Key Points
Diagnosis of TCL often relies on PCR-based TCR clonality testing, which displays low specificity and requires high-quality DNA.
Targeted NGS and T-cell clonality assessment are complementary techniques that should be applied in hard front-line diagnosis.
Abstract
Immunomorphological diagnosis of T-cell lymphoma (TCL) may be challenging, especially on needle biopsies. Multiplex polymerase chain reaction (PCR) assays to assess T-cell receptor (TCR) gene rearrangements are now widely used to detect T-cell clones and provide diagnostic support. However, PCR assays detect only 80% of TCL, and clonal lymphocyte populations may also appear in nonneoplastic conditions. More recently, targeted next-generation sequencing (t-NGS) technologies have been deployed to improve lymphoma classification. To the best of our knowledge, the comparison of these techniques’ performance in TCL diagnosis has not been reported yet. In this study, 82 TCL samples and 25 nonneoplastic T-cell infiltrates were divided into 2 cohorts (test and validation) and analyzed with both multiplex PCR and t-NGS to investigate TCR gene rearrangements and somatic mutations, respectively. The detection of mutations appeared to be more specific (100.0%) than T-cell clonality assessment (41.7%-45.5%), whereas no differences were observed in terms of sensitivity (95.1%-97.4%). Furthermore, t-NGS provided a reliable basis for TCL diagnosis in samples with partially degraded DNA that was impossible to assess with PCR. Finally, although multiplex PCR assays appeared to be less specific than t-NGS, both techniques remain complementary, as PCR recovered some t-NGS negative cases.
Introduction
T-cell lymphomas (TCL) account for less than 10% of all cases of non-Hodgkin lymphomas, but they represent the most aggressive lymphoid tumors.1 In a recent survey from the French Lymphopath Network, angioimmunoblastic T-cell lymphoma (AITL) and peripheral T-cell lymphoma not otherwise specified (PTCL NOS) were the most frequent subtypes, but also some of the most challenging tumors to diagnose.2 Distinguishing neoplastic from reactive T-cell infiltrates on the basis of immunomorphological criteria is getting increasingly difficult as ever-smaller biopsies are nowadays being performed. Given these difficulties, molecular techniques, such as multiplex polymerase chain reaction (PCR) assays to assess T-cell receptor (TCR) gene rearrangements are now widely used in daily practice. However, only 80% of TCL are found to show a PCR-detectable clone.3 In addition, some clonal lymphocyte populations may also appear in nonneoplastic situations such as infections, immunodeficiency, and autoimmune disorders. More recently, the emergence of next-generation sequencing (NGS), allowing for broad molecular profiling of tumors, has generated extensive additional information relevant for both malignancy diagnosis and tumor classifications.4-7 To the best of our knowledge, the diagnostic performance of targeted-NGS (t-NGS) techniques to assess T-cell malignancy has not yet been evaluated in routine practice.
Here, we compared the diagnostic performance of PCR-based clonality assessment to the detection of mutations through t-NGS techniques in a series of lymph node samples suspected of TCL.
Methods
A total of 107 formalin-fixed, paraffin-embedded samples with suspected noncutaneous TCL diagnosis on histopathological examination were collected in the Pathology Department of the Cancer University Institute of Toulouse. These samples were divided into 2 groups: a retrospective test cohort including samples collected from January 2018 to March 2020 (N = 57) and a prospective validation cohort, containing samples collected from March 2020 to December 2020 (N = 50). All samples were assessed through both multiplex PCR assay and t-NGS using a panel of 69 genes involved in lymphomagenesis. For each case, clinical data were retrieved to support the final diagnosis (supplemental Table 1). Additional methodology is provided in supplemental Methods and supplemental Table 2.
Results and discussion
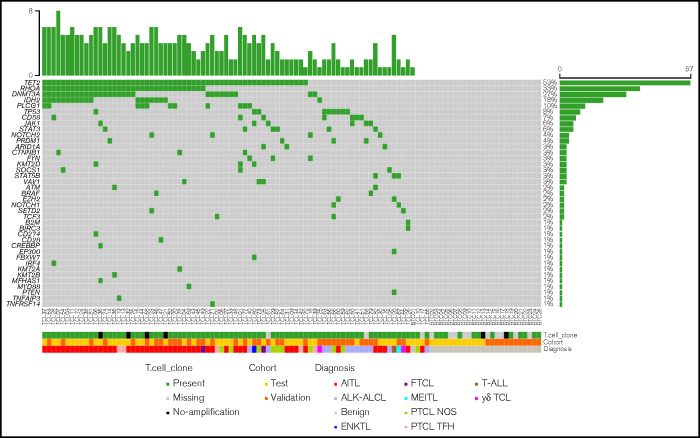
After clinicopathological correlations, TCL diagnosis was confirmed in 44/57 (77.2%) and 38/50 (76.0%) cases included in the test and validation cohorts, respectively, with a majority of AITL followed by PTCL NOS (supplemental Table 1). The remaining 13/57 (test cohort) and 12/50 (validation cohort) samples were considered nonneoplastic T-cell infiltrates (supplemental Table 1). The results provided by multiplex PCR and t-NGS are reported in Figure 1 and the related diagnostic performances in Table 1. The variants detected through t-NGS are detailed in supplemental Table 3.
Figure 1.
Targeted next-generation sequencing (t-NGS) and multiplex PCR-based clonality results of 82 T-cell lymphoma samples and 25 benign T-cell infiltrates divided into test (N = 57) and validation (N = 50) cohorts. Each column of this plot represents an individual case. Mutated genes are labeled in green and are sorted according to their mutation frequencies; the percentage of mutated cases is provided on the far right. For visualization purposes, only genes mutated in ≥1 case are displayed. Tracks at the bottom of the plot provide data on T-cell clone detection through multiplex PCR assays, on the cohort the case belongs to and on its diagnosis. Color codes are indicated in the key. AITL, angioimmunoblastic T-cell lymphoma; ALK-ALCL, ALK-negative anaplastic large cell lymphoma; ENKTL, extranodal NK/T-cell lymphoma nasal type; FTCL, follicular T-cell lymphoma; γδTCL, γδ T-cell lymphoma; MEITL, monomorphic epitheliotropic intestinal T-cell lymphoma; PTCL NOS, peripheral T-cell lymphoma not otherwise specified; PTCL TFH, peripheral T-cell lymphoma with T-follicular helper phenotype; T-ALL, T-cell acute lymphoblastic leukemia/lymphoma.
Table 1.
Diagnostic performances of PCR-based T-cell clonality assessment and mutation detection through targeted NGS (t-NGS) in T-cell lymphoma diagnosis
| Diagnosis | Diagnostic performances | ||||||
|---|---|---|---|---|---|---|---|
| TCL | BTC | Se (%) | Sp (%) | Accuracy (%) | |||
| (95% CI) | (95% CI) | (95% CI) | |||||
| Test cohort (N = 57) |
PCR | + | 39 | 7 | 95.1 (83.5-99.4) |
41.7 (15.2-72.3) |
83.0 (70.2-91.9) |
| − | 2 | 5 | |||||
| NC | 3 | 1 | |||||
| t-NGS | + | 42 | 0 | 95.5 (84.5-99.4) |
100.0 (75.3-100.0)* |
96.5 (87.9-99.6) |
|
| − | 2 | 13 | |||||
| NC | 0 | 0 | |||||
| Validation cohort (N = 50) |
PCR | + | 35 | 6 | 94.6 (81.8-99.3) |
45.5 (16.7-76.6) |
83.3 (69.8-92.5) |
| − | 2 | 5 | |||||
| NC | 1 | 1 | |||||
| t-NGS | + | 37 | 0 | 97.4 (86.2-99.9) |
100.0 (73.5-100.0)* |
98.0 (89.4-99.9) |
|
| − | 1 | 12 | |||||
| NC | 0 | 0 | |||||
1-sided 97.5% CI.
BTC, benign T-cell infiltrate; NC, noncontributory; Se, sensitivity; Sp, specificity; TCL, T-cell lymphoma.
First, considering the results provided by each molecular technique separately, a T-cell clone was detected through multiplex PCR assays in 39/41 (test cohort) and 35/37 (validation cohort) confirmed TCL samples with amplifiable DNA. However, a T-cell clone was also found in 7/12 (test cohort) and 6/11 (validation cohort) reactive T-cell infiltrates, leading to sensitivities of 95.1% (95% confidence interval [CI], 83.5-99.4) and 94.6% (95% CI, 81.8-99.3) and specificities of 41.7% (95% CI, 15.2-72.3) and 45.5% (95% CI, 16.7-76.6) in the test and validation cohorts, respectively. In 6 of 107 samples (5.6%), DNA quality was unsuitable to amplify TCR genes, leading to noncontributory multiplex PCR results. Then, according to the t-NGS approach, mutations in genes taking part in our lymphoma panel were detected in 42/44 (test cohort) and 37/38 (validation cohort) TCL samples and in none of the 25 reactive T-cell infiltrates assessed. Thus, t-NGS showed sensitivities of 95.5% (95% CI, 84.5-99.4) and 97.4% (95% CI, 86.2-99.9) in the test and validation cohorts, respectively, with 100% specificity in both of them (97.5% CI, 75.3-100.0 and 73.5-100.0) (Table 1). PCR and t-NGS techniques therefore showed similar performances in terms of sensitivity, with relative sensitivities of t-NGS on T-cell clonality of 1 (95% CI, 0.93-1.07) and 1.03 (95% CI. 0.93-1.13) in the test and validation cohorts, respectively. However, t-NGS appeared to be more specific than T-cell clonality with relative specificities of 2.4 (95% CI, 1.23-4.69) and 2.2 (95% CI, 1.15-4.20), respectively.
Second, the 4 TCL samples with nonamplifiable DNA through multiplex PCR assays were found to show some common TCL mutations (eg, mutations in RHOA, IDH2, PTEN, PLCG1)4,8 detected with t-NGS. In addition, 2 of the 3 t-NGS-negative TCL specimens had a PCR-detectable T-cell clone and only 1 case remained negative using both techniques. Taken together, our results strongly suggest that both molecular techniques should be combined to optimize atypical T-cell infiltrate diagnosis. However, the detection of a characteristic TCL mutation with t-NGS in a sample may be sufficient to assert malignancy and thus avoid clonality PCR analysis in more than 90% of cases. Similarly, a recent study focusing on screening for the RHOA (G17V) mutation has demonstrated the relevance of mutation detection for the early diagnosis of PTCL with T-follicular helper (TFH) phenotype in specimens that showed no evidence of lymphoma after combined histological and clonality analyses.9
Regarding our t-NGS approach, the pathogenicity of the mutations detected in samples with a negative PCR-based clonality might be questionable and raises the issue of clonal hematopoiesis (CH). Some of the genes included in our lymphoma panel have been shown to be related to CH (DNMT3A, EZH2, JAK2, TET2, TP53).10 Of interest, the 6 cases harboring negative clonality and positive t-NGS carried some mutations in major genes involved in lymphomagenesis, such as RHOA (G17V), IDH2 (R172), PTEN, and PLCG1,4,8 which have never been reported in CH to our knowledge. Thus, these 6 cases were considered to show true-positive t-NGS results. Furthermore, in our case series, the mutational profiles related to PTCL subtypes were consistent with the literature: the IDH2 (R172) mutation was found in AITL cases (N = 18/47) only and the RHOA (G17V) mutation was exclusive to PTCL cases with TFH phenotype (N = 34/54).5,11
Importantly, the 4 cases with nonamplifiable TCR genes but positive t-NGS indicated a difference regarding applicability between these molecular techniques. Within these cases, the failure to amplify TCR genes during multiplex PCR assays was related to the poor quality of the extracted DNA. Indeed, although BIOMED multiplex PCR assays require relatively large amounts of high-quality DNA,12 t-NGS techniques tolerate low input and variable DNA quality. Thus, t-NGS techniques should be prioritized over multiplex PCR assays for samples with a low quality or low quantity of extracted DNA.
Regarding the limitations of the study, the frequency of clonality in reactive cases was surprisingly high in both the test and validation cohorts (N = 7/13 and 6/12, respectively). This might be explained by the fact that, in practice, t-NGS is more commonly performed on clonality-positive T-cell infiltrates. In addition, further studies including cutaneous specimens would be of interest before extrapolating these results to cutaneous TCL.
In conclusion, our results confirm, in 2 independent cohorts, the major importance of molecular techniques in TCL diagnosis. Although the relevance of TCR gene rearrangement studies has long been proven,13,14 our study is the first to highlight that the mutational profile provides a reliable basis for T-cell malignancy diagnosis in routine histopathology. Despite the fact that T-cell clonality assessment by PCR appears to be less specific and requires higher quality DNA than t-NGS, both techniques remain complementary because PCR recovers some t-NGS-negative cases. Accordingly, we suggest a testing strategy using t-NGS as the primary test, followed by TCR clonality analysis if negative (supplemental Figure 1). Finally, NGS-based clonality assays are emerging in clinical laboratories,15-17 and the design of a single t-NGS test encompassing both clonal rearrangements of TCR genes and mutational status of target genes should represent an attractive alternative to conventional multiplex PCR in the near future.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgment
The authors thank Samira Icher for her help in collecting cases, as well as the different technicians who helped with the preparation of the patients’ samples and performed the NGS, and Ana Darquier for proofreading the manuscript.
Authorship
Contribution: P.B. is the principal investigator of this study and has full access to all of the included data and takes responsibility for its integrity and accuracy of its analysis; C.S., P.G., S.P., L.O., and L.Y. collected data; C.S., S.M.E., C.L., and L.L. performed data analysis; D.G. and F.E. contributed reagents, materials, and data analysis (NGS); B.C. performed statistical analysis; P.B. and C.S. wrote the manuscript; and all the coauthors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pierre Brousset, Department of Pathology, Cancer Institute University of Toulouse-Oncopole and Labex TOUCAN, 1 avenue Irène Joliot-Curie, 31059 Toulouse CEDEX 9, France; e-mail: brousset.pierre@iuct-oncopole.fr.
References
- 1.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laurent C, Baron M, Amara N, et al. Impact of expert pathologic review of lymphoma diagnosis: study of patients from the French Lymphopath Network. J Clin Oncol. 2017;35(18):2008-2017. [DOI] [PubMed] [Google Scholar]
- 3.Tan BT, Warnke RA, Arber DA.. The frequency of B- and T-cell gene rearrangements and Epstein-Barr virus in T-cell lymphomas: a comparison between angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified with and without associated B-cell proliferations. J Mol Diagn. 2006;8(4):466-475, quiz 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandell RF, Boddicker RL, Feldman AL.. Genetic landscape and classification of peripheral T cell lymphomas. Curr Oncol Rep. 2017;19(4):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watatani Y, Sato Y, Miyoshi H, et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019;33(12):2867-2883. [DOI] [PubMed] [Google Scholar]
- 6.Moffitt AB, Dave SS.. Clinical applications of the genomic landscape of aggressive non-Hodgkin lymphoma. J Clin Oncol. 2017;35(9):955-962. [DOI] [PubMed] [Google Scholar]
- 7.Yao W-Q, Wu F, Zhang W, et al. Angioimmunoblastic T-cell lymphoma contains multiple clonal T-cell populations derived from a common TET2 mutant progenitor cell. J Pathol. 2020;250(3):346-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017; 49(8):1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobson R, Du PY, Rásó-Barnett L, et al. Early detection of T-cell lymphoma with T follicular helper phenotype by RHOA mutation analysis [published online ahead of print 11 February 2021]. Haematologica. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman RL, Busque L, Levine RL.. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell. 2018;22(2):157-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Lee D, Brimer T, Hussaini M, Sokol L.. Genomics of peripheral T-cell lymphoma and its implications for personalized medicine. Front Oncol. 2020;10:898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Dongen JJM, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257-2317. [DOI] [PubMed] [Google Scholar]
- 13.Bertness V, Kirsch I, Hollis G, Johnson B, Bunn PA Jr. T-cell receptor gene rearrangements as clinical markers of human T-cell lymphomas. N Engl J Med. 1985;313(9):534-538. [DOI] [PubMed] [Google Scholar]
- 14.Mahe E, Pugh T, Kamel-Reid S.. T cell clonality assessment: past, present and future [published correction appears in J Clin Pathol. 2018;71(7):660]. J Clin Pathol. 2018;71(3):195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho CC, Tung JK, Zehnder JL, Zhang BM.. Validation of a next-generation sequencing-based T-cell receptor gamma gene rearrangement diagnostic assay: transitioning from capillary electrophoresis to next-generation sequencing. J Mol Diagn. 2021;23(7):805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schumacher JA, Duncavage EJ, Mosbruger TL, Szankasi PM, Kelley TW.. A comparison of deep sequencing of TCRG rearrangements vs traditional capillary electrophoresis for assessment of clonality in T-Cell lymphoproliferative disorders. Am J Clin Pathol. 2014;141(3):348-359. [DOI] [PubMed] [Google Scholar]
- 17.Greiner TC, Bagg A, Langerak AW.. Transitioning from T-cell clonality testing to high-throughput sequencing [published correction appears in J Mol Diagn. 2021;23(9):1205]. J Mol Diagn. 2021;23(7):781-783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.