Abstract
Colorectal cancer (CRC) is a common malignant tumour of human digestive tract. The high mortality rate of CRC is closely related to the limitations of existing treatments. Thus, there is an urgent need to search for new anti-CRC agents. In this work, twenty novel coumarin-dithiocarbamate derivatives (IDs) were designed, synthesized and evaluated in vitro. The results suggest that the most active compound ID-11 effectively inhibited the proliferation of CRC cell lines while shown little impact on normal colon epithelial cells. Mechanism studies revealed that ID-11 displayed bromodomain-containing protein 4 inhibitory activity, and induced G2/M phase arrest, apoptosis as well as decreased the expression levels of the key genes such as c-Myc and Bcl-2 in CRC cell lines. Moreover, the ADMET properties prediction results shown that ID-11 possess well metabolic characteristics without obvious toxicities. Our data demonstrated that compound ID-11 may be a promising anti-CRC agent and deserved for further development.
Keywords: Colorectal cancer, coumarin-dithiocarbamate derivatives, bromodomain-containing protein 4, antitumor activities
1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide. In 2018, nearly 1.8 million new cases occurred and 881 000 patients died1. It was predicted that the number of deaths for both colon and rectal cancer would increase by 60.0 and 71.5% in all countries until 2035, respectively2. Like other types of cancer, CRC is the result of accumulation of genetic and epigenetic mutations in specific oncogenes and/or tumour suppressor genes. Besides familial CRC (about 10–30%), CRC often occurred in patients with a defined hereditary syndrome (about 5%), sporadic adenomatous colorectal polyps (about 75%) and extensive ulcerative colitis (about 4%)3. Currently, surgical section combined with radiotherapy and chemotherapy still be the main treatments for CRC. However, drug resistance and the severe side effects of these therapies are inevitable, the quality of life of patients would be severely affected. Although some new treatments such as immunotherapy have emerged in recent years, the high costs of which would be a burden for most patients4–7. Therefore, exploring for new anti-CRC agents is of great significance.
The bromodomain-containing protein 4 (BRD4) is one of the most important member of BET (bromodomain and extra-terminal domain) family of transcriptional regulatory proteins, which specially recognise acetylated lysine residues on histones with its bromodomains and recruits transcriptional regulatory complexes to acetylated chromatin.8 Previous studies have shown that the expression disorder of BRD4 is associated with the progression of many cancers including CRC8,9. As BRD4 affects the transcription process of some key oncogenes such as c-Myc, inhibiting the activity of BRD4 may produce potential anticancer effect10. Compounds such as (+)-JQ111, which specifically designed as a BRD4 inhibitor, had been proved to own powerful anticancer activities. Thus, targeting BRD4 represents a promising therapeutic strategy against CRC.
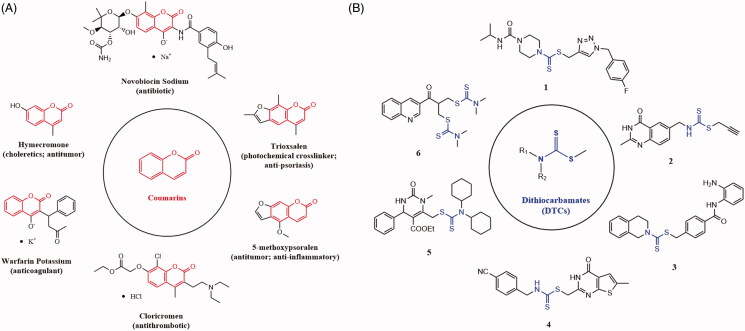
Over the past few decades, natural products play an important role in the discovery of anticancer drugs12–14. Coumarin (benzopyran-2-one, Figure 1) is a classic natural product original from plants, bacteria and fungi15,16. The scaffold of coumarin had been widely used in drug design, especially in anticancer drugs17–19 (Figure 1). Actually, the targets of different anticancer coumarins vary much from each other, mainly involved carbonic anhydrase (isoforms IX and XII)20–23, STAT324, tubulin25, topoisomerase II26, and BRD427, etc. Dithiocarbamates (DTCs) is another important pharmacophore, which often be incorporated into the structure of antitumor agents, such as compound 128 (against MCF-7, MGC-823, SMMC-7721 and EC-9706 cell lines), compound 229 (against A549, MCF-7, Hela, HT-29 and HCT-116 cell lines), compound 330 (against A375, MGC-803, Hela, SMMC-7721, H1299 and HCT-116 cell lines), compound 431 (against A549 and HCT-116 cell lines), compound 532 (against leukaemia, non-small cell lung cancer, melanoma, etc.) and compound 633 (against H460, HepG2, MCF-7 cell lines, etc.) (Figure 1). Likewise, anticancer agents containing DTCs fragment also have different targets, including carbonic anhydrase (isoforms IX and XII)34,35, VEGFR-236 and FAK37, etc.
Figure 1.
Drugs containing coumarin core (A) and antitumor agents containing dithiocarbamate fragment (B).
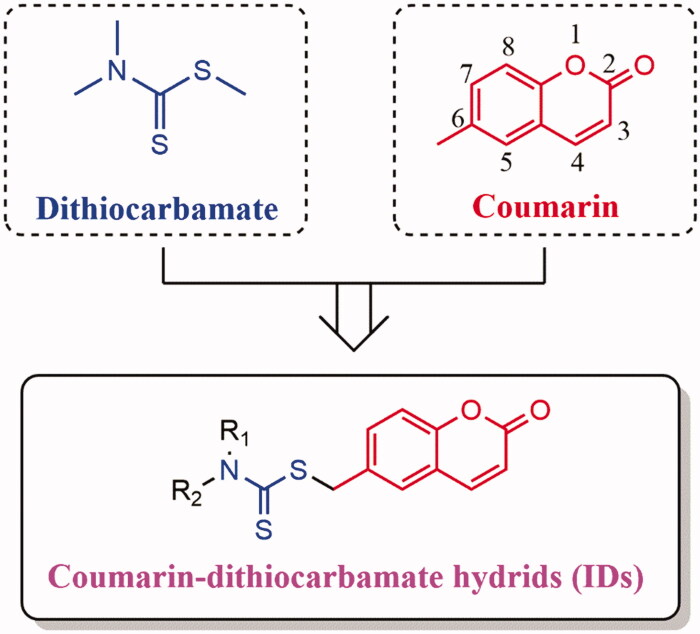
To our knowledge, there are few reports about the hybrids between coumarins and DTCs as potential anti-CRC agents. In this study, we focus on the modification of position C-6 of coumarin scaffold and hybridized with the DTCs. A series of novel coumarin-dithiocarbamate derivatives (Figure 2), which we named IDs uniformly, were designed and synthesized. Herein, we reported the discovery of these IDs as potential anti-CRC agents and explored their underlying anticancer mechanisms including BRD4 inhibition assay, the effects on cell cycle, cell apoptosis as well as the expression levels of c-Myc and Bcl-2 in CRC cell lines in vitro. In addition, the physicochemical properties of the most active agent were also investigated.
Figure 2.
Design strategy of the novel coumarin-dithiocarbamate derivatives (IDs).
2. Materials and methods
2.1. Chemistry
All chemicals were purchased from Aladdin (Shanghai, China) and Energy Chemical (Shanghai, China) and used without further purification. Reaction progress was monitored by thin layer chromatography (TLC) on precoated silica gel GF254 plates (Qingdao Haiyang Chemical Plant, Qingdao, China) and visualised under UV light (254 nm). 1H NMR and 13 C NMR spectra were recorded on Bruker AVANCE III 400 or 600 spectrometer with DMSO-d6 or CDCl3 as the solvent, and the chemical shifts (d) given in parts per million (ppm) relative to tetramethylsilane (TMS) as an internal standard. Coupling constants (J) were reported in hertz, and splitting patterns were used as follows: s, singlet; br. s, broad singlet; d, doublet; dd, doublet of doublets; t, triplet; m, multiplet. High resolution mass spectrometry (HRMS) spectra were recorded on SHIMADZU LCMS-IT-TOF with an electrospray ion source (ESI). Melting point was measured on an SGW®X-4A micro melting point apparatus (Shanghai INESA Physico-Optical Instrument Co., Ltd, China).
2.2. General procedures
2.2.1. Synthesis of intermediate 6-(bromomethyl)-2H-chromen-2-one (9)
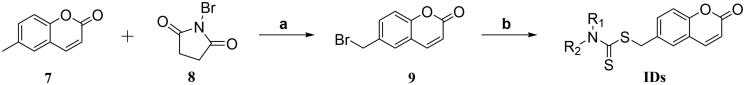
6-Methyl-2H-chromen-2-one (7, 1.00 g, 6.24 mmol) and 2,2′-dimethyl-2,2′-azodipropionitrile (102 mg, 0.624 mmol) was dissolved in CCl4 (25 ml), and then the 1-bromopyrrolidine-2,5-dione (8, 1.11 g, 6.24 mmol) was added in batches at room temperature within 15 min. After that, the mixture was transferred, stirred and refluxed at 90 °C for another 10 h until the reaction finished. Then the CCl4 was removed under reduced pressure, and the residue containing 6-(bromomethyl)-2H-chromen-2-one (9) was used in next step directly without further purification.
2.2.2. General procedures for the preparation of final products ID-1 to ID-20
First, dimethylamine (28 μL, 0.557 mmol) and KOH (31 mg, 0.557 mmol) was dissolved in dry ethanol (10 ml), and the mixture was stirred at room temperature for 20 min. Then, CS2 (33 μL, 0.557 mmol) was added and stirred for another 30 min. After that, intermediate 9 (100 mg, 0.418 mmol) was added slowly and continue to stir at room temperature for another 24 h. After the reaction finished, ethanol was removed under reduced pressure, and the residue was extracted with ethyl acetate/H2O (v/v, 100 ml) for three times. The ethyl acetate layer was collected, concentrated, and purified by silica gel column chromatography (PE: EA = 5: 1) to afford the title product ID-1.
Compounds ID-6, ID-7, ID10, ID-12, ID-15, and ID-20 were prepared with a similar procedure which described for the synthesis of compound ID-1. For the synthesis of compounds ID-2 ∼ ID-5, ID-8, ID-9, ID-13, ID-14, and ID-16 ∼ ID-19, in which the solvent was dioxane/H2O instead of dry ethanol and the alkali was K2CO3 instead of KOH. For the synthesis of compound ID-11, actone was used as the solvent and K3PO4 was used as the alkali.
2.2.2.1. (2-Oxo-2H-chromen-6-yl)methyl dimethylcarbamodithioate (ID-1)
White powder. Yield: 71%. m.p: 151.6–152.5 °C. 1H NMR (400 MHz, CDCl3) (ppm): 7.67 (1H, d, J = 9.2 Hz, H-4), 7.54 (1H, d, J =7.2 Hz, H-7), 7.53 (1H, s, H-5), 7.26 (1H, d, J = 6.4 Hz, H-8), 6.41 (1H, d, J = 9.2 Hz, H-3), 4.61 (2H, s, –SCH2–), 3.55 (3H, s, –N(CH3)2), 3.36 (3H, s, –N(CH3)2). 13 C NMR (100 MHz, CDCl3) (ppm): 195.9, 160.7, 153.2, 143.3, 133.3, 132.8, 128.3, 118.7, 117.0, 116.8, 45.6, 41.5, 41.0. HRMS (ESI, m/z) calcd. for C13H13NO2S2 (M + H)+ 280.0460, found 208.0461.
2.2.2.2. (2-Oxo-2H-chromen-6-yl)methyl diethylcarbamodithioate (ID-2)
White powder. Yield: 50%. m.p: 100.1–101.0 °C. 1H NMR (400 MHz, DMSO-d6) (ppm): 8.04 (1H, d, J =9.6 Hz, H-4), 7.73 (1H, d, J =2.4 Hz, H-5), 7.62 (1H, dd, J1=8.8 Hz, J2=2.4 Hz, H-7), 7.35 (1H, d, J =8.4 Hz, H-8), 6.48 (1H, d, J = 9.6 Hz, H-3), 4.58 (2H, s, –SCH2–), 3.96 (2H, q, J = 6.8 Hz, –N(CH2CH3)2), 3.72 (2H, q, J =6.8 Hz, –N(CH2CH3)2), 1.21–1.15 (6H, m, –N(CH2CH3)2). 13 C NMR (100 MHz, DMSO-d6) (ppm): 193.0, 159.8, 152.6, 144.0, 133.0, 132.7, 128.6, 118.5, 116.4 × 2, 49.1, 46.5, 39.6, 12.3, 11.3. HRMS (ESI, m/z) calcd. for C15H17NO2S2 (M + Na)+ 330.0593, found 330.0599.
2.2.2.3. (2-Oxo-2H-chromen-6-yl)methyl dipropylcarbamodithioate (ID-3)
Yellowish powder. Yield: 60%. m.p: 52.3–53.2 °C. 1H NMR (400 MHz, CDCl3) (ppm): 7.65 (1H, d, J = 9.6 Hz, H-4), 7.51–7.48 (2H, m, H-5, H-7), 7.20 (1H, d, J = 8.4 Hz, H-8), 6.35 (1H, d, J = 9.6 Hz, H-3), 4.55 (2H, s, –SCH2–), 3.86 (2H, t, –N(CH2CH2CH3)2), 3.56 (2H, t, –N(CH2CH2CH3)2), 1.76–1.63 (4H, m, –N(CH2CH2CH3)2), 0.88 (6H, t, –N(CH2CH2CH3)2). 13 C NMR (100 MHz, CDCl3) (ppm): 194.8, 160.6, 153.1, 143.3, 133.3, 132.8, 128.4, 118.6, 116.9, 116.7, 57.0, 54.4, 40.7, 20.7, 19.6, 11.1 × 2. HRMS (ESI, m/z) calcd. for C17H21NO2S2 (M + Na)+ 358.0906, found 358.0918.
2.2.2.4. (2-Oxo-2H-chromen-6-yl)methyl dibutylcarbamodithioate (ID-4)
Yellowish liquid. Yield: 70%. 1H NMR (400 MHz, CDCl3) (ppm): 7.69 (1H, d, J =9.6 Hz, H-4), 7.55–7.53 (2H, m, H-5, H-7), 7.24 (1H, d, J =8.4 Hz, H-8), 6.40 (1H, d, J =9.6 Hz, H-3), 4.60 (2H, s, –SCH2–), 3.95 (2H, t, J = 8.0 Hz, –N(CH2CH2CH2CH3)2), 3.64 (2H, t, J = 8.0 Hz, –N(CH2CH2CH2CH3)2), 1.74–1.67 (4H, m, –N(CH2CH2CH2CH3)2), 1.38–1.31 (4H, m, –N(CH2CH2CH2CH3)2), 0.96–0.91 (6H, m, –N(CH2CH2CH2CH3)2). 13 C NMR (100 MHz, CDCl3) (ppm): 194.6, 160.6, 153.2, 143.3, 133.3, 132.9, 128.4, 118.7, 116.9, 116.7, 55.2, 52.5, 40.7, 29.3, 28.3, 20.0 × 2, 13.8, 13.7. HRMS (ESI, m/z) calcd. for C19H25NO2S2 (M + Na)+ 386.1219, found 386.1222.
2.2.2.5. (2-Oxo-2H-chromen-6-yl)methyl azetidine-1-carbodithioate (ID-5)
White powder. Yield: 56%. m.p: 144.0–145.0 °C. 1H NMR (400 MHz, CDCl3) (ppm): 7.66 (1H, d, J =9.6 Hz, H-4), 7.55–7.53 (2H, m, H-5, H-7), 7.25 (1H, d, J =9.2 Hz, H-8), 6.40 (1H, d, J = 9.6 Hz, H-3), 4.59 (2H, s, –SCH2–), 4.30 (2H, t, J =7.6 Hz, H-a), 4.17 (2H, t, J =7.6 Hz, H-c), 2.43–2.35 (2H, m, H-b). 13 C NMR (100 MHz, CDCl3) (ppm): 192.8, 160.7, 153.2, 143.2, 133.9, 132.7, 128.2, 118.7, 117.0, 116.8, 54.8, 53.1, 38.9, 15.5. HRMS (ESI, m/z) calcd. for C14H13NO2S2 (M + Na)+ 314.0280, found 314.0281.
2.2.2.6. (2-Oxo-2H-chromen-6-yl)methyl 3-hydroxyazetidine-1-carbodithioate (ID-6)
White powder. Yield: 55%. m.p: 137.8–138.7 °C. 1H NMR (400 MHz, DMSO-d6) (ppm): 8.04 (1H, d, J = 9.6 Hz, H-4), 7.70 (1H, d, J =1.2 Hz, H-5), 7.61 (1H, dd, J1 = 8.4 Hz, J2 = 1.6 Hz, H-7), 7.35 (1H, d, J = 8.4 Hz, H-8), 6.48 (1H, d, J = 9.6 Hz, H-3), 5.94 (1H, d, J = 6.4 Hz, –N(CH2)2CHOH), 4.58–4.52 (3H, m, –SCH2–, –N(CH2)2CHOH), 4.39 (2H, m, –N(CH2)2CHOH), 3.90–3.84 (2H, m, –N(CH2)2CHOH). 13 C NMR (100 MHz, DMSO-d6) (ppm): 191.7, 159.8, 152.6, 144.0, 133.8, 132.6, 128.4, 118.5, 116.4 × 2, 64.4, 62.5, 59.8, 38.2. HRMS (ESI, m/z) calcd. for (M + Na)+ 330.0229, found 330.0232.
2.2.2.7. (2-Oxo-2H-chromen-6-yl)methyl pyrrolidine-1-carbodithioate (ID-7)
White powder. Yield: 50%. m.p: 110.2–111.1 °C. 1H NMR (400 MHz, CDCl3) (ppm): 7.67 (1H, d, J =9.6 Hz, H-4), 7.56–7.54 (2H, m, H-5, H-7), 7.25 (1H, d, J = 9.2 Hz, H-8), 6.40 (1H, d, J = 9.6 Hz, H-3), 4.63 (2H, s, –SCH2–), 3.93 (2H, t, J =6.8 Hz, –NCH2CH2CH2CH2N–(pyrrolidine)), 3.63 (2H, t, J =6.8 Hz, –NCH2CH2CH2CH2N–(pyrrolidine)), 2.10–2.03 (2H, m, –NCH2CH2CH2CH2N–(pyrrolidine)), 2.01–1.94 (2H, m, –NCH2CH2CH2CH2N–(pyrrolidine)). 13 C NMR (100 MHz, CDCl3) (ppm): 191.5, 160.7, 153.2, 143.3, 133.7, 132.8, 128.3, 118.7, 117.0, 116.8, 55.3, 50.6, 39.8, 26.0, 24.2. HRMS (ESI, m/z) calcd. for C15H15NO2S2 (M + Na)+ 328.0436, found 328.0444.
2.2.2.8. (2-Oxo-2H-chromen-6-yl)methyl piperidine-1-carbodithioate (ID-8)
Yellowish powder. Yield: 70%. m.p: 130.1–131.0 °C. 1H NMR (400 MHz, CDCl3) (ppm): 7.67 (1H, d, J =9.6 Hz, H-4), 7.56–7.53 (2H, m, H-5, H-7), 7.26 (1H, d, J =8.0 Hz, H-8), 6.40 (1H, d, J = 9.6 Hz, H-3), 4.62 (2H, s, –SCH2–), 4.28 (2H, br, H-a), 3.86 (2H, br, H-e), 1.69 (6H, br, H-b, H-c, H-d). 13 C NMR (100 MHz, CDCl3) (ppm): 194.3, 160.7, 153.2, 143.3, 133.3, 132.9, 128.4, 118.7, 117.0, 116.8, 53.2, 51.4, 40.7, 26.0, 25.4, 24.2. HRMS (ESI, m/z) calcd. for C16H17NO2S2 (M + Na)+ 342.0593, found 342.0585.
2.2.2.9. (2-Oxo-2H-chromen-6-yl)methyl azepane-1-carbodithioate (ID-9)
White powder. Yield: 66%. m.p: 72.6–73.5 °C. 1H NMR (400 MHz, CDCl3) (ppm): 7.65 (1H, d, J = 9.6 Hz, H-4), 7.53–7.50 (2H, m, H-5, H-7), 7.22 (1H, d, J = 8.0 Hz, H-8), 6.37 (1H, d, J = 9.6 Hz, H-3), 4.58 (2H, s, –SCH2–), 4.15 (2H, t, J = 6.0 Hz, H-a), 3.83 (2H, t, J = 6.0 Hz, H-f), 1.84–1.79 (4H, m, H-b, H-e), 1.23–1.20 (2H, m, H-c), 0.85–0.78 (2H, m, H-d). 13 C NMR (100 MHz, CDCl3) (ppm): 195.0, 160.6, 153.2, 143.3, 133.4, 132.8, 128.4, 118.7, 116.9, 116.7, 55.8, 52.8, 40.6, 27.3, 26.6, 26.5, 26.1. HRMS (ESI, m/z) calcd. for C17H19NO2S2 (M + Na)+ 356.0749, found 356.0751.
2.2.2.10. (2-Oxo-2H-chromen-6-yl)methyl morpholine-4-carbodithioate (ID-10)
White powder. Yield: 57%. m.p: 135.4–136.3 °C. 1H NMR (600 MHz, DMSO-d6) (ppm): 8.04 (1H, d, J =9.6 Hz, H-4), 7.73 (1H, s, H-5), 7.63 (1H, dd, J1=8.4 Hz, J2=1.8 Hz, H-7), 7.35 (1H, d, J = 9.0 Hz, H-8), 6.48 (1H, d, J =9.0 Hz, H-3), 4.63 (2H, s, –SCH2–), 4.22 (2H, br, –N(CH2CH2)2O–(morpholine)), 3.90 (2H, br, -N(CH2CH2)2O-(morpholine)), 3.65 (4H, br, –N(CH2CH2)2O–(morpholine)). 13 C NMR (125 MHz, DMSO-d6) (ppm): 197.8, 162.9, 155.8, 147.1, 136.0, 135.9, 131.8, 121.7, 119.5, 119.5, 68.6 × 2, 54.4, 53.3, 43.1. HRMS (ESI, m/z) calcd. for C15H15NO3S2 (M + Na)+ 344.0386, found 344.0391.
2.2.2.11. (2-Oxo-2H-chromen-6-yl)methyl 1H-imidazole-1-carbodithioate (ID-11)
Yellow powder. Yield: 68%. m.p: 139.1–140.0 °C. 1H NMR (400 MHz, DMSO-d6) (ppm): 8.64 (1H, s, H-b), 8.05 (1H, d, J =9.6 Hz, H-4), 8.01 (1H, s, H-5), 7.82 (1H, s, H-e), 7.71 (1H, d, J = 8.4 Hz, H-7), 7.40 (1H, d, J =8.4 Hz, H-8), 7.17 (1H, s, H-d), 6.50 (1H, d, J =9.6 Hz, H-3), 4.78 (2H, s, –SCH2–). 13 C NMR (100 MHz, DMSO-d6) (ppm): 197.8, 159.7, 153.0, 143.8, 135.9, 133.0, 131.7, 130.6, 129.0, 118.7, 118.3, 116.6 × 2, 39.6. HRMS (ESI, m/z) calcd. for C14H10N2O2S2 (M + Na)+ 325.0076, found 325.0070.
2.2.2.12. (2-Oxo-2H-chromen-6-yl)methyl 4-methylpiperazine-1-carbodithioate (ID-12)
Yellowish powder. Yield: 53%. m.p: 124.3–125.2 °C. 1H NMR (400 MHz, DMSO-d6) (ppm): 8.03 (1H, d, J = 9.6 Hz, H-4), 7.73 (1H, d, J = 2.0 Hz, H-5), 7.62 (1H, dd, J1=8.8 Hz, J2=2.0 Hz, H-7), 7.35 (1H, d, J =8.4 Hz, H-8), 6.48 (1H, d, J =9.6 Hz, H-3), 4.61 (2H, s, –SCH2–), 4.23 (2H, br, –N(CH2CH2)2N–(piperazine)), 3.87 (2H, br, –N(CH2CH2)2N–(piperazine)), 2.38 (4H, br, –N(CH2CH2)2N–(piperazine)), 2.19 (3H, s, –NCH3). 13 C NMR (100 MHz, DMSO-d6) (ppm): 194.1, 159.8, 152.6, 143.9, 132.9, 132.8, 128.7, 118.5, 116.4 × 2, 53.9 × 2, 50.9, 49.5, 45.0 × 2. HRMS (ESI, m/z) calcd. for C16H18N2O2S2 (M + Na)+ 357.0702, found 357.0709.
2.2.2.13. (2-Oxo-2H-chromen-6-yl)methyl 4-isopropylpiperazine-1-carbodithioate (ID-13)
Yellowish powder. Yield: 58%. m.p: 73.6–74.5 °C. 1H NMR (400 MHz, DMSO-d6) (ppm): 8.04 (1H, d, J = 9.6 Hz, H-4), 7.73 (1H, d, J = 2.0 Hz, H-5), 7.63 (1H, dd, J1=8.8 Hz, J2=2.4 Hz, H-7), 7.35 (1H, d, J =8.4 Hz, H-8), 6.48 (1H, d, J =9.6 Hz, H-3), 4.61 (2H, s, –SCH2–), 4.21 (2H, br, –N(CH2CH2)2N–CH(CH3)2), 3.86 (2H, br, –N(CH2CH2)2N-CH(CH3)2), 2.71–2.65 (1H, m, –N(CH2CH2)2N–CH(CH3)2), 2.50–2.49 (4H, m, –N(CH2CH2)2N–CH(CH3)2), 0.96 (3H, s, –N(CH2CH2)2N–CH(CH3)2), 0.95 (3H, s, –N(CH2CH2)2N–CH(CH3)2). 13 C NMR (100 MHz, DMSO-d6) (ppm): 193.7, 159.8, 152.6, 143.9, 132.9, 132.8, 128.7, 118.5, 116.4 × 2, 53.4, 51.5, 50.1, 47.7, 47.6 × 2, 18.0 × 2. HRMS (ESI, m/z) calcd. for C18H22N2O2S2 (M + H)+ 363.1195, found 363.1201.
2.2.2.14. (2-Oxo-2H-chromen-6-yl)methyl 4-(tert-butyl)piperazine-1-carbodithioate (ID-14)
Yellowish powder. Yield: 62%. m.p: 159.4–160.3 °C. 1H NMR (400 MHz, DMSO-d6) (ppm): 8.04 (1H, d, J =9.6 Hz, H-4), 7.73 (1H, d, J = 2.0 Hz, H-5), 7.62 (1H, dd, J1=8.8 Hz, J2=2.4 Hz, H-7), 7.35 (1H, d, J =8.4 Hz, H-8), 6.48 (1H, d, J =9.6 Hz, H-3), 4.60 (2H, s, –SCH2–), 4.18 (2H, br, –N(CH2CH2)2N–C(CH3)3), 3.84 (2H, br, –N(CH2CH2)2N–C(CH3)3), 2.55 (4H, br, –N(CH2CH2)2N–C(CH3)3), 1.00 (9H, s, –N(CH2CH2)2N–C(CH3)3). 13 C NMR (100 MHz, DMSO-d6) (ppm): 193.5, 159.8, 152.6, 143.9, 133.0, 132.8, 128.7, 118.5, 116.4 × 2, 53.3, 51.7, 50.4, 45.4, 45.3, 45.2, 25.5 × 3. HRMS (ESI, m/z) calcd. for C19H24N2O2S2 (M + H)+ 377.1352, found 377.1349.
2.2.2.15. (2-Oxo-2H-chromen-6-yl)methyl 4–(2-hydroxyethyl)piperazine-1-carbodithioate (ID-15)
Yellowish powder. Yield: 43%. m.p: 120.9–121.8 °C. 1H NMR (400 MHz, DMSO-d6) (ppm): 8.04 (1H, d, J =9.6 Hz, H-4), 7.73 (1H, d, J = 2.0 Hz, H-5), 7.62 (1H, dd, J1=8.4 Hz, J2=2.0 Hz, H-7), 7.35 (1H, d, J =8.4 Hz, H-8), 6.48 (1H, d, J =9.6 Hz, H-3), 4.61 (2H, s, –SCH2–), 4.47 (1H, t, J =4.8 Hz, –NCH2CH2OH), 4.22 (2H, br, –N(CH2CH2)2N–(piperazine)), 3.87 (2H, br, –N(CH2CH2)2N–(piperazine)), 3.50 (2H, q, J = 5.6 Hz, –NCH2CH2OH), 2.50 (4H, br, –N(CH2CH2)2N–(piperazine)), 2.42 (2H, t, J = 6.0 Hz, –NCH2CH2OH). 13 C NMR (100 MHz, DMSO-d6) (ppm): 193.9, 159.8, 152.6, 143.9, 132.9, 132.8, 128.7, 118.5, 116.4 × 2, 59.4, 58.4, 52.5, 52.4, 51.0, 49.6, 39.5. HRMS (ESI, m/z) calcd. for C17H20N2O3S2 (M + Na)+ 387.0808, found 387.0808.
2.2.2.16. (2-Oxo-2H-chromen-6-yl)methyl 4-cyclohexylpiperazine-1-carbodithioate (ID-16)
Yellowish powder. Yield: 68%. m.p: 106.9–107.8 °C. 1H NMR (400 MHz, DMSO-d6) (ppm): 8.04 (1H, d, J = 9.6 Hz, H-4), 7.73 (1H, d, J = 2.0 Hz, H-5), 7.62 (1H, dd, J1=8.4 Hz, J2=2.0 Hz, H-7), 7.35 (1H, d, J =8.8 Hz, H-8), 6.48 (1H, d, J =9.6 Hz, H-3), 4.60 (2H, s, –SCH2–), 4.19 (2H, br, H-a), 3.84 (2H, br, H-d), 2.56–2.54 (5H, m, H-b, H-c, H-e), 1.73–1.70 (4H, m, H-f, H-j), 1.56–1.53 (1H, m, H-h), 1.22–1.00 (5H, m, H-g, H-h, H-i). 13 C NMR (100 MHz, DMSO-d6) (ppm): 193.7, 179.3, 159.8, 152.6, 143.9, 132.9, 132.8, 128.7, 118.5, 116.4, 62.2, 51.5, 50.2, 48.2, 48.1, 48.0, 29.4, 28.2, 25.7, 25.1 × 2. HRMS (ESI, m/z) calcd. for C21H26N2O2S2 (M + H)+ 403.1508, found 403.1509.
2.2.2.17. (2-Oxo-2H-chromen-6-yl)methyl 4–(4-hydroxyphenyl)piperazine-1-carbodithioate (ID-17)
Yellowish powder. Yield: 55%. m.p: 195.5–196.4 °C. 1H NMR (400 MHz, DMSO-d6) (ppm): 8.91 (1H, s, –OH), 8.04 (1H, d, J = 9.6 Hz, H-4), 7.74 (1H, d, J = 2.0 Hz, H-5), 7.64 (1H, dd, J1=8.4 Hz, J2=2.0 Hz, H-7), 7.36 (1H, d, J =8.4 Hz, H-8), 6.80 (2H, d, J =8.8 Hz, H-f, H-g), 6.66 (2H, d, J =8.8 Hz, H-e, H-h), 6.48 (1H, d, J =9.6 Hz, H-3), 4.63 (2H, s, –SCH2–), 4.36 (2H, br, H-a), 4.05–3.99 (2H, m, H-d), 3.05 (4H, br, H-b, H-c). 13 C NMR (100 MHz, DMSO-d6) (ppm): 194.2, 159.8, 152.6, 151.4, 143.9, 143.1, 132.9, 132.8, 128.7, 118.5, 118.3 × 2, 116.4 × 2, 115.4 × 2, 51.0, 50.9, 49.7 × 2, 39.5. HRMS (ESI, m/z) calcd. for C21H20N2O3S2 (M–H)- 411.0843, found 411.0860.
2.2.2.18. (2-Oxo-2H-chromen-6-yl)methyl 4–(4-cyanophenyl)piperazine-1-carbodithioate (ID-18)
Yellowish powder. Yield: 55%. m.p: 181.5–182.4 °C. 1H NMR (400 MHz, DMSO-d6) (ppm): 8.04 (1H, d, J = 9.6 Hz, H-4), 7.74 (1H, d, J = 1.6 Hz, H-5), 7.64 (1H, dd, J1=8.4 Hz, J2=1.6 Hz, H-7), 7.60 (2H, d, J =8.8 Hz, H-f, H-g), 7.36 (1H, d, J =8.4 Hz, H-8), 6.94 (2H, d, J =8.8 Hz, H-e, H-h), 6.48 (1H, d, J = 9.6 Hz, H-3), 4.65 (2H, s, –SCH2–), 4.35 (2H, br, H-a), 4.06 (2H, br, H-d), 3.55 (4H, t, J =5.2 Hz, H-b, H-c). 13 C NMR (100 MHz, DMSO-d6) (ppm): 194.4, 159.8, 152.6, 151.9, 143.9, 133.3 × 2, 132.9, 132.8, 128.7, 119.9, 118.5, 116.4, 116.4, 113.3 × 2, 98.1, 50.2, 50.1, 48.6, 48.5, 44.9. HRMS (ESI, m/z) calcd. for C22H19N3O2S2 (M + Na)+ 444.0811, found 444.0827.
2.2.2.19. (2-Oxo-2H-chromen-6-yl)methyl 4-phenylpiperidine-1-carbodithioate (ID-19)
White powder. Yield: 66%. m.p: 115.6–116.5 °C. 1H NMR (400 MHz, CDCl3) (ppm): 7.68 (1H, d, J = 9.6 Hz, H-4), 7.58–7.56 (2H, m, H-5, H-7), 7.33–7.27 (3H, m, H-g, H-i, H-8), 7.24–7.18 (3H, m, H-f, H-h, H-j), 6.42 (1H, d, J = 9.2 Hz, H-3), 5.75 (1H, br, H-a), 4.72–4.64 (3H, m, –SCH2–, H-a), 3.26–3.16 (2H, m, H-e), 2.92–2.84 (1H, m, H-c), 2.00–1.96 (2H, m, H-b), 1.80 (2H, br, H-d). 13 C NMR (100 MHz, CDCl3) (ppm): 194.9, 160.6, 153.3, 144.2, 143.2, 133.2, 132.9, 128.6 × 2, 128.4, 126.8, 126.7 × 2, 118.7, 117.0, 116.9, 52.6, 50.8, 42.5, 40.8, 33.2, 32.6. HRMS (ESI, m/z) calcd. for C22H21NO2S2 (M + Na)+ 418.0906, found 418.0910.
2.2.2.20. (2-Oxo-2H-chromen-6-yl)methyl 4–(4-methylpiperazin-1-yl)piperidine-1-carbodithioate (ID-20)
Orange powder. Yield: 66%. m.p: 95.0–96.0 °C. 1H NMR (400 MHz, DMSO-d6) (ppm): 8.04 (1H, d, J =9.6 Hz, H-4), 7.73 (1H, d, J =2.0 Hz, H-5), 7.62 (1H, dd, J1=8.4 Hz, J2=2.4 Hz, H-7), 7.35 (1H, d, J =8.8 Hz, H-8), 6.48 (1H, d, J =9.6 Hz, H-3), 4.59 (2H, d, J = 4.0 Hz, –SCH2–), 3.36–3.23 (2H, m, –CH(CH2CH2)N–(piperidine)), 2.58–2.53 (1H, m, –CH(CH2CH2)N–(piperidine)), 2.52–2.49 (2H, m, –CH(CH2CH2)N–(piperidine)), 2.45 (4H, br, –N(CH2CH2)2N–CH3–(piperazine)), 2.30 (4H, br, –N(CH2CH2)2N–CH3–(piperazine)), 2.14 (3H, s, –N(CH2CH2)2N–CH3), 1.867–1.839 (2H, m, –CH(CH2CH2)N–(piperidine)), 1.38–1.34 (2H, m, –CH(CH2CH2)N–(piperidine)). 13 C NMR (100 MHz, DMSO-d6) (ppm): 193.2, 159.8, 152.6, 144.0, 133.0, 132.8, 128.7, 118.5, 116.4 × 2, 59.7, 54.9 × 2, 48.3 × 3, 45.5 × 2, 39.8, 28.0, 27.5. HRMS (ESI, m/z) calcd. for C21H27N3O2S2 (M + H)+ 418.1617, found 418.1630.
2.3. Biological assay
2.3.1. Cell lines and cell culture
Human colorectal cancer cell lines including RKO, SW620 and SW480 were purchased from Chinese Academy of Sciences, a typical cell library culture preservation committee (Shanghai, China). Normal human colon epithelial cell line NCM460 was purchased from ATCC (Washington, DC, USA). RPMI-1640 medium (12633020), foetal bovine serum (FBS, 26170043), penicillin-streptomycin (10378016) and phosphate buffered saline (PBS, 70011044) were purchased from Gibco (Eggenstein, Germany). 5-Fluorouracil (5-FU, E0201010050, purity ≥98%) was purchased from Energy Chemical (Shanghai, China). Dimethyl sulfoxide (DMSO, 41639) was obtained from Sigma-Aldrich (St. Louis, MO). 3–(4,5-dimethylthiazol-2-yl)-5–(3-carboxymethoxyphenyl)-2–(4-sulfophenyl)-2H-tetrazolium (MTS, G1111) was purchased from Promega (Madison, WI, USA). All cell lines were cultured in RPMI-1640 medium supplemented with 10% FBS, penicillin (100 units/mL) and streptomycin (100 mg/mL) at 37 °C in a humidified atmosphere containing 5% CO2.
2.3.2. MTS assay
Cell viability was measured via MTS assay. RKO, SW620, SW480 and NCM460 cells were seeded in 96-well plates at a density of 5000 cells per well for 24 h. After treatment with the test compounds at indicated concentration for another 48 h or 72 h at 37 °C, MTS solution was added to each well and incubated for another 30 min at 37 °C. Then, the absorbance at 490 nm was measured by a BioTek Synergy H4 microplate Reader (Vermont, USA). The IC50 values were calculated with Graphpad 7.0 software packages.
2.3.3. Binding affinity of target compounds to BRD4(BD1) and BRD4(BD2) by HTRF assay
In brief, compounds were serially diluted in Echo plate according to the plate map and DMSO’s final fraction is 0.1%. Then, transferring compounds and DMSO to 384-well assay plate by Echo. After that, 2× Protein and Peptide Mix was added to the assay plate and 2× Detection Mix was then added to assay plate and shook for 30 s. After incubating the plate at room temperature for 1.5 h, the HTRF signals (Excitation wavelength at 340 nm, Emission wavelength at 615 & 665 nm) was read on EnVision (PerkinElmer, UK). (+)-JQ1 (BPS Bioscience, Cat. No. 27402) was used as a positive control. BRD4-1 (RD-11–157) and BRD4-2 (RD-11–158) were purchased from Reaction Biology Corp. (PENN, USA). For Curve fitting, the inhibition values was calculated in Excel using Equation (1): Inhibition %=(Max-Signal)/(Max-Min)*100, and the IC50 values was calculated in XL-Fit using Equation (2): Y = Bottom + (Top-Bottom)/(1+(IC50/X)*HillSlope), Y is %inhibition and X is compound concentration.
2.3.4. Colony formation
SW620 and SW480 cells were seeded in 6-well plates (1000 cells per well) and cultured in RPMI-1640 medium containing 10% FBS overnight. After the cells treatment with DMSO or compound ID-11 (4, 8, 16, and 32 μM) for 24 h, the culture medium was replaced by fresh RPMI-1640 medium (containing 10% FBS) and continue culturing for another 10 days. Then the cells were washed with PBS and fixed with methanol for 15 min and stained with 0.1% crystal violet for another 30 min. Finally, the cells were washed with running water, air-dried and visualised by photographing.
2.3.5. Cell cycle analysis
Cells were seeded in 6-well plates (40,000 cells per well) and cultured in RPMI-1640 medium containing 10% FBS overnight. Then the cells were exposed to DMSO or compound ID-11 (8 or 16 μM) for 24 h. After that, cells were collected, washed with cold PBS for twice and fixed in 75% ethanol overnight at 4 °C. Subsequently, ethanol was removed and suspended in PBS for twice, and then RNase A and propidium iodide (PI) (Multi Sciences, Hangzhou, China) was added and keep in the darkness for 30 min. Finally, the percentages of cells in different cell cycle phases were measured using a FACS Calibur flow cytometer (Bectone Dickinson, San Jose, CA, USA).
2.3.6. Cell apoptosis analysis
Cell apoptosis was determined using Annexin V-FITC/PI Apoptosis Detection Kit (BD Biosciences, New Jersey, USA) in according with manufacturer's instruction. Briefly, cells were cultured in 6-well plates (40 000 cells per well) till reaching logarithmic growth phase and then treatment with DMSO or compound ID-11 (8 or 16 μM) for 24 h. Afterwards, the cells were harvested, washed with PBS for twice and stained with 100 μL of the mixture of Annexin V/FITC and PI in binding buffer (v/v, 5: 5: 100) in the darkness for 30 min. Another 200 μL binding buffer was added before detection. The apoptotic cells were then analysed by FACS Calibur flow cytometry immediately.
2.3.7. Western blot assay
Cells were cultured in 6-well plates at a density of 40,000 cells per well overnight, and then exposed to selected compounds at different concentrations for 24 h. Subsequently, the cells were lysed in lysate buffer containing 50 mM Tris-HCl (PH 6.8), 2% SDS, 0.1% bromophenol blue, 1.5% DTT, 10% glycerol. The total protein extracts were boiled, sonicated, separated by 10–12% SDS-PAGE gels and transferred to polyvinylidene fluoride (PVDF) membranes. Then the membranes were blocked with 5% non-fat milk at room temperature for 1 h and incubated with primary antibodies at 4 °C overnight. After that, membranes were washed with 1× TBST for three times and incubated with secondary antibody at room temperature for 2 h. Finally, after washing with 1× TBST for three times, the bolts were visualised by an enhanced chemiluminescence kit (Beyotime, China) using the Amersham Imager 600 system (GE Healthcare Life Science, Shanghai, China).
2.3.8. RNA isolation and quantitative RT-PCR
Cells (50,000 cells per well) incubated in 6-well plates were treated with compound ID-11 for 24 h. After that, the total RNA was extracted using Total RNA Extraction Reagent (Invitrogen, USA) according to the manufacturer’s instructions and quantified with NanoDrop 2000 (Nanodrop, USA). Then the cDNA was synthesized from total RNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA). The SYBR Green Master reagents (Applied Biosystems, USA) was used to determine the relative expression of mRNA level by quantitative real-time PCR (qRT-PCR). The primer sequences are shown below.
| Gene | Primer sequence (5’-3’) |
|---|---|
| c-Myc Forward | GCCTCAGAGTGCATCGAC |
| c-Myc Reverse | TCCACAGAAACAACATCG |
| Bcl-2 Forward | GTGTGTGGAGAGCGTCAACC |
| Bcl-2 Reverse | CTTGTGGCCCAGATAGGCA |
| Actin Forward | ACTCTTCCAGCCTTCCTTCC |
| Actin Reverse | CGTCATACTCCTGCTTGCTG |
2.3.9. Molecular docking
The cocrystal structure of BRD4 with (+)-JQ1 (PDB: 3MXF) was downloaded from Protein Data Bank (https://www.rcsb.org/) and the whole molecular docking process were carried out in the Ledock software (https://www.lephar.com/). The receptor was prepared using the LePro module with default parameters and a receptor grid was defined as the ligand-binding site based on the cocrystallised ligand and an enclosed box that was in similar in size to the cocrystallised ligand were used to fit the compounds to be docked. The ligand was prepared using ChemBio3D Ultra 14.0 and minimised to the lowest energy. Before the docking study started, the native ligand (+)-JQ1 was re-docked into the binding site using the same set of parameters as described above. The RMSD of the best-docked pose (binding energy=‒8.66 kcal/mol) was 0.628 Å, thus the docking method using Ledock was suitable. Then the docking work between the receptor and ligand was conducted in the LeDock module and the best docking pose was selected as the dominant conformation. The final docking results were processed with PyMOL software (Version 2.1.0).
2.3.10. Molecular properties prediction of compound ID-11
Molecular properties including absorption, distribution, metabolism and excretion (ADME), toxicity and drug-likeness property of compound ID-11 were predicted at related available website (https://preadmet.bmdrc.kr/).
2.3.11. Statistic analysis
Data were presented as mean ± standard deviation (SD). One-way ANOVA was used to analyse the statistics by GraphPad Prism 7.0 packages. p < 0.05 was considered as statistically significance.
3. Results and discussion
3.1. Chemistry
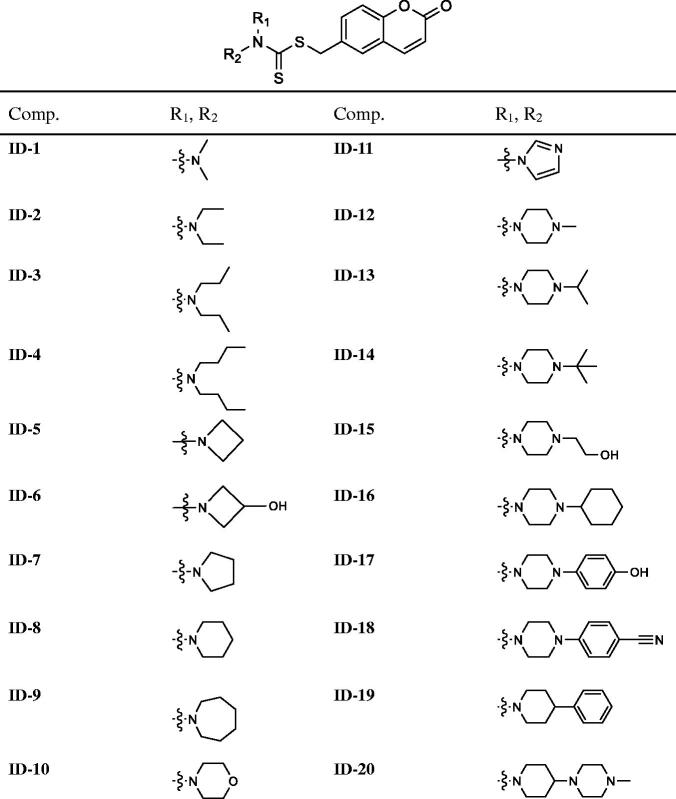
The overall synthetic routes of novel coumarin-dithiocarbamate derivatives (IDs) were shown in Scheme 1. Intermediate 6-(bromomethyl)-2H-chromen-2-one (9) was prepared via the reaction between 6-methyl-2H-chromen-2-one (7) and 1-bromopyrrolidine-2,5-dione (8), and the bromination reaction was catalysed by 2,2′-dimethyl-2,2′-azodipropionitrile (AIBN) with a moderate yield. Then, the final products ID-1, ID-6, ID-7, ID10, ID-12, ID-15 and ID-20 could be obtained via the reactions between intermediate 9, CS2 and appropriate secondary amines in dry ethanol and KOH conditions. Compounds ID-2 ∼ ID-5, ID-8, ID-9, ID-13, ID-14 and ID-16 ∼ ID-19 were prepared with a similar procedure which described for the synthesis of compound ID-1, and the solvent was dioxane/H2O instead of dry ethanol and the alkali was K2CO3 instead of KOH. Especially, for the synthesis of compound ID-11, actone was used as the solvent and K3PO4 was used as the alkali. The chemical structures of novel coumarin-dithiocarbamate derivatives (IDs) were shown in Table 1. All final products were confirmed by 1H NMR, 13 C NMR, and HRMS spectra.
Scheme 1.
Synthetic routes of novel coumarin-dithiocarbamate derivatives (IDs). Reagents and Conditions: (a) CCl4, AIBN (cat.), 90 °C, reflux, 10 h; (b) appropriate secondary amines, CS2, alkali (KOH, K2CO3 or K3PO4), solvent (ethanol, dioxane/H2O (1/4, v/v) or actone), rt, 12–24 h.
Table 1.
Structures of novel coumarin-dithiocarbamate derivatives (IDs).
|
3.2. In vitro antiproliferative activities of IDs
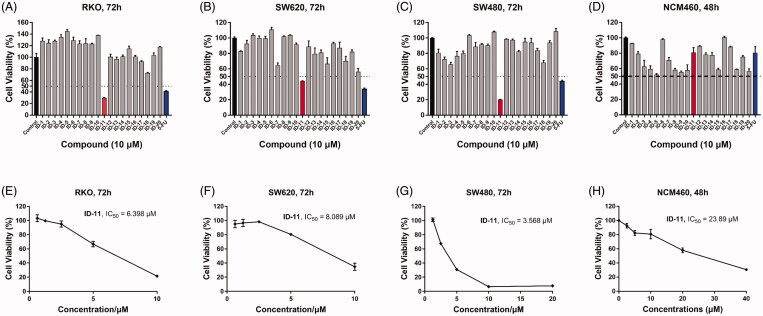
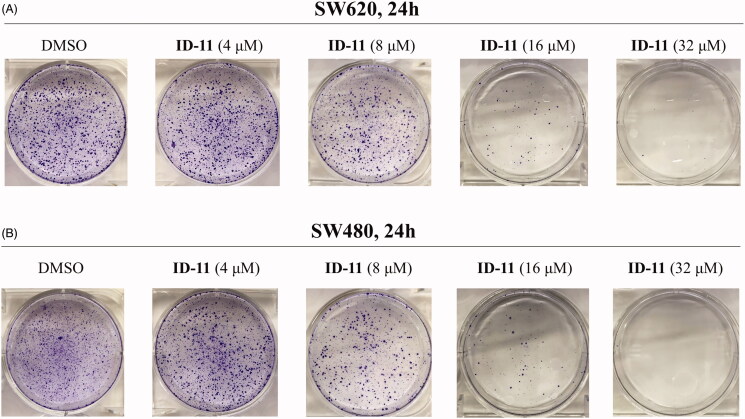
To investigate the in vitro tumour cell growth inhibitory activities of all newly designed compounds and explore the structure-activity relationships (SAR), three different kinds of human colorectal cancer cell lines, including RKO, SW620 and SW480 were applied. As 5-Fluorouracil (5-FU) is a first-line drug for the treatment of CRC38, so we chose it as a positive control in the in vitro antiproliferative assay. As shown in Figure 3(A–C), after treatment with the tested compounds (ID-1 ∼ ID-20) at 10 μM for 72 h, the cell viabilities of RKO, SW620 and SW480 cells were all over 50% apart from compound ID-11 (R1, R2= imidazole) treatment group, the cell viabilities of these three CRC cell lines after ID-11 treatment were 29.43, 43.94, and 20.18%, respectively. At the same condition, the cell viabilities of which were 40.73, 33.57, and 43.87% for 5-FU treatment, respectively. The MTS assay results indicated that, when the R1, R2 positions replaced by C1–C4 linear alkyl (ID-1 to ID-4), four-membered ring to seven-membered ring secondary amines (ID-5 to ID-9), morpholine (ID-10), substituted piperazine or piperidine secondary amines (ID-12 to ID-20) will weaken antiproliferative activities against these cell lines. Next, we measured the IC50 values of ID-11 towards these cancer cell lines. As illustrated in Figure 3(E–G), the IC50 values of ID-11 against RKO, SW620 and SW480 cells were 6.398, 8.809, and 3.568 μM, respectively. Furthermore, we also evaluated the cytotoxicity of these newly designed compounds towards NCM460 cell line (normal human colon epithelial cell line). As shown in Figure 3(D), after incubation with these IDs and 5-FU for 48 h at 10 μM, the viabilities of NCM460 cells were all exceed 50% (80.84% for ID-11 treatment and 80.36% for 5-FU treatment). We also measured the IC50 value of ID-11 against NCM460 cells, and the IC50 value of which was 23.89 μM after ID-11 incubation (Figure 3(H)). We next evaluated the long-term effects on the colony formation of CRC cell lines with the treatment of the most active compound ID-11. As shown in Figure 4(A,B), compound ID-11 effectively inhibited the colony formation of both SW620 and SW480 cells in a concentration dependent way (from 0 to 32 μM). The above results indicated that these IDs (especially for ID-11) have potential growth inhibitory activities towards CRC cell lines in vitro while shown little impact on normal human colon epithelial cells. In view of the excellent antiproliferative activities of ID-11 against CRC cell lines, ID-11 was chosen as the representative compound for further anticancer mechanism investigations.
Figure 3.
The inhibitory activities of IDs against different human CRC cell lines and normal human colon epithelial cells. (A–C) RKO, SW620 and SW480 cells were treated with IDs (ID-1 ∼ ID-20) and 5-FU for 72 h at 10 μM and then cell viability was determined by MTS assay. (D) NCM460 cells were treated with IDs (ID-1 ∼ ID-20) and 5-FU for 48 h at 10 μM and then cell viability was determined by MTS assay. (E-G) RKO, SW620 and SW480 cells were treated with ID-11 for 72 h at variable concentrations (20, 10, 5, 2.5, 1.25, 0.625 μM) and then cell viability determined by MTS assay, finally the IC50 values were calculated. (H) NCM460 cells were treated with ID-11 for 48 h at variable concentrations and then cell viability determined by MTS assay, finally the IC50 values were calculated.
Figure 4.
Effects on colony formation of different CRC cell lines by treatment of compound ID-11. (A) SW620 cells. (B) SW480 cells. Cells after treating with ID-11 (0, 4, 8, 16, and 32 μM) for 24 h, continue to cultivate with fresh culture medium for 10 days and then fixed, stained, air-dried and photographed.
3.3. BRD4 inhibitory assay
Growing evidences have shown that the abnormality of BRD4 is closely related to the development of CRC8. We want to know whether the antiproliferative activities of these IDs towards CRC cell lines was associated with the inhibition of BRD4, so the HTRF assay was performed. In the HTRF assay, reference compound (+)-JQ111 was selected as positive control. The results were shown in Table 2. Among all the twenty IDs, at the concentration of 10 μM, only compound ID-11 shown potential inhibitory activity against BRD4 (BD1), the inhibitory rate of which was 68.9%. In the same conditions, (+)-JQ1 could strongly inhibit both BRD4 (BD1) and BRD4 (BD2), the inhibitory rate of (+)-JQ1 against BRD4 (BD1) and BRD4 (BD2) were all exceed 98%. We next measured the IC50 value of ID-11 against BRD4 (BD1), and the results shown that ID-11 inhibited BRD4 (BD1) at μM level, the IC50 value of which was 7.8 μM.
Table 2.
BRD4 inhibitory activities of IDs in vitro.
| Comp. | (Inhibitory rate at 10 μMa) or IC50 (μM)b |
cLogPc | |
|---|---|---|---|
| BRD4 (BD1) | BRD4 (BD2) | ||
| ID-1 | (2.2%) | (1.5%) | 2.02 |
| ID-2 | (4.3%) | (5.1%) | 3.08 |
| ID-3 | (1.6%) | (3.3%) | 4.14 |
| ID-4 | (4.2%) | (3.1%) | 5.20 |
| ID-5 | (6.2%) | (5.8%) | 2.10 |
| ID-6 | (15.0%) | (8.7%) | 1.32 |
| ID-7 | (9.2%) | (7.1%) | 2.66 |
| ID-8 | (7.8%) | (8.2%) | 3.21 |
| ID-9 | (6.3%) | (5.0%) | 3.77 |
| ID-10 | (27.2%) | (19.6%) | 1.70 |
| ID-11 | (68.9%) | (35.7%) | 2.67 |
| 7.8 ± 2.2 | |||
| ID-12 | (35.5%) | (26.5%) | 2.26 |
| ID-13 | (14.4%) | (10.8%) | 3.10 |
| ID-14 | (12.8%) | (16.6%) | 3.50 |
| ID-15 | (40.2%) | (33.3%) | 1.69 |
| ID-16 | (20.1%) | (12.9%) | 4.08 |
| ID-17 | (39.0%) | (27.7%) | 3.03 |
| ID-18 | (11.8%) | (8.4%) | 3.52 |
| ID-19 | (6.5%) | (3.2%) | 4.62 |
| ID-20 | (36.6%) | (29.3%) | 1.04 |
| (+)-JQ1 | (98.9%) | (99.5%) | 4.82 |
| 0.06 ± 0.01 | 0.03 ± 0.01 | ||
aAll results were measured three times.
bData were shown as mean ± SD value of three independent experiments.
ccLogP values were calculated using ChemDraw 14.0.
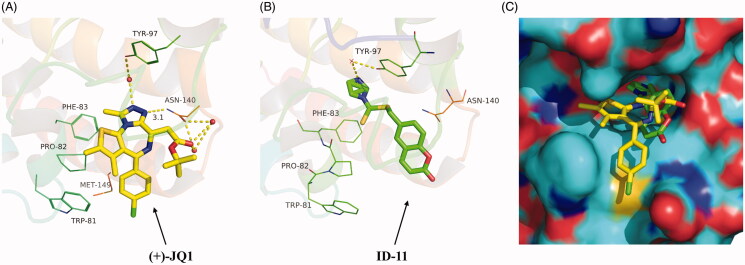
Based on the inhibitory activity of compound ID-11 against BRD4 in vitro, we next explored the potential binding mode of ID-11 with BRD4. As shown in Figure 5(A,B), comparing the best docking conformation of ID-11 with BRD4 (binding energy = ‒6.59 kcal/mol) and the cocrystal structure of (+)-JQ1 with BRD4, the N atom of imidazole in ID-11 formed a hydrogen bond with Tyr-97 residue through a water molecule, which was similar as the binding mode of (+)-JQ1 with BRD4, while (+)-JQ1 formed more complex interactions with BRD4, in which the N atom on triazole of (+)-JQ1 formed another hydrogen bond with ASN-140 (the distance is 3.1 Å) and the carbonyl oxygen of (+)-JQ1 also formed a hydrogen bond with ASN-140 through a water molecule. In addition, (+)-JQ1 could also form hydrophobic interactions between the chlorobenzene moiety in (+)-JQ1 with BRD4. The abundant interactions of (+)-JQ1 with BRD4 maybe explained the reason why the inhibitory activity of (+)-JQ1 against BRD4 was much stronger than ID-11. As shown in Figure 5(C), we also found that ID-11 buried more deeply than (+)-JQ1 in BRD4 binding pocket.
Figure 5.
Proposed binding mode of ID-11 with BRD4. (A) Crystal structure of BRD4 bound to (+)-JQ1 (yellow stick, in which carbon atoms in yellow, chlorine atom in green, sulphur atom in orange yellow, oxygen atoms in red and nitrogen atoms in blue) (PDB ID: 3MXF); (B) Docking conformation of ID-11 (green stick, in which carbon atoms in green, sulphur atom in orange yellow, oxygen atoms in red and nitrogen atoms in blue) with BRD4 (PDB ID: 3MXF); (C) Superimposition of (+)-JQ1 (yellow stick) and ID-11 (green stick) in their putative bioactive conformations with BRD4 (PDB ID: 3MXF).
3.4. Compound ID-11 induced G2/M cell cycle arrest in human colorectal cancer cells
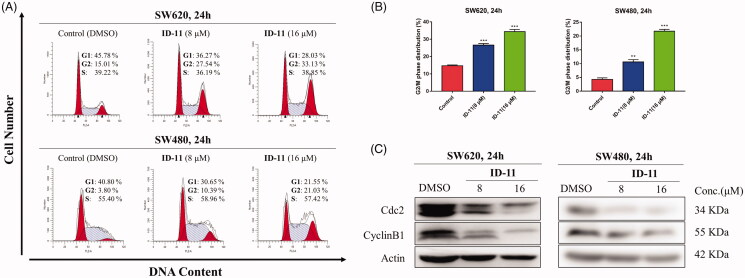
Control of cell cycle is important for mediating the equilibrium between cell proliferation and death. Moreover, blockade of the cell cycle regarded as an effective strategy in cancer therapy39. In order to explore the compound ID-11 whether could induce cell cycle arrest in human colorectal cancer cells, flow cytometry assay was performed. As shown in Figure 6(A,B), SW620 and SW480 cells were both arrested at G2/M phase, and the percentage of cell cycle distribution of G2/M phase increased in a concentration-dependent manner (from 15.01 to 33.13% for SW620 cells, and from 3.80 to 21.03% for SW480 cells) after treatment with compound ID-11 from 0 to 16 μM for 24 h.
Figure 6.
Compound ID-11 induced G2/M phase arrest in CRC cell lines. (A) SW620 and SW480 cells were incubated with ID-11 for 24 h at concentration of 0, 8, and 16 μM. Cells were harvested and stained with PI and then cell cycle distributions were measured by flow cytometry. (B) Histogram of G2/M phase distributions. (C) The expression levels of G2/M phase related proteins were determined by western blot assay. Actin was used as the internal control. Error bar original from three independent experiments. (**p < 0.01; ***p < 0.001 vs. control).
Previous studies had revealed that Cdc2 protein kinase plays an important role in cell cycle transition of eukaryotic cells, which was regulated by other factors, such as cyclin B1 and Cdc25c proteins40. Thus, western bolt assay was performed to detect the expression level of several cell cycle related proteins in human colorectal cancer cells at the presence or absence of compound ID-11. As shown in Figure 6(C), the expression levels of cdc2 and cyclin B1 in SW620 and SW480 cells were both decreased in a concentration-dependent way after exposed to compound ID-11 from 0 to 16 μM for 24 h.
3.5. Compound ID-11 induced apoptosis in human colorectal cancer cells
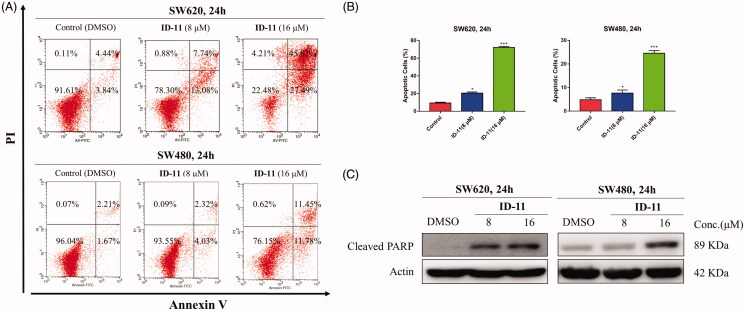
Annexin V-FITC/PI staining assay was performed by flow cytometry to investigate whether the antiproliferative potency of compound ID-11 was consistent with the induction of apoptosis. As shown in Figure 7(A,B), the total apoptosis rate in SW620 and SW480 cells were both dose-dependently increased (from 8.28 to 73.31% for SW620 cells, and from 3.88 to 23.23% for SW480 cells) after incubated with compound ID-11 from 0 to 16 μM for 24 h.
Figure 7.
Compound ID-11 induced apoptosis in CRC cell lines. (A) SW620 and SW480 cells were incubated with ID-11 for 24 h at concentration of 0, 8, and 16 μM. Cells were harvested and stained with PI/Annexin V and then cell distributions were measured by flow cytometry. (B) Histogram of apoptotic cells. (C) The expression levels of apoptosis-related proteins were determined by western blot assay. Actin was used as the internal control. Error bar original from three independent experiments. (*p < 0.05; ***p < 0.001 vs. control).
The process of cell apoptosis was regulated by some vital proteins including, anti-apoptotic proteins (e.g. Bcl-2 and Bcl-xl), pro-apoptotic proteins (e.g. Bax and Bad) and caspase family, etc.41. To explore whether the apoptosis effect induced by compound ID-11 in SW620 and SW480 cells was correlated to the influence on apoptosis-related proteins, western blot assay was performed. The results showed that, the expression level of cleaved PARP was increased in a dose-dependent way in both SW620 and SW480 cells after treatment with compound ID-11 from 0 to 16 μM for 24 h (Figure 7(C)).
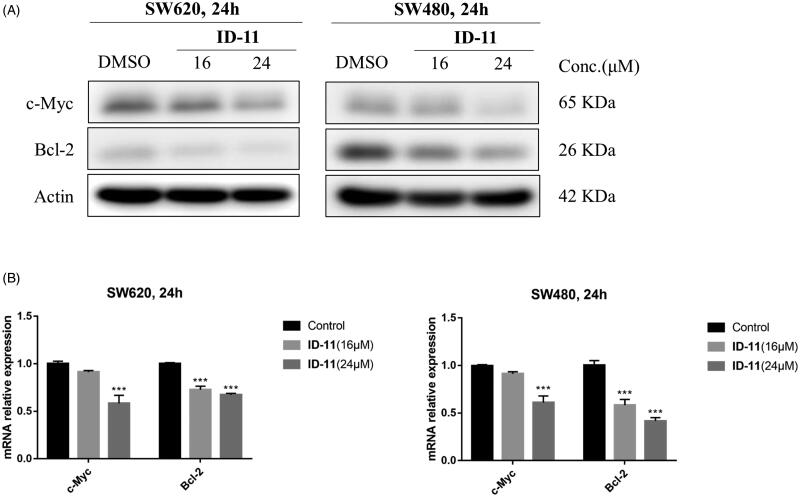
3.6. Effects on c-Myc and bcl-2 protein and mRNA expression
Previous study shown that the inhibition of BRD4 activity usually affect the expression of some genes such as c-Myc, Bcl-2 and CDK6, etc.42. Considering the BRD4 inhibitory activity and the ability to induce apoptosis of compound ID-11 against CRC cell lines, we next measured the protein and mRNA expression levels of c-Myc and Bcl-2 after incubating with ID-11 in CRC cell lines. As shown in Figure 8(A), ID-11 treatment group decreased the protein expression levels of c-Myc and Bcl-2 in a concentration dependent way (from 0 to 24 μM) in both SW620 and SW480 cells. Meanwhile, the mRNA relative expression levels of c-Myc and Bcl-2 were also decreased in a concentration dependent way (from 0 to 24 μM) (Figure 8(B)) in both SW620 and SW480 cells.
Figure 8.
Effects on c-Myc and Bcl-2 protein and mRNA expression in CRC cell lines by treatment of compound ID-11. (A) The expression levels of proteins c-Myc and Bcl-2 in SW620 and SW480 cells. Cells after treating with ID-11 (0, 16, and 24 μM) for 24 h, proteins were extracted and the relative expression levels were determined by western blot assay. (B) The mRNA expression levels of c-Myc and Bcl-2 in SW620 and SW480 cells. Cells after treating with ID-11 (0, 16, and 24 μM) for 24 h, the total RNA was extracted, quantified and the mRNA levels of c-Myc and Bcl-2 were determined by qRT-PCR.
3.7. Molecular properties of compound ID-11
The proper physicochemical properties of a compound are essential for its druggability. Based on the screening results in vitro, we further evaluated the physicochemical properties of compound ID-11. As shown in Table 3, we predicted the ADMET and drug-likeness properties of ID-11, the results indicated that ID-11 has a high human intestinal absorption rate and plasma protein binding rate without P-glycoprotein inhibitory activity. We also found that ID-11 follows Lipinski’ rule of five and shown little toxicity. These prediction results shown that ID-11 owns proper physicochemical properties and well druggability.
Table 3.
Molecular properties prediction of compound ID-11a.
| ADME |
Toxicity |
Drug-likeness |
|||
|---|---|---|---|---|---|
| ID | Value | ID | Value | ID | Value |
| BBB | 0.256131 | algae_at | 0.0613152 | CMC_like_Rule | Qualified |
| Buffer_solubility_mg_L | 268.239 | Ames_test | Mutagen | CMC_like_Rule_Violation_Fields | – |
| Caco2 | 28.7315 | Carcino_Mouse | Negative | CMC_like_Rule_Violations | 0 |
| CYP_2C19_inhibition | Inhibitor | Carcino_Rat | Negative | Lead-like_Rule_Violation_Fields | AlopP98_value |
| CYP_2C9_inhibition | Inhibitor | daphnia_at | 0.0456393 | Lead_like_Rule | Violated |
| CYP_2D6_inhibition | Non | hERG_inhibition | Medium_risk | Lead_like_Rule_Violations | 1 |
| CYP_2D6_substrate | Non | medaka_at | 0.00500794 | MDDR_like_Rule | Mid-structure |
| CYP_3A4_inhibition | Inhibitor | minnow_at | 0.004834 | MDDR_like_Rule_Violation_Fields | No_Rotatable_bonds |
| CYP_3A4_substrate | Substrate | TA100_10RLI | Positive | MDDR_like_Rule_Violations | 1 |
| HIA | 99.553241 | TA100_NA | Positive | Rule_of_Five | Suitable |
| MDCK | 16.196 | TA1535_10RLI | Negative | Rule_of_Five_Violation_Fields | – |
| Pgp_inhibition | Non | TA1535_NA | Positive | Rule_of_Five_Violations | 0 |
| Plasma_Protein_Binding | 89.406977 | WDI_like_Rule | In 90% cut-off | ||
| Pure_water_solubility_mg_L | 47.2348 | WDI_like_Rule_Violation_Fields | – | ||
| Skin_Permeability | −3.7303 | WDI_like_Rule_Violations | 0 | ||
| SKlogD_value | 1.986330 | ||||
| SKlogP_value | 1.986330 | ||||
| SKlogS_buffer | −3.052010 | ||||
| SKlogS_pure | −3.806270 | ||||
aThe predicted results were performed at: https://preadmet.bmdrc.kr/.
4. Conclusions
In this work, we provided a series of novel coumarin-dithiocarbamate derivatives (IDs) and found that compound ID-11 shown the best antiproliferative activities towards several human CRC cell lines including RKO, SW620 and SW480 cells, while shown little effect on the normal human colon epithelial cells. Further investigations of the anticancer mechanism indicated that ID-11 has potential BRD4 inhibitory activity, as well as induced apoptosis and G2/M phase arrest in CRC cells. In addition, ID-11 also decreased the expression levels of related genes such as c-Myc and Bcl-2. Furthermore, the prediction results of the physicochemical properties of ID-11 demonstrated that it may be a promising anti-CRC leading agent and deserved for further development.
Supplementary Material
Funding Statement
This work was supported by the China State Key Research [No. 2016YEF0200503] and National Natural Science Foundation of China [No. 21532005] as well as National Natural Science Foundation of China [No. 81761138047].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68: 394–424. [DOI] [PubMed] [Google Scholar]
- 2.Araghi M, Soerjomataram I, Jenkins M, et al. Global trends in colorectal cancer mortality: projections to the year 2035. Int J Cancer 2019; 144: 2992–3000. [DOI] [PubMed] [Google Scholar]
- 3.Kanth P, Grimmett J, Champine M, et al. Hereditary colorectal polyposis and cancer syndromes: a primer on diagnosis and management. Am J Gastroenterol 2017;112:1509–25. [DOI] [PubMed] [Google Scholar]
- 4.Ganesh K, Stadler ZK, Cercek A, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol 2019;16:361–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauer R, Becker H, Hohenberger W, German Rectal Cancer Study Group, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med 2004;351:1731–40. [DOI] [PubMed] [Google Scholar]
- 6.Bruheim K, Guren MG, Skovlund E, et al. Late side effects and quality of life after radiotherapy for rectal cancer. Int J Radiat Oncol Biol Phys 2010;76:1005–11. [DOI] [PubMed] [Google Scholar]
- 7.Van der Jeught K, Xu HC, Li YJ, et al. Drug resistance and new therapies in colorectal cancer. World J Gastroenterol 2018;24:3834–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donati B, Lorenzini E, Ciarrocchi A.. BRD4 and cancer: going beyond transcriptional regulation. Mol Cancer 2018;17:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White ME, Fenger JM, Carson WE. 3rd,. Emerging roles of and therapeutic strategies targeting BRD4 in cancer. Cell Immunol 2019;337:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otto C, Schmidt S, Kastner C, et al. Targeting bromodomain-containing protein 4 (BRD4) inhibits MYC expression in colorectal cancer cells. Neoplasia 2019;21:1110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature 2010;468:1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues T, Reker D, Schneider P, et al. Counting on natural products for drug design. Nat Chem 2016;8:531–41. [DOI] [PubMed] [Google Scholar]
- 13.Newman DJ, Cragg GM.. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 2016;79:629–61. [DOI] [PubMed] [Google Scholar]
- 14.Matulja D, Wittine K, Malatesti N, et al. Marine natural products with high anticancer activities. Curr Med Chem 2020;27:1243–307. [DOI] [PubMed] [Google Scholar]
- 15.Vazquez-Rodriguez S, Matos MJ, Borges F, et al. Bioactive coumarins from marine sources: origin, structural features and pharmacological properties. Curr Top Med Chem 2015;15:1755–66. [DOI] [PubMed] [Google Scholar]
- 16.Costa TM, Tavares LBB, de Oliveira D.. Fungi as a source of natural coumarins production. Appl Microbiol Biotechnol 2016;100:6571–84. [DOI] [PubMed] [Google Scholar]
- 17.Miura K, Satoh M, Kinouchi M, et al. The use of natural products in colorectal cancer drug discovery. Exp Opin Drug Disc 2015;10:411–26. [DOI] [PubMed] [Google Scholar]
- 18.Kaur M, Kohli S, Sandhu S, et al. Coumarin: a promising scaffold for anticancer agents. Anticancer Agents Med Chem 2015;15:1032–48. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Xu Z.. Coumarin-containing hybrids and their anticancer activities. Eur J Med Chem 2019;181:111587. [DOI] [PubMed] [Google Scholar]
- 20.Supuran CT. Carbonic anhydrase inhibitors as emerging agents for the treatment and imaging of hypoxic tumors. Expert Opin Investig Drugs 2018;27:963–70. [DOI] [PubMed] [Google Scholar]
- 21.Supuran CT. Coumarin carbonic anhydrase inhibitors from natural sources. J Enzyme Inhib Med Chem 2020;35:1462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Supuran CT. How many carbonic anhydrase inhibition mechanisms exist? J Enzyme Inhib Med Chem 2016;31:345–60. [DOI] [PubMed] [Google Scholar]
- 23.Fois B, Distinto S, Meleddu R, et al. Coumarins from Magydaris pastinacea as inhibitors of the tumour-associated carbonic anhydrases IX and XII: isolation, biological studies and in silico evaluation. J Enzyme Inhib Med Chem 2020;35:539–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai G, Yu W, Song D, et al. Discovery of fluorescent coumarin-benzo[b]thiophene 1, 1-dioxide conjugates as mitochondria-targeting antitumor STAT3 inhibitors. Eur J Med Chem 2019;174:236–51. [DOI] [PubMed] [Google Scholar]
- 25.Govindaiah P, Dumala N, Mattan I, et al. Design, synthesis, biological and in silico evaluation of coumarin-hydrazone derivatives as tubulin targeted antiproliferative agents. Bioorg Chem 2019;91:103143. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Ren ZL, Wang W, et al. Novel coumarin-pyrazole carboxamide derivatives as potential topoisomerase II inhibitors: Design, synthesis and antibacterial activity. Eur J Med Chem 2018;157:81–7. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Gu L, Wang B, et al. Discovery of novel coumarin derivatives as potent and orally bioavailable BRD4 inhibitors based on scaffold hopping. J Enzyme Inhib Med Chem 2019;34:808–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan YC, Zheng YC, Li XC, et al. Design, synthesis and antiproliferative activity studies of novel 1,2,3-triazole-dithiocarbamate-urea hybrids. Eur J Med Chem 2013;64:99–110. [DOI] [PubMed] [Google Scholar]
- 29.Cao SL, Wang Y, Zhu L, et al. Synthesis and cytotoxic activity of N-((2-methyl-4(3H)-quinazolinon-6-yl)methyl)dithiocarbamates. Eur J Med Chem 2010;45:3850–7. [DOI] [PubMed] [Google Scholar]
- 30.Xie R, Li Y, Tang P, et al. Design, synthesis and biological evaluation of novel 2-aminobenzamides containing dithiocarbamate moiety as histone deacetylase inhibitors and potent antitumor agents. Eur J Med Chem 2018;143:320–33. [DOI] [PubMed] [Google Scholar]
- 31.Yang CR, Peng B, Cao SL, et al. Synthesis, cytotoxic evaluation and target identification of thieno[2,3-d]pyrimidine derivatives with a dithiocarbamate side chain at C2 position. Eur J Med Chem 2018;154:324–40. [DOI] [PubMed] [Google Scholar]
- 32.Mostafa AS, Selim KB.. Synthesis and anticancer activity of new dihydropyrimidinone derivatives. Eur J Med Chem 2018;156:304–15. [DOI] [PubMed] [Google Scholar]
- 33.Li RD, Wang HL, Li YB, et al. Discovery and optimization of novel dual dithiocarbamates as potent anticancer agents. Eur J Med Chem 2015;93:381–91. [DOI] [PubMed] [Google Scholar]
- 34.Carta F, Aggarwal M, Maresca A, et al. Dithiocarbamates: a new class of carbonic anhydrase inhibitors. Crystallographic and kinetic investigations. Chem Commun 2012;48:1868–70. [DOI] [PubMed] [Google Scholar]
- 35.Supuran CT. Structure-based drug discovery of carbonic anhydrase inhibitors. J Enzyme Inhib Med Chem 2012;27:759–72. [DOI] [PubMed] [Google Scholar]
- 36.Zeidan MA, Mostafa AS, Gomaa RM, et al. Design, synthesis and docking study of novel picolinamide derivatives as anticancer agents and VEGFR-2 inhibitors. Eur J Med Chem 2019;168:315–29. [DOI] [PubMed] [Google Scholar]
- 37.Su Y, Li R, Ning X, et al. Discovery of 2,4-diarylaminopyrimidine derivatives bearing dithiocarbamate moiety as novel FAK inhibitors with antitumor and anti-angiogenesis activities. Eur J Med Chem 2019;177:32–46. [DOI] [PubMed] [Google Scholar]
- 38.Vodenkova S, Buchler T, Cervena K, et al. 5-Fluorouracil and other fluoropyrimidines in colorectal cancer: past, present and future. Pharmacol Ther 2020;206:107447. [DOI] [PubMed] [Google Scholar]
- 39.Wu J, Yi J, Wu Y, et al. 3, 3'-Dimethylquercetin inhibits the proliferation of human colon cancer RKO cells through inducing G2/M cell cycle arrest and apoptosis. Anticancer Agents Med. Chem 2019;19:402–9. [DOI] [PubMed] [Google Scholar]
- 40.Draetta G, Beach D.. The mammalian cdc2 protein kinase: mechanisms of regulation during the cell cycle. J Cell Sci Suppl 1989;12:21–7. [DOI] [PubMed] [Google Scholar]
- 41.Bruckheimer EM, Cho SH, Sarkiss M, et al. The Bcl-2 gene family and apoptosis. Adv Biochem Eng Biotechnol 1998;62:75–105. [DOI] [PubMed] [Google Scholar]
- 42.Dawson MA, Prinjha RK, Dittmann A, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011; 478:529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.