Abstract
In the current study, new thienopyrimidine conjugates bearing 1,2,3-triazole core and different sugar moieties have been designed and synthesized by Cu(I)-catalysed click dipolar cycloaddition. The cytotoxic activity of the synthesised conjugates 2, 5, 7, and 13–18 was studied against HCT-116 and MCF-7 cell lines by the MTT assay. The triazole glycosides 16 and 18 provided significant cytotoxic activities against HCT-116 cell lines comparable to that of doxorubicin and other studied compounds. The cytotoxic behaviour against MCF-7 exhibited that all the investigated compounds were more potent than doxorubicin. Moreover, all screened targets were evaluated against mutant EGFR kinase type L858R and the results revealed that the acetylated 1,2,3-triazole glycosides 13–18 exhibited excellent EGFR inhibitory activity in comparison with gefitinib. Furthermore, molecular modelling studies were performed to investigate the binding affinity of the most active compounds to EGFR enzyme.
Keywords: Click chemistry, thienopyrimidines, glycosides, anticancer, EGFR
1. Introduction
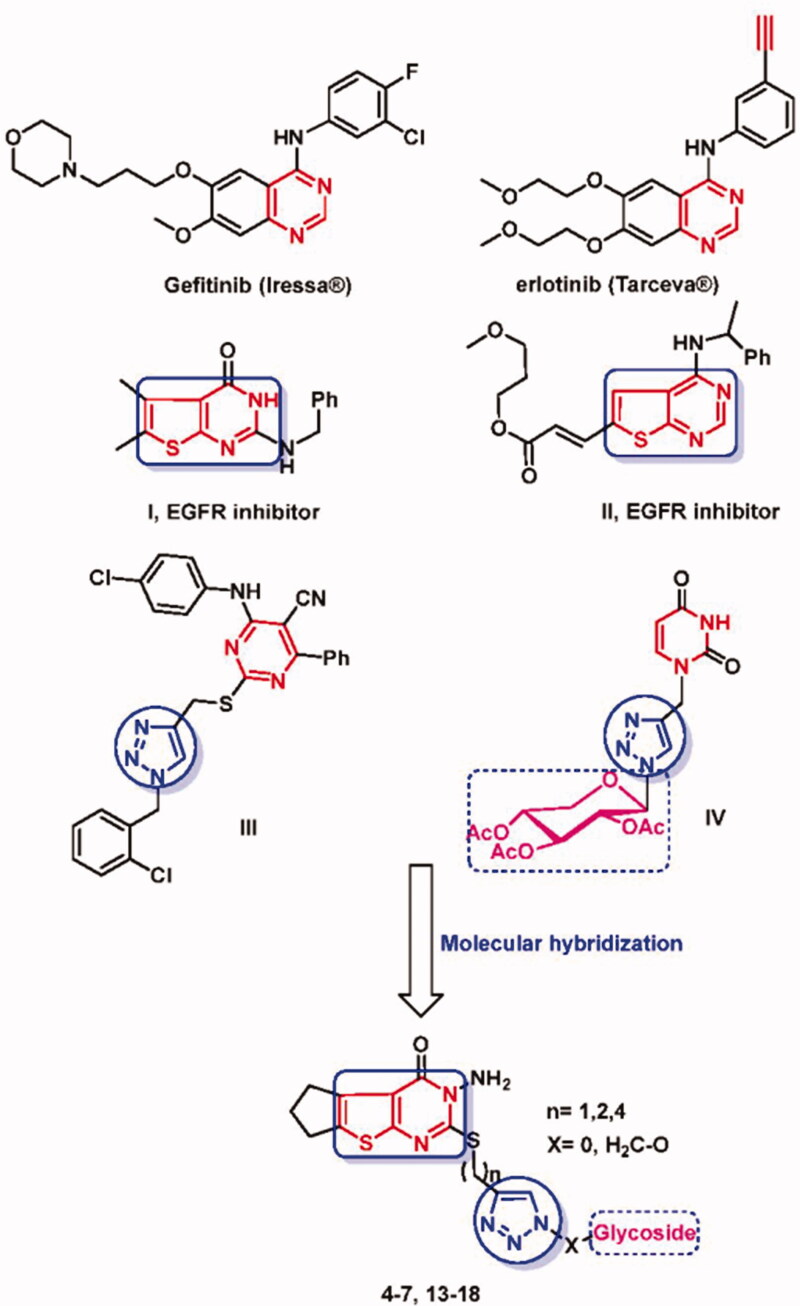
Cancer causes a significant percentage of all human deaths (7.9 million), and it is expected to rise to 12 million deaths per year by 20301. Consequently, discovering new anti-cancer candidates has become a key aim in recent scientific research. Research strategies involving enzyme inhibition pathways are the most followed approaches in cancer drug research. Kinases have been revealed as a group of the most frequent targets of proteins in the field of anticancer drug discovery due to their importance in the regulation of cellular pathways. Research for discovering inhibitors of these targets has found considerable interest owing to their outstanding role in the treatment of cancer and various diseases2–4. Among the most widely investigated, receptor tyrosine kinase (RTK) is the epidermal growth factor (EGF) family which is also called erythroblastic B (ErbB) receptors due to its various developmental, physiological functions in human and for triggering cancer5. Disruption of the EGFR signalling system, both by antagonistic effects on EGFR binding sites, particularly on the extracellular domain of the receptor or by inactivating intracellular tyrosine kinase, can potentially control (inhibit) the growth EGFR-mediated tumours and show some improvement in the patient's condition. Therefore, EGFR illustrates a valuable role in cancer drug discovery and its inhibitors are now clinically available as gefitinib (Iressa®)6 and erlotinib (Tarceva®)7 that have been approved and ratified for the treatment of gastrointestinal stromal tumours and lung cancer (Figure 1).
Figure 1.
Reported examples of anticancer agents and structural modification to design new thienopyrimidines linked triazole glycosides.
Based on click strategy, synthetic approaches provide efficient pathways for rapid and mild synthesis of bioactive leads, as possible candidates with enzyme inhibitory actions. The Cu (I)-catalysed 1,3-dipolar cycloaddition of functionalised azide and alkyne substrates which affords 1,4-disubstituted 1,2,3-triazole applying mild conditions exhibits superb selectivity and good yields8. One of the most exciting features of click cycloaddition is the unique structural attributes of the yielded 1,2,3-triazole motifs which are known to be present in a large number of compounds owing to their anticancer9, and antiviral10,11 activities as well as their ability of being potent pharmacophores12,13. Triazole-amended structures possessing potent anticancer activity were prepared by applying the catalysed azide-alkyne click cycloaddition14. Besides, the exceptional plane stability to metabolic biotransformation, the aromatic nature of the triazole core as well as the characterised dipole moment and H-bonding formation, gained the triazole motif more advantages for being a good linker group15,16. On the other hand, incorporation of more than one pharmacophore in one hybrid structure (known as molecular hybridisation) has been found to be an efficient tool for designing new compounds with promising activities17. Thieno[2,3-d]pyrimidine system that was afforded through bioisosterism of gefitinib, erlotinib, is an outright ingredient in different cancer chemotherapeutic agents18–20 possessing distinct anti-cancer activity via enzymes inhibition mechanism21–25. Reported thienopyrimidines, I and II displayed promising anticancer and kinase inhibitory activity against EGFR26,27 (Figure 1). Furthermore, compounds possessing thienopyrimidine motif were reported with interesting bioactivities such as anticancer28,29, antiviral30, antibacterial31, and anti-inflammatory32 activities. Different glycosyl heterocycles have been synthesised and revealed potential anticancer and enzyme inhibition activities providing essential roles in biological systems, and glycosides possessing the 1,2,3-triazole core, e.g. III and IV33–37 are an essential feature of such compounds (Figure 1).
As a consequence of the above findings, it is assumed that novel hybrid structures possessing the three pharmacophoric systems could be of biological interest. We have continuing research interest for discovering potent and selective anticancer compounds38,39 by the synthesis of new heterocyclic glycosides with modified base and/or glycon constituent. Owing to the significance of the above findings, in the current study, a number of molecular hybrids of thienopyrimidine linked triazole glycosides were synthesised and studied for their anticancer activity in addition to docking studies and EGFR kinase inhibition investigation.
2. Results and discussion
2.1. Chemistry
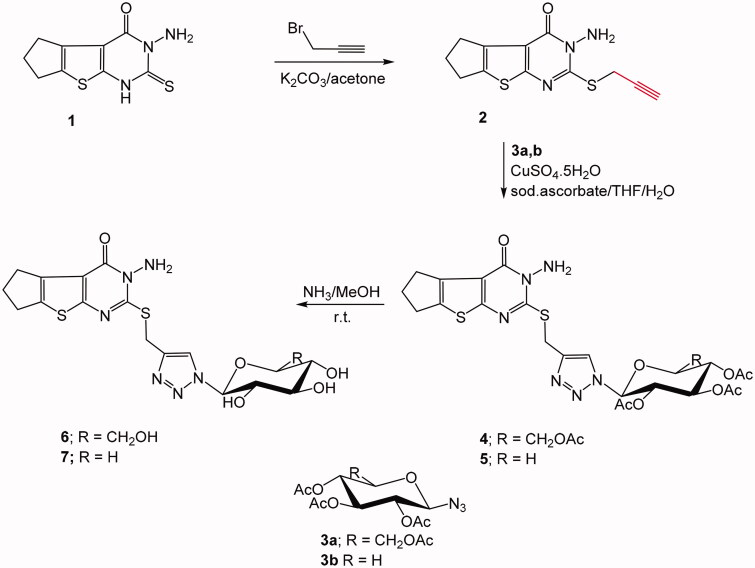
In the current investigation, two new targeted 1,2,3-triazole linked thienopyrimidine motif glycol-conjugates were synthesised via catalysed click dipolar cycloaddition strategy. The first is a 1,2,3-triazole glycoside in which the carbohydrate moiety is directly attached to the triazole motif via a C-N1 linkage. The starting acetylenic functionalised thienopyrimidine compound 2 was afforded by reaction of 3-amino-2-thioxothieno[2,3-d]pyrimidin-4-one derivative 140 and propargyl bromide. The terminal acetylenic compound was allowed to react with two glycosyl azides; definitely tetra-O-acetyl-β-d-gluco- and tri-O-acetyl-β-d-xylopyranosyl azides (3a, b), under click dipolar cycloaddition conditions which lead to the formation of the targeted 1,2,3-glycoside derivatives 4 and 5, respectively in 65–60% yield. Generation of the required Cu (I) catalyst species was performed by means of using sodium ascorbate and copper sulphate, via in situ reduction of copper (II) salt. Tetrahydrofuran/water mixture (3:1) was revealed as the best solvent system after studying a number of single and mixed solvent systems (Scheme 1).
Scheme 1.
Synthesis of triazole glycosides based thienopyrimidine system.
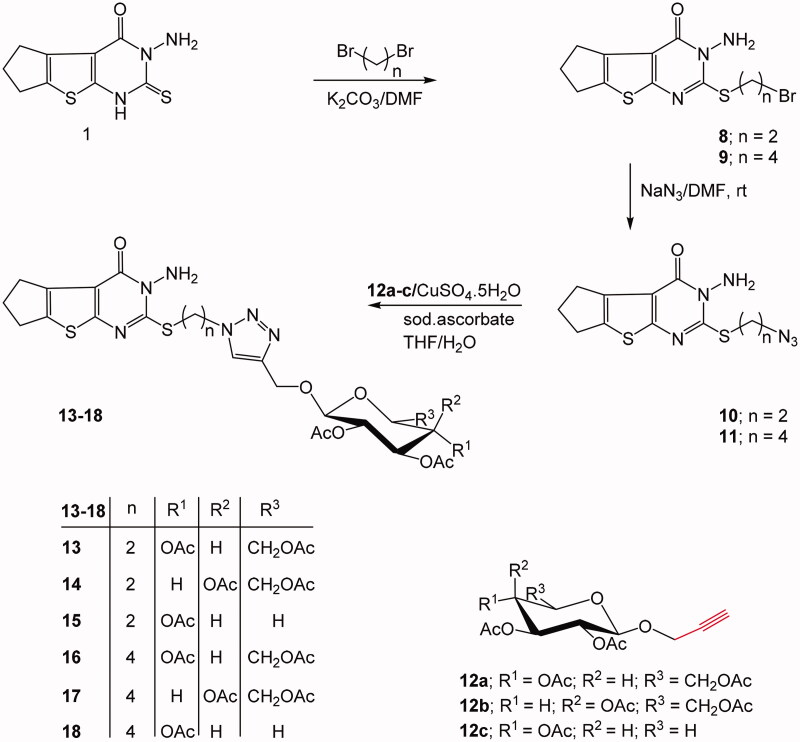
On the other hand, the second thienopyrimidine bridged triazole glycoside derivatives exhibited the 1,2,3-triazole core linked to five and six-carbon sugar moieties since the sugar is attached to the 1,2,3-triazole ring through an oxymethylenic linkage. In such strategy, the azide function was provided into the thienopyrimidine system by halo-alkylation of 3-amino-2-thioxothieno[2,3-d]pyrimidin-4-one (1) by means of 1,2-dibroalkyl compounds; namely 1,2-dibromoethane or 1,4-dibromobutane, which produced the bromoalkyl derivatives 8 and 9, respectively in good yield. The latter bromoalkyl compounds were then reacted with sodium azide and afforded the required azide compound 10 and 11, respectively (Scheme 2).
Scheme 2.
Synthesis of C-linked glycosyl triazoles.
The formed azide compounds were allowed to react with acetylated acetylenic glucose, galactose, or xylose in a copper catalysed dipolar cycloaddition in THF/H2O (3:1) solvent and afforded the targeted glycosyl 1,2,3-triazoles incorporating thienopyrimidine system 13–18.
2.2. Cytotoxic activity
The cytotoxic activities were studied in vitro against HCT-116 (human colorectal carcinoma) and MCF-7 (human breast adenocarcinoma) cell lines using the MTT assay. The results of the cytotoxicity of the screened compounds against the two carcinoma cell lines were compared to that of doxorubicin used as a reference drug in the current investigation. Their IC50 values were calculated and depicted in Table 1. It has been generally observed that most of the tested compounds revealed higher activities against MCF-7 than HCT-116 cell lines. The data shown in Table 1 against HCT-116, indicated that compounds 16, 17, and 18 demonstrated the most potent cytotoxic activities (IC50=8.1 ± 0.8, 8.4 ± 1.1, and 8.3 ± 1.2 µM, respectively) comparable to that of the reference drug, doxorubicin (IC50=7.8 ± 0.7 µM). The remaining derivatives were slightly less active against such cancer cell line (IC50 ranged from 8.6 ± 0.9 to 8.9 ± 1.1) compared to the later three compounds. On the other hand, all the investigated synthesised thienopyrimidine compounds were active towards MCF-7 cell lines (IC50 ranged from 2.1 ± 0.3 to 5.3 ± 0.8) in comparison with doxorubicin (IC50=6.7 ± 0.9 µM). Moreover, the descending order of activity was as follows 2>5>18>13>7>14>15>17>16.
Table 1.
The IC50 values of compounds 2, 5, 7, and 13–18 against HCT-116 and MCF-7 cancer cells according to the MTT assay.
| Compound no. | IC50 (µM)±SD |
|
|---|---|---|
| HCT-116 | MCF-7 | |
| 2 | 8.6 ± 1.9 | 2.1 ± 0.3 |
| 5 | 8.6 ± 0.9 | 2.9 ± 0.4 |
| 7 | 8.7 ± 1.5 | 4.3 ± 0.8 |
| 13 | 8.7 ± 1.6 | 4.2 ± 0.5 |
| 14 | 8.8 ± 1.1 | 4.5 ± 0.7 |
| 15 | 8.9 ± 1.1 | 5.0 ± 0.9 |
| 16 | 8.1 ± 0.8 | 5.3 ± 0.8 |
| 17 | 8.4 ± 1.1 | 5.2 ± 0.9 |
| 18 | 8.3 ± 1.2 | 3.9 ± 0.7 |
| Doxorubicin | 7.8 ± 0.7 | 6.7 ± 0.9 |
Correlation of the afforded results with the structural features of the screened derivatives indicated that the attachment of certain glycosyl moieties to the thienopyrimidine ring system resulted in enhanced cytotoxic activities against the HCT-116 cell line and the activity was raised in compounds such as 16–18. For the activity results against MCF-7 cancer cells, the most potent compounds were the terminal acetylenic thienopyrimidine derivative which does not incorporate sugar part. It has also been observed the 1,2,3-triazole glycosides based thienopyrimidine system possessing a linker of four methylene (CH2) groups revealed higher activities against HCT-116 cells than their analogues with shorter linker (two CH2 groups). Furthermore, it has also been found that the glycosyl-1,2,3-triazole derivatives of the thienopyrimidine system with cyclic glucopyranosyl unit, either acetylated or free hydroxyl moiety, were found more potent against HCT-116 cell line than their analogues with xylopyranosyl fragments. In addition, glycosyl-1,2,3-triazole 18 incorporating the xylopyranosyl moiety showed relatively higher cytotoxic activity against the MCF-7 cancer cell lines more than the gluco- and galactopyranosyl analogues.
2.3. Kinase inhibitory activity
The RTKs involving epidermal growth factor receptor (EGFR) and its three related proteins (the ERBB family) have been shown to play essential roles cancerous conditions in addition to their role in normal physiological conditions41. The investigated compounds were studied for their inhibitory activity against mutant EGFR kinase type L858R. The resulted IC50 values are displayed in Table 2 in comparison with the reference drug, gefitinib (IC50=0.014 ± 0.18 µM). According to the revealed results, the acetylated 1,2,3-triazole glycosides 13–18 exhibited excellent EGFR inhibition with IC50 in the range of 0.010 ± 0.14 to 0.020 ± 0.23 µM. Furthermore, the glycosyl-1,2,3-triazole 18 incorporating the acetylated xylopyranosyl moiety displayed the most potent activity (IC50=0.010 ± 0.14 µM) among this series of studied compounds. It is proposed that the direct attachment of the 1,2,3-triazolyl and glycopyranosyl moieties in the synthesised glycosides 5 and 7 resulted in drastic decrease in the inhibitory activity (IC50=0.130 ± 1.45 and 0.170 ± 0.43 µM, respectively) due to the absence of the separating linker. Additionally, compound 2 was found to be of weak inhibitory activity of EGFR enzyme (IC50=0.500 ± 1.00 µM).
Table 2.
Inhibitory evaluation of the tested compounds against EGFRL858R.
| Compound no. | IC50 (mean ± SEM) (µM) |
|---|---|
| EGFR | |
| Gefitinib | 0.014 ± 0.18 |
| 2 | 0.500 ± 1.00 |
| 5 | 0.130 ± 1.45 |
| 7 | 0.170 ± 0.43 |
| 13 | 0.015 ± 0.81 |
| 14 | 0.013 ± 0.22 |
| 15 | 0.017 ± 0.30 |
| 16 | 0.014 ± 0.12 |
| 17 | 0.020 ± 0.23 |
| 18 | 0.010 ± 0.14 |
IC50: compound concentration required to inhibit the enzyme activity by 50%; SEM: standard error mean; each value is the mean of three values.
2.4. Molecular docking study
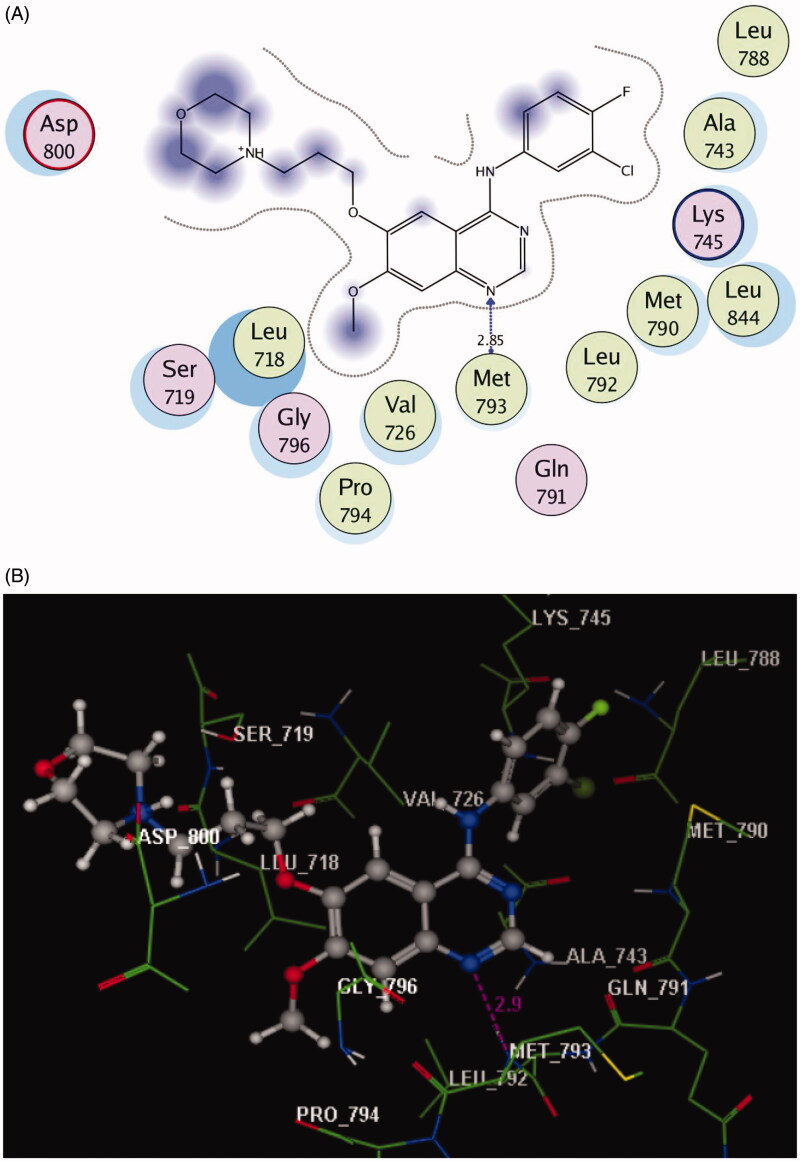
Based on the results of kinase inhibitory assessment, we have been promoted to perform a molecular docking study of the studied compounds 2, 5, 7, and 13–18 against EGFR kinase by using MOE software version 2008.10. The binding site of EGFR (PDB ID: 3UG2) with its inhibitor gefitinib was retrieved from PDB bank http://www.rcsb.org/pdb. Redocking of the reference ligand showed that the root mean square difference (RMSD) between the top docking pose and original crystallographic geometry of the cocrystallised ligand, gefitinib was 0.87 ˚A with a perfect superimposition between them (Figure 2(C)).
Figure 2.
(A) 2D molecular interactions of gefitinib with amino acids of the EGFR enzyme pocket (PDB code: 3UG2). (B) 3D molecular interactions of gefitinib with amino acids of the EGFR enzyme pocket (PDB code: 3UG2). (C) 3D representation of the superimposition of the docking pose (yellow) and the co-crystallised (red) of gefitinib with an RMSD of 0.87 Å.
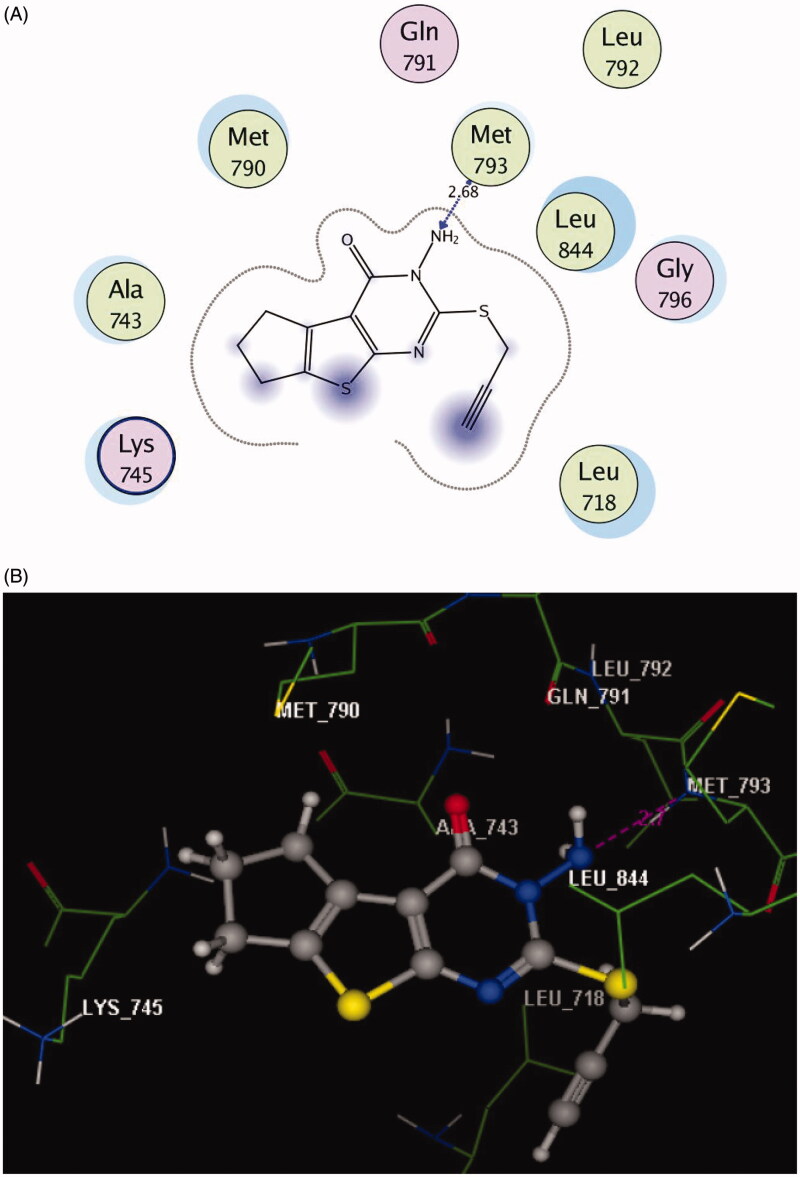
As reported in the docking profile of gefitinib bearing a quinazoline core, the N1 formed hydrogen bond acceptor with the backbone of Met793 (distance: 2.85 Å) (Figure 2(A,B)). Moreover, the whole structure was inserted nicely within the active site of EGFR through hydrophobic interactions with the essential amino acids Leu718, Ser719, Val726, Ala743, Lys745, Leu788, Met790, Gln791, Leu792, Pro794, Gly796, Asp800, and Leu84442.
The data of docking scores (kcal/mol) and interactive binding residues of the studied compounds 2, 5, 7, and 13–18 are depicted in Table 3. It can be noticed that all derivatives displayed variable and promising energy scores ranging from −7.30 to −9.20 kcal/mol and bind to the key amino acid Met793 like the original ligand, gefitinib.
Table 3.
Docking results of the compounds 2, 5, 7, and 13–18 with EGFR kinase (PDB code: 3UG2) using MOE software version 2008.10.
| Compound | Docking score (kcal/mol) | Amino acid residues (bond length, Å) | Atoms of compound | Type of bond |
|---|---|---|---|---|
| Gefitinib | –8.15 | Met793(2.85) | N1(Quinazoline) | H-acc |
| 2 | –7.30 | Met793(2.68) | N(NH2) | H-acc |
| 5 | –8.22 | Met793(1.54); | H(NH2); | H-don |
| Ser719(2.37) | O(CO)(OCOCH3) | H-acc | ||
| 7 | –7.62 | Met793(1.60); | H(NH2); | H-don |
| Ser719(2.24) | O(OH) | H-acc | ||
| 13 | –8.40 | Met793(1.51); | H(NH2); | H-don |
| Lys745(2.33) | O(CO)(OCOCH3) | H-acc | ||
| 14 | –8.25 | Met793(1.65); | H(NH2); | H-don |
| Lys745(2.27) | O(CO)(OCOCH3) | H-acc | ||
| 15 | –7.92 | Met793(1.55); | H(NH2); | H-don |
| Lys745(2.22) | O(CO)(OCOCH3) | H-acc | ||
| 16 | –8.65 | Met793(1.50); | H(NH2); | H-don |
| Lys745(2.40) | O(CO)(OCOCH3) | H-acc | ||
| 17 | –8.36 | Met793(1.66); | H(NH2); | H-don |
| Lys745(2.55) | O(CO)(OCOCH3) | H-acc | ||
| 18 | –9.20 | Met793(1.50); | H(NH2); | H-don |
| Lys745(2.43) | O(CO)(OCOCH3) | H-acc |
For example, the lowest biologically active derivative 2 anchored with EGFR binding site via hydrogen bond interaction between the N of the amino group at p-3 of cyclopenta[4,5]thieno[2,3-d]pyrimidinone with Met793 (distance: 2.68 Å) (Figure 3).
Figure 3.
(A) 2D molecular interactions of compound 2 with amino acids of the EGFR enzyme pocket (PDB code: 3UG2). (B) 3D molecular interactions of compound 2 with amino acids of the EGFR enzyme pocket (PDB code: 3UG2).
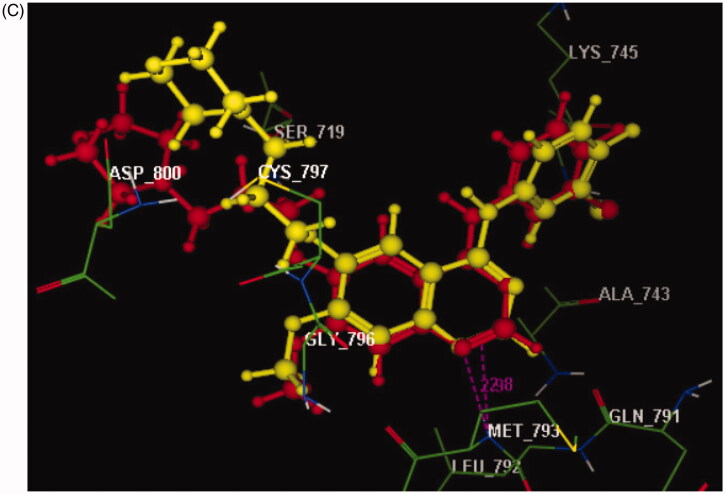
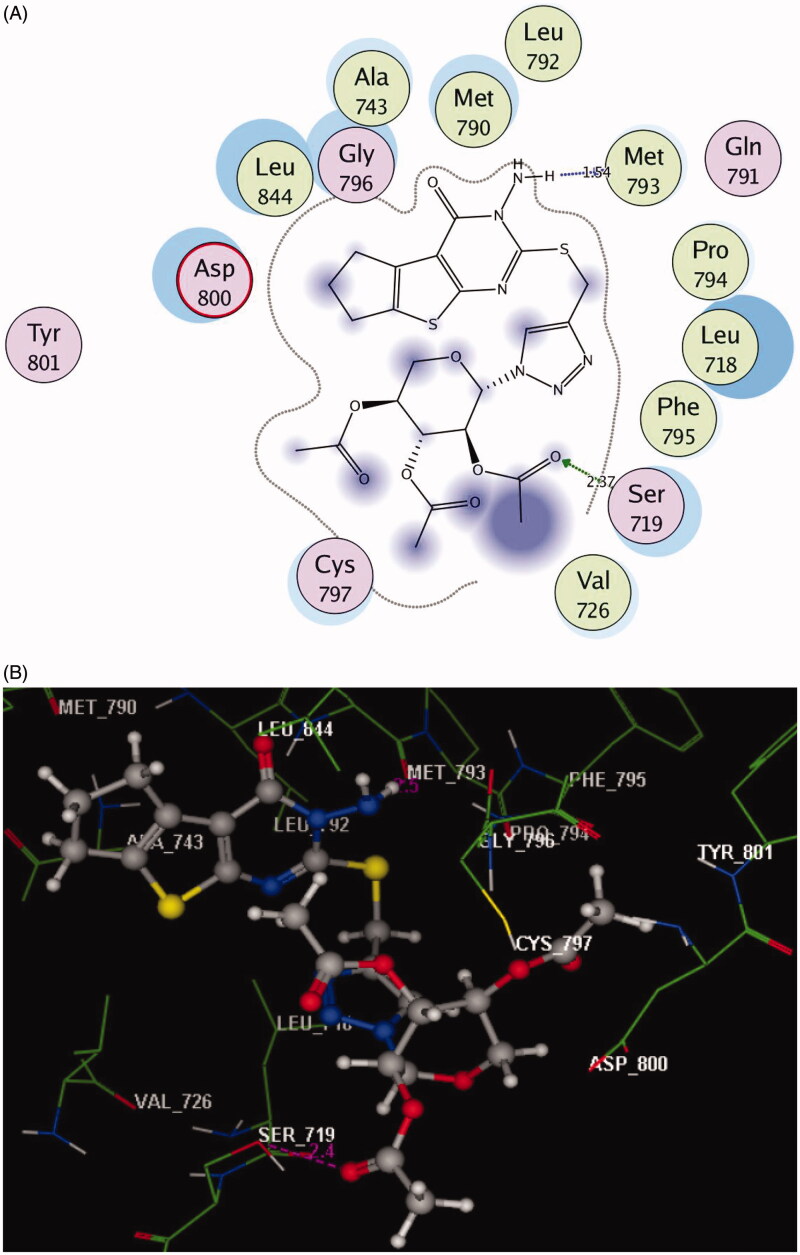
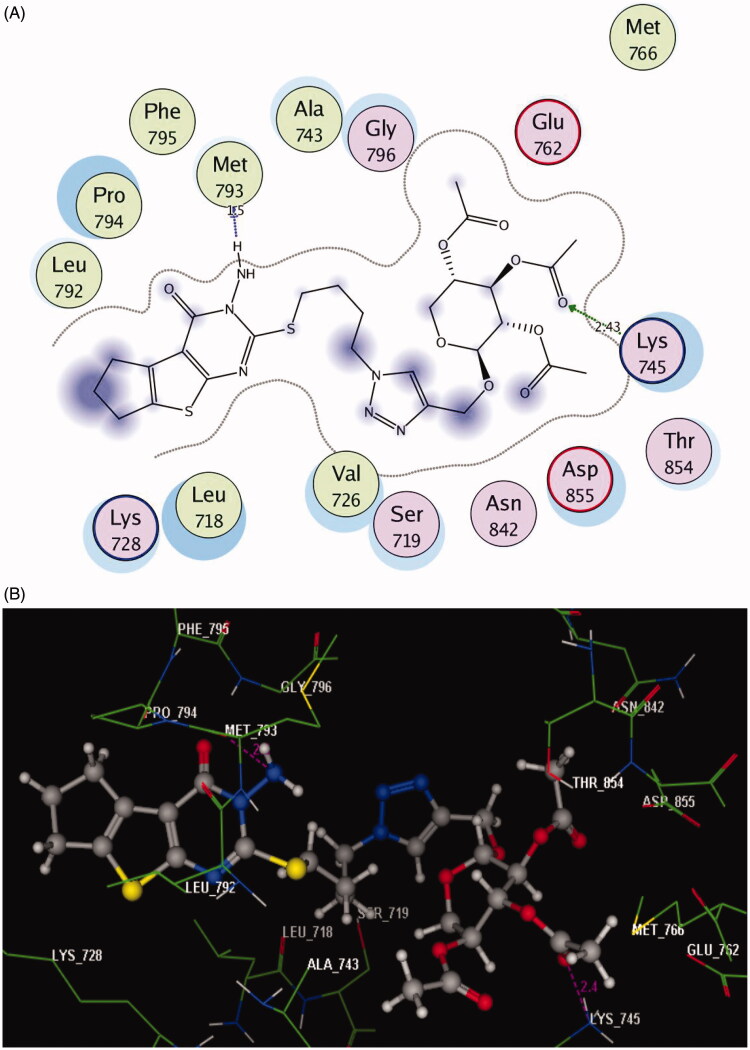
Incorporation of the 3-amino-2-thioxothieno[2,3-d]pyrimidin-4-one nucleus with 1,2,3-triazole and acetylated glycoside scaffolds gave the chance for more fitting within the ATP pocket through different interactions. As observed in Figures 4 and 5, the oxygen of the acetyl group established extra H-bond acceptors with the sidechain of Ser719 in the triazole-N-glycoside 5 (with moderate inhibitory activity and bearing 1C distance chain) and with the sidechain of Lys745 in triazole-oxymethyl-glycoside 18 (with the highest inhibitory activity and bearing 4C distance chain) (distance: 2.37 and 2.43 Å, respectively).
Figure 4.
(A) 2D molecular interactions of compound 5 with amino acids of the EGFR enzyme pocket (PDB code: 3UG2). (B) 3D molecular interactions of compound 5 with amino acids of the EGFR enzyme pocket (PDB code: 3UG2).
Figure 5.
(A) 2D molecular interactions of compound 18 with amino acids of the EGFR enzyme pocket (PDB code: 3UG2). (B) 3D molecular interactions of compound 18 with amino acids of the EGFR enzyme pocket (PDB code: 3UG2).
Overall, the docking study, along with in vitro EGFR results, confirmed that the presence of triazole, oxymethyl, and β-d-glycosyl moieties greatly contribute to the chain elongation and the enhancement of the inhibitory activities of these 3-aminocyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one-based compounds through contributing in formation of extra significant H-bonds inside the ATP-binding cavity.
3. Conclusions
A series of novel 3-aminocyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one hybrids linked with 1,2,3-triazol and different glycosides were designed, synthesised and evaluated for their cytotoxic activity against two cancer cell lines HCT-116 and MCF-7 by the MTT assay. Regarding to HCT-116 cell lines, the triazole glycosides 16 and 18 revealed the highest cytotoxic activity in comparison with doxorubicin, while all the studied derivatives displayed excellent activity against MCF-7. The newly synthesised compounds 2, 5, 7, and 13–18 were investigated in vitro for their suppression activity against mutant EGFR kinase type L858R (Table 3). The acetylated 1,2,3-triazole glycosides 13–18 illustrated excellent EGFR inhibitory activity in comparison with gefitinib. Additionally, the molecular docking studies represented the binding orientations of the promising compounds in the active pocket of EGFR and it could be deduced that the 3-aminocyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one nucleus attached to acetylated 1,2,3-triazole-oxymethylglycoside moiety improved the binding strength within the active site through H-bonding with the key amino acids Met793 and Lys745. The previous findings may provide promising templates in the anticancer field.
4. Experimental
4.1. Chemistry
All melting points were measured using a Reichert Thermovar apparatus and are uncorrected. Yields listed are of isolated compounds. The IR spectra were recorded on a Perkin-Elmer model 1720 FTIR spectrometer for KBr disc (Waltham, MA). Routine NMR measurements were made on a Bruker AC-300 or DPX-300 spectrometer (Billerica, MA). Chemical shifts were reported in δ scale (ppm) relative to TMS as a reference standard and the coupling constants J values are given in Hz. The progress of the reactions was monitored by TLC using aluminium silica gel plates 60 F245. IR, 1H NMR, 13C NMR, and elemental analyses were performed at the Micro analytical centre at the Faculty of Science, Cairo University, Cairo, Egypt. The starting compound 3-amino-2-thioxo-1,2,3,5,6,7-hexahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (1) was synthesised as previously reported40. mp 253–255 °C.
4.1.1. 3-Amino-2-(prop-2-yn-1-ylthio)-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (2)
To a mixture of aminothienopyrimidine derivative 1 (1.195 g, 5 mmol) with propargyl bromide (0.594 g, 5 mmol) and K2CO3 (0.691 g, 5 mmol) in acetone (15 ml), the reaction mixture was stirred at room temperature for 14 h. The reaction mixture poured in into ice-water; the solid obtained was filtered off, dried and recrystallised from DMF to give the corresponding derivatives 2.
Yield 75%, mp 181–182 °C, IR (KBr, υ, cm−1): 3391, 3296 (NH2), 2218 (C–C alkyne), 1686 (C═O); 1H NMR (DMSO-d6, δ ppm): 2.38–2.40 (m, 2H, CH2), 2.51 (t, 2H, J= 1.8 Hz, CH2), 2.90 (t, 2H, J= 7.2 Hz, CH2), 3.29 (s, 1H, CH), 3.83 (s, 2H, CH2), 5.78 (br s, 2H, NH2 exchangeable with D2O). Anal. Calcd. for C12H11N3OS2 (277.36): C, 51.97; H, 4.00; N, 15.15. Found: C, 51.80; H, 4.07; N, 15.02.
4.1.2. General procedure synthesis of 1,2,3-triazole acetylated N-glycosides derivatives (4, 5)
To a well stirred solution of the terminal acetylenic derivative 2 (0.554 g, 2.0 mmol) in a mixture of THF–H2O (1:2, 15 ml) was added the azido-sugar (2,3,4,6-tetra-O-acetyl-d-glucopyranosyl or 2,3,4-tri-O-acetyl-d-xylopyranosyl azide) (2.0 mmol). Sodium ascorbate (0.08 g, 0.4 mmol) and few drops of diisopropylethylamine (DIPEA) followed by CuSO4·5H2O (0.4 mmol, 0.11 g) were then added. The mixture was stirred at room temperature overnight (TLC, petroleum ether–ethyl acetate (4:1)). Extraction of the organic compound layer was performed by shaking the mixture two times for 5 min with ethyl acetate. The organic layers were combined, dried over Na2SO4 and the solvent evaporated. Purification by column chromatography (hexane/ethyl acetate, 5:1, as the eluent) gave the title products.
4.1.3. 3-Amino-2-(((1-(2,3,4,6-tetra-O-acetyl-β-d-glucopyranosyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3,5,6,7-tetrahydro-4H-cyclopenta [4,5]thieno[2,3-d]pyrimidin-4-one (4)
Yield: 65%, mp 151–152 °C. IR (KBr, υ, cm−1): 3440 (NH2), 1751 (C═O), 1668 (C═O); 1H NMR (DMSO-d6, δ ppm): 1.94, 1.95, 1.97, 2.01 (4s, 12H, 4CH3), 2.27–2.34 (m, 2H, CH2), 2.39 (t, 2H, J = 1.8 Hz, CH2), 2.89 (t, 2H, J = 7.2 Hz, CH2), 3.82 (s, 2H, CH2), 4.05–4.09 (m, 1H, H5′), 4.30 (dd, 1H, J = 11.2, 3.7 Hz, H6′), 4.31 (dd, 1H, J = 8.2, 6.4 Hz, H6″), 5.14–4.19 (m, 1H, H4′), 5.50–5.57 (m, 1H, H3′), 5.73 (t, 1H, J= 8.2 Hz, H2′), 5.77 (br s, 2H, NH2 exchangeable with D2O), 6.32 (d, 1H, J= 8.7 Hz, H1′), 8.26 (s, 1H, triazole-H); 13C NMR (DMSO-d6, δ ppm): 20.69, 20.82, 20.89, 26.05, 27.72, 29.06, 29.51, 39.35, 67.98, 68.20, 70.47, 72.56, 73.67, 84.22, 115.99, 123.01, 136.67, 139.42, 144.01, 157.64, 159.42; 166.47, 196.80, 169.98, 170.43, 170.47. Anal. Calcd. for C26H30N6O10S2 (650.68): C, 47.99; H, 4.65; N, 12.92. Found: C, 47.70; H, 4.51; N, 13.06.
4.1.4. 3-Amino-2-(((1-(2,3,4-tri-O-acetyl-β-d-xyloopyranosyl)-1H-1,2, 3-triazol-4-yl)methyl)thio)-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (5)
Yield: 60%, mp 155–156 °C. IR (KBr, υ, cm−1): 3432 (NH2), 1741 (C═O), 1666 (C═O), 1H NMR (DMSO-d6, δ ppm): 1.97, 1.98, 2.01 (3s, 9H, 3CH3), 2.26–2.31 (m, 2H, CH2), 2.48 (t, 2H, J= 1.8 Hz, CH2), 2.88 (t, 2H, J= 7.2 Hz, CH2), 4.00 (s, 2H, CH2), 4.29 (dd, 1H, J= 3.7, 10.3 Hz, H5′), 4.31 (dd, 1H, J= 10.2, 6.8 Hz, H5″), 4.97–5.03 (m, 1H, H4′), 5.25–5.30 (m, 1H, H3′), 5.57 (t, 1H, J= 8.2 Hz, H2′), 5.73 (br s, 2H, NH2 exchangeable with D2O), 6.30 (d, 1H, J= 8.7 Hz, H1′), 8.25 (s, 1H, triazole-H), 13C NMR (DMSO-d6, δ ppm). Anal. Calcd. for C23H26N6O8S2 (578.62): C, 47.74; H, 4.53; N, 14.52. Found: C, 47.60; H, 4.57; N, 14.46.
4.1.5. General procedure synthesis of 3-amino-2-(((1-(β-d-glycopyranosyl)-1H-1,2,3-triazol-4-yl)methyl)thio)-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (6, 7)
A solution of the acetylated glycoside compound 4 or 5 (0.5 g) in a saturated methanolic ammonia (20 ml) was stirred at room temperature for 7 h. Upon completion of the deacetylation process (TLC, petroleum ether–hexane, 2:1), the solvent was evaporated under reduced pressure at 40 °C to give a residue, which was triturated with diethyl ether (25 ml) to afford a solid which was filtered off, dried and crystallised from ethanol to give 6 or 7, respectively.
4.1.6. 3-Amino-2-(((β-d-glucopyranosyl)-1H-1,2,3-triazol-4-yl)methyl) thio)-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (6)
Yield: 55%, mp 180–182 °C. IR (KBr, υ, cm−1): 3450–3421 (OH), 3419 (NH2), 1669 (C═O), 1H NMR (DMSO-d6, δ ppm): 2.30–2.38 (m, 2H, CH2), 2.47 (t, 2H, J= 1.8 Hz, CH2), 2.93 (t, 2H, J= 7.2 Hz, CH2), 4.09 (s, 2H, CH2), 4.10–4.19 (m, 2H, H6′,6″), 4.27–4.31 (m, 1H, H5″), 4.34–4.43 (m, 2H, H4′,3′), 4.71–4.75 (m, 1H, OH), 4.97–5.17 (m, 2H, OH. H2′), 5.49–5.60 (m, 2H, 2OH), 5.75 (br s, 2H, NH2 exchangeable with D2O), 5.88 (d, 1H, J= 8.2 Hz, H1′), 8.36 (s, 1H, triazole-H). Anal. Calcd. for C18H22N6O6S2 (482.53): C, 44.81; H, 4.60; N, 17.42. Found: C, 44.69; H, 4.66; N, 17.51.
4.1.7. 3-Amino-2-(((1-(β-d-xylopyranosyl)-1H-1,2,3-triazol-4-yl)methyl) thio)-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (7)
Yield: 60%, mp 189–190 °C. IR (KBr, υ, cm−1): 3448–3419 (OH), 3429 (NH2), 1665 (C═O), 1H NMR (DMSO-d6, δ ppm): 2.28–2.36 (m, 2H, CH2), 2.50 (t, 2H, J= 1.8 Hz, CH2), 2.95 (t, 2H, J= 7.2 Hz, CH2), 4.11 (s, 2H, CH2), 4.31–4.38 (m, 2H, H5′,5″), 4.39–4.48 (m, 2H, H4′,3′), 4.49–4.53 (m, 1H, OH), 4.98–5.07 (m, 2H, OH. H2′), 5.43–5.40 (m, 1H, OH), 5.72 (br s, 2H, NH2 exchangeable with D2O), 5.85 (d, 1H, J= 8.2 Hz, H1′), 8.34 (s, 1H, triazole-H) Anal. Calcd. for C17H20N6O5S2 (452.50): C, 45.12; H, 4.46; N, 18.57. Found: C, 44.88; H, 4.51; N, 18.41.
4.1.8. General procedure synthesis of 3-amino-2-((4-bromoalkyll)thio)-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (8, 9)
To a solution of the aminothienopyrimidine derivative 1 (1.195 g, 5 mmol) in DMF (15 ml) was added to anhydrous K2CO3 (0.691 g, 5 mmol) and the mixture was stirred at room temperature for (1 h). 1,2-Dibromoethane or 1,2-dibromobutane (5 mmol) was slowly added under vigorous stirring, and the resulting mixture was stirred at room temperature for further 10 h. Water (100 ml) was added.
4.1.9. 3-Amino-2-((2-bromoethyl)thio)-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (8)
Yield: 75%, mp 169–170 °C. IR (KBr, υ, cm−1): 3442 (NH2), 1665 (C═O), 1H NMR (DMSO-d6, δ ppm): 2.30–2.38 (m, 2H, CH2), 2.59 (t, 2H, J= 2.8 Hz, CH2), 2.98 (t, 2H, J= 7.2 Hz, CH2), 3.38 (t, 2H, J= 7.2 Hz, CH2), 3.65 (t, 2H, J= 7.2 Hz, CH2), 5.89 (br s, 2H, NH2 exchangeable with D2O); MS: m/z (%)=346 (M+, 20). Anal. Calcd. for C11H12BrN3OS2 (346.26): C, 38.16; H, 3.49; N, 12.14. Found: C, 38.05; H, 3.45; N, 12.28.
4.1.10. 3-Amino-2-((4-bromobutyl)thio)-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (9)
Yield 77%, mp 161–162 °C. IR (KBr, υ, cm–1): 3439 (NH2), 1666 (C═O), 1H NMR (DMSO-d6, δ ppm): 2.09–2.18 (m, 2H, CH2), 2.42–2.53 (m, 2H, CH2), 2.55 (m, 2H, CH2), 2.90 (t, 2H, J= 7.2 Hz, CH2), 2.93 (t, 2H, J= 6.9 Hz, CH2), 3.17 (t, 2H, J= 6.9 Hz, CH2), 3.41 (t, 2H, J= 6.6 Hz, CH2), 5.77 (br s, 2H, NH2 exchangeable with D2O). Anal. Calcd. for C13H16BrN3OS2 (374.32): C, 41.71; H, 4.31; N, 11.23. Found: C, 41.52; H, 4.38; N, 11.14.
4.1.11. General procedure synthesis of 3-amino-2-((azidoethyl)thio)-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (10, 11)
A mixture of bromoalkyl derivatives 8 or 9 (3 mmol) and sodium azide (0.292 g, 4.5 mmol) in DMF (15 ml) was stirred at room temperature for (14 h). The reaction mixture was poured into ice-water; the solid obtained was filtered off, dried and recrystallised from DMF–water (1:1) to give the corresponding derivatives 10 and 11, respectively.
4.1.12. 3-Amino-2-((2-azidoethyl)thio)-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (10)
Yield: 75%, mp 189–190 °C. IR (KBr, υ, cm−1): 3316, 3205 (NH2), 2102 (N3), 1668 (C═O), 1H NMR (DMSO-d6, δ ppm), 2.17–2.23 (m, 2H, CH2), 2.52 (t, 2H, J = 2.8 Hz, CH2), 2.91 (t, 2H, J = 7.2 Hz, CH2), 3.15 (t, 2H, J = 6.8 Hz, CH2), 3.37 (t, 2H, J = 6.8 Hz CH2), 5.71 (br s, 2H, NH2 exchangeable with D2O). MS: m/z (%)= 309 (M+, +1, 14). Anal. Calcd. for C11H12N6OS2 (308.38): C, 42.84; H, 3.92; N, 27.25. Found: C, 42.70; H, 3.85; N, 27.34.
4.1.13. 3-Amino-2-((4-azidobutyl)thio)-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (11)
Yield: 77%, mp 186–187 °C. IR (KBr, υ, cm−1): 3296, 3191 (NH2), 2101 (N3), 1680 (C═O); 1H NMR (DMSO-d6, δ ppm): 2.25–2.33 (m, 2H, CH2), 2.39–2.48 (m, 2H, CH2), 2.50 (m, 2H, CH2), 2.89 (t, 2H, J= 7.2 Hz, CH2), 2.91 (t, 2H, J= 6.9 Hz, CH2), 3.05 (t, 2H, J= 6.8 Hz, CH2), 3.38 (t, 2H, J= 6.8 Hz, CH2), 5.69 (br s, 2H, NH2 exchangeable with D2O). Anal. Calcd. for C13H16N6OS2 (336.43): C, 46.41; H, 4.79; N, 24.98. Found: C, 46.15; H, 4.71; N, 25.08.
4.1.14. General procedure synthesis of acetylated 1,2,3-triazole glycosides (13–18)
Sodium ascorbate (0.08 g, 0.4 mmol) and few drops of DIPEA were added sequentially to a mixture of the respective propargyl sugars 12a, 12b, or 12c (2 mmol) and azido thienopyrimidine derivatives 10 or 11 (2 mmol) in the solvent mixture of THF–H2O (2:1; 15 ml). CuSO4·5H2O (0.11 g, 0.4 mmol,) was then added and the mixture was stirred at 60 °C for (6 h) (TLC, petroleum ether–EtOAc, 3:1). Ethyl acetate (30 ml) was added followed by shaking the reaction content for 5 min then the organic layer was separated, washed with water and the solvent was evaporated. The residue was purified by column chromatography (petroleum ether–EtOAc, 4:1) to afford the triazole-O-linked glycosyl derivatives 13–18.
4.1.15. 3-Amino-2-(ethylylthio)-{2-[4-({[2,3,4,6-tetra-O-acetyl-β-d-glucopyranosyl] oxy}methyl)-1H-1,2,3-triazol-1-yl]ethoxy}-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d] pyrimidin-4-one (13)
Yield: 65%, mp 159–160 °C. IR (KBr, υ, cm−1): 3434 (NH2), 1745 (C═O), 1666 (C═O), 1H NMR (DMSO-d6, δ ppm): 1.93, 1.96, 1.98, 2.00 (4s, 12H, 4CH3), 2.36–2.47 (m, 2H, CH2), 2.86 (t, 2H, J= 1.8 Hz, CH2), 3.00 (t, 2H, J= 7.2 Hz, CH2), 3.29 (t, 2H, J= 6.8 Hz, CH2), 3.35 (t, 2H, J= 6.8 Hz CH2), 4.02–4.05 (m, 1H, H5′), 4.11 (dd, 1H, J= 3.3, 10.8 Hz, H6′), 4.64 (dd, 1H, J= 11.3, 3.3 Hz, H6″), 4.81–4.87 (m, 1H, H4′), 5.19 (s, 2H, CH2), 5.21 (dd, 1H, J= 8.4, 9.2 Hz, H3′), 5.41 (t, 1H, J= 9.3 Hz, H2′), 5.65 (br s, 2H, NH2 exchangeable with D2O), 5.71 (d, 1H, J= 9.8 Hz, H1′), 8.27 (s, 1H, triazole-H); 13C NMR (DMSO-d6, δ ppm): 20.70, 20.81, 20.98, 21.06, 21.09, 21.14, 21.22, 21.48, 56.32, 68.50, 68.92, 69.80, 71.05, 71.21, 72.42, 99.03, 115.73, 124.63, 129.11, 139.41, 143.33, 169.77, 169.96, 170.22; 170.48, 170.53, 170.74, 172.44. Anal. Calcd. for C28H34N6O11S2 (694.73):C, 48.41; H, 4.93; N, 12.10. Found: C, 48.28; H, 4.83; N, 12.19.
4.1.16. 3-Amino-2-(ethylthio)-{2-[4-({[2,3,4,6-tetra-O-acetyl-β-d-galactopyranosyl]oxy}methyl)-1H-1,2,3-triazol-1-yl]ethoxy}-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (14)
Yield: 72%, mp 156–157 °C. IR (KBr, υ, cm−1): 3429 (NH2), 1742 (C═O), 1665 (C═O), 1H NMR (DMSO-d6, δ ppm): 1.95, 1.96, 1.97, 2.01 (4s, 12H, 4CH3), 2.33–2.38 (m, 2H, CH2), 2.47 (t, 2H, J= 2.8 Hz, CH2), 2.90 (t, 2H, J= 7.2 Hz, CH2), 3.30 (t, 2H, J= 6.9 Hz, CH2), 3.35 (t, 2H, J= 6.6 Hz, CH2), 3.97–4.14 (m, 1H, H5′), 4.26 (dd, 1H, J= 3.3, 10.8 Hz, H6′), 4.29 (dd, 1H, J= 11.3, 3.3 Hz, H6″), 4.50–4.82 (m, 1H, H4′), 4.94 (s, 2H, CH2), 5.13 (dd, 1H, J= 8.4, 9.2 Hz, H3′), 5.50 (t, 1H, J= 9.30 Hz, H2′), 5.60 (br s, 2H, NH2 exchangeable with D2O), 6.20 (d, 1H, J= 9.8 Hz, H1′), 8.26 (s, 1H, triazole-H), 13C NMR (DMSO-d6, δ ppm): 20.87, 20.95, 21.11, 21.49, 25.68, 27.69, 29.06, 29.40, 56.10, 68.96, 69.05, 70.87, 71.02, 71.58, 71.74, 100.96, 107.29, 124.55, 136.34, 139.41, 143.42, 157.63, 162.00, 166.00; 169.44, 169.94, 170.00, 172.44. Anal. Calcd. for C28H34N6O11S2 (694.73): C, 48.41; H, 4.93; N, 12.10. Found: C, 48.19; H, 4.95; N, 11.98.
4.1.17. 3-Amino-2-(ethylthio)-{2-[4-({[2,3,4-tri-O-acetyl-β-d-xylopyranosyl]oxy}methyl)-1H-1,2,3-triazol-1-yl]ethoxy}-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (15)
Yield: 71%, mp 153–154 °C. IR (KBr, υ, cm−1): 3441 (NH2), 1746 (C═O), 1669 (C═O), 1H NMR (DMSO-d6, δ ppm): 1.96, 1.97, 1.99 (3s, 9H, 3CH3), 2.34–2.48 (m, 2H, CH2), 2.87 (t, 2H, J= 2.8 Hz, CH2), 3.01 (t, 2H, J= 7.2 Hz, CH2), 3.31 (t, 2H, J= 6.9 Hz, CH2), 3.36 (t, 2H, J= 6.6 Hz, CH2), 4.11 (dd, 1H, J= 3.3, 10.8, Hz, H5″), 4.17 (dd, 1H, J= 11.3, 3.3 Hz, H5″), 4.40–4.58 (m, 1H, H4′), 4.71 (s, 2H, CH2), 4.87 (dd, 1H, J= 8.4, 9.2 Hz, H3′), 5.23 (t, 1H, J= 9.3 Hz, H2′), 5.68 (br s, 2H, NH2 exchangeable with D2O), 5.69 (d, 1H, J= 9.8 Hz, H1′), 8.30 (s, 1H, triazole-H). Anal. Calcd. for C25H30N6O9S2 (622.67): C, 48.22; H, 4.86; N, 13.50. Found: C, 48.05; H, 4.92; N, 13.36.
4.1.18. 3-Amino-2-(butylthio)-{2-[4-({[2,3,4,6-tetra-O-acetyl-β-d-glucopyranosyl] oxy}methyl)-1H-1,2,3-triazol-1-yl]ethoxy}-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno-[2,3-d]pyrimidin-4-one (16)
Yield: 68%, mp 163–164 °C. IR (KBr, υ, cm−1): 3442 (NH2), 1749 (C═O), 1667 (C═O), 1H NMR (DMSO-d6, δ ppm): 1.53–1.58 (m, 2H, CH2), 1.93, 1.95, 1.97, 1.99 (4s,12H, 4CH3), 2.15–2.42 (m, 2H, CH2), 2.48 (t, 2H, J= 1.8 Hz, CH2), 2.74 (t, 2H, J= 7.2 Hz, CH2), 2.84 (t, 2H, J= 6.9 Hz, CH2), 3.16 (t, 2H, J= 6.9 Hz, CH2), 3.35 (t, 2H, J= 6.6 Hz CH2), 3.93 (m, 1H, H5′), 4.11 (dd, 1H, J= 3.3, 10.8, Hz, H6′), 4.14 (dd, 1H, J= 11.3, 3.3 Hz, H6″), 4.64–4.68 (m, 1H, H4′), 4.70 (s, 2H, CH2), 4.89 (dd, 1H, J= 8.4, 9.20 Hz, H3′), 5.33 (t, 1H, J= 9.30 Hz, H2′), 5.65 (br s, 2H, NH2 exchangeable with D2O), 5.68 (d, 1H, J= 9.8 Hz, H1′), 8.25 (s, 1H, triazole-H), 13C NMR (DMSO-d6, δ ppm): 20.28, 20.69, 20.82, 20.89, 26.05, 27.00, 27.72, 29.06, 29.51, 31.98, 56.67, 66.57, 67.98, 68.20, 70.47, 72.28, 72.42, 99.22, 115.99, 123.01, 136.67, 139.42, 144.01, 157.64, 166.47, 169.69, 169.80, 169.98, 170.43, 170.47. Anal. Calcd. for C30H38N6O11S2 (722.79): C, 49.85; H, 5.30; N, 11.63. Found: C, 49.59; H, 5.37; N, 11.52.
4.1.19. 3-Amino-2-(butylthio)-{2-[4-({[2,3,4,6-tetra-O-acetyl-β-d-galactopyranosyl] oxy}methyl)-1H-1,2,3-triazol-1-yl]ethoxy}-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (17)
Yield: 70%, mp 159–160 °C. IR (KBr, υ, cm−1): 3433 (NH2), 1751 (C═O), 1666 (C═O), 1H NMR (DMSO-d6, δ ppm): 1.61–1.66 (m, 2H, CH2), 1.98, 1.99, 2.00, 2.01 (4s,12H, 4CH3), 2.39–2.48 (m, 2H, CH2), 2.50 (t, 2H, J= 1.8 Hz, CH2), 2.83 (t, 2H, J= 7.2 Hz, CH2), 2.94 (t, 2H, J= 6.9 Hz, CH2), 3.01 (t, 2H, J= 6.9 Hz, CH2), 3.36 (t, 2H, J = 6.6 Hz, CH2), 4.11 (m, 1H, H5′), 4.15 (dd, 1H, J= 3.3, 10.8, Hz, H6′), 4.19 (dd, 1H, J= 11.3, 3.3 Hz, H6″), 4.52–4.54 (m, 1H, H4′), 4.62 (s, 2H, CH2), 4.74 (dd, 1H, J= 8.4, 9.2 Hz, H3′), 5.25 (t, 1H, J= 9.3 Hz, H2′), 5.67 (br s, 2H, NH2 exchangeable with D2O), 5.70 (d, J= 9.8 Hz, 1H, H1′), 8.29 (s, 1H, triazole-H). Anal. Calcd. for C30H38N6O11S2 (722.79): C, 49.85; H, 5.30; N, 11.63. Found: C, 49.69; H, 5.35; N, 11.50.
4.1.20. 3-Amino-2-(butylthio)-{2-{2-[4-({[2,3,4-tri-O-acetyl-β-d-xylopyranosyl]oxy}methyl)-1H-1,2,3-triazol-1-yl]ethoxy}-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one (18)
Yield: 75%, mp 159–160 °C. IR (KBr, υ, cm−1), 3429 (NH2), 1750 (C═O), 1668 (C═O), 1H NMR (DMSO-d6, δ ppm): 1.62–1.64 (m, 2H, CH2), 1.93, 1.95, 1.99 (3s, 9H, 3CH3), 2.35–2.42 (m, 2H, CH2), 2.48 (t, 2H, J= 1.8 Hz, CH2), 2.85 (t, 2H, J= 7.2 Hz, CH2), 2.87 (t, 2H, J= 6.9 Hz, CH2), 3.01 (t, 2H, J= 6.9 Hz, CH2), 3.37 (t, 2H, J= 6.6 Hz, CH2), 4.20 (dd, 1H, J= 3.3, 10.8, Hz, H5″), 4.38 (dd, 1H, J= 11.3, 3.3 Hz, H5″), 4.60 (m, 1H, H4′), 4.72 (s, 2H, CH2), 4.88 (dd, 1H, J= 8.4, 9.2 Hz, H3′), 5.13 (t, 1H, J= 9.30 Hz, H2′), 5.67 (br s, 2H, NH2 exchangeable with D2O), 5.81 (d, 1H, J= 9.80 Hz, H1′), 8.09 (s, 1H, triazole-H), 13C NMR (DMSO-d6, δ ppm): 23.69, 25.68, 27.68, 28.80, 29.06, 29.40, 29.51, 30.24, 30.28, 49.31, 68.56, 68.92, 71.07, 71.29, 72.49, 99.03, 115.73, 124.63, 129.11, 139.41, 143.33, 157.63, 160.05, 169.43; 169.72, 169.96, 171.51. Anal. Calcd. for C27H34N6O9S2 (650.72): C, 49.84; H, 5.27; N, 12.92. Found: C, 49.72; H, 5.20; N, 13.02.
4.2. In vitro cytotoxic activity
HCT-116 (human colorectal carcinoma) and MCF-7 (human breast adenocarcinoma) cell lines were purchased from the American Type Culture Collection (Rockville, MD) and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated foetal bovine serum (FBS), 100 U ml−1 penicillin, and 100 U ml−1 streptomycin. The cells were grown at 37 °C in a humidified atmosphere of 5% CO2.
Cytotoxicity activity against HCT-116 and MCF-7 was estimated by the 3-[4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay. This test is based on MTT cleavage by mitochondrial dehydrogenases from viable cells43,44. Cells were placed in a 96-well sterile microplate (5 × 104 cells well−1) and incubated at 37 °C in serum-free media containing dimethyl sulphoxide (DMSO) and either a series of various concentrations of each compound or doxorubicin (positive control) for 48 h before the MTT assay. After incubation, the media were removed and 40 µl MTT (2.5 mg ml−1) was added to each well. Incubation was resumed for an additional 4 h. The purple formazan dye crystals were solubilised with 200 µl DMSO. Absorbance was measured at 590 nm in a Spectra Max Paradigm Multi-Mode microplate reader (Molecular Devices, LLC, San Jose, CA). Relative cell viability was expressed as the mean percentage of viable cells compared to the untreated control cells.
All experiments were conducted in triplicate and repeated on three different days. All values were reported as mean ± SD. The IC50 values were determined by SPSS Inc. probit analysis (IBM Corp., Armonk, NY).
4.3. Kinase inhibitory activity
The in vitro enzyme inhibition assessment was carried out in confirmatory diagnostic unit, Vacsera (Giza, Egypt). The evaluation performed profiling of the tested compounds against mutant EGFR kinase type L858R using gefitinib as a reference and more details were provided in the supplementary data. The in vitro enzyme inhibition assessment was carried out in confirmatory diagnostic unit, Vacsera (Giza, Egypt). The master mixture (6 μl 5× kinase buffer + 1 μl ATP (500 μM)+1 μl 50× PTK substrate + 17 μl water) was prepared then, 25 μl to every well was added. Five microlitres of inhibitor solution of each well labelled as “Test Inhibitor” was added. However, for the “Positive Control” and “Blank”, 5 μl of the same solution without inhibitor (inhibitor buffer) was added. Three millilitres of 1× kinase buffer by mixing 600 μl of 5× kinase buffer with 2400 μl water was prepared. Hence, 3 ml of 1× kinase buffer became sufficient for 100 reactions. To the wells designated as “Blank”, 20 μl of 1× kinase buffer was added. EGFR enzyme on ice was thawed. Upon first thaw, briefly the tube containing enzyme was spun to recover full content of the tube. The amount of EGFR required for the assay and dilute enzyme to 1 ng/μl with 1× kinase buffer was calculated. Moreover, the remaining undiluted enzyme in aliquots was stored at –80 °C. The reaction was initiated by adding 20 μl of diluted EGFR enzyme to the wells designated “Positive Control” and “Test Inhibitor Control”, after that it was incubated at 30 °C for 40 min. After the 40 minutes’ reaction, 50 μl of Kinase-Glo Max reagent was added to each well and the plate was covered with aluminium foil and incubated at room temperature for 15 min. Luminescence was measured using the microplate reader.
4.4. Molecular docking study
Docking analysis was performed using molecular operating environment (MOE, 10.2008) software according to the previously reported method42,45. The three-dimensional X-ray structure of EGFR (PDB code: 3UG2)46 was obtained from the Protein Data Bank through the Internet. The structure of the EGFR enzyme was prepared using Protonate 3D protocol in MOE with the default options. The co-crystallised ligand, gefitinib was used to define the active site for molecular docking. First, the co-crystallised inhibitor was re-docked into the assigned active EGFR enzyme and the root-mean-square deviation value was evaluated. Then, the molecular docking procedure was done for the newly synthesised compounds 2, 5, 7, and 13–18 into ATP binding site of EGFR. The docking protocol was done using Triangle Matcher placement method and London dG scoring function. All minimisations were achieved with MOE until an RMSD gradient of 0.05 kcal.mol−1 Å–1 with MMFF94x force field and the partial charges were automatically calculated.
Supplementary Material
Disclosure statement
The authors declare that they have no conflicts of interest to disclose.
References
- 1.World Health Organization. Cancer: data and country profiles; 2020. Available from: http://www.who.int/cancer/country/en/.
- 2.Krishnamurty R, Maly DJ.. Biochemical mechanisms of resistance to small-molecule protein kinase inhibitors. ACS Chem Biol 2010;5:121–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu P, Nielsen TE, Clausen MH.. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol Sci 2015;36:422–39. [DOI] [PubMed] [Google Scholar]
- 4.Wu P, Nielsen TE, Clausen MH.. Small-molecule kinase inhibitors: an analysis of FDA-approved drugs. Drug Discov Today 2016;21:5–10. [DOI] [PubMed] [Google Scholar]
- 5.Wieduwilt MJ, Moasser MM.. The epidermal growth factor receptor family: biology driving targeted therapeutics. Cell Mol Life Sci 2008;65:1566–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakeling AE, Guy SP, Woodburn JR, et al. ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res 2002;62:5749–54. [PubMed] [Google Scholar]
- 7.Moyer JD, Barbacci EG, Iwata KK, et al. Induction of apoptosis and cell cycle arrest by CP-358,774, an inhibitor of epidermal growth factor receptor tyrosine kinase. Cancer Res 1997;57:4838–48. [PubMed] [Google Scholar]
- 8.Ameen MA, Ahmed EK, El Malah T, et al. Click chemistry based synthesis of novel architectures bearing sugar unit at the pyridothienopyrimidines. J Heterocycl Chem 2015;52:1093–8. [Google Scholar]
- 9.Kashmiri L, Pinki Y.. Recent advancements in 1,4-disubstituted-1H-1,2,3-triazoles as potential anticancer agents, anti-cancer agents. Med Chem 2018;18:21–37. [DOI] [PubMed] [Google Scholar]
- 10.El-Sayed WA, Khalaf HS, Mohamed SF, et al. Synthesis and antiviral activity of 1,2,3-triazole glycosides based substituted pyridine via click cycloaddition. Russ J Gen Chem 2017;87:2444–53. [Google Scholar]
- 11.Li J, Zheng M, Tang W, et al. Syntheses of triazole-modified zanamivir analogues via click chemistry and anti-AIV activities. Bioorg Med Chem Lett 2006;16:5009–13. [DOI] [PubMed] [Google Scholar]
- 12.Kolb HC, Sharpless KB.. The growing impact of click chemistry on drug discovery. Drug Discovery Today 2003;8:1128–37. [DOI] [PubMed] [Google Scholar]
- 13.Bourne Y, Kolb HC, Radic Z, et al. Freeze-frame inhibitor captures acetylcholinesterase in a unique conformation. Proc Natl Acad Sci U S A 2004;101:1449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahad MA, Hussein HE, Mohamed NB, et al. Synthesis and cytotoxic activity of new 1,3,4-thiadiazole thioglycosides and 1,2,3-triazolyl-1,3,4-thiadiazole N-glycosides. Molecules 2019;24:3738–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dondoni A, Marra A.. C-glycoside clustering on calix[4]arene, adamantane, and benzene scaffolds through 1,2,3-triazole linkers. J Org Chem 2006;71:7546–75. [DOI] [PubMed] [Google Scholar]
- 16.Hotha S, Kashyap S.. "Click chemistry" inspired synthesis of pseudo-oligosaccharides and amino acid glycoconjugates. J Org Chem 2006;71:364–7. [DOI] [PubMed] [Google Scholar]
- 17.Decker M, Design of hybrid molecules for drug development. London (UK): Elsevier Ltd.; 2017. [Google Scholar]
- 18.Elrazaz EZ, Serya RAT, Ismail NSM, et al. Thieno[2,3-d]pyrimidine based derivatives as kinase inhibitors and anticancer agents. Fut J Pharm Sci 2015;1:33–41. [Google Scholar]
- 19.Wasfy AAF, Hassan AA, Khattab RR, et al. Synthesis of some new thioglycosides derived from thieno[2,3-d]pyrimidine derivatives and their anticancer and antioxidant activity. Res J Pharm Biol Chem Sci 2018;9:77–88. [Google Scholar]
- 20.Kandeel MM, Mounir AA, Refaat HM, Kassab AE.. Synthesis of thieno[2,3-d]pyrimidines, thieno[2,3-d]triazinones and thieno[2,3-e]diazepinones of anticipated anti-cancer activity. J Chem Res 2012;36:105–10. [Google Scholar]
- 21.Porter JR, Fritz CC, Depew KM.. Discovery and development of Hsp90 inhibitors: a promising pathway for cancer therapy. Curr Opin Chem Biol 2010;14:412–20. [DOI] [PubMed] [Google Scholar]
- 22.Janin YL. ATPase inhibitors of heat-shock protein 90 second season. Drug Discov Today 2010;15:335–42. [DOI] [PubMed] [Google Scholar]
- 23.Horiuchi TH, Nagata M, Kitagawa M, et al. Discovery of novel thieno[2,3-d]pyrimidin-4-yl hydrazone-based inhibitors of cyclin D1-CDK4: synthesis, biological evaluation and structure–activity relationships. Part 2. Bioorg Med Chem 2009;17:7850–60. [DOI] [PubMed] [Google Scholar]
- 24.Pédeboscq S, Gravier D, Casadebaig F, et al. Synthesis and study of antiproliferative activity of novel thienopyrimidines on glioblastoma cells. Eur J Med Chem 2010;45:2473–9. [DOI] [PubMed] [Google Scholar]
- 25.Hasler WL. Serotonin and the GI tract. Curr Gastroenterol Rep 2009;11:383–91. [DOI] [PubMed] [Google Scholar]
- 26.Shyyka O, Pokhodylo N, Finiuk N, et al. Anticancer activity evaluation of new thieno [2, 3-d] pyrimidin-4 (3H)-ones and thieno [3, 2-d] pyrimidin-4 (3H)-one derivatives. Sci Pharm 2018;86:28. [DOI] [PubMed] [Google Scholar]
- 27.Bysting F, Bugge S, Sundby E, Hoff BH.. Investigation of Heck coupling on 6-bromo [2, 3-d] thienopyrimidines for construction of new EGFR inhibitor lead structures. RSC Adv 2017;7:18569–77. [Google Scholar]
- 28.El-Sayed WA, Hemat SK, Dalia AAO, et al. Synthesis and anticancer activity of new pyrazolyl and oxadiazolyl glycosides based on theinopyrimidine nucleus and their acyclic analogs. Acta Pol Pharm Drug Res 2017;74:1739–51. [Google Scholar]
- 29.Amawi H, Karthikeyan C, Pathak R, et al. Thienopyrimidine derivatives exert their anticancer efficacy via apoptosis induction, oxidative stress and mitotic catastrophe. Eur J Med Chem 2017;138:1053–65. [DOI] [PubMed] [Google Scholar]
- 30.Khattab RR, Hassan AA, Kutkat OM, et al. Synthesis and antiviral activity of novel thieno[2,3-d]pyrimidine hydrazones and their C-nucleosides. Russ J Gen Chem 2019;89:1707–17. [Google Scholar]
- 31.Mulla JAS, Khazi MIA, Panchamukhi SI, et al. Synthesis and pharmacological evaluation of novel thienopyrimidine and triazolothienopyrimidine derivatives. Med Chem Res 2014;23:3235–43. [Google Scholar]
- 32.El-Sayed NNE, Abdelaziz MA, Wardakhan WW, Mohareb RM.. The Knoevenagel reaction of cyanoacetylhydrazine with pregnenolone: synthesis of thiophene, thieno[2,3-d]pyrimidine, 1,2,4-triazole, pyran and pyridine derivatives with anti-inflammatory and anti-ulcer activities. Steroids 2016;107:98–111. [DOI] [PubMed] [Google Scholar]
- 33.El Ashry ESH, El Kilany Y.. Acyclonucleosides: part 3, tri- tetra-, and pentaseco-nucleosides. Adv Heterocycl Chem 1998;69:129–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Y, Pang LP, Wang B, et al. Design and synthesis of novel 1,2,3-triazole-pyrimidine hybrids as potential anticancer agents. Eur J Med Chem 2014;86:368–80. [DOI] [PubMed] [Google Scholar]
- 35.Mohamed AM, Al-Qalawi HRM, El-Sayed WA, et al. Anticancer activity of newly synthesized triazolopyrimidine derivatives and their nucleoside analogs. Acta Pol Pharm Drug Res 2015;72:307–18. [PubMed] [Google Scholar]
- 36.Adel AH, Ibrahim FN, Amira KFS, et al. Synthesis, docking studies into CDK-2 and anticancer activity of new derivatives based pyrimidine scaffold and their derived glycosides. Mini Rev Med Chem 2019;19:1093–110. [DOI] [PubMed] [Google Scholar]
- 37.Halay E, Ay E, Şalva E, et al. Syntheses of 1,2,3-triazole-bridged pyranose sugars with purine and pyrimidine nucleobases and evaluation of their anticancer potential. Nucleosides Nucleotides Nucleic Acids 2017;36:598–619. [DOI] [PubMed] [Google Scholar]
- 38.El-Sayed WA, El-Essawy FA, Ali OM, et al. Synthesis and antiviral evaluation of new 2,5-disbstituted 1,3,4-oxadiazole derivatives and their acyclic nucleoside analogs. Monatsh Chem 2010;141:1021–8. [Google Scholar]
- 39.El-Sayed WA, Abdel-Rahman AAH, Ramiz MMM.. Anti-hepatitis B virus activity of new N4-beta-d-glycoside pyrazolo [3,4-d]pyrimidine derivatives. Z Naturforsch C J Biosci 2009;64:323–8. [DOI] [PubMed] [Google Scholar]
- 40.Hassan AA, Khattab RR, Wasfy AAF, et al. Convenient one pot synthesis of thieno[2′,3′:4,5]pyrimido[1,2-b]-[1,2,4,5] tetrazines. J Het Chem 2018;55:907–12. [Google Scholar]
- 41.Mitsudomi T, Yatabe Y.. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J 2010;277:301–8. [DOI] [PubMed] [Google Scholar]
- 42.Asmaa FK, Eman MHA, Dina SK, et al. Design, synthesis and anticancer activity of new thiazole-tetrazole or triazole hybrid glycosides targeting CDK-2 via structure-based virtual screening. Mini Rev Med Chem 2019;19:933–48. [DOI] [PubMed] [Google Scholar]
- 43.Emam AN, Loutfy SA, Mostafa AA, et al. Cyto-toxicity, biocompatibility and cellular response of carbon dots–plasmonic based nano-hybrids for bioimaging. RSC Adv 2017;7:23502–14. [Google Scholar]
- 44.Flefel EM, El-Sayed WA, Mohamed AM, et al. Synthesis and anticancer activity of new 1-thia-4-azaspiro[4,5]decane, their derived thiazolopyrimidine and 1,3,4-thiadiazole thioglycosides. Molecules 2017;22:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amr AE-GE, Abo-Ghalia MH, Moustafa GO, et al. Synthesis and docking studies of novel macrocyclic pentapeptides as anticancer multi-targeted kinase inhibitors. Molecules 2018;23:2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshikawa S, Kukimoto-Niino M, Parker L, et al. Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor. Oncogene 2013;32:27–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.