Abstract
Background and Aims
Although genome-wide association studies have identified many loci that influence smoking behaviors, much of the genetic variance remains unexplained. We characterized the genetic architecture of four smoking behaviors using single nucleotide polymorphism (SNP) heritability (). This is an estimate of narrow-sense heritability specifically estimating the proportion of phenotypic variation due to causal variants (CVs) tagged by SNPs.
Design
Partitioned analysis of smoking behavior traits.
Setting
UK Biobank.
Participants
UK Biobank participants of European ancestry. The number of participants varied depending on the trait, from 54 792 to 323 068.
Measurements
Smoking initiation, age of initiation, cigarettes per day (CPD; count, log-transformed, binned and dichotomized into heavy versus light) and smoking cessation with imputed genome-wide SNPs.
Findings
We estimated that, in aggregate, approximately 18% of the phenotypic variance in smoking initiation was captured by imputed SNPs [, standard error (SE) = 0.01] and 12% [SE = 0.02] for smoking cessation, both of which were more than twice the previously reported estimates. Estimated age of initiation (, SE = 0.01) and binned CPD (, SE = 0.01) were substantially below published twin-based h2 of 50%. CPD encoding influenced estimates, with dichotomized CPD . There was no evidence of dominance genetic variance for any trait.
Conclusion
A biobank study of smoking behavior traits suggested that the phenotypic variance explained by SNPs of smoking initiation, age of initiation, cigarettes per day and smoking cessation is modest overall.
Keywords: Dominance variance, genetic architecture, h2SNP heritability, nicotine, smoking
INTRODUCTION
Cigarette smoking is a leading cause of premature death world-wide [1], and many smokers struggle to quit, despite interest and numerous attempts [2]. Although smoking prevalence has decreased in recent decades due to public health efforts [3], rates of alternative forms of nicotine use (e.g. vaping) have grown rapidly during this time [4], demonstrating a pressing need to characterize the underlying biology of nicotine use and smoking to reduce subsequent premature death.
A key aspect of that underlying biology is the genetic architecture [5] of smoking behaviors, including the relative contribution of rare versus common variants, functional annotation of associated loci and characterization of the neutral and selective forces shaping that architecture. Numerous [6-10] twin, adoption and family studies have demonstrated that up to 50% of the variance in nicotine dependence and individual smoking behaviors, such as quantity, is attributable to genetic influences. Recent genome-wide association studies (GWAS) have improved our understanding of this genetic basis by identifying more than 200 conditionally independent loci associated with these traits to date [11-17]. This genetic signal is enriched in loci that influence the epigenome and within specific brain regions, such as the hippocampus, providing a more nuanced interpretation of specific class(es) of variants, candidate brain regions and potential causal mechanisms that influence smoking [11]. Together, this body of work strongly indicates a highly polygenic architecture to smoking behaviors. Nonetheless, significantly associated loci collectively explain only a small proportion of the family-based genetic variance, leaving many additional loci undiscovered and the majority of the genetic variance unexplained.
While additional common variants of very small effect are likely to be identified as sample sizes grow, some of the unexplained variability probably arises from uncommon and rare variants (MAF < 0.01), although their relative contribution is uncertain. The most recent large GWAS [11,15] of smoking behaviors and nicotine dependence, using GWAS summary statistics-based linkage disequilibrium (LD) score regression (LDSC), estimate the SNP-based heritability [i.e. , the proportion of the total phenotypic due to causal variants (CVs) tagged by single nucleotide polymorphisms (SNPs)] [18] due to common variants as 0.05–0.09 across traits [19]. LD score regression, although computationally efficient and attractive in the ability to partition SNP heritability into functional annotations, is unable to assess the contribution of rare variation [19-21]. A related exome sequencing study [22] estimated that rare coding variants explained approximately 1–2% of the phenotypic variance. However, given that the majority of identified associations are intergenic [11], exome-based studies are unlikely to identify most rare variants influencing these behaviors. Thus, the rare-variant contribution to smoking behaviors may yet be substantial when assessed with methods that can account for the aggregated influence of common and rare variation.
Additionally, the contribution of non-additive genetic variance to these smoking behaviors is poorly understood. Twin-based studies have typically evaluated ACE models [9], which estimate additive genetic (A), common environment (C) and unique environment (E) variances using twin correlations, implicitly assuming zero dominance genetic variance. Alternatively, ADE models can be used to estimate dominance variance (D) rather than common environment variance. Often, ACE models are chosen over ADE models because of improved fit. However, because such studies are based only on correlations of monozygotic and dizygotic twins, they cannot estimate both simultaneously, although both sources may, in fact, influence trait variance [23]. Extended twin kinship models can estimate dominance genetic variance and shared environmental effects simultaneously, and the only such model to evaluate smoking initiation found no evidence of dominance genetic variance [24]. To our knowledge, only one estimate of SNP-based dominance genetic variance () has been reported in addition to previous estimates of smoking behaviors, which found of smoker status indistinguishable from zero [25]. Furthermore, the allele frequency spectrum and contribution of functional annotations related to LD, allele frequency, recombination and related genomic features for smoking behaviors has not been fully explored. The only published work has examined a single trait, smoking status, finding contributions of low-LD and -MAF variants consistent with negative or purifying selection [21,26]. One study applied partitioned approaches to evaluate tissue-specific effects, with results indicating that genes expressed in the cerebellum are enriched in their contribution to nicotine dependence [15]. Whether these same patterns exist for other smoking behaviors, such as quantity of use or cessation, is unknown.
A comprehensive evaluation of the frequency spectrum, the influence of dominance genetic variance and the contributions of functional annotations is needed to provide a more complete picture of the genetic architecture underlying complex smoking behaviors. Here, we use recently developed methods [18,20,21,25,27-29] to evaluate these heritable contributions and characterize the genetic architecture of four smoking behaviors: smoking initiation (whether an individual has ever been a regular smoker), age of initiation of regular smoking, cigarettes per day (CPD, evaluated with different data encodings) and smoking cessation. These four behaviors represent a cross-section of the full spectrum from experimentation to dependence [30-32], and have been evaluated in recent GWAS [11,16,17].
METHODS
Phenotype and genetic data sets
Using the UK Biobank [33] full release, we assessed the same four smoking phenotypes as the GSCAN project [11], defined identically (final sample sizes after quality control; see below): (1) smoking initiation (n = 323 068), defined as whether an individual had ever in their life-time been a regular smoker by having smoked more than 100 cigarettes during one’s life-time; (2) age of smoking initiation (n = 122 200), defined as the age at which an individual began smoking regularly (UK Biobank data fields 3426 and 2867); (3) CPD (n = 116 258), defined as a five-bin variable based on responses for the number of cigarettes smoked per day (fields 2887, 3456 and 6183); and (4) smoking cessation (n = 160 390), defined as individuals who were not current smokers but had been regular smokers at one point (fields 1239 and 1249). The latter three phenotypes required an individual to be a current or former regular smoker. Genome-based restricted maximum likelihood (GREML) variance estimation (see below) was limited by available RAM (1 Tb) on a single compute node; therefore, we analyzed the smoking initiation and smoking cessation data using three and two separate, equally sized subsamples, respectively, and meta-analyzed the results using inverse-variance weighting. Age of initiation and CPD were each analyzed in a single analysis. In addition to the binned CPD metric used in recent genetic association meta-analyses [11], we examined the influence of CPD scale on estimates, which we previously found to influence association effect sizes [34]. We evaluated raw CPD count, log-transformed CPD, √CPD, CPD(2/3) and dichotomized CPD (heavy versus light) using four different sets of CPD cut-offs for heavy and light smoker definitions [we applied the following heavy (H) and light (L) cut-offs of CPD: (a) H: > 20, L: ≤ 10; b) H: > 30, L: ≤ 10; (c) H: > 40, L ≤ 5; (d) median CPD of 20 (H: > 20, L: ≤ 20; Supporting information, Fig. S1]. Final sample sizes for the different CPD encodings are presented in the Supporting information. Together, these phenotypes, initiation, age of initiation, CPD and cessation, encompass key aspects of nicotine dependence [35].
The UK Biobank release included ~97 M imputed variants using both the Haplotype Reference Consortium (HRC) and 1000 Genomes + UK10K reference panels [33]. We removed individuals with mismatched self-reported and genetic sex, ∣Fhet∣ ≥ 0.2, and/or no phenotypic information. We restricted our analyses to bi-allelic SNPs with minor allele frequency (MAF) ≥ 0.0001, imputation INFO score ≥ 0.3, Hardy–Weinberg equilibrium test (HWE) P-value ≥ 10−10 and variant missingness ≤ 0.02 using plink1.9 [36], yielding 22 982 114 SNPs. The choice of INFO score threshold was based on previous results demonstrating that variants with relatively poor imputation still contribute to estimates [27], although will be underestimated relative to the true SNP-heritability as a result of error during imputation [18]. We note that while we used the HRC-imputed UK Biobank full release, imputation to larger and more diverse samples, such as TOPMed [37], would probably improve estimates. We identified individuals of European ancestry using principal components analysis using flashpca [38] from a set of MAF- and LD-pruned array markers (plink2 command: --maf 0.05 --indep-pairwise 50 5 0.2), retaining those whose scores on the first four PCs fell within the range of the UK Biobank-identified individuals of European ancestry (UK Biobank data field ID 22006). We identified unrelated individuals using GCTA version 1.91.3 [39] with an initial relatedness cut-off of < 0.05. After observing differences between REML- and Haseman–Elston-based variance estimators (see below), we applied relatedness thresholds of 0.02, 0.03, 0.04 and 0.05 to assess the potential for environmental effects confounding rare variation. Because sample size varied for each of the four phenotypes, we applied these relatedness thresholds for each phenotype separately. All sample sizes are presented in Supporting information, Tables S1-S3.
Variance estimation
We estimated genetic variance in unrelated individuals using a set of genetic relatedness matrices (GRMs) partitioned by MAF- and individual marker LD-stratified bins (LDMS-I), which provides the most robust estimates of genetic variance across the allelic frequency spectrum in imputed data [18] and can be used in a GREML (GCTA [39]) or moment-matching framework, such as phenotype correlation–genotype correlation (PCGC) regression [40,41]. These analyses were not pre-registered, and are therefore exploratory. We used both GCTA and PCGC (for binary traits) to estimate variances accounted for by GRMs (described next), and included the following as fixed-effect covariates: sex (UK Biobank field ID 31), age (21003), age2, Townsend deprivation index (189), educational attainment (6138), genotyping batch (22 000), scores of the first 10 world-wide principal components (22 009) and scores of the first 10 principal components of the retained individuals of European ancestry estimated, as described above.
We estimated using six LDMS-I-partitioned GRMs. We calculated LD scores for all imputed markers (GCTA: --ld-score-region 200). We stratified markers into four MAF intervals [(0.0001, 0.001), (0.001, 0.01), (0.01, 0.05) and (≥ 0.05)]. For the two more common MAF bins, we further stratified SNPs into low and high individual SNP LD score bins based on median LD score within MAF bins. We did not LD stratify the two more rare MAF bins because there is (1) low variation in LD for low MAF SNPs (most SNPs have low LD), (2) limited power to differentiate across LD bins of SNPs of low MAF and (3) inclusion of more GRMs required more memory than available. Because of incomplete data across all four phenotypes, we estimated all GRMs for each set of unrelated individuals for each phenotype separately.
To estimate dominance genetic variance, we included a dominance genetic relatedness matrix [25] for each data set (GCTA: --make-bin-d) using all markers with MAF > 0.01. We did not partition the dominance matrix by MAF or LD due to the practical limitations noted above.
For binary traits (smoking initiation, smoking cessation and heavy/light CPD), we converted observed scale estimates to the liability scale using within-sample trait prevalence and the conversion of Lee et al. [42].
Finally, we evaluated the influence of possible confounding from the ascertainment of samples genotyped on the UK Biobank Lung Exome Variant Evaluation (BiLEVE) chip versus Axiom array and the relatedness threshold used, i.e. potential environmental confounding and cryptic relatedness. To assess the influence of the relatedness threshold, we applied progressively lower relatedness thresholds (0.02, 0.03, 0.04 and 0.05), then estimating as above. To assess the influence of ascertainment of heavy smokers for samples genotyped on the BiLEVE versus Axiom chip, we ran our GREML-LMDS-I models after excluding all BiLEVE batches and compared estimates to those with all individuals included. Because of the relatively small number of individuals genotyped on the BiLEVE array, we are unable to run the same 6-GRM models with only those samples, but comparison to the full model provides an assessment of possible influence. Resulting sample sizes across thresholds are presented in Supporting information, Tables S1-S3.
Functional annotation and tissue- and cell type-specific expression heritability enrichment
We used LD score regression to estimate partitioned for functional annotations [20]. We applied the baseline + LD model [21] to assess functional annotations such as LD, allele frequency and age, recombination rate and related annotations, and the possible role of purifying selection. We applied a Bonferroni cut-off either within traits (P < 0.00052, as suggestive) or across all traits (P < 0.00013) to identify significant LDSC regression coefficients.
We also used LD score regression to estimate partitioned for tissue- and cell type-specific gene expression patterns [20]. As previously detailed in applying this method for nicotine dependence [15], we used 205 tissues and cell types with publicly available gene expression data as measured via RNA-sequencing (53 human tissues/cell types from GTEx [43]) or microarrays [152 human and mouse tissues/cell types from the Data-driven Expression Prioritized Integration for Complex Traits (DEPICT) [44,45] tool]. We used Bonferroni correction to identify tissues/cell types with suggestive gene expression patterns within each trait (P < 2.4 × 10−4, α = 0.05/205) and to declare significance based on additional correction for testing all four traits (P < 6.1 × 10−5).
RESULTS
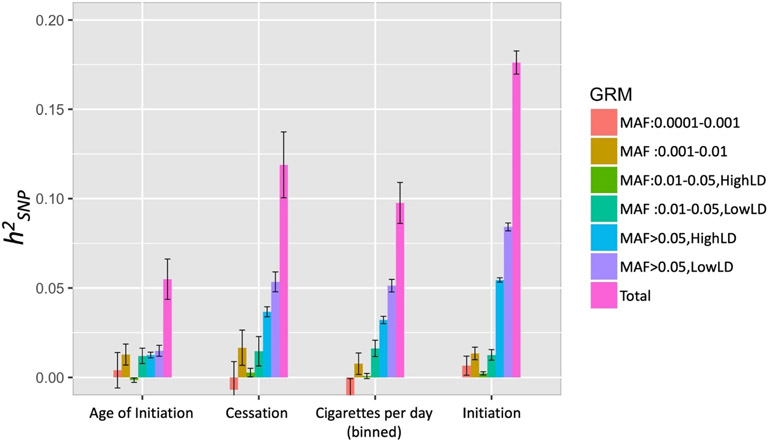
Using GREML-LDMS-I with unrelated individuals, we estimated smoking initiation (standard error [SE]) = 0.176 (0.007), smoking cessation , cigarettes per day and age of initiation (Fig. 1, Supporting information, Table S1). MAF- and LD-partitioned heritability estimates differed across traits. Common variants (MAF > 0.05) contributed substantially to all traits, particularly common variants with relatively low LD (Fig. 1). Uncommon variants (MAF = 0.01–0.05) with low LD, but not high LD, contributed to all traits. Alternatively, rare (MAF < 0.01) variants contributed significantly only to smoking initiation and age of initiation and very rare (MAF < 0.001) variants did not contribute significantly to any trait, as their 95% confidence intervals (CIs) overlapped zero, although we note that overlapping CIs are only one measure of significance, and that others, such as likelihood ratio tests, may still indicate statistical significance.
Figure 1.
estimates (± standard error) across four smoking behaviors, partitioned using GREML-LDMS-I. Note that twin-based estimates are approximately 50% across these smoking traits.
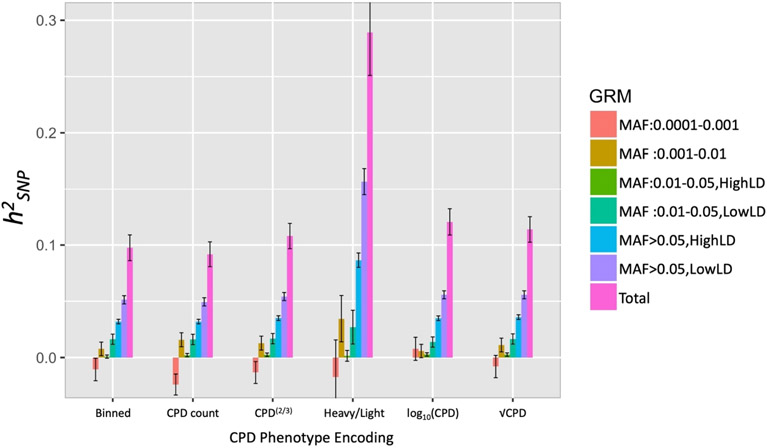
Notably, we estimated significantly different (non-overlapping 95% CI) total and binned for different CPD encodings. Total ranged from 0.092 (0.011) for the raw CPD count to 0.289 (0.038) when CPD was dichotomized into heavy (CPD > 20)/light (CPD ≤ 10) smokers (Fig. 2 and Supporting information, Figs S2-S3, Tables S1-S3). All dichotomized CPD total estimates (except using the median) were > 0.2. We found differences in partitioned estimates across CPD scale, such that common variants (MAF > 0.05) contributed to substantially higher of heavy/light CPD than the other CPD encodings. The more rare (MAF = 0.001–0.01) variant contribution was also higher, although the smaller sample size of the dichotomized data led to larger SEs.
Figure 2.
estimates (± standard error) of cigarettes per day (CPD) using different phenotype encodings, partitioned using GREML-LDMS-I. Heavy versus light is dichotomized with light: CPD ≤ 10 and heavy: CPD > 20; estimated shown on the liability scale using a prevalence of 0.42.
We estimated the contribution of dominance variance. For all traits, the 95% CI of estimates overlapped zero (Supporting information, Table S4).
The relatedness threshold strongly influenced estimated when using PCGC, but not when using GREML (Supporting information, Tables S1-S3, Figs S2-S6). Specifically, the PCGC estimates were considerably higher than GREML estimates when applying a relatedness < 0.05 cut-off with smoking initiation and smoking cessation, but dropped and had overlapping 95% CIs at lower relatedness thresholds. The higher estimates when using PCGC with relatedness < 0.05 were driven by a much greater contribution of rare variant (MAF < 0.0001; Supporting information, Figs S5-S6).
When we excluded the batches genotyped on the BiLEVE array, we found qualitatively similar estimates across the four smoking behaviors and the six GRMs (Supporting information, Fig. S7, Table S1).
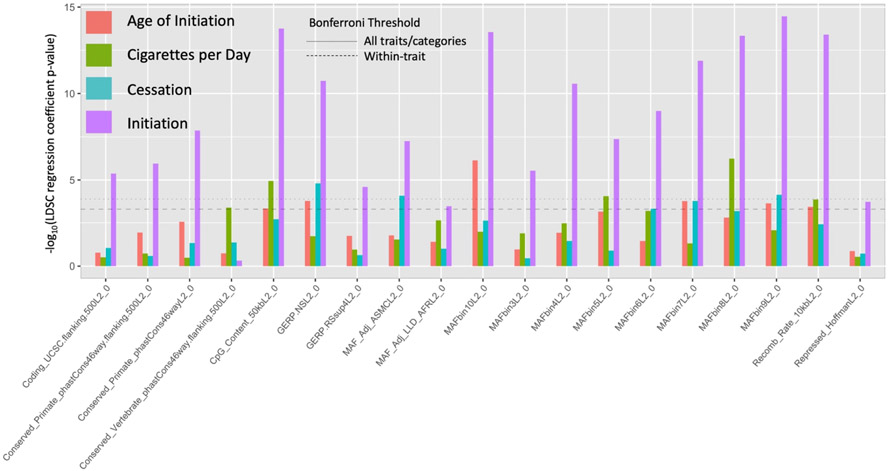
We applied partitioned LDSC to assess contribution of functional annotations and the role of LD and selective constraint in smoking behaviors. Across smoking behaviors we found that SNPs that were highly conserved, that had lower MAF-adjusted LD or lower MAF quantiles (MAF > 0.001 in Liu et al. [11]) and that were in areas of high cytosine–phosphate–guanine (CpG) content and low recombination rate contributed significantly to heritable genetic variation (Supporting information, Figs S3 and S7, Table S5).
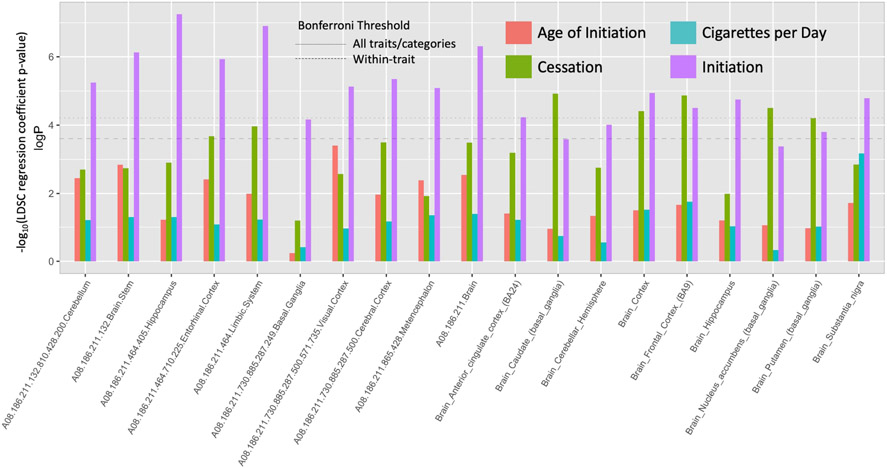
The tissue- and cell type-specific expression analysis of partitioned LDSC identified cortical and nucleus accumbens (NAcc) regions as significantly contributing to heritable variation for smoking initiation and/or cessation, among others (Fig. 4, Supporting information, Table S6); these results indicate that genes spanning SNPs associated with initiation and cessation are enriched for expression specifically in the frontal cortex, for example, compared to other tissues. Even broader evidence was observed for initiation, with significant enrichment for heritability for genes expressed in 14 total brain tissues including hippocampus, cerebellum and substantia nigra. No tissues or cell types showed significant enrichment for age of initiation or CPD, and genes specifically expressed in non-brain tissues, including lung tissue, did not significantly contribute to heritable variation for any smoking behavior (Supporting information, Table S6).
Figure 4.
Partitioned linkage disequilibrium score (LDSC) regression coefficient P-values for all annotations with at least one significant coefficient across all traits using the cell type and tissue-specific expression model. See Supporting information, Table S6 for all annotations.
DISCUSSION
We estimated and across four key smoking behaviors, and partitioned variance according to frequency (rare versus common), functional annotation and gene expression. Our estimates are more than double the previously reported [11] LDSC-based and single-component GREML-based estimates for smoking initiation (0.18 versus 0.08 and 0.12) and smoking cessation (0.12 versus 0.05 and 0.06), but are nearly identical for binned CPD (0.1). Our estimate of age of smoking initiation is nearly identical to the LDSC-based estimate, but is much lower than the previous single-component GREML estimate of 0.11. The difference in age of initiation may be due to including all variants in a single GRM when the causal variants are relatively common [18]. Partitioned estimates of common, well-tagged variants are similar to the LDSC-based estimates [11] across all four traits, consistent with expectations, as LDSC estimates variance due to common, well-tagged variants [18,19]. The higher estimates for smoking initiation and cessation results from larger contributions of low-LD and low-frequency variants (MAF < 0.01), suggesting that for these traits a non-trivial portion of the genetic variance is due to more rare variants and those that are poorly tagged by surrounding SNPs. This contribution is probably underestimated in the current study, given that these sites are typically poorly imputed even using large reference panels such as HRC, which leads to a downward bias in estimates [18,27].
Alternative CPD encodings led to different estimates, wherein total for dichotomized heavy/light smoker status was more than twice that of other encodings. This may be explained by one or more possible phenomena that occur after restricting the analyses to phenotypic extremes, i.e. removing the center of the distribution. First, the extremes of the CPD distribution may be capturing a phenotype more closely approximating physical dependence on nicotine. Tolerance and withdrawal may index severity of nicotine dependence [46], a construct for which we do not have formal diagnoses, but which is highly heritable. In such a case, while lacking other important aspects of the clinical presentation such as craving or loss of control, the dichotomized heavy/light phenotype is comparing individuals who may find overnight abstinence less aversive and start smoking later in the day, and endorse lower levels of nicotine dependence (light) to those who meet criteria for severe nicotine dependence (heavy), whereas the standard continuous CPD encoding includes intermediate levels of smoking heaviness that may or may not correlate with clinical presentations of nicotine dependence. Our GREML-based estimate of common, well-tagged (~0.09) is approximately the same as one recently reported LDSC-based estimate of nicotine dependence [15], consistent with this hypothesis. Alternatively, the dichotomized phenotype may reflect lower environmental variance and result in higher if, for example, environmental effects such as reduced access to cigarettes or regular use of nicotine replacement therapy lead to intermediate values of CPD where higher values would otherwise be observed, or if intermediate values of CPD are intrinsically noisier. Such differences in variance cannot be tested when either trait is dichotomous, because the liability underlying the dichotomous trait must be assumed to have unit variance. Future work may distinguish between these two possibilities, and determine whether variants that contribute to heavy/light CPD and other smoking behaviors examined here also contribute to nicotine dependence liability or severity.
We found no evidence of dominance genetic variance for any phenotype, although we note that the power to detect is lower relative to [25], and therefore we may be limited by our sample size to detect low, but non-zero . Our findings are consistent with those of Zhu et al. [25], who reported low across 79 traits and for one smoking phenotype: smoking status. We conclude, therefore, that for the four smoking phenotypes in the current study, dominance genetic variance probably contributes little or not at all to the phenotypic variance. We note that dominance effects of individual alleles, when the allele frequency is low, will primarily contribute to the estimated additive genetic variance (i.e. ) [47]. Alternatively, interactions between, rather than within, loci may lead to epistatic genetic variation underlying smoking behaviors, and such effects could not be tested using the current approach.
We identified several functional annotations related to LD, MAF and sequence conservation that significantly contribute to (Fig. 3, Supporting information, Table S5). In addition, GREML-LDMS-I analyses identified higher contribution of poorly tagged variants relative to well-tagged variants within the same MAF range across all four traits, and also identified nominally significant (95% CI > 0) contribution of rare variants (MAF < 0.01) for smoking initiation, raw CPD count and age of initiation. Across the four traits analyzed, rare variants accounted for between 10 and 20% of total (Supporting information, Table S1). This suggests a role of low-frequency SNPs in low LD with surrounding regions, consistent with purifying and background selection acting to remove mutations with deleterious effects. Given that tobacco use in high concentrations, such as found in cigarettes, is evolutionarily novel for humans, it is unlikely that negative selection acted directly on these smoking behaviors, but rather mutations that today influence nicotine-related behaviors may have pleiotropic effects on other traits that were subject to negative selection across evolutionary time [26].
Figure 3.
Partitioned linkage disequilibrium score (LDSC) regression coefficient P-values for all annotations with at least one significant coefficient across all traits using the LD+ baseline model. See Supporting information, Fig. S5 and Table S5 for all annotations.
We also found significant heritable contribution of genes with tissue-specific gene expression across the brain to smoking initiation and cessation, although the specific regions were only partly overlapping. Tissues with a strong evidence base for their involvement in addiction processes via dopamine and neuronal transmission (cortical, NAcc and substantia nigra tissues [48-51]) were found to significantly contribute to along with addiction-relevant tissues as indicated via neuroimaging studies (amygdala, caudate basal ganglia and frontal cortex [52-54]). These results support functional follow-up studies, for instance, in animal models or drug target studies of nicotine addiction, focused on genes expressed in frontal cortex and NAcc as key addiction-relevant tissues. Further, these results highlight the potential relevance of other brain regions for smoking behaviors, such as cerebellum [55], that was found to have the most significant contribution for nicotine dependence [15]. Most of the significant LDSC associations were observed for smoking initiation, which has the largest GWAS sample size of the four smoking phenotypes. This suggests that as discovery GWAS sample size increases for the other smoking behaviors, additional tissues may be implicated and highlight phenotype-specific neural circuits and brain regions thought to be involved in the different stages of addiction [56,57]. Nonetheless, some of the implicated tissues may also reflect gene expression correlations across tissues rather than direct involvement, and perturbations in addiction-associated gene expression are probably pervasive across the brain.
Cumulatively, our results point to a highly polygenic nature of these four smoking behaviors, consistent with purifying selection, and highlight the roles of key brain regions and the possible influence of rare variation for at least some smoking behaviors. Genes expressed within these regions that harbor rare variants may be useful to target in detailed sequencing or functional studies, particularly if such genes could be targeted by repurposed therapies [58].
Our estimates are still considerably lower than twin-based estimates, which range from 50 to 80% for dependence, smoking initiation and quantity of use [6-10], suggesting that additional still-missing heritability remains. This is unlikely to be explained by common causal variants, which are well-tagged in current imputation reference panels and from which we expect little downward bias in estimates [18]. Further work will be required to fully characterize non-additive genetic variance, such as epistasis or gene–environment interaction. Regardless, rare variants are a probable source of the still-missing heritability. The SE of the most rare MAF partitions were substantially larger than the common variant partition SE, indicating that increased sample size will improve the precision of estimates of rare-variant contribution. Overall, estimates are still generally low compared with those attributable to common variants, and even with large reference panels such as the HRC rare variants are expected to be poorly imputed, resulting in downwardly biased [18,27]. Further work through deep sequencing of large samples [37] or using those deeply sequenced individuals as an improved imputation reference panel is needed to obtain less-biased estimates of rare-variant , For example, height and body mass index (BMI) estimates using whole genome sequencing have approached twin-based heritability estimates; rare variants account for a substantial proportion of the heritability [59].
Beyond the limitation of rare variant imputation, our study highlights several key issues in estimation. First, although we used the largest relatively homogeneous sample available, even larger samples will be needed for more precise estimation of rare variant contribution, as demonstrated by the much smaller SE of estimates of traits with larger sample sizes. Secondly, estimates are sensitive to the estimation method, i.e. H–E regression-based versus GREML, which may be due to how environmental confounding differentially influences estimates across methods. GREML-based estimates were relatively stable across relatedness thresholds (Supporting information, Table S1). However, PCGC-based estimates were quite sensitive to relatedness thresholds, being much higher than GREML-based estimates at a.05 threshold and declining with lower thresholds. Although a full assessment of performance of estimators is beyond the scope of this study, it will be important to assess the potential for environmental confounding. As with the possibility of rare variant–environment confounding in GWAS [60], environmental confounding is particularly relevant to estimates of rare variant , because very rare variants are more likely to be shared by individuals sharing recent common ancestors and who may therefore be more likely to share environmental influences. Models that incorporate environmental sharing of families, partners and close relatives or geography (e.g. [61,62]) are a possible avenue to address confounding. To this effect, we note that a full extended twin-family design found a lower and possibly sex-dependent estimate of common additive genetic variance, as well as strong environmental influences [24]. Finally, the UK Biobank is not a random sample of the United Kingdom and, importantly, has a lower proportion of smokers and higher educational attainment than a random sample of the UK population would have, which introduces the possibility of collider bias [63]. When we excluded the individuals run on the BiLEVE array (which were ascertained for smoking-relevant traits [64]), we found no evidence that such ascertainment biased our estimates, but we cannot rule out the possibility that other factors could lead to biases.
In conclusion, although our estimates of the four different smoking behaviors were generally modest, they are higher than previously published estimates for smoking initiation and cessation, emphasize contributions of multiple brain tissues with specific gene expression profiles and indicate that additional genetic variance may be explained by low- and rare-frequency variants, which may be due to the impact of purifying selection on genes involved in these highly polygenic traits. Quantity of use, as measured by CPD, may also be modestly heritable, but as it depends on the encoding of the variable, additional characterization of the phenotype and its relationship with nicotine dependence is required. All estimates will be improved by the use of complete whole genome sequencing of large numbers of individuals or the use of larger, more diverse imputation panels [37], including the contribution of rare variants to smoking behaviors.
Supplementary Material
Figure S1 Distribution of raw CPD count, and different transformations. Note that 1 pack is approximately 20CPD. For dichotomized CPD, we applied the following Heavy (H) and Light (L) cutoffs of CPD: a) H: >20, L: <= 10; b) H: >30, L: <= 10; c) H: >40, L: <= 5; d) Median CPD of 20 (H: >20, L: <=20).
Figure S2 Cigarettes per day (CPD) (+/− SE), at four different relatedness thresholds and with different CPD transformations as a continuous variable.
Figure S3 Cigarettes per day (CPD) (+/− SE), at four different relatedness thresholds and with CPD dichotomized using the indicated thresholds for low and high CPD.
Figure S4 Age of initiation (+/− SE), at four different relatedness thresholds.
Figure S5 Smoking cessation (+/− SE), at four different relatedness thresholds, and using two different estimation methods.
Figure S6 Smoking initiation (+/− SE), at four different relatedness thresholds, and using two different estimation methods.
Figure S7 (+/− SE) estimates for the four smoking behaviors shown in Fig. 1 of the main text, after exclusion of those samples genotyped on the UK BiLEVE genotyping array, using a relatedness threshold of 0.05. Comparison of these results to those in Fig. 1 show estimates are qualitatively similar after excluding those batches, which were oversampled for heavy smokers from the full UK Biobank sample based on FEV1 phenotypes.
Figure S8 Partitioned LDSC regression coefficient p-values for all annotations across all traits.
Table S1 Estimates of partitioned h2SNP in 6 MAF & LD-stratified bins, and the total h2SNP estimate, across the four traits (and four CPD encodings) using different relatedness cutoffs (0.02, 0.03, 0.04 & 0.05) using GREML. For binary traits, we applied two different estimation methods (GREML & PCGC), and present estimates on the liability scale. Estimates for CPD use the binned encoding, matching GSCAN (Liu et al. 2019); for other CPD encodings, see Tables S2-3. Prevlance for the cessation phenotype is given as the proportion of current smokers. Models that indicate “NoBiLEVE” were run with the genotyping batches using the BiLEVE array excluded from the analysis.
Table S2 Estimates of partitioned h2SNP in 6 MAF & LD-stratified bins, and the total h2SNP estimate, across different continous CPD encodings using different relatedness cutoffs (0.02, 0.03, 0.04 & 0.05) using GREML.
Table S3 Estimates of partitioned h2SNP in 6 MAF & LD-stratified bins, and the total h2SNP estimate, across differet dichotomous CPD encodings using different relatedness cutoffs (0.02, 0.03, 0.04 & 0.05). We applied two different estimation methods (GREML & PCGC), and report estimates on the liability scale using the in-sample prevalence estimates. Designations of heavy (H) and light (L) smokers are indicated in the “Dichotomous Encoding” column, where, for example, “L10H20” indicates the CPD cutoff values of light (CPD < = 10) and heavy (CPD > 20) smokers. Median CPD was 20 (L: CPD < = 20, H:CPD > 20).
Table S4 Estimated variance explained including dominance and additive GREML-LDMS-I-paritioned GRMs. Relatedness < 0.05 for all analyses.
Table S5 LDSC-based estimates regression coefficients for annotation categories from the baseline+LD model. † = Bonferroni correction applied for all traits and categories, * = Bonferroni correction applied for all categories within-traits separately.
Table S6 LDSC-based estimates of regression coefficients for annotation categories from the cell type- and tissue-specific expression model. † = Bonferroni correction applied for all traits and categories, * = Bonferroni correction applied for all categories within-traits separately.
Acknowledgements
This work was supported by National Institute of Mental Health R01 MH100141-06 [Principal Investigator (PI): M.C.K.]; National Institute on Drug Abuse R01 DA 044283, National Institute on Drug Abuse R01 DA 037904, National Human Genome Research Institute R01 HG 008983 (PI: S.I.V), and National Institute on Drug Abuse R01 DA042090 (PI: D.B.H.); and the Institute for Behavioral Genetics. We thank John Hewitt, Jerry Stitzel, Charles Hoeffer, Laura Saba, Christian Hopfer, Naomi Wray and Peter Visscher for helpful discussion and comments. This research was conducted using the UK Biobank Resource (application number 1665). We thank the UK Biobank and the participants of the UK Biobank.
Footnotes
Declaration of interests
None.
Supporting Information
Additional supporting information may be found online in the Supporting information section at the end of the article.
References
- 1.US Department of Health and Human Services Health Consequences of Smoking—50 Years of Progress. A Report of the Surgeon General. Atlanta, GA: Centers for Disease Control and Prevention; 2014. [Google Scholar]
- 2.Centers For Disease Control and Prevention Quitting smoking among adults—United States, 2001–2010. Morb Mort Wkly Rep 2011; 60: 1513–9. [PubMed] [Google Scholar]
- 3.Van Meijgaard J, Fielding JE Estimating benefits of past, current, and future reductions in smoking rates using a comprehensive model with competing causes of death. Prev Chronic Dis 2012E122; 10.5888/pcd9.110295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen KA, Ambrose BK, Gentzke AS, Apelberg BJ, Jamal A, King BA Notes from the field: use of electronic cigarettes and any tobacco product among middle and high school students—United States, 2011–2018. Morb Mort Wkly Rep 2018; 67: 1276–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timpson NJ, Greenwood CMT, Soranzo N, Lawson DJ, Richards JB Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet 2017; 19: 110–24. [DOI] [PubMed] [Google Scholar]
- 6.Haberstick BC, Ehringer MA, Lessem JM, Hopfer CJ, Hewitt JK Dizziness and the genetic influences on subjective experiences to initial cigarette use. Addiction 2011; 106: 391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haberstick BC, Zeiger JS, Corley RP, Hopfer CJ, Stallings MC, Rhee SH, et al. Common and drug-specific genetic influences on subjective effects to alcohol, tobacco and marijuana use. Addiction 2011; 106: 215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaprio J Genetic epidemiology of smoking behavior and nicotine dependence. COPD 2009; 6: 304–6. [DOI] [PubMed] [Google Scholar]
- 9.Rose RJ, Broms U, Korhonen T, Dick DM, Kaprio J Genetics of smoking behavior. In: Kim Y-K, editor. Handbook of Behavior Genetics. New York, NY: Springer; 2009, pp. 411–32. [Google Scholar]
- 10.Kendler KS, Schmitt E, Aggen SH, Prescott CA, Virginia V Genetic and environmental influences on alcohol, caffeine, cannabis, and nicotine use from early adolescence to middle adulthood. Arch Gen Psychiatry 2008; 65: 674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 2019; 51: 237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tobacco and, Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 2010; 42: 441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock DB, Guo Y, Reginsson GW, Gaddis NC, Lutz SM, Sherva R, et al. Genome-wide association study across European and African American ancestries identifies a SNP in DNMT3B contributing to nicotine dependence. Mol Psychiatry 2018; 23: 1911–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hancock DB, Wang JC, Gaddis NC, Levy JL, Saccone NL, Stitzel JA, et al. A multiancestry study identifies novel genetic associations with CHRNA5 methylation in human brain and risk of nicotine dependence. Hum Mol Genet 2015; 24: 5940–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quach BC, Bray MJ, Gaddis NC, Liu M, Palviainen T, Minica CC, et al. Expanding the genetic architecture of nicotine dependence and its shared genetics with multiple traits. Nat Commun 2020; 11: 5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matoba N, Akiyama M, Ishigaki K, Kanai M, Takahashi A, Momozawa Y, et al. GWAS of smoking behaviour in 165,436 Japanese people reveals seven new loci and shared genetic architecture. Nat Hum Behav 2019; 3: 471–7. [DOI] [PubMed] [Google Scholar]
- 17.Erzurumluoglu AM, Liu M, Jackson VE, Barnes DR, Datta G, Melbourne CA, et al. Meta-analysis of up to 622,409 individuals identifies 40 novel smoking behaviour associated genetic loci. Mol Psychiatry 2020; 25: 2392–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans LM, Tahmasbi R, Vrieze SI, Abecasis GR, Das S, Gazal S, et al. Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat Genet 2018; 50: 737–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47: 291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 2015; 47: 1228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gazal S, Finucane HK, Furlotte NA, Loh PR, Palamara PF, Liu X, et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat Genet 2017; 49: 1421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brazel DM, Jiang Y, Hughey JM, Turcot V, Zhan X, Gong J, et al. Exome Chip meta-analysis fine maps causal variants and elucidates the genetic architecture of rare coding variants in smoking and alcohol use. Biol Psychiatry 2019; 85: 946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller MC, Coventry WL Quantifying and addressing parameter indeterminacy in the classical twin design. Twin Res Hum Genet 2012; 8: 201–13. [DOI] [PubMed] [Google Scholar]
- 24.Maes HH, Morley K, Neale MC, Kendler KS, Heath AC, Eaves LJ, et al. Cross-cultural comparison of genetic and cultural transmission of smoking initiation using an extended twin kinship model. Twin Res Hum Genet 2018; 21: 179–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Z, Bakshi A, Vinkhuyzen AA, Hemani G, Lee SH, Nolte IM, et al. Dominance genetic variation contributes little to the missing heritability for human complex traits. Am J Hum Genet 2015; 96: 377–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoech AP, Jordan DM, Loh PR, Gazal S, O’Connor LJ, Balick DJ, et al. Quantification of frequency-dependent genetic architectures in 25 UK biobank traits reveals action of negative selection. Nat Commun 2019; 10: 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AAE, Lee SH, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet 2015; 47: 1114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans LM, Keller MC Using partitioned heritability methods to explore genetic architecture. Nat Rev Genet 2018; 19: 185–185. [DOI] [PubMed] [Google Scholar]
- 29.Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 2018; 50: 621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pihl RO Personality disorders, behavioral disinhibition, and addiction: a commentary. Biol Psychiatry 2007; 62: 551–2. [DOI] [PubMed] [Google Scholar]
- 31.Palmer RH, Knopik VS, Rhee SH, Hopfer CJ, Corley RC, Young SE, et al. Prospective effects of adolescent indicators of behavioral disinhibition on DSM-IV alcohol, tobacco, and illicit drug dependence in young adulthood. Addict Behav 2013; 38: 2415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hicks BM, Iacono WG, McGue M Index of the transmissible common liability to addiction: heritability and prospective associations with substance abuse and related outcomes. Drug Alcohol Depend 2012; 123: S18–S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK biobank resource with deep phenotyping and genomic data. Nature 2018; 562: 203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adjangba C, Border R, Romero Villela PN, Ehringer MA, Evans LM Little evidence of modified genetic effect of rs16969968 on heavy smoking based on age of onset of smoking. Nicotine Tob Res 2020; 10.1093/ntr/ntaa229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO The Fagerstrom test for nicotine dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict 1991; 86: 1119–27. [DOI] [PubMed] [Google Scholar]
- 36.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. bioRxiv 2019; 10.1101/563866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abraham G, Inouye M Fast principal component analysis of large-scale genome-wide data. PLOS ONE 2014; 9:e93766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, Lee SH, Goddard ME, Visscher PM GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011; 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golan D, Lander ES, Rosset S Measuring missing heritability: inferring the contribution of common variants. Proc Natl Acad Sci USA 2014; 111: E5272–E5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weissbrod O, Flint J, Rosset S Estimating SNP-based heritability and genetic correlation in case–control studies directly and with summary statistics. Am J Hum Genet 2018; 103: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee SH, Yang J, Chen GB, Ripke S, Stahl EA, Hultman CM, et al. Estimation of SNP heritability from dense genotype data. Am J Hum Genet 2013; 93: 1151–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.GETx Consortium Genetic effects on gene expression across human tissues. Nature 2017; 550: 204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pers TH, Karjalainen JM, Chan Y, Westra HJ, Wood AR, Yang J, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun 2015; 6: 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fehrmann RS, Karjalainen JM, Krajewska M, Westra HJ, Maloney D, Simeonov A, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet 2015; 47: 115–25. [DOI] [PubMed] [Google Scholar]
- 46.Haberstick BC, Timberlake D, Ehringer MA, Lessem JM, Hopfer CJ, Smolen A, et al. Genes, time to first cigarette and nicotine dependence in a general population sample of young adults. Addiction 2007; 102: 655–65. [DOI] [PubMed] [Google Scholar]
- 47.Falconer DS, Mackay TFC Introduction to quantitative genetics Essex. England: Longman; 1996. [Google Scholar]
- 48.Hyman SE, Malenka RC, Nestler EJ Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci 2006; 29: 565–98. [DOI] [PubMed] [Google Scholar]
- 49.Nestler EJ Is there a common molecular pathway for addiction? Nat Neurosci 2005; 8: 1445–9. [DOI] [PubMed] [Google Scholar]
- 50.Del Arco A, Mora F Prefrontal cortex-nucleus accumbens interaction: in vivo modulation by dopamine and glutamate in the prefrontal cortex. Pharmacol Biochem Behav 2008; 90: 226–35. [DOI] [PubMed] [Google Scholar]
- 51.Grace AA, Floresco SB, Goto Y, Lodge DJ Regulation of firing of dopaminergic neurons and control of goal-directed behaviors. Trends Neurosci 2007; 30: 220–7. [DOI] [PubMed] [Google Scholar]
- 52.Bonelli RM, Cummings JL Frontal-subcortical circuitry and behavior. Dialogues Clin Neurosci 2007; 9: 141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feil J, Sheppard D, Fitzgerald PB, Yucel M, Lubman DI, Bradshaw JL Addiction, compulsive drug seeking, and the role of frontostriatal mechanisms in regulating inhibitory control. Neurosci Biobehav Rev 2010; 35: 248–75. [DOI] [PubMed] [Google Scholar]
- 54.Yuan K, Yu D, Bi Y, Li Y, Guan Y, Liu J, et al. The implication of frontostriatal circuits in young smokers: a resting-state study. Hum Brain Mapp 2016; 37: 2013–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miquel M, Vazquez-Sanroman D, Carbo-Gas M, Gil-Miravet I, Sanchis-Segura C, Carulli D, et al. Have we been ignoring the elephant in the room? Seven arguments for considering the cerebellum as part of addiction circuitry. Neurosci Biobehav Rev 2016; 60: 1–11. [DOI] [PubMed] [Google Scholar]
- 56.Koob GF, Volkow ND Neurocircuitry of addiction. Neuropsychopharmacology 2010; 35: 217–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koob GF, Volkow ND Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry 2016; 3: 760–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.So HC, Chau CK, Chiu WT, Ho KS, Lo CP, Yim SH, et al. Analysis of genome-wide association data highlights candidates for drug repositioning in psychiatry. Nat Neurosci 2017; 20: 1342–9. [DOI] [PubMed] [Google Scholar]
- 59.Wainschtein P, Jain DP, Yengo L, Zheng Z, Cupples LA, Shadyab AH, et al. Recovery of trait heritability from whole genome sequence data. bioRxiv 2019; 10.1101/588020. [DOI] [Google Scholar]
- 60.Mathieson I, McVean G Differential confounding of rare and common variants in spatially structured populations. Nat Genet 2012; 44: 243–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xia C, Amador C, Huffman J, Trochet H, Campbell A, Porteous D, et al. Pedigree- and SNP-associated genetics and recent environment are the major contributors to anthropometric and cardiometabolic trait variation. PLOS Genet 2016; 12: e1005804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heckerman D, Gurdasani D, Kadie C, Pomilla C, Carstensen T, Martin H, et al. Linear mixed model for heritability estimation that explicitly addresses environmental variation. Proc Natl Acad Sci USA 2016; 113: 7377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Munafo MR, Tilling K, Taylor AE, Evans DM, Davey SG Collider scope: when selection bias can substantially influence observed associations. Int J Epidemiol 2018; 47: 226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wain LV, Shrine N, Miller S, Jackson VE, Ntalla I, Artigas MS, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK biobank. Lancet Respir Med 2015; 3: 769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Distribution of raw CPD count, and different transformations. Note that 1 pack is approximately 20CPD. For dichotomized CPD, we applied the following Heavy (H) and Light (L) cutoffs of CPD: a) H: >20, L: <= 10; b) H: >30, L: <= 10; c) H: >40, L: <= 5; d) Median CPD of 20 (H: >20, L: <=20).
Figure S2 Cigarettes per day (CPD) (+/− SE), at four different relatedness thresholds and with different CPD transformations as a continuous variable.
Figure S3 Cigarettes per day (CPD) (+/− SE), at four different relatedness thresholds and with CPD dichotomized using the indicated thresholds for low and high CPD.
Figure S4 Age of initiation (+/− SE), at four different relatedness thresholds.
Figure S5 Smoking cessation (+/− SE), at four different relatedness thresholds, and using two different estimation methods.
Figure S6 Smoking initiation (+/− SE), at four different relatedness thresholds, and using two different estimation methods.
Figure S7 (+/− SE) estimates for the four smoking behaviors shown in Fig. 1 of the main text, after exclusion of those samples genotyped on the UK BiLEVE genotyping array, using a relatedness threshold of 0.05. Comparison of these results to those in Fig. 1 show estimates are qualitatively similar after excluding those batches, which were oversampled for heavy smokers from the full UK Biobank sample based on FEV1 phenotypes.
Figure S8 Partitioned LDSC regression coefficient p-values for all annotations across all traits.
Table S1 Estimates of partitioned h2SNP in 6 MAF & LD-stratified bins, and the total h2SNP estimate, across the four traits (and four CPD encodings) using different relatedness cutoffs (0.02, 0.03, 0.04 & 0.05) using GREML. For binary traits, we applied two different estimation methods (GREML & PCGC), and present estimates on the liability scale. Estimates for CPD use the binned encoding, matching GSCAN (Liu et al. 2019); for other CPD encodings, see Tables S2-3. Prevlance for the cessation phenotype is given as the proportion of current smokers. Models that indicate “NoBiLEVE” were run with the genotyping batches using the BiLEVE array excluded from the analysis.
Table S2 Estimates of partitioned h2SNP in 6 MAF & LD-stratified bins, and the total h2SNP estimate, across different continous CPD encodings using different relatedness cutoffs (0.02, 0.03, 0.04 & 0.05) using GREML.
Table S3 Estimates of partitioned h2SNP in 6 MAF & LD-stratified bins, and the total h2SNP estimate, across differet dichotomous CPD encodings using different relatedness cutoffs (0.02, 0.03, 0.04 & 0.05). We applied two different estimation methods (GREML & PCGC), and report estimates on the liability scale using the in-sample prevalence estimates. Designations of heavy (H) and light (L) smokers are indicated in the “Dichotomous Encoding” column, where, for example, “L10H20” indicates the CPD cutoff values of light (CPD < = 10) and heavy (CPD > 20) smokers. Median CPD was 20 (L: CPD < = 20, H:CPD > 20).
Table S4 Estimated variance explained including dominance and additive GREML-LDMS-I-paritioned GRMs. Relatedness < 0.05 for all analyses.
Table S5 LDSC-based estimates regression coefficients for annotation categories from the baseline+LD model. † = Bonferroni correction applied for all traits and categories, * = Bonferroni correction applied for all categories within-traits separately.
Table S6 LDSC-based estimates of regression coefficients for annotation categories from the cell type- and tissue-specific expression model. † = Bonferroni correction applied for all traits and categories, * = Bonferroni correction applied for all categories within-traits separately.