Abstract
In various malaria-endemic regions, the appearance of resistance has precluded the use of pyrimidine-based antifolate drugs. Here, a three-step fragment screening was used to identify new non-pyrimidine Plasmodium falciparum dihydrofolate reductase (PfDHFR) inhibitors. Starting from a 1163-fragment commercial library, a two-step differential scanning fluorimetry screen identified 75 primary fragment hits. Subsequent enzyme inhibition assay identified 11 fragments displaying IC50 in the 28-695 μM range and selectivity for PfDHFR. In addition to the known pyrimidine, three new anti-PfDHFR chemotypes were identified. Fragments from each chemotype were successfully co-crystallized with PfDHFR, revealing a binding in the active site, in the vicinity of catalytic residues, which was confirmed by molecular docking on all fragment hits. Finally, comparison with similar non-hit fragments provides preliminary input on available growth vectors for future drug development.
Keywords: Malaria, Plasmodium falciparum, fragment-based screening, dihydrofolate reductase, small molecule inhibitors
Introduction
In the fight against malaria, antifolates were once regarded as safe, efficient drugs against the Plasmodium spp. parasite. However, the appearance of resistance-inducing mutations in the folate biosynthesis enzymes such as dihydrofolate reductase (DHFR) and dihydropteroate synthase (DHPS) in the parasite has precluded the use of the existing antifolate drugs1. Although the recent anti-malarial drugs are geared towards exploiting different drug targets, such as P-type Na+-ATPases transporter (PfATP4), V-type H+-ATPase transporter, Phosphatidylinositol 4-kinase (PfPI4K) or dihydroorotate dehydrogenase (PfDHODH)2, PfDHFR remains attractive in light of the fact that known three-dimensional structures enable rational drug design against the WT and mutant parasite strains3–5.
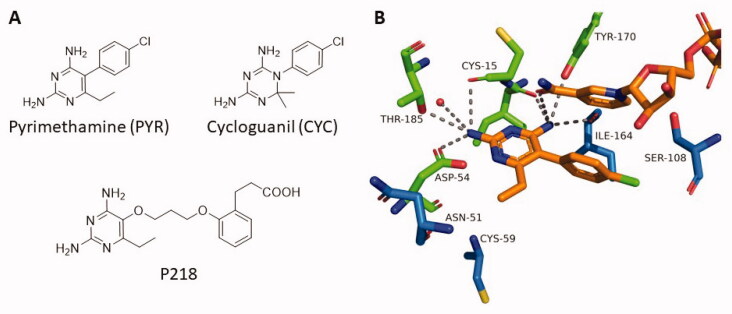
Historically, antifolates have been developed by mimicking the enzymes natural substrates. As such, the two DHFR-targeting drugs pyrimethamine (PYR) and cycloguanil (CYC) share a similar 2,4-diamino pyrimidine/triazine scaffold mimicking the 2-amino 4-oxo-pteridine core of DHF (Figure 1(A)). A series of PfDHFR co-crystal structures with antifolate derivatives (e.g., PDB 3QGT6, 3UM87, 1J3K4, 4DP38) generated by our team have enabled elucidation of the role of each functional group in ligand binding (Figure 1). This combination of interactions makes of these antifolates excellent WT PfDHFR inhibitors, with Ki in the (sub-) nanomolar range. However, their efficiency dramatically decreases against PfDHFR variants carrying S108N mutation such as the double mutant (DM) (C59R + S108N) and quadruple mutant (QM) (DM + N51I + I164L) PfDHFR due to steric clash of the pCl-phenyl of PYR and CYC with S108N4.
Figure 1.
(A) Structure of some Plasmodium antifolates. (B) Binding mode of PYR in WT PfDHFR (PDB 3QGT). Resistance-induced mutated residues appear in blue.
In 2012, our group discovered the compound P218 which displays nanomolar range in vitro activities for P. falciparum strains carrying WT and QM PfDHFR8. The compound was recently found to display favourable safety, tolerability and pharmacokinetics in first-in-human clinical trial9. P218 was developed through the combination of the 2,4-diaminopyrimidine moiety, with a flexible linker carrying a phenyl ethylcarboxylate group, taking advantage of the conserved R122 to give P218 a second distant anchoring point (Figure 1(A)). The design of P218 demonstrated the utility of new chemotypes to target specific residues in the PfDHFR active site. By combining a careful deconstruction of long-known inhibitors with the incorporation of new chemotypes, it became possible to develop new-generation antifolates that by-pass the drug resistance hurdle.
In the last decade, the fragment-based screening (FBS) strategy has emerged as a powerful method to identify new chemotypes for drug discovery and development. Techniques used in FBS, which typically include differential scanning fluorimetry (DSF), surface plasmon resonance (SPR), NMR and X-ray crystallography10,11 are simple, robust and reproducible. FBS strength resides in an efficient sampling of the chemical space, as even small fragment libraries can encompass high levels of diversity. Interactions between fragment hits and their protein targets are also of higher quality, which eases downstream lead molecule development and optimisation. For these reasons, FBS is widely used in both pharmaceutical industry and academia10. Although it has been successfully applied to other infectious diseases12–16, FBS has only been scarcely used against malaria drug targets17,18.
Following this idea, we have employed the FBS strategy to identify new chemotypes for inhibitor development against WT PfDHFR. Upon PYR and CYC drug pressure, multiple mutations of PfDHFR have appeared to preclude the binding of these inhibitors, decreasing their binding affinity for QM by several hundred to thousand folds as compared to WT. Several examples have shown that, using the same chemotypes, new inhibitors (such as m-Cl derivatives of PYR and CYC, WR99210 and P218) could be rationally designed to avoid steric clash with S108N and display similar binding affinity for WT and QM enzymes4,8,19–21. Following this idea, we proposed to identify new active fragments that could interact with both WT and QM. Using WT PfDHFR has practical advantages, as it is more stable, well-characterised, and easier to target with low-affinity fragments compared to the mutant enzymes. For these reasons, we chose to focus our efforts on the fragment-based high-throughput screening of WT PfDHFR. Once the fragment hits identified and characterised, the structural specificities of both WT and QM variants can be integrated in the drug design process, using the extensive structural information available to design larger drug candidates4,6–8.
Here, a commercial fragment library was screened using differential scanning fluorimetry (DSF) and fragment hits were further validated by enzymatic assay and X-ray crystallography to identify several active fragments against WT PfDHFR.
Materials and methods
Chemicals
SYPRO Orange was purchased from Invitrogen and PYR and NADPH were purchased from Sigma Aldrich. DHA22, P2188, P3023, P4519, P2423, L48 and L5 (manuscript in preparation) were synthesised following previously described procedures. Compounds were numbered as follows: the P series corresponds to previously reported PfDHFR inhibitor prototypes developed by our group. The L compounds are reaction intermediates that were used here as controls. Commercial fragments were numbered following the random order provided by the supplier.
Fragment library selection and preparation
The BIONET Premium fragment library was purchased from Key Organics, containing 1163 fragments. Fragments were received as dried powder/oils, and stock solutions were prepared by dissolving ca. 3 mg in DMSO to 200 mM. For primary screening, quadruplex fragment mixtures were prepared by mixing equal volumes of four fragments, providing 50 mM solutions. For secondary screening of hit mixtures, individual fragment solutions were prepared by four-fold dilutions of the 200 mM stock in DMSO. For fragment hits, pKa values were predicted using MarvinSketch 20.13.
Protein expression and purification
PfDHFR and HsDHFR expression was completed following a previously published procedure24. In brief, E. coli BL21(DE3) pLysS cells carrying the plasmid encoding for WT-PfDHFR and HsDHFR, respectively, were grown at 37 °C until reaching an OD600 ca. 0.8. Protein expression was induced by addition of 0.4 mM IPTG and cells were cultured overnight at 20 °C. Cells were harvested and cell pellet was stored at −20 °C. For purification, cells were thawed on ice and lysed by French Press. Lysate was then clarified by centrifugation and applied to a methotrexate (MTX) affinity column. The column was washed overnight, and protein was eluted using dihydrofolate (DHF). In a second purification step, protein was concentrated and applied to a Q-Sepharose ion-exchange column. Protein was eluted using a gradient of KCl. Fractions containing protein were concentrated and their purity was assessed by SDS-PAGE. Protein concentration was measured using Bradford assay.
Differential scanning fluorimetry
Prior to screening, DSF parameters such as protein and SYPRO Orange concentration, and buffer composition and heating rate were optimised to give the best-defined thermal denaturation peak. The most suitable conditions were found to be 10 μM PfDHFR, 8x SYPRO Orange in Phosphate buffer pH 7.2 20 mM containing 50 mM KCl, 0.1 mM EDTA and 20% glycerol.
In a typical DSF experiment, a master mix containing buffer, PfDHFR and 8x SYPRO Orange dye were prepared in a microcentrifuge tube. Ligands of interest were dispensed in low-profile 96-well plates (Bio-Rad) to 1 mM final concentration, and the master mix was dispensed to a final volume of 50 μL per well. In these conditions, DMSO content was maintained constant at 2%. The microplate was sealed with adhesive film and mixed by shaking for 2 min at 800 rpm at RT. The microplate was then submitted to a DSF run on a CFX96 RT-PCR (Bio-Rad). DSF program was designed by starting with a 3 min equilibration phase at 30 °C, followed by a temperature gradient of 1 °C/min, recording fluorescence every 0.5 °C. Fluorescence was recorded using the FRET channel (λexc = 450–490 nm, λem = 560–580 nm). Curves were fitted using the Precision Melt Analysis software (Bio-Rad).
For the primary screening, DSF chromatograms were recorded for PfDHFR in the presence of quadruplex fragment mixtures containing 4 fragments at 50 mM each. The experiment was set as described above, using 1 μL quadruplex mixtures in a final volume of 50 μL (1 mM final fragment concentration). For each experiment, a DMSO negative control was included. Every experiment was run in triplicate and the average ΔTm was considered. The standard deviation of the negative control was calculated and a threshold of twice the standard deviation was applied to define hits.
For secondary screening of each hit mixture obtained from the quadruplex screen, individual fragment stocks were prepared to a final concentration of 50 mM in DMSO. DSF experiments were then run for each individual fragment as described above. For fragments displaying a significant ΔTm, the experiment was run in triplicate. The standard deviation of the negative control was calculated and a threshold of twice the standard deviation was applied to define hits.
PfDHFR activity assay
PfDHFR activity was assessed following a previously published procedure24. In brief, a master mix was prepared by mixing DHF and NADPH in activity buffer (Tris pH 7.2 50 mM, β-mercaptoethanol 75 mM, BSA 1 mg/mL) to a final concentration of 100 μM. In a 96-well plate, 2 μL of fragments in DMSO were dispensed, followed by 178 μL of the master mix. The reaction was initiated by addition of 20 μL of enzyme followed by immediate mixing. NADPH consumption was monitored by recording absorbance at 340 nm for 80 s. Curve was fitted using a linear regression function and % activity was calculated by comparison with a DMSO control. For IC50 determination, data points corresponding to variable inhibitor concentrations were plotted in a semi-logarithmic scale, and curve was fitted using the Hill equation.
Molecular docking
Molecular docking experiments were conducted using Autodock 4 program. PDB file 3QGT was edited to remove water molecules and co-crystallized PYR ligand. The protein receptor file was then generated using Autodock Tools 1.5.6. Ligand files were obtained by drawing 2 D structure using ChemDraw software, and 3D structure was built and energy-minimized using Avogadro software Universal Force Field. Area of interest was defined using a grid encompassing the pterin binding pocket of the PfDHFR active site. Rigid molecular docking was performed using Autodock 425 using default parameters. Results were processed by clustering data and considering the lowest energy cluster and/or the most populated cluster. Structural graphics were drawn with PyMOL26.
X-ray crystallography
Co-crystallization of purified WT PfDHFR-TS (15 mg/mL) with 2–5 mM each of the compound of interest, NADPH and dUMP was performed using a previously published procedure [4]. Data were collected using SC XRD series: D8 venture Bruker at NSTDA Characterisation and Testing Service Centre (NCTC). Data were processed using PROTEUM3 software. Structure model was built by MOLREP27, using PDB 1J3I as a template and refined by REFMAC528 in CCP4. The model building was done using COOT29. Structural graphics were drawn with PyMOL26. Co-crystal structures with fragments 263, 820 and 148 were deposited in the PDB database under accession numbers 7CTY, 7CTW and 7CTZ.
Results
Validation of DSF for PfDHFR inhibitor screening
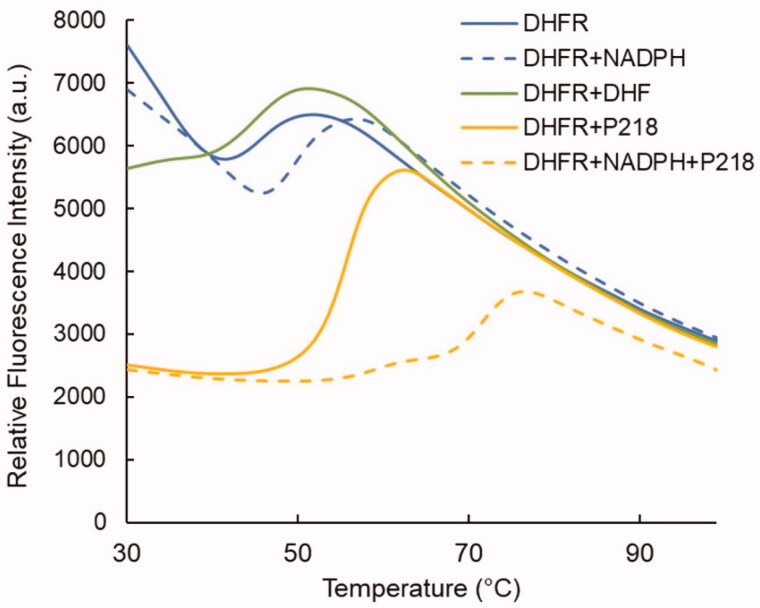
Among the available primary fragment screening methods, DSF was selected for its low cost and ease of use30. In order to validate its efficacy to detect PfDHFR binders, DSF was first tested on DHFR substrates, as well as a series of known inhibitors. While dihydrofolate (DHF), and tetrahydrofolate (THF) showed minor melting temperature variations, NADPH showed a notable +4 °C stabilisation (Figure 2). This result was further confirmed in the presence of inhibitors. When tested on the apo PfDHFR, the P218 inhibitor displayed a + 10 °C stabilising effect. This effect increases to +23.1 °C in the presence of NADPH. The same effect is observed with other pyrimidine-based inhibitors and confirms that NADPH is needed to pre-organize the pterin binding site, so that inhibitors can bind in their most favourable configuration, as reported for DHFR from other organisms31,32.
Figure 2.
DSF curves obtained for PfDHFR in the presence of its substrate, cofactor, and P218 inhibitor.
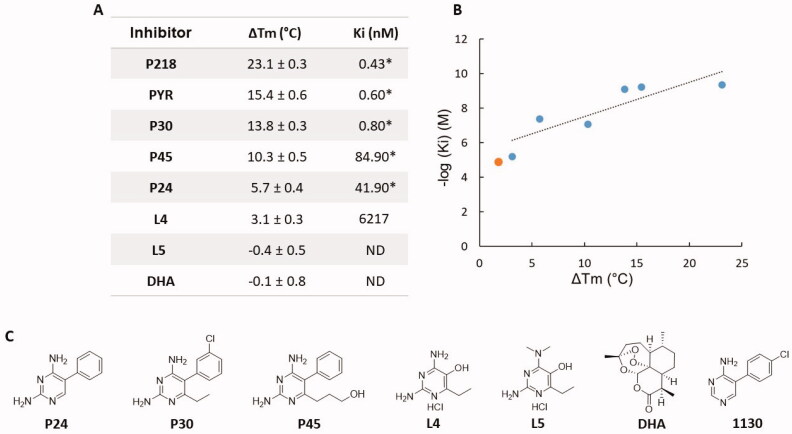
As a preliminary assessment of DSF for PfDHFR ligand detection, PfDHFR inhibitors displaying variable activity were assayed in the presence of NADPH. All inhibitors displayed measurable ΔTm, except for L5 (synthetic precursor of the 2,4-diaminopyrimidine series). Dihydroartemisinin (DHA) was added as a negative control, as an antimalarial that does not target folate biosynthesis enzymes. ΔTm results obtained for the different inhibitors appear in Figure 3. Although the correlation between the ΔTm and the Ki values is not strictly linear (r2 = 0.756, Figure 3(B)), a consistent trend is observed. This is consistent with the complex nature of inhibitor binding, as the sum of variable enthalpic vs. entropic contributions that results in an apparent stabilising/destabilizing effect33. In this view, it is interesting to note that when comparing P218 and PYR, the presence of the carboxylate chelating group does not involve a drastic change of Ki, while a strong structure stabilising effect is observed by DSF with ΔTm of 23.1 vs 15.1 °C for P218 and PYR, respectively. The technique also shows sensitivity to small structural changes in the pyrimidine-based inhibitor series. This result demonstrates the ability of DSF to detect fragments with a Ki as high as micromolar (L4). However, as ΔTm is a function of the enthalpic vs. entropic nature of the binding, and Ki represents completion in binding between substrate and inhibitor, one cannot extrapolate this to all chemotypes33.
Figure 3.
(A) ΔTm and inhibition constants of the different inhibitors tested. Data noted * are from8,19. (B) Correlation between ΔTm and inhibition constants of the inhibitors (blue). Linear trendline appears as black dashed line (r2 = 0.756). Fragment 1130 (orange) was added for comparison. (C) Structure of the inhibitors. ND: Not detected.
Fragment library screening
A DSF primary screening of PfDHFR was undertaken using the BIONET Premium library containing 1163 fragments. This library was chosen over other commercial ones as it is rule-of-three compliant and was curated from any PAINS and aggregating compounds. All fragments are also soluble at 200 mM in DMSO and display high diversity.
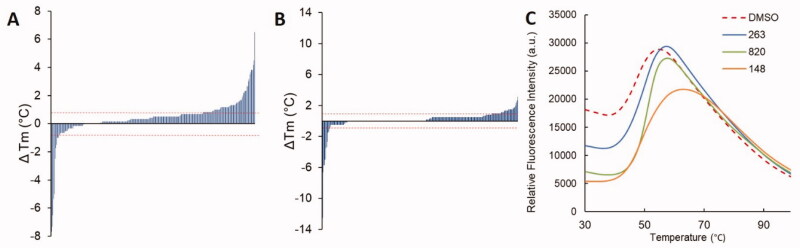
Fragments were first screened against PfDHFR as mixtures of 4 at 1 mM in the presence of NADPH (Figure 4(A)). In these conditions, a Tm of 47.26 ± 0.4 °C was measured for PfDHFR in the presence of DMSO as a control. Considering the multiple thermodynamic factors influencing the ΔTm, it seems virtually impossible to apply a definitive threshold that would encompass all potential fragment binders. We thus decided, as others before us, to consider our hit threshold as twice the standard deviation of the DMSO control, relying on the experimental technique limitations. Fragments were thus considered as hits when their individual ΔTm exceeded ± 0.8 °C. Out of 291 quadruplex mixtures tested, 87 exceeded the set criterion, with ΔTm ranging from −7.66 °C to +6.5 °C. The fragments composing these mixtures were thus examined individually, affording a total of 75 fragment hits (6.4% hit rate) with ΔTm spanning between −12.5 °C and +5.2 °C (Figure 4(B)). Among these, 14 fragments displayed a negative Tm. The overall 6.4% hit rate matches the results obtained in typical fragment screening campaigns.
Figure 4.
Distributions of fragment hits ΔTm for quadruplex mixtures screening (A) and individual fragment screening (B). The ± 0.8 °C threshold applied appears as red dashed lines. C. Representative examples of DSF curves obtained.
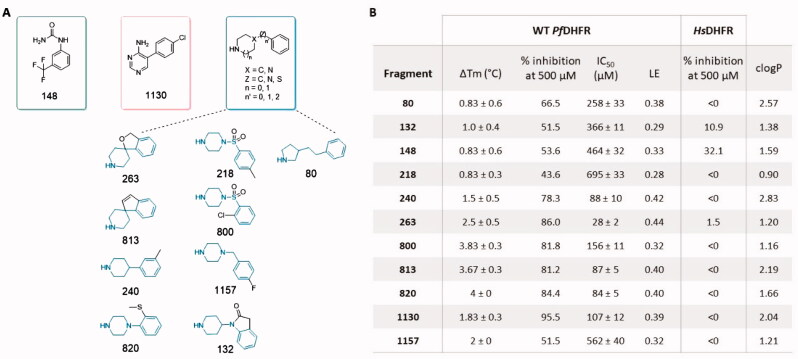
In order to validate these primary hits, the 75 primary fragment hits were subjected to a DHFR inhibition assay. As shown in Figure 5, 11 fragments displayed 43–95% PfDHFR inhibition at 500 μM (i.e., 0.8% of the library). These fragments were grouped by tentative chemotypes.
Figure 5.
(A) Fragment hits structures grouped by tentative chemotypes. (B) Corresponding experimental data obtained for the fragment hits. clogP values were calculated using DataWarrior43. LE: Ligand Efficiency.
The fragment hits found display IC50 values in the 28 − 695 µM range. Among the 11 active fragments, 5 were found to inhibit WT PfDHFR with 78 − 84% inhibition at 500 µM, with corresponding IC50 values of 28 − 156 µM. Fragment 263 exhibited the best overall inhibition, while fragment 1130 showed the best inhibition at 500 µM. Little correlation was observed between the ΔTm and the ability to inhibit the enzyme (ri2 = 0.41). As such, fragments 80, 148 and 218 all display a ΔTm of 0.83 °C, close to the limit of detection, while showing enzyme inhibition varying by several folds (IC50 PfDHFR of 258, 464 and 695 µM, respectively). All the active fragment hits display a positive ΔTm. In order to evaluate enzyme selectivity, inhibition of the Homo sapiens (Hs) DHFR was also measured for the 11 active hits in the same conditions. All fragments were found inactive against HsDHFR, showing overall selectivity for the Pf enzyme (Figure 5(B)).
Fragment 1130 (5–(4-chlorophenyl)pyrimidin-4-amine) is the only compound related to the 2,4-diaminopyrimidine-based antifolate series. Its core structure relates to PYR but lacks one aromatic amino group and the 6-ethyl side substituent. Our previous work demonstrated that these groups are necessary to promote direct interaction with the protein, through Cys15, Asp54 and Thr185 (Figure 1) 8. The absence of these two groups in 1130 results in a loss of affinity by four orders of magnitude compared to PYR, in line with our DSF results (Figure 3). In addition, it should be noted that in a retrosynthetic approach, fragments L4 and 1130 complement each other into PYR. This illustrates the end goal of fragment-based drug discovery in which the merging of medium affinity fragments leads to a compound with dramatically improved affinity34,35. This result also suggests that the p-Cl phenyl ring (present in 1130) and the amino and ethyl substituents (present in L4) equally contribute to the high binding affinity of PYR.
The main chemotype unravelled by this screening was defined as bicyclic fragments with a heteroalkyl cyclic ring, a linker, and a phenyl ring with or without substituent. This group includes four 4-phenyl-piperidines (fragments 132, 240, 263, and 813), four 4-phenyl-piperazines (fragments 218, 800, 820 and 1157), and one phenyl-pyrrolidine (fragment 80). These compounds with high pKa values (7.16 < pKa<11.43) are in their protonated form at physiological pH, which might result in poor cell penetration due to their positive charge. Fragments 263 and 813 differ by the presence of a fused 5-member ring that rigidifies the system and prevents free rotation of the two rings.
Finally, fragment 148 is the only monocyclic fragment, constituted of a phenyl ring bearing urea and a trifluoromethyl substituents in 1,3 position. It is neutral at physiological pH, which could ease future lead design.
Crystallography of selected fragments complexed with PfDHFR-TS
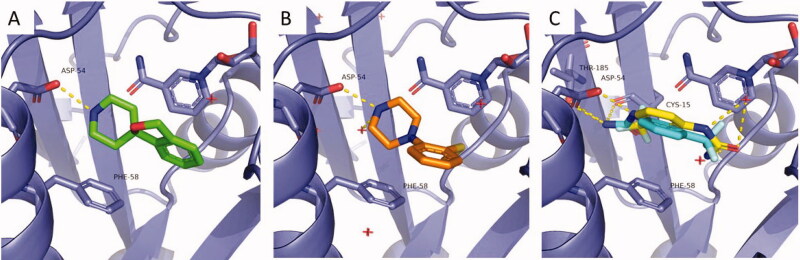
Three representative active fragments belonging to each chemotype, fragments 148 (phenyl urea), 820 (piperazine) and 263 (piperidine) were selected for co-crystallization with PfDHFR-TS (the native bifunctional form of the protein) based on their highest DHFR inhibitory activity at 500 µM. Despite an overall low fragment occupancy, crystal structures were successfully obtained for each complex at resolutions between 2.70 − 2.85 Å. All crystals showed the expected orthorhombic arrangement (P212121), with two protein chains per asymmetric unit. All the data collection and refinement statistics appear in Table S1. Very little variation was observed between the protein backbone of three structures (Figure S1).
All three fragments were confirmed to bind into PfDHFR active site, occupying the space delimitated by Asp54, Phe58, and NADPH normally occupied by the pteridine moiety of DHF. Fragments 263 and 820 display a similar binding mode, with a H-bond mediated interaction with Asp54 (Figure 6(A,B)). As the secondary amines are in their protonated form at physiological pH, the interaction is likely to be strengthened through charge pairing. For the two fragments, the phenyl ring is stacked in the apolar region defined by NADPH and Phe58, however the ring positioning differs between the two complexes. In 263, the phenyl ring is positioned 4.7 Å from the NADPH nicotinamide ring and forms a π-π interaction, while in 820, the phenyl ring is coplanar to the Phe58 phenyl ring. This could be the origin of the higher inhibitory potential of fragment 263, as its binding mode might prevent substrate access to the catalytic Asp54 as well as to the NADPH cofactor.
Figure 6.
Crystal structures for fragment 263 (PDB 7CTY) (A), fragment 820 (PDB 7CTW) (B) and fragment 148 (PDB 7CTZ) in conformation A (cyan) and conformation B (yellow) (C). Interacting residues and NADPH appear as sticks, polar contacts appear as yellow dash lines.
Due to its similarly sized substituents in 1,3 position of the phenyl ring, the electron density for fragment 148 displays a symmetrical shape that is compatible with two ligand orientations. Because crystallographic evidence did not allow conclusion on a preferred conformation, both were proposed in the crystal structure (Figure 6(C)). In the first conformation, the urea moiety interacts with the protein via a double H-bond mediated interaction with Asp54. Polar contacts are also observed with Thr185 side chain hydroxy group and Cys15 backbone carbonyl. The phenyl ring is equidistant from Phe58 and nicotinamide ring of NADPH, with the trifluoromethyl substituent pointing towards Ser108. In the second conformation, the trifluoromethyl substituent is facing Asp54 and Cys15, while the urea moiety forms polar contact with Ile164 backbone carbonyl and with a water molecule. The phenyl ring is tilted towards Phe58.
From a strictly chemical perspective, the first conformation would appear as more likely, as the set of interactions observed resembles the interactions of the DHF substrate. However, at this stage, it is not possible to conclude from crystallographic data which conformation is occurring, or if the two binding modes are present simultaneously.
Molecular docking analyses of selected fragment hits
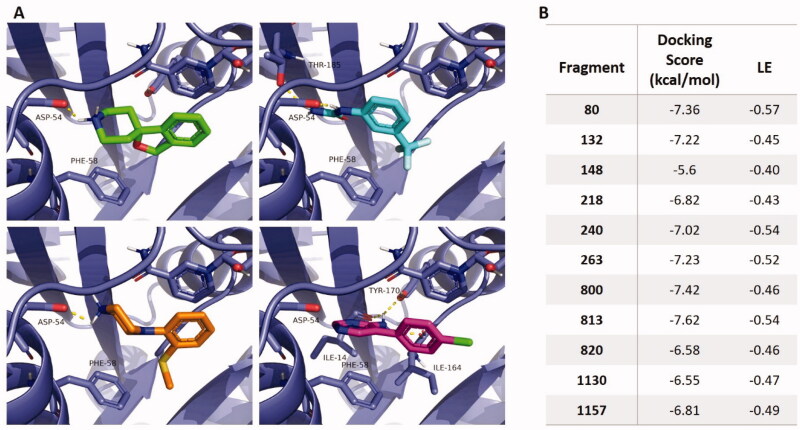
As a complement to crystallographic data, molecular docking experiments were conducted to study binding mode variations for all the active fragment hits. The PfDHFR structure (PDB 3QGT) was used, from which PYR and water molecules were removed, while bound NADPH was conserved, and docking parameters were defined to encompass the whole catalytic pocket. Docking scores and ligand efficiencies (LE), corresponding to the calculated binding energy per non-hydrogen atom, appear in Figure 7(B).
Figure 7.
(A) Detail of molecular docking results for fragments 263 (green), 820 (orange), 148 (cyan) and 1130 (pink). Interacting residues and NADPH appear as sticks. (B) Corresponding docking scores and ligand efficiencies (LE) for each fragment.
Along the phenyl piperidine/piperazine series, molecular docking suggests a binding mode consistent with the crystal structure obtained for fragments 263 and 820, with an interaction between the fragment protonated secondary amine and Asp54. Although an important flexibility is conferred by the aliphatic ring, the aromatic rings tend to align with the NADPH nicotinamide ring for all fragments, except for the extended ring of fragment 132 (Figure 7(A) and Figure S2).
The docking pose of fragment 148 is consistent with the first conformation proposed in the crystal structure, with a coordination of the urea to Asp54 via two parallel H-bonds, while the trifluoromethyl substituent is pointing towards Ile164 and Ser108. The phenyl ring is tilted compared to the crystal structure to be coplanar with NADPH. The comparatively low docking score obtained could be attributable to a poor estimation of the trifluoromethyl contribution by the software’s scoring function (Figure 7(B)).
Expectedly, molecular docking on fragment 1130 provided a similar binding pose as co-crystallized PYR, with a slightly twisted conformation of the rings, possibly due to the absence of interaction with Asp54 (Figure 7(A)). Its docking score is comparatively lower than for the phenyl piperidine/piperazine derivatives (Figure 7(B)).
Discussion
PfDHFR fragment-based screening and re-discovery of PYR by fragment-based approach
Starting from a 1163 fragments library, our screening workflow enabled us to identify 11 fragments displaying inhibitory activity on WT PfDHFR. Compared to other fragment-based screening studies, ours can be considered successful, as it is common to see campaigns yielding fragments that bind their target too weakly to detect any measurable inhibition34.
Out of 11 active fragment hits, only one fragment resembles the pyrimidine-based inhibitor series. This constitutes a validation of FBS applied to PfDHFR, as FBS would have allowed discovery of PYR and CYC, if they had not already been developed. More importantly, the fact that besides 1130, 10 other unrelated fragments showed similar (or better) inhibitory properties confirmed the success of the FBS approach to identify novel anti-PfDHFR chemotypes.
New anti PfDHFR chemotypes
Among the fragment hits identified, the largest part is constituted of 4-phenyl-piperidines and 4-phenyl-piperazines. Two successive studies from 2005 and 2012 demonstrated the antimalarial effect of mono- and di-substituted piperazines on both chloroquine-sensitive and resistant P. falciparum strains36,37. A structure-activity relationship (SAR) study showed that the presence of a free amino group was essential to guarantee the inhibition, prompting the authors to assume of the existence of an unknown protein target. In this study, we provide evidence that both phenyl piperidine and piperazine derivatives target the active site of PfDHFR. This is supported by the crystal structure obtained for fragments 820 and 263 showing electron density in the PfDHFR active site.
Looking closely at the structure of these fragment hits, and as importantly at the non-hits, allows us to build a preliminary SAR study along this fragment series (Figure S3). Consistently with previous observation37, we note that none of the piperazine fragments with protected secondary amine present in the library appeared as a hit by DSF. 2-phenyl piperazine derivatives (i.e., asymmetric piperazines bearing the phenyl ring linked to an aliphatic carbon) were also negative.
Although hits display variable linker lengths, the shortest ones seem favourable for inhibition. For the piperazines, the introduction of an angle between the rings is also a beneficial feature (fragments 2, 132, 263, 813), probably promoting a favourable alignment with NADPH. Finally, the nature and position of the phenyl ring substituents seem to be of crucial importance. At this stage, it cannot be concluded whether these variations are due to steric constraints or to a precise tuning of the electronic properties of the phenyl ring, and it seems clear that a more systematic SAR study will be key for future lead development.
A second chemotype identified is the [3-(trifluoromethyl)phenyl]urea (fragment 148). This is in line with the work from Rastelli et al. who identified (thio)urea derivatives as a pharmacophore of interest by computational screen on PfDHFR38. In their study, PfDHFR inhibition was confirmed experimentally for seven derivatives, showing Ki values in the low μM range. Here, our fragment-based screening was able to further minimise the chemical group responsible for interaction. Importantly, none of the substituted ureas, either through methylation (fragments 894, 895, 959, 960) or cyclisation (fragments 361, 369), showed any interaction or inhibition towards PfDHFR (Figure S4). This supports the first conformation proposed in the crystal structure, showing that the presence of both the primary and the secondary amine are involved in the interaction through two parallel hydrogen bonds and two polar contacts. An equilibrium with the tautomeric form of the urea could also be envisaged.
Opportunities for lead development
For the two new chemotypes identified in this screening campaign, novel opportunities appear in terms of growth vectors. Using piperidines or piperazines offers a chance to evolve the fragment in three dimensions, taking advantage of the ring flexibility, which cannot be done with the planar 2,4-diaminopyrimidine. This series of fragment hits also accommodates a variety of phenyl ring substituents, enabling functionalization in different directions. The high LE values measured for these fragments make them promising starting points for lead development, provided that LE can be maintained through the design process. Another important parameter to consider for future drug design is the clogD value. Because they are positively charged at physiological pH, piperazines and piperidines are commonly used to improve drugs water solubility39,40. However, this may come with a decrease in cell penetration and/or tissue distribution. Design will have to include a strategy to increase lipophilicity of future lead candidates to reach a suitable clogD range.
The potential of phenyl urea fragment 148 resides in the positioning of its phenyl ring. Because the urea moiety is relatively small, the phenyl ring is positioned about 2 Å deeper into the active site compared to PYR and is facing Phe58. This reduces the risk of steric clash with S108N, but also provides an opportunity to target this residue, by incorporation of a suitable functional group. Complete SAR studies will be needed to elucidate the role of the trifluoro substituent. Functionalization in ortho or para position could also be envisaged, as suggested by the results obtained with fragment 327 (Figure S4).
Another promising feature of these new chemotypes is their selectivity, as they do not show inhibition against HsDHFR. This contrasts with the 2,4-diaminopyrimidine-based scaffolds, that do bind HsDHFR and for which careful optimisation was necessary to obtain selective compounds. Starting from chemotypes with high selectivity would surely ease the drug design process, although a constant monitoring of selectivity factors should be observed.
Finally, it should be noted that the three active chemotypes identified through this screening were found to bind the enzyme active site, in close proximity with the Asp54 and NADPH. This residue is known to be crucial, both for substrate binding and catalytic activity, and it is not surprising to find that μM-range binders can impede catalytic activity. However, for portions of the active site with more limited contribution to substrate binding, a μM-range binding event might not result in detectable inhibition. For this reason, the absence of enzyme inhibition does not make the 64 other fragment hits irrelevant, as they might still bind other portions of the active site, or in other sites. Although this study has focussed on the fragment hits displaying promising inhibitory properties, a complete structural characterisation of the remaining primary fragment hits using X-rays crystallography and molecular docking would provide valuable information for future design.
Conclusions
In the context of antifolate resistance, development of new generation antifolates based on novel chemotypes is warranted. Although the fragment-based screening approach has been successfully applied to DHFR enzymes in various bacteria and protozoa41,42, no similar study has yet been reported in the malaria parasite Plasmodium falciparum. Here, we describe the first fragment-based screening on PfDHFR. In addition to the widely used diaminopyrimidine, three new chemotypes displaying μM-range PfDHFR inhibition were identified. Most of the 11 active fragment hits showed high selectivity for the target enzyme compared to the human isoform, highlighting the efficiency and specificity of the fragment-based strategy.
The crystal structures obtained for both phenyl piperazine and phenyl urea chemotypes confirm a binding in the enzyme active site, similarly to DHF substrate and pyrimidine-based antifolates. These scaffolds offer new possibilities in terms of growth vectors, enabling exploration of novel regions of the active site. Future work will be devoted to the investigation of these growth vectors through a structure-activity relationship study, paying attention to the selectivity profile towards the antifolate-resistant enzyme variants. These results could pave the way for the discovery of new lead molecules against P. falciparum DHFR and give a fresh start to antifolate drug development.
Supplementary Material
Acknowledgements
We thank the NSTDA Characterization and Testing Service Center (NCTC) for SC XRD series: D8 venture, and thank Waraporn Pinyo for helping with the D8 Venture system. We thank Netnapa Charoensetakul for the preparation of compounds L4 and L5.
Funding Statement
This research was supported by grants from the Medicines for Malaria Venture (MMV), BIOTEC, NSTDA’s Cluster and Program Management (P1450883) and NSTDA’s Researcher Chair Grant (P1850116).
Disclosure statement
The authors report no conflict of interest.
References
- 1.Yuthavong Y, Kamchonwongpaisan S, Leartsakulpanich U, et al. Folate metabolism as a source of molecular targets for antimalarials. Future Microbiol 2006;1:113–25. [DOI] [PubMed] [Google Scholar]
- 2.Tse EG, Korsik M, Todd MH.. The past, present and future of anti-malarial medicines. Malar J 2019;18:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuthavong Y. Basis for antifolate action and resistance in malaria. Microbes Infect 2002;4:175–82. [DOI] [PubMed] [Google Scholar]
- 4.Yuvaniyama J, Chitnumsub P, Kamchonwongpaisan S, et al. Insights into antifolate resistance from malarial DHFR-TS structures. Nat Struct Biol 2003;10:357–65. [DOI] [PubMed] [Google Scholar]
- 5.Yuthavong Y, Yuvaniyama J, Chitnumsub P, et al. Malarial (Plasmodium falciparum) dihydrofolate reductase-thymidylate synthase: structural basis for antifolate resistance and development of effective inhibitors. Parasitology 2005;130:249–59. [DOI] [PubMed] [Google Scholar]
- 6.Vanichtanankul J, Taweechai S, Yuvaniyama J, et al. Trypanosomal dihydrofolate reductase reveals natural antifolate resistance. ACS Chem Biol 2011;6:905–11. [DOI] [PubMed] [Google Scholar]
- 7.Vanichtanankul J, Taweechai S, Uttamapinant C, et al. Combined spatial limitation around residues 16 and 108 of Plasmodium falciparum dihydrofolate reductase explains resistance to cycloguanil. Antimicrob Agents Chemother 2012;56:3928–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuthavong Y, Tarnchompoo B, Vilaivan T, et al. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci USA 2012;109:16823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chughlay MF, Rossignol E, Donini C, et al. First-in-human clinical trial to assess the safety, tolerability and pharmacokinetics of P218, a novel candidate for malaria chemoprotection. Br J Clin Pharmacol 2020;86:1113–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erlanson DA, de Esch IJP, Jahnke W, et al. Fragment-to-Lead Medicinal Chemistry Publications in 2018. J Med Chem 2020;63:4430–44. [DOI] [PubMed] [Google Scholar]
- 11.Bancet A, Raingeval C, Lomberget T, et al. Fragment linking strategies for structure-based drug design. J Med Chem 2020;63:11420–35. [DOI] [PubMed] [Google Scholar]
- 12.Chhabra S, Dolezal O, Hattarki M, et al. Fragment screening on Staphylococcus aureus HPPK – a folate pathway target. Aust J Chem 2013;66:1537–43. [Google Scholar]
- 13.Benmansour F, Trist I, Coutard B, et al. Discovery of novel dengue virus NS5 methyltransferase non-nucleoside inhibitors by fragment-based drug design. Eur J Med Chem 2017;125:865–80. [DOI] [PubMed] [Google Scholar]
- 14.Christopeit T, Leiros H-KS.. Fragment-based discovery of inhibitor scaffolds targeting the metallo-β-lactamases NDM-1 and VIM-2. Bioorg Med Chem Lett 2016;26:1973–7. [DOI] [PubMed] [Google Scholar]
- 15.Hudson SA, McLean KJ, Surade S, et al. Application of fragment screening and merging to the discovery of inhibitors of the mycobacterium tuberculosis cytochrome P450 CYP121. Angew Chem Int Ed Engl 2012;51:9311–6. [DOI] [PubMed] [Google Scholar]
- 16.Liu M, Quinn RJ.. Fragment-based screening with natural products for novel anti-parasitic disease drug discovery. Expert Opin Drug Discov 2019;14:1283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vu H, Roullier C, Campitelli M, et al. Plasmodium gametocyte inhibition identified from a natural-product-based fragment library. ACS Chem Biol 2013;8:2654–9. [DOI] [PubMed] [Google Scholar]
- 18.Vu H, Pedro L, Mak T, et al. Fragment-based screening of a natural product library against 62 potential malaria drug targets employing native mass spectrometry. ACS Infect Dis 2018;4:431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamchonwongpaisan S, Quarrell R, Charoensetakul N, et al. Inhibitors of multiple mutants of Plasmodium falciparum dihydrofolate reductase and their antimalarial activities. J Med Chem 2004;47:673–80. [DOI] [PubMed] [Google Scholar]
- 20.Saepua S, Sadorn K, Vanichtanankul J, et al. 6-Hydrophobic aromatic substituent pyrimethamine analogues as potential antimalarials for pyrimethamine-resistant Plasmodium falciparum. Bioorg Med Chem 2019;27:115158. [DOI] [PubMed] [Google Scholar]
- 21.Kamchonwongpaisan S, Charoensetakul N, Srisuwannaket C, et al. Flexible diaminodihydrotriazine inhibitors of Plasmodium falciparum dihydrofolate reductase: binding strengths, modes of binding and their antimalarial activities. Eur J Med Chem 2020; 195:112263. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Cui K, Lu W, et al. Synthesis and antimalarial activity of novel dihydro-artemisinin derivatives. Molecules 2011;16:4527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarnchompoo B, Sirichaiwat C, Phupong W, et al. Development of 2,4-diaminopyrimidines as antimalarials based on inhibition of the S108N and C59R + S108N mutants of dihydrofolate reductase from pyrimethamine-resistant Plasmodium falciparum. J Med Chem 2002;45:1244–52. [DOI] [PubMed] [Google Scholar]
- 24.Chitnumsub P, Yavaniyama J, Vanichtanankul J, et al. Characterization, crystallization and preliminary X-ray analysis of bifunctional dihydrofolate reductase-thymidylate synthase from Plasmodium falciparum. Acta Crystallogr D Biol Crystallogr 2004;60:780–3. [DOI] [PubMed] [Google Scholar]
- 25.Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 2009;30:2785–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The PyMOL Molecular Graphics System, Version 2.4.1 Schrödinger , LLC; 2010. [Google Scholar]
- 27.Vagin A, Teplyakov A.. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 2010;66:22–5. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov GN, Skubák P, Lebedev AA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 2011;67:355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, et al. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010;66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mortenson PN, Erlanson DA, de Esch IJP, et al. Fragment-to-Lead Medicinal Chemistry Publications in 2017: miniperspective. J Med Chem 2019;62:3857–72. [DOI] [PubMed] [Google Scholar]
- 31.Sawaya MR, Kraut J.. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry 1997;36:586–603. [DOI] [PubMed] [Google Scholar]
- 32.Rod TH, Radkiewicz JL, Brooks CL.. Correlated motion and the effect of distal mutations in dihydrofolate reductase. Proc Natl Acad Sci USA 2003;100:6980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simeonov A. Recent developments in the use of differential scanning fluorometry in protein and small molecule discovery and characterization. Expert Opin Drug Discov 2013;8:1071–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Montfort RLM, Workman P, Lamoree B, et al. Current perspectives in fragment-based lead discovery (FBLD). Essays Biochem 2017;61:453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray CW, Rees DC.. Opportunity knocks: organic chemistry for fragment-based drug discovery (FBDD). Angew Chem Int Ed Engl 2016;55:488–92. [DOI] [PubMed] [Google Scholar]
- 36.Molyneaux C-A, Krugliak M, Ginsburg H, et al. Arylpiperazines displaying preferential potency against chloroquine-resistant strains of the malaria parasite Plasmodium falciparum. Biochem Pharmacol 2005;71:61–8. [DOI] [PubMed] [Google Scholar]
- 37.Ibezim E, Duchowicz PR, Ortiz EV, et al. QSAR on aryl-piperazine derivatives with activity on malaria. Chemom Intell Lab Syst 2012;110:81–8. [Google Scholar]
- 38.Rastelli G, Pacchioni S, Sirawaraporn W, et al. Docking and database screening reveal new classes of Plasmodium falciparum dihydrofolate reductase inhibitors. J Med Chem 2003;46:2834–45. [DOI] [PubMed] [Google Scholar]
- 39.Walker MA. Novel tactics for designing water-soluble molecules in drug discovery. Expert Opin Drug Discov 2014;9:1421–33. [DOI] [PubMed] [Google Scholar]
- 40.Rathi AK, Syed R, Shin H-S, et al. Piperazine derivatives for therapeutic use: a patent review (2010-present). Expert Opin Ther Pat 2016;26:777–97. [DOI] [PubMed] [Google Scholar]
- 41.Shelke RU, Degani MS, Raju A, et al. Fragment discovery for the design of nitrogen heterocycles as Mycobacterium tuberculosis dihydrofolate reductase inhibitors. Arch Pharm (Weinheim) 2016;349:602–13. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz V, Czyzyk DJ, Valhondo M, et al. Novel allosteric covalent inhibitors of bifunctional Cryptosporidium hominis TS-DHFR from parasitic protozoa identified by virtual screening. Bioorg Med Chem Lett 2019;29:1413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sander T, Freyss J, von Korff M, et al. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 2015;55:460–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.