Abstract
Poly(ADP‐ribose) polymerase 1 (PARP1) is a key mediator of various forms of DNA damage repair and plays an important role in the progression of several cancer types. The enzyme is activated by binding to DNA single-strand and double-strand breaks. Its contribution to chromatin remodeling makes PARP1 crucial for gene expression regulation. Inhibition of its activity with small molecules leads to the synthetic lethal effect by impeding DNA repair in the treatment of cancer cells. At first, PARP1 inhibitors (PARPis) were developed to target breast cancer mutated cancer cells. Currently, PARPis are being studied to be used in a broader variety of patients either as single agents or in combination with chemotherapy, antiangiogenic agents, ionizing radiation, and immune checkpoint inhibitors. Ongoing clinical trials on olaparib, rucaparib, niraparib, veliparib, and the recent talazoparib show the advantage of these agents in overcoming PARPi resistance and underline their efficacy in targeted treatment of several hematologic malignancies. In this review, focusing on the crucial role of PARP1 in physiological and pathological effects in myelodysplastic syndrome and acute myeloid leukemia, we give an outline of the enzyme’s mechanisms of action and its role in the pathophysiology and prognosis of myelodysplastic syndrome/acute myeloid leukemia and we analyze the available data on the use of PARPis, highlighting their promising advances in clinical application.
Introduction
Poly(ADP-ribose) polymerases (PARPs) are a family of enzymes that use the oxidized form of nicotinamide adenine dinucleotide (NAD+) to transfer ADP-ribose to other proteins (poly ADP-ribosylation). The family consists of at least 18 enzymes that are encoded by different genes and all share a conserved catalytic domain.1 Therefore, some isoforms, such as poly(ADP-ribose) polymerase 1 (PARP1), are most known for their implication in different cellular processes. PARP1 is an ∼113 kDa nuclear protein and the first member of the PARP family identified. It is connected to a plethora of cellular procedures such as DNA repair, transcriptional and posttranscriptional regulation of gene expression, control of protein degradation, and cell death.2 PARP1 is overexpressed in many cancers such as testicular and other germ cell tumors, neuroblastoma, malignant lymphoma, Ewing sarcoma, breast cancer, and colon cancer.3-5 It also contributes to progression of endometrial cancer, breast cancer (BRCA)-mutated ovarian cancer, and BRCA-mutated serous ovarian cancer.6 Although, most studies focus on the DNA damage detection and repair effects of the molecule, PARP1 has more recently been studied in the context of the regulation of chromatin structure and transcription and linked to DNA methylation, imprinting, and chromosome organization.7-9
Evaluating the DNA damage response (DDR) machinery as a therapeutic target is a promising strategy for designing novel pharmaceutical agents. DDR in hematological malignancies has been extensively studied but not fully understood. It has been reported that the BRCA1 expression level was found reduced in acute myelogenous leukemia (AML) samples. When AML was treated with DNA damaging agents, the loss of BRCA1 function led to the accumulation of genomic alterations, and even to synthetic lethality.10 Consequently, it is important to understand the roles of PARP1 in DNA repair to proceed to developing successful therapeutic regimens for treating different cancers.11 PARP inhibitors (PARPis) are already available and have shown significant benefits in a variety of malignancies. A study trying to illustrate the importance of oncogenic transcription factors for AML progression12 demonstrated for the first time a potential utility of PARPi-induced lethality for leukemia, driven by AML1-ETO and PML-RARa. AML cells with low expression of key members of the DDR pathway, such as Rad51, ATM, BRCA1, and BRCA2, displayed obvious sensitivity to PARPis.12 Furthermore, they showed that combining a PARPi with a GSK3 inhibitor proved an effective therapeutic strategy for PARPi-resistant AML. In this review, we give an outline of PARP1 mechanism of action7 (Figure 1), focusing on its crucial role in physiological and pathological effects in hematologic malignancies. Moreover, we analyze the available data on the use of PARPis in AML and/or myelodysplastic syndromes (MDS).
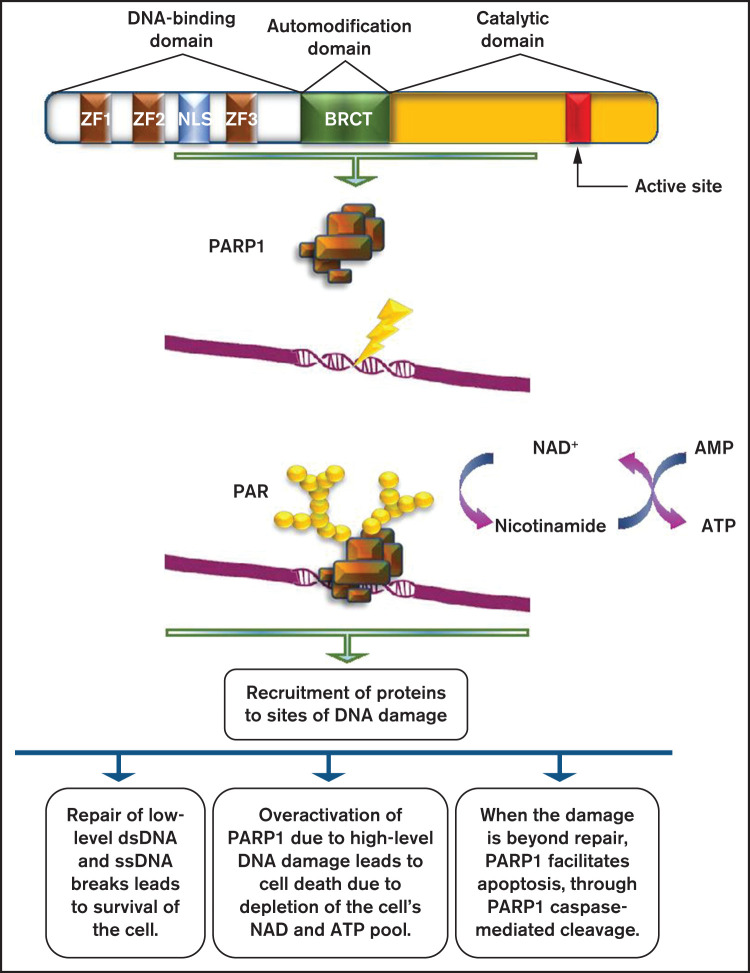
Figure 1.
Role of PARP1 in DNA damage repair. PARP1 consists of a DNA-binding domain with 3 zinc finger motifs, an automodification domain that contains the BRCA1 C terminus (BRCT) domain, and a carboxy-terminal catalytic domain, which contains the active site of the enzyme. PARP1 is usually activated by DNA damage occurring as a result of the DNA damage response. The net result of its activation is the production of PAR chains, with the use of nicotinamide adenine dinucleotide (NAD+) as substrate. PARylation results in the recruitment of several proteins with multiple roles on DNA damage repair.
PARP1 biochemical activities
In human cells, normal metabolic activities as well as environmental factors can lead to DNA damage, resulting in a range from 20,000 to 1 million individual molecular damages per cell per day.13,14 Consequently, the DNA repair machine responds to lesions by activating apoptosis.15 When normal repair processes fail to respond, nonrepairable DNA damage may arise, including double-strand breaks (DSBs) of DNA. Activation of PARP1 is a usual event in the DDR. Posttranscriptionally, PARPs attach to a polymer termed poly(ADP-ribose) (PAR), to PARP1 itself, and to other histone and nonhistone proteins within a dynamic process called poly(ADP ribosyl)ation (PARylation).16,17 DNA damage may produce extensive auto PARylation, resulting in the inhibition of PARP1 binding to DNA and to a decrease of its catalytic activities.6 Several kinds of DNA damage may cause rapid recruitment of PARP1 to the sites of damage through DNA binding. Single base alternations or nucleotide damage are the most common type of DNA damage. They can modify base-pairing nature and result in spontaneous mutations that are strongly associated with cancer predisposition. Single-strand breaks (SSBs) are rapidly detected and bound by PARP1.9,10 The rate of SSB repair (SSBR) is likely to be increased with the use of the molecule. SSBR activation could be mediated by the induction of accumulation of SSBR components such as DNA ligase 3 (LIG3), DNA polymerase β and bifunctional polynucleotide kinase 3′-phosphatase and/or their stabilization at SSBs,18 thus inducing the repair process. DNA-damaging agents, such as ionizing and ultraviolet radiation or impairments from by-products of the cell own metabolism are among the most common causes of DSB production.19 DDR proteins such as the ataxia telangiectasia mutated (ATM) kinase target DSBs. ATM consists of PAR-binding domains, and the stimulation of PAR activity depends on its interaction with them.20 However, there are other critical pathways for the activation of the DDR because PARP1-lack of function only delays the recruitment of certain proteins but does not completely suspend it.
Parthanatos
PARP1 function ranges from supporting survival to inducing death. Parthanatos (from the Greek Θανατoς, “death”), a form of programmed cell death, shares cytological and morphological features with apoptosis and necrosis, but in contrast to apoptosis, it is the result of a caspase-independent molecular mechanism.21,22 It is mediated by the pathologically high levels of PARP1 activity. The main event is the translocation of the apoptosis-inducing factor from mitochondria to the nucleus,23 leading to dissociation of histones and DNA degradation. PAR toxicity depends on the length and complexity of the PAR polymer. PAR polymers with a size greater than 60 ADP-ribose units are more toxic than less complex polymers. Delivery of >60 ADP-ribose polymer kills cells in a dose-dependent manner. The toxicity of PAR polymers is emphasized in studies of poly-(ADP) ribose glycohydrolase (PARG). PARG is responsible for encoding four isoforms, the nuclear 110-kDa isoform, the cytoplasmic 102-kDa and 99-kDa isoforms (detected only in humans), and the 59-kDa isoform (60 kDa in mice).24 Different sizes of PAR polymer can bind to proteins through either covalent or noncovalent binding. That several PARPs but only 2 PARGs have been identified so far makes it clear that the polymers produced by the PARP family members are structurally distinct, a fact that plays a role in the different functions of the several PARPs. Noncovalent binding of PAR to proteins is stable, whereas in vitro PAR binding is resistant to strong acids, chaotropes, detergents, and high salt concentrations. Limitation of PARP1 overactivation affects apoptosis-inducing factor release from the mitochondria, indicating that there is a nuclear-mitochondrial interaction that occurs in PARP1-mediated cell death. Because parthanatos subtends PARP1-mediated cell death through the actions of the PAR polymer, the next steps involve the identification of specific PAR targets and the designation of how PAR binding leads to cytotoxicity.
Methylation
Epigenetics is defined as heritable changes in gene expression that do not go along with changes in DNA sequence.25 Hypermethylation of CpG islands in gene promoters, histone deacetylation, and genome hypomethylation are common events in tumorigenesis. DNA methylation and histone deacetylation may be affected by hypomethylating agents (HMAs) and histone deacetylase inhibitors.26 PARP1 is related to DNA methylation and is correlated to gene expression silencing. Based on the enzyme’s involvement and regulating gene expression, a study reported its association with chromatin structure modification. The activity of enzyme was inhibited by a histone variant, thus leading to reactivation of a gene on the inactive X chromosome, establishing the participation of PARP1 in silencing conservation.27 Inhibitors of PARP1 have shown to act synergistically with HMAs. They inhibit DNA methylation by inactivating and trapping DNA methyltransferase (DNMT) to DNA, facilitating the repair of damaged DNA by the base excision repair machinery. PARP1 may affect methylation by regulating the expression of the DNA methyltransferase-1 (DNMT1) gene. Binding to the gene promoter and protecting it from methylation, it leads to induction of the DNMT1 activation.28 Inhibition of DNMT1 expression causes hypomethylation of the whole genome. This correlation of PARP1 with methylation is a key point in DNA repair, since hypermethylation is a major event in the pathogenesis of MDS and AML, and hypomethylating agents are an effective treatment option for these patients.
Targeting genetic abnormalities in MDS and AML
Gene mutations that are likely associated with AML can be classified into 9 distinct functional categories: the signaling genes (FLT3, KIT), the DNA-methylation-associated genes (DNMT3A, TET2, IDH1/2), the chromatin-modifying genes (ASXL1, EZH2, MLL), the nucleophosmin encoding gene (NPM1), the myeloid transcription factor genes (CEBPA, RUNX1), the transcription factor fusion genes (PML-RARA, RUNX1-RUNX1T1, CBFB-MYH11), the tumor suppressor genes (TP53, WT1), the spliceosome–complex genes (SF3B1, SRSF2, U2AF1), and the cohesin-complex genes (SMC1, SMC3, STAG2, RAD21). NPM1 and biallelic CEBPA mutations are associated with a better prognosis. In contrast, patients with mutations of FLT3-ITD, RUNX1, ASXL1, and TP53 are considered to present with a worse prognosis.29 That genomic alterations are present in AML and other hematological malignancies allows the presumption that these DNA changes might have as an origin the aberrant DSB repair.30-34 As an example, predisposition to develop MDS and AML is evident in patients with Fanconi anemia; a chromosomal disorder known for the mutation of homologous recombination (HR) components (BRCA2 [FANCD1], FANCO [RAD51C], FANCJ [BRIP1], and FANCN [PALB2]).35 Gene mutations in Fanconi anemia have been identified in therapy-related AML and MDS, predisposing to several malignancies.36,37
Patients with high-risk MDS have few treatment options and are often not eligible for intensive chemotherapy (ICT) because of comorbidities. They are treated with less aggressive therapies, including low-dose cytarabine and HMAs. ICT has been reported to induce a complete remission in ∼50% of higher-risk MDS patients, which is significantly lower than that of AML patients. The duration of response is usually short, and patients with adverse karyotypes or TP53 mutations have lower response rates.38 ICT can be used as a bridge to transplantation for patients failing 5-azacytidine (AZA).39
Venetoclax, is an orally selective inhibitor of the B-cell lymphoma 2 protein that, when acting synergistically with AZA, increases both response and prolonged survival as it was observed in a phase III trial.40,41 Indeed, prior trials trying to evaluate the efficacy of their combination comparing to venetoclax alone in AML patients, showed promising results.42,43 This combination therapy, which led to an astonishing complete remission rate in the venetoclax plus HMA cohort, resulted in its approval by the US Food and Drug Administration (FDA) in 2018.44 In a retrospective study with high-risk MDS patients, it was shown that an HMA plus venetoclax leads to high response rates.45 However, the addition of venetoclax to HMAs is associated with potent myelosuppression because of the bone marrow dysplasia and cytopenias, thus needing further investigation of this combination in clinical trials.41 At the time being, a phase I and 2 phase II trials evaluating venetoclax in combination with AZA in treatment-naïve higher risk MDS (NCT02942290), AML elderly patients with no previous treatment (NCT03466294) and MRD+ patients after AML/MDS allogeneic hematopoietic stem cell transplantation (NCT04809181) are under investigation.46-48
Glasdegib is a potent oral inhibitor of the Hedgehog signaling pathway, producing rapid and complete tumor regression as a single agent or in combination with chemotherapy, reduced expression of key leukemia stem cell regulators, and decreased leukemia stem cell populations in patient-derived AML cells.49-51
Lenalidomide is widely used in MDS. The combination of lenalidomide and AZA has significant clinical activity and can induce cytogenetic responses in patients with del(5q). However, controlled studies have failed to show a clinical benefit of adding lenalidomide to azacytidine in mixed cohorts of all cytogenetic subtypes. Guadecitabine is a novel azanucleoside, with clinical activity in refractory patients but the usefulness of this drug needs to be investigated in larger trials.52,53
Immune checkpoint inhibitors as monotherapy seem to have modest clinical activity in MDS, although there are limited published data.54,55 Studies have evaluated the clinical usefulness of combining AZA with immune checkpoint inhibitors.56
Midostaurin, enasidenib, and ivosidenib, 3 kinase inhibitors that are orally bioavailable, were approved in 2017 and 2018 for the treatment of AML. Midostaurin has been approved for targeting the FLT3-mutated AML combined with the “7 plus 3” regimen.57 Gilteritinib, a small molecule type I FLT3 inhibitor is being studied both in newly diagnosed AML and in refractory or relapsed AML. It was found to have efficacy alone or in combination with chemotherapy or azacytidine, compared with azacytidine as monotherapy.58 Enasidenib (isocitrate dehydrogenase 2 [IDH2] inhibitor) and ivosidenib (IDH1 inhibitor) have been approved for the treatment of adults with IDH1- or IDH2-mutated relapsed/refractory AML.59,60 Moreover, BCR-ABL1 kinase inhibitors have been approved for targeting the rare subtype of BCR-ABL1+ AML. Published preclinical data regarding the development of PARPis are listed and analyzed in Table 1.
Table 1.
Preclinical data on the development of PARPi
| Reference | Genetics/studied parameters | Disease | Sensitivity to PARPi/results |
|---|---|---|---|
| Esposito et al, 2015 | Synthetic lethality of oncogenic transcription for leukemia treatment | AML | Sensitivity to PARPis of AML cells with low expression of members of the DDR pathway. AML cells driven by repressive transcription factors, including AML1-ETO and PML-RARα fusion oncoproteins, are sensitive to PARPis. Sensitivity to PARPis of AML cells with low expression of Rad51, ATM, BRCA1, and BRCA2. Genetic or pharmacological inhibition of HOXA9 impairs DDR and sensitizes MML leukemia to PARPis. |
| Zampieri et al, 2009 | DNA methylation | Ovarian cancer, breast cancer | PARPis with HMAs lead to synergistic inhibition. Inactivation and trapping of DNMT to DNA facilitates the role of BER machinery. |
| Lord et al, 2017 | Synthetic lethality, DNA repair | Ovarian cancer, breast cancer | PARPis trap PARP1 on DNA, preventing autoPARylation and PARP1 release from the site of damage of BRCA-mutant cells. |
| Boussios et al, 2012 | Synthetic lethality, DNA repair | Ovarian cancer, breast cancer | Tumors carrying mutations in BRCA1/2 implicated in homologous repair deficiency are particularly sensitive to PARPis. |
| Meng et al, 2014 | Apoptosis, knockdown of PARP1 and/or PARP2 | AML | Synergistic action of PARPi with death ligands results in enhanced expression of DR5 and Fas and sensitivity to treatment with multiple death ligands (agonistic anti-Fas antibody, recombinant human TRAIL, and agonistic anti-DR5 antibody). |
| Faraoni et al, 2018 | Apoptosis resistance, modulation of FAS and TRAIL receptors | AML | AML ΒΜ samples express FAS and DR5 transcripts at lower levels than normal BM. Apoptosis triggered by olaparib is associated with a dose-dependent up-regulation |
| Maifrede et al, 2017 | PARP1 knockdown | AML bearing MLL translocations | inhibitors of PARP1 enhance the therapeutic effect of cytotoxic drugs against MLL leukemias. |
| Molenaar et al, 2018 | Correlation of IDH1/IDH2 mutations to DNA damage and responses to PARPis | AML | IDH1/2MUT cells are sensitive to PARPi as monotherapy or/and in combination with DNA-damaging agents. Concomitant administration of IDH1/2MUT inhibitors during cytotoxic therapy decrease the efficacy of both agents in IDH1/2MUT AML. |
| Faraoni et al, 2014 | Αpoptosis, in vitro sensitivity to olaparib | AML | Olaparib induced cell death in the majority of AML samples (88%) and tested cell lines. Olaparib preferentially killed leukemic blasts and did not affect the viability of normal BM and CD34− peripheral blood cells. |
| Nieborowska-Skorska et al, 2017 | DNA repair | MPN | PARPi combination with ruxolitinib-mediated inhibition of DSB repair and/or hydroxyurea causes accumulation of lethal DSBs, resulting in elimination of MPN cells. |
| Patel et al, 2019 | DNA repair, genomic instability | MPN | In veliparib and busulfan treated SET2 and HEL cells, veliparib decreased busulfan’s IC50. Combination treatment of SET2 cells caused G2M arrest in 53% of cells, compared with 30% with veliparib alone and 35% with busulfan alone. |
| Muvarak et al, 2016 | DNA damage-related binding between DNMTs and PARP1 | AML, breast cancer | Combining DNMTi and PARPi (talazoparib) increases tight binding of PARP1 in chromatin, frequency of DSBs, and synergistic cytotoxicity while it decreases clonogenicity |
| Zhao et al, 2017 | Synthetic lethality | AML driven by MLL fusion proteins | Combining olaparib with DNMT inhibitor induce cell-cycle block and apoptosis. Olaparib can sensitize MLL leukemic cells to both DNMT inhibitors and chemotherapy agents. |
AML-ETO, acute myeloid leukemia-eight twenty-one oncoprotein; ATM, ataxia-telangiectasia mutated; BER, base excision repair; BM, bone marrow; DR5, death receptor 5; FAS, FS-7-associated surface antigen; HEL, hevein-like preprotein; IC50, half-maximal inhibitory concentration; MML, myelomastocytic leukemia; PML-RARα, promyelocytic leukemia/retinoic acid receptor α; TRAIL, tumor necrosis factor-related apoptosis inducing ligand; SET2, nucleosomal histone H3-selective methyltransferase.
PARPis
Synthetic lethal targeted therapy
Almost 15 years ago, 2 groups described the “synthetic lethal (SL) interaction” between PARP inhibition and BRCA1 or BRCA2 mutations, suggesting a new treatment of patients with BRCA-mutant tumors.61,62 SL as a concept was initially introduced a century ago by geneticists to describe the situation in which a defect in either 1 of 2 genes has little impact on the cell/organism, but the combination of defects in both genes results in death.63 It is quite important to achieve a better understanding of what causes/triggers an SL interaction, which are the factors that determine the robustness of SL interactions, and how these interactions can be predicted. For example, it has been suggested that protein–protein interaction networks are used to identify robust SL interaction effects associated with different pairs of genes.64 Moreover, proteins with similar functions seem to share SL interactions, leading to computational approaches that identify SL relationships.65
Clinical development of PARPi
PARPis are small molecules that compete with the oxidized form of nicotinamide adenine dinucleotide (NAD+) at the catalytic pocket of PARPs66 (mainly PARP1 and PARP2) (Figure 2), resulting in the inhibition of DNA repair enzymatic activities regulated by PARylation.67 Some PARPis can trap PARP1 and PARP2 on the so-called endogenous DNA breaks.68,69 Currently, PARPis such as olaparib, rucaparib, and niraparib are approved by the FDA and the European Medicine Agency for the treatment of ovarian cancer, whereas veliparib is in the latest stage of clinical development.70 Talazoparib was approved by the FDA for the treatment of metastatic germline BRCA1/2-mutated breast cancer in October 2018. They have been studied both as single agents and in combination with chemotherapy, antiangiogenic agents, and ionizing radiation.71 The PARP trapping depends on a toxic allosteric effect that varies among PARPis (talazoparib > niraparib > olaparib = rucaparib > veliparib).72 The mechanisms involved in PARPi cytotoxicity vary depending on the type of the defective DNA repair pathway and on the status of tumor cell proliferation.56
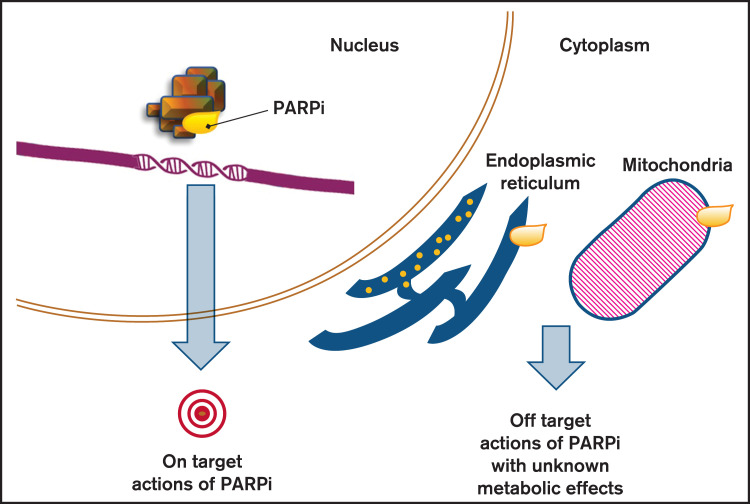
Figure 2.
Effects of PARP inhibition. PARPis are able to create their own interaction network with proteins outside the nucleus, beside the sole blockage of PARP (on target action of PARPis). By inducing signaling pathways and impacting secondary proteins, they can affect cell functions and cause metabolic responses that constitute the off-target actions of PARPi.
In addition to DNA repair, PARPs are able to control cellular functions that are important for tumor survival by regulating the chromatin structure and modifying the signaling molecules that interfere with gene transcription.73-77 Studies performed in primary AML blasts78 and tumor cell lines79,80 revealed that the PARPi cytotoxic effect could be due to upregulation of death receptor 5 (DR5) and FS-7-associated surface antigen (FAS) death receptors that respectively requires the activation of SP1 and NF-KB. As a result, leukemia cells are sensitized to produce endogenously Fas ligand (FASL) and TNF-related apoptosis inducing ligand (TRAIL). In conclusion, PARPis can cause tumor cytotoxicity via several pathways and mechanisms that may be different from those associated with DNA damage sensing and repair.81
The clinical studies initially focused on hereditary tumors such as breast, prostate, ovarian, and pancreatic cancers. These tumors are associated with pathogenic mutations in the BRCA genes.82-84 Olaparib was approved in 2014 as maintenance therapy for recurrent, BRCA-mutated ovarian cancer, or for the treatment of BRCA-mutated advanced ovarian cancer (after 3 or more lines of chemotherapy).85 In 2018, olaparib was finally approved by the FDA in first-line maintenance setting of BRCA-mutated advanced ovarian cancer. Niraparib and rucaparib have shown promising results when used as maintenance treatment of recurrent platinum-sensitive ovarian cancer after completion of platinum-based chemotherapy.86,87
Olaparib and talazoparib were recently approved (2018) for HER2-negative metastatic breast cancer in patients with deleterious/suspected deleterious germline mutations of BRCA genes (after at least 2 lines of chemotherapy).88,89 Some other PARPis that are being investigated in clinical trials are veliparib, pamiparib, fluzoparib, and 2X-121.90-92
PARPi activity as monotherapy in AML and MDS
Almost a decade ago, it was suggested that PARPi therapy could be beneficial for MDS and AML. More specifically, microsatellite instability was associated with downregulation of HR repair genes in MDS/AML. In fact, mutations in CtIP (high-risk MDS) and in CtIP and meiotic recombination protein 11 (primary AML samples) had as a result the leukemia cells to be sensitive to PARPis.93 Additionally, it was reported that samples from myeloproliferative neoplasms (MPN) patients, with a tendency to evolve to AML, were more sensitive to olaparib and veliparib compared with normal bone marrow cells. Additionally, myeloblasts with nonmutated JAK2 were more sensitive to PARPi than JAK2-V617F mutated samples.94 This effect was the result of JAK2-V617F ability to activate transcription through signal transducer and activator of transcription and to inhibit apoptosis.95
The t(8;21) (RUNX1-RUNX1T1) is a repeated chromosomal translocation in AML. A relevant study showed that transformed RUNX1-RUNX1T1 primary hematopoietic cells (mice) were very sensitive to olaparib and veliparib compared with bone marrow cells. PARPi had little effect in the presence of the t(9;11) (p21; q23)(mixed lineage leukemia [MLL]-MLLT3≈MLL-AF9) and t(1;19)(TCF3-PBX1≈E2A-PBX) translocation.12 Another study demonstrated that olaparib inhibited the proliferation and quiescence of MLL-AF9 AML stem cells proposing that it is possible for PARPi to be used for MLL-rearranged leukemia.96
IDH1/2 mutations in primary AML cells demonstrated induction of HR defects and decrease in ATM expression that renders AML cells sensitive to PARPi. The IDH1/2 inhibitors can protect cells against PARPi because they restore ATM expression and decrease DNA damage. Thus, combining PARPi with IDH1/2 inhibitors is better to be avoided in IDH1/2 mutated AML.97
The levels of PARP1 protein were considered to affect tumor response to PARPis. PARP1 and PARP298-100 were studied in primary AML and MDS.101,102 However, the expression of PARP1/2 was not directly associated with sensitivity to PARPis. High levels of the MPL proto-oncogene thrombopoietin receptor (upregulated in RUNX1/RUNXT1 AML) correlate with PARP expression.87,103 MPL stimulation by thrombopoietin results in the activation of AKT1 and ERK1/2 pathways leading to proliferation and apoptosis resistance of leukemia cells.83 PARP1 is an important mediator of translocation common in AML (via the Alt-EJ pathway).104 However, because data for MDS and AML treated with PARPi are still scarce, some concerns have arisen regarding delayed adverse events from PARPi. An observational clinical study (NCT04326023) with 178 enrolled patients, using the VigiBase database, tried to investigate potent adverse events related to PARPis. It was observed that PARPis increased the risk of MDS and AML vs placebo treatment, suggesting that these adverse events need further evaluation.105 Currently, a phase II clinical trial (NCT03953898) compares the efficacy of olaparib in IDH mutated MDS and refractory/relapsed (R/R) AML patients to that of standard chemotherapy.106 Alongside, an ongoing phase I pilot study (NCT03974217) examines if mutations in the cohesin complex can represent a therapeutic target for talazoparib, a new experimental drug, in this subset of patients.107 Results of clinical trials on the use of PARPis in several malignancies are presented and analyzed in Table 2.
Table 2.
Results from clinical trials of PARPi in several malignancies
| Clinical trial (EudraCT number)/reference | Disease | Therapy | Potential mechanism | Results |
|---|---|---|---|---|
| NCT00753545 L edermann et al, 2014 | Ovarian cancer | Olaparib 400 mg BID vs placebo | Synthetic lethality DNA repair |
BRCA+: median PFS significantly longer in the olaparib group OS not significantly different between groups Serious adverse events reported in 25 (18%) patients under olaparib and 11 (9%) under placebo. |
| NCT01891344 Swisher et al, 2017 | Ovarian cancer | Rucaparib 600 mg PO, BID |
Homologous recombination deficiency DNA methylation Genomic LOH |
BRCA+: PFS longer in high LOH Grade ≥3 treatment-related adverse events: anemia (45 [22%] patients) and elevations in ALT/AST (25 [12%]) |
| NCT01618136 Plummer et al, 2020 | Ovarian cancer, B-cell malignancies, malignant solid tumors, triple-negative breast cancer, advanced melanoma | E7449 50-800 mg QD, PO E7449 plus TMZ QD, PO E7449 plus carboplatin and paclitaxel QD, PO |
PARP-DNA trapping Synthetic lethality |
Antitumor activity of E7449 in 13 patients, durable in 8. The 2X-121 DRP identified patients achieving PR and durable SD. E7449: good tolerability, promising antitumor activity and significant concentration-dependent PARP inhibition. |
| NCT043260230 Morice et al, 2020 | MDS, AML (review of randomized controlled trials) | Olaparib, rucaparib, niraparib, talazoparib, veliparib | PARPis significantly increased the risk of MDS and AML compared with placebo treatment with no between-study heterogeneity. | |
| NCT03953898 Reference number106 | R/R AML, MDS | Olaparib | Inhibition of cancer cell growth by blocking enzymes needed for cell growth. | No results Still recruiting |
| NCT03974217 Reference number107 | Leukemia | Talazoparib | Synthetic lethality (leukemia cells with a mutation in cohesin may be dependent on PARP activity to survive; when inhibiting PARP with talazoparib the leukemia cells die) | No results Still recruiting |
| NCT00588991 Reference number122 | R/R AML, high-risk MDS, aggressive myeloproliferative disorder | Veliparib Carboplatin Topotecan Hydrochloride |
DNA repair cytotoxicity of multiple classes of chemotherapy drugs, including topoisomerase I inhibitors and platinating agents. | No results/not recruiting Veliparib/topotecan/carboplatin combination warrants further investigation |
| NCT04207190 Reference number123 | R/R AML | Gemtuzumab- ozogamicin, talazoparib, talazoparib tosylate | PARP1 trapping Potential ability of talazoparib to enhance levels of DNA damage induced by GO therapy. |
Still recruiting |
| NCT02878785 Reference number124 | Untreated AML, R/R AML | Decitabine, talazoparib | SSB repair | Active, not recruiting Suggestion that talazoparib will increase the effects of decitabine in leukemia cells |
BID, twice per day; DRP, drug response predictor; EudraCT, European Union Drug Regulating Authorities Clinical Trials Database; GO, gemtuzumab ozogamicin; LOH, loss of heterozygosity; OS, overall survival; PFS, progression-free survival; PO (per os), oral administration; PR, partial response; QD, once a day; SD, stable disease.
Candidate biomarkers of sensitivity to PARPi
Finally, special attention has been devoted to the discovery of predictive biomarkers focusing on the identification of patients that are likely to benefit from PARPi. Knowledge of the homologous recombination deficiency (HRD) status seems to be crucial for treatment decision. PARPis selectively target HRD, especially in tumors from patients bearing the deleterious mutations of BRCA1 and 2. Up to 55% of sporadic epithelial ovarian cancer are homologous recombination repair-deficient for several reasons, including BRCA1 promoter methylation, meiotic recombination protein 11 mutation and BRCA2-interacting transcriptional suppressor overexpression.108,109 Reliable candidate biomarkers for the identification of patients with defects in the HR pathway would expedite the clinical development of PARPi-based therapies. BRCA1/2 loss-of-function mutation impairs homologous recombination repair and induces PARP hyperactivation reflected by an increased abundance of PARs.110,111 HRD may occur without BRCA mutation in the context of “BRCAness.”112 BRCA2-interacting transcriptional suppressor genes and ETS fusion genes that are reported in different tumors inhibit BRCA2.111,113 In ovarian cancer, the importance of platinum sensitivity, which seems to be one of the most credible biomarkers is distinct, but because this information is not to our knowledge at the start of chemotherapy, we need to rely more on the HRD and BRCA status to identify patients that are likely to respond to PARPis.114 In AML, despite the important efforts in understanding the molecular basis of the disease, there are still more to be elucidated. A study performed in AML cell lines observed a direct correlation of RAD51 and an inverse correlation of γH2AX with olaparib half-maximal inhibitory concentration.100 Additionally, the PARPi caused a dose-dependent increase in the number of γH2AX+ cells, which suggested a possible sensitivity to olaparib.100 These data support the role of the repair proteins RAD51 and γH2AX as potential markers of sensitivity of AML cells to olaparib. In conclusion, multiple molecular signatures in combination with a personalized approach should be considered for the prediction of patients who may benefit from PARPi therapy.
PARPi-based combination therapies
Combination therapies evaluate the effects of PARPi interaction with agents that target genetic alterations present in AML/MDS and MPN. Agents with lower toxicity in comparison with chemotherapy are better to be combined with PARPis. The JAK2-V617F mutation is known for unrestrained HR activity resulting in genetic instability. It is the most frequent genetic alteration in MPN and ruxolitinib (JAK1/2 inhibitor) has been approved for the treatment myelofibrosis and polycythemia vera in patients resistant to hydroxyurea.115 Ruxolitinib treatment sensitizes MPN cells to SL caused by talazoparib.116 The JAK2-V617F MPN model was also studied for the combination of a PARPi with busulfan (component of regimens of allo-HSCT).117 It was assumed that combining busulfan with a PARPi might lead to increased efficacy. More precisely, when busulfan and veliparib were combined, the result was synergistic cytotoxicity in mutated MPN cells.118
FLT3-ITD-mutated AML is characterized by increased genomic instability (through Alt-EJ repair factors, LIG3, and PARP1), associated with induction of MYC expression.119-121 Additionally, when FLT3-ITD cells are mutated, they present an increase in ROS production and evidence of interchromosomal HR which may result in loss of heterozygosity, which is common in myeloid malignancies.122,123 When quizartinib, a tyrosine kinase inhibitor, inhibits FLT3-ITD, there is a decrease in HR and NHEJ protein expression, an increase in DSB formation and sensitization of FLT3-ITD+ AML cells to olaparib and talazoparib. More precisely, although quizartinib mainly exhibits cytostatic effects in proliferating cells,124 PARPis exhibit cytotoxic effects in NHEJ-deficient AML cells. This is an important conclusion, because therapeutic options for AML usually fail to eliminate quiescent drug-refractory leukemia stem cells. As a result, many patients relapse after allo-HSCT.125
Another case of combination therapy is PARPi synergizing with the AZA and decitabine used in MDS and AML. In this case, there is reactivation of hypermethylated tumor suppressor genes involved in the differentiation, transformation, and induction of DNA damage. Some groups have shown that low doses of 5-azacitidine or decitabine together with talazoparib or olaparib have as a result synergistic cytotoxicity in AML cells (because of increased DNA damage and delayed DNA repair).126,127
Finally, with the PARPis being under investigation in vitro, in vivo, and in clinical trials as monotherapy or in combination with chemotherapy, progress in their development has led to a better understanding of the contribution of different mechanisms of action and to improvement of their therapeutic potential.128 However, because of the concerning potential adverse events associated with MDS/AML, especially with the BRCA-mutated patients, further investigations should be carried out for prolonged toxicity. An active phase I clinical trial (NCT00588991) is trying to study the side effects and best dose of veliparib combined with chemotherapy in patients with high-risk MDS, R/R AML, or aggressive MPN to address the optimal inhibition of cancer cell growth.129 To date, talazoparib is being studied in a phase I/Ib clinical trial in combination with gemtuzumab ozogamicin in CD33+ patients with R/R AML, to assess if the addition of talazoparib in the existing therapy increases its efficacy.130 In a phase II interventional trial (NCT02878785), talazoparib is given in combination with decitabine, to find out if these 2 agents work better together than decitabine alone for the treatment of AML.131
Conclusion
To date, thanks to novel technologies, accumulating facts from preclinical studies point out PARPi as promising therapies entering many clinical trials. Because of the different profiles of each agent, patient-specific factors should be taken into consideration when deciding which inhibitor to use as the preferable option.132 Research and development of novel targets related to other PARP family members may also contribute to the revealing of new drugs and the further comprehension of the biological and clinical role of PARPi.93 A recent study suggested that the positive correlation of PARP1, PARP2, PARP3, and TRPM2 genes in physiological cells, is disturbed in patients with AML.133 Consequently, the need for further research of the mutual expression and regulation of different PARP family members is mandatory to identify possible biomarkers, provide more perspectives for optimal PARPi-based combination outline, and outspread their therapeutic outlook.
Authorship
Contribution: All authors participated in designing the review article; C.-N.K. and D.T. participated in drafting the manuscript; P.T.D. and N.-A.V. contributed in the critical review of the article; and all authors have read and approved the manuscript.
Conflict-of-interest disclosure: P.T.D. reports personal fees for presentations for Roche, Celgene, and Novartis. N.-A.V. reports investigational grants and personal fees for presentations and advisory roles from Celgene/Genesis Pharma. The remaining authors declare no competing financial interests.
Correspondence: Christina-Nefeli Kontandreopoulou, Hematology Unit, First Department of Internal Medicine, National and Kapodistrian University of Athens, “Laikon” General Hospital, Athens 11527, Greece; e-mail: elina_knt@hotmail.com.
References
- 1.Morales J, Li L, Fattah FJ, et al. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr. 2014;24(1):15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vyas S, Matic I, Uchima L, et al. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. 2014;5(1):4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mego M, Cierna Z, Svetlovska D, et al. PARP expression in germ cell tumours. J Clin Pathol. 2013;66(7):607-612. [DOI] [PubMed] [Google Scholar]
- 4.Newman EA, Lu F, Bashllari D, Wang L, Opipari AW, Castle VP.. Alternative NHEJ pathway components are therapeutic targets in high-risk neuroblastoma. Mol Cancer Res. 2015;13(3):470-482. [DOI] [PubMed] [Google Scholar]
- 5.Tomoda T, Kurashige T, Moriki T, Yamamoto H, Fujimoto S, Taniguchi T.. Enhanced expression of poly(ADP-ribose) synthetase gene in malignant lymphoma. Am J Hematol. 1991;37(4):223-227. [DOI] [PubMed] [Google Scholar]
- 6.Manchana T, Phoolcharoen N, Tantbirojn P.. BRCA mutation in high grade epithelial ovarian cancers. Gynecol Oncol Rep. 2019;29(29):102-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray Chaudhuri A, Nussenzweig A.. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18(10):610-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajawat J, Shukla N, Mishra DP.. Therapeutic targeting of poly(ADP-ribose) polymerase-1 (PARP1) in cancer: current developments, therapeutic strategies, and future opportunities. Med Res Rev. 2017;37(6):1461-1491. [DOI] [PubMed] [Google Scholar]
- 9.Krishnakumar R, Kraus WL.. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39(1):8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Topatana W, Juengpanich S, et al. Development of synthetic lethality in cancer: molecular and cellular classification. Signal Transduct Target Ther. 2020;5(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satoh MS, Lindahl T.. Role of poly (ADP-ribose) formation in DNA repair. Nature. 1992;26(6367):356-358. [DOI] [PubMed] [Google Scholar]
- 12.Esposito MT, Zhao L, Fung TK, et al. Synthetic lethal targeting of oncogenic transcription factors in acute leukemia by PARP inhibitors. Nat Med. 2015;21(12):1481-1490. [DOI] [PubMed] [Google Scholar]
- 13.Lodish H, Berk A, Matsudaira P, et al. Molecular Biology of the Cell. 5th ed. New York: WH Freeman; 2004:963 [Google Scholar]
- 14.Vilenchik MM, Knudson AG.. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci USA. 2003;100(22):12871-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Amours D, Sallmann FR, Dixit VM, Poirier GG.. Gain-of-function of poly(ADP-ribose) polymerase-1 upon cleavage by apoptotic proteases: implications for apoptosis. J Cell Sci. 2001;114(Pt 20):3771-3778. [DOI] [PubMed] [Google Scholar]
- 16.Piao L, Fujioka K, Nakakido M, Hamamoto R.. Regulation of poly(ADP-ribose) polymerase 1 functions by post-translational modifications. Front Biosci. 2018;23(23):13-26. [DOI] [PubMed] [Google Scholar]
- 17.Wei H, Yu X.. Functions of PARylation in DNA damage repair pathways. Genomics Proteomics Bioinformatics. 2016;14(3):131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher AE, Hochegger H, Takeda S, Caldecott KW.. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007;27(15):5597-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F.. Poly (ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp Cell Res. 2014;329(1):18-25. [DOI] [PubMed] [Google Scholar]
- 20.Rondeau S, Vacher S, De Koning L, et al. ATM has a major role in the double-strand break repair pathway dysregulation in sporadic breast carcinomas and is an independent prognostic marker at both mRNA and protein levels. Br J Cancer. 2015;112(6):1059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fatokun AA, Dawson VL, Dawson TM.. Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. Br J Pharmacol. 2014;171(8):2000-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.David KK, Andrabi SA, Dawson TM, Dawson VL.. Parthanatos, a messenger of death. Front Biosci. 2009;14(14):1116-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Kim NS, Haince JF, et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci Signal. 2011;4(167):ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassa PO, Haenni SS, Elser M, Hottiger MO.. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70(3):789-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miranda TB, Jones PA.. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213(2):384-390. [DOI] [PubMed] [Google Scholar]
- 26.Valdez BC, Li Y, Murray D, et al. Combination of a hypomethylating agent and inhibitors of PARP and HDAC traps PARP1 and DNMT1 to chromatin, acetylates DNA repair proteins, down-regulates NuRD and induces apoptosis in human leukemia and lymphoma cells. Oncotarget. 2017;9(3):3908-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nusinow DA, Hernández-Muñoz I, Fazzio TG, Shah GM, Kraus WL, Panning B.. Poly (ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem. 2007;282(17):12851-12859. [DOI] [PubMed] [Google Scholar]
- 28.Zampieri M, Passananti C, Calabrese R, et al. Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PLoS One. 2009;4(3):e4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ley TJ, Miller C, Ding L, et al. ; Cancer Genome Atlas Research Network . Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaymes TJ, Mufti GJ, Rassool FV.. Myeloid leukemias have increased activity of the nonhomologous end-joining pathway and concomitant DNA misrepair that is dependent on the Ku70/86 heterodimer. Cancer Res. 2002;62(10):2791-2797. [PubMed] [Google Scholar]
- 31.Rassool FV, Gaymes TJ, Omidvar N, et al. Reactive oxygen species, DNA damage, and error-prone repair: a model for genomic instability with progression in myeloid leukemia? Cancer Res. 2007;67(18):8762-8771. [DOI] [PubMed] [Google Scholar]
- 32.Zhao L, So CW.. PARP-inhibitor-induced synthetic lethality for acute myeloid leukemia treatment. Exp Hematol. 2016;44(10):902-907. [DOI] [PubMed] [Google Scholar]
- 33.Takagi M. DNA damage response and hematological malignancy [published correction appears in Int J Hematol. 2017;106(3):450-453]. Int J Hematol. 2017;106(3):345-356. [DOI] [PubMed] [Google Scholar]
- 34.Delia D, Mizutani S.. The DNA damage response pathway in normal hematopoiesis and malignancies. Int J Hematol. 2017;106(3):328-334. [DOI] [PubMed] [Google Scholar]
- 35.Taniguchi T, D’Andrea AD.. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107(11):4223-4233. [DOI] [PubMed] [Google Scholar]
- 36.Voso MT, Fabiani E, Zang Z, et al. Fanconi anemia gene variants in therapy-related myeloid neoplasms. Blood Cancer J. 2015;5(7):e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Churpek JE, Marquez R, Neistadt B, et al. Inherited mutations in cancer susceptibility genes are common among survivors of breast cancer who develop therapy-related leukemia. Cancer. 2016;122(2):304-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadia TM, Jain P, Ravandi F, et al. TP53 mutations in newly diagnosed acute myeloid leukemia: clinicomolecular characteristics, response to therapy, and outcomes. Cancer. 2016;122(22):3484-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murakami K, Ueno H, Okabe T, et al. Bridging-to-transplant with azacitidine for myelodysplastic syndrome and acute myeloid leukemia, reduces the incidence of acute graft-versus-host disease. Hematol Rep. 2017;9(2):7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ball BJ, Famulare CA, Stein EM, et al. Venetoclax and hypomethylating agents (HMAs) induce high response rates in MDS, including patients after HMA therapy failure. Blood Adv. 2020;4(13):2866-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiNardo C, Jonas B, Pullarkat V, et al. A randomized, double-blind, placebo-controlled study of venetoclax with azacitidine vs azacitidine in treatment-naïve patients with acute myeloid leukemia ineligible for intensive therapy-viale-a. Available at: https://library.ehaweb.org/eha/2020/eha25th/303390/courtney.dinardo.a.randomized.doubleblind.placebocontrolled.study.of.html. Accessed 6 June 2020.
- 42.DiNardo CD, Pratz KW, Letai A, et al. Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: a non-randomised, open-label, phase 1b study. Lancet Oncol. 2018;19(2):216-228. [DOI] [PubMed] [Google Scholar]
- 43.DiNardo CD, Pratz K, Pullarkat V, et al. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood. 2019;133(1):7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia JS. Prospects for venetoclax in myelodysplastic syndromes. Hematol Oncol Clin North Am. 2020;34(2):441-448. [DOI] [PubMed] [Google Scholar]
- 45.Azizi A, Ediriwickrema A, Dutta R, et al. Venetoclax and hypomethylating agent therapy in high risk myelodysplastic syndromes: a retrospective evaluation of a real-world experience. Leuk Lymphoma. 2020;61(11):2700-2707. [DOI] [PubMed] [Google Scholar]
- 46.A Study Evaluating Venetoclax in Combination With Azacitidine in Subjects With Treatment-Naïve Higher-Risk Myelodysplastic Syndromes (MDS). Available at: https://clinicaltrials.gov/ct2/show/NCT02942290. Accessed 26 July 2021.
- 47.Azacitidine and Venetoclax Induction Therapy With Venetoclax Maintenance in the Elderly With AML. Available at: https://clinicaltrials.gov/ct2/show/NCT03466294. Accessed 26 July 2021.
- 48.Azacitidine in Combination With Venetoclax Treatment for MRD Positive Post Allo-HSCT AML/MDS Patients. Available at: https://clinicaltrials.gov/ct2/show/NCT04809181. Accessed 26 July 2021.
- 49.Kent A, Vasu S, Schatz D, et al. Glasdegib as maintenance therapy for patients with AML and MDS patients at high risk for postallogeneic stem cell transplant relapse. Blood Adv. 2020;4(13):3102-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolska-Washer A, Robak T.. Glasdegib in the treatment of acute myeloid leukemia. Future Oncol. 2019;15(28):3219-3232. [DOI] [PubMed] [Google Scholar]
- 51.Cortes JE, Dombret H, Merchant A, et al. Glasdegib plus intensive/nonintensive chemotherapy in untreated acute myeloid leukemia: BRIGHT AML 1019 Phase III trials. Future Oncol. 2019;15(31):3531-3545. [DOI] [PubMed] [Google Scholar]
- 52.Daher-Reyes GS, Merchan BM, Yee KWL.. Guadecitabine (SGI-110): an investigational drug for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Expert Opin Investig Drugs. 2019;28(10):835-849. [DOI] [PubMed] [Google Scholar]
- 53.Kantarjian HM, Roboz GJ, Kropf PL, et al. Guadecitabine (SGI-110) in treatment-naive patients with acute myeloid leukaemia: phase 2 results from a multicentre, randomised, phase 1/2 trial. Lancet Oncol. 2017;18(10):1317-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berger R, Rotem-Yehudar R, Slama G, et al. Phase I safety and pharmacokinetic study of CT-011, a humanized antibody interacting with PD-1, in patients with advanced hematologic malignancies. Clin Cancer Res. 2008;14(10):3044-3051. [DOI] [PubMed] [Google Scholar]
- 55.Zeidan A, Zeidner JF, Duffield A, et al. Stabilization of myelodysplastic syndromes (MDS) following hypomethylating agent (HMAs) failure using the immune checkpoint inhibitor ipilimumab: a phase I trial. Blood. 2015;126(33):1666. [Google Scholar]
- 56.Liao D, Wang M, Liao Y, Li J, Niu T.. A review of efficacy and safety of checkpoint inhibitor for the treatment of acute myeloid leukemia. Front Pharmacol. 2019;10:609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lancet JE, Uy GL, Cortes JE, et al. CPX-351 (cytarabine and daunorubicin) liposome for injection versus conventional cytarabine plus daunorubicin in older patients with newly diagnosed secondary acute myeloid leukemia. J Clin Oncol. 2018;36(26):2684-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larrosa-Garcia M, Baer MR.. FLT3 inhibitors in acute myeloid leukemia: current status and future directions. Mol Cancer Ther. 2017;16(6):991-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang X, Li W, Li X, Bai H, Zhang Z. . Current status and future prospects of PARP inhibitor clinical trials in ovarian cancer. Cancer Manag Res. 2019;11:4371–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stone RM, Mandrekar SJ, Sanford BL, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med. 2017; 377(5):454-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035): 917-921. [DOI] [PubMed] [Google Scholar]
- 62.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase [published correction appears in Nature. 2007;447(7142):346]. Nature. 2005;434(7035):913-917. [DOI] [PubMed] [Google Scholar]
- 63.Stein EM, DiNardo CD, Pollyea DA, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130(6):722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DiNardo CD, Stein EM, de Botton S, et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N Engl J Med. 2018; 378(25):2386-2398. [DOI] [PubMed] [Google Scholar]
- 65.Ashworth A, Lord CJ, Reis-Filho JS.. Genetic interactions in cancer progression and treatment. Cell. 2011;145(1):30-38. [DOI] [PubMed] [Google Scholar]
- 66.Doig CL, Lavery GG.. PARP inhibitors: staying on target? Cell Chem Biol. 2016;23(12):1442-1443. [DOI] [PubMed] [Google Scholar]
- 67.Faraoni I, Graziani G.. Role of BRCA mutations in cancer treatment with poly(ADP-ribose) polymerase (PARP) inhibitors. Cancers (Basel). 2018;10(12):487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lord CJ, Ashworth A.. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355(6330):1152-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ström CE, Johansson F, Uhlén M, Szigyarto CA-K, Erixon K, Helleday T.. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 2011;39(8):3166-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang X, Li W, Li X, Bai H, Zhang Z.. Current status and future prospects of PARP inhibitor clinical trials in ovarian cancer. Cancer Manag Res. 2019; 11(11):4371-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boussios S, Abson C, Moschetta M, et al. Poly (ADP-ribose) polymerase inhibitors: talazoparib in ovarian cancer and beyond. Drugs R D. 2020;20(2):55-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murai J, Huang SY, Das BB, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72(21):5588-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas A, Murai J, Pommier Y.. The evolving landscape of predictive biomarkers of response to PARP inhibitors. J Clin Invest. 2018;128(5): 1727-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kraus WL, Lis JT.. PARP goes transcription. Cell. 2003;113(6):677-683. [DOI] [PubMed] [Google Scholar]
- 75.Gibson BA, Kraus WL.. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13(7): 411-424. [DOI] [PubMed] [Google Scholar]
- 76.Muthurajan UM, Hepler MRD, Hieb AR, et al. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc Natl Acad Sci USA. 2014;111(35):12752-12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martin KA, Cesaroni M, Denny MF, Lupey LN, Tempera I.. Global transcriptome analysis reveals that poly(ADP-ribose) polymerase 1 regulates gene expression through EZH2. Mol Cell Biol. 2015;35(23):3934-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meng XW, Koh BD, Zhang J-S, et al. Poly(ADP-ribose) polymerase inhibitors sensitize cancer cells to death receptor-mediated apoptosis by enhancing death receptor expression. J Biol Chem. 2014;289(30):20543-20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hottiger MO. Nuclear ADP-ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu Rev Biochem. 2015;84(1): 227-263. [DOI] [PubMed] [Google Scholar]
- 80.Faraoni I, Aloisio F, De Gabrieli A, et al. The poly(ADP-ribose) polymerase inhibitor olaparib induces up-regulation of death receptors in primary acute myeloid leukemia blasts by NF-κB activation. Cancer Lett. 2018;423:127-138. [DOI] [PubMed] [Google Scholar]
- 81.Shah GM, Robu M, Purohit NK, Rajawat J, Tentori L, Graziani G.. PARP inhibitors in cancer therapy: magic bullets but moving targets. Front Oncol. 2013;3:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Campbell J, Ryan CJ, Brough R, et al. Large-scale profiling of kinase dependencies in cancer cell lines. Cell Rep. 2016;14(10):2490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Young JH, Marcotte EM.. Predictability of genetic interactions from functional gene modules. G3 (Bethesda). 2017;7(2):617-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15(8):852-861. [DOI] [PubMed] [Google Scholar]
- 85.Swisher EM, Lin KK, Oza AM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18(1):75-87. [DOI] [PubMed] [Google Scholar]
- 86.Gupta S, Nag S, Aggarwal S, Rauthan A, Warrier N.. Maintenance therapy for recurrent epithelial ovarian cancer: current therapies and future perspectives – a review. J Ovarian Res. 2019;12(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gogineni V, Morand S, Staats H, et al. Current ovarian cancer maintenance strategies and promising new developments. J Cancer. 2021;12(1):38-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oza AM, Tinker AV, Oaknin A, et al. Antitumor activity and safety of the PARP inhibitor rucaparib in patients with high-grade ovarian carcinoma and a germline or somatic BRCA1 or BRCA2 mutation: integrated analysis of data from Study 10 and ARIEL2. Gynecol Oncol. 2017;147(2):267-275. [DOI] [PubMed] [Google Scholar]
- 89.Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33(3):244-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xie H, Wang W, Xia B, Jin W, Lou G.. Therapeutic applications of PARP inhibitors in ovarian cancer. Biomed Pharmacother. 2020;127:110204. [DOI] [PubMed] [Google Scholar]
- 91.Plummer R, Dua D, Cresti N, et al. First-in-human study of the PARP/tankyrase inhibitor E7449 in patients with advanced solid tumours and evaluation of a novel drug-response predictor. Br J Cancer. 2020;123(4):525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Curtin NJ, Szabo C.. Poly(ADP-ribose) polymerase inhibition: past, present and future. Nat Rev Drug Discov. 2020;19(10):711-736. [DOI] [PubMed] [Google Scholar]
- 93.Gaymes TJ, Mohamedali AM, Patterson M, et al. Microsatellite instability induced mutations in DNA repair genes CtIP and MRE11 confer hypersensitivity to poly (ADP-ribose) polymerase inhibitors in myeloid malignancies. Haematologica. 2013;98(9):1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pratz KW, Koh BD, Patel AG, et al. Poly (ADP-ribose) polymerase inhibitor hypersensitivity in aggressive myeloproliferative neoplasms. Clin Cancer Res. 2016;22(15):3894-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mesa RA, Tefferi A, Lasho TS, et al. Janus kinase 2 (V617F) mutation status, signal transducer and activator of transcription-3 phosphorylation and impaired neutrophil apoptosis in myelofibrosis with myeloid metaplasia. Leukemia. 2006;20(10):1800-1808. [DOI] [PubMed] [Google Scholar]
- 96.Maifrede S, Martinez E, Nieborowska-Skorska M, et al. MLL-AF9 leukemias are sensitive to PARP1 inhibitors combined with cytotoxic drugs. Blood Adv. 2017;1(19):1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Molenaar RJ, Radivoyevitch T, Nagata Y, et al. IDH1/2 mutations sensitize acute myeloid leukemia to PARP inhibition and this is reversed by IDH1/2-mutant inhibitors. Clin Cancer Res. 2018;24(7):1705-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang L, Cai W, Zhang W, et al. Inhibition of poly(ADP-ribose) polymerase 1 protects against acute myeloid leukemia by suppressing the myeloproliferative leukemia virus oncogene. Oncotarget. 2015;6(29):27490-27504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pashaiefar H, Yaghmaie M, Tavakkoly-Bazzaz J, et al. The association between parp1 and lig3 expression levels and chromosomal translocations in acute myeloid leukemia patients. Cell J. 2018;20(2):204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Faraoni I, Compagnone M, Lavorgna S, et al. BRCA1, PARP1 and γH2AX in acute myeloid leukemia: role as biomarkers of response to the PARP inhibitor olaparib. Biochim Biophys Acta. 2015;1852(3):462-472. [DOI] [PubMed] [Google Scholar]
- 101.Diamantopoulos P, Zervakis K, Zervakis P, et al. Poly (ADP-ribose) polymerase 1 mRNA levels strongly correlate with the prognosis of myelodysplastic syndromes. Blood Cancer J. 2017;7(2):e533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Diamantopoulos PT, Kontandreopoulou CN, Symeonidis A, et al. ; Hellenic MDS Study Group . Bone marrow PARP1 mRNA levels predict response to treatment with 5-azacytidine in patients with myelodysplastic syndrome. Ann Hematol. 2019;98(6):1383-1392. [DOI] [PubMed] [Google Scholar]
- 103.Rauch PJ, Ellegast JM, Widmer CC, et al. MPL expression on AML blasts predicts peripheral blood neutropenia and thrombocytopenia. Blood. 2016;128(18):2253-2257. [DOI] [PubMed] [Google Scholar]
- 104.Wray J, Williamson EA, Singh SB, et al. PARP1 is required for chromosomal translocations. Blood. 2013;121(21):4359-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morice PM, Leary A, Dolladille C, et al. Myelodysplastic syndrome and acute myeloid leukaemia in patients treated with PARP inhibitors: a safety meta-analysis of randomised controlled trials and a retrospective study of the WHO pharmacovigilance database. Lancet Haematol. 2020;8(2):E122-E134. [DOI] [PubMed] [Google Scholar]
- 106.Using the Anticancer Drug Olaparib to Treat Relapsed/Refractory Acute Myeloid Leukemia or Myelodysplastic Syndrome With an Isocitrate Dehydrogenase (IDH) Mutation. Available at: https://www.clinicaltrials.gov/ct2/show/NCT03953898. Accessed 26 July 2021.
- 107.Talazoparib for Cohesin-Mutated AML and MDS With Excess Blasts. Available at: https://www.clinicaltrials.gov/ct2/show/NCT03974217. Accessed 26 July 2021.
- 108.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma [published correction appears in Nature. 2012;490(7419):298]. Nature. 2011;474(7353):609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hughes-Davies L, Huntsman D, Ruas M, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115(5):523-535. [DOI] [PubMed] [Google Scholar]
- 110.Michels J, Vitale I, Saparbaev M, Castedo M, Kroemer G.. Predictive biomarkers for cancer therapy with PARP inhibitors. Oncogene. 2014;33(30):3894-3907. [DOI] [PubMed] [Google Scholar]
- 111.Hodgson DR, Dougherty BA, Lai Z, et al. Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br J Cancer. 2018;119(11):1401-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Turner N, Tutt A, Ashworth A.. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4(10):814-819. [DOI] [PubMed] [Google Scholar]
- 113.Cousineau I, Belmaaza A.. EMSY overexpression disrupts the BRCA2/RAD51 pathway in the DNA-damage response: implications for chromosomal instability/recombination syndromes as checkpoint diseases. Mol Genet Genomics. 2011;285(4):325-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mirza MR, Coleman RL, González-Martín A, et al. The forefront of ovarian cancer therapy: update on PARP inhibitors [published correction in Ann Oncol. 2021;32(8):1066-1067]. Ann Oncol. 2020;31(9):1148-1159.32569725 [Google Scholar]
- 115.Bose P, Verstovsek S.. JAK2 inhibitors for myeloproliferative neoplasms: what is next? Blood. 2017;130(2):115-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nieborowska-Skorska M, Maifrede S, Dasgupta Y, et al. Ruxolitinib-induced defects in DNA repair cause sensitivity to PARP inhibitors in myeloproliferative neoplasms. Blood. 2017;130(26):2848-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sharma P, Shinde SS, Damlaj M, et al. Allogeneic hematopoietic stem cell transplant in adult patients with myelodysplastic syndrome/myeloproliferative neoplasm (MDS/MPN) overlap syndromes. Leuk Lymphoma. 2017;58(4):872-881. [DOI] [PubMed] [Google Scholar]
- 118.Patel PR, Senyuk V, Rodriguez NS, et al. Synergistic cytotoxic effect of busulfan and the PARP inhibitor veliparib in myeloproliferative neoplasms. Biol Blood Marrow Transplant. 2019;25(5):855-860. [DOI] [PubMed] [Google Scholar]
- 119.Tobin LA, Robert C, Rapoport AP, et al. Targeting abnormal DNA double-strand break repair in tyrosine kinase inhibitor-resistant chronic myeloid leukemias. Oncogene. 2013;32(14):1784-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fan J, Li L, Small D, Rassool F.. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: implications for genomic instability and therapy. Blood. 2010;116(24):5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Muvarak N, Kelley S, Robert C, et al. c-MYC generates repair errors via increased transcription of alternative-NHEJ factors, LIG3 and PARP1, in tyrosine kinase-activated leukemias. Mol Cancer Res. 2015;13(4):699-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sallmyr A, Fan J, Datta K, et al. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: implications for poor prognosis in AML. Blood. 2008;111(6):3173-3182. [DOI] [PubMed] [Google Scholar]
- 123.Gaymes TJ, Mohamedali A, Eiliazadeh AL, Darling D, Mufti GJ.. FLT3 and JAK2 mutations in acute myeloid leukemia promote interchromosomal homologous recombination and the potential for copy neutral loss of heterozygosity. Cancer Res. 2017;77(7):1697-1708. [DOI] [PubMed] [Google Scholar]
- 124.Taylor SJ, Dagger SA, Thien CBF, Wikstrom ME, Langdon WY.. Flt3 inhibitor AC220 is a potent therapy in a mouse model of myeloproliferative disease driven by enhanced wild-type Flt3 signaling. Blood. 2012;120(19):4049-4057. [DOI] [PubMed] [Google Scholar]
- 125.Nieborowska-Skorska M, Sullivan K, Dasgupta Y, et al. Gene expression and mutation-guided synthetic lethality eradicates proliferating and quiescent leukemia cells. J Clin Invest. 2017;127(6):2392-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Muvarak NE, Chowdhury K, Xia L, et al. Enhancing the cytotoxic effects of PARP inhibitors with DNA demethylating agents – a potential therapy for cancer. Cancer Cell. 2016;30(4):637-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhao L, So CWE.. PARPi potentiates with current conventional therapy in MLL leukemia. Cell Cycle. 2017;16(20):1861-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ricks TK, Chiu HJ, Ison G, et al. Successes and challenges of PARP inhibitors in cancer therapy. Front Oncol. 2015;5:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Veliparib and Topotecan With or Without Carboplatin in Treating Patients With Relapsed or Refractory Acute Leukemia, High-Risk Myelodysplasia, or Aggressive Myeloproliferative Disorders. Available at: https://www.clinicaltrials.gov/ct2/show/NCT00588991. Accessed 26 July 2021.
- 130.Talazoparib and Gemtuzumab Ozogamicin for the Treatment of CD33 Positive Relapsed or Refractory Acute Myeloid Leukemia. Available at: https://www.clinicaltrials.gov/ct2/show/NCT04207190. Accessed 26 July 2021.
- 131.Decitabine and Talazoparib in Untreated AML and R/R AML (1565GCC). Available at: https://www.clinicaltrials.gov/ct2/show/NCT02878785. Accessed 26 July 2021.
- 132.Hennes ER, Dow-Hillgartner EN, Bergsbaken JJ, Piccolo JK.. PARP-inhibitor potpourri: a comparative review of class safety, efficacy, and cost. J Oncol Pharm Pract. 2020;26(3):718-729. [DOI] [PubMed] [Google Scholar]
- 133.Gil-Kulik P, Dudzińska E, Radzikowska-Büchner E, et al. Different regulation of PARP1, PARP2, PARP3 and TRPM2 genes expression in acute myeloid leukemia cells. BMC Cancer. 2020;20(1):435. [DOI] [PMC free article] [PubMed] [Google Scholar]