Abstract
Deep learning is transforming the analysis of biological images, but applying these models to large datasets remains challenging. Here we describe the DeepCell Kiosk, cloud-native software that dynamically scales deep learning workflows to accommodate large imaging datasets. To demonstrate the scalability and affordability of this software, we identified cell nuclei in 106 1-megapixel images in ~5.5 h for ~US$250, with a cost below US$100 achievable depending on cluster configuration. The DeepCell Kiosk can be downloaded at https://github.com/vanvalenlab/kiosk-console; a persistent deployment is available at https://deepcell.org/.
While deep learning is an increasingly popular approach to extracting quantitative information from biological images, its limitations substantially hinder its widespread adoption. Chief among these limitations are the requirements for expansive sets of training data and computational resources. Here we sought to overcome the latter limitation. While deep learning methods have remarkable accuracy for a range of image-analysis tasks, including classification1, segmentation2–4 and object tracking5,6, they have limited throughput even with GPU acceleration. For example, even when running segmentation models on a GPU, typical inference speeds on megapixel-scale images are in the range of 5–10 frames per second, limiting the scope of analyses that can be performed on images in a timely fashion. The necessary domain knowledge and associated costs of GPUs pose further barriers to entry, although recent software packages7–11 have attempted to solve these two issues. While cloud computing has proven effective for other data types12–15, scaling analyses to large imaging datasets in the cloud while constraining costs is a considerable challenge.
To meet this need, we have developed the DeepCell Kiosk (Fig. 1a). This software package takes in configuration details (user authentication, GPU type, etc.) and creates a cluster on Google cloud that runs predefined deep learning–enabled image-analysis pipelines. This cluster is managed by Kubernetes, an open-source framework for running software containers (software that is bundled with its dependencies so it can be run as an isolated process) across a group of servers. An alternative way to view Kubernetes is as an operating system for cloud computing. Data are submitted to the cluster through a web-based front-end, a command line tool, or an ImageJ plugin. Once submitted, they are placed in a database where the specified image analysis pipeline can pick up the dataset, perform the desired analysis and make the results available for download. Results can be visualized by a variety of visualization software tools16,17.
Fig. 1 |. Architecture and performance of the DeepCell Kiosk.
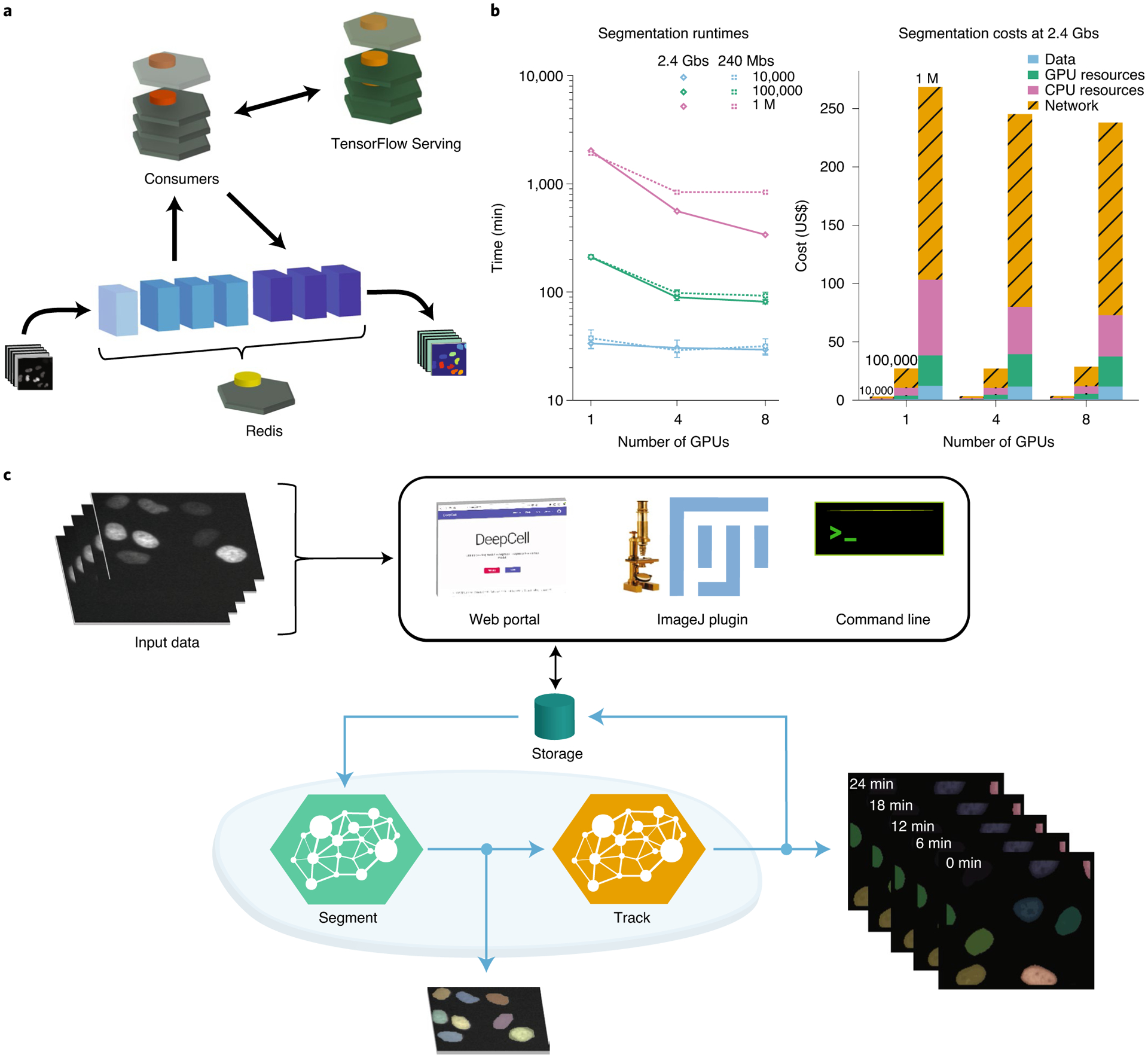
a, Data flow through the DeepCell Kiosk. Images submitted to a running Kiosk cluster are entered into a queue. Once there, images are processed by objects called consumers, which execute all the computational operations in a given pipeline. Consumers perform most computations themselves but access deep learning by submitting data to TensorFlow Serving and capturing the result. This separation allows conventional operations and deep learning operations to occur on different types of nodes, which is essential for efficient resource allocation. After processing, the result is returned to the queue, ready to be downloaded by the user. b, Left, benchmarking of inference speed demonstrating scaling to large imaging datasets (see the Supplementary Information for details). Throughput is ultimately limited by data transfer speed. right, an analysis of cost demonstrating affordability. network costs are incurred by inter-zone network traffic in multi-zone clusters, which are more stable and scale faster than their single-zone counterparts. This cost can be avoided when users configure a single-zone cluster. Benchmarking was performed once for 1 million (M) image runs and in triplicate otherwise. Error bars, when present, represent the s.d. of replicate measurements. c, The DeepCell Kiosk enables construction and scaling of multi-model pipelines. For example, a live-cell imaging consumer can access deep learning models for both segmentation and tracking. The live-cell imaging consumer places the frames for a movie into the queue for a segmentation consumer, where they are processed in parallel. Once segmented, the images are processed by a tracking consumer to link cells together over time and to construct lineages. The results, which consist of label images of segmented and tracked cells as well as a JSOn file describing mother–daughter relationships, are uploaded to a cloud bucket, from which they can be downloaded by the user.
To ensure that image analysis pipelines can be run efficiently on this cluster, we made two software design choices. First, image analysis pipelines access trained deep learning models through a centralized model server in the cluster. This strategy enables the cluster to efficiently allocate resources, as the various computational steps of a pipeline (deep learning and conventional computer vision operations) are run on the most appropriate hardware. Second, the DeepCell Kiosk treats hardware as a resource that can be allocated dynamically, as opposed to a fixed resource. This conceptual shift means that cluster size scales to meet data analysis demand: small datasets lead to small clusters, while large datasets lead to large clusters. This feature, which has previously been demonstrated for other biological data types13, is made possible by our use of Kubernetes, which enables us to scale analyses to large datasets, reducing analysis time while constraining costs (Fig. 1b). A full description of the DeepCell Kiosk’s software architecture, scaling policies, and benchmarking is provided in the Supplementary Information.
Given the wide range of image analysis problems that deep learning can now solve, we designed the DeepCell Kiosk with the flexibility to work with arbitrary collections of deep learning models and image analysis pipelines. Pipelines can exploit multiple deep learning models—and even other pipelines—while still leveraging the scaling ability provided by Kubernetes. For example, we constructed a pipeline (Fig. 1c) that pairs deep learning models for cell segmentation and cell tracking5 to analyze live-cell imaging data. Additional examples are described in the Supplementary Information, and documentation on creating custom pipelines is available at https://deepcell-kiosk.readthedocs.io.
In conclusion, the DeepCell Kiosk enables cost-effective scaling of customizable deep learning–enabled image analysis pipelines to large imaging datasets. The dynamic nature of our scaling enables individual users to process large datasets. Further, it allows a single cluster to serve an entire community of researchers—much like BLAST18 has rendered sequence alignment accessible to the scientific community through a web portal. At the cost of network latency, software ecosystems like ImageJ can access complete deep learning–enabled pipelines hosted by such a cluster through a plugin. While the data analyzed here came from in vitro cell culture, our emphasis on deep learning makes it possible for our software to analyze tissue data, including spatial genomics data19. We believe that, by reducing costs and time to analysis, this work should accelerate the rate of biological discovery and change the relationship that biologists have with imaging data. Further, by focusing on deployment, this work highlights aspects of the performance of deep learning models beyond accuracy (such as inference speed) that substantially impact their utility. Finally, this work highlights the growing definition of the term ‘software’. Just as deep learning has made data a part of the software stack, Kubernetes can do the same for hardware. Jointly developing data, code, and computing should substantially improve the performance of scientific software crafted in the era of Software 2.0.
Methods
Data collection.
Cells were imaged with either a Nikon Ti-E or Nikon Ti2 fluorescence microscope controlled by either Micro-Manager or Nikon Elements. Data were processed using the scientific computing stack for Python.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We thank numerous colleagues including A. Anandkumar, M. Angelo, J. Bois, I. Brown, A. Butkovic, L. Cai, I. Camplisson, M. Covert, M. Elowitz, J. Freeman, C. Frick, L. Geontoro, A. Ho, K. Huang, K. C. Huang, G. Johnson, L. Keren, D. Litovitz, D. Macklin, U. Manor, S. Patel, A. Raj, N. Pelaez Restrepo, C. Pavelchek, S. Shah and M. Thomson for helpful discussions and contributing data. We gratefully acknowledge support from the Shurl and Kay Curci Foundation, the Rita Allen Foundation, the Paul Allen Family Foundation through the Allen Discovery Center at Stanford University, the Rosen Center for Bioengineering at Caltech, Google Research Cloud, Figure 8’s AI For Everyone award, and a subaward from NIH U24-CA224309-01.
Footnotes
Code availability
We used Kubernetes and TensorFlow, along with the scientific computing stack for Python. A persistent deployment of the software described can be accessed at https://deepcell.org/. All source code, including version requirements and explicit usage, is under a modified Apache license and is available at https://github.com/vanvalenlab. Detailed instructions are available at https://deepcell-kiosk.readthedocs.io.
Competing interests
The authors have filed a provisional patent for the described work; the software described here is available under a modified Apache license and is free for non-commercial uses.
Supplementary information is available for this paper at https://doi.org/10.1038/s41592-020-01023-0.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41592-020-01023-0.
Data availability
All data that were used to generate the figures in this paper are available at https://deepcell.org/data and at https://github.com/vanvalenlab/deepcell-tf under the deepcell.datasets module.
References
- 1.Ouyang W et al. Analysis of the Human Protein Atlas Image Classification competition. Nat. Methods 16, 1254–1261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falk T et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Van Valen DA et al. Deep learning automates the quantitative analysis of individual cells in live-cell imaging experiments. PLoS Comput. Biol 12, e1005177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt U, Weigert M, Broaddus C & Myers G Cell detection with star-convex polygons. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018 (eds. Frangi AF et al.) 265–273 (Springer International Publishing, 2018). [Google Scholar]
- 5.Moen E et al. Accurate cell tracking and lineage construction in live-cell imaging experiments with deep learning. Preprint at bioRxiv 10.1101/803205 (2019). [DOI] [Google Scholar]
- 6.Anjum S & Gurari D CTMC: Cell Tracking with Mitosis Detection dataset challenge. in Proc. IEEE/CVF Conf. Computer Vision and Pattern Recognition (CVPR) Workshops 982–983 (2020). [Google Scholar]
- 7.Gómez-de-Mariscal E et al. DeepImageJ: a user-friendly plugin to run deep learning models in ImageJ. Preprint at bioRxiv 10.1101/799270 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Ouyang W, Mueller F, Hjelmare M, Lundberg E & Zimmer C ImJoy: an open-source computational platform for the deep learning era. Nat. Methods 16, 1199–1200 (2019). [DOI] [PubMed] [Google Scholar]
- 9.von Chamier L et al. ZeroCostDL4Mic: an open platform to simplify access and use of deep-learning in microscopy. Preprint at bioRxiv 10.1101/2020.03.20.000133 (2020). [DOI] [Google Scholar]
- 10.McQuin C et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 16, e2005970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haberl MG et al. CDeep3M—Plug-and-Play cloud-based deep learning for image segmentation. Nat. Methods 15, 677–680 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emami Khoonsari P et al. Interoperable and scalable data analysis with microservices: applications in metabolomics. Bioinformatics 35, 3752–3760 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novella JA et al. Container-based bioinformatics with Pachyderm. Bioinformatics 35, 839–846 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capuccini M et al. On-demand virtual research environments using microservices. PeerJ Comput. Sci 5, e232 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters K et al. PhenoMeNal: processing and analysis of metabolomics data in the cloud. GigaScience 8, giy149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sofroniew N et al. napari/napari: 0.3.5. Zenodo 10.5281/zenodo.3900158 (2020). [DOI] [Google Scholar]
- 18.Altschul SF et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keren L et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that were used to generate the figures in this paper are available at https://deepcell.org/data and at https://github.com/vanvalenlab/deepcell-tf under the deepcell.datasets module.


