ABSTRACT
We report a draft genome assembly of the causal agent of tomato vascular wilt, Fusarium oxysporum f. sp. lycopersici isolate 59, obtained from the Andean region in Colombia.
ANNOUNCEMENT
Fusarium oxysporum f. sp. lycopersici is a soilborne fungus belonging to the F. oxysporum species complex (FOSC). F. oxysporum f. sp. lycopersici causes fusarium wilt in tomato (Solanum lycopersicum), which often leads to significant yield losses (1, 2). F. oxysporum f. sp. lycopersici isolate 59 was isolated from root and stem tissue from a wilted tomato plant grown in the Andean region of Colombia (3). Isolate 59 was classified as F. oxysporum f. sp. lycopersici race 2, using PCR markers for phylogenetic analysis (3).
For whole-genome sequencing, fungal hyphae from a 6-day-old culture (Czapek-Dox medium) were collected and lyophilized overnight. High-molecular-weight (HMW) DNA was extracted using a modified phenol-chloroform/isoamyl alcohol method (4). For Nanopore sequencing, a library was prepared using the ligation sequencing kit (SQK-LSK109) according to the manufacturer’s instructions (Oxford Nanopore Technologies, Oxford, UK) using 1 μg HMW DNA. The long-fragment buffer (LFB) supplied in the kit was used to enrich long DNA fragments of >3 kb. An R9.4.1 flow cell (Oxford Nanopore Technologies) was loaded and run for 24 h. Base calling was performed using Guppy version 4.0.21 within MinKNOW (Oxford Nanopore Technologies). Illumina sequencing was performed using a fungal sample collected as previously described. Total DNA was isolated using the cetyltrimethylammonium bromide (CTAB) protocol (5). DNA (350 ng/μL) was used for library preparation with the Nextera DNA Flex library preparation kit in dual index format (Illumina, Inc., San Diego, CA, USA) according to the manufacturer’s instructions. The library was sequenced in paired-end format on the Illumina HiSeq 4000 sequencing system (Macrogen, South Korea).
The quality of the Nanopore and Illumina reads was assessed via NanoPlot version 1.30.1 (6) and FastQC version 11.7 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), respectively. A total of 1,742,231 raw reads were generated from the Nanopore sequencing. Approximately 16 million 151-bp paired-end reads were obtained from the Illumina sequencing. The resulting long reads were first processed using Porechop version 0.2.4 to divide chimeric sequences (https://github.com/rrwick/Porechop) (7); then, the reads were filtered by length and quality using Filtlong version 0.2.0 (https://github.com/rrwick/Filtlong). The N50 length of the Nanopore reads was 9.569 kbp. A total of 885,847 filtered reads were assembled using the de novo long-read assembler Shasta version 0.1.0 (8). The sequenced short reads were processed by first removing residual adapters and poor quality reads using Trim Galore version 0.6.5 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Reads shorter than 100 bp were filtered using the FASTX-Toolkit version 0.0.14 (fastx_trimmer; http://hannonlab.cshl.edu/fastx_toolkit). The de novo assembly Nanopore and Illumina reads were polished using Racon version 1.4.13 (9) and Pilon (10), respectively. Whole-genome assembly was carried out using a hybrid de novo assembly approach, incorporating Nanopore long reads and Illumina short reads.
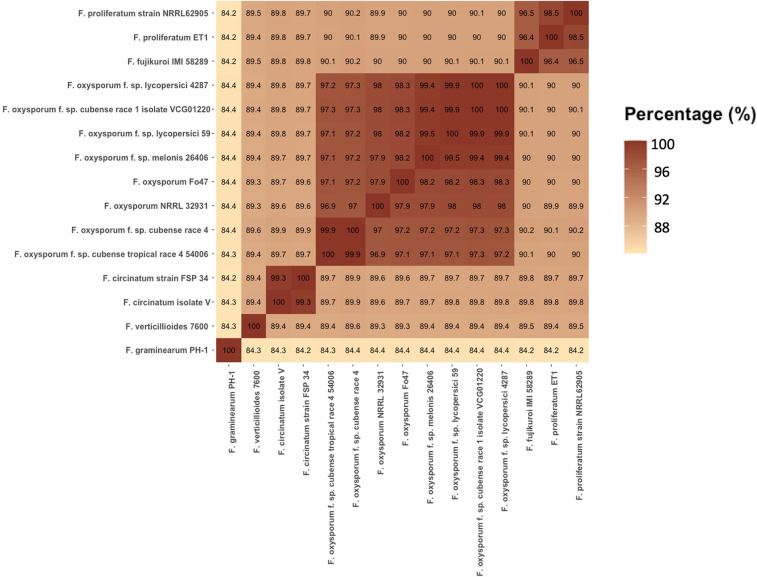
A summary of assembly statistics was generated using BBMap version 38.90 (11), and the assembly completeness was evaluated using the Benchmarking Universal Single-Copy Orthologs (BUSCO) version 4.0.6 software (12) (Table 1). PYANI version 0.2.10 was used to calculate the average nucleotide identity (ANI) and relatedness measures of whole-genome comparisons among Fusarium species (13) (Fig. 1). The draft assembly (combining long reads and Illumina short reads) has a total size of 54.2 Mb and a coverage of approximately 75.5×. The completeness of the assembly was calculated using BUSCO with the Hypocreales_odb10 lineage gene data set; the analysis showed that 4,441 out of 4,494 BUSCO markers were found, and only a few duplicated or missing BUSCO orthologs were identified (Table 1).
TABLE 1.
Comparison of assembly statistics of Fusarium oxysporum f. sp. lycopersici isolates
| Characteristic | Data for F. oxysporum f. sp. lycopersici strain: |
||
|---|---|---|---|
| 59 | 4287 | 4287 | |
| Accession no. (database) | PRJNA756266 (BioProject) | GCF_000149955.1 (GenBank assembly) | GCA_003315725.1 (GenBank assembly) |
| Sequencing method | Oxford Nanopore + Illumina | Sanger | PacBio + Illumina |
| Total length (Gbp) | 5.36 | 6.1 | 5.39 |
| No. of contigs | 361 | 1,362 | 504 |
| Coverage (×) | 75.5 | 6.5 | 76 |
| Assembly size (Mb) | 54.2 | 59.9 | 53.9 |
| Longest contig (bp) | 6,457,141 | 5,700,000 | |
| % GC | 47.67 | 48.4 | 47.7 |
| Contig N50 (bp) | 3,035,620 | 95,416 | 1,338,693 |
| Contig L50 | 7 | 184 | 11 |
| Complete BUSCOs (%) | 99.60 | 97.70 | 99.90 |
| Total no. of BUSCOs | 4,494 | 4,494 | 2,294 |
| No. of duplicate BUSCOs | 37 | 40 | 34 |
| No. of fragmented BUSCOs | 0 | 24 | 1 |
| No. of missing BUSCOs | 7 | 78 | 7 |
| Reference | This study | 15 | 14 |
FIG 1.
Heatmap table of the average nucleotide identity (ANI) values generated from a pairwise comparison of 15 Fusarium isolates. An ANI score greater than 95% between two genomes indicates that they are the same species. The genomes of the Fusarium isolates were downloaded from NCBI: F. oxysporum f. sp. cubense race 4 (GenBank accession number GCA_000350365.1), F. circinatum strain FSP 34 (GCA_000497325.3), F. oxysporum f. sp. melonis 26406 (GCA_002318975.1), F. oxysporum Fo47 (GCA_013085055.1), F. circinatum isolate V (GCA_013168815.1), F. oxysporum f. sp. cubense race 1 isolate VCG01220 (GCA_016802225.1), F. proliferatum strain NRRL62905 (GCA_900029915.1), F. verticillioides 7600 (GCF_000149555.1), F. oxysporum f. sp. lycopersici 4287 (GCA_003315725.1), F. graminearum PH-1 (GCF_000240135.3), F. oxysporum f. sp. cubense tropical race 4 strain 54006 (GCF_000260195.1), F. oxysporum NRRL 32931 (GCF_000271745.1), F. proliferatum ET1 (GCF_900067095.1), F. fujikuroi IMI 58289 (GCF_900079805.1).
The results of this study will contribute to building a more robust phylogenetic framework that will guide inquiries concerning the evolution of important traits in the FOSC group.
Data availability.
The described genome assembly is available in GenBank under BioProject accession number PRJNA756266. The Illumina and Oxford Nanopore reads are deposited at the Sequence Read Archive (SAR) under accession numbers SRX11976571 and SRX11976570, respectively. F. oxysporum f. sp. lycopersici strain 59 was registered in the National Collections Registry (RNC129) and was collected under AGROSAVIA permit framework number 1466 from 2014, updated by resolution 04039 on 19 July 2018.
ACKNOWLEDGMENTS
This study was funded by (i) Colombian Ministry of Agriculture grant number 1000941 (TV19) under a collaborative project with Universidad de los Andes, (ii) the U.S. Department of Agriculture’s Agriculture and Food Research Initiative grant FLA-PLP-006039, and (iii) the Research Office of the University of Florida’s Institute for Food and Agricultural Science.
Contributor Information
Mauricio Soto-Suarez, Email: msoto@agrosavia.co.
Antonis Rokas, Vanderbilt University.
REFERENCES
- 1.FAOSTAT. Crops and livestock products. http://www.fao.org/faostat/en/#data/QCL. Accessed 22 September 2021.
- 2.Srinivas C, Nirmala Devi D, Narasimha Murthy K, Mohan CD, Lakshmeesha TR, Singh B, Kalagatur NK, Niranjana SR, Hashem A, Alqarawi AA, Tabassum B, Abd Allah EF, Chandra Nayaka S. 2019. Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: biology to diversity—a review. Saudi J Biol Sci 26:1315–1324. doi: 10.1016/j.sjbs.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmona SL, Burbano-David D, Gómez MR, Lopez W, Ceballos N, Castaño-Zapata J, Simbaqueba J, Soto-Suárez M. 2020. Characterization of pathogenic and nonpathogenic Fusarium oxysporum isolates associated with commercial tomato crops in the Andean region of Colombia. Pathogens 9:70. doi: 10.3390/pathogens9010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chavarro‐Carrero EA, Vermeulen JP, Torres D, Usami T, Schouten HJ, Bai Y, Seidl MF, Thomma BPHJ. 2021. Comparative genomics reveals the in planta‐secreted Verticillium dahliae Av2 effector protein recognized in tomato plants that carry the V2 resistance locus. Environ Microbiol 23:1941–1958. doi: 10.1111/1462-2920.15288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffith GW, Shaw DS. 1998. Polymorphisms in Phytophthora infestans: four mitochondrial haplotypes are detected after PCR amplification of DNA from pure cultures or from host lesions. Appl Environ Microbiol 64:4007–4014. doi: 10.1128/AEM.64.10.4007-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Coster W, D'Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom 3:e000132. doi: 10.1099/mgen.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shafin K, Pesout T, Lorig-Roach R, Haukness M, Olsen HE, Bosworth C, Armstrong J, Tigyi K, Maurer N, Koren S, Sedlazeck FJ, Marschall T, Mayes S, Costa V, Zook JM, Liu KJ, Kilburn D, Sorensen M, Munson KM, Vollger MR, Monlong J, Garrison E, Eichler EE, Salama S, Haussler D, Green RE, Akeson M, Phillippy A, Miga KH, Carnevali P, Jain M, Paten B. 2020. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat Biotechnol 38:1044–1053. doi: 10.1038/s41587-020-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaser R, Sovic I, Nagarajan N, Sikic M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. LBNL-7065E. LBNL, Berkeley, CA. [Google Scholar]
- 12.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 13.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 14.Ayhan DH, López-Díaz C, Di Pietro A, Ma L-J. 2018. Improved assembly of reference genome Fusarium oxysporum f. sp. lycopersici strain Fol4287. Microbiol Resour Announc 7:e00910-18. doi: 10.1128/MRA.00910-18. [DOI] [PMC free article] [PubMed]
- 15.Ma L-J, Van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, Di Pietro A, Dufresne M, Freitag M, Grabherr M, Henrissat B, Houterman PM, Kang S, Shim W-B, Woloshuk C, Xie X, Xu J-R, Antoniw J, Baker SE, Bluhm BH, Breakspear A, Brown DW, Butchko RAE, Chapman S, Coulson R, Coutinho PM, Danchin EGJ, Diener A, Gale LR, Gardiner DM, Goff S, Hammond-Kosack KE, Hilburn K, Hua-Van A, Jonkers W, Kazan K, Kodira CD, Koehrsen M, Kumar L, Lee Y-H, Li L, Manners JM, Miranda-Saavedra D, Mukherjee M, Park G, Park J, Park S-Y, Proctor RH, Regev A, Ruiz-Roldan MC, et al. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464:367–373. doi: 10.1038/nature08850. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The described genome assembly is available in GenBank under BioProject accession number PRJNA756266. The Illumina and Oxford Nanopore reads are deposited at the Sequence Read Archive (SAR) under accession numbers SRX11976571 and SRX11976570, respectively. F. oxysporum f. sp. lycopersici strain 59 was registered in the National Collections Registry (RNC129) and was collected under AGROSAVIA permit framework number 1466 from 2014, updated by resolution 04039 on 19 July 2018.