Abstract
Background
Cellular prion protein (PrPC) is a cell surface GPI-anchored protein, usually known for its role in the pathogenesis of human and animal prionopathies. However, increasing knowledge about the participation of PrPC in prion pathogenesis contrasts with puzzling data regarding its natural physiological role. PrPC is expressed in a number of tissues, including at high levels in the nervous system, especially in neurons and glial cells, and while previous studies have established a neuroprotective role, conflicting evidence for a synaptic function has revealed both reduced and enhanced long-term potentiation, and variable observations on memory, learning, and behavior. Such evidence has been confounded by the absence of an appropriate knock-out mouse model to dissect the biological relevance of PrPC, with some functions recently shown to be misattributed to PrPC due to the presence of genetic artifacts in mouse models. Here we elucidate the role of PrPC in the hippocampal circuitry and its related functions, such as learning and memory, using a recently available strictly co-isogenic Prnp0/0 mouse model (PrnpZH3/ZH3).
Results
We performed behavioral and operant conditioning tests to evaluate memory and learning capabilities, with results showing decreased motility, impaired operant conditioning learning, and anxiety-related behavior in PrnpZH3/ZH3 animals. We also carried in vivo electrophysiological recordings on CA3-CA1 synapses in living behaving mice and monitored spontaneous neuronal firing and network formation in primary neuronal cultures of PrnpZH3/ZH3 vs wildtype mice. PrPC absence enhanced susceptibility to high-intensity stimulations and kainate-induced seizures. However, long-term potentiation (LTP) was not enhanced in the PrnpZH3/ZH3 hippocampus. In addition, we observed a delay in neuronal maturation and network formation in PrnpZH3/ZH3 cultures.
Conclusion
Our results demonstrate that PrPC promotes neuronal network formation and connectivity. PrPC mediates synaptic function and protects the synapse from excitotoxic insults. Its deletion may underlie an epileptogenic-susceptible brain that fails to perform highly cognitive-demanding tasks such as associative learning and anxiety-like behaviors.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12915-021-01203-0.
Keywords: Cellular prion protein, Epilepsy, Hippocampus, Behavior, Anxiety, LTP
Background
Cellular prion protein (PrPC) is a cell surface glycosylphosphatidylinositol (GPI) anchored protein expressed in several tissues with high levels in the nervous system. It is expressed ubiquitously in the brain, especially in neurons and glial cells [1–5]. PrPC is known for its crucial role in the pathogenesis of human and animal prionopathies [6, 7]. In these diseases, PrPC is transformed into a misfolded β-sheet-rich isoform, the infectious prion protein (PrPSc) [6]. Increasing knowledge about the participation of PrPC in prion pathogenesis contrasts with puzzling data regarding its natural physiological role/s [8–10]. Indeed, this controversy was also strengthened by the absence, until a few years ago, of an appropriate Prnp0/0 mouse model without PrPC protein, with high breeding capability to dissect biological relevance in specific processes [9, 11–13].
PrPC has been previously described as neuroprotective, mainly by using loss-of-function approaches [14–16], while in other studies, PrPC overexpression was associated with increased susceptibility to neurotoxicity and cell death [15, 17–19]. This might mean that Prnp levels should be constrained to a certain level to develop their natural functions [11, 15]. This balance is altered in several injuries and neurodegenerative processes, presenting changes in mRNA and protein expression, for example, in Alzheimer’s disease, dementia with Lewy bodies, some tauopathies [20, 21], human prionopathies (e.g., sporadic Creutzfeldt Jakob Disease (sCJD) [22]), and multiple sclerosis [23].
As indicated, numerous studies have explored the physiological roles of PrPC in vivo using Prnp0/0 mice. However, it was later demonstrated that some physiological functions were unfortunately misattributed to PrPC due to genetic artifacts generated during the production of the most commonly used knock-out model, still in use in some laboratories: the Zürich I (PrnpZH1/ZH1) mouse [24]. This mouse displayed a mixed background (B6129: C57BL6/J + 129Sv) and was further backcrossed with C57BL/6 J mice to generate the B6.129 mouse line [19, 25], and with FVB mice to generate the FVBN-PrnpZH1/ZH1 model [25] in order to reduce the 129/Sv-associated genes. However, the generated lines were systematically confounded by the Prnp-linked loci polymorphic region containing numerous 129/Sv-associated “flanking genes” (FG) close to Prnp deletion [26, 27]. In fact, after crossing PrnpZH1/ZH1 mice with C57BL/6 J for more than ten generations to reduce FG, a remnant of ≈ 2–5% of the 129/Sv genome markers still persisted in B6.129 mice [26, 28, 29]. Genome analysis of these models revealed that the number of FG in the chromosome 2 regions where Prnp is localized is 62, related to different functions (i.e., cancer, depression, anxiety, among others [26]). Thus, we consider that most of the physiological phenotypes attributed to the Prnp absence or overexpression, using these mouse models carrying FG, need to be revaluated and confirmed in other FG-free models. As an example, in previous studies, we and others demonstrated that these FGs masked the real neuroprotective function of PrPC against kainate (KA) administration in vivo [19, 30–32]. Although a full description of the FG-associated effects in a null Prnp background is not available, one of these FGs is the signal regulatory protein alpha (Sirpα), an important regulator of several innate immune functions [33]. Although prion disease evolution is not modified in Sirpα0/0 mice [34], it has been clearly demonstrated that Sirpα is mainly responsible for a phagocytic function previously attributed to PrPC [26, 27]. The number of functions misattributed to PrPC increased when a recent study described a substrain-related dependence of Cu(I)-ATPase activity among Prnp0/0 mice related to the 129/Sv FGs and not PrPC [35].
In neurons, PrPC is transported along axons [36] and enriched at the synaptic terminal, where it has been described that it interacts with some subunits of the glutamate receptors (e.g., NMDA-R, GluK2/GluK3, GluN2D, or mGluR5), modulating their activity, and with anchoring proteins such as PSD-95 [37–39]. However, due to the different mouse strains used and some experimental differences, the involvement of PrPC in neurotransmission is still elusive. For example, different studies reported reduced [40–43], normal [25], or enhanced [19, 44] long-term potentiation (LTP) in Prnp0/0 mice compared to wild-type mice. Following these descriptions, the consequences of Prnp absence in memory, learning, and behavior lead to variable results in studies using mice carrying FGs [24, 45–48] or not [42, 49].
In the present study, we focused our attention on reexamining some PrPC functions associated with neurotransmission, learning, and behavior, taking advantage of a recently generated Prnp0/0 mouse model: the Zürich 3 (PrnpZH3/ZH3) [50]. This co-isogenic mouse was generated in a pure C57BL/6 J background using TALEN technology [50] and it is resistant to prion infection [51]. Here we performed a set of behavioral tests to analyze PrnpZH3/ZH3 mouse activity, learning, and memory capabilities. In addition, basic synaptic functions, KA-mediated excitability, and LTP induction were evaluated electrophysiologically in alert behaving mice. Finally, PrPC roles during neuronal differentiation and activity were also evaluated in primary cortical cultures. Results indicate that adult PrnpZH3/ZH3 mice display reduced activity and anxiety-like behavior. They also fail to acquire different instrumental learning tasks. In addition, our experiments show that hippocampal CA3-CA1 PrnpZH3/ZH3 synapse fails to induce LTP, most likely due to an exacerbated endogenous excitability, further corroborated in vivo after KA administration. Lastly, our results are sustained by the observed alteration in the expression patterns of several genes associated with neuronal system function and synaptic protein-protein interactions in the PrnpZH3/ZH3 hippocampus by an RNAseq analysis and its RT-qPCR validation.
Results
The absence of PrPC has been related to deficiencies in behavior, learning, and memory in several mouse models with different results [24, 42, 52, 53]. In order to evaluate the implication of PrPC in systemic behavioral tasks, we took advantage of the new knock-out model, the PrnpZH3/ZH3 mouse. First, we analyzed the nest building capacity between PrnpZH3/ZH3 vs. Prnp+/+ mice as an indicator of mouse welfare. In contrast to Schmitz et al., where they used PrnpZH1/ZH1 mice (reviewed in [54]), results showed a slightly increased but not significantly nest-building capacity in PrnpZH3/ZH3 compared to controls (PrnpZH3/ZH3 = 3.81 ± 0.64, n = 7 vs. Prnp+/+ = 3.00 ± 0.41, n = 7; mean ± S.E.M., p = 0.15; Mann-Whitney U non-parametric test) (Additional file 1: Fig. S1), suggesting that the two genotypes had similar welfare conditions [55].
Reduced activity, increased thigmotaxis, and anxiety-related behavior in PrnpZH3/ZH3 mice
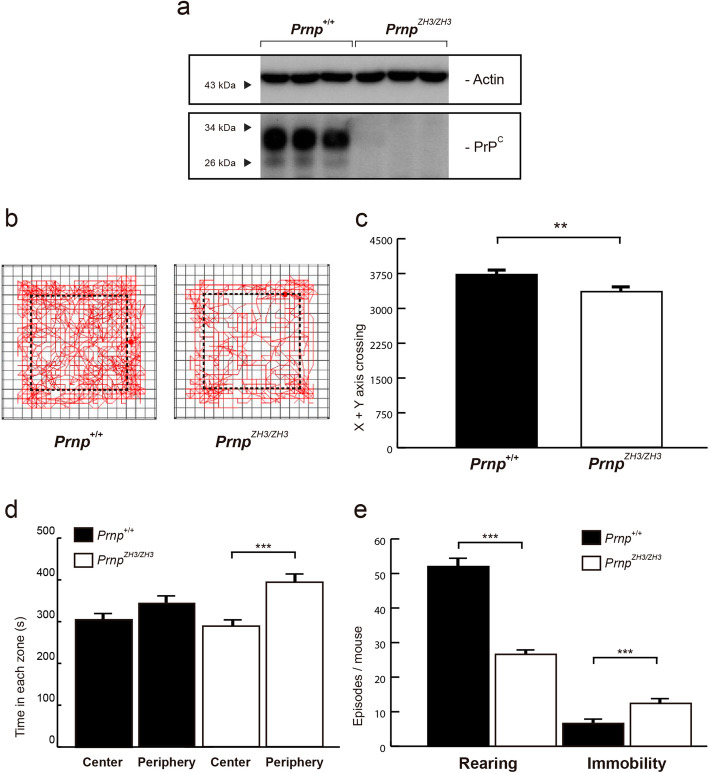
First, we performed the open field test to measure the general locomotor activity levels, anxiety, and willingness in knock-out mice (Fig. 1). Prnp+/+ and PrnpZH3/ZH3 mice (n = 49 for each genotype) were individually placed (in rounds of two animals in parallel) in the open field arena for 15 min, and their activity was evaluated on the X-Y-Z axes (Fig. 1b). PrnpZH3/ZH3 mice showed significantly reduced displacement in the field (Prnp+/+ = 3725 ± 93 a.u. vs. PrnpZH3/ZH3 = 3370 ± 95 a.u.; **p = 0.009; Mann-Whitney U non-parametric test) (Fig. 1c). Anxiety and stress increased the thigmotaxis behavior and the natural aversion to exploring the inner square of the field during the test [56]. Thus, to evaluate anxiety-like behavior in the PrnpZH3/ZH3 mice, we measured this thigmotaxis performance as the time spent in the center (inner region of the field) vs. the periphery (outer region of the field) for each mouse (Fig. 1b). Prnp+/+ mice spent the same amount of time in the two regions, while PrnpZH3/ZH3 animals remained significantly more time in the periphery close to the walls, suggesting an apprehension of the center of the field that reflects an anxiety-like behavior (Prnp+/+: Center = 303.3 ± 14.5 vs. Periphery = 345.7 ± 18,0; and PrnpZH3/ZH3: Center = 288.2 ± 15.1 vs. Periphery = 394.3 ± 20.3; mean ± S.E.M., p = 0.071 and ***p < 0.001 respectively; Mann-Whitney U non-parametric test) (Fig. 1d). Stressed behavior was also assessed by counting the number of rearing and immobility episodes during the test. PrnpZH3/ZH3 mice displayed significantly fewer rearing episodes (Prnp+/+ = 52 ± 2 vs. PrnpZH3/ZH3 = 26.6 ± 1.7; mean ± S.E.M., ***p < 0,001; Mann-Whitney U non-parametric test) and more immobility episodes (Prnp+/+ = 6.6 ± 1.2 vs. PrnpZH3/ZH3 = 12.4 ± 0.9; mean ± S.E.M., ***p < 0.001; Mann-Whitney U non-parametric test) confirming an anxiety-like behavior (Fig. 1e).
Fig. 1.
PrnpZH3/ZH3 mice show reduced activity and anxiety-related behavior. a Immunoblot analysis of PrPC expression in Prnp+/+ and PrnpZH3/ZH3 mice forebrain. Actin is used as a loading control. b Representative images of Prnp+/+ and PrnpZH3/ZH3 mouse exploratory behavior in the open field test. The dotted line delineates the center and the periphery of the arena. c Mouse activity in the open field test represented as the number of lines crossed in the X + Y-axis. d Time spent (s) by the mice in the center and periphery of the open field arena. e Number of rearing and immobility episodes displayed by Prnp+/+ and PrnpZH3/ZH3 mice during the open field test. In total, 98 animals (n = 49 for each genotype) were tested individually, in rounds of two animals in parallel. Data are presented as mean ± S.E.M. **p < 0.01 and ***p < 0.001, Mann-Whitney U non-parametric test. Source data and individual data values are available in Additional file 10
PrnpZH3/ZH3 mice and PrnpZH1/ZH1 failed to acquire instrumental learning tasks
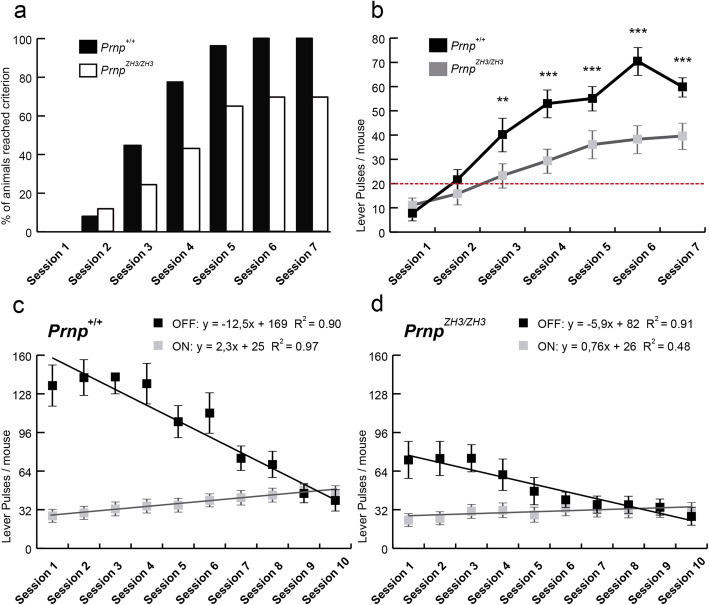
Our next goal was to examine the capabilities of PrnpZH3/ZH3 mice in performing highly demanding learning tasks. Instrumental learning capabilities were tested with operant conditioning in the Skinner box (n = 49 for each genotype) (Fig. 2). Collected data were compared to those obtained using PrnpZH1/ZH1 mice (Additional file 2: Fig. S2). Thirty-one percent of PrnpZH3/ZH3 mice did not reach the learning criterion (to obtain ≥ 20 pellets for two consecutive sessions) at the end of the training session. In contrast, all Prnp+/+ mice (100%) meet the selected criterion from the 6th session (Fig. 2a). Similarly, in a second set of experiments using PrnpZH1/ZH1 mice (Prnp+/+ = 10 and PrnpZH1/ZH1 = 10), 50% of PrnpZH1/ZH1 mice failed to reach the criterion, but 80% of wild-type mice reached it at the end of the sessions (Additional file 2: Fig. S2a). These results strongly suggest that mice lacking Prnp (both ZH1 and ZH3 backgrounds) present evident instrumental learning deficiencies. Also, Prnp+/+ mice pressed the lever significantly more times from session 3 onwards than PrnpZH3/ZH3 animals (Session 1: Prnp+/+ = 7.8 ± 1.3 vs. PrnpZH3/ZH3 = 11.12 ± 1.9, p > 0.99; Session 2: Prnp+/+ = 21.6 ± 2.51 vs. PrnpZH3/ZH3 = 15.8 ± 2.2, p > 0.99; Session 3: Prnp+/+ = 40.2 ± 5.1 vs. PrnpZH3/ZH3 = 23.3 ± 3.2, **p = 0.0023; Session 4: Prnp+/+ = 53.0 ± 3.9 vs. PrnpZH3/ZH3 = 29.4 ± 3.1, ***p < 0.0001; Session 5: Prnp+/+ = 55.1 ± 3.3 vs. PrnpZH3/ZH3 = 36.1 ± 3.9, ***p = 0.0003; Session 6: Prnp+/+ = 70.5 ± 4.1 vs. PrnpZH3/ZH3 = 38.3 ± 3.9 ***p < 0.0001; Session 7: Prnp+/+ = 59.9 ± 2.3 vs PrnpZH3/ZH3 = 39.6 ± 3.5, ***p < 0.0001; two-way ANOVA + Bonferroni’s multiple comparisons test) (Fig. 2b). However, as observed in the open field test, PrnpZH3/ZH3 mice presented considerable inactive behaviors (Fig. 1). To distinguish the reduction of activity from real learning deficits, 44 mice (Prnp+/+ = 24 and PrnpZH3/ZH3 = 20) were subjected to a more complex operant conditioning paradigm, the light ON/light OFF task (see “Material and methods”). As expected, the total number of lever pulses during the OFF period was higher in the Prnp+/+ mice and drastically reduced along with sessions (Session 1 = 134.8 ± 13.3 vs. Session 10 = 39.7 ± 5.2) (Fig. 2c). In parallel, Prnp+/+ mice increased the number of pulses in the ON period (Session 1 = 26.9 ± 1.8 vs. Session 10 = 45.7 ± 2.8) (Fig. 2c). In contrast, PrnpZH3/ZH3 mice showed a reduced decrease of pulses in the OFF period (Session 1 = 73.2 ± 11.7 vs. Session 10 = 26.5 ± 3.9) and an incipient increase in the ON period (Session 1 = 23.4 ± 1.6 vs. Session 10 = 31.1 ± 2.9) (Fig. 2d). Learning capacity, measured as the difference in the curve slope during ON or OFF periods, was drastically reduced in PrnpZH3/ZH3 mice (OFF: Prnp+/+ = − 12.5, R2 = 0.90; PrnpZH3/ZH3 = − 5.9, R2 = 0.92; ON: Prnp+/+ = 2.3, R2 = 0.96; PrnpZH3/ZH3 = 0.8, R2 = 0.48) (Fig. 2c,d). These differences show that PrnpZH3/ZH3 mice failed to learn to avoid OFF periods and push the lever during the ON periods, indicating that PrPC seems to be necessary to properly acquire instrumental learning tasks. A similar study was developed using PrnpZH1/ZH1 mice, and the task accuracy ratio ((lever presses during light ON − lever presses during light OFF)/(total number of lever presses)) was evaluated. At the end of the experiment (sessions 7 and 8), the PrnpZH1/ZH1 mice showed lower values than wild-type mice (session 7: Prnp+/+ = 0.31 vs. PrnpZH1/ZH1 = − 0.02; session 8: Prnp+/+ = 0.54 vs. PrnpZH1/ZH1 = 0.1), reinforcing the notion that the absence of PrPC decreases the instrumental learning goals in mutant mice (Additional file 2: Fig. S2b).
Fig. 2.
Impairments in the acquisition of an instrumental learning task in PrnpZH3/ZH3 mice. a Percentage of mice reaching the selected criterion (to collect ≥ 20 food pellets for two consecutive days) in the successive training sessions. b Lever presses of Prnp+/+ and PrnpZH3/ZH3 mice during the fixed ratio (1:1) conditioning paradigm. The test was performed daily for seven consecutive days. c,d Lever presses of Prnp+/+ (c) and PrnpZH3/ZH3 (d) mice during the ON/OFF training paradigm. Lines represent best linear fits for lever presses evoked during light ON (gray) and light OFF (black) periods. Equations corresponding to regression lines are illustrated in c and d, including R2 coefficients. Data are presented as a percentage in a, and as mean ± S.E.M. in b–d. **p < 0.01 and ***p < 0.001, two-way ANOVA + Bonferroni’s multiple comparisons test. Source data and individual data values are available in Additional file 10
To test for possible deficiencies in motor coordination and balance that could also affect in the results from the Skinner box and the open field tests, we compared the performances of both Prnp+/+ and PrnpZH3/ZH3 mice in the accelerating rotarod test. After a training session, the mice latency to fall from the rods was tested for five sessions during two consecutive days. In the first day, the PrnpZH3/ZH3 mice displayed a significantly lower latency just in the first two session compared to the control mice (Additional file 3: Fig. S3a). However, from the third run, their performance was not significantly different (Session 1: Prnp+/+ = 48.51 ± 3.34 vs. PrnpZH3/ZH3 = 24.48 ± 7.56, *p = 0.014; Session 2: Prnp+/+ = 56.65 ± 6.56 vs. PrnpZH3/ZH3 = 27.30 ± 8.03, **p = 0.007; Session 3: Prnp+/+ = 61.49 ± 4.69 vs. PrnpZH3/ZH3 = 41.11 ± 8.20, p = 0.121; Session 4: Prnp+/+ = 55.71 ± 2.82 vs. PrnpZH3/ZH3 = 37.00 ± 6.45, p = 0.189; Session 5: Prnp+/+ = 54.30 ± 1.73 vs. PrnpZH3/ZH3 = 38.39 ± 6.44, p = 0.380; mean ± S.E.M., two-way ANOVA + Bonferroni’s multiple comparisons test). In the second day, no significant difference was observed (Session 1: Prnp+/+ = 45.61 ± 5.59 vs. PrnpZH3/ZH3 = 31.95 ± 8.16; Session 2: Prnp+/+ = 53.54 ± 3.25 vs. PrnpZH3/ZH3 = 44.99 ± 7.00; Session 3: Prnp+/+ = 53.67 ± 4.68 vs. PrnpZH3/ZH3 = 47.23 ± 8.03; Session 4: Prnp+/+ = 59.91 ± 5.48 vs. PrnpZH3/ZH3 = 48.16 ± 9.06; Session 5: Prnp+/+ = 52.64 ± 3.35 vs. PrnpZH3/ZH3 = 59.01 ± 9.76; in all the sessions p > 0.89, mean ± S.E.M.; two-way ANOVA + Bonferroni’s multiple comparisons test) (Additional file 3: Fig. S3b). These results indicate similar motor capacities in both groups, but PrnpZH3/ZH3 mice needed more trials to reach steady measurements for the same task compared to Prnp+/+ mice, suggesting that the knock-out mice have deficits in acquiring instrumental learning as we also observe with the Skinner test (Fig. 2), but not any motor impairment.
Finally, episodic memory was evaluated with the object recognition test (Additional file 4: Fig. S4). In our experiments, most of the PrnpZH3/ZH3 mice interacted with the objects for just a few seconds (Additional file 4: Fig. S4c-d). Therefore, we ruled out this approach due to PrnpZH3/ZH3 mouse inactivity, related to the anxiety-like behavior we observed in the open field test. This inactivity led to reduced interactions with the objects that rendered the learning results unreliable. To further support increased anxiety levels in the object recognition test, the fecal bodies left in the arena after the habituation session were counted by the observer once the test subject was removed. PrnpZH3/ZH3 mice exhibited significant increase in fecal bodies present when compared to wild-type mice (PrnpZH3/ZH3 = 4.76 ± 0.56 vs. Prnp+/+ = 1.29 ± 0.52. mean ± S.E.M., *** p < 0.001, Mann-Whitney U non-parametric test) (Additional file 4: Fig. S4b). These results also correlate with the thigmotaxis levels measured in the PrnpZH3/ZH3 mice and indicate that the knock-out mice showed increased emotionality and anxiety compared to their wild-type counterparts. This was in contrast to what was observed using PrnpZH1/ZH1mice, where the test could be performed. In the habituation session, knock-out mice showed a significant decrease in rearing episodes (Prnp+/+ = 43.21 ± 10.01 vs. PrnpZH1/ZH1 = 15.7 ± 2.8, mean ± S.E.M., ***p < 0.001, Mann-Whitney U non-parametric test) (Additional file 4: Fig. S4e). No changes were observed in the training phase (Additional file 4: Fig. S4f), although the PrnpZH1/ZH1 showed a tendency (p = 0.053) to explore the second object less time ((second − first) / total) compared to the wild-type mice (Prnp+/+ = 0.3 ± 0.15 vs. PrnpZH1/ZH1 = − 0.17 ± 0.15, mean ± S.E.M.) (Additional file 4: Fig. S4g).
Increased paired-pulse facilitation at high intensities in PrnpZH3/ZH3 Schaffer collateral pathway
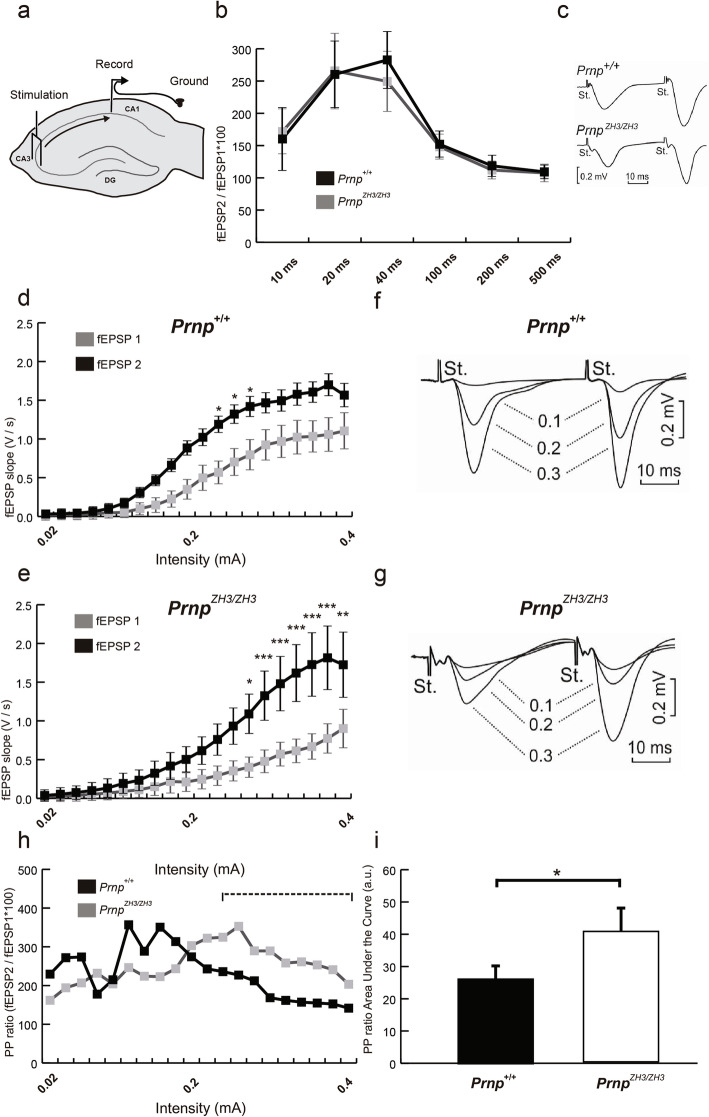
PrPC has been described as a regulator of glutamatergic neurotransmission in the hippocampus [19, 37]. As an example, PrPC has been shown to inhibit NMDAr containing the NR2D subunit [37], or the GluR6/7 receptor [38], see also [57] for review. Therefore, we analyzed the activation of the well-characterized hippocampal Schaffer collateral pathway (CA3-CA1 synapses). Stimulating and recording electrodes were permanently implanted in the CA3 and CA1 regions, allowing us to record and quantify the evoked fEPSPs in living behaving mice (Fig. 3a). First, we evaluated the putative synaptic facilitation or depression evoked at CA3-CA1 synapses by paired-pulse stimulation of the ipsilateral Schaffer collaterals (Fig. 3b). Paired-pulse stimuli were presented to Prnp+/+ (n = 27) and PrnpZH3/ZH3 (n = 24) mice at different inter-stimulus intervals (from 10 to 500 ms). As already reported for CA3-CA1 synapses [58], this approach generates a higher fEPSP from the second stimulus (fEPSP2) than from the first (fEPSP1) at short intervals due to presynaptic facilitation. In our experiments, as can be observed in the representative examples of fEPSP1 and fEPSP2 (Fig. 3c), no differences were observed between Prnp+/+ and PrnpZH3/ZH3 facilitation (Fig. 3b), suggesting that PrPC does not participate in presynaptic mechanisms related to synaptic facilitation at the least at the selected intensities (2 × Threshold; ≈ 0.2 mA).
Fig. 3.
CA3-CA1 synapses in PrnpZH3/ZH3 mice show enhanced excitability. a Schematic representation of electrodes implanted in mouse dorsal hippocampus. Two stimulation electrodes are implanted in the Schaffer collateral pathway in the CA3 region and two recording electrodes in the CA1 stratum radiatum. b Effects of the paired-pulse stimulation of the Schaffer collateral pathway at increasing inter-stimulus intervals (10, 20, 40, 100, 200, 500 ms). Data are presented as the percentage of increase of fEPSP2 in relation to fEPSP1 (fEPSP2/fEPSP1 × 100). c The inset illustrates representative examples of fEPSPs (averaged 5 times) evoked by paired pulses (40 ms of inter-pulse interval) of similar intensities (2 × Threshold; ≈ 0.2 mA) in Prnp+/+ and PrnpZH3/ZH3 mice. d,e Input/output curves of fEPSPs (V/s) in CA1 after the presentation of paired-pulses of increasing intensities in the CA3 area (0.02 mA to 0.4 mA) of Prnp+/+ (d) and PrnpZH3/ZH3 (e) mice. f,g The insets show representative recordings of fEPSPs evoked in Prnp+/+ (f) and PrnpZH3/ZH3 (g) mice by paired pulses (40 ms of inter-pulse interval) of similar intensities (0.1 mA, 0.2 mA, and 0.3 mA). h Paired-pulse ratio (fEPSP2 / fEPSP1 × 100) of data illustrated in d, e, and i area under the curve (a.u.) of PP ratio from 0.24 mA to 0.4 intensities. Data are presented as mean ± S.E.M; p < 0.05, **p < 0.01, and ***p < 0.001, two-way ANOVA + Bonferroni’s multiple comparisons test. Source data and individual data values are available in Additional file 10
Next, we analyzed the consequences of PrPC deficiency in hippocampal synaptic excitability at a large range of stimulus intensities (Fig. 3d–g). The slope of fEPSP facilitation evoked by paired-pulse (40 ms inter-stimulus interval) stimulation was measured at increasing intensities (from 0.02 to 0.4 mA). In Prnp+/+ mice (n = 14), fEPSP1 and fEPSP2 increased steadily more or less in parallel after 0.18 mA stimulation, reaching asymptotic values at 0.32 mA (Fig. 3d,e). fESPS2 was significantly greater in three stimulation intensities before arriving at the asymptotic values showing synaptic facilitation (0.20 mA: fESPS1 = 0.35 ± 0.1 V/s and fEPSP2 = 0.88 ± 0.2 V/s, p = 0.124; 0.22 mA: fESPS1 = 0.50 ± 0.1 V/s and fEPSP2 = 1.02 ± 0.2 V/s, p = 0.144; 0.24 mA: fESPS1 = 0.57 ± 0.1 V/s and fEPSP2 = 1.19 ± 0.2 V/s, *p = 0.030; 0.26 mA: fESPS1 = 0.70 ± 0.1 V/s and fEPSP2 = 1.32 ± 0.2 V/s, *p = 0.031; 0.28 mA: fESPS1 = 0.80 ± 0.1 V/s and fEPSP2 = 1.42, ± 0.2 V/s, *p = 0.028, 0.30 mA: fESPS1 = 0.92 ± 0.1 V/s and fEPSP2 = 1.47, ± 0.2 V/s, p = 0.111; 0.32 mA: fESPS1 = 0.97 ± 0.2 V/s and fEPSP2 = 1.50, ± 0.3 V/s, p = 0.192; 0.34 mA: fESPS1 = 1.02 ± 0.2 V/s and fEPSP2 = 1.58, ± 0.3 V/s, p = 0.172; mean ± S.E.M.; two-way ANOVA + Bonferroni’s multiple comparisons test). From 0.26 mA stimulation, fEPSP1 and fEPSP2 were statistically equal in Prnp+/+ mice; thus, there was no synaptic facilitation at high intensities (Fig. 3d,f). This phenomenon has been described as a putative protective mechanism in high-intensity insults to maintain hippocampal homeostasis [58]. In contrast, in PrnpZH3/ZH3 mice (n = 15), fEPSP1 and fEPSP2 did not increase in parallel, showing an increased facilitation to paired-pulse presentations, and therefore suggesting the absence of this protective mechanism (Fig. 3e,g). fEPSP2 was significantly greater than fEPSP1 at higher intensities (0.24 mA: fESPS1 = 0.293 ± 0.1 V/s and fEPSP2 = 0.76 ± 0.1 V/s, p = 0.36; 0.26 mA: fESPS1 = 0.35 ± 0.1 V/s and fEPSP2 = 0.93 ± 0.2 V/s, p = 0.069; 0.28 mA: fESPS1 = 0.4 ± 0.1 V/s and fEPSP2 = 1.09 ± 0.2 V/s, *p = 0.010; 0.30 mA: fESPS1 = 0.48 ± 0.1 V/s and fEPSP2 = 1.33 ± 0.3 V/s, ***p = 0.0008; 0.32 mA: fESPS1 = 0.57 ± 0.1 V/s and fEPSP2 = 1.5 ± 0.3 V/s, ***p = 0.0002; 0.34 mA: fESPS1 = 0.61 ± 0.1 V/s and fEPSP2 = 1.62 ± 0.3 V/s, ***p < 0.0001; 0.36 mA: fESPS1 = 0.67 ± 0.1 V/s and fEPSP2 = 1.73 ± 0.3 V/s, ***p < 0.0001; 0.38 mA: fESPS1 = 0.77 ± 0.1 V/s and fEPSP2 = 1.81 ± 0.4 V/s, ***p < 0.0001 0.40 mA: fESPS1 = 0.9 ± 0.2 V/s and fEPSP2 = 1.73 ± 0.4 V/s, **p = 0.004; mean ± S.E.M.; two-way ANOVA + Bonferroni’s multiple comparisons test). Prnp+/+ fEPSP1 increased steadily to greater asymptotic values than PrnpZH3/ZH3 fEPSP1 (from 0.3 mA stimulation ≈ 50% increased), but fEPSP2 were almost equal (fEPSP1: Prnp+/+ ≈ 1.0 V/s; PrnpZH3/ZH3 ≈ 0.7 V/s and fEPSP2: Prnp+/+ ≈ 1.5 V/s; PrnpZH3/ZH3 ≈ 1.6 V/s). Consequently, the increase in fEPSP1 related to fEPSP2 was ≈ 50% in Prnp+/+ individuals, but > 140% in their PrnpZH3/ZH3 counterparts.
Exacerbation of synaptic facilitation was clearly observed with the paired-pulse (PP) ratio (fEPSP2/fEPSP1 × 100). At high intensities, the PP ratio was larger in PrnpZH3/ZH3 connections (Fig. 3h). The area under the curve (AUC) from 0.24 mA intensity was significantly lower in the Prnp+/+ (Prnp+/+ = 26.07 ± 4.0 a.u vs. PrnpZH3/ZH3 = 40.75 ± 7.1 a.u., mean ± S.E.M., *p = 0.04; Mann-Whitney U non-parametric test) (Fig. 3i). These results suggest that PrPC regulates neuronal excitability or, perhaps, synaptic homeostasis at high-intensity stimulations, hinting at a neuroprotective role.
High-frequency stimulation evoked epileptic seizures in PrnpZH3/ZH3 Schaffer collaterals but failed to increase LTP
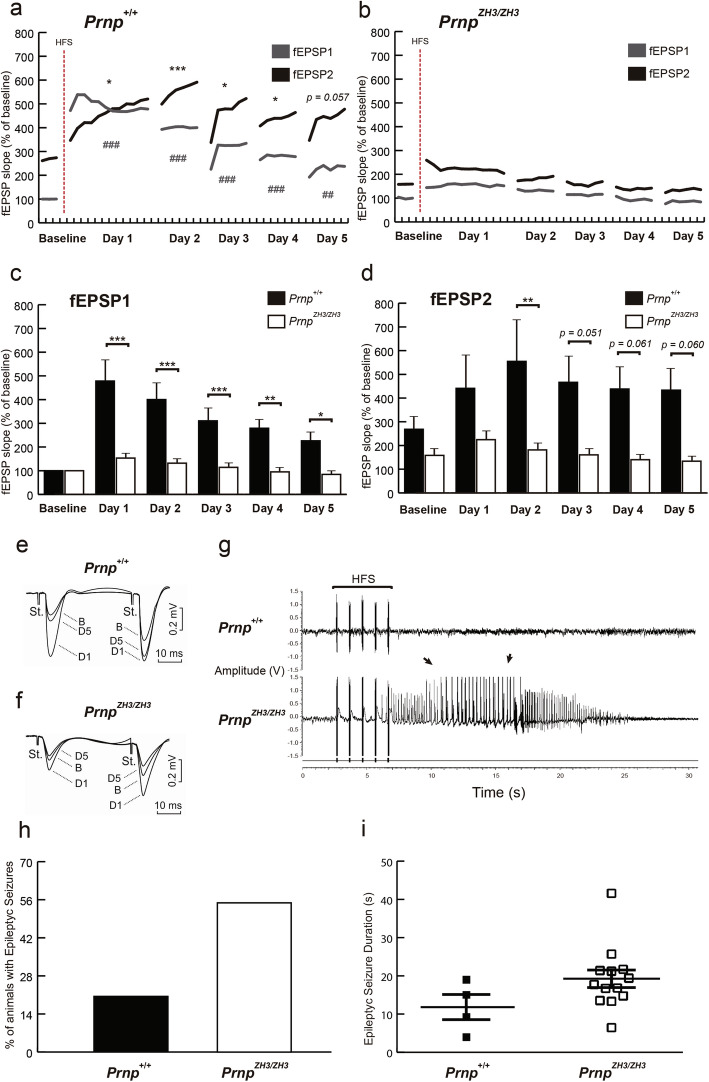
As indicated, several studies reported differing data on LTP in Prnp0/0 mice (see “Background”). So, to analyze LTP in PrnpZH3/ZH3 mice, we performed an LTP induction protocol based on high-frequency stimulation (HFS) in 40 mice (Prnp+/+ = 20 and PrnpZH3/ZH3 = 20) (Fig. 4). First, the baseline fEPSPs were recorded for 15 min, evoked by double pulses at an inter-stimulus interval of 40 ms. Afterward, the HFS protocol was presented. This consisted of five trains (200 Hz, 100 ms) of pulses (1/s) presented six times (1/min). Recordings were maintained for 60 min immediately after the HFS and repeated 30 min daily for 4 days from the HFS presentation session. Prnp+/+ displayed significant LTP for both pulses (Fig. 4a). fESPS1 and fESPS2 recordings were significantly larger than the baseline after the HFS, and this potentiation lasted for the 5 days of recording sessions (Fig. 4e). As expected, HFS reduced paired-pulsed facilitation on the first day [58]. However, from the second day, facilitation recovered steadily but with a range of increase from 350 to 150% with respect to fEPSP1 baseline (baseline: fEPSP1 = 100%; fEPSP2 = 268.2 ± 43.1%; day 1: fEPSP1 = 478.3 ± 78.4%, ***p < 0,001; fEPSP2 = 441.1 ± 140.2%, *p = 0,040; day 2: fEPSP1 = 399.9 ± 65.0%, ***p < 0,001; fEPSP2 = 554.8 ± 164.2%, ***p < 0,001; day 3: fEPSP1 = 310.4 ± 47.0%, ***p < 0,001; fEPSP2 = 466.3 ± 106.7%, *p = 0,012; day 4: fEPSP1 = 279.2 ± 36.0%, ***p < 0,001; fEPSP2 = 438.1 ± 90.7%, *p = 0,046; day 5: fEPSP1 = 226.25 ± 29.4%, **p = 0,0014; fEPSP2 = 433.14 ± 87.7%, p = 0,057; mean ± S.E.M.; two-way ANOVA + Bonferroni’s multiple comparisons test) (Fig. 4a). In contrast, in PrnpZH3/ZH3 mice, LTP induction was virtually absent and paired-pulsed facilitation was maintained (≈ 60%) from the first day (e.g., baseline: EPSP1 = 100%; fEPSP2 = 158.4 ± 23.0%; day 1: fEPSP1 = 153.43 ± 15.7%; fEPSP2 = 224.3 ± 39.8%; day 3: fEPSP1 = 114.2 ± 11.9%; fEPSP2 = 160.4 ± 23.8%; in all the sessions p > 0.6; mean ± S.E.M.; two-way ANOVA + Bonferroni’s multiple comparisons test) (Fig. 4b,f).
Fig. 4.
LTP is not induced at CA3-CA1 synapses of PrnpZH3/ZH3 mice, and the HFS presentation generates epileptic seizures. a,b Evolution of fEPSP1 evoked in the CA1 region by paired-pulsed stimulation of Schaffer collaterals for the Prnp+/+ (a) and PrnpZH3/ZH3 (b) mice after the HFS session. Data are presented as the percentage of increase from baseline. Significant differences with baseline values are presented for fEPSP1 (#) and fEPSP2 (*) recordings in Prnp+/+ mice. c,d fEPSP mean slopes from Prnp+/+ (c) and PrnpZH3/ZH3 (d) mice before and after the HFS session. Data are presented as the percentage of increase from baseline values. e,f The insets show representative recordings (averaged 5 times) of fEPSPs evoked in Prnp+/+ (e) and PrnpZH3/ZH3 (f) mice by paired pulses (40 ms of inter-pulse interval) of similar intensities (2 × Threshold; ≈ 0.2 mA). g Representative examples of long (30 s) recordings carried out after an HFS stimulation protocol in Prnp+/+ and PrnpZH3/ZH3 Schaffer collaterals. Note the presence of a hippocampal seizure in the PrnpZH3/ZH3 mouse (arrows). h Percentage of mice that presented epileptic seizures following HFS presentations. i Seizure duration (s) following HFS. Data are presented as Mean ± S.E.M. *p < 0.05, **p < 0.01, ***p < 0.001, ##p < 0.01 and ###p < 0.001, two-way ANOVA + Bonferroni’s multiple comparisons test and Mann-Whitney U non-parametric test. Source data and individual data values are available in Additional file 10
In addition, Prnp+/+ presented significantly larger fEPSP1 than PrnpZH3/ZH3 mice (day 1: Prnp+/+ = 478.3 ± 78.3% vs. PrnpZH3/ZH3 = 153.4 ± 15.7%, ***p < 0,001; day 2: Prnp+/+ = 399.90 ± 65.0% vs. PrnpZH3/ZH3 = 131.77 ± 12.3%, ***p < 0,001; day 3: Prnp+/+ = 310.42 ± 47.0% vs. PrnpZH3/ZH3 = 114.22 ± 11.9%, ***p = 0.0009; day 4: Prnp+/+ = 279.20 ± 36.05% vs. PrnpZH3/ZH3 = 95.06 ± 9.26%, **p = 0.0021 day 5: Prnp+/+ = 226.3 ± 29.4% vs. PrnpZH3/ZH3 = 84.7 ± 8.0%, *p = 0.034; mean ± S.E.M.; two-way ANOVA + Bonferroni’s multiple comparisons test) (Fig. 4c). Following the same tendency, in PrnpZH3/ZH3 mice, fEPSP2 was also smaller than in Prnp+/+ (baseline: Prnp+/+ = 268.24 ± 43.1% vs. PrnpZH3/ZH3 = 158.4 ± 23.0%, p > 0.99; day 1: Prnp+/+ = 441.1 ± 140.2% vs. PrnpZH3/ZH3 = 224.3 ± 39.8%, p = 0.370; day 2: Prnp+/+ = 554.80 ± 164.2% vs. PrnpZH3/ZH3 = 181.4 ± 25.9%, **p = 0.0083; day 3: Prnp+/+ = 466.3 ± 106.7% vs. PrnpZH3/ZH3 = 160.39 ± 23.8%, p = 0,051; day 4: Prnp+/+ = 438.12 ± 90.7% vs. PrnpZH3/ZH3 = 139.73 ± 19.9%, p = 0,062; day 5: Prnp+/+ = 433.1 ± 87.7% vs. PrnpZH3/ZH3 = 133.53 ± 18.0%, p = 0.06; mean ± S.E.M.; two-way ANOVA + Bonferroni’s multiple comparisons test) (Fig. 4d). These results indicate that LTP increased fEPSPs in Prnp+/+ connections but not in PrnpZH3/ZH3 ones. These results were surprising and were not in accordance with previous publications in which PrnpZH1/ZH1 mice showed even exacerbated LTP [19]. In an attempt to explain these results, we checked the in situ registers in detail during the HFS protocol (Fig. 4g). We observed that 55% of the PrnpZH3/ZH3 mice suffered from epileptic seizures due to HFS in contrast to 20% of the Prnp+/+ mice (Fig. 4h). The PrnpZH3/ZH3 epileptic crises tended to be longer (although not statistically significantly) than those suffered by Prnp+/+ mice (Prnp+/+ = 12.08 ± 3.3 s; PrnpZH3/ZH3 = 19.55 ± 2.5 s, mean ± S.E.M., p = 0.12; Mann-Whitney U non-parametric test) (Fig. 4i). We postulate that this exacerbated excitability in PrnpZH3/ZH3 synapse impaired a proper LTP generation. The HFS may bring about an aberrant synaptic activation (even generating epileptic seizures) that enables activation of the molecular mechanisms needed for LTP induction. Therefore, as published with chemoconvulsants models [28, 31], PrPC might exert protection against electrically induced seizures.
To gain insight into the gene expression patterns altered in the PrnpZH3/ZH3 mice, RNA-seq was performed from the hippocampus region of 8 Prnp+/+ and 8 PrnpZH3/ZH3 animals. The sequencing data have been deposited at the Gene Expression Omnibus (GEO) with accession code: GSE189691. (Matamoros-Angles, A; Hervera, A; Soriano, J; Martí, E; Carulla, P, Llorens, F; Nuvolone, M; Aguzzi, A; Ferrer I; Gruart, A; Delgado-García, JM; Del Río, JA. RNA sequencing of hippocampus of Prnp+/+ and PrnpZH3/ZH3 animals. https://identifiers.org/geo:GSE189691). Around 700 genes showed alterations in their expression profile (323 upregulated and 390 downregulated in PrnpZH3/ZH3 compared to Prnp+/+) (Additional files 5 and 6: Table S1 and Table S2). According to pathway analysis in Reactome v77, the main alterations related to brain functions were the downregulation of genes associated with the “MECP2 regulates neuronal receptors and channels” (10 genes; padj = 1.06 E-07), the “neuronal system” (28 genes; padj. = 0.002), and “protein-protein interaction at synapses” (10 genes; padj. = 8.65 E−04) (Additional file 7: Fig. S5), all in line with the previous behavioral and electrophysiological findings. Among the dysregulated genes, we validated some by RT-qPCR that could explain the phenotype shown by the PrnpZH3/ZH3 mice: including the downregulation of glutamate ionotropic receptor NMDA type subunit 2B (Grin2b), the Gamma-Aminobutyric Acid Type A Receptor Subunit Rho2 (Gabrr2), the Potassium Voltage-Gated Channels: Subfamily J Member 2 (Kcnj2) and 6 (Kcnj6), Subfamily A Member 1 (Kcna1), and Subfamily Q member 3 (Kcnq3) (Additional file 7: Fig. S5).
Enhanced susceptibility to KA-induced seizures in PrnpZH3/ZH3 mice correlates with neuronal death in the hippocampus
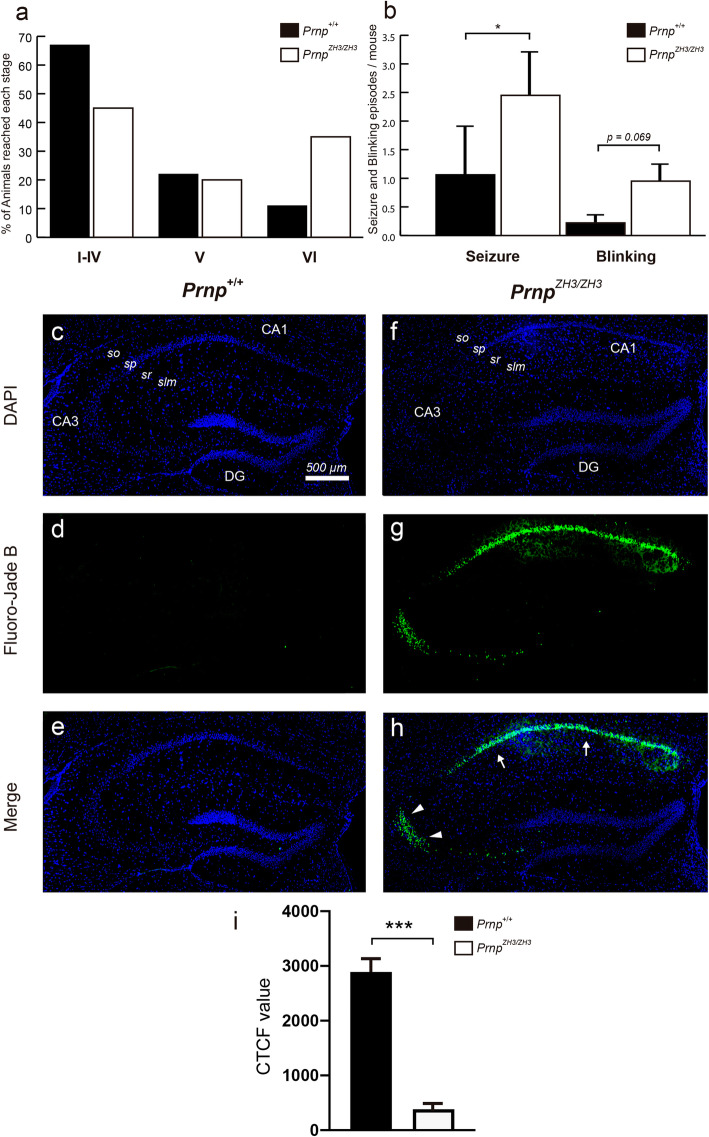
Next, we aimed to explore whether the absence of PrPC in the PrnpZH3/ZH3 mice increased their susceptibility to epileptic seizures following KA (i.p.) injections, as reported in PrnpZH1/ZH1 mice (B6129 and B6.129 backgrounds) [28] (Fig. 5). All experiments were carried out on a blind basis, and two different researchers carried out data evaluation (see “Methods” section). Three consecutive injections of KA (10 mg/kg b.w.) were administrated at intervals of 30 min. The epileptic behavior was monitored for 3 h and was categorized into six stages according to its severity (Fig. 5a). Results indicate that 67% of Prnp+/+ mice did not suffer any severe epileptic episodes (stages I–IV). Only 22% and 11% of wild-type mice reached stages V and VI, respectively. In contrast, 55% of PrnpZH3/ZH3 mice suffered severe epileptic episodes, 20% reaching stage V and 35% at stage VI (Fig. 5a and Additional file 8: Movie. S1). Moreover, PrnpZH3/ZH3 mice presented more seizures and blinking episodes per animal than Prnp+/+ individuals (Seizure: PrnpZH3/ZH3 = 2.45 ± 0.74 vs. Prnp+/+ = 1.06 ± 0.83; p = 0.019; Blinking: PrnpZH3/ZH3 = 0.95 ± 0.29 vs. Prnp+/+ = 0.22 ± 0.13; mean ± S.E.M. p = 0.069; Mann-Whitney U non-parametric test) (Fig. 5b).
Fig. 5.
PrnpZH3/ZH3 mice are more susceptible to KA-induced epilepsy correlating with increased neuronal death in CA1 and CA3 pyramidal layers. a Percentage of mice reaching stage I–IV, V, or VI epileptic phenotype after KA administration (10 mg/kg). b Number of seizures and blinking episodes presented by Prnp+/+ and PrnpZH3/ZH3 mice for 3 h after KA administration. c–h Photomicrographs showing the pattern of neurodegeneration with Fluoro-Jade B staining seven days after KA treatment in Prnp+/+ (c–e) and PrnpZH3/ZH3 (f–h) mouse hippocampus. Nuclei are stained with DAPI (c, f). Dying cells (d, g, stained with Fluoro-Jade B) are located in the pyramidal cell layer of CA1 (arrows) and CA3 (arrowheads) areas. i Graph illustrating the analysis of the CTCF values in the CA1-3 pyramidal layer of PrnpZH3/ZH3 and wild-type mice (see “Methods” for details). Data are presented as a percentage in a and as mean ± S.E.M, in b and i; *p < 0.05 and ***p < 0.001, Mann–Whitney U non-parametric test. Abbreviations: so, stratum oriens; sp, stratum pyramidale; sr, stratum radiatum; slm, stratum lacunosum-moleculare; DG, dentate gyrus. The scale bar in c is also representative for d–h. Source data and individual data values are available in Additional file 10
Additionally, we evaluated neuronal damage after KA-induced epilepsy with Fluoro-Jade B (Fig. 5c–h). PrnpZH3/ZH3 mice showed relevant numbers of labeled cells in the pyramidal layer of the CA1 and CA3 (Fig. 5c–e), while no signal was observed in Prnp+/+ sections (Fig. 5f–h). Indeed, CTCF analysis of Fluoro-Jade B labeling in the pyramidal layer of the CA1 and CA3 (see “Methods” section for details) demonstrated statistical differences between PrnpZH3/ZH3 vs wild-type mice: CTCF value for PrnpZH3/ZH3 = 2893 ± 349.3 vs. wild-type = 380 ± 84.05; mean ± S.E.M., *** p < 0,001; Mann–Whitney U non-parametric test (Fig. 5i). These results corroborated the absence of Prnp to generate an exacerbated synaptic excitability in the hippocampal region that increases susceptibility to electrical and KA-induced seizures, causing neuronal death in the CA1 and CA3 regions of the pyramidal layer of the hippocampus proper.
Neuronal PrnpZH3/ZH3-derived cultures show reduced bursting and impairment network formation in vitro
PrPC has also been described as a regulator of neurogenesis and neuronal differentiation in vitro and in vivo (see [6–8] for reviews). Furthermore, defects in neuronal network connectivity and maturation are related to epilepsy [59]. Consequently, we tested whether PrnpZH3/ZH3 increased excitability might be due to changes in the neuronal differentiation inducing aberrant connectivity and an immature neuronal network. To analyze this, calcium imaging was performed in primary cortical cultures (n = 10 in both genotypes) from Prnp+/+ and PrnpZH3/ZH3 mouse embryos (E16.5–E17.5) expressing the GECI indicator GCaMP6f under the neuronal syntaxin promoter [60], allowing us to record calcium traces of the same neuronal population after 8, 11, 13, and 15 days in vitro (DIV) (Fig. 6).
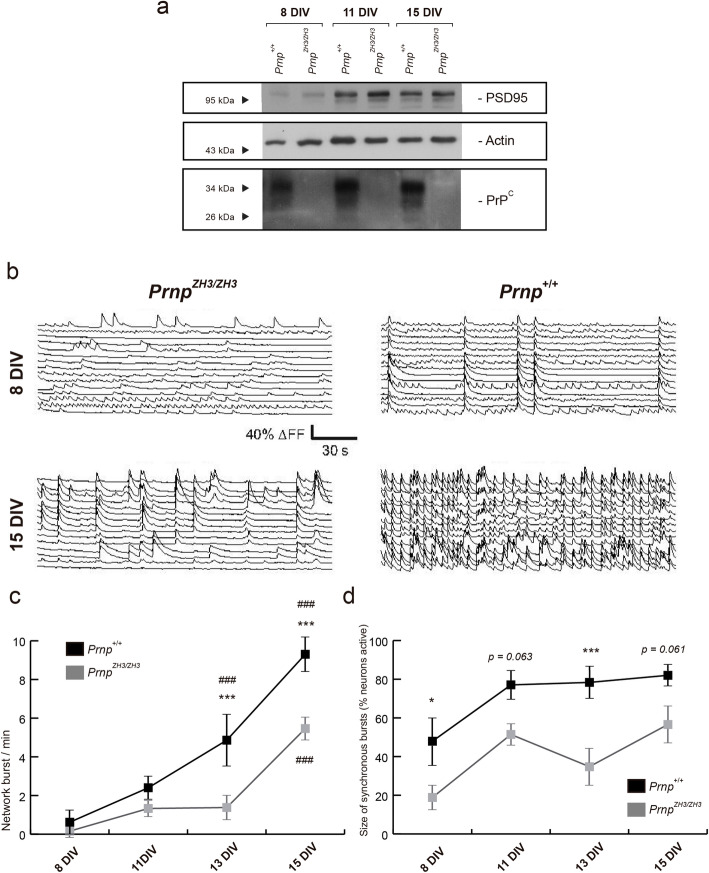
Fig. 6.
Reduced bursting and network formation in neuronal PrnpZH3/ZH3-derived cultures. a Immunoblot analysis of PrPC expression and PSD95 in Prnp+/+ and PrnpZH3/ZH3 derived primary neuronal cultures after 8, 11, and 15 DIV. Note the absence of PrPC in the PrnpZH3/ZH3 cultures and the same PSD95 expression in each DIV. Actin is used as a loading control. b Representative examples of neuronal traces at 8 and 15 DIV in the Prnp+/+ and PrnpZH3/ZH3 primary cultures. Note the asynchrony in the PrnpZH3/ZH3 culture. c Evolution of network bursting in Prnp+/+ and PrnpZH3/ZH3 neuronal cultures from 8 to 15 DIV. Data are presented as the mean of bursts/min ± S.E.M. d Evolution of size of synchronous bursts from 8 to 15 DIV. Data are presented as the mean percentage of active neurons ± S.E.M. Asterisks (*) indicate significant differences between Prnp+/+ and PrnpZH3/ZH3 bursting. Number sign (#) indicates significant differences with the respective baseline bursting at 8 DIV. *p < 0.05, ***p < 0.001 and ###p < 0.001, two-way ANOVA + Bonferroni’s multiple comparisons test. Source data and individual data values are available in Additional file 10
Prnp+/+ and PrnpZH3/ZH3 cultures displayed the same number of collective bursts/min at 8 and 11 DIV (Fig. 6c). After that, a delay in PrnpZH3/ZH3 neuron activity was observed compared to controls. Prnp+/+ cultures increased the number of bursts/min significantly at 13 DIV; however, PrnpZH3/ZH3 cultures needed two additional days, at 15 DIV (Fig. 6b,c). Moreover, Prnp+/+ neurons exhibited significantly more bursts/min at 13 DIV and 15 DIV than PrnpZH3/ZH3 ones and overall the latter showed a reduced firing interval along development (8 Div: Prnp+/+ = 0.62 ± 0.4 vs. PrnpZH3/ZH3 = 0.16 ± 0.1 bursts/min; 13 Div: Prnp+/+ = 4.86 ± 1.1 vs. PrnpZH3/ZH3 = 1.38 ± 0.4; ***p < 0.001; 15 Div: Prnp+/+ = 9.30 ± 0.7 vs. PrnpZH3/ZH3 = 5.46 ± 0.4; mean ± S.E.M. ***p < 0.001; ANOVA + Bonferroni’s multiple comparisons test) (Fig. 6c). The PrnpZH3/ZH3 cultures also showed a reduction in the size of synchronous bursts. In the Prnp+/+ cultures, around 80% of the neurons showed synchronic activity at 11, 13, and 15 DIV, while this value was around 50% in PrnpZH3/ZH3 cultures (Fig. 6d). These results demonstrate that collective bursting is reduced and delayed in PrnpZH3/ZH3 cultures, suggesting that Prnp expression is necessary for network formation and maturation.
Discussion
PrPC has been associated with several physiological functions using in vivo approaches; however, the consequences of PrPC deletion in behavior and cognition have not been extensively evaluated [9]. There are some studies about Prnp0/0 mouse behavior, motor capabilities, and learning performance, but the results are not conclusive, especially after the description of the so-called FG in the background of the PrnpZH1/ZH1 model that masks specific PrPC roles [26]. For example, concerning KA susceptibility, a clear decrease is observed in mice expressing a lower percentage of 129/Sv-associated polymorphisms (B6.129) compared to the B6129 original strain of the PrnpZH1/ZH1 mice with a higher percentage of 129/Sv genome [28]. Here we assess the consequences of the absence of PrPC in behavior and neurotransmission using the new strictly co-isogenic mouse model PrnpZH3/ZH3 [50]. However, another relevant aspect of these studies is the age of the analyzed mice, since physiological differences in the absence of Prnp have been described in association with age for PrnpZH1/ZH1 [45] or FVB/N-PrnpZH1/ZH1 [61]. Thus, in our study, we only used and compared results obtained from mice of 3–5 months of age. Concerning nest-building behavior, our results suggest similar behavior in PrnpZH3/ZH3 and wild-type mice. This contrasts with previously reported results [42], but it has been largely demonstrated that this capacity is dependent on mouse background [62, 63].
Our results also reveal that PrnpZH3/ZH3 mice display reduced activity in the open field test. The reduced rearing exploration and peripheral preference in the arena, and the high defecation rate observed in PrnpZH3/ZH3 mice, suggest an anxiety-like behavior. Indeed, PrnpZH3/ZH3 showed higher thigmotaxis than wild-type mice (see [64] for technical details). Our data correlate with those reported by Schmitz et al. [45] and Lobao-Soares et al., both using 3-month-old PrnpZH1/ZH1 mice, illustrating reduced mobility between the inner and outer regions of the open field and increased defecation in PrnpZH1/ZH1 mice [65]. However, they are in contrast to Nico et al. [66], where they found no differences, and with Gadotti et al. [52], that showed 10-week-old PrnpZH1/ZH1 mice displayed increased crossing in open-field test although thigmotaxis changes were not analyzed in the study. Although technical details could also play a role (i.e., handling of the mice, the initial position of the mice in the field, geometry of the field), we think that these discrepancies reinforce the relevance of the homogenous genetic background in our study vs. the others. In fact, the differences we observed also extend to the rotarod test. Our results showed no motor alternations in the PrnpZH3/ZH3 mice, but they need more sessions to learn the task compared to controls. Previous publications also showed any motor alterations in two different knock-out strains [45, 65, 67]. However, Nazor et al. observed changes just in mice older than 95 days [61].
In our experiments, PrnpZH3/ZH3 mice also failed to achieve instrumental learning in the Skinner box. These results are similar to those observed in PrnpZH1/ZH1 mice (B6.129 background, 2–5% 129/Sv markers, [28]). Striking differences in motility between wild-type and knock-out mice were identified using this approach. However, learning capacity based on the ON/OFF paradigm confirmed the operant conditioning deficiencies, a type of associative learning.
Following our results, alterations in locomotor activity and increased latency to initiate exploration were previously reported in other Prnp0/0 mouse models [42, 47]. Anxiety-related behavior [54], depressive tendencies [52], and alterations in spatial memory and learning have also been described [42, 45]. In contrast, Bueler et al. reported no alterations in PrnpZH1/ZH1 behavior [24]. This disparity in results might be explained by the age of the animals used in the studies. Bueler and collaborators performed the test with 2- and 3-month-old mice, which could potentially uncover the behavior impairment as it was reported to be an age-dependent decline in other publications using the PrnpZH1/ZH1 model [45, 68]. Another study also showed no deficits in Prnp0/0 mouse behavior, where Prnp was conditionally deleted at 12 or 16 months, and the results included no alterations in the Morris water maze or object recognition test [53]. We reported behavioral deficits in Prnp0/0 mouse models with some discrepancies with the previous works, most likely related to age-related sampling and the mice’s background.
Glutamate neurotransmission is, in large part, responsible for cortical signaling, and its impairment has been related to behavioral deficits [69]. PrPC has been described as a regulator of glutamate synapses [13]. Even glutamate inhibition with an NMDA antagonist (MK-801) ameliorates depressive-like behavior in Prnp0/0 mice [52]. Thus, our next step was to study glutamate connectivity to better understand behavioral alterations. The hippocampal Schaffer collaterals and their implication in operant conditioning, spatial learning, and anxiety-related behavior were evaluated as a well-defined model of glutamate circuitry [70].
The paired-pulse facilitation test did not show differences between PrnpZH3/ZH3 and Prnp+/+ animals. Therefore, PrPC deletion did not alter synaptic facilitation, at least in the living behaving mouse model, at the least at relatively low stimulation intensities (Fig. 3b). These results may be explained by the fact that synaptic facilitation is mainly a presynaptic phenomenon [71], and PrPC has been related to postsynaptic neurotransmission mechanisms [37, 38]. These differences in pre- and post-synaptic mechanisms would explain the different results collected from PrnpZH3/ZH3 mice and their littermate controls in paired-pulse ratio and LTP tests.
Nevertheless, PrnpZH3/ZH3 mice displayed increased synaptic facilitation at high intensities, a fact not observed in controls. This could represent a sort of compensatory phenomenon for their evident LTP deficits. In addition, PrnpZH3/ZH3 mice presented an increased susceptibility to KA-induced seizures. This epileptogenic phenotype may explain our results on anxiety behavior in PrnpZH3/ZH3 mice. Comorbid anxiety disorders affect patients with epilepsy [72], and cognitive decline has been described in epileptic animal models [73]. Moreover, increased excitability was previously reported, especially in susceptibility to KA, NMDA, and pentylenetetrazol (PTZ) insults [30, 31].
However, contradictory results were published by other groups, who described an elevated epileptic threshold in Prnp0/0 hippocampal slices treated with bicuculline, zero-magnesium conditions, and PTZ [74], and also normal neurotransmission-associated parameters compared with wild-type mice [25]. Both studies recorded hippocampal slices of the FVB/N-Prnp0/0 model, a mouse with a triple mixture background (FVB/129Sv/C57BL6) which carried the FG [26]. Different susceptibility to KA-induced seizures among mice with different backgrounds has been described; even the FVB background has been shown as highly susceptible to epilepsy [75, 76]. Our results using PrnpZH3/ZH3 animals in KA susceptibility were similar to those previously obtained with the other available Prnp0/0 co-isogenic mouse model, the Edinburgh 129/Ola [28]. Therefore, we postulate that these contradictory results published about the excitability of Prnp0/0 synapse are likely associated with the mouse backgrounds, the FG effect, and the experimental approach. However, the ZH3 mice results demonstrate that PrPC indeed protects against KA-induced epilepsy.
Our results show that the presentation of HFS protocols causes epileptic seizures in most PrnpZH3/ZH3 mice but fails to generate significant LTP at the hippocampus. Some controversial results have been published about the implication of PrPC in LTP generation. Different experimental approaches (hippocampal slices or in vivo experiments) and a mixture of mouse models with distinct backgrounds were used, generating non-comparable data (i.e., [19, 25, 40, 42]). Here, using ZH3 mice, we hypothesize that the absence of PrPC ends in LTP induction failure due to exacerbated synaptic excitability, although we cannot rule out putative GABAergic disinhibition. It is convincingly demonstrated that severe epileptic seizures cause neuronal death, which hampers LTP generation. Moreover, non-severe epileptic seizures generate similar molecular and synaptic changes to LTP [77, 78]. This suggests that non-severe HFS-induced seizures somehow saturate the postsynaptic terminal, over-activating LTP-induction mechanisms that reduce LTP production capacity by HFS. Additionally, PrPC has also been described as interacting with key elements required for LTP-related mechanisms AMPA or NMDA receptors [37, 39, 79, 80], see also [57] for review.
The presented data show that Prnp0/0 hippocampal synapse is highly excitable and epileptogenic. Alterations in brain connectivity due to developmental alterations, traumas, or infections contribute to this neuromodulation imbalance [59]. In order to assess whether the epileptic phenotype displayed by PrnpZH3/ZH3 animals came from neuronal connectivity alterations, we studied bursting and network formation in PrnpZH3/ZH3-derived primary neuronal cultures. Relevant PrPC expression in vitro is observed from 4 to 5 DIV [81]. Our results indicate that PrnpZH3/ZH3 cultures did not mature or connect properly; they displayed asynchronous and very low bursting compared to wild-type cultures. These results suggest that the absence of PrPC causes a delay in neuronal maturation, but more relevantly in neural network formation and function. In fact, the role of PrPC-mediated signals in neuritogenesis has been demonstrated [82–84]. However, to our knowledge, this is the first description of network alterations due to the absence of PrPC. However, and as indicated by Benvegnu and coworkers [85], there are gene expression changes during the development of FVB/N Prnp0/0 and wild-type hippocampus. Hence, changes in ion conductance or channel receptor expression might be involved in this delay. In this line, electrophysiological experiments with biochemical characterization might confirm the basis of the delay in maturation in vitro.
Finally, the gene ontology analysis of the RNAseq of PrnpZH3/ZH3 and Prnp+/+ mouse hippocampus showed downregulation of genes associated with the neuronal system and protein-protein interaction at synapses, fitting the phenotype we observed in the knock-out mice. The significant downregulation of Gabrr2 and Grin2b in PrnpZH3/ZH3 mice would produce dysregulation in the excitatory/inhibitory balance, increasing the excitability of the system, as we describe with the KA and the HFS analyses. The alteration of the inhibitory neurotransmission was already shown in Prnp0/0 models since they were susceptible to suffering from epileptic crises (see references above). PrPC has been widely described as a regulator of glutamatergic neurotransmission and its receptors, as we show here with the Grin2b. Moreover, mutations at Grin2b have recently been related to a rare brain disease, the GRIN2B-related neurodevelopmental disorder that causes intellectual disability, autism-spectrum-like behavior, epilepsy, and, sometimes, locomotor deficiencies as well [86, 87]. Therefore, the altered expression of Grin2b might contribute to the behavior and learning deficits observed in the PrnpZH3/ZH3 mice.
Conclusions
In conclusion, our study points that the absence of PrPC impairs neuronal network formation and connectivity, producing enhanced susceptibility to excitotoxicity insults such as HFS and KA exposure. This epileptogenic circuitry seems to impair highly cognitive-demanding functions such as associative learning, and it produces anxiety-like behavior.
Methods
Animals
Adult C57BL/6 J mice (Prnp+/+) were purchased from Charles River Laboratories (Paris, France). PrnpZH3/ZH3 mice line was provided by A. Aguzzi (Switzerland) (see [50] for details). PrnpZH1/ZH1 mice [24] were purchased from the European Mouse Mutant Archive (EMMA, Monterotondo, Italy). A total of 185 adult (3–5 months old) male mice (ZH3: Prnp+/+ = 81 and PrnpZH3/ZH3 = 84; ZH1: Prnp+/+ = 10 and PrnpZH1/ZH1 = 10) were used in the present study. In ZH1 mouse experiments, null PrnpZH1/ZH1 and control mice (Prnp+/+) were obtained by crossing heterozygous Prnp+/ZH1 mice to obtain a mixed background (B6.129). It is well described that behavior and neural physiology are different between male and female rodent models due to several hormone and non-hormone-derived reasons [88]. Thus, we used only males in order to establish an equivalent group comparable with previous publications. All experiments were performed following the protocols and guidelines of the Ethical Committee for Animal Experimentation (CEEA) of the University of Barcelona. CEEA of the University of Barcelona approved the protocol for using animals in this study (CEEA approval #276/16 and 141/15). Behavioral and electrophysiological studies were performed following the guidelines of the European Union Council (2010/276:33-79/EU) and current Spanish regulations (BOE 34:11370-421, 2013) for the use of laboratory animals in chronic experiments. Experiments were also approved by the Ethics Committee for Animal Care and Handling of the Pablo de Olavide University (UPO-JA 06/03/2018/025).
Immunoblotting
Proteins from brain tissue lysates or primary cortical neurons were extracted using RIPA buffer with protease and phosphatase inhibitor cocktails (Roche). Total lysates were obtained by 30 s of centrifugation at 4 °C. The protein concentration of the lysates was quantified using Pierce BCA Protein Assay Kit (Thermo Scientific). Then, 10–50 μg of proteins were loaded to SDS-PAGE gels and transferred and transferred to nitrocellulose membranes for 1 h. Membranes were blocked with Tris-buffered solution with 0.1% tween, 5% skimmed milk, and 2% of FBS for 1 h at room temperature (RT) and incubated with PSD95 (1:1.000, MAB1598; Millipore), PrP (1:500; 6H4; Thermo) or Actin (1:20.000; MAB1501; Millipore) antibodies at 4 °C O/N. Following HRP-linked secondary antibody (Dako) incubation for 1 h at RT, membranes were developed with ECL substrate (Thermo).
Behavioral studies
A total of 147 animals were used in these sets of experiments (ZH3: Prnp+/+ = 63 and PrnpZH3/ZH3 = 64; ZH1: Prnp+/+ = 10 and PrnpZH1/ ZH1 = 10). Mice were housed alone in boxes on a 12/12 h light/dark cycle with constant ambient temperature (21 ± 1 °C). Water and food were provided ad libitum except for the instrumental learning tests (see below).
Nest building
For this test, a total of 14 mice (3 months old) were used (PrnpZH3/ZH3 = 7 and Prnp+/+ = 7). On the first day of testing, one piece of tissue paper (36 × 12 cm) was placed in the cage to facilitate nest building (Additional file 1: Fig. S1). The presence and the quality of each nest were photo-documented and evaluated the following day according to a modified 5-point scale using the method described by Deacon [89]. Two different blinded researchers evaluated the nest generated by each mouse. Data are presented as the mean ± S.E.M. in (Additional file 1: Fig. S1). The statistical analysis was performed with the Mann-Whitney non-parametric test (GraphPad Prism 8 software).
Open field test
In this test, PrnpZH1/ZH1 mice were not used since detailed studies were already developed using this model [45, 54]. In our experiments, mice (Prnp+/+ = 49 and PrnpZH3/ZH3 = 49) were placed in a square open field altimeter box (35 × 35 × 25 cm, Cibertec, Madrid, Spain). The field had a grid (16 × 16 cm) of infrared lasers on the XY-axis and one on the Z-axis. Locomotor activity was measured for 15 min in mice with the MUX-XYZ16L software. Mice were placed in the box’s periphery for 15 min for two consecutive days, and their behavior was recorded. The first day was considered a training session to reduce mouse anxiety associated with manual handling, and the data analyzed and displayed in the manuscript corresponded to the second session. The system inferred mouse activity by counting laser intersections. For anxiety-related behavior measurement, the center (inner) square of the field (10 × 10 cm) was considered as the central zone and the rest of the square as the peripheral (outer) zone [56] (see Fig. 1a). For quantification and to distinguish motility from exploratory behavior, it was considered that a mouse spent time in one of the regions (center vs. periphery) if it remained in the region at least 3 s. Rearing episodes were considered when the animal stood up for at least 3 s, and immobility episodes if immobile for an additional 3 s. Obtained data were analyzed, and the sum of the crossed X- and the Y-axes are presented together to show total mouse mobility in the experiments. The time spent in the maze periphery zones measures thigmotaxis or wall-hugging behavior and indicates anxiety-related behavior [56]. Data are presented as the mean ± S.E.M. The statistical analysis was performed with a T-test or Mann-Whitney U non-parametric test (GraphPad Prism 8 software). The asterisks indicate significant differences: **p < 0.01 and ***p < 0.001. The arena and the walls were cleaned with soap and ethanol between trials to remove olfactory cues between experiments.
Operant conditioning tests
The instrumental learning tests were performed as described in previous studies of our group [90]. Six Skinner boxes were used simultaneously (12.5 × 13.5 × 18.5 cm; MED Associates, St. Albans, VT, USA). Each Skinner box was housed in a sound-attenuating cubicle (90 × 55 × 60 cm) constantly exposed to white noise (± 45 dB) and dim light (Cibertec, S.A, Madrid, Spain). The boxes had a trough to receive food pellets (Noyes formula P; 45 mg; Sandown Scientific, Hampton, UK) by pressing a lever. Before the test, mouse food availability was monitored for 7 days to reduce initial mouse weight to 85%. First, mice (ZH3: Prnp+/+ = 49 and PrnpZH3/ZH3 = 49; and ZH1: Prnp+/+ = 10 and PrnpZH1/ZH1 = 10) were trained to press the lever to receive food pellets in a fixed-ratio (1:1) schedule. Seven daily sessions (20 min/each) were held. The boxes were cleaned with soap and ethanol (30%) between trials. Obtaining ≥ 20 pellets for two consecutive sessions was defined as the criterion to assume the learning criteria achievement. Following this first operant conditioning test, we increased the paradigm complexity to test the mice in a more demanding cognitive task for an additional 10 days. Only animals that met the learning criterion were tested (ZH3: Prnp+/+ = 24 and PrnpZH3/ZH3 = 20; and ZH1: Prnp+/+ = 8 and PrnpZH1/ZH1 = 5). The paradigm consisted of light (ON period) and dark periods (OFF period) randomly distributed during the session. The light was provided by a small light bulb located over the lever. During the ON period (20 s), lever presses were reinforced with food pellets at a ratio of 1:1. During the OFF period, lever presses were not rewarded and were penalized by adding ten additional seconds (20 ± 10 s) to the next ON period. The number of lever presses during the different conditioning paradigms was monitored and recorded with the MED-PC program (MED Associates, St. Albans, VT, USA). Statistical analysis was carried out using two-way ANOVA with repeated measures and Bonferroni’s multiple comparisons test (GraphPad Prism 8 software). Asterisks indicate significant differences: **p < 0.01; and ***p < 0.001. Data are presented as the mean ± S.E.M. or as a percentage (as indicated in each figure).
Rotarod test
For this test, a total of 15 mice of 4 months were used (PrnpZH3/ZH3 = 8 and Prnp+/+ = 7). Motor performance was tested using an accelerating rotarod. Mice were pre-trained to the task to reach a minimum of 30 s performance at 5 rpms on the 1st day of testing. In each training run, animals were placed on the rods at an initial speed of 5 rpm for 30 s. After that, the testing consisted of 5 consecutive trials with 15-min inter-trial intervals. Each trial consisted of 30 s at 5 rpm followed by 5 rpm increases every 15 s with a cut-off of 5 min. Results are expressed as the mean latency of animals to fall from the rod ± S.E.M. The statistical analysis was performed with the two-way ANOVA + Bonferroni’s multiple comparisons test (GraphPad Prism 8 software).
Object recognition test
The object recognition test was performed in a homemade arena (30 × 25 × 20 cm), as described [91]. A total of 23 Prnp+/+ and 24 PrnpZH3/ZH3 mice were analyzed. Additionally, 6 PrnpZH1/ZH1 mice were also used with 7 Prnp+/+ counterparts. The test consisted of four phases of 10 min/each. First, animals were habituated to the field without any object (habituation session). One hour later, two identical plastic objects were placed in the center of the arena for the training session. A short-term memory test was performed 2–3 h later by changing one of the objects (see Additional File 4: Fig. S4). ZH1 mouse mobility was expressed as the number of rearing episodes during the habituation session. The arena and the objects were cleaned with soap and 30% ethanol between trials to remove olfactory cues. Mouse behavior was recorded with a video camera placed over the arena, and these recordings were used to measure the exploratory behavior blindly. Sniffing and gently touching the objects were counted as exploratory behavior. To further support increased anxiety levels in the PrnpZH3/ZH3 mice, fecal bodies left in the maze during the habituation session were counted by the observer once the test subject was removed since it has been demonstrated that highly emotional animals exhibit increased defecation [56]. Statistical analysis was performed with the Mann-Whitney U non-parametric test (GraphPad Prism 8 software). Data are presented as the mean ± S.E.M. or as a percentage (indicated in each figure).
Mouse surgery
A total of 98 adult male (3-5 months) mice were implanted with stimulating and recording electrodes (Prnp+/+ = 49 and PrnpZH3/ZH3 = 49). Four of them died during surgery, and 33 mice were excluded because of the inability to obtain reliable and clean recordings. Thus, the experiments were performed with 61 mice (Prnp+/+ = 31 and PrnpZH3/ZH3 = 30). Surgery was performed as described in [19, 92]. Mice were deeply anesthetized with ketamine (35 mg/kg) and xylazine (2 mg/kg), and electrodes were aimed at the right dorsal hippocampus. Two recording electrodes were implanted in the stratum radiatum of the CA1 area (2.2 mm caudal to Bregma, 1.2 mm lateral, and 1.3 mm ventral), and two stimulating electrodes were implanted in the Schaffer collateral pathway of the CA3 region (1.5 mm posterior to Bregma, 2 mm lateral, and 1.3 mm ventral). Electrodes were made of 50 μm Teflon-coated tungsten wires (Advent Research, Eynsham, UK). Electrode localizations were checked according to the field excitatory postsynaptic potential (fEPSP) profile evoked by a single stimulation. A silver wire was fixed to the skull as ground. All the wires were soldered to a six-pin socket (RS Amidata, Madrid, Spain) fixed to the skull with dental cement. Recordings were started at a minimum of 1 week after the surgery.
Electrophysiology recordings
Animals were consecutively recorded in groups of six individuals since they reach the total number of animals used in each experiment. Each animal was placed in a small plastic cubicle (5 × 5 × 10 cm) inside a large Faraday box (30 × 30 × 20 cm). fEPSPs were recorded with a high impedance probe (2 × 1012 Ω, 10 pF) using differential amplification at a bandwidth of 0.1 Hz–10 kHz (P511, Grass-Telefactor, West Warwick, RI, USA). For each experiment, artefactual recordings were discarded. The stimulation intensity threshold of each animal was set with paired-pulse stimulations at 40 ms of inter-stimulus interval. The stimulus intensity was set to 40–60% of the amount necessary to evoke a suturing response. These intensity values were used for all the experiments.
Paired-pulse stimulation
For synaptic facilitation experiments, 51 mice (Prnp+/+ = 27 and PrnpZH3/ZH3 = 24) were stimulated at Schaffer collaterals with a pair of pulses at different inter-stimulus intervals (10, 20, 40, 100, 200, and 500 ms) at 2 × threshold intensities (≈ 0.2 mA). Threshold values were previously defined for each mouse. As classically defined, threshold values were determined as the intensity evoking fEPSP responses in 50% of the cases. For all the inter-pulse intervals, the stimulations were repeated ten times. Data are represented as the mean percentage increases of fEPSP2 from fEPSP1 recordings (fEPSP2 / fEPSP1 × 100) ± S.E.M.
Input/output curves
Schaffer collaterals of 29 mice (Prnp+/+ = 14 and PrnpZH3/ZH3 = 15) were stimulated with paired pulses at 20 increasing intensities (from 0.02 to 0.4 mA, increased in steps of 0.02 mA) at 40 ms of inter-stimulus interval. For all the selected intensities, the stimulations were repeated ten times. Data are represented as the mean of fEPSP slopes (V/s) ± S.E.M. The same data are presented as the mean of paired-pulsed ratio (PP ratio) ± S.E.M. PP ratio is the percentage of the increase of the fEPSP2 from fEPSP1 recordings (fEPSP2 / fEPSP1 × 100). The area under the curve (AUC) was calculated from the PP ratio of all the animals using GraphPad Prism 8 software. Statistical analysis was performed using the Mann-Whitney-Wilcoxon non-parametric test or two-way ANOVA + Bonferroni’s multiple comparisons test (GraphPad Prism 8 software). The asterisks indicate significant differences: *p < 0.05; **p < 0.01; and ***p < 0.001 in the figure.
Long-term potentiation experiments
For long-term potentiation experiments, 40 mice (Prnp+/+ = 20 and PrnpZH3/ZH3 = 20) were stimulated at Schaffer collaterals. In a first experimental step, fEPSP baseline values were evoked and recorded for 15 min, with paired-pulse stimulus presented every 20 s (40 ms inter-stimulus). Stimulus intensities were selected to evoke fEPSPs of about 0.2–0.3 mV of amplitude (see insets in Fig. 4a,b). Next, LTP was evoked with a high-frequency stimulation (HFS) protocol. HFS consisted of five trains of pulses at a rate of 1/s (200 Hz, 100 ms) with the same intensity as the baseline recording. The HFS was repeated six times at intervals of 1 min. After the HFS protocol, fEPSPs were recorded, as for baseline, for 1 h. The following 4 days, the recordings were repeated for 30 min. fEPSPs and 1 V rectangular pulses corresponding to stimulus presentations were saved on a PC using an analog/digital converter (CED 1401 Plus, Cambridge, England). Data were analyzed offline using Spike2 and Signal 5.04 software with homemade representation programs [58]. Collected recordings were represented and analysed offline with the help of commercial (Spike 2 and Signal 5.04) programs following procedures described elsewhere. The slope of collected fEPSPs was computed as its first derivative (volts/s). Five successive fEPSs were averaged and the mean value of the slope was determined. Data are presented as the mean of the percentage compared to the baseline ± S.E.M. The statistical analysis was performed using two-way ANOVA + Bonferroni’s multiple comparisons test (GraphPad Prism 8 software). The asterisks and symbols indicate significant differences: *p < 0.05; **p < 0.01; and ***p < 0.001; ##p < 0.01; and ###p < 0.001.
KA-induced epilepsy and seizure analysis
Adult (3–4 months old) male mice were used for these sets of experiments (Prnp+/+ = 18 and PrnpZH3/ZH3 = 20) essentially as described in [28]. A KA (Sigma-Aldrich, Darmstadt, Germany) solution was freshly prepared for each experiment in 0.1 M phosphate buffer. Mice were injected with KA (10 mg/kg b.w.) three times: at 0 min, 30 min, and 60 min. After the first injection, mice were placed in clean boxes (1–3 mice/box). The presence of epileptic seizures was monitored in situ and recorded with a video camera for 3 h after drug administration. Seizure severity was scored in grades following the following criteria: grade I-II: hypoactivity and immobility; grade III-IV: hyperactivity and scratching; grade V: loss of balance control and intermittent convulsions; grade VI: continuous seizures and bouncing activity (also reported as blinking episodes or “pop-corn” behavior). Data are presented as the mean ± S.E.M. or as a percentage (as indicated in each figure). Statistical analysis was performed with the Mann-Whitney U non-parametric test (GraphPad Prism 8 software). The asterisk indicates significant differences: *p < 0.05 in the figure.
RNAseq
Hippocampi were extracted, flash frozen on dry ice, and RNA was harvested using RNAEasy Mini kit (Qiagen). Libraries were prepared using the TruSeq Stranded mRNA Sample Prep Kit v2 according to the manufacturer’s protocol. Briefly, 500 ng of total RNA was used for poly(A)-mRNA selection using Oligo (dT) magnetic beads and subsequently fragmented to approximately 300 bp. cDNA was synthesized using reverse transcriptase (SuperScript II, Invitrogen) and random primers. The second strand of the cDNA incorporated dUTP in place of dTTP. Double-stranded DNA was further used for library preparation. dsDNA was subjected to A-tailing and ligation of the barcoded Truseq adapters. All purification steps were performed using AMPure XP Beads. Library amplification was performed with PCR using the primer cocktail supplied in the kit. Final libraries were analyzed using Agilent DNA 1000 chip to estimate the quantity, check the size distribution, and then quantified by qPCR using the KAPA Library Quantification Kit (KapaBiosystems, Merck, Darmstadt, Germany) before amplification Illumina’s cBot. Libraries were sequenced 1 × 50 bp on Illumina’s HiSeq 2500.
The quality of the fast files was checked using the FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). An estimation of ribosomal RNA in the raw data was obtained with riboPicker [93]. Reads were aligned with the STAR mapper [94] to release M14 of the Mus musculus Gencode version of the genome (GRMm38/mm10 assembly) (https://www.gencodegenes.org/mouse/release_M14.html). A raw count of reads per gene was also obtained with STAR (-quantMode TranscriptomeSAM GeneCounts option). The R/Bioconductor package DESeq2 [95, 96] was used to assess differential expression between experimental groups (Wald statistical test + false discovery rate correction). Prior to processing the differential expression analysis, genes for which the sum of raw counts across all samples was less than two were discarded. Deregulated genes with a padj < 0.05 were used to disclose relevant pathway alterations in the REACTOME v77 pathway database gene expression. The analysis has been done just with the protein-coding genes. The gene difference was considered biologically relevant if they are upregulated or downregulated with a fold change of > 1.2 or < 0.85, respectively. A pathway was considered relevant if it was related to neuronal and/or cerebral functions, showed significance (padj < 0.05) and contained more than 10 deregulated genes. The sequencing data have been deposited at the Gene Expression Omnibus (GEO) with accession code: GSE189691 (Matamoros-Angles, A; Hervera, A; Soriano, J; Martí, E; Carulla, P, Llorens, F; Nuvolone, M; Aguzzi, A; Ferrer I; Gruart, A; Delgado-García, JM; Del Río, JA. RNA sequencing of hippocampus of Prnp+/+ and PrnpZH3/ZH3 animals. https://identifiers.org/geo:GSE189691).
RT-qPCR
For RT-qPCR validations, cDNA was obtained with a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) following the supplier’s instructions. RT-qPCR reactions contained 4.5 μL cDNA and mixed with 0.5 μL 20X TaqMan Gene Expression Assays and 5 μL of 2X TaqMan Universal PCR Master Mix (Applied Biosystems) for a final volume of 10 μL. TaqMan probes used were as follows: Grin2b Mm00433820_m1, Gabrr2 Mm00433507_m1, Kacnj6 Mm01215650_m1, Kcna1 Mm00439977_s1, Kcnj2 Mm00434616_m1, Kcnq3 Mm00548884_m1 (Applied Biosystems). Actb Mm02619580_g1 and Aars Mm00507627_m1 were used as endogenous controls. The assay was performed using technical duplicates per sample in 384-well optical plates with ABI Prism 7900 Sequence Detection system (Applied Biosystems, Life Technologies) following the supplier’s parameters: 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The Sequence Detection Software (SDS version 2.2.2, Applied Biosystems) was used for data processing. It was further analyzed with the ∆∆Ct method, which consists of obtaining ∆Ct by normalizing each target gene to its endogenous control, followed by subtracting the mean-∆Ct of the control group samples to each ∆Ct to obtain ∆∆Ct values, and finally using these ∆∆Ct values as the negative exponent with base 2, thereby obtaining fold change per sample.
Fluoro-Jade B staining
Mice were perfused 7 days after the KA administration with 4% paraformaldehyde (PFA) dissolved in 0.1 M phosphate buffer (pH = 7.3–7.4). Brains were dissected and postfixed overnight with the same fixative solution. The following day, they were cryoprotected in 0.1 M phosphate buffer containing 30% sucrose (w/v). After freezing in dry ice, 50-μm-thick coronal sections were obtained with a freezing microtome (Leica, Wetzlar, Germany). Sections containing the dorsal hippocampus were selected and rinsed in 0.1 M phosphate buffer and mounted on gelatin-coated slides. The slides containing sections were dried at 37 °C overnight. The following day, they were heated at 50 °C for 45 min before staining to improve adhesion. The staining started with pretreatment for 3 min in absolute alcohol, followed by 1 min in 70% ethanol and 1 min in deionized water. After that, they were oxidized in a solution of 0.06% KMnO4 for 15 min. Following three rinses (2 min/each) in deionized water, they were incubated in a solution of 0.001% Fluoro-Jade B (Chemicon, Temecula, CA, USA) containing 0.05% of DAPI in 0.1% acetic acid for 30 min. Finally, sections were rinsed in deionized water (3 min), dehydrated with ethanol, cleared with xylene, and coverslipped with EukittTM (Merck, Darmstadt, Germany). Considering that wild-type mice did not displayed Fluoro-Jade B labeled cells in the CA1-CA3 regions after KA treatments and only a very few background could be seen at high magnification and long exposures times (> 500 ms), the Fluoro-Jade B fluorescence in the pyramidal layer of the dorsal hippocampal region (4 sections of each mouse, n = 4 mice per genotype) was photo-documented using an Olympus BX61 epifluorescence microscope equipped with a cooled DP12L camera (Hamburg, Germany). Photomicrographs were obtained using a × 40 objective with identical time exposure between preparations from each wild-type and respective knock-out mouse. No modifications were applied to the obtained pictures. Changes in Fluoro-Jade B labeling were determined by analyzing the corrected total cell fluorescence (CTCF) values (see Matamoros-Angles et al. [97], for details) in the pyramidal layer of hippocampal CA1-3 regions of four mice of each genotype, taking a region of interest of 200 × 100 μm centered in the pyramidal layer, and 4–5 different sections per animals were analyzed and averaged. Data were expressed as mean ± S.E.M. The statistical analysis of the obtained data was performed using Mann-Whitney U non-parametric test in GraphPad Prism 8 software. A value of ***p < 0.001 was considered statistically significant in the CTCF analysis.
Primary cortical cultures of PrnpZH3/ZH3 and wild-type mice
Primary cortical cultures were fashioned from E16.5–E17.5 Prnp+/+ and PrnpZH3/ZH3 mouse embryos, as explained elsewhere [98]. Brains were removed from the skull and rinsed in cold Hank’s balanced salt solution (HBSS) containing glucose (6.5 mg/ml). The meninges were removed, and the cortical lobes were isolated. Tissue pieces were treated with trypsin for 15 min at 37 °C. After the addition of horse serum followed by centrifugation, cells were isolated mechanically with a polished glass pipette after treatment with 0.025% DNAse for 10 min at 37 °C. One million cells were plated on a 35-mm diameter glass-bottom gridded culture dish (Ibidi, Martinsried, Germany) previously coated with poly-d-lysine (Sigma-Aldrich). NeurobasalTM medium supplemented with 6.5 mg/ml glucose, 2 mM glutamine, penicillin/streptomycin, 5% of horse serum, and B27 was used as a culture medium (all from Invitrogen-Thermo Fisher Scientific, MA, USA). As Prnp0/0-derived cells are sensitive to serum removal [99], after 24 h, the serum was reduced to 2.5%. The medium was changed every 2 days. Horse serum was entirely removed on the eighth day of culture.
Calcium imaging in neuronal culture
Primary cortical neurons were infected 24 h after seeding with AAV9-Synapsin-GCaMP6f [60] (Watertown, MA, USA). In our cultures, the genetically encoded calcium indicators started to express 3–4 days after infection. Calcium changes in GCaMP6f-expressing neurons were recorded at 8, 11, 13, and 15 days in vitro (DIV) using an Olympus IX71 inverted microscope (Olympus, Hamburg, Germany), equipped with an ORCA-Flash 4.0 camera (Hamamatsu Photonics, Japan). During recording, the cells were maintained in a microscope stage incubator at 5% CO2 and 37 °C (Okolab S.R.L., Italy). The same region of the culture was recorded throughout the days following the culture dish grid references. Images (1024 × 1024 pixels) were captured using a × 20 objective and 470 nm wavelength (CoolLED’s pE-300white, Delta Optics, Madrid, Spain) every 100 ms for 8–10 min using the CellSensTM software (Olympus) or the Micro-Manager Open Source Microscopy Software (https://micro-manager.org). Exposure levels and frequency were maintained between cultures and evaluation days. GCaMP6f activity was measured in four different identified squares of each culture dish during these 4 days.
Neuronal activity traces, spike events, and network bursts
The recordings were analyzed offline using two MATLABTM toolboxes: NETCAL (www.itsnetcal.com) [100, 101] and NeuroCa [102]. In NeuroCa, an automatic analysis was performed afterward to corroborate obtained NETCAL results. Using NETCAL, a highly contrasted image of the recording’s average fluorescence was created, and regions of interest (ROIs) were automatically detected as those objects with a circular shape whose brightness was over a preset threshold. NETCAL and NeuroCa software-rendered a similar number of ROIs and calcium traces. About 400 ROIs, uniformly covering the field of view, were typically identified per recording. The average fluorescence Fi (t) in each ROI i along the recording was then extracted, corrected from global drifts and artifacts, and finally normalized as (Fi (t) − F(0,i)) / F(0,i) = fi (t), where F0,i is the background fluorescence of the ROI. The time series of fi (t) was analyzed with NETCAL to infer neuronal activation timing using the Schmitt trigger method. Our analysis used + 2 S.E.M. of the baseline noise as the high threshold, + 1.5 S.E.M. as the low threshold, and 200 ms as the minimum event length. Calcium traces were calculated, and raster plots of network activity were then constructed by representing the trains of detected neuronal spikes over time. Next, network bursts were analyzed to quantify the ability of the neuronal networks to exhibit collective dynamics, i.e., the collective activation of a group of neurons in a short time window. Bursts were investigated using two approaches. In the first approach, raster plots of spike events were scanned to detect collective occurrences in which at least 5% of the neurons in the network fired synchronously within a 500-ms window. This threshold of 5% was set to disregard random activations that coincided in time. In the second approach, the fluorescence time series of all neurons in the network were averaged. The resulting trace was analyzed with the Schmitt trigger method to detect sufficiently strong fluorescence peaks associated with bursting episodes. Both approaches produced consistent results. Although the detected bursts contained a different number of participating neurons, this information was disregarded in the present analysis and treated later. The total number of detected network bursts divided by the recording duration reflected the culture’s activity and was indicated as bursts/min.
The fraction of active neurons in the network was calculated as follows. All detected ROIs were assigned as neurons. After inferring the spike trains, those neurons exhibiting at least two spikes along the recording were considered active, and their number NA was set as a proxy of the healthy population in the neuronal network. The average fraction of active neurons in each condition was then determined as f = NA/NT, where NT is the total number of detected ROIs.
At least ten videos of each genotype/day from different culture plates were consecutively analyzed. Data are presented as the mean of network burst/min ± S.E.M. Statistical analysis was performed using two-way ANOVA + Bonferroni’s multiple comparisons test (GraphPad Prism 8 software). Asterisks indicate significant differences between Prnp+/+ and PrnpZH3/ZH3 cultures at a given DIV: *p < 0.05 and ***p < 0.001. The # symbols indicate significant differences between a given DIV with the initial value at 8 DIV: ###p < 0.001.
Statistical analysis
All statistical analysis was performed with GraphPad PRISM 8 (GraphPad Software, USA). Unless otherwise stated, data is plotted as the mean ± SEM. All experiments were performed three times unless specified. Normality of the distributions was checked via Shapiro-Wilk test. All tests performed were two-sided, and adjustments for multiple comparisons and/or significantly different variances (Fisher’s F) applied were indicated. All data analysis was performed blind to the experimental group by two independent experimenters. Unless otherwise stated, sample size was chosen in order to ensure a power of at least 0.8, with a type I error threshold of 0.05, in view of the minimum effect size that was expected.
Supplementary Information
Additional file 1: Figure S1. PrnpZH3/ZH3 mice showed similar nest-building behavior to wild-type mice. a, Representative images of nests constructed by Prnp+/+ (left) and PrnpZH3/ZH3 mice (right). b, Quantification illustrating the mean of the nest score (see Material and Methods for details). Data are presented as mean ± S.E.M. Source data and individual data values are available in Additional file 10.
Additional file 2: Figure S2. PrnpZH1/ZH1 mice failed to acquire an instrumental learning test in the Skinner box test using a fixed ratio (1:1) schedule. a, Percentage of Prnp+/+ and PrnpZH1/ZH1 mice reached the selected criterion during the training sessions. b, Task accuracy ((lever presses during light ON – lever presses during the light OFF) / Total number of lever presses) during the light ON / light OFF conditioning paradigm. Data are presented as a percentage in a and as mean ± S.E.M. in b. Source data and individual data values are available in Additional file 10.
Additional file 3: Figure S3. PrnpZH3/ZH3 mice have deficits in acquiring instrumental learning but not motor impairment in the Rotarod test. After a training session in the Rotarod, mice were tested two consecutive days with 5 sessions in each. Latency of mice to fall (sec.) from the Rotarod in the first day (a) or in the second (b). Data are presented as mean ± S.E.M.. *p < 0.05 and **p < 0.01, two-way ANOVA + Bonferroni’s multiple comparisons test. Source data and individual data values are available in Additional file 10.
Additional file 4: Figure S4. Stressed-like behavior in PrnpZH3/ZH3 mice impairing object recognition test performance. PrnpZH1/ZH1 mice also showed stressed-like behavior and failed to acquire short-term memory. a, Scheme of object recognition protocol that consisted of 3 sessions (10 min each). First, mice were habituated in the empty arena. One hour later, they were placed again in the arena with two identical objects for the training session. Finally, 2-3 h later, they were once more placed in the arena, replacing one object with a novel one for the short-term memory test. Fecal bodies (PrnpZH3/ZH3) and rearing episodes (PrnpZH1/ZH1) were counted in the habituation session as an indicator of animal stress. b, Number of fecal bodies generated per animal (Prnp+/+ and PrnpZH3/ZH3) during the habituation session. Data are presented as mean ± S.E.M. c-d Time that Prnp+/+ (c), and PrnpZH3/ZH3 (d) mice interacted with the objects in the short-term test. Data are presented as the percentage of mice interacting with the object in each time interval (0 to 35 s). e, Number of rearing episodes in the habituation session (Prnp+/+ and PrnpZH1/ZH1). f-g, Relative time that the two exposed objects are explored to the total time invested. Object 1 (O1) and object 1’ (O1’) in the training session (f) and number, and object 1 (O1) and object 2 (O2) in the short-term memory test (g). Data are presented as percentages in C-D and as mean ± S.E.M. in b, e, f, g. *p < 0.05 and ***p < 0.001, Mann-Whitney U non-parametric test. Source data and individual data values are available in Additional file 10.
Additional file 5: Table S1. List of the protein-coding significantly downregulated genes in PrnpZH3/ZH3 hippocampus compared to Prnp+/+.
Additional file 6: Table S2. List of the protein-coding significantly upregulated genes in PrnpZH3/ZH3 hippocampus compared to Prnp+/+.
Additional file 7: Figure S5. Gene ontology of the genes significantly downregulated and upregulated in PrnpZH3/ZH3 hippocampus compared to Prnp+/+. a, Volcano plot of the protein-coding genes analyzed in the RNAseq (~ 16.000). In the X-axis is plotted the Fold change (log2) and in the Y-axis the padj (-log10). The red line separates the significantly expressed genes (padj < 0,05). The blue lines indicate the 0,85 and 1,2 fold changes. b, Gene ontology analysis of the downregulated and upregulated genes with the Reactome software (see Materials and Methods for details). Validation of the main genes altered by RT-qPCR: Grin2b (c), Gabrr2 (d), Kcnj6 (e), Kcna1 (f), Kcnj2 (g), and Kcnq3 (h). Data are presented as mean ± S.E.M. *p < 0.05 and ***p < 0.001. Source data and individual data values are available in Additional file 10.
Additional file 8: Movie S1. Representative movie of Prnp+/+ (bottom and up-left) and PrnpZH3/ZH3 (middle and up-right) mouse behavior after KA (middle and bottom) and PBS (top) administration. Note the blinking and seizure episodes suffered by the PrnpZH3/ZH3 mice treated with KA (labeled with asterisks at the beginning of the video). Source data and individual data values are available in Additional file 10.
Additional file 9: Figure S6. Images of the original, uncropped blots for Figs. 1A and 6A.
Additional file 10. Excel file 1. File providing individual data values for all figures from the manuscript.
Acknowledgements
The authors thank Tom Yohannan for editorial advice and Miriam Segura-Feliu, María Sánchez-Enciso, and José M. González-Martín for their technical help.
Abbreviations
- AMPA
Alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- ANOVA
Analysis of variance
- AUC
Area under the ROC curve
- CA3-CA1
Cornnu ammonis 3 and 1 of the hippocampus.
- CEEA
Ethical Committee for Animal Experimentation
- CTCF
Corrected total cell fluorescence
- fEPSP
Field excitatory postsynaptic potential
- FG
Flanking genes
- GECI
Genetically encoded calcium indicator
- GLUK2
Glutamate ionotropic receptor kainate type subunit 2
- GPI
Glycosylphosphatidylinositol
- HFS
High-frequency stimulation
- KA
Kainate
- LTP
Long-term potentiation
- mGluR5
Metabotropic glutamate receptor 5
- MK-801
NMDA antagonist
- NMDA-R
N-methyl-d-aspartate receptor
- PFA
Paraformaldehyde
- Prnp
Cellular prion protein gene
- PrPC
Cellular prion protein
- PSD-95
Post synaptic density protein of 95 kD
- PTZ
Pentylenetetrazol
- ROI
Region of interest
- sCJD
Sporadic Creutzfeldt-Jakob disease
- SEM
Standard deviation of the mean
- TALEN
Transcription activator-like effector nuclease
Authors’ contributions
A M-A, A.H., J.S., P.C., E.M., and F.L. performed most of the experiments. A M-A. and A.H. participated in all the experiments, performing and designing, and contributed equally to this project. P.C performed the behavior experiments. J.S. analyzed and interpreted the calcium imaging data. E.M. and F.L. planned and organized the RNAseq and performed the gene ontology analysis. A.G. and J.M D-G developed the electrophysiological experiments. J.A.D.R., A.G., J.M. D.-G., A.A., M.N., and I.F designed and supervised the study. J.M. D.-G., and J.A.D.R analyzed the results. J.A.D.R. and J.M. D.-G. wrote the draft and circulated it among the authors. The authors read and approved the final manuscript.
Funding
This research was supported by PRPSEM Project with ref. RTI2018-099773-B-I00 from MCINN/AEI/10.13039/501100011033/ FEDER “Una manera de hacer Europa”, the CERCA Programme, and the Commission for Universities and Research of the Department of Innovation, Universities, and Enterprise of the Generalitat de Catalunya (SGR2017-648), CIBERNED (CMED2018-2) to JADR and IF. The project leading to these results received funding from the “la Caixa” Foundation (ID 100010434) under the agreement LCF/PR/HR19/52160007 and the María de Maeztu Unit of Excellence (Institute of Neurosciences, University of Barcelona) MDM-2017-0729 to JADR. JS was supported by FIS2016-78507-C2-2-P from (MCIU/FEDER/AEI), SGR2017-1061 from the Generalitat de Catalunya, and the European Union’s Horizon 2020 research and innovation program under the grant agreement No. 713140 (MESOBRAIN). Support was also received from MINECO (BFU2017-82375-R), and Junta de Andalucía (BIO-122, UPO-1250734, and P18-FR-823) grants to AG and JMDG. F.LL. was supported by Instituto Carlos III (grant PI19-00144). A.M-A. was supported by the Tatiana Pérez de Guzmán el Bueno Foundation.
Availability of data and materials
All data generated or analysed during this study are included in this published article and additional information files. RNAseq data have been deposited at the Gene Expression Omnibus (GEO) with accession code: GSE189691. (Matamoros-Angles, A; Hervera, A; Soriano, J; Martí, E; Carulla, P, Llorens, F; Nuvolone, M; Aguzzi, A; Ferrer I; Gruart, A; Delgado-García, JM; Del Río, JA. RNA sequencing of hippocampus of Prnp+/+ and PrnpZH3/ZH3 animals. https://identifiers.org/geo:GSE189691). Source data for all figures have been provided in Additional file 10. Uncropped blots from Figs. 1 and 6 are available in Additional file 9.
Declarations
Ethics approval and consent to participate
All experiments were performed following the protocols and guidelines of the Ethical Committee for Animal Experimentation (CEEA) of the University of Barcelona. CEEA of the University of Barcelona approved the protocol for using animals in this study (CEEA approval #276/16 and 141/15). Behavioral and electrophysiological studies were performed following the guidelines of the European Union Council (2010/276:33-79/EU) and current Spanish regulations (BOE 34:11370-421, 2013) for the use of laboratory animals in chronic experiments. Experiments were also approved by the Ethics Committee for Animal Care and Handling of the Pablo de Olavide University (UPO-JA 06/03/2018/025).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A. Matamoros-Angles and A. Hervera contributed equally to this work.
Contributor Information
J. M. Delgado-García, Email: jmdelgar@upo.es
J. A. Del Río, Email: jadelrio@ibecbarcelona.eu
References
- 1.Barmada S, Piccardo P, Yamaguchi K, Ghetti B, Harris DA. GFP-tagged prion protein is correctly localized and functionally active in the brains of transgenic mice. Neurobiol Dis. 2004;16(3):527–537. doi: 10.1016/j.nbd.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Moser M, Colello RJ, Pott U, Oesch B. Developmental expression of the prion protein gene in glial cells. Neuron. 1995;14(3):509–517. doi: 10.1016/0896-6273(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 3.Adle-Biassette H, Verney C, Peoc'h K, Dauge MC, Razavi F, Choudat L, Gressens P, Budka H, Henin D. Immunohistochemical expression of prion protein (PrPC) in the human forebrain during development. J Neuropathol Exp Neurol. 2006;65(7):698–706. doi: 10.1097/01.jnen.0000228137.10531.72. [DOI] [PubMed] [Google Scholar]
- 4.Tremblay P, Bouzamondo-Bernstein E, Heinrich C, Prusiner SB, DeArmond SJ. Developmental expression of PrP in the post-implantation embryo. Brain Res. 2007;1139:60–67. doi: 10.1016/j.brainres.2006.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailly Y, Haeberle AM, Blanquet-Grossard F, Chasserot-Golaz S, Grant N, Schulze T, Bombarde G, Grassi J, Cesbron JY, Lemaire-Vieille C. Prion protein (PrPc) immunocytochemistry and expression of the green fluorescent protein reporter gene under control of the bovine PrP gene promoter in the mouse brain. J Comp Neurol. 2004;473(2):244–269. doi: 10.1002/cne.20117. [DOI] [PubMed] [Google Scholar]
- 6.Prusiner SB, DeArmond SJ. Prion diseases and neurodegeneration. Ann Rev Neurosci. 1994;17(1):311–339. doi: 10.1146/annurev.ne.17.030194.001523. [DOI] [PubMed] [Google Scholar]
- 7.Aguzzi A. Molecular pathology of prion diseases. Vox Sang. 2000;78(Suppl 2):25. [PubMed] [Google Scholar]
- 8.Linden R, Martins VR, Prado MA, Cammarota M, Izquierdo I, Brentani RR. Physiology of the prion protein. Physiol Rev. 2008;88(2):673–728. doi: 10.1152/physrev.00007.2007. [DOI] [PubMed] [Google Scholar]
- 9.Wulf MA, Senatore A, Aguzzi A. The biological function of the cellular prion protein: an update. BMC Biol. 2017;15(1):34. doi: 10.1186/s12915-017-0375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Legname G. Elucidating the function of the prion protein. PLoS Pathog. 2017;13(8):e1006458. doi: 10.1371/journal.ppat.1006458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.del Rio JA, Gavin R. Functions of the cellular prion protein, the end of Moore's law, and Ockham's razor theory. Prion. 2016;10(1):25–40. doi: 10.1080/19336896.2015.1126038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steele AD, Lindquist S, Aguzzi A. The prion protein knockout mouse: a phenotype under challenge. Prion. 2007;1(2):83–93. doi: 10.4161/pri.1.2.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watts JC, Bourkas MEC, Arshad H. The function of the cellular prion protein in health and disease. Acta Neuropathol. 2018;135(2):159–178. doi: 10.1007/s00401-017-1790-y. [DOI] [PubMed] [Google Scholar]
- 14.Roucou X, Gains M, LeBlanc AC. Neuroprotective functions of prion protein. J Neurosci Res. 2004;75(2):153–161. doi: 10.1002/jnr.10864. [DOI] [PubMed] [Google Scholar]
- 15.Gavin R, Lidon L, Ferrer I, Del Rio JA. The quest for cellular prion protein functions in the aged and neurodegenerating brain. Cells. 2020;9(3). 10.3390/cells9030591. [DOI] [PMC free article] [PubMed]
- 16.Chiarini LB, Freitas AR, Zanata SM, Brentani RR, Martins VR, Linden R. Cellular prion protein transduces neuroprotective signals. EMBO J. 2002;21(13):3317–3326. doi: 10.1093/emboj/cdf324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paitel E, Sunyach C, Alves da Costa C, Bourdon JC, Vincent B, Checler F. Primary cultured neurons devoid of cellular prion display lower responsiveness to staurosporine through the control of p53 at both transcriptional and post-transcriptional levels. J Biol Chem. 2004;279(1):612–618. doi: 10.1074/jbc.M310453200. [DOI] [PubMed] [Google Scholar]
- 18.Paitel E, Fahraeus R, Checler F. Cellular prion protein sensitizes neurons to apoptotic stimuli through Mdm2-regulated and p53-dependent caspase 3-like activation. J Biol Chem. 2003;278(12):10061–10066. doi: 10.1074/jbc.M211580200. [DOI] [PubMed] [Google Scholar]
- 19.Rangel A, Madronal N, Gruart A, Gavin R, Llorens F, Sumoy L, Torres JM, Delgado-Garcia JM, Del Rio JA. Regulation of GABA(A) and glutamate receptor expression, synaptic facilitation and long-term potentiation in the hippocampus of prion mutant mice. PloS one. 2009;4(10):e7592. doi: 10.1371/journal.pone.0007592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goetzl EJ, Peltz CB, Mustapic M, Kapogiannis D, Yaffe K. Neuron-derived plasma exosome proteins after remote traumatic brain injury. J Neurotrauma. 2020;37(2):382–388. doi: 10.1089/neu.2019.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leng B, Sun H, Zhao J, Liu Y, Shen T, Liu W, Liu X, Tan M, Li F, Zhang J, Li Z. Plasma exosomal prion protein levels are correlated with cognitive decline in PD patients. Neurosci Lett. 2020;723:134866. doi: 10.1016/j.neulet.2020.134866. [DOI] [PubMed] [Google Scholar]
- 22.Llorens F, Ansoleaga B, Garcia-Esparcia P, Zafar S, Grau-Rivera O, Lopez-Gonzalez I, Blanco R, Carmona M, Yague J, Nos C, et al. PrP mRNA and protein expression in brain and PrP(c) in CSF in Creutzfeldt-Jakob disease MM1 and VV2. Prion. 2013;7(5):383–393. doi: 10.4161/pri.26416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scalabrino G, Veber D, De Giuseppe R, Roncaroli F. Low levels of cobalamin, epidermal growth factor, and normal prions in multiple sclerosis spinal cord. Neuroscience. 2015;298:293–301. doi: 10.1016/j.neuroscience.2015.04.020. [DOI] [PubMed] [Google Scholar]
- 24.Büeler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, DeArmond SJ, Prusiner SB, Aguet M, Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356(6370):577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 25.Lledo PM, Tremblay P, DeArmond SJ, Prusiner SB, Nicoll RA. Mice deficient for prion protein exhibit normal neuronal excitability and synaptic transmission in the hippocampus. Proc Natl Acad Sci U S A. 1996;93(6):2403–2407. doi: 10.1073/pnas.93.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nuvolone M, Kana V, Hutter G, Sakata D, Mortin-Toth SM, Russo G, Danska JS, Aguzzi A. SIRPalpha polymorphisms, but not the prion protein, control phagocytosis of apoptotic cells. J Exper Med. 2013;210(12):2539–2552. doi: 10.1084/jem.20131274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Almeida CJ, Chiarini LB, da Silva JP, ES PM, Martins MA, Linden R. The cellular prion protein modulates phagocytosis and inflammatory response. J Leukoc Biol. 2005;77(2):238–246. doi: 10.1189/jlb.1103531. [DOI] [PubMed] [Google Scholar]
- 28.Carulla P, Llorens F, Matamoros-Angles A, Aguilar-Calvo P, Espinosa JC, Gavin R, Ferrer I, Legname G, Torres JM, del Rio JA. Involvement of PrP(C) in kainate-induced excitotoxicity in several mouse strains. Sci Rep. 2015;5(1):11971. doi: 10.1038/srep11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sparkes RS, Simon M, Cohn VH, Fournier RE, Lem J, Klisak I, Heinzmann C, Blatt C, Lucero M, Mohandas T, et al. Assignment of the human and mouse prion protein genes to homologous chromosomes. Proc Natl Acad Sci U S A. 1986;83(19):7358–7362. doi: 10.1073/pnas.83.19.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rangel A, Burgaya F, Gavin R, Soriano E, Aguzzi A, Del Rio JA. Enhanced susceptibility of Prnp-deficient mice to kainate-induced seizures, neuronal apoptosis, and death: role of AMPA/kainate receptors. J Neurosci Res. 2007;85(12):2741–2755. doi: 10.1002/jnr.21215. [DOI] [PubMed] [Google Scholar]
- 31.Walz R, Amaral OB, Rockenbach IC, Roesler R, Izquierdo I, Cavalheiro EA, Martins VR, Brentani RR. Increased sensitivity to seizures in mice lacking cellular prion protein. Epilepsia. 1999;40(12):1679–1682. doi: 10.1111/j.1528-1157.1999.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 32.Striebel JF, Race B, Pathmajeyan M, Rangel A, Chesebro B. Lack of influence of prion protein gene expression on kainate-induced seizures in mice: studies using congenic, coisogenic and transgenic strains. Neuroscience. 2013;238:11–18. doi: 10.1016/j.neuroscience.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown EJ, Frazier WA. Integrin-associated protein (CD47) and its ligands. Trends Cell Biol. 2001;11(3):130–135. doi: 10.1016/S0962-8924(00)01906-1. [DOI] [PubMed] [Google Scholar]
- 34.Nuvolone M, Paolucci M, Sorce S, Kana V, Moos R, Matozaki T, Aguzzi A. Prion pathogenesis is unaltered in the absence of SIRPalpha-mediated "don't-eat-me" signaling. PloS One. 2017;12(5):e0177876. doi: 10.1371/journal.pone.0177876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adao-Novaes J, Valverde RHF, Landemberger MC, Silveira MS, Simoes-Pires EN, Lowe J, Linden R. Substrain-related dependence of Cu(I)-ATPase activity among prion protein-null mice. Brain Res. 2020;1727:146550. doi: 10.1016/j.brainres.2019.146550. [DOI] [PubMed] [Google Scholar]
- 36.Encalada SE, Szpankowski L, Xia CH, Goldstein LS. Stable kinesin and dynein assemblies drive the axonal transport of mammalian prion protein vesicles. Cell. 2011;144(4):551–565. doi: 10.1016/j.cell.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khosravani H, Zhang Y, Tsutsui S, Hameed S, Altier C, Hamid J, Chen L, Villemaire M, Ali Z, Jirik FR, Zamponi GW. Prion protein attenuates excitotoxicity by inhibiting NMDA receptors. J Cell Biol. 2008;181(3):551–565. doi: 10.1083/jcb.200711002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carulla P, Bribián A, Rangel A, Gavín R, Ferrer I, Caelles C, Del Río JA, Llorens F. Neuroprotective role of PrPC against kainate-induced epileptic seizures and cell death depends on the modulation of JNK3 activation by GluR6/7-PSD-95 binding. Mol Biol Cell. 2011;22(17):3041–3054. doi: 10.1091/mbc.e11-04-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferreira DG, Temido-Ferreira M, Vicente Miranda H, Batalha VL, Coelho JE, Szego EM, Marques-Morgado I, Vaz SH, Rhee JS, Schmitz M, et al. Alpha-synuclein interacts with PrP(C) to induce cognitive impairment through mGluR5 and NMDAR2B. Nat Neurosci. 2017;20(11):1569–1579. doi: 10.1038/nn.4648. [DOI] [PubMed] [Google Scholar]
- 40.Collinge J, Whittington MA, Sidle KC, Smith CJ, Palmer MS, Clarke AR, Jefferys JG. Prion protein is necessary for normal synaptic function. Nature. 1994;370(6487):295–297. doi: 10.1038/370295a0. [DOI] [PubMed] [Google Scholar]
- 41.Manson JC, Hope J, Clarke AR, Johnston A, Black C, MacLeod N. PrP gene dosage and long term potentiation. Neurodegeneration. 1995;4(1):113–114. doi: 10.1006/neur.1995.0014. [DOI] [PubMed] [Google Scholar]
- 42.Criado JR, Sanchez-Alavez M, Conti B, Giacchino JL, Wills DN, Henriksen SJ, Race R, Manson JC, Chesebro B, Oldstone MB. Mice devoid of prion protein have cognitive deficits that are rescued by reconstitution of PrP in neurons. Neurobiol Dis. 2005;19(1-2):255–265. doi: 10.1016/j.nbd.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Curtis J, Errington M, Bliss T, Voss K, MacLeod N. Age-dependent loss of PTP and LTP in the hippocampus of PrP-null mice. Neurobiol Dis. 2003;13(1):55–62. doi: 10.1016/S0969-9961(03)00017-2. [DOI] [PubMed] [Google Scholar]
- 44.Maglio LE, Martins VR, Izquierdo I, Ramirez OA. Role of cellular prion protein on LTP expression in aged mice. Brain Res. 2006;1097(1):11–18. doi: 10.1016/j.brainres.2006.04.056. [DOI] [PubMed] [Google Scholar]
- 45.Schmitz M, Greis C, Ottis P, Silva CJ, Schulz-Schaeffer WJ, Wrede A, Koppe K, Onisko B, Requena JR, Govindarajan N, Korth C, Fischer A, Zerr I. Loss of prion protein leads to age-dependent behavioral abnormalities and changes in cytoskeletal protein expression. Mol Neurobiol. 2014;50(3):923–936. doi: 10.1007/s12035-014-8655-3. [DOI] [PubMed] [Google Scholar]
- 46.Lipp HP, Stagliar-Bozicevic M, Fischer M, Wolfer DP. A 2-year longitudinal study of swimming navigation in mice devoid of the prion protein: no evidence for neurological anomalies or spatial learning impairments. Behav Brain Res. 1998;95(1):47–54. doi: 10.1016/S0166-4328(97)00209-X. [DOI] [PubMed] [Google Scholar]
- 47.Coitinho AS, Roesler R, Martins VR, Brentani RR, Izquierdo I. Cellular prion protein ablation impairs behavior as a function of age. Neuroreport. 2003;14(10):1375–1379. doi: 10.1097/00001756-200307180-00019. [DOI] [PubMed] [Google Scholar]
- 48.Fainstein N, Dori D, Frid K, Fritz AT, Shapiro I, Gabizon R, Ben-Hur T. Chronic progressive neurodegeneration in a transgenic mouse model of prion disease. Front Neurosci. 2016;10:510. doi: 10.3389/fnins.2016.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manson JC, Clarke AR, Hooper ML, Aitchison L, McConnell I, Hope J. 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol Neurobiol. 1994;8(2-3):121–127. doi: 10.1007/BF02780662. [DOI] [PubMed] [Google Scholar]
- 50.Nuvolone M, Hermann M, Sorce S, Russo G, Tiberi C, Schwarz P, Minikel E, Sanoudou D, Pelczar P, Aguzzi A. Strictly co-isogenic C57BL/6 J-Prnp-/- mice: A rigorous resource for prion science. J Exper Med. 2016;213(3):313–327. doi: 10.1084/jem.20151610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nuvolone M, Sorce S, Paolucci M, Aguzzi A. Extended characterization of the novel co-isogenic C57BL/6 J Prnp(-/-) mouse line. Amyloid. 2017;24(sup1):36–37. doi: 10.1080/13506129.2017.1289913. [DOI] [PubMed] [Google Scholar]
- 52.Gadotti VM, Bonfield SP, Zamponi GW. Depressive-like behaviour of mice lacking cellular prion protein. Behav Brain Res. 2012;227(2):319–323. doi: 10.1016/j.bbr.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 53.Salazar SV, Gallardo C, Kaufman AC, Herber CS, Haas LT, Robinson S, Manson JC, Lee MK, Strittmatter SM. Conditional deletion of Prnp rescues behavioral and synaptic deficits after disease onset in transgenic Alzheimer's disease. J Neurosci. 2017;37(38):9207–9221. doi: 10.1523/JNEUROSCI.0722-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmitz M, Zafar S, Silva CJ, Zerr I. Behavioral abnormalities in prion protein knockout mice and the potential relevance of PrP(C) for the cytoskeleton. Prion. 2014;8(6):381–386. doi: 10.4161/19336896.2014.983746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jirkof P. Burrowing and nest building behavior as indicators of well-being in mice. J Neurosci Methods. 2014;234:139–146. doi: 10.1016/j.jneumeth.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 56.Seibenhener ML, Wooten MC. Use of the open field maze to measure locomotor and anxiety-like behavior in mice. J Vis Exp. 2015;96(96):e52434. doi: 10.3791/52434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steinert JR. Prion protein as a mediator of synaptic transmission. Commun Integr Biol. 2015;8(4):e1063753. doi: 10.1080/19420889.2015.1063753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madroñal N, Gruart A, Delgado-García JM. Differing presynaptic contributions to LTP and associative learning in behaving mice. Front Behav Neurosci. 2009;3:7. doi: 10.3389/neuro.08.007.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stafstrom CE, Carmant L. Seizures and epilepsy: an overview for neuroscientists. In: Cold Spring Harbor perspectives in medicine. vol. 5. New York: Cold Spring Harbor Laboratory Press; 2015. [DOI] [PMC free article] [PubMed]
- 60.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nazor KE, Seward T, Telling GC. Motor behavioral and neuropathological deficits in mice deficient for normal prion protein expression. Biochim Biophys Acta. 2007;1772(6):645–653. doi: 10.1016/j.bbadis.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bult A, Lynch CB. Nesting and fitness: lifetime reproductive success in house mice bidirectionally selected for thermoregulatory nest-building behavior. Behav Genet. 1997;27(3):231–240. doi: 10.1023/A:1025610130282. [DOI] [PubMed] [Google Scholar]
- 63.Gaskill BN, Karas AZ, Garner JP, Pritchett-Corning KR. Nest building as an indicator of health and welfare in laboratory mice. J Vis Exp. 2013;82(82):51012. doi: 10.3791/51012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simon P, Dupuis R, Costentin J. Thigmotaxis as an index of anxiety in mice. Influence of dopaminergic transmissions. Behav Brain Res. 1994;61(1):59–64. doi: 10.1016/0166-4328(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 65.Lobao-Soares B, Walz R, Carlotti CG, Jr, Sakamoto AC, Calvo F, Terzian AL, da Silva JA, Wichert-Ana L, Coimbra NC, Bianchin MM. Cellular prion protein regulates the motor behaviour performance and anxiety-induced responses in genetically modified mice. Behav Brain Res. 2007;183(1):87–94. doi: 10.1016/j.bbr.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 66.Nico PB, de-Paris F, Vinade ER, Amaral OB, Rockenbach I, Soares BL, Guarnieri R, Wichert-Ana L, Calvo F, Walz R, et al. Altered behavioural response to acute stress in mice lacking cellular prion protein. Behav Brain Res. 2005;162(2):173–181. doi: 10.1016/j.bbr.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 67.Janner DR, de Lima EV, da Silva RT, Clarke JR, Linden R. Dissociation of genotype-dependent cognitive and motor behavior in a strain of aging mice devoid of the prion protein. Behav Brain Res. 2021;411:113386. doi: 10.1016/j.bbr.2021.113386. [DOI] [PubMed] [Google Scholar]
- 68.Rial D, Duarte FS, Xikota JC, Schmitz AE, Dafre AL, Figueiredo CP, Walz R, Prediger RD. Cellular prion protein modulates age-related behavioral and neurochemical alterations in mice. Neuroscience. 2009;164(3):896–907. doi: 10.1016/j.neuroscience.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 69.Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. 2013;14(9):609–625. doi: 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anacker C, Hen R. Adult hippocampal neurogenesis and cognitive flexibility — linking memory and mood. Nat Rev Neurosci. 2017;18(6):335–346. doi: 10.1038/nrn.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jackman SL, Regehr WG. The mechanisms and functions of synaptic facilitation. Neuron. 2017;94(3):447–464. doi: 10.1016/j.neuron.2017.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kanner AM. Anxiety disorders in epilepsy: the forgotten psychiatric comorbidity. Epilepsy Currents. 2011;11(3):90–91. doi: 10.5698/1535-7511-11.3.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Medel-Matus JS, Shin D, Sankar R, Mazarati A. Kindling epileptogenesis and panic-like behavior: their bidirectional connection and contribution to epilepsy-associated depression. Epilepsy Behav E&B. 2017;77:33–38. doi: 10.1016/j.yebeh.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ratte S, Vreugdenhil M, Boult JK, Patel A, Asante EA, Collinge J, Jefferys JG. Threshold for epileptiform activity is elevated in prion knockout mice. Neuroscience. 2011;179:56–61. doi: 10.1016/j.neuroscience.2011.01.053. [DOI] [PubMed] [Google Scholar]
- 75.McKhann GM, 2nd, Wenzel HJ, Robbins CA, Sosunov AA, Schwartzkroin PA. Mouse strain differences in kainic acid sensitivity, seizure behavior, mortality, and hippocampal pathology. Neuroscience. 2003;122(2):551–561. doi: 10.1016/S0306-4522(03)00562-1. [DOI] [PubMed] [Google Scholar]
- 76.Royle SJ, Collins FC, Rupniak HT, Barnes JC, Anderson R. Behavioural analysis and susceptibility to CNS injury of four inbred strains of mice. Brain Res. 1999;816(2):337–349. doi: 10.1016/S0006-8993(98)01122-6. [DOI] [PubMed] [Google Scholar]
- 77.Meador KJ. The basic science of memory as it applies to epilepsy. Epilepsia. 2007;48(Suppl 9):23–25. doi: 10.1111/j.1528-1167.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 78.Reid IC, Stewart CA. Seizures, memory and synaptic plasticity. Seizure. 1997;6(5):351–359. doi: 10.1016/S1059-1311(97)80034-9. [DOI] [PubMed] [Google Scholar]
- 79.You H, Tsutsui S, Hameed S, Kannanayakal TJ, Chen L, Xia P, Engbers JD, Lipton SA, Stys PK, Zamponi GW. Abeta neurotoxicity depends on interactions between copper ions, prion protein, and N-methyl-D-aspartate receptors. Proc Natl Acad Sci U S A. 2012;109(5):1737–1742. doi: 10.1073/pnas.1110789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watt NT, Taylor DR, Kerrigan TL, Griffiths HH, Rushworth JV, Whitehouse IJ, Hooper NM. Prion protein facilitates uptake of zinc into neuronal cells. Nat Commun. 2012;3(1):1134. doi: 10.1038/ncomms2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gavin R, Urena J, Rangel A, Pastrana MA, Requena JR, Soriano E, Aguzzi A, Del Rio JA. Fibrillar prion peptide PrP(106-126) treatment induces Dab1 phosphorylation and impairs APP processing and Abeta production in cortical neurons. Neurobiol Dis. 2008;30(2):243–254. doi: 10.1016/j.nbd.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 82.Pantera B, Bini C, Cirri P, Paoli P, Camici G, Manao G, Caselli A. PrPc activation induces neurite outgrowth and differentiation in PC12 cells: role for caveolin-1 in the signal transduction pathway. J Neurochem. 2009;110(1):194–207. doi: 10.1111/j.1471-4159.2009.06123.x. [DOI] [PubMed] [Google Scholar]
- 83.Loubet D, Dakowski C, Pietri M, Pradines E, Bernard S, Callebert J, Ardila-Osorio H, Mouillet-Richard S, Launay JM, Kellermann O, Schneider B. Neuritogenesis: the prion protein controls beta1 integrin signaling activity. FASEB J. 2012;26(2):678–690. doi: 10.1096/fj.11-185579. [DOI] [PubMed] [Google Scholar]
- 84.Amin L, Nguyen XT, Rolle IG, D'Este E, Giachin G, Tran TH, Serbec VC, Cojoc D, Legname G. Characterization of prion protein function by focal neurite stimulation. J Cell Sci. 2016;129(20):3878–3891. doi: 10.1242/jcs.183137. [DOI] [PubMed] [Google Scholar]
- 85.Benvegnu S, Roncaglia P, Agostini F, Casalone C, Corona C, Gustincich S, Legname G. Developmental influence of the cellular prion protein on the gene expression profile in mouse hippocampus. Physiol Genom. 2011;43(12):711–725. doi: 10.1152/physiolgenomics.00205.2010. [DOI] [PubMed] [Google Scholar]
- 86.Platzer K, Yuan H, Schutz H, Winschel A, Chen W, Hu C, Kusumoto H, Heyne HO, Helbig KL, Tang S, et al. GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects. J Med Genet. 2017;54(7):460–470. doi: 10.1136/jmedgenet-2016-104509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Platzer K, Lemke JR. GRIN2B-related neurodevelopmental disorder. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, LJH B, Stephens K, Amemiya A, editors. GeneReviews((R)). Seattle (WA): University of Washington; 1993. [PubMed]
- 88.Cahill L. Why sex matters for neuroscience. Nat Rev Neurosci. 2006;7(6):477–484. doi: 10.1038/nrn1909. [DOI] [PubMed] [Google Scholar]
- 89.Deacon RM. Assessing nest building in mice. Nat Protoc. 2006;1(3):1117–1119. doi: 10.1038/nprot.2006.170. [DOI] [PubMed] [Google Scholar]
- 90.Madronal N, Lopez-Aracil C, Rangel A, del Rio JA, Delgado-Garcia JM, Gruart A. Effects of enriched physical and social environments on motor performance, associative learning, and hippocampal neurogenesis in mice. PloS One. 2010;5(6):e11130. doi: 10.1371/journal.pone.0011130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clarke JR, Cammarota M, Gruart A, Izquierdo I, Delgado-García JM. Plastic modifications induced by object recognition memory processing. Proc Natl Acad Sci U S A. 2010;107(6):2652–2657. doi: 10.1073/pnas.0915059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Medrano-Fernandez A, Delgado-Garcia JM, Del Blanco B, Llinares M, Sanchez-Campusano R, Olivares R, Gruart A, Barco A. The epigenetic factor CBP is required for the differentiation and function of medial ganglionic eminence-derived interneurons. Mol Neurobiol. 2018;56(6):4440–4454. doi: 10.1007/s12035-018-1382-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmieder R, Lim YW, Edwards R. Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics. 2012;28(3):433–435. doi: 10.1093/bioinformatics/btr669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12(2):115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matamoros-Angles A, Gayosso LM, Richaud-Patin Y, di Domenico A, Vergara C, Hervera A, Sousa A, Fernandez-Borges N, Consiglio A, Gavin R, et al. iPS cell cultures from a Gerstmann-Straussler-Scheinker patient with the Y218N PRNP mutation recapitulate tau pathology. Mol Neurobiol. 2018;55(4):3033–3048. doi: 10.1007/s12035-017-0506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Urrea L, Segura-Feliu M, Masuda-Suzukake M, Hervera A, Pedraz L, Garcia Aznar JM, Vila M, Samitier J, Torrents E, Ferrer I, et al. Involvement of cellular prion protein in alpha-synuclein transport in neurons. Mol Neurobiol. 2018;55(3):1847–1860. doi: 10.1007/s12035-017-0451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nishimura T, Sakudo A, Hashiyama Y, Yachi A, Saeki K, Matsumoto Y, Ogawa M, Sakaguchi S, Itohara S, Onodera T. Serum withdrawal-induced apoptosis in ZrchI prion protein (PrP) gene-deficient neuronal cell line is suppressed by PrP, independent of Doppel. Microbiol Immunol. 2007;51(4):457–466. doi: 10.1111/j.1348-0421.2007.tb03920.x. [DOI] [PubMed] [Google Scholar]
- 100.Teller S, Tahirbegi IB, Mir M, Samitier J, Soriano J. Magnetite-amyloid-beta deteriorates activity and functional organization in an in vitro model for Alzheimer's disease. Sci Rep. 2015;5(1):17261. doi: 10.1038/srep17261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Orlandi JG, Stetter O, Soriano J, Geisel T, Battaglia D. Transfer entropy reconstruction and labeling of neuronal connections from simulated calcium imaging. PloS One. 2014;9(6):e98842. doi: 10.1371/journal.pone.0098842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jang MJ, Nam Y. NeuroCa: integrated framework for systematic analysis of spatiotemporal neuronal activity patterns from large-scale optical recording data. Neurophotonics. 2015;2(3):035003. doi: 10.1117/1.NPh.2.3.035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. PrnpZH3/ZH3 mice showed similar nest-building behavior to wild-type mice. a, Representative images of nests constructed by Prnp+/+ (left) and PrnpZH3/ZH3 mice (right). b, Quantification illustrating the mean of the nest score (see Material and Methods for details). Data are presented as mean ± S.E.M. Source data and individual data values are available in Additional file 10.
Additional file 2: Figure S2. PrnpZH1/ZH1 mice failed to acquire an instrumental learning test in the Skinner box test using a fixed ratio (1:1) schedule. a, Percentage of Prnp+/+ and PrnpZH1/ZH1 mice reached the selected criterion during the training sessions. b, Task accuracy ((lever presses during light ON – lever presses during the light OFF) / Total number of lever presses) during the light ON / light OFF conditioning paradigm. Data are presented as a percentage in a and as mean ± S.E.M. in b. Source data and individual data values are available in Additional file 10.
Additional file 3: Figure S3. PrnpZH3/ZH3 mice have deficits in acquiring instrumental learning but not motor impairment in the Rotarod test. After a training session in the Rotarod, mice were tested two consecutive days with 5 sessions in each. Latency of mice to fall (sec.) from the Rotarod in the first day (a) or in the second (b). Data are presented as mean ± S.E.M.. *p < 0.05 and **p < 0.01, two-way ANOVA + Bonferroni’s multiple comparisons test. Source data and individual data values are available in Additional file 10.
Additional file 4: Figure S4. Stressed-like behavior in PrnpZH3/ZH3 mice impairing object recognition test performance. PrnpZH1/ZH1 mice also showed stressed-like behavior and failed to acquire short-term memory. a, Scheme of object recognition protocol that consisted of 3 sessions (10 min each). First, mice were habituated in the empty arena. One hour later, they were placed again in the arena with two identical objects for the training session. Finally, 2-3 h later, they were once more placed in the arena, replacing one object with a novel one for the short-term memory test. Fecal bodies (PrnpZH3/ZH3) and rearing episodes (PrnpZH1/ZH1) were counted in the habituation session as an indicator of animal stress. b, Number of fecal bodies generated per animal (Prnp+/+ and PrnpZH3/ZH3) during the habituation session. Data are presented as mean ± S.E.M. c-d Time that Prnp+/+ (c), and PrnpZH3/ZH3 (d) mice interacted with the objects in the short-term test. Data are presented as the percentage of mice interacting with the object in each time interval (0 to 35 s). e, Number of rearing episodes in the habituation session (Prnp+/+ and PrnpZH1/ZH1). f-g, Relative time that the two exposed objects are explored to the total time invested. Object 1 (O1) and object 1’ (O1’) in the training session (f) and number, and object 1 (O1) and object 2 (O2) in the short-term memory test (g). Data are presented as percentages in C-D and as mean ± S.E.M. in b, e, f, g. *p < 0.05 and ***p < 0.001, Mann-Whitney U non-parametric test. Source data and individual data values are available in Additional file 10.
Additional file 5: Table S1. List of the protein-coding significantly downregulated genes in PrnpZH3/ZH3 hippocampus compared to Prnp+/+.
Additional file 6: Table S2. List of the protein-coding significantly upregulated genes in PrnpZH3/ZH3 hippocampus compared to Prnp+/+.
Additional file 7: Figure S5. Gene ontology of the genes significantly downregulated and upregulated in PrnpZH3/ZH3 hippocampus compared to Prnp+/+. a, Volcano plot of the protein-coding genes analyzed in the RNAseq (~ 16.000). In the X-axis is plotted the Fold change (log2) and in the Y-axis the padj (-log10). The red line separates the significantly expressed genes (padj < 0,05). The blue lines indicate the 0,85 and 1,2 fold changes. b, Gene ontology analysis of the downregulated and upregulated genes with the Reactome software (see Materials and Methods for details). Validation of the main genes altered by RT-qPCR: Grin2b (c), Gabrr2 (d), Kcnj6 (e), Kcna1 (f), Kcnj2 (g), and Kcnq3 (h). Data are presented as mean ± S.E.M. *p < 0.05 and ***p < 0.001. Source data and individual data values are available in Additional file 10.
Additional file 8: Movie S1. Representative movie of Prnp+/+ (bottom and up-left) and PrnpZH3/ZH3 (middle and up-right) mouse behavior after KA (middle and bottom) and PBS (top) administration. Note the blinking and seizure episodes suffered by the PrnpZH3/ZH3 mice treated with KA (labeled with asterisks at the beginning of the video). Source data and individual data values are available in Additional file 10.
Additional file 9: Figure S6. Images of the original, uncropped blots for Figs. 1A and 6A.
Additional file 10. Excel file 1. File providing individual data values for all figures from the manuscript.
Data Availability Statement
All data generated or analysed during this study are included in this published article and additional information files. RNAseq data have been deposited at the Gene Expression Omnibus (GEO) with accession code: GSE189691. (Matamoros-Angles, A; Hervera, A; Soriano, J; Martí, E; Carulla, P, Llorens, F; Nuvolone, M; Aguzzi, A; Ferrer I; Gruart, A; Delgado-García, JM; Del Río, JA. RNA sequencing of hippocampus of Prnp+/+ and PrnpZH3/ZH3 animals. https://identifiers.org/geo:GSE189691). Source data for all figures have been provided in Additional file 10. Uncropped blots from Figs. 1 and 6 are available in Additional file 9.