SUMMARY
Viruses rearrange host membranes to support different entry steps. Polyomavirus simian virus 40 (SV40) reorganizes the endoplasmic reticulum (ER) membrane to generate focus structures that enable virus ER-to-cytosol escape, a decisive infection step. The molecular architecture of the ER exit site that might illuminate why it is ideally suited for membrane penetration is unknown. Here 3D focused ion beam scanning electron microscopy (FIB-SEM) reconstruction reveals that the ER focus structure consists of multi-tubular ER junctions where SV40 preferentially localizes, suggesting that tubular branch points are virus ER-to-cytosol penetration sites. Functional analysis demonstrates that lunapark—an ER membrane protein that typically stabilizes three-way ER junctions—relocates to the ER foci, where it supports focus formation, leading to SV40 ER escape and infection. Our results reveal how a virus repurposes the activity of an ER membrane protein to form a virus-induced ER substructure required for membrane escape and suggest that ER tubular junctions are vulnerable sites exploited by viruses for membrane penetration.
In brief
How viruses escape a host membrane during entry to cause infection is unclear. Here Bagchi et al. reveal polyomavirus SV40 hijacks the activity of lunapark, an endoplasmic reticulum (ER) membrane protein, to generate virus-induced multi-tubular ER junctions. These ER substructures represent vulnerable sites where the virus penetrates the ER membrane.
Graphical Abstract

INTRODUCTION
Viruses are the etiologic agents for many devastating human diseases. To cause infection, they must deliver their nucleic acid across a cellular membrane into the cytosol (or nucleus) of their host cell. For enveloped viruses surrounded by a lipid bilayer, this requires fusion of the viral and host membranes, resulting in delivery of the viral particle across the limiting membrane (Helenius and Moss, 2013). In contrast, nonenveloped viruses cannot invade host cells by a fusion mechanism, because they lack a lipid bilayer. How they cross a biological membrane therefore remains mysterious (Moyer and Nemerow, 2011; Tsai, 2007).
Polyomaviruses (PyVs) are nonenveloped viruses responsible for many human diseases (DeCaprio and Garcea, 2013), ranging from BK PyV-associated nephropathy and hemorrhagic cystitis caused by the BK PyV to an aggressive skin cancer called Merkel cell carcinoma (MCC) caused by the human Merkel cell PyV (Arora et al., 2012). Because the prototype PyV simian virus 40 (SV40) is genetically and structurally similar to human PyVs, elucidating the cellular mechanism of SV40 entry has illuminated human PyV infections (Tsai and Qian, 2010). Structurally, SV40 is composed of 72 pentamers of the coat protein VP1 that encloses its DNA genome (Liddington et al., 1991; Stehle et al., 1996), with each pentamer encasing an internal hydrophobic protein VP2 or VP3 (Chen et al., 1998). Three forces stabilize the viral architecture: the VP1 C terminus invades a neighboring VP1 pentamer to provide inter-pentamer support, the presence of intra- and inter-pentamer disulfide bonds stabilizes the viral structure, and calcium ions bound to the virus strengthen its arrangement (Liddington et al., 1991; Stehle et al., 1996). When assembled, SV40 is a spherical particle approximately 50 nm in diameter.
To infect cells, SV40 undergoes receptor-mediated endocytosis, reaching the endosome (Engel et al., 2011) and then the endoplasmic reticulum (ER) (Kartenbeck et al., 1989). Here the virus penetrates the ER membrane to escape into the cytosol (Geiger et al., 2011; Inoue and Tsai, 2011), from which it transports into the nucleus to cause infection (Greber and Kasamatsu, 1996; Yamada and Kasamatsu, 1993). ER-to-cytosol membrane penetration of SV40—a decisive infection step—continues to be enigmatic, although a better understanding of this process is emerging. Specifically, when SV40 reaches the ER from the plasma membrane, ER redox enzymes disrupt the viral disulfide bonds and unfold the VP1 C-terminal arms (Inoue et al., 2015; Schelhaas et al., 2007; Walczak and Tsai, 2011). These reactions partially destabilize SV40, exposing the hydrophobic proteins VP2 and VP3 (Daniels et al., 2006; Rainey-Barger et al., 2007). This in turn generates a hydrophobic particle that inserts into the ER membrane (Daniels et al., 2006; Geiger et al., 2011; Rainey-Barger et al., 2007). Once inserted in the membrane, SV40 remodels the ER to create suborganellar structures—called the ER foci—which function as the viral ER-to-cytosol membrane penetration site (Bagchi et al., 2015; Geiger et al., 2011; Ravindran et al., 2015, 2017; Walczak et al., 2014). In the final step, the membrane-inserted virus is ejected across the ER foci into the cytosol by cytosolic extraction machinery en route to the nucleus for infection (Dupzyk and Tsai, 2018; Dupzyk et al., 2017; Ravindran et al., 2015; Walczak et al., 2014). Despite this model, major gaps in our understanding of ER escape by SV40 remain. In particular, what is the molecular architecture of the ER foci that might explain why this structure is ideally suited for viral membrane penetration, and how does SV40 exploit host factors to construct this ER exit site?
We used super-resolution focused ion beam scanning electron microscopy (FIB-SEM) followed by 3D reconstruction to visualize the molecular arrangement of the virus-induced ER foci. Strikingly, our results reveal that the ER focus structure consists of a network of multiple ER tubules coalescing at distinct sites. SV40 preferentially localizes at these multi-tubular ER junctions, where it appears partially exposed to the cytosol, suggesting that the tubular branch points are likely virus ER-to-cytosol membrane penetration sites. We identify that the host factor lunapark (LNP)—an ER membrane protein that typically stabilizes three-way ER junctions—reorganizes to the ER foci, where it stabilizes focus formation to enable SV40 ER membrane escape and infection. Our findings demonstrate how a virus repurposes the function of an ER membrane protein to promote the formation of an ER substructure required during entry. They also suggest that ER tubular junctions—thought to be particularly destabilized within the ER membranous network—are vulnerable sites well suited for viral membrane penetration.
RESULTS
Identification of the SV40-induced focus structure by confocal and electron microscopy
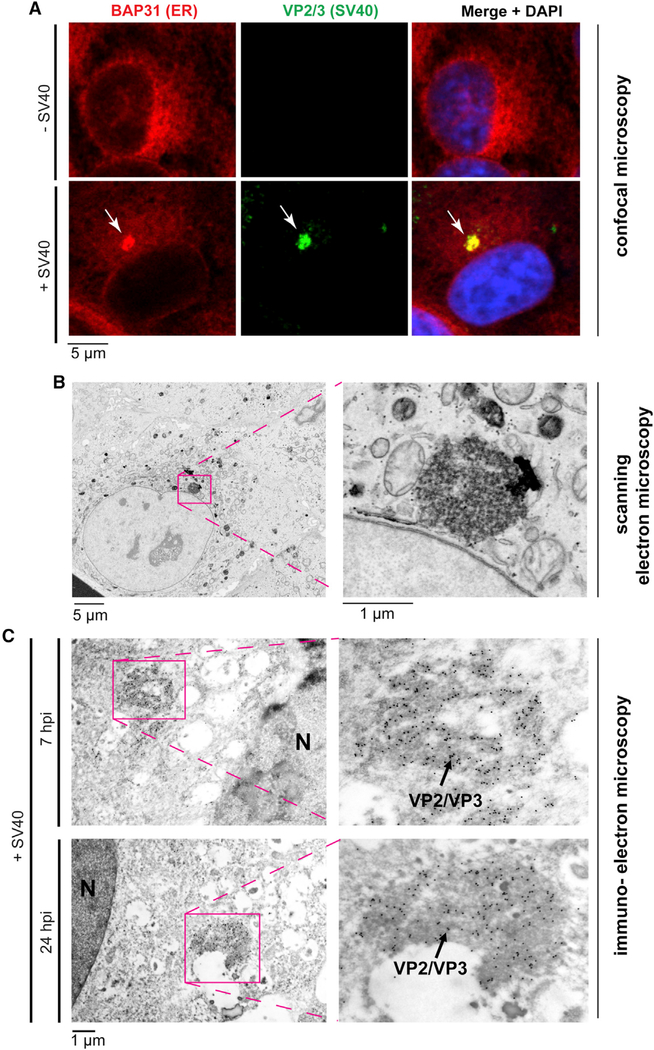
We used FIB-SEM to visualize the molecular architecture of the SV40-induced focus structure. As a first step, we identified this viral ER escape site by conventional scanning electron microscopy (SEM). Using standard confocal microscopy, a virus-induced mature foci can typically be recognized by the characteristic appearance of a perinuclear punctate structure (approximating 2 μm in diameter) that stains positive for the BAP31 ER membrane protein starting 7–8 h post-infection (hpi) during SV40 entry in CV-1 cells (Figure 1, first column, bottom panel; a 7 hpi sample is shown) (Walczak et al., 2014); we refer this as a large and intact focus structure. The formation of two large intact foci can be observed, although less frequently (<20%). As previously reported (Bagchi et al., 2015), VP2/VP3-exposed SV40—representing the membrane penetration-competent viral particle—also accumulates at the ER foci (Figure 1A, second column, bottom panel). Importantly, when a similar SV40-infected CV-1 cell sample was subjected to SEM, a perinuclear electron-dense remodeled ER structure (also approximating 2 μm in diameter) was found in the virus-infected cells (Figure 1B, left panel); this structure was not found in uninfected cells. An enlarged view of this virus-triggered structure reveals the presence of many particles that likely correspond to SV40 (Figure 1B, right panel). Immunoelectron microscopy using a VP2/VP3 antibody revealed that the electron-dense structure of electron microscopy (EM) images (observed in cells infected with SV40 for 7 or 24 h) are SV40 positive (Figure 1C), consistent with the idea that this electron-dense EM structure corresponds to the SV40-positive foci.
Figure 1. Identification of the SV40-induced focus structure by confocal and electron microscopy.
(A) Mock-infected CV-1 cells or CV-1 cells infected with SV40 for 7 h were fixed and stained with the indicated antibodies and analyzed by confocal microscopy.
(B) CV-1 cells infected with SV40 for 7 h were fixed and analyzed by scanning electron microscopy.
(C) Immunogold-stained transmission electron microscopy (TEM) images of SV40-induced foci in CV-1 cells at 7 and 24 hpi. 18-nm gold particles label SV40 VP2/VP3 at the focus structure. N, nucleus.
Virus-induced foci consist of multi-tubular ER junctions with SV40 localized at the junctions
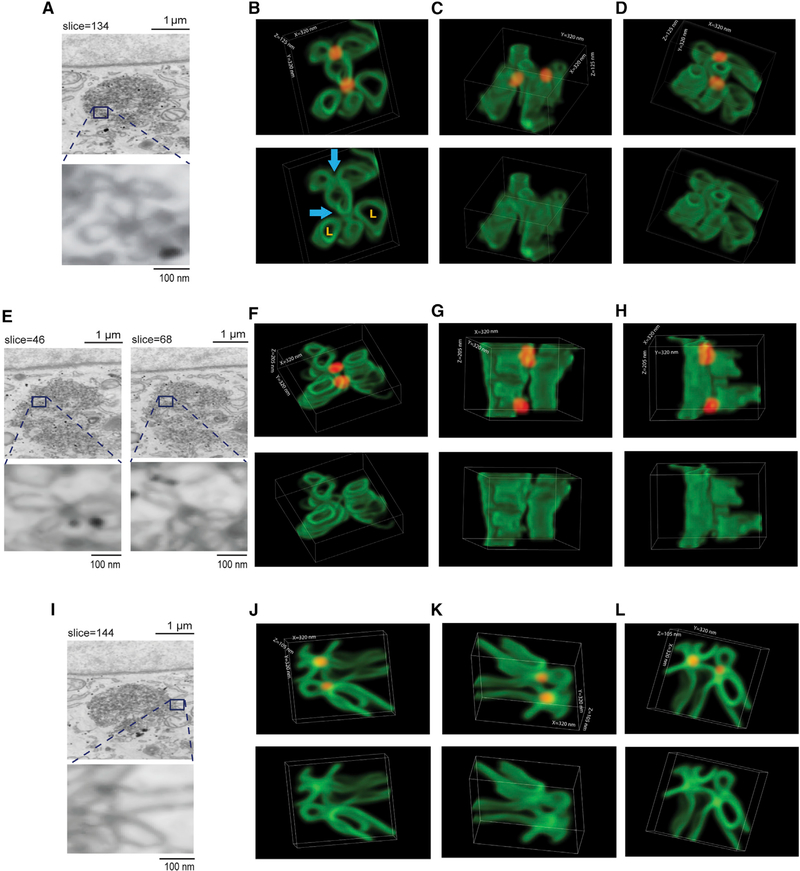
The SV40-induced focus structure was then subjected to FIB-SEM analysis. A representative image (obtained at slice 134) is shown in Figure 2A (top). When this image is enlarged, we identified repeating flower-like structures, each containing a spherical and dense object approximating 50 nm—consistent with SV40—located at the center (Figure 2A, bottom). The petal of this flower-like structure has a luminal cavity with a diameter slightly longer than the 50 nm SV40 particle, in agreement with the reported 60- to 90-nm diameter of the ER lumen (Georgiades et al., 2017).
Figure 2. Virus-induced foci consist of multi-tubular ER junctions with SV40 localized at the junctions.
(A) FIB-SEM image of an ER focus (obtained at slice 134) induced by SV40 in CV-1 cells at 7 hpi.
(B–D) 3D reconstruction of the zoomed-in focus ultrastructure shown in (A) from different angles, where slice 134 serves as the mid-point.
(E) FIB-SEM image of an ER focus (obtained at slices 46 and 68) induced by SV40 in CV-1 cells at 7 hpi.
(F–H) 3D reconstruction of the zoomed-in focus ultrastructure shown in (E) from different angles, starting at slice 46.
(I) FIB-SEM image of an ER focus (obtained at slice 144) induced by SV40 in CV-1 cells at 7 hpi.
(J–L) 3D reconstruction of the zoomed-in focus ultrastructure shown in (I) from different angles, with slice 144 serving as the mid-point.
The green structures mark the ER, and the red structures represent SV40. The blue arrows identify the ER tubular junctions. L, ER lumen.
When this flower-like structure is subjected to 3D FIB-SEM reconstruction (by serial sectioning, with slice 134 serving as the mid-point), a striking 3D image is revealed: from the top view (Figure 2B, top), each petal of the flower-like structure corresponds to the rim of an individual ER tubule (green) enclosing a defined luminal cavity, whereas the center of this structure is a junctional site between the ER tubules where SV40 (red) is localized. For clarity, Figure 2B (bottom) is presented without SV40 to highlight the multi-tubular ER junctions (blue arrows), as well as the ER luminal cavity (marked by an orange L). Figures 2C and 2D show the same structure except from different angles, where the ER tubules are more evident. A corresponding video of this structure is presented in Video S1.
When a set of two flower-like structures from a different z plane within the same ER foci (Figure 2E, obtained at slices 46 and 68) is reconstructed by FIB-SEM (via serial sectioning starting at slice 46), a similar architecture is observed: multiple ER tubules coalescing at distinct junctional sites where SV40 is located (Figure 2F, top view; Figures 2G and 2H, different angles). In this sample, SV40 appears to be partially exposed to the cytosol (Figure 2G, top), suggesting that ER tubular junctions function as SV40 membrane escape sites. Video S2 is the corresponding video of this structure. Figures 2I–2L represent yet another example of SV40 found at junctional sites between many ER tubules within the same ER foci but from a different z plane; the corresponding video of this structure is presented in Video S3.
To corroborate these findings, we used different ER foci from a different cell and found that SV40 is again located at similar multi-tubular ER junctional sites (Figure S1), in agreement with the results presented in Figure 2. Collectively, these 3D FIB-SEM reconstruction images uncover the molecular architecture of the SV40-induced foci, revealing a striking virus-induced ER-derived architecture devised to support virus membrane penetration.
LNP promotes SV40 infection
What host factors exploited by SV40 might promote the formation of the multi-tubular ER junctions within the ER foci? LNP is an ER membrane protein that typically stabilizes three-way ER junctions, resulting in the characteristic reticular weblike ER morphology (Chen et al., 2012, 2015; Wang et al., 2016). We reasoned that because the SV40-induced multi-tubular ER junctional sites are reminiscent of the typical three-way ER junctions, LNP might support the stable formation of the multi-tubular ER junctions, which allows LNP to promote ER escape and infection of SV40.
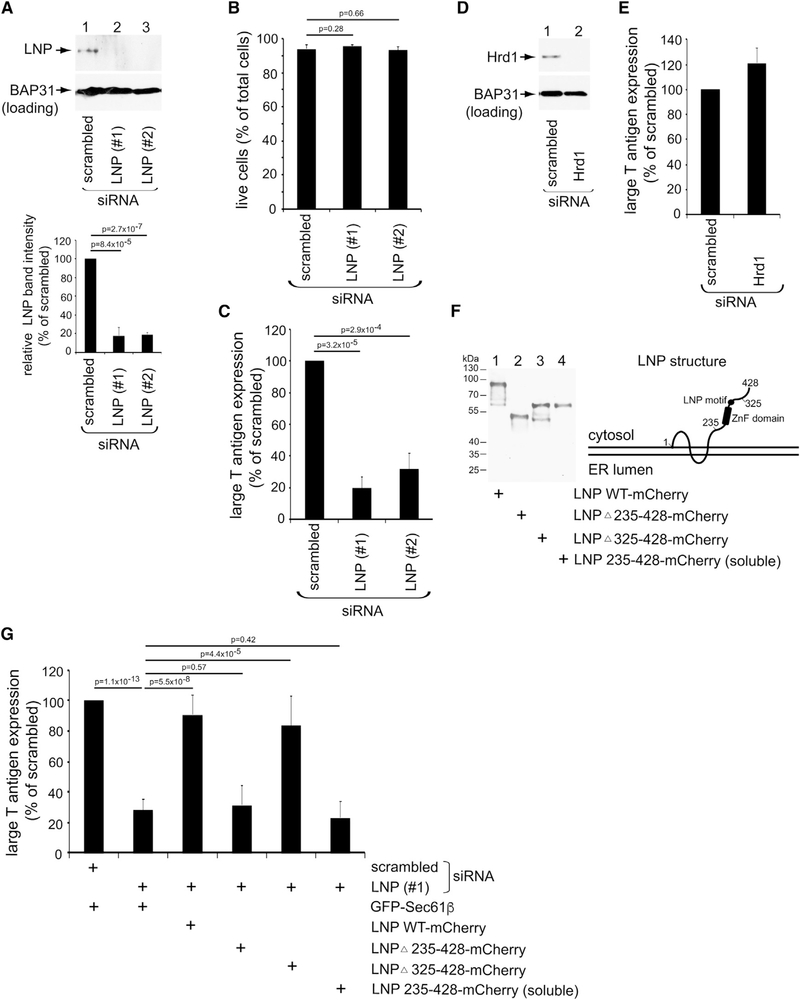
We first asked whether LNP is important during SV40 infection. CV-1 cells were transfected with the control, small interfering RNA (siRNA) (scrambled), or either of two siRNAs targeting LNP (#1 or #2). As expected, endogenous LNP decreased markedly in cells transfected with LNP siRNA compared with control cells (Figure 3A, top panel; quantified in bar graph), with no effect on cell viability (Figure 3B). To measure infection under LNP knockdown (KD), we scored for the presence of the large T antigen, a virally encoded protein that is expressed only when the incoming viral particle successfully reaches the nucleus. Importantly, we found that LNP KD results in a pronounced block in large T antigen expression (Figure 3C), indicating that LNP promotes SV40 infection. As a control, KD of the Hrd1 ER membrane protein (Figure 3D) did not impair SV40 infection (Figure 3E).
Figure 3. LNP facilitates SV40 infection.
(A) CV-1 cells were transfected with the indicated siRNA for 48 h and lysed with 1% Triton X-100, and the resulting extract was subjected to SDS-PAGE and immunoblotting with the indicated antibodies. The relative LNP band intensity is quantified in the graph below by using FIJI/ImageJ (NIH) software. The data were normalized to scrambled siRNA. The values represent means and standard deviations (SDs) of the results of three independent experiments. A two-tailed t test was used.
(B) CV-1 cells transfected with the indicated siRNA for 72 h were treated with trypan blue after transfection to stain dead cells. The live cells were quantified relative to the total cells to check the cell viability. Data represent the mean ± SD of three independent experiments.
(C) Large T antigen-positive nuclei were scored in SV40-infected CV-1 cells transfected with the indicated siRNAs. The data were normalized to the scrambled siRNA, with the values representing means and SDs (n ≥ 3).
(D) CV-1 cells were transfected with the indicated siRNA for 48 h and lysed with 1% Triton X-100, and the resulting extract was subjected to SDS-PAGE and immunoblotting with the indicated antibodies.
(E) Large T antigen-positive nuclei were scored in SV40-infected CV-1 cells transfected with the indicated siRNAs. The data were normalized to the scrambled siRNA, with the values representing means and SDs (n ≥ 3).
(F) (Diagram) Structure of LNP transmembrane protein in the ER membrane. The C-terminal domain of LNP in the cytosol contains the zinc-finger (ZnF) domain and LNP motif. The indicated LNP constructs were expressed in CV-1 cells at 24 h post-transfection. LNP 235–428-mCherry is a cytosolic soluble protein.
(G) CV-1 cells were transfected with the indicated siRNA for 24 h before transfection with the indicated plasmids for 30 h. The cells were then infected with SV40, fixed, and immunostained with anti-large T antigen antibodies. The percentage of large T antigen-positive cells was determined only in cells (≥600) expressing the indicated GFP- or mCherry-tagged protein by immunofluorescence microscopy. The data were normalized to the scrambled siRNA, with the values representing means and SDs (n ≥ 3).
To verify the integrity of the LNP siRNAs, cells transfected with the scrambled or LNP (#1) siRNA were cotransfected with either the GFP-Sec61β control plasmid or a siRNA-resistant LNP wild-type-mCherry (LNP WT-mCherry) construct. Only cells expressing GFP-Sec61β or LNP WT-mCherry were analyzed for infection. Using this method, we found that expression of LNP WT-mCherry (Figure 3F, bottom, lane 1) in LNP-depleted cells largely restored the block in large T antigen expression because of LNP KD (Figure 3G, compare the third and second bars). These results indicate that the impaired SV40 infection because of transfection with the LNP siRNA is caused by loss of endogenous LNP and is not the result of unintended off-target effects, unambiguously establishing LNP as playing a critical function during SV40 infection.
Structurally, LNP is a two-pass transmembrane protein with both the N and the C termini facing the cytosol (Figure 3F, diagram). The cytosolic domain of LNP contains a zinc-finger (ZnF) domain that mediates LNP-LNP dimerization followed by a LNP motif (Casey et al., 2015; Wang et al., 2016). The LNP Δ235–428 deletion mutant (lacking the ZnF domain and LNP motif) fused to mCherry (Figure 3F, lane 2; LNP Δ235–428-mCherry) was previously shown to not stabilize typical three-way ER junctions, because it does not mobilize to this structure (Wang et al., 2016). We find that this LNP mutant does not rescue SV40 infection in LNP-depleted cells (Figure 3G, compare the fourth and second bars). By contrast, the LNP Δ325–428-mCherry deletion mutant, which does contain the ZnF domain and LNP motif (Figure 3F, lane 3; LNP Δ325–428-mCherry) and can stabilize typical three-way ER junctions (Wang et al., 2016), largely restored the block in SV40 infection under LNP KD (Figure 3G, compare the fifth and second bars). These findings suggest that the membrane-stabilizing function of LNP is critical during SV40 infection. Not surprisingly, although the cytosolic soluble LNP 235–428-mCherry mutant contains the ZnF domain and LNP motif (Figure 3F, lane 4; LNP 235–428-mCherry), its inability to rescue SV40 infection in LNP-depleted cells (Figure 3G, compare the sixth and second bars) demonstrates that proper anchoring of LNP’s ZnF domain and LNP motif to the ER membrane is essential for their function.
LNP is required for ER-to-cytosol escape of SV40
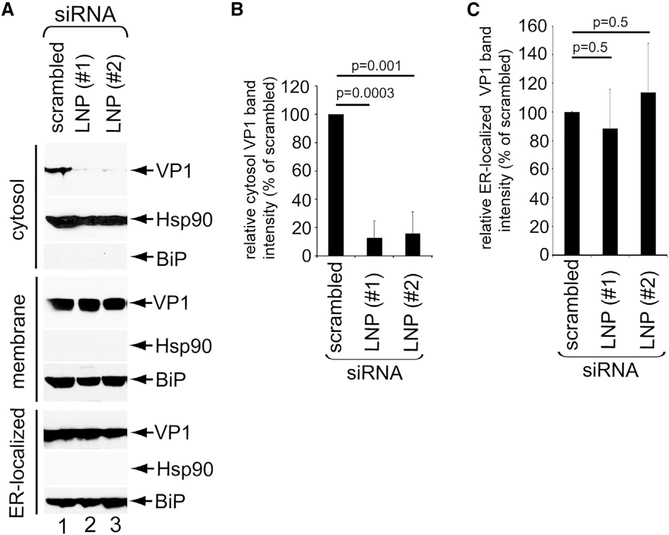
Because LNP lies at the interface of the ER and cytosol, we suspected that this ER membrane protein supports ER-to-cytosol transport of SV40 to promote infection. To monitor cytosol arrival of SV40, we used an established cell-based, semi-permeabilized ER-to-cytosol transport assay (Geiger et al., 2011; Inoue and Tsai, 2011). In this assay, CV-1 cells (transfected with scrambled, LNP #1, or LNP #2 siRNA) were incubated with SV40, harvested, and treated to a low concentration of digitonin to permeabilize the plasma membrane without impairing internal membranes. Cells were then centrifuged to generate two fractions: a supernatant fraction that contains cytosolic proteins and virus that reaches the cytosol (cytosol fraction) and a pellet fraction that harbors membranes including the ER, as well as virus associated with membranes (membrane fraction). The integrity of the fractionation procedure can be monitored in the release of cytosol-localized heat shock protein 90 (Hsp90) to the cytosolic fraction and the pelleting of ER-resident BiP with the membrane fraction (Figure 4A).
Figure 4. LNP is required for ER-to-cytosol escape of SV40.
(A) CV-1 cells infected with SV40 (multiplicity of infection [MOI] 5) and transfected with the indicated siRNA were subjected to the ER-to-cytosol transport assay (see STAR Methods). The ER luminal protein BiP served as the membrane marker, whereas cytosolic Hsp90 served as the cytosol marker.
(B) Relative VP1 band intensities in the cytosol fraction from (A) were determined by using FIJI/ImageJ software. The data were normalized to scrambled siRNA. The values represent means and SD of the results of three independent experiments. A two-tailed t test was used.
(C) Relative VP1 band intensities in ER-localized fraction from (A) were determined by using FIJI/ImageJ software. A two-tailed t test was used.
Using this assay, we found that depletion of LNP (using either siRNA) markedly reduced the SV40 VP1 level in the cytosol compared with control cells (Figure 4A, top panel; the VP1 band intensity is quantified in Figure 4B). As a control, we asked whether KD of LNP reduced arrival of SV40 to the ER from the cell surface using an established biochemical fractionation protocol that isolates ER-localized SV40 from infected cells (Inoue and Tsai, 2011), and we found that it did not (Figure 4A, 7th panel; quantified in Figure 4C). Hence, our data demonstrate that LNP promotes cytosol arrival of SV40 from the ER.
SV40 triggers LNP reorganization to the ER focus structure to stabilize this ER exit site
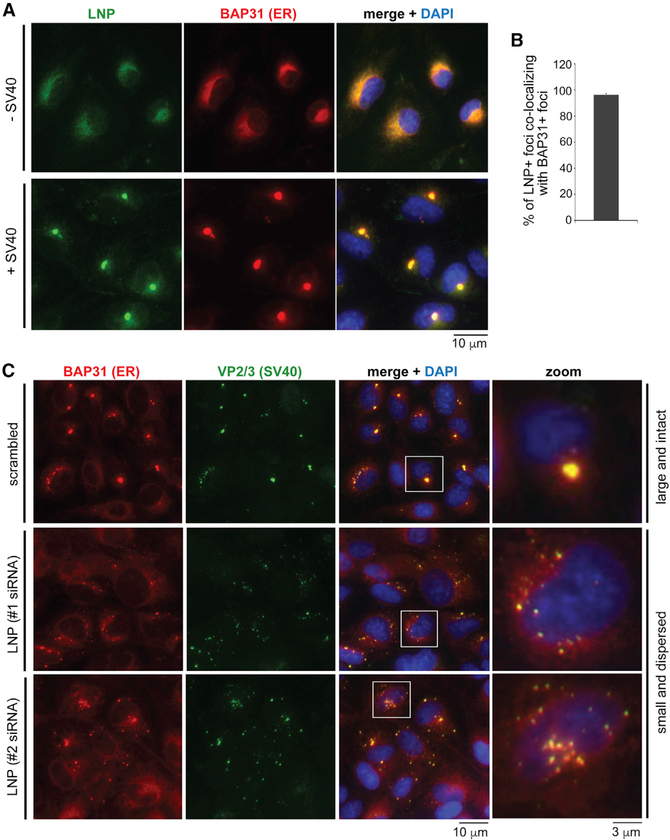
We hypothesize that LNP reorganizes into the ER focus structure to promote the stable formation of this exit site to support SV40 ER-to-cytosol escape (Figure 4) and infection (Figure 3). Consistent with this idea, we found that LNP relocates to the BAP31-positive ER foci (Figure 5A); quantification revealed that essentially all BAP31-positive foci contain the LNP-positive puncta (Figure 5B). In addition, no LNP-positive foci were found to be BAP31-puncta negative. Thus, LNP mobilizes to the SV40-induced ER focus structure. These confocal images were taken at 16 hpi, even though the ER focus structure begins to form around 6–7 hpi (Walczak et al., 2014). When the ER foci are observed at 16 hpi by EM, the highly intense electron-dense signal of the focus structure precludes us from clearly delineating any structure within the ER foci. By contrast, the relatively lower electron-dense signal of the ER foci at 7 hpi by EM enables us to identify the flower-like structure within the ER foci (Figures 1 and 2).
Figure 5. SV40 triggers LNP reorganization into the ER focus structure to stabilize this ER exit site.
(A) CV-1 cells were immunostained with LNP (green), ER marker BAP31 (red), and DAPI (blue) with or without SV40 infection (MOI 5) for 16 h. The BAP31 focus structure represents the SV40 ER exit site.
(B) Quantification of the percentage of LNP foci co-localizing with BAP31 foci in (A). More than 400 cells were counted in each experiment, and three independent experiments were performed.
(C) CV-1 cells transfected with the indicated siRNA were infected with SV40 for 16 h. Cells were fixed; immunostained for BAP31 (red), SV40 VP2/3 (green), and DAPI (blue); and analyzed by immunofluorescence microscopy. Scale bar is 10 μm. The far-right (zoom) panels show 3.3× enlargements of the boxed focus regions.
Analogous to its role in stabilizing a three-way ER junction (Chen et al., 2012, 2015; Wang et al., 2016), we postulate that LNP supports the stable formation of the viral focus structure; the inability of the LNP stabilization-defective mutant to rescue SV40 infection under LNP KD is consistent with this idea (Figure 3D). To support this hypothesis, we monitored appearance of the ER focus structure in SV40-infected cells at 16 hpi. Although an obvious large and intact ER focus (approximating 2 μm) can be found in control cells (Figure 5C, top row [zoom]), it appeared infrequently under LNP KD. Instead, multiple small and dispersed focus structures appeared in the LNP-depleted cells (Figure 5C, second and third rows [zoom]). When these small and dispersed foci were examined by SEM, flower-like structures in the foci—found in the large ER focus (Figure 2)—were absent (Figure S2). These findings suggest that LNP supports stable assembly of the large and intact ER focus that is critical for the formation of the flower-like structures.
Depletion of LNP blunts the formation of the large and intact ER focus
To mechanistically probe how LNP promotes the stable formation of this viral ER exit site, we envision two scenarios to account for the observation that small and dispersed foci, not the large and intact focus, are found under LNP KD. In the first scenario, the small and dispersed foci seen under LNP KD could reflect disassembly of a large and intact focus after this structure has formed; in this scenario, LNP stabilizes the large and intact focus once it is formed. In the second scenario, the appearance of small and dispersed foci under LNP KD might reflect the inability of multiple small foci to fuse and form the large and intact focus; in this case, LNP stabilizes an intermediate focus structure after fusion of multiple small foci.
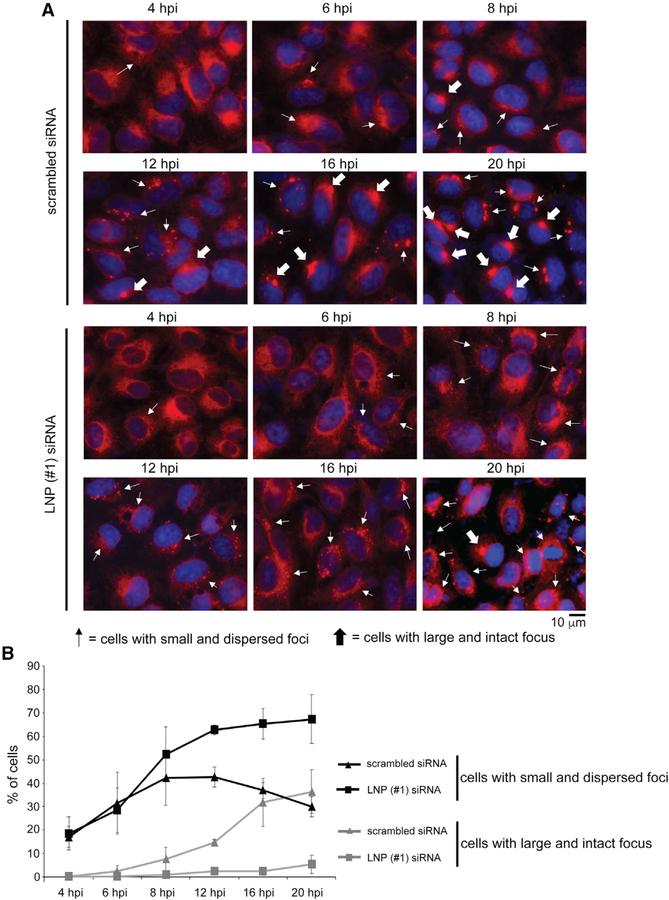
To clarify which scenario is appropriate, we performed a time course infection experiment (4–20 hpi) and quantified the percentage of control and LNP KD cells containing either the large and intact focus or the small and dispersed foci. In control cells, the percentage of cells harboring small and dispersed foci steadily increased up to 12 hpi, followed by a gradual decrease after this time point (Figure 6A; quantified in Figure 6B). In parallel, the percentage of cells harboring the large and intact focus increased throughout this time course experiment (Figure 6A; quantified in Figure 6B). Importantly, between 12 and 16 hpi, the decrease in the percentage of cells with small and dispersed foci corresponds to a sharp increase in the percentage of cells with the large and intact focus, suggesting that fusion of the multiple dispersed foci to generate the large and intact focus occurs efficiently within this time frame.
Figure 6. Depletion of LNP blunts formation of the large and intact ER focus.
(A) CV-1 cells transfected with scrambled siRNA or LNP (#1) siRNA were infected with SV40 and, at the indicated time point, fixed, stained with BAP31 (red) and DAPI (blue), and analyzed by epifluorescence microscopy. Cells with small and dispersed foci are marked as thin arrows, and cells with large and intact focus are marked as thick arrows.
(B) Quantification of the percentage of cells with small and dispersed foci, as well as the percentage of cells with the large and intact focus, under the scrambled or LNP KD condition from (A). At least 200 cells were analyzed for each condition, and three independent biological replicates were performed. The error bar represents the standard deviation of the results of three independent experiments.
By contrast, in LNP-depleted cells, the percentage of cells with small and dispersed foci steadily increased throughout the time points, yet a very low percentage of cells harboring the large and intact focus appeared throughout this time course study (Figure 6A; quantified in Figure 6B). Thus, without LNP, the small and dispersed foci persist, failing to fuse and generate the large and intact focus. These findings are consistent with the second scenario, in which LNP stabilizes the fusion intermediate of the small and dispersed foci that leads to the formation of the large and intact focus.
DISCUSSION
A remarkable discovery in the field of virus-host interaction is that viruses remodel the host ER to create ER-derived structures that support distinct entry steps during virus infection (Inoue and Tsai, 2013). In the case of SV40, this polyomavirus triggers the formation of an ER-derived focus structure that promotes virus ER-to-cytosol membrane escape, a critical infection step (Bagchi et al., 2015; Geiger et al., 2011; Ravindran et al., 2015, 2017; Walczak et al., 2014). We and another group have reported a succession of findings that strongly support the idea that the ER foci act as the ER-to-cytosol exit sites of SV40, including the following: (1) proper focus formation is required to promote SV40 ER membrane escape to the cytosol and infection; (2) membrane penetration-competent SV40 preferentially localizes to the ER foci; (3) the ER foci are formed before cytosol entry of the virus; (4) select ER membrane proteins that promote SV40 ER membrane escape reorganize to the ER foci, whereas those that are dispensable in this penetration process do not; (5) perturbing cytosolic factors that extract SV40 from the ER into the cytosol trap the virus in the ER foci; and (6) the ER foci are physically surrounded by cytosolic disassembly machinery that disassembles SV40 after ER membrane escape (Bagchi et al., 2015; Dupzyk et al., 2017; Geiger et al., 2011; Ravindran et al., 2015, 2017; Spriggs et al., 2020; Walczak et al., 2014).
Despite these insights, the molecular architecture of the membrane penetration structure that might explain why it is suited for virus membrane transport remains unknown. To address this, we used super-resolution FIB-SEM followed by 3D reconstruction. This combinatorial approach offers the opportunity to capture an image in a cell in 3D with high resolution and has provided important information about virus-host interactions, such as in the case of HIV and coronavirus infections (Baena et al., 2021). Using this strategy, we discovered that within the ER foci, SV40 is located at the multi-tubular ER junctions. The junction-localized virus can be seen to be partially exposed to the cytosol, suggesting that these tubular junctions are membrane escape sites. Creating the multi-tubular junctions is an ingenious strategy devised by the virus, because typical three-way ER branch sites are thought to be unstable within the ER tubular network (Georgiades et al., 2017)—their structural vulnerability ideally suits them for virus membrane penetration. We believe that this virus-induced structure has not been observed in other virus infection events.
How does SV40 promote fusion of many ER tubules to generate the multi-tubular ER junctions in the ER foci? The virus-triggered multi-tubular ER junction is reminiscent of typical three-way ER junctions that give rise to the classic weblike reticular ER morphology (Chen et al., 2013; Wang and Rapoport, 2019). In this latter case, several ER membrane proteins, including atlastins (Chen et al., 2012; Wang et al., 2016) and LNP (Chen et al., 2015), play key roles in establishing the three-way ER junction. Specifically, although atlastins initially trigger tubule-tubule fusion to generate the three-way ER junction (Chen et al., 2012; Wang et al., 2016), LNP is subsequently recruited to the three-way ER branch site, where it stabilizes this structure (Chen et al., 2015).
Therefore, we asked whether LNP reorganizes to the SV40-induced foci to assist in the formation of this structure and found that it did. A LNP mutant that is unable to stabilize a typical three-way ER junction (Wang et al., 2016) cannot promote SV40 infection. This indicates that the junction-stabilizing activity of LNP is critical during SV40 entry, consistent with our hypothesis that LNP stabilizes the multi-tubular ER junction when it is recruited to this site. Mechanistically, our analysis further suggests that LNP participates in the maturation of the large and intact focus structure. LNP likely does so by stabilizing a transient fusion intermediate that forms after the coalescence of multiple small focus structures and then becomes the large and intact focus. This model agrees with our previous report demonstrating that the formation of the large and intact focus results from fusion of multiple small foci (Ravindran et al., 2017). At present, a role of the atlastins during SV40 ER focus formation is unknown and requires future investigation.
In summary, our study unveils the molecular arrangement of a virus-induced ER-derived subcellular structure responsible for promoting viral ER escape, a critical infection step. It also identifies a host component whose function is exploited to assist in the formation of this structure. Virus-induced structures derived from the host ER are known for supporting different viral entry steps, such as in the cases of flavivirus and coronavirus infections in which ER-derived organelles are created to support virus replication (Paul and Bartenschlager, 2015). Our discovery of an ER-derived architecture—multi-tubular ER junctions—that is designed to support virus membrane escape thus expands the repertoire of structures that can be created from the complex ER membranous network.
Limitations of the study
In the FIB-SEM 3D analysis of the multi-tubular ER junctions, the precise structural features of the membrane-penetrating SV40 remain unclear. Are VP2 and VP3 exposed in the penetrating virus? Higher imaging techniques are required to address this question. Another caveat of our study is incomplete understanding regarding how the multi-tubular ER junctions are generated. What additional cellular components might act with LNP to promote the formation of this virus-induced ER-derived membrane penetration structure? Finally, a conceptual limitation is the broader implication of our results: Do other pathogens trigger the formation of a similar membrane escape structure to support their infection cycle?
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Billy Tsai (btsai@umich.edu).
Materials availability
Further information and requests for reagents and resources should be directed to and will be fulfilled by the Lead Contact.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and Reagents
CV-1 (male) cells were obtained from ATCC, cultured at 37°C under 5% CO2 in complete Dulbecco’s modified Eagle’s medium (DMEM), containing 10% fetal bovine serum (FBS), 10 U/ml penicillin, and 10 μg/ml streptomycin (GIBCO, Waltham, MA). DMEM, Opti-MEM, and 0.25% trypsin-EDTA were purchased from Thermo Fisher Scientific (Waltham, MA). FBS was purchased from R&D systems (Minneapolis, MN). Lipofectamine RNAiMax and ProLong Diamond Antifade Mountant with DAPI was purchased from Thermo Fisher Scientific (Waltham, MA). Phosphate-buffered saline (PBS) (1X) was purchased from GIBCO (Waltham, MA). Triton X-100, digitonin, and phenylmethylsulfonyl fluoride (PMSF) were purchased from Millipore Sigma (St. Louis, MO). Fugene HD was purchased from Promega (Madison, WI). Lunapark (Novus Biologicals, Centennial, CO), SV40 large T antigen (Santa Cruz Biotechnology, Santa Cruz, CA), SV40 VP2/3 and VP1 (Abcam, Cambridge, MA), Hsp90 (Santa Cruz Biotechnology, Santa Cruz, CA), BiP (Abcam, Cambridge, MA), and BAP31 (Thermo Fisher, Waltham, MA) antibodies were purchased from the indicated companies. The mCherry antibody is a generous gift from the lab of Dawen Cai (University of Michigan, Ann Arbor, MI). Details of the cell lines and reagents are listed in the Key resources table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-BAP31 | Thermo Fisher Scientific | Cat# MA3-002; RRID: AB_325095 |
| Anti-Lunapark | Novus Biologicals | Cat# NBP1-80637; RRID: AB_11013558 |
| Anti-SV40 large T antigen | Santa Cruz Biotechnology | Cat# sc-147; RRID: AB_628305 |
| Anti-Hsp90 | Santa Cruz Biotechnology | Cat# sc-13119; RRID: AB_675659 |
| Anti-SV40 VP1 antibody | Abcam | Cat# ab53977, RRID: AB_946338 |
| Anti-BiP | Abcam | Cat# ab21685; RRID: AB_2119834 |
| Anti-SV40 VP2/3 | Abcam | Cat# ab53983; RRID: AB_946339 |
| Anti-mCherry | University of Michigan | N/A |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| SV40 | Lab stored | Inoue and Tsai, 2011 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| DMEM | Thermo Fisher Scientific | Cat# 11995 |
| Fetal bovine serum | R&D system | Cat# S11150 |
| Trypsin-EDTA (0.25%) | Thermo Fisher Scientific | Cat# 25300054 |
| Fugene HD | Promega | Cat# E2311 |
| Lipofectamine RNAiMax | Thermo Fisher Scientific | Cat# 13778150 |
| Penicillin-Streptomycin | GIBCO | Cat# 15140-122 |
| ProLong Diamond Antifade Mountant with DAPI | Thermo Fisher Scientific | Cat# P36941 |
| Digitonin | Millipore Sigma | Cat# 300410-1GM |
| Triton X-100 | Millipore Sigma | Cat# 45-93443 |
| Phenylmethylsulfonyl fluoride | Millipore Sigma | Cat# BCCD4192 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| CV-1 | ATCC | Cat# CCL-70 |
|
| ||
| Recombinant DNA | ||
|
| ||
| LNP-WT-mCherry | Gift from Tom Rapoport (Harvard Medical School) | N/A |
| LNP-Δ235-428-mCherry | Gift from Tom Rapoport (Harvard Medical School) | N/A |
| LNP-Δ325-428-mCherry | Gift from Tom Rapoport (Harvard Medical School) | N/A |
| LNP-235-428-mCherry | Gift from Tom Rapoport (Harvard Medical School) | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| FIJIa | NIH | N/A |
| Avizo 9.2.0. | Thermo Fisher Scientific | N/A |
| FluoRender | Scientific Computing and Imaging Institute, Utah | N/A |
METHOD DETAILS
Preparation of Virus
WT SV40 was prepared using the OptiPrep gradient system. Briefly, SV40-infected or viral genome-transfected CV-1 cells were lysed in a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, and 0.5% Brij58 at 4°C for 30 min and centrifuged at 16,100 g for 10 min. The supernatant was loaded onto a discontinuous 20% and 40% OptiPrep gradient and centrifuged at 49,500 rpm for 2 h at 4°C in an SW 55Ti rotor. A viral particle fraction between 20% and 40% OptiPrep was collected with a needle.
Widefield Epifluorescence and Confocal Microscopy
CV-1 cells were grown in 12-well plate with coverslips. For knockdown studies, cells were reverse transfected with the desired siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific) at the time of cell seeding. SV40-infected or mock-infected cells washed with PBS followed by fixation with 4% formaldehyde at room temperature were then permeabilized using 0.2% Triton X-100, and blocked by 5% milk with 0.2% Tween. Primary antibodies were incubated for 1 h at room temperature or overnight at 4°C, followed by incubation of fluorescent-conjugated secondary antibodies for 2 h at room temperature. Coverslips were mounted with ProLong Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific) for widefield epifluorescence and confocal microscopy. Images were taken using inverted epifluorescence microscope (Nikon Eclipse TE2000-E) equipped with 100X oil immersion objective (N.A. 1.4), Sola lumencore light engine and Photometrics CoolSnap HQ camera, Nikon N-SIM E in confocal mode with CFI SR HP Apochromat TIRF 60XC Oil immersion objective (NA 1.49), LU-NV series laser unit, and ORCA-Flash 4.0 sCMOS camera (Hamamatsu Photonics K.K.). NIS-Elements AR software was used to take images. FIJI distribution of ImageJ (Schindelin et al., 2012; Schneider et al., 2012) was used for image processing, analyses, and assembly.
Immuno-EM
CV-1 cells were infected with SV40 for 7h and 24 h, fixed chemically (with 3% paraformaldehyde and 0.2% glutaraldehyde), dehydrated with ethanol, and embedded in London Resin (LR) white (Electron Microscopy Sciences, PA) at 55°C for 48 hours. The thin sections (70 nm in thickness) were cut using an ultramicrotome with a diamond knife. The sections were collected onto bare nickel grids. The sections are then stained immunochemically with primary antibodies raised against antigens exposed on the surface of the sections. The primary antibodies are visualized by staining immunochemically with secondary antibodies raised against the species and isotype of the primary antibodies, conjugated to colloidal gold particles. The immunochemically stained sections are then contrast stained with 0.5% uranyl acetate (Electron Microscopy Sciences, PA) to reveal the ultrastructure of the cells followed by carbon evaporation by Leica EM ACE600 high vacuum coater and are finally viewed by TEM (JEOL JEM 1400 Plus).
Scanning Electron Microscopy (SEM) and Focused Ion Beam-Scanning Electron Microscopy (FIB-SEM)
CV-1 cells were grown in 35 mm MatTek grid dishes, infected with SV40 (MOI 50) and fixed chemically [2.5% glutaraldehyde (GA) in 0.1M sodium cacodylate buffer (CB)] at 7 hours post infection, stained (1% OsO4, 1% uranyl acetate, Walton’s lead aspartate), dehydrated with ethanol, and embedded in Durcupan ACM Epoxy resin (Electron Microscopy Sciences, PA) for SEM or FIB-SEM study. Tescan MIRA3 field emission SEM was used for the SEM study, and Thermofisher Helios 650 nanolab focused ion beam SEM was used for the FIB-SEM study. The pixel resolution used for FIB-SEM was 5 nm and each slice represents 5 nm size. FIB-SEM images are then aligned and cropped using Avizo 9.2.0. software followed by a non-local means filter used on the dataset to remove noise and the contrast was inverted. Structures of interest within the ER-focus were segmented using FIJI distribution of ImageJ followed by 3D rendering using FluoRender software. FIB-SEM was performed at the University of Michigan core facility.
siRNA and Plasmids
Allstar negative-control siRNA (QIAGEN, Hilden, Germany) was used as a scrambled control siRNA. Two siRNAs specific to human lunapark were synthesized by Millipore-Sigma (St. Louis, MO), and the target sequences are: siRNA #1, UACGUUGGCACUGGUACGAAU; siRNA #2, GAAACUUACAAGACGGCUA. Target sequence for Hrd1 siRNA is GGAGACUGCCACUACAGUUGUdTdT and synthesized by Millipore-Sigma (St. Louis, MO). CV-1 cells were reverse transfected with each siRNA at 40 nM using Lipofectamine RNAi Max (Thermo Fisher Scientific) and incubated for 48 h prior to SV40 infection. All LNP constructs were generous gifts from Tom Rapoport (Harvard Medical School, Boston, MA) and has been previously described (Wang et al., 2016).
Cell Viability Assay
CV-1 cells were seeded with approximately 1.5 × 105 cells per well in 12-well plates and reverse transfected with 40 nM of the indicated siRNA for 72 hr. Cells were then trypsinized, resuspended in PBS, and diluted at a 1:1 ratio in 0.4% Trypan Blue Solution (Thermo Fisher). The solution was loaded on a hemocytometer and the number of blue-staining cells was quantified relative to the number of total cells. Three biological replicates were performed for a total of ~250 cells per condition. Bars represent the means ± SD of three independent experiments.
SV40 Infection and ER-foci Formation Assay
CV-1 cells were seeded and transfected with 40 nM LNP siRNA or scrambled siRNA (along with Opti-MEM and RNAiMax) onto coverglass in 12-well plates at 37°C for 48 h before infection with SV40. For knockdown-rescue experiments, after 24 h of siRNA transfection as described above, the CV-1 cells were washed and transfected with mCherry tagged LNP constructs or GFP-Sec61β using Fugene and Opti-MEM for 30 h. Cells were then infected with SV40 [multiplicity of infection (MOI), 0.5] for 20 h and scored for large T antigen expression, or at MOI of 5 for 16 h to monitor BAP31-positive or VP2/3-positive ER-foci formation. The infected cells were fixed with 4% paraformaldehyde (Alfa Aesar) for 15 min at room temperature followed by permeabilization with 0.2% Triton X-100 in PBS for 5 min. Cells were then immunostained with primary antibody diluted in blocking buffer containing 5% milk in TBST (Tris-buffered saline with 0.02% Tween 20) for 1 h at room temperature and then washed four times with blocking buffer. Cells were then incubated with a fluorescent dye conjugated secondary antibody for 30 min and then washed three times with blocking buffer, PBS, and water before air drying and mounting on glass slides using ProLong Diamond Antifade Mountant. Images were taken using an inverted epifluorescence microscope (Nikon Eclipse TE2000-E, Melville, NY) equipped with 10x, 40x, and 60x 1.40 NA objectives and standard DAPI (blue), FITC (green) and TRITC (red) filter cubes. Images were processed using the Fiji (NIH). In each experiment, > 8,000 cells were scored for large T antigen-positive cells. For knockdown-rescue studies, > 200 cells expressing GFP- or mCherry-tagged construct were scored under each condition, and at least three independent experiments were performed.
ER-arrival and ER-to-cytosol membrane transport assays
The ER-to-cytosol transport assay was performed as described previously (Inoue and Tsai, 2011). Briefly, 6 × 105 CV-1 cells were seeded in a 6-cm dish and the cells were transfected with 40 nM LNP siRNA or scramble siRNA for 48 h. The cells were then infected with purified SV40 (MOI 5) for 16 h, semi-permeabilized using 0.1% digitonin for 10 min, and then centrifuged. The resulting supernatant fraction represented the cytosol fraction, while the pellet fraction represented the membrane fraction. The membrane fraction was further treated with 1% Triton X-100 and subjected to centrifugation − the resulting supernatant contained ER-localized SV40. The different fractions generated by this assay were subjected to SDS-PAGE followed by western blotting using the indicated antibodies. The VP1 band intensity was quantified using FIJI/ImageJ software and normalized to the Hsp90 or BiP loading control band.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data obtained from at least three independent experiments (biological replicates) were combined for statistical analyses. Results were analyzed using Student two-tailed t test. Data are represented as the mean values and error bar represents standard deviation (SD) (n ≥ 3) where indicated. p < 0.05 was considered to be significant.
Supplementary Material
Highlights.
SV40-induced foci consist of multi-tubular ER junctions with virus at the junctions
SV40 triggers reorganization of the lunapark ER membrane protein to the ER foci
Lunapark promotes SV40 infection and is required for ER-to-cytosol escape of SV40
Depletion of lunapark blunts formation of the large and intact ER focus
ACKNOWLEDGMENTS
We thank Chelsey Spriggs (University of Michigan) for critical review of this manuscript. B.T. is supported by the National Institutes of Health (RO1AI064296). We also thank Aaron Taylor (University of Michigan) and Allen Hunter (University of Michigan) for their assistance in the FIB-SEM experiment.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110077.
REFERENCES
- Arora R, Chang Y, and Moore PS (2012). MCV and Merkel cell carcinoma: a molecular success story. Curr. Opin. Virol 2, 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena V, Conrad R, Friday P, Fitzgerald E, Kim T, Bernbaum J, Berensmann H, Harned A, Nagashima K, and Narayan K (2021). FIB-SEM as a Volume Electron Microscopy Approach to Study Cellular Architectures in SARS-CoV-2 and Other Viral Infections: A Practical Primer for a Virologist. Viruses 13, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi P, Walczak CP, and Tsai B (2015). The endoplasmic reticulum membrane J protein C18 executes a distinct role in promoting simian virus 40 membrane penetration. J. Virol 89, 4058–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey AK, Chen S, Novick P, Ferro-Novick S, and Wente SR (2015). Nuclear pore complex integrity requires Lnp1, a regulator of cortical endoplasmic reticulum. Mol. Biol. Cell 26, 2833–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XS, Stehle T, and Harrison SC (1998). Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J 17, 3233–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Novick P, and Ferro-Novick S (2012). ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat. Cell Biol 14, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Novick P, and Ferro-Novick S (2013). ER structure and function. Curr. Opin. Cell Biol 25, 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Desai T, McNew JA, Gerard P, Novick PJ, and Ferro-Novick S (2015). Lunapark stabilizes nascent three-way junctions in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 112, 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R, Rusan NM, Wadsworth P, and Hebert DN (2006). SV40 VP2 and VP3 insertion into ER membranes is controlled by the capsid protein VP1: implications for DNA translocation out of the ER. Mol. Cell 24, 955–966. [DOI] [PubMed] [Google Scholar]
- DeCaprio JA, and Garcea RL (2013). A cornucopia of human polyomaviruses. Nat. Rev. Microbiol 11, 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupzyk A, and Tsai B (2018). Bag2 Is a Component of a Cytosolic Extraction Machinery That Promotes Membrane Penetration of a Nonenveloped Virus. J. Virol 92, e00607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupzyk A, Williams JM, Bagchi P, Inoue T, and Tsai B (2017). SGTA-Dependent Regulation of Hsc70 Promotes Cytosol Entry of Simian Virus 40 from the Endoplasmic Reticulum. J. Virol 91, e00232–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel S, Heger T, Mancini R, Herzog F, Kartenbeck J, Hayer A, and Helenius A (2011). Role of endosomes in simian virus 40 entry and infection. J. Virol 85, 4198–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, and Helenius A (2011). BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat. Cell Biol 13, 1305–1314. [DOI] [PubMed] [Google Scholar]
- Georgiades P, Allan VJ, Wright GD, Woodman PG, Udommai P, Chung MA, and Waigh TA (2017). The flexibility and dynamics of the tubules in the endoplasmic reticulum. Sci. Rep 7, 16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber UF, and Kasamatsu H (1996). Nuclear targeting of SV40 and adenovirus. Trends Cell Biol 6, 189–195. [DOI] [PubMed] [Google Scholar]
- Helenius A, and Moss B (2013). Virus entry–an unwilling collaboration by the cell. Curr. Opin. Virol 3, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, and Tsai B (2011). A large and intact viral particle penetrates the endoplasmic reticulum membrane to reach the cytosol. PLoS Pathog 7, e1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, and Tsai B (2013). How viruses use the endoplasmic reticulum for entry, replication, and assembly. Cold Spring Harb. Perspect. Biol 5, a013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Dosey A, Herbstman JF, Ravindran MS, Skiniotis G, and Tsai B (2015). ERdj5 Reductase Cooperates with Protein Disulfide Isomerase To Promote Simian Virus 40 Endoplasmic Reticulum Membrane Translocation. J. Virol 89, 8897–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartenbeck J, Stukenbrok H, and Helenius A (1989). Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol 109, 2721–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, and Harrison SC (1991). Structure of simian virus 40 at 3.8-A resolution. Nature 354, 278–284. [DOI] [PubMed] [Google Scholar]
- Moyer CL, and Nemerow GR (2011). Viral weapons of membrane destruction: variable modes of membrane penetration by non-enveloped viruses. Curr. Opin. Virol 1, 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, and Bartenschlager R (2015). Flaviviridae Replication Organelles: Oh, What a Tangled Web We Weave. Annu. Rev. Virol 2, 289–310. [DOI] [PubMed] [Google Scholar]
- Rainey-Barger EK, Magnuson B, and Tsai B (2007). A chaperone-activated nonenveloped virus perforates the physiologically relevant endoplasmic reticulum membrane. J. Virol 81, 12996–13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran MS, Bagchi P, Inoue T, and Tsai B (2015). A Non-enveloped Virus Hijacks Host Disaggregation Machinery to Translocate across the Endoplasmic Reticulum Membrane. PLoS Pathog 11, e1005086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran MS, Engelke MF, Verhey KJ, and Tsai B (2017). Exploiting the kinesin-1 molecular motor to generate a virus membrane penetration site. Nat. Commun 8, 15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelhaas M, Malmström J, Pelkmans L, Haugstetter J, Ellgaard L, Grünewald K, and Helenius A (2007). Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131, 516–529. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs CC, Badieyan S, Verhey KJ, Cianfrocco MA, and Tsai B (2020). Golgi-associated BICD adaptors couple ER membrane penetration and disassembly of a viral cargo. J. Cell Biol 219, e201908099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle T, Gamblin SJ, Yan Y, and Harrison SC (1996). The structure of simian virus 40 refined at 3.1 A resolution. Structure 4, 165–182. [DOI] [PubMed] [Google Scholar]
- Tsai B (2007). Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol 23, 23–43. [DOI] [PubMed] [Google Scholar]
- Tsai B, and Qian M (2010). Cellular entry of polyomaviruses. Curr. Top. Microbiol. Immunol 343, 177–194. [DOI] [PubMed] [Google Scholar]
- Walczak CP, and Tsai B (2011). A PDI family network acts distinctly and coordinately with ERp29 to facilitate polyomavirus infection. J. Virol 85, 2386–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CP, Ravindran MS, Inoue T, and Tsai B (2014). A cytosolic chaperone complexes with dynamic membrane J-proteins and mobilizes a nonenveloped virus out of the endoplasmic reticulum. PLoS Pathog 10, e1004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, and Rapoport TA (2019). Reconstituting the reticular ER network—mechanistic implications and open questions. J. Cell Sci 132, jcs227611. [DOI] [PubMed] [Google Scholar]
- Wang S, Tukachinsky H, Romano FB, and Rapoport TA (2016). Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. eLife 5, e18605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, and Kasamatsu H (1993). Role of nuclear pore complex in simian virus 40 nuclear targeting. J. Virol 67, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.