Abstract
Gains and losses at chromosome 3p12–21 are common in breast tumors and associated with patient outcomes. We hypothesized that the LRIG1 gene at 3p14.1, whose product functions in ErbB-family member degradation, is a critical tumor modifier at this locus. We analyzed 971 stage I/II breast tumors using Affymetrix Oncoscan™ molecular inversion probe arrays that include 12 probes located within LRIG1. Copy number results were validated against gene expression data available in the public database. By partitioning the LRIG1 probes nearest exon 12/13, we confirm a breakpoint in the gene and show that gains and losses in the sub-regions differ by tumor and patient characteristics including race/ethnicity. In analyses adjusted for known prognostic factors, loss of LRIG1 was independently associated with risk of any relapse (HR, 1.90; 95% CI, 1.32–2.73), relapse ≥ 5 y (HR, 2.39; 95% CI, 1.31–4.36), and death (HR, 1.55; 95% CI, 1.11–2.16). Analyses of copy number across chromosome 3, as well as expression data from pooled, publically available datasets, corroborated the hypothesis of an elevated and persistent risk among cases with loss of or low LRIG1. We concluded that loss/low expression of LRIG1 is an independent risk factor for breast cancer metastasis and death in stage I/II patients. Increased hazard in patients with loss/low LRIG1 persists years after diagnosis, suggesting that LRIG1 is acting as a critical suppressor of tumor metastasis and an early clinical indicator of risk for late recurrences in otherwise low-risk patients.
Keywords: breast cancer, relapse, death, LRIG1, early stage
INTRODUCTION
The human LRIG1 gene at chromosome 3p14.1 encodes a ‘leucine-rich repeats and immunoglobulin-like domains-1’ protein(1) that negatively regulates the ERRB2 gene product, HER2, and other oncogenic receptor tyrosine kinases, including EGFR, HER3 and 4, MET (hepatocyte growth factor receptor), and RET (rearranged during transfection)(2–5). The LRIG1 protein is expressed in most tissues analyzed(6) and limits the size of epithelial progenitor cell populations by promoting the degradation of members of the ErbB family(7–9). In mouse models, deletion results in epidermal hyperplasia(10, 11) and expansion of base columnar cells in the intestinal crypt(9).
We previously analyzed LRIG1 copy number (CN) by in situ hybridization and showed increased CN at exon 12/13 in 34% of 73 breast tumors(12, 13). Moreover, we observed co-incident ERBB2 gene amplification in tumors with gain of exon 12/13 probe signal. In another study, LRIG1 expression was observed to be down-regulated in HER2-overexpressing tumors(4). Recently, high levels of LRIG1 expression were observed in estrogen receptor (ER)+ breast tumors with functional evidence for direct induction of LRIG1 expression by estrogen that was antagonized by HER2(14). Using pooled results from public expression array data, Krig et al. reported that LRIG1 overexpression in breast tumors was associated with lower risk of relapse, though analyses were not adjusted for clinical covariates(14). These findings contrast with reports of chromosome 3 losses between p12 and p21 common in breast cancer, and Staaf et al. showed that loss of 3p13, adjusted for patient and clinical characteristics including ERBB2 amplification, was independently prognostic for recurrence and breast cancer death(15).
To evaluate the clinical importance and correlates of the LRIG1 chromosomal region, we applied high-resolution molecular inversion probe (MIP) analysis in a cohort of early-stage breast tumors with long-term follow-up to characterize the effect of LRIG1 gene dose on patient outcomes, considering amplification of ERBB2.
PATIENTS AND METHODS
Patients and specimen characteristics.
Patients from the Early Stage Breast Cancer Repository, a cohort of 2327 women with stage I/II breast cancer, were treated at the University of Texas MD Anderson Cancer Center between 1985 and 2000. Details of the cohort have been reported, including methodology for medical record abstraction for clinical, pathological, and treatment covariates(16). Of 2327 women, 1003 cases were Texas residents and had primary tumor tissue with >80% tumor cells from formalin-fixed, paraffin-embedded tissue. For CN analyses, cases were matched ~2:1 for non-Hispanic white (NHW)-to-Hispanic and NHW-to-black women, controlling for stage and age of diagnosis. Case status was last updated in 2007 with a median follow-up time of 8.9 y.
ER and progesterone receptor (PR) status were determined as described(17). Intrinsic subtypes were defined as follows: Luminal A: ER+ and/or PR+ and <15% Ki-67; Luminal B: ER+ and/or PR+ and Ki-67 ≥15%; and triple-negative breast cancer (TNBC): ER–, PR, and HER2–. HER2+ tumors were defined based on ERRB2 gene CN by MIP array with a threshold of >2.3 for gain.
Assay methods.
Details of Affymetrix Oncoscan™ Version 1 results with clinical and histopathological patient characteristics have been reported(17). Briefly, MIP probes are hybridized to genomic DNA and split into two tubes containing paired nucleotide mixes (triphosphates of adenine + thymine or cytosine + guanine). In the presence of DNA polymerase and ligase, MIP probes circularize with their complementary nucleotide. Allele discrimination is enzymatically derived and highly specific, allowing multiplexed assays. Of 1003 case samples, 971 passed quality control and included.
We followed two approaches to compute CN at LRIG1. Pre-processing included application of the Nexus™ Copy Number Segmentation algorithm to each of the samples. We extracted CN information from 11 of 12 probes that passed quality control (call rate >90%, relative standard deviation <0.4) located within the start and stop locations of LRIG1 assembly UCSC hg18 (NCBI Build 36.1); 16.9% of calls were removed as outliers. To quantify CN, we identified the segment that contained the start-end locations of LRIG1 (chromosome 3: 66,511,911–66,633,535), with each sample having distinct start and stop sites. The CN assigned to the LRIG1 ‘locus’ is the mean value of that region as reported by Nexus Copy Number. Thus, the per-tumor locus that includes LRIG1 is large [median (mean) length of 12.05 (14.16) mb] relative to the LRIG1 locus (122 kb). The segmentation yielded a median (mean) number of probes/segment of 1183 (13). The distributions of segment lengths and probes are illustrated in Supplemental Figures 1A–B.
In the second case, we focused the location of our previous LRIG1 probe, which was positioned at 66,527,771–66,531,685 (intron 10 spanning exon 11). The MIP probes located at 66,515,866 and 66,532,949 were closest to the exons. To reduce noise generated from using single exon-flanking probes, we calculated CN from all evaluable probes 5’ and 3’ of exon 12. For these ‘intra-gene’ analyses, the median of the first five probes 5’ of exon 12 are designated LRIG11–5, and the median of the last six probes located 3’ of exon 13 are designated LRIG16–11 (Supplemental Figure 2). To account for the inherent noise, normal contamination, and possible effects of mosaisicm on CN determination, we applied a threshold for CN gain in LRIG1 as >2.3 and loss as <1.7, as reported previously. To determine the impact of the threshold selection on any observed associations, we conducted a series of sensitivity analyses across a range of CN thresholds for the determination of LRIG1 gain or loss. The application of stringent thresholds under an assumption of pure tumor cells produced the magnitude and direction of the association though our power was reduced. The results of the sensitivity analyses indicate that the associations were not sensitive to the selection of thresholds.
Statistical analyses.
We used Fisher’s exact test to examine the association between LRIG1 CN variation and ERBB2 status or intrinsic subtype. The Spearman’s rank correlation coefficient was used to assess the correlation between the logarithm of CN at the ERBB2 locus and the LRIG1 locus. We conducted the Kruskal-Wallis test for differences in LRIG1 CN across the different intrinsic subtypes and Cox proportional-hazards regression for survival analysis. For patient outcomes of any relapse (local and distant, n=252), a clinical-only multivariate Cox model was built. In the first step, all covariates shown in Table 1 were included; then stepwise selection was performed to minimize Akaike Information Criterion (AIC), which resulted in a reduced clinical model with diagnosis age, tumor size, and nodal status. These three selected covariates were included in multivariate Cox models with various outcomes (recurrence, distant metastasis, and overall survival).
Table 1.
Clinical characteristics of breast cancer patients (N = 971), by LRIG1 copy number imbalance status
| Characteristic* | Loss (%) | Normal (%) | Gain (%) | p value |
|---|---|---|---|---|
| No. of patients (n=971) | 86 (8.9) | 847 (87.2) | 38 (3.9) | |
| Median follow-up time (y) | 10.26 | 9.50 | 10.26 | |
| Age at diagnosis (y) | ||||
| Mean (SD) | 52.3 (11.8) | 54.5 (12.7) | 55.6(13.6) | |
| <50 (n=394) | 37 (9.4) | 343 (87.1) | 14 (3.6) | 0.88 |
| ≥50 (n=555) | 48 (8.6) | 485 (87.4) | 22 (4) | |
| Race/ethnicity | ||||
| Non-Hispanic white (n=715) | 55 (7.7) | 633 (88.5) | 27 (3.8) | 0.03 |
| Black (n=125) | 16 (12.8) | 100 (80) | 9 (7.2) | |
| Hispanic (n=123) | 15 (12.2) | 106 (86.2) | 2 (1.6) | |
| ER status | ||||
| Positive (n=666) | 49 (7.4) | 590 (88.6) | 27 (4.1) | 0.07 |
| Negative (n=293) | 35 (11.9) | 248 (84.6) | 10 (3.4) | |
| HER2 status | ||||
| Negative (n=768) | 61 (7.9) | 679 (88.4) | 28 (3.6) | 0.09 |
| Positive (n=203) | 25 (12.3) | 168 (82.8) | 10 (4.9) | |
| HER2/ER status | ||||
| HER2+/ER+ (n=115) | 15 (13.0) | 93 (80.9) | 7 (6.1) | 0.67 |
| HER2+/ER– (n=84) | 9 (10.7) | 72 (85.7) | 3 (3.6) | |
| Pathologic stage | ||||
| I (n=304) | 27 (8.9) | 268 (88.2) | 9 (3) | 0.61 |
| II (n=662) | 58 (8.8) | 575 (86.9) | 29 (4.4) | |
| Nuclear grade† | ||||
| I (n=92) | 9 (9.8) | 81 (88) | 2 (2.2) | 0.11 |
| II (n=477) | 35 (7.3) | 426 (89.3) | 16 (3.4) | |
| III (n=336) | 40 (11.9) | 279 (83) | 17 (5.1) | |
| Tumor size (cm) | ||||
| < 2 (n=566) | 48 (8.5) | 500 (88.3) | 18 (3.2) | 0.30 |
| ≥ 2 (n=369) | 36 (9.8) | 315 (85.4) | 18 (4.9) | |
| Lymph node status | ||||
| Negative (n=565) | 54 (9.6) | 489 (86.5) | 22 (3.9) | 0.74 |
| Positive (n=383) | 31 (8.1) | 338 (88.3) | 14 (3.7) | |
| Tumor subtype# | ||||
| Luminal A (n=373) | 18 (4.8) | 343 (92) | 12 (3.2) | 0.005 |
| Luminal B (n=145) | 14 (9.7) | 125 (86.2) | 6 (4.1) | |
| HER2+ (n=203) | 25 (12.3) | 168 (82.8) | 10 (4.9) | |
| TNBC (n=174) | 24 (13.8) | 144 (82.8) | 6 (3.4) | |
| Radiation therapy | ||||
| Yes (n=410) | 41 (10) | 354 (86.3) | 15 (3.7) | 0.63 |
| No (n=535) | 44 (8.2) | 470 (87.9) | 21 (3.9) | |
| Chemotherapy | ||||
| None (n=480) | 41 (8.5) | 420 (87.5) | 19 (4) | 0.94 |
| Anthracycline (n=323) | 30 (9.3) | 282 (87.3) | 11 (3.4) | |
| Anthracycline/taxane (n=114) | 8 (7) | 101 (88.6) | 5 (4.4) | |
| Endocrine therapy | ||||
| Yes (n=422) | 33 (7.8) | 368 (87.2) | 21 (5) | 0.16 |
| No (n=522) | 51 (9.8) | 456 (87.4) | 15 (2.9) |
Abbreviations: SD, standard deviation; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer.
Numbers do not add up to column totals due to missing values for the individual factors.
Tumor subtype was determined using ER, PR, Ki67, and HER2 as defined in Materials and Methods.
Nuclear grade was determined by the modified Black’s method.
RESULTS
LRIG1, patient and tumor characteristics.
Of 971 stage I/II breast tumors, 3.9% had LRIG1 gene-level gains, whereas 8.9% showed losses (Table 1). The overall distribution of LRIG1 CN status (loss, normal, or gain) differed significantly by tumor subtype (p=0.005) and by race/ethnicity (p=0.03). LRIG1 loss was more common in TNBC (13.8%) and HER2+ (12.3%) tumors than Luminal B (9.7%) and Luminal A (4.8%) tumors. Median LRIG1 CN differed significantly among the four subtypes (p<0.0001; Supplemental Figure 3). Furthermore, CN loss was more prevalent in black (12.8%) and Hispanic (12.2%) cases than NHWs (7.7%); distribution reflects a higher proportion of TNBC and HER2+ tumors in these patients (Supplemental Table 1). There were no overall differences in LRIG1 CN status by tumor stage, diagnosis age, nuclear grade, tumor size, lymph node status, or treatment (yes/no or type; Table 1). Further, unlike previous smaller studies of more-advanced tumors, CN gains in the LRIG1 gene were not significantly associated with tumor subtypes, including HER2+ tumors.
LRIG1 CN 5’ and 3’ to exon 12 and clinicopathologic characteristics.
We previously hypothesized a putative breakpoint in LRIG1 with gain of exon 11 co-incident with ERRB2 amplification(13). Using MIP data in the LRIG1 locus, we conducted two statistical tests comparing log-CN values of two regions for all possible 11-probe partitions. Both tests produced a minimum p-value for comparing the regions 5’ versus 3’ to exon 12 independent of any a priori fitting (p=2.04−46 and 3.36−49 for a Wilcoxon rank-sum test and a t-test, respectively). These results support our prior hypothesis of a common breakpoint in LRIG1 occurring near the exon 12/13 junction.
Overall, partitioned data of the smaller intra-gene events in tumors reflect those using the whole probe set (Supplemental Table 2). For the loci 5’ of exon 12 (LRIG11–5), 20.9% of cases had gains and 14.9% showed loss. Losses in the 5’ region were significantly more common in black (20.8%) and Hispanic (20.3%) cases than in NHW patients (13%). Gains in the region did not differ significantly by patient or tumor characteristics with the exception that gains of LRIG11–5 were ~3-fold higher in ER+/HER2+ tumors than ER–/HER2+ tumors (27.8% versus 11.9%, respectively, p=0.008). Events at the 3’ partitioned loci (LRIG16–11) were slightly less frequent than those occurring 5’ to exon 12 with 10.0 and 19.8% of tumors showing gains or losses, respectively. CN in the region 3’ of exon 12 showed significant differences by race/ethnicity, ER status, and treatment. Specifically, loss of this region was more common among black (28%) and Hispanic (22%) cases than NHW (18.2%) (p=0.04). Gains in this region were more common in ER+ (11.3%) than ER– (7.5%) tumors (p = 0.08), with losses more common in ER– than ER+ tumors (23.9 versus 18.0%, respectively p=0.04).
LRIG1 CN and relapse risk.
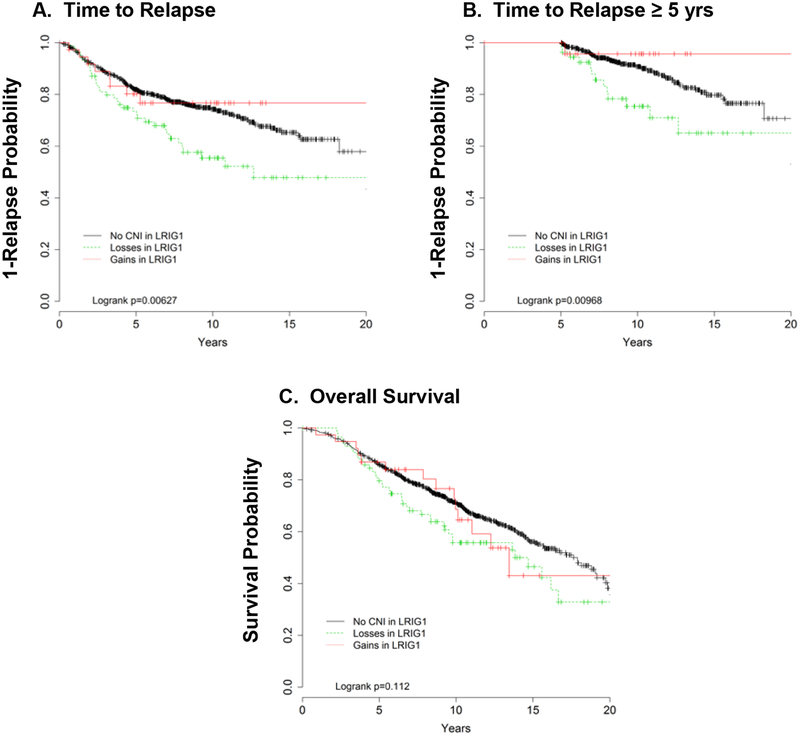
In univariate analyses, LRIG1 loss was significantly associated with time-to-relapse, with a non-significant trend for lower overall survival (Figure 1A), for both early (<5 y) and late (≥5 y) events (Figure 1B). The 5 year cut-point was selected based on the clinical relevance of this time point for patients and the arbitrary consideration of ‘late’ recurrences being those that occur after 5 years. For example, when we excluded recurrences that occurred <5 y after diagnosis, the probability of relapse was 46% in patients with LRIG1 loss compared with only 28% in patients with normal CN. However, when we applied the scaled Schoenfeld residuals, plotted against time, there was no statistically significant time trend of the effect of LRIG1 gain or loss (p values for linear trend = 0.23 for losses). If anything, we find that LRIG1 loss shows the biggest effect between 2 and 7 years from diagnosis and remains independent of the other clinicopathological characteristics. When adjusted for diagnosis age, tumor size, and nodal status associated relapse risk, LRIG1 loss was significantly associated with recurrence (HR, 1.91; 95% CI, 1.33–2.74), distant metastasis (2.10, 95% CI, 1.43–3.09) and overall mortality (HR, 1.55; 95% CI, 1.11–2.16; Table 2). The increased hazard remained even after forcing in treatment and tumor subtype (data not shown). While the effect of LRIG1 loss was generally similar in treated and untreated cases, risk of relapse was stronger and more immediate in untreated patients (Supplemental Table 3). However, we observed no association between LRIG1 gain and relapse or overall mortality (data not shown).
Figure 1.
Kaplan Meier curves for unadjusted analyses of time to relapse and overall survival by LRIG1 tumor copy number.
Table 2.
Multivariate Cox proportional hazards models for risk of recurrence, distant metastasis and overall survival for copy number imbalance in LRIG1
| Patient or tumor characteristic | Any Recurrence | Distant Metastasis | Overall Survival | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | HR (95% CI) | p value | |
| Age, diagnosis (y) | ||||||
| < 50 | Reference | Reference | Reference | |||
| ≥ 50 | 1.60 (1.24–2.07) | 0.0003 | 1.58 (1.19–2.09) | 0.002 | 0.69 (0.55–0.87) | 0.002 |
| Tumor size (cm) | ||||||
| < 2 | Reference | Reference | Reference | |||
| ≥ 2 | 1.71 (1.33–2.22) | < 0.0001 | 1.99 (1.50–2.64) | < 0.0001 | 1.59 (1.28–1.98) | < 0.0001 |
| Lymph node | ||||||
| Negative | Reference | Reference | Reference | |||
| Positive | 1.62 (1.26–2.10) | 0.0002 | 1.85 (1.39–2.46) | < 0.0001 | 1.39 (1.12–1.74) | 0.003 |
| LRIG1 | ||||||
| Copy normal | Reference | Reference | Reference | |||
| Loss | 1.91 (1.33–2.74) | 0.0004 | 2.10 (1.43–3.09) | < 0.0001 | 1.55 (1.11–2.16) | 0.009 |
| Gain | 0.87 (0.43–1.78) | 0.71 | 0.94 (0.44–2.01) | 0.87 | 0.97 (0.56–1.71) | 0.93 |
Abbreviations: HR, hazard ratio; CI, confidence interval
Note: The clinical covariates shown above were chosen using a stepwise selection procedure to minimize Akaike Information Criterion.
Chromosome 3 CN analyses support LRIG1 as a major driver event at 3p12–21.
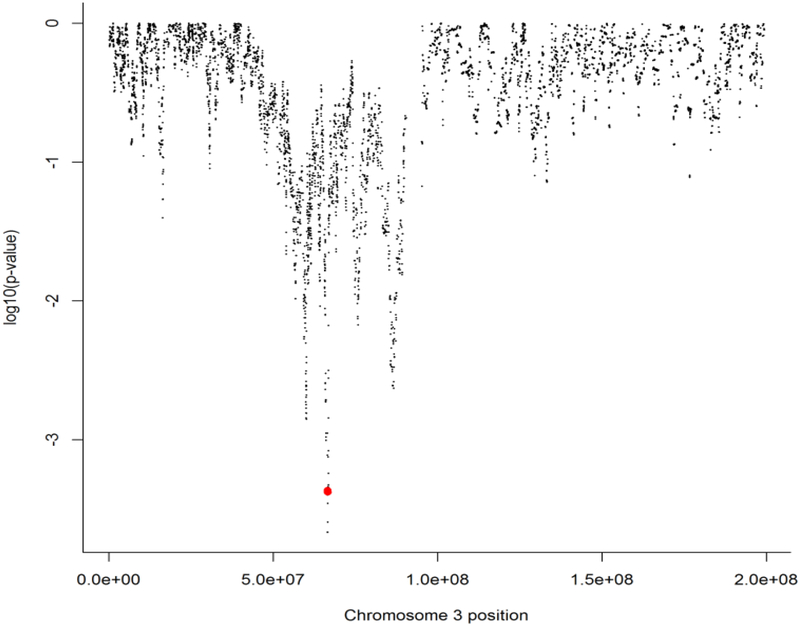
Gains and losses at chromosome 3p12–21 are common and heterogeneous across breast tumors. To investigate the contribution of LRIG1 to the associations found for chromosome 3 overall, we first generated a high-resolution noise-reduced profile for each sample, applying the quantification method used for LRIG1 in a priori hypothesis testing to the rest of chromosome 3, and assigning a CN value to each chromosome 3 probe (n=17,557) for each sample. Since we were interested in LRIG1 CN losses, we conservatively designated loss for each probe of the smoothed data as CN <–1.7 and fit a Cox model adjusted for age, tumor size, and nodal status. We calculated log10 p-values for the association of loss with relapse across chromosome 3 and a small (178 kb) region that includes LRIG1 and SLC25A26 (cytogenic band at 3p14), which is positively associated with disease recurrence (Figures 2A–B). In addition, we computed the average overlap with LRIG1 for each probe (Supplemental Figure 4). The only probes showing low p-values for association with recurrence were those with high LRIG1 overlap. We also used percent agreement of the loss/no-loss calls for each probe with the LRIG1 loss/no-loss calls and found that the few probes with very high (>99%) correlation with LRIG1 loss showed small p-values for recurrence, and all probes belonged to either LRIG1 or SLC25A26.
Figure 2.
The log10 p-values for association with any recurrence across chromosome 3. A small region that includes LRIG1 (red dots) on the short arm of chromosome 3 is associated with recurrence. The length of the region is narrow and it only includes LRIG1 and its close neighbor SLC25A26.
Association between LRIG1 loss and recurrence is independent of loss at fragile histidine triad.
Deletions on 3p are thought to arise as a consequence of frequent breaks due to the presence of a common fragile site encompassed by FHIT (fragile histidine triad), a set of highly unstable genomic regions at 3p14.2(13, 14) FHIT is abnormally transcribed in 30% of breast tumors(18) and has been associated with poor outcomes(19). In our dataset, FHIT loss was highly correlated with LRIG1 loss (R2=0.55; p<0.0001). In order to eliminate FHIT as an explanation for our observed associations between LRIG1 CN and patient outcomes, we conducted a multivariate model adjusted for FHIT loss; LRIG1 loss remained significantly associated with relapse risk (HR, 2.01; 95% CI, 1.15–3.52; p=0.01).
LRIG1 gene expression and patient outcomes.
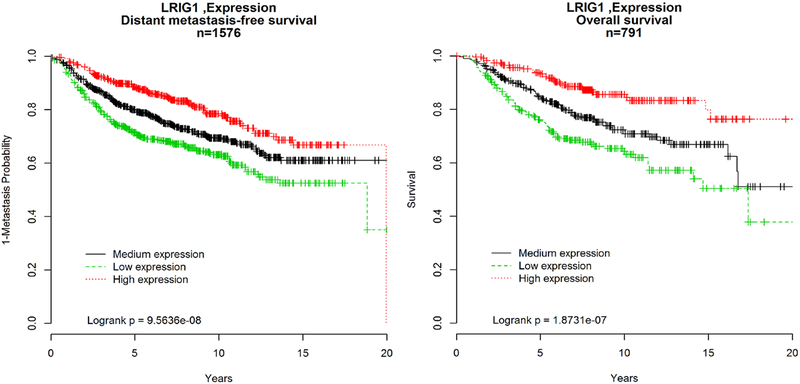
For further evidence that LRIG1 CN may explain the previous association between losses of 3p12–121 and worse patient outcomes (5), we evaluated 18 publically available gene expression datasets with breast cancer outcomes, yielding 1576 samples with information on distant metastasis-free survival (DMFS) and 791 with overall survival (Supplemental Methods). The risk of DMFS and death were significantly higher in breast cancer patients whose tumors expressed low LRIG1 levels compared with medium or high expression (Figure 3). Importantly, as with the CN analyses, the risk persists beyond 5 y after diagnosis. Further, to rule out a contribution from SLC25A26, we tested the association between SLC25A26 gene expression and patient outcomes using publically available data. Though the sample set is smaller due to differences in probe availability of platforms for SLC25A26, unlike its neighbor LRIG1, we observed no evidence for an association between SLC25A26 expression level and DMFS or overall mortality (Supplemental Figure 5). In contrast, low LRIG1expression levels were positively associated with higher rates of distant metastasis and mortality, even in the smaller sample set.
Figure 3.
Kaplan Meier curves for unadjusted analyses of A. time to distant metastasis free survival and B. overall survival by level of LRIG1 gene expression. --- low, --- medium, --- high.
DISCUSSION
Our findings support those of Staaf et al.(15), which show that genomic loss near 3p13 in breast tumors is an independent risk factor for relapse and poor survival. Our results strongly implicate LRIG1 loss as the major driver event in this region, with localized deletion or low LRIG1 gene expression significantly associated with distant metastasis and breast cancer death. Importantly, the increased hazard persists in these patients beyond 5 y post-diagnosis, suggesting the loss may be a strong indicator of late events in otherwise low-risk patients.
The higher frequency of loss in black and Hispanic women was positively associated with higher LRIG1 loss in TNBC, HER2+, and Luminal B tumors; subtypes that are disproportionally higher in these patients (Supplemental Table 2). In contrast to our previous report(13), we observed no significant association between LRIG1-specific CN imbalances and ERBB2 amplification, though specific probe losses were significantly more common among ER– tumors, and gains were more common in ER+ disease. Among HER2+ tumors, gains in LRIG1 were more common among ER+/HER2+ than HER2+/ER– tumors, consistent with an overall higher frequency of gains in ER+ versus ER– tumors and losses in ER– versus ER+ tumors. Given the complex pattern of splicing in LRIG1, additional analyses of expressed transcripts may provide further insight and risk stratification among patient populations.
Our data confirm previous reports of amplification in the LRIG1 gene region(9); however, we were unable to replicate our prior observation(13) of concomitant gains in LRIG1 and ERBB2 amplification. Similar to Krig et al.(14), we observed a significant association between gains and ER+ tumors and non-significantly better patient outcomes (data not shown). Our findings support those of Miller et al.(4), which suggest LRIG1 loss is more common in tumor subtypes with disturbances in ErbB family members (i.e., ERBB2 amplification in ER–/HER2+ tumors and EGFR in TNBCs). Analyses of probes flanking exon 12/13 favor our original hypothesis for a breakpoint in the gene, possibly reflecting localized fragility involving FHIT that destabilizes the region (76 of 86 tumors bearing LRIG1 loss also show FHIT loss).
LRIG1 protein has been shown to oppose MET synergy with HER2 in cellular invasion and to negatively regulate other oncogenic receptor tyrosine kinases in the ErbB family, including EGFR, HER3 and 4, MET, and RET(4). These functional data support a key role of LRIG1 as a tumor suppressor gene important in limiting tumor invasion, a putative mechanism that aligns with our finding of greater metastasis risk in cases with LRIG1 loss.
In our previous study, we showed increased CN at exon 12/13 in LRIG1 in 34% of 73 breast tumors (12, 13). Moreover, we observed co-incident ERBB2 gene amplification in tumors with gain of exon 12/13 probe signal. In contrast, LRIG1 expression was reported to be down-regulated in HER2-overexpressing tumors(4). There are several possible explanations for differences between our earlier results and the present study. First, the Oncoscan array did not provide any probes at the exact genomic location of our previous FISH probe and confidence with few MIP probes is limited, thus the results are not directly comparable. Second, the prior results analyzed a smaller and more selected set of cases including patients with larger and more-advanced tumors.
In conclusion, our results provide strong evidence that LRIG1 is the tumor suppressor gene on chromosome 3 near p13–14 whose loss is a critical driver event in breast cancer metastasis that is independent of stage and tumor subtype. Though LRIG1 loss is proportionally higher in TNBC and HER2+ tumors, loss in otherwise low-risk cases (e.g., Luminal A) may partly explain late relapse events in these patients. Along with efforts to understand the mechanistic impact of LRIG1 loss on degradation of ErbB family members and control of tumor stem cells in the breast, prospective efforts that combine gene expression and CN determination of LRIG1 are needed to confirm the clinical value of LRIG1 expression status for patient-risk stratification.
Supplementary Material
Contributor Information
Patricia A. Thompson, Department of Cellular and Molecular Medicine, The University of Arizona Cancer Center, 1515 Campbell Avenue, Tucson, Arizona 85724.
Ingrid Ljuslinder, Department of Radiation Sciences, Umeå University, Sweden.
Spyros Tsavachidis, Dan L. Duncan Center Center, Baylor College of Medicine
Abenaa Brewster, Department of Clinical Cancer Prevention, The University of Texas MD Anderson Cancer Center
Aysegul Sahin, Department of Pathology, The University of Texas MD Anderson Cancer Center
Håkan Hedman, Department of Radiation Sciences, Umeå University, Sweden
Roger Henriksson, Department of Radiation Sciences, Umeå University, Sweden
Melissa L. Bondy, Department of Pediatrics, Baylor College of Medicine, One Baylor Plaza, MS:305, Houston, TX 77030.
Beatrice S. Melin, Department of Radiation Sciences, Umeå University, Sweden.
REFERENCES
- 1.Nilsson J, Vallbo C, Guo D, Golovleva I, Hallberg B, Henriksson R, et al. Cloning, characterization, and expression of human LIG1. Biochem Biophys Res Commun 2001;284(5):1155–61. [DOI] [PubMed] [Google Scholar]
- 2.Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J 2004;23(16):3270–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL 3rd, et al. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem 2004;279(45):47050–6. [DOI] [PubMed] [Google Scholar]
- 4.Miller JK, Shattuck DL, Ingalla EQ, Yen L, Borowsky AD, Young LJ, et al. Suppression of the negative regulator LRIG1 contributes to ErbB2 overexpression in breast cancer. Cancer Res 2008;68(20):8286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci 2008;28(1):39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nilsson J, Starefeldt A, Henriksson R, Hedman H. LRIG1 protein in human cells and tissues. Cell Tissue Res 2003;312(1):65–71. [DOI] [PubMed] [Google Scholar]
- 7.Wong VW, Stange DE, Page ME, Buczacki S, Wabik A, Itami S, et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol 2012;14(4):401–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 2012;149(1):146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ordonez-Moran P, Huelsken J. Lrig1: a new master regulator of epithelial stem cells. EMBO J 2012;31(9):2064–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, et al. Lrig1 Expression Defines a Distinct Multipotent Stem Cell Population in Mammalian Epidermis. Cell Stem Cell 2009;4(5):427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki Y, Miura H, Tanemura A, Kobayashi K, Kondoh G, Sano S, et al. Targeted disruption of LIG-1 gene results in psoriasiform epidermal hyperplasia. FEBS Letters 2002;521(1–3):67–71. [DOI] [PubMed] [Google Scholar]
- 12.Ljuslinder I, Malmer B, Golovleva I, Thomasson M, Grankvist K, Hockenstrom T, et al. Increased copy number at 3p14 in breast cancer. Breast Cancer Res 2005;7(5):R719–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ljuslinder I, Golovleva I, Henriksson R, Grankvist K, Malmer B, Hedman H. Co-incidental increase in gene copy number of ERBB2 and LRIG1 in breast cancer. Breast Cancer Res 2009;11(3):403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krig SR, Frietze S, Simion C, Miller JK, Fry WHD, Rafidi H, et al. Lrig1 Is an Estrogen-Regulated Growth Suppressor and Correlates with Longer Relapse-Free Survival in ERα-Positive Breast Cancer. Molecular Cancer Research 2011;9(10):1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staaf J, Jonsson G, Ringner M, Vallon-Christersson J, Grabau D, Arason A, et al. High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer. Breast Cancer Research 2012;12(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brewster AM, Do KA, Thompson PA, Hahn KM, Sahin AA, Cao Y, et al. Relationship between epidemiologic risk factors and breast cancer recurrence. J Clin Oncol 2007;25(28):4438–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson PA, Brewster AM, Kim-Anh D, Baladandayuthapani V, Broom BM, Edgerton ME, et al. Selective Genomic Copy Number Imbalances and Probability of Recurrence in Early-Stage Breast Cancer. PLoS ONE 2011;6(8):e23543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pekarsky Y, Palamarchuk A, Huebner K, Croce CM. FHIT as Tumor Suppressor: Mechanisms and Therapeutic Opportunities. Cancer Biology & Therapy 2002;1(3):232–236. [DOI] [PubMed] [Google Scholar]
- 19.Ginestier C, Bardou V-J, Popovici C, Charafe-Jauffret E, Bertucci F, Geneix J, et al. Loss of FHIT protein expression is a marker of adverse evolution in good prognosis localized breast cancer. International Journal of Cancer 2003;107(5):854–862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.