journal.pone.0123875. 10:e0123875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore T.M., Lin A.J., Strumwasser A.R., Cory K., Whitney K., Ho T., Ho T., Lee J.L., Rucker D.H., Nguyen C.Q., Yackly A., Mahata S.K., Wanagat J., Stiles L., Turcotte L.P., Crosbie R.H., Zhou Z. Mitochondrial Dysfunction Is an Early Consequence of Partial or Complete Dystrophin Loss in mdx Mice. Front Physiol. 2020:11:690. doi: 10.3389/fphys.2020.00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vila M.C., Rayavarapu S., Hogarth M.W., Van der Meulen J.H., Horn A., Defour A., Takeda S., Brown K.J., Hathout Y., Nagaraju K., Jaiswal J.K. Mitochondria mediate cell membrane repair and contribute to Duchenne muscular dystrophy. Cell Death Differ. 2017;24:330–342. doi: 10.1038/cdd.2016.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arahata K., Ishiura S., Ishiguro T., Tsukahara T., Suhara Y., Eguchi C., Ishihara T., Nonaka I., Ozawa E., Sugita H. Immunostaining of skeletal and cardiac muscle surface membrane with antibody against Duchenne muscular dystrophy peptide. Nature. 1988;333:861–863. doi: 10.1038/333861a0. [DOI] [PubMed] [Google Scholar]
- 72.Suminaga R., Takeshima Y., Wada H., Yagi M., Matsuo M. C-terminal truncated dystrophin identified in skeletal muscle of an asymptomatic boy with a novel nonsense mutation of the dystrophin gene. Pediatr Res. 2004;56:739–743. doi: 10.1203/01.PDR.0000142734.46609.43. [DOI] [PubMed] [Google Scholar]
- 73.Hoffman E.P., Brown R.H.,, jr, Kunkel L.M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]