Abstract
Amyloid-induced neurodegeneration plays a central role in Alzheimer’s disease (AD) pathogenesis. Here, we show that telomerase reverse transcriptase (TERT) haploinsufficiency decreases BDNF and increases amyloid-β (Aβ) precursor in murine brain. Moreover, prior to disease onset, the TERT locus sustains accumulation of repressive epigenetic marks in murine and human AD neurons, implicating TERT repression in amyloid-induced neurodegeneration. To test the impact of sustained TERT expression on AD pathobiology, AD mouse models were engineered to maintain physiological levels of TERT in adult neurons, resulting in reduced Aβ accumulation, improved spine morphology, and preserved cognitive function. Mechanistically, integrated profiling revealed that TERT interacts with β-catenin and RNA polymerase II at gene promoters and upregulates gene networks governing synaptic signaling and learning processes. These TERT-directed transcriptional activities do not require its catalytic activity nor telomerase RNA. These findings provide genetic proof-of-concept for somatic TERT gene activation therapy in attenuating AD progression including cognitive decline.
Editor summary:
The authors link neuronal expression of the Telomerase Reverse Transcriptase (TERT) to amyloid pathology and cognitive dysfunction in preclinical models of Alzheimer’s disease.
Introduction
Alzheimer’s disease (AD) is the most common type of dementia and is characterized by the accumulation of neurotoxic protein aggregates in the brain, eventually leading to progressive neuronal loss and decline of learning, memory, and other cognitive functions 1. While multiple mechanisms contribute to AD pathobiology, the amyloid precursor protein (APP) and its neurotoxic metabolite amyloid-β (Aβ) play key roles in disease pathology and cognitive dysfunction. Progressive Aβ accumulation, which appears early in the disease, provokes a complex pathological cascade of neuroinflammation, neuronal and synaptic loss and tau pathology 2,3. Correspondingly, in AD patients, Aβ levels correlate more positively with the degree of disease progression and cognitive decline than the number and extent of extracellular plaques 4–6. Many factors can influence the trajectory of AD including neurotrophic factors 7 such as brain-derived neurotrophic factor (BDNF), which supports memory formation and storage via modulation of synaptic plasticity and protects against Aβ-induced neurotoxicity 8. Accumulating clinical and preclinical studies have documented reduced BDNF levels in AD-affected brain areas and BDNF administration can attenuate neuronal atrophy and synaptic loss in various AD models 9,10. Together, these insights have spurred efforts to identify drugs designed to reduce Aβ and/or preserve neuronal survival and function.
Telomerase, a specialized ribonucleoprotein complex, is responsible for the maintenance of telomere length and genome integrity. TERT is the catalytic subunit of the telomerase complex, but also exhibits telomere-independent functions that are not dependent on its reverse transcriptase activity or TERC yet can impact gene regulation, chromatin organization, and stress responses 11. Mice engineered to experience telomere dysfunction exhibit premature aging phenotypes 12,13 including neurodegeneration 14; moreover, aging human brains with neurodegenerative disease also show signs of genotoxic stress in neurons 15. In TERT deficient mice with severe telomere dysfunction and neurodegeneration, TERT reactivation reverses microcephaly, restores neuron stem cell pools and newborn neuron formation, promotes oligodendrocyte maturation and myelination, and improves learning and memory 16. Curiously, in these TERT reactivation studies, we also observed that post-mitotic cells such as cardiomyocytes and hepatocytes showed improved function, suggesting that TERT-induced rejuvenation may involve mechanisms unrelated to its classical telomere synthesis function during cell division 82. Of relevance to this study, TERT can exert neuroprotective effects following various insults in rodent and human neurons 17–19 and specifically enhance neuronal resistance against pathological tau, which is a downstream mediator of Aβ toxicity 17,20. In this study, we explored whether and how modest changes in the level of TERT expression might also influence the molecular and cellular biology of neurons of the normal mouse brain and regulate pathophysiological processes driving neurodegenerative diseases such as AD.
Results
Tert insufficiency induces APP and reduces BDNF in mouse brain.
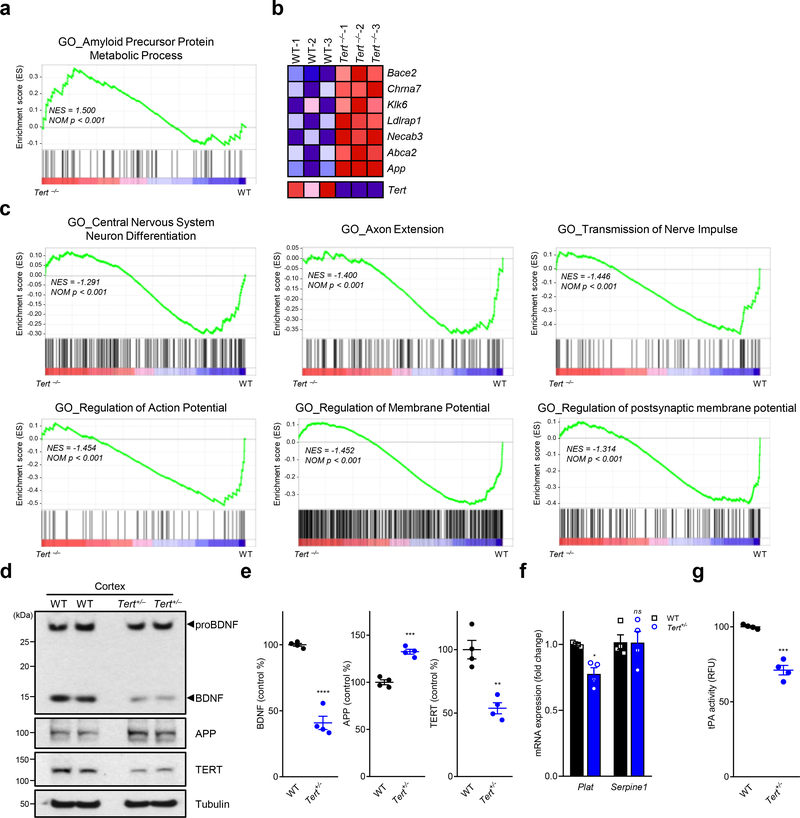
Telomere dysfunction has been reported to elicit clinical features of premature aging and systemic degenerative phenotypes in adult mice that can be reversed by enforced Tert expression 16,21. In auditing differentially expressed genes in the brains of adult Tert+/+, Tert+/− and Tert−/− mice, we observed that the transcriptome of first-generation (G1) Tert−/− brains (with intact telomeres) showed enrichment of APP metabolic process signature relative to Tert+/+ controls (Fig. 1a). Tert deficiency also showed elevated expression of genes encoding strong AD genetic risk factors (APP and ABCA2), AD- and aging-related proteins (CHRNA7, KLK6, LDLRAP1 and NECAB3) and APP processing enzyme (BACE2) (Fig. 1b). Conversely, Tert deficiency was associated with suppression of neural pathway signatures, including neuron differentiation, axon extension, transmission of nerve pulse, and regulation of action/membrane potentials, in the mouse brain (Fig. 1c). These findings prompted a survey of the cerebral cortex of adult Tert+/− haploinsufficient mice with an emphasis on amyloid proteins and neurotrophic factors involved in AD pathogenesis 8. Strikingly, we observed that the adult cerebral cortex of Tert+/− mice possessed reduced levels of TERT and less mature BDNF, yet comparable precursor proBDNF levels (Fig. 1d, e). Since proBDNF is converted to the mature BDNF form by an extracellular proteolytic system, such as tissue-type plasminogen activator (tPA)-plasmin cascade, after its release from neurons 22, we examined tPA activity in Tert+/− brains, revealing a significant decrease in tPA proteolytic activity as well as a decrease in expression of the Plat gene encoding tPA (Fig. 1f, g). Tert haploinsufficiency was also associated with increased amyloid precursor protein (APP) (Fig. 1d, e). Thus, modest changes in TERT levels can alter the expression of genes involved in brain health and AD pathogenesis.
Fig. 1. Tert insufficiency enhances APP and reduces mature BDNF levels in mouse brains.
a, Gene set enrichment analysis (GSEA) plots showing relative upregulation of APP metabolic process genes in the cerebral cortex of Tert–/– (TertLSL/LSL) G1 mouse brain relative to wildtype controls (n = 3 per group). b, RNA-Seq heat map of genes involved in APP metabolic process in wildtype and Tert–/– mouse brain (p < 0.001). c, GSEA plots showing downregulated Gene Ontology pathways in the hippocampus of Tert–/– mouse brain relative to wildtype controls. GSEA in a-c was based on the two-sided Kolmogorov-Smirnov statistic, and nominal p values unadjusted for multiple comparisons were calculated from 1,000 iterations of permutation on sample labels. d, Immunoblots for the indicated endogenous proteins in wildtype (C57BL/6) or Tert heterozygous Tert+/– (Tert+/LSL) mouse cerebral cortex. A tubulin was used as a loading control. e, Quantitative comparison of BDNF, APP and TERT levels in the immunoblots (n = 4 per group; p < 0.001, p = 0.001, p = 0.0016, respectively, two-tailed unpaired t-test). f, mRNA levels of the tPA gene Plat and the PAI-1 gene Serpine1 in the neurons isolated from mouse hippocampus and cerebral cortex (n = 4 per group; p = 0.0306, p = 0.9993, respectively, two-way ANOVA with Sidak’s multiple comparisons test). g, tPA proteolytic activity in the mouse hippocampus and cerebral cortex (n = 4 per group; p = 0.0001, two-tailed unpaired t-test). Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant.
Neuronal Tert downregulation via H3K9me3 heterochromatin.
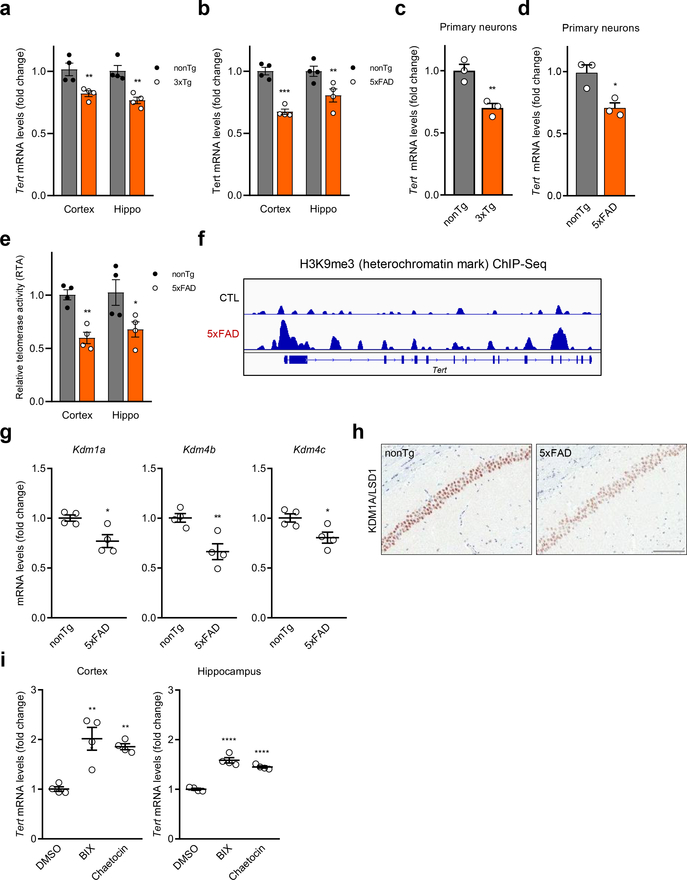
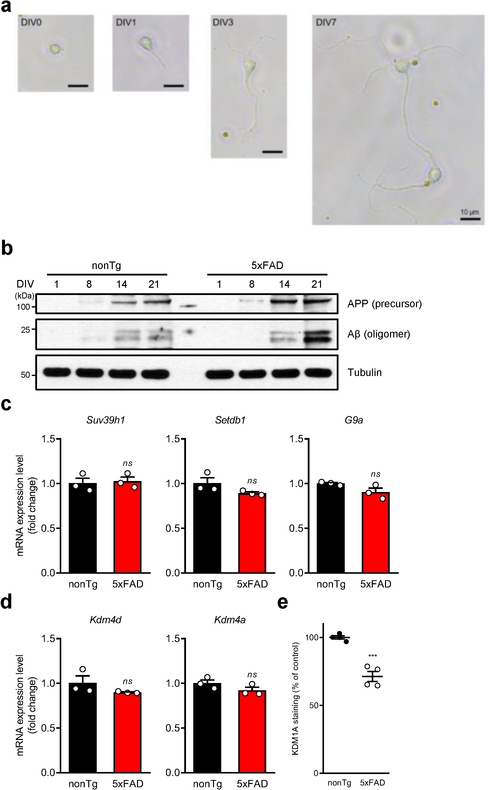
These observations prompted assessment of the relationship of TERT levels and amyloid pathology in two distinct and well-established mouse models of AD, 3xTg-AD 23 and 5xFAD 24. Cortical and hippocampal tissues of 2- to 3-month-old 3xTg-AD and 5xFAD mice, possessing minimal signs of neuropathology 23,24, showed a significant reduction in Tert expression, relative to age- and gender-matched controls (Fig. 2a, b). Similarly, Tert mRNA levels were further reduced in primary cortical and hippocampal neuron cultures isolated from 3xTg-AD and 5xFAD brains versus controls at 14 days in vitro (DIV) at a point coinciding with maturation of the synaptic network and accumulation of intraneuronal Aβ oligomers (Fig. 2c, d and Extended Data Fig. 1a, b). Correspondingly, telomerase activity was lower in freshly isolated cortical and hippocampal neurons from 5xFAD brain relative to wildtype controls (Fig. 2e). Consistent with decreased Tert gene expression, the Tert gene body and promoter region exhibited an increase in the repressive epigenetic mark, H3K9me3, in cultured 5xFAD mouse neurons at 14 DIV relative to controls (Fig. 2f). Analysis of expression of histone methyltransferases and demethylases revealed that H3K9 demethylases Kdm1a, Kdm4b and Kdm4c were significantly downregulated in the neurons of mouse AD brains relative to controls (Fig. 2g, h and Extended Data Fig. 1c–e). To confirm H3K9me3-mediated repression of Tert, 5xFAD mice were treated with the histone methyltransferase inhibitors chaetocin and BIX-01294, shown previously to reduce H3K9 methylation marks in the brain 25,26. Both small-molecule inhibitors de-repressed Tert gene expression in the cortex and hippocampus of the 5xFAD brains (Fig. 2i). These findings, together with previous work showing that this epigenetic mark accumulates at gene bodies and is critical for transcriptional repression of neuronal genes 27, raised the possibility that soluble Aβ may negatively regulate Tert gene expression via altered expression of H3K9 demethylases or methyltransferases in AD mouse neurons.
Fig. 2. Downregulation of Tert via H3K9me3-dependent heterochromatin in two distinct AD mouse models.
a, Tert mRNA levels in the cortex and hippocampus of 3xTg-AD and wildtype control (B6;129S) mice (n = 4, 3-month-old; nonTg vs. 3xTg-AD: p = 0.007, p = 0.0017, respectively, two-way ANOVA with Sidak’s multiple comparisons test). b, Tert mRNA levels in the cortex and hippocampus of 5xFAD and wildtype littermate control (B6;SJL) mice (n = 4, 2~3-month-old; nonTg vs. 5xFAD: p = 0.0001, p = 0.0074, respectively, two-way ANOVA with Sidak’s multiple comparisons test). c, Tert mRNA levels in primary neurons isolated from 3xTg-AD and control mice at DIV 14 (n = 3, p = 0.0089, two-tailed unpaired t-test). d, Tert mRNA levels in primary neurons isolated from 5xFAD and control mice at DIV 14 (n = 3, p = 0.0198, two-tailed unpaired t-test). e, Telomerase activity in cortical and hippocampal neurons isolated from 5xFAD and control mice (n = 4, 2~3-month-old; nonTg vs. 5xFAD: p = 0.0067, p = 0.0173, respectively, two-way ANOVA with Sidak’s multiple comparisons test). f, Representative view of H3K9me3 repressive histone mark occupancy in Tert gene of 5xFAD and control mouse primary neurons at DIV 14. g, mRNA levels of histone demethylase Kdm1a, Kdm4b, and Kdm4c genes in cortical and hippocampal neurons of 5xFAD and wildtype littermate control mice (n = 4, 2~3-month-old; p = 0.0178, p = 0.0094, p = 0.0289, respectively, two-tailed unpaired t-test). h, Representative images of KDM1A immunostaining in the CA1 hippocampal subfield of 5xFAD and wildtype littermate control mice (2~3-month-old). Experiments were repeated three times independently with similar results. Scale bar, 100 μm. i, Tert mRNA levels in the cortex and hippocampus of 5xFAD mice treated with chaetocin or BIX-01294 (n = 4, 2~3-month-old; DMSO vs. inhibitor: cortex: p = 0.0015, p = 0.0036, respectively, hippocampus: p < 0.0001, p < 0.0001, respectively, two-way ANOVA with Tukey’s multiple comparisons test). Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
TERT activation alleviates amyloid pathology.
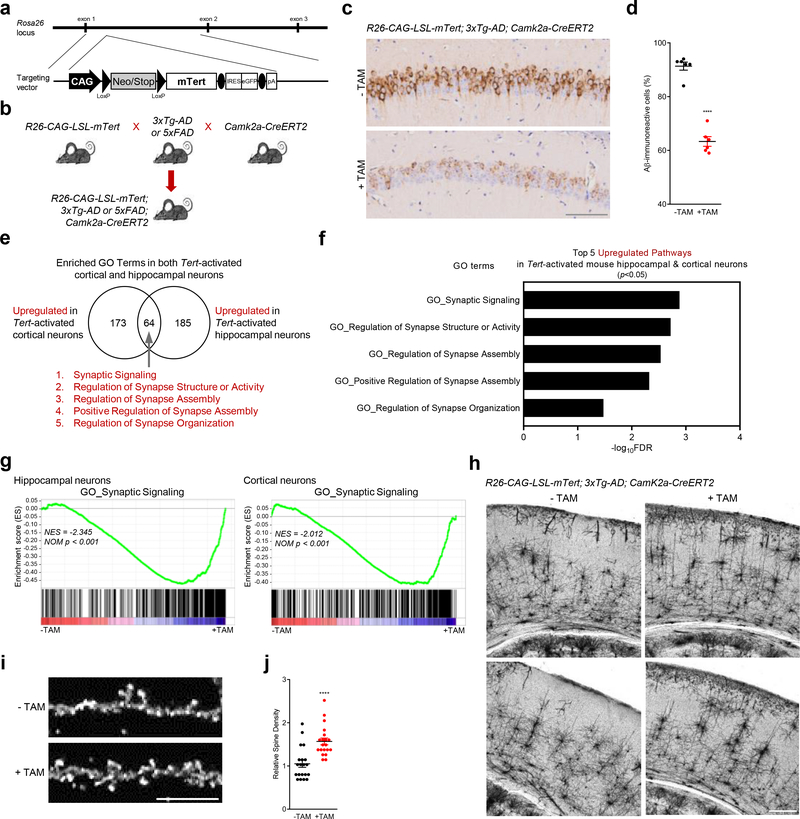
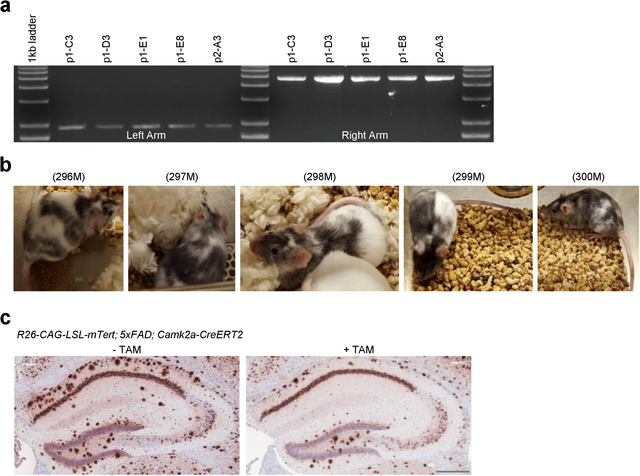
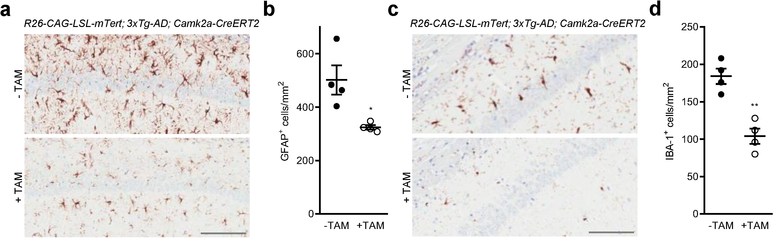
As repression of Tert gene expression occurs early in the accumulation of amyloid in the AD mouse, we next tested whether increased Tert gene expression in AD neurons could ameliorate or prevent amyloid pathophysiology. To that end, we inserted into the Rosa 26 locus a Cre-inducible Tert knock-in allele consisting of the ubiquitously expressed CAG promoter, followed by the loxP-flanked stop cassette and mouse Tert open reading frame (R26-CAG-LSL-mTert) (Fig. 3a and Extended Data Fig. 2a, b). To selectively drive Tert expression in neuronal populations of the AD mouse models, we incorporated a neuron-specific Cre allele which is under the control of the calcium/calmodulin-dependent protein kinase type II alpha promoter (Camk2a-CreERT2) 28,29 (Fig. 3b). Brain amyloid deposition was monitored in these TERT-AD models following Tert activation, revealing a striking decline in Aβ accumulation in the hippocampus of R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mouse model (Fig. 3c, d; note: tamoxifen was administered at 2 to 3 months of age when intracellular Aβ begins to accumulate in the brain). Similar amyloid load reduction was observed in the R26-CAG-LSL-mTert; 5xFAD; Camk2a-CreERT2 model (Extended Data Fig. 2c). We further examined whether TERT induction could influence AD-associated neuroinflammation, characterized by persistent activation of astrocytes and microglia 30. Consistent with the reduction in Aβ accumulation, we observed a significant decrease in the immunoreactivity of astrocytes and microglia in the brains of Tert-activated R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice (Extended Data Fig. 3a–d).
Fig. 3. TERT activation not only alleviates amyloid pathology but enhances synaptic pathways and neural networks in AD mouse model.
a, Targeting strategy for the generation of CAG-LSL-mTert-IRES-eGFP-polyA knock-in mice. b, Breeding strategy of R26-CAG-LSL-mTert with 3xTg-AD or 5xFAD and Camk2a-CreERT2 mice. c, Aβ immunostaining in the CA1 hippocampal subfield of adult (8-month-old) control and Tert-activated R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice. Scale bar, 100 μm. d, Quantitative comparison of Aβ-immunoreactive pyramidal neurons in the CA1 regions (n = 6 per group, 8-month-old; p < 0.0001, two-way ANOVA with Tukey’s multiple comparisons test). e, Venn diagram showing intersections of upregulated biological processes based on the RNA-Seq results from Tert-activated cortical and hippocampal neurons of R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice compared with control groups. f, Top 5 overlapping pathways upregulated in both Tert-activated cortical and hippocampal neurons. g, Gene Set Enrichment Analysis (GSEA) plots showing relative upregulation of synaptic signaling genes in Tert-activated cortical and hippocampal neurons by comparison with control neurons. GSEA was based on the two-sided Kolmogorov-Smirnov statistic, and nominal p values unadjusted for multiple comparisons were calculated from 1,000 iterations of permutation on sample labels. h, Representative images of Golgi-stained cortical neurons from aged (18 months) control and Tert-activated R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice (n = 4 independent biological replicates per group). Experiments were repeated four times independently with similar results. Scale bar, 250 μm. i, High magnification of dendritic spines in impregnated pyramidal cortical neurons of aged control and Tert-activated R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice. Scale bar, 10 μm. j, Quantification of dendritic spine density (n = 5 dendrites per mouse, n = 4 mice per group, 18-month-old; p < 0.0001, two-tailed unpaired t-test). Data are mean ± s.e.m. ****P < 0.0001.
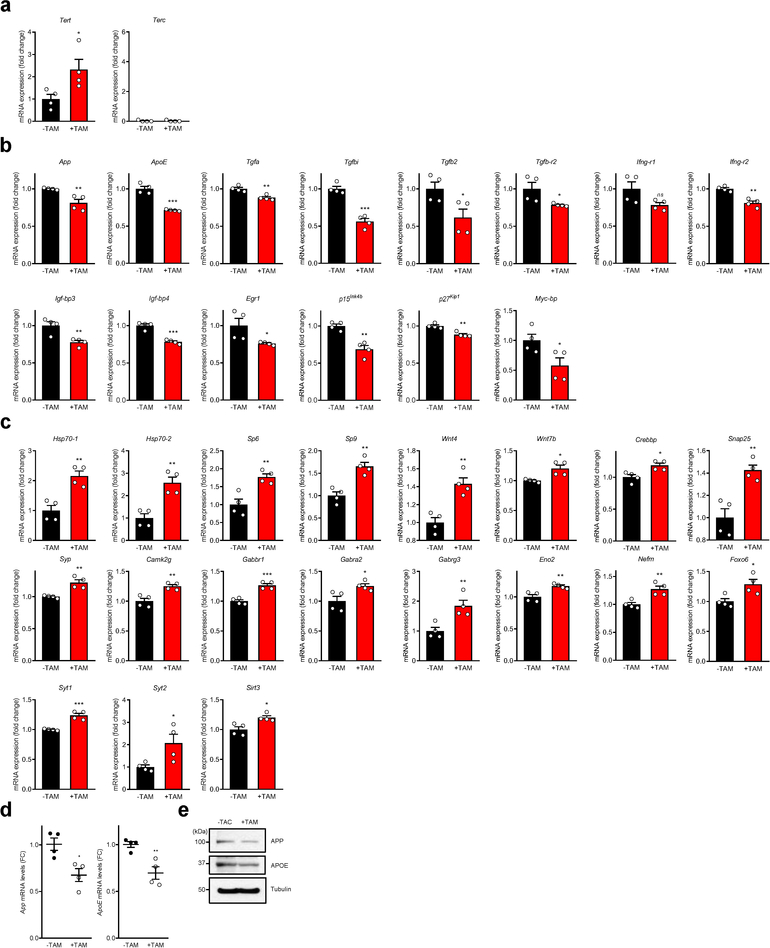
To further explore the impact of a modest increase in TERT levels in neurons, genome-wide RNA sequencing (RNA-Seq) was performed on cortical and hippocampal neurons isolated from R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice. This profile confirmed increased Tert gene expression in isolated neurons of our model upon tamoxifen treatment, and no changes in expression of the Terc gene which encodes the telomerase RNA component (Extended Data Fig. 4a). Moreover, Tert levels in our model system were well within the physiological range and comparable to those present during differentiation of mouse neurons and far less than those present in neural stem cells (Extended Data Fig. 5a–c). Computational analysis revealed that Tert induction in cortical and hippocampal AD neurons correlated with activation of pathways associated with synaptic signaling, nervous system development, neuron projection/differentiation, synaptic transmission/plasticity, and cognition/learning/memory (Fig. 3e, f and Extended Data Fig. 6). Gene set enrichment analysis (GSEA) also showed that genes involved in synaptic signaling were globally upregulated in both neuronal populations after Tert induction (Fig. 3g). Examination of expression of genes integral to AD biology revealed that Tert induction resulted in significant reduction in the expression of App (amyloid-β precursor protein) and ApoE (apolipoprotein E, a strong genetic risk factor for AD) genes (Extended Data Fig. 4b, d, e). Notably, Tert induction increased expression of the Hsp70 genes which encode a molecular chaperone that can reduce Aβ-induced cellular toxicity and effectively protect neurons in various AD animal models 31,32 and whose functional interaction with telomerase has been shown in cancer cells 33 (Extended Data Fig. 4c). Moreover, multiple aging- and/or AD-associated molecules, including inflammatory biomarkers (Ifng and Tgfb) and their receptors (Ifng-r and Tgfb-r), senescence-associated genes (Igf-bp3/4 and Egr1), cell cycle inhibitors (p15Ink4b and p27Kip1) and an oncogenic molecule (Mycbp) were also downregulated in Tert-activated AD neurons (Extended Data Fig. 4b). Conversely, transcription factors (Sp6/9 and Foxo6), transcriptional coactivators and axonal attractants (Wnt4 and Wnt7b), chromatin-modifying and transcription cofactor for memory formation (Crebbp), synaptic proteins (Snap-25, Syp and Camk2g), neurotransmitter receptors (Gabbr1, Gabra2 and Gabrg3) and neuron-specific markers (Eno2 and Nefm) were significantly upregulated in Tert-activated neurons (Extended Data Fig. 4c). Together, these unbiased transcriptomic analyses show that modest Tert induction in neurons can impact the expression of a large group of genes in postmitotic neurons in vivo that are strongly linked to AD pathobiology and central to synapse formation and neuronal activity and health.
Synapse loss and dysfunction are major correlates of cognitive decline in AD 2,34,35. To test whether induction of neuronal Tert could lead to protection against synaptic and network dysfunction in the AD brain, we examined neuronal morphology in vivo using Golgi-Cox staining. We observed that Tert induction was associated with increased neuronal complexity and density of dendritic spines in the aged cerebral cortex of Tert-activated R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice relative to controls (Fig. 3h–j). We conclude that elevated Tert levels in neurons activate synaptic signaling cascades and reduce spine shrinkage and synaptic loss in neurons of the mouse AD brain.
TERT enhances learning pathway networks in human neurons.
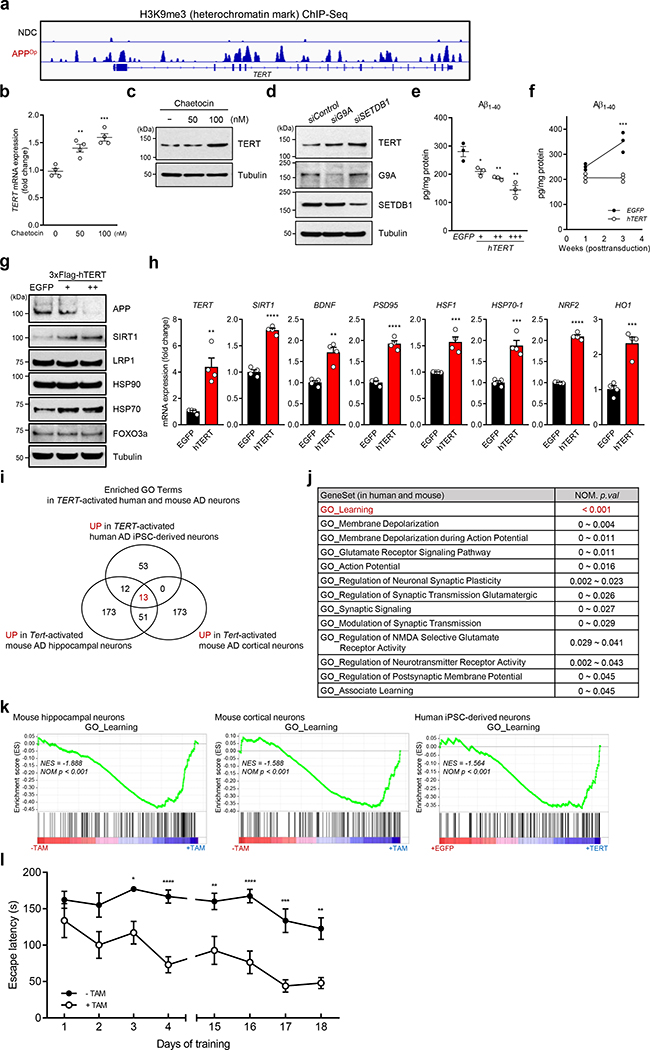
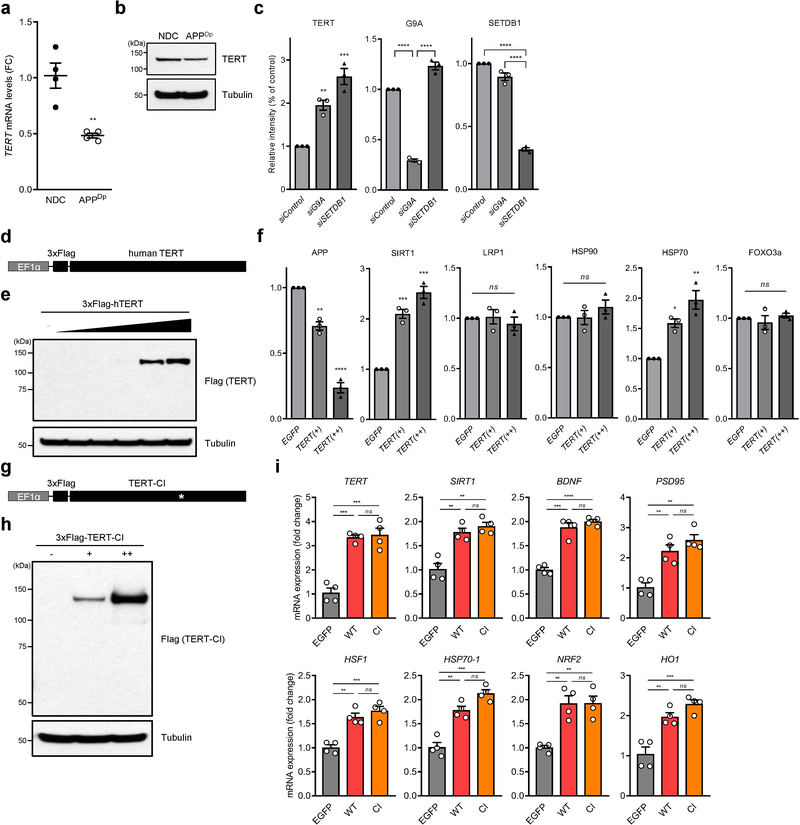
These murine observations prompted assessment of the biological impact of TERT induction in a well-established induced pluripotent stem cells (iPSCs) model generated from AD patient harboring genomic APP duplication 36. Mirroring the murine findings (Fig. 2f), APP duplication (APPDp) was associated with decreased expression of TERT coincident with enrichment of the heterochromatic mark H3K9me3 in the TERT gene locus of iPSC-derived human neurons relative to non-demented control (NDC) (Fig. 4a and Extended Data Fig. 7a, b). Accordingly, a H3K9 methyltransferase inhibitor restored human TERT mRNA and protein levels in iPSC-derived neurons (Fig. 4b, c). Moreover, the genetic knockdown of two key H3K9 methyltransferases, G9A and SETDB1, also de-repressed TERT suppression (Fig. 4d and Extended Data Fig. 7c).
Fig. 4. TERT activation alleviates amyloid pathology in human iPSC-derived neurons and ameliorates learning and memory deficits in AD.
a, H3K9me3 repressive histone mark occupancy in TERT gene of neurons from APPDp patient- and non-demented control (NDC) individual-derived iPSCs. b, c, TERT mRNA levels (b, n = 4; 0 vs. each concentration: p = 0.0038, p = 0.0005, respectively, two-way ANOVA with Tukey’s multiple comparisons test) and TERT protein levels (c) in chaetocin-treated human iPSC-derived APPDp neurons. d, TERT protein levels in human iPSC-derived APPDp neurons treated with siRNAs targeting histone methyltransferase genes, G9A or SETDB1 (n = 3 independent biological replicates per group). e, f, Aβ1–40 levels measured by ELISA in EGFP- or TERT-transduced iPSC-derived APPDp neurons (n = 3; EGFP vs. hTERT: p = 0.0251, p = 0.007, p = 0.0049, respectively, two-tailed unpaired t-test (e) and week 3: p = 0.0001, two-way ANOVA with Sidak’s multiple comparisons test (f)). g, Immunoblots for the indicated endogenous proteins in EGFP- or TERT-transduced iPSC-derived APPDp neurons (n = 3 independent biological replicates per group). h, Relative gene expression by qRT-PCR in EGFP- or TERT-transduced iPSC-derived APPDp neurons (n = 4; EGFP vs. hTERT: p = 0.0025, p < 0.0001, p = 0.0013, p < 0.0001, p = 0.001, p = 0.0005, p < 0.0001, p = 0.0006, respectively, two-tailed unpaired t-test). i, Venn diagram showing intersections of upregulated biological processes based on three (3) independent RNA-Seq results from Tert-activated mouse cortical and hippocampal neurons of R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice (n = 4 for each group) and TERT-activated human iPSC-derived APPDp neurons (n = 3) compared with each control group (all p < 0.05). j, List of 13 overlapping pathways upregulated in all Tert-activated mouse cortical and hippocampal AD neurons and TERT-activated human iPSC-derived APPDp neurons. k, GSEA plots showing relative upregulation of learning-related genes in Tert-activated cortical and hippocampal AD neurons and TERT-activated human iPSC-derived APPDp neurons by comparison with each control group. GSEA in i-k was based on the two-sided Kolmogorov-Smirnov statistic, and nominal p values unadjusted for multiple comparisons were calculated from 1,000 iterations of permutation on sample labels. l, Escape latency of aged (22~26 months) control and Tert-activated R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice in the Barnes maze over training days (n = 9 for each group; day 3: p = 0.0251, day 4: p < 0.0001, day 15: p = 0.0078, day 16: p < 0.0001, day 17: p = 0.0001, day 18: p = 0.0022, two-way ANOVA with Sidak’s multiple comparisons test). Experiments in d and g were repeated three times independently with similar results. Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We next measured the impact of TERT induction on Aβ accumulation in human iPSC-derived neurons infected with lentiviral vectors expressing TERT or EGFP. Similar to the murine studies, TERT induction resulted in a significant dose- and time-dependent reduction of intracellular Aβ accumulation in human iPSC-derived APPDp neurons as measured by sandwich ELISA (enzyme-linked immunosorbent assay) (Fig. 4e, f and Extended Data Fig. 7d, e). Furthermore, TERT induction not only decreased APP protein levels, but also triggered activation of the anti-aging gene (SIRT1), molecular chaperone and stress sensor genes (HSP70–1 and HSF1), synaptic plasticity-related genes (BDNF and PSD-95), and antioxidant genes (NRF2 and HO1) (Fig. 4g, h and Extended Data Fig. 7f), which are known to be crucial in reducing Aβ processing and cytotoxicity as well as improving synaptic plasticity and memory formation in the adult brain 31,37–39. Furthermore, the catalytically inactive TERT mutant also led to the upregulation of these genes (Extended Data Fig. 7g–i), indicating that TERT’s transactivation function is independent of its catalytic activity.
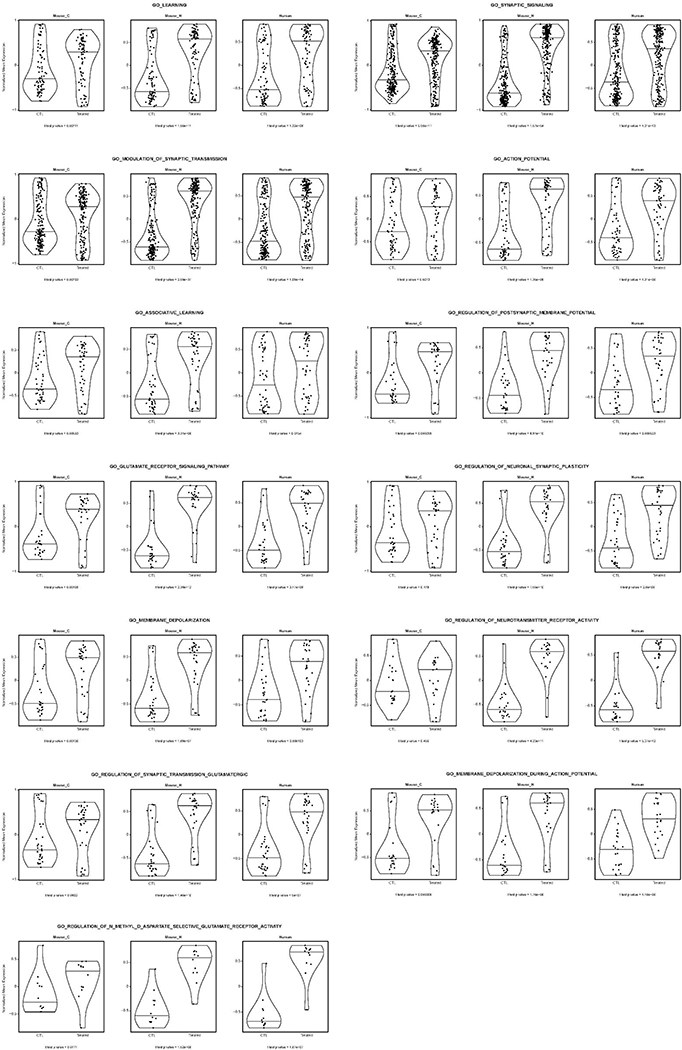
To gain insight into the functional significance of neuronal TERT activation in AD, we intersected RNA-Seq transcriptional profiles and pathway analysis of mouse AD cortical neurons, mouse AD hippocampal neurons, and human iPSC-derived APPDp neurons. This integrative cross-species analysis of the neuronal TERT activation network identified overlap of multiple neuron-specific pathways (Fig. 4i) with learning process as the most significantly enriched pathway (all p < 0.001) followed by membrane depolarization, glutamate receptor signaling, action potential, and synaptic signaling as the downstream consequences of TERT activation (Fig. 4j and Extended Data Fig. 8). All the enrichment profiles from three groups displayed a high level of concordant regulation of gene sets involved in learning processes in mouse and human AD neurons (Fig. 4k), suggesting that TERT regulates critical disease-associated pathways in the AD brain.
Since TERT induces dendritic spine formation on the cellular and tissue levels as well as activates learning process genes at the molecular level, we next investigated whether TERT activation could ameliorate the learning deficits of AD models in vivo. To that end, spatial learning and memory were assessed in the R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 model versus AD controls. While AD controls showed impaired acquisition of spatial learning at old age on the Barnes maze, age- and gender-matched Tert-activated AD mice achieved significant prevention of learning and memory dysfunction, as indicated by a reduction in latencies to enter the escape hole (Fig. 4l). These findings, which align with above cellular and molecular data, indicate that Tert activation attenuates age-associated learning impairment of AD mice.
TERT as a novel transactivation component in AD neurons.
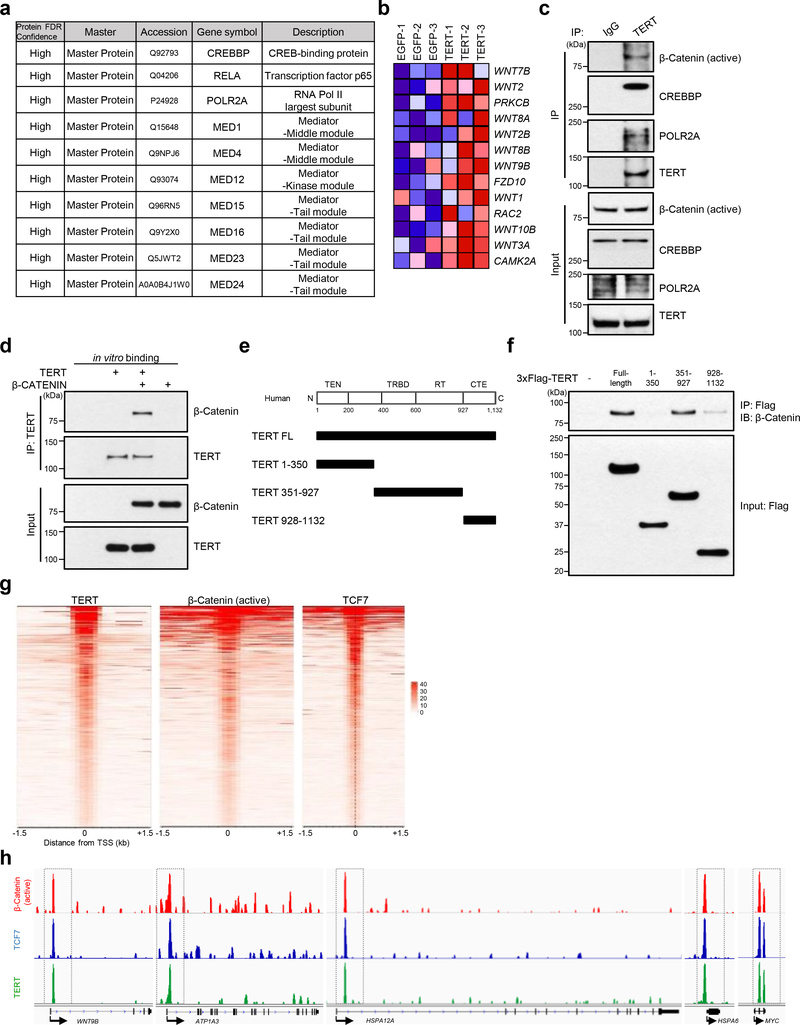
To identify the mechanistic basis of TERT activation and gene regulation, we conducted proteome-wide analysis of potential interaction partners of TERT in neurons. Characterization of TERT-containing protein complexes by mass spectrometry identified transcriptional regulators CREB-binding protein (CREBBP) and RELA, RNA polymerase II’s largest and catalytic subunit POLR2A, and multiple Mediator complex subunits (MED1, 4, 12, 15, 16, 23, 24) which link transcriptional regulators to RNA polymerase II in human iPSC-derived neurons (Fig. 5a). RNA-Seq also revealed that various WNTs and WNT pathway components were elevated in TERT-activated AD neurons (Fig. 5b) which gains added significance in light of the known neuroprotective action of WNT signaling in neurodegenerative disease 40; moreover, previous work in proliferating cell systems has established the interconnectedness of WNT/β-Catenin signaling and TERT in the context of mouse cells 41. These observations prompted us to assess whether endogenous human TERT in postmitotic neurons physically interact with transcriptional regulatory complexes containing β-Catenin, a pivotal player in the transduction of WNT signaling. Co-immunoprecipitation assays indeed confirmed that neuronal TERT protein physically interacts with the activated nuclear form of β-Catenin as well as CREBBP and POLR2A at endogenous levels in human iPSC-derived neurons (Fig. 5c). To provide further evidence for the binding specificity of TERT, we next employed an in vitro system using purified recombinant human proteins. Using human TERT or β-Catenin proteins that were first expressed in a human cell line and purified using an independent affinity resin, we conducted in vitro binding assays, showing that TERT binds to β-Catenin (Fig. 5d). To define the TERT protein domains responsible for this interaction, we performed pull-down assays with human TERT fragments, demonstrating that full-length or the TERT fragment containing the RNA-binding and reverse transcriptase (RT) motif binds to β-Catenin (Fig. 5e, f). Thus, TERT interacts with β-Catenin both in vivo and in vitro.
Fig. 5. TERT interacts with β-Catenin-containing transactivation complex and is recruited to the promoters of specific target genes in AD neurons.
a, List of TERT-interacting proteins identified by mass spectrometry in human iPSC-derived APPDp neurons. b, RNA-Seq heat map of WNT signaling pathway genes in EGFP- and TERT-transduced human iPSC-derived APPDp neurons (n = 3). c, Co-immunoprecipitation of endogenous β-Catenin (active), CREBBP, POLR2A, and TERT from human iPSC-derived APPDp neurons. d, In vitro binding assay between human TERT and β-Catenin. e, Schematic of full-length and truncated forms of human TERT. The upper panel represents four domains of TERT protein as the TERT N-terminal (TEN) domain, the TERT-specific RNA-binding domain (TRBD), the reverse transcriptase (RT) domain and the C-terminal extension (CTE)/thumb domain. f, Co-immunoprecipitation of endogenous β-Catenin with the full-length and truncated forms of 3xFlag-TERT. g, ChIP-Seq density heat maps of TERT, β-Catenin (active) and TCF7 across the gene promoters of human iPSC-derived APPDp neurons. h, Chromatin-state maps showing β-Catenin (active), TCF7 and TERT binding peaks for the WNT9B, ATP1A3, HSPA12A, HSPA6, and MYC locus, as determined by ChIP-Seq. Experiments in c, d and f were repeated three times independently with similar results.
To further assess a possible global enrichment of the association of TERT and β-Catenin/TCF7 on the genomic level, the genome-wide distribution of TERT and β-Catenin/TCF7 was determined in human iPSC-derived neurons by ChIP-Seq using specific antibodies. Both TERT and β-Catenin as well as TCF7, which is a transcription complex partner, predominantly occupied the transcription start sites (TSS) of gene promoters (Fig. 5g). We also determined that TERT-binding sites were occupied by both β-Catenin and TCF7 at the promoter regions of highly relevant genes including a WNT family member, WNT9B; a Na+/K+-ATPase catalytic subunit, ATP1A3 (one of 5 overlapping genes upregulated in both TERT-activated human and mouse neurons in our study [Supplementary Table 1]); HSP70 family members HSPA12A and HSPA6; and a positive feed-forward regulator of TERT, MYC (Fig. 5h), suggesting that the recruitment of TERT to these promoters amplifies their transcriptional activation in postmitotic neurons.
Discussion
In this study, we observed that Tert haploinsufficiency affected the expression of genes integral to AD pathogenesis and that neurons from amyloid-based AD models exhibited early epigenetic repression of neuronal TERT expression. We established that physiological increases in TERT levels result in a marked reduction of Aβ levels in neurons in the brains of two AD mouse models and in cultured human iPSC-derived neurons harboring genomic APP duplication. Mechanistically, TERT induces the β-Catenin/TCF7 complex to up-regulate key neuronal genes governing synaptic signaling and learning pathways and protecting neuron health in both mouse and human neurons. Notably, neuronal TERT expression improved dendritic spine formation and cognitive function in aging AD mouse models. Together, this work highlights somatic TERT activation as a potential disease modification strategy for AD.
Cross-species transcriptomic profiles revealed that TERT activation in neurons upregulates genes promoting learning, action potential, and synaptic signaling pathways as well as neuronal protection in the setting of toxic Aβ accumulation in human and mouse AD models (Fig. 4). Our work aligns with observations that TERT protein can exert potent neuroprotective effects against oxidative damage/pathological Tau in human postmortem AD brains 17 and Aβ-peptide-induced apoptosis of cultured embryonic neurons 42. It also provides additional mechanistic insights that TERT can reinforce the survival and functioning of neurons through enhancement of HSP70-mediated neuroprotection, NRF2/HO1-regulated antioxidant defense, and BDNF-induced cell survival system. Current evidence supports the involvement of repressive histone modifications in the TERT gene and associated downregulation of genes governing neuronal transcriptional homeostasis, memory formation and brain function 43,44. Inhibition of histone methyltransferases, which represses transcription in terminally differentiated cells 45, can induce dendritic spine formation in the hippocampus and restore learning and memory function in aged animals 46. Conversely, loss of histone demethylase KDM1A/LSD1 can result in widespread neuronal cell death and severe neurodegeneration 47. These collective insights prompt speculation that TERT serves as a key target of disease-associated epigenetic dysregulation of gene networks critical to neuronal health in the aging AD brain.
Our finding of the TERT/β-Catenin intersection in AD pathobiology gains significance in light of previous work showing that disruption of WNT/β-Catenin signaling has been implicated in the selective vulnerability of neurological disorders, including AD, autism, and schizophrenia 48–50. Along these lines, it is intriguing that several single nucleotide polymorphisms (SNPs) have been identified in the TERT locus from autism spectrum disorder patients 51 as well as schizophrenia patients 52 -- thus our findings encourage the study of TERT biology in these diseases. With respect to AD, our findings of the physical association of TERT and the β-Catenin/TCF transcription complex, and the TERT enhancement of β-Catenin/TCF transcriptional activity in AD neurons (Extended Data Fig. 9) point to important roles for TERT and WNT signaling in the progression of AD disease. At the same time, while WNT surfaced as the top TERT-modulated gene network, we acknowledge that additional neuronal pathways, such as NMDA receptor-dependent signaling cascades and calcium signaling pathway, are also influenced by TERT activation. Furthermore, our functional findings of the beneficial impact of TERT expression also rationalizes why mature neurons would sustain TERT expression in the postmitotic state. Finally, our data do not exclude the possibility that TERT plays additional roles including cooperation with transcriptional regulatory factors and/or functions in other subcellular compartments, such as mitochondria 53,54.
Emerging evidence has suggested that TERT is a potent protector from brain aging and age-associated pathologies 16,17,53,55–57. Telomerase plays a critical role in aging and age-related diseases and its activation has been proposed as a potential therapeutic for human aging and age-related degenerative diseases 16,55,56,58. While the role of TERT in these processes has focused primarily on its classical functions in telomere synthesis and end protection in proliferative cells, there is a growing appreciation that TERT also functions in post-mitotic tissues via modulation of gene expression 14–19. Our findings provide a deeper understanding of how TERT and its transcriptional regulatory networks contribute to neuronal health in the setting of amyloid pathology and cognitive deficits in both mouse and human models of AD at the molecular level. Our genetic and mechanistic study justifies the testing of various somatic TERT therapies such as exosome-mediated TERT mRNA, small molecule activators of TERT gene expression, and/or small molecules decreasing H3K9 methyl marks at the TERT locus as potential therapeutic options for AD patients.
Methods
Animals.
All animal procedures used in this study were reviewed and approved by the University of Texas MD Anderson Cancer Center Institutional Animal Care and Use Committee. Tert deficient LSL-mTert mice generated by inserting a transcriptional stopper cassette flanked by loxP sites (LSL) in the endogenous Tert locus to prevent the expression of Tert gene were previously described 59. Homozygous 3xTg-AD 23, its non-transgenic control B6129SF2/J, hemizygous 5xFAD 24, and Camk2a-CreERT2 28 mice were purchased from the Jackson Laboratory. To generate Cre-inducible Tert knock-in allele, we used a previously established knock-in strategy 60. Linearized CAG-LSL-mTert-IRES-EGFP-pA construct was targeted into C57BL/6-derived JM8F6 embryonic stem (ES) cells by electroporation, and the Rosa26 locus was modified by targeted insertion of ubiquitously expressed CAG promoter, followed by a lox-STOP-lox cassette-controlled mTert gene (CAG-LSL-mTert). Positive ES clones were identified by Long Range PCR (New England Biolabs) using the following primers: left arm 5ʹ-GGT CGT GTG GTT CGG TGT CTC TTT-3ʹ and 5ʹ-ATG GGC TAT GAA CTA ATG ACC CCG-3ʹ right arm 5ʹ- CAC TAC CAG CAG AAC ACC CCC ATC-3ʹ and 5ʹ-GTG CCA CTA GTA CCA ACA GCC TCT-3ʹ. PCR products were sent to sequencing to confirm the correct recombination and karyotyping. Eventually, two independent clones were identified, injected into C57BL/6 albino blastocysts to generate chimeric mice. The chimeric mice from each clone produced germline transmission. To generate our TERT-AD model, the resulting mice were mated to 3xTg-AD or 5xFAD mice, then subsequently bred with Camk2a-CreERT2 strain. This TERT-AD model enabled spatial (neuron-specific) and temporal (tamoxifen-inducible) control of Tert gene expression in two independent and widely studied AD (3xTg-AD and 5xFAD) mouse models. Mice used in this study were of both genders. All animals were housed in pathogen-free, ambient temperature (21–23 °C), 45–55% humidity, and 12-h dark/light cycle conditions, and cared for in accordance with the International Association for Assessment and Accreditation of Laboratory Animal Care policies and certification.
Primary mouse neuronal culture.
Primary mouse cortical and hippocampal neuronal cultures were prepared from E18.5 ~ E19.5 embryos from 3xTg-AD, its non-transgenic control B6129SF2/J, 5xFAD and its wildtype littermate control mice using methods and procedures as previously described 61,62. The brains were removed from the mouse embryos, and the hippocampi and cortices were separated using sterile instruments in CMF-HBSS. The hippocampal and cortical tissues were dissociated using Neural Tissue Dissociation Kits (MACS), and the neuronal populations were isolated by Neuron Isolation Kit (MACS), according to the manufacturer’s protocols. The isolated neuronal cells were plated in poly-D-lysine-coated plates with Neuronal Plating Medium (Eagle’s MEM (ATCC), 10% FBS (Invitrogen), 1x penicillin/streptomycin (Invitrogen)) for an hour, and then incubated with Neuronal Maintenance Medium (Neurobasal medium (Invitrogen), 1x GlutaMax (Invitrogen), 1x B27 (Invitrogen), 1x penicillin/streptomycin (Invitrogen)). Half of the medium was replaced with fresh Neuronal Maintenance Medium every 3 days. These primary neurons were maintained in culture for up to about 4 weeks.
Human iPSC-derived NPC culture.
For human AD neural culture, neural progenitor cells (NPCs) 63 derived from induced pluripotent stem cells (iPSCs) which were previously derived from familial Alzheimer’s disease patients harboring genomic duplication of APP gene (APPDp) or from non-demented control individual 36 were used and maintained in neural stem cell maintenance medium (Millipore) supplemented with 20 ng/ml bFGF (Peprotech). For neuronal differentiation as previously described 64,65, NPCs were plated onto poly-L-ornithine (PLO)/laminin-coated plates, and incubated in neural differentiation media (DMEM/F12 (Invitrogen), 1x GlutaMax (Invitrogen), 1x N2 (Invitrogen), 1x B27 (Invitrogen), 20 ng/ml BDNF (Peprotech), 20 ng/ml GDNF (Peprotech), 1 μM dibutyryl-cyclic AMP (Sigma), and 0.2 μM ascorbic acid (Sigma). iPSCs-derived neurons were differentiated for 1~4 months.
Isolation of adult mouse neuron and neural stem cells.
Adult mouse neurons were isolated using the density gradient procedures, as previously described 66. The mice were euthanized, and the cortices and hippocampi were separately dissected from adult mouse brains in HABG (HibernateA (BrainBits) / B27 (Invitrogen) / GlutaMax (Invitrogen) medium). The hippocampi and cortices were dissociated with fire-polished glass pasteur pipettes and Neural Tissue Dissociation Kit (MACS), according to the manufacturer’s protocol. The cell suspensions were placed on the top of the prepared OptiPrep density gradient (Sigma), and the neuronal cells were separated by centrifugation. After additional washing-centrifugation to remove debris, the pellet of isolated neurons was proceeded to examine the expression profiles. Neural stem cells (NSCs) from the subventricular zone (SVZ) of the adult mouse brain were isolated using methods and procedures as previously described 67.
RNA sequencing and analysis.
Total RNA was extracted from collected samples using TRIzol Reagent (Invitrogen), according to the manufacturer’s protocol. For mouse samples, both mouse hippocampal and cortical neurons were freshly isolated from 7- or 8-month-old Tert+/+, Tert–/– (TertLSL/LSL), or R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice eight days after the final tamoxifen or vehicle administration, as previously described 66. For human samples, differentiated neurons derived from APPDp NPCs were prepared eight days after EGFP or TERT lentiviral transduction. RNA quantity was determined to be optimal for each sample before further processing. For each RNA seq, purified RNA was amplified to construct Illumina sequencing libraries using standard mRNA-Seq guide (the PE protocol), and the libraries were sequenced on Illumina HiSeq 4000 instrument. Gene-level quantification was implemented with htseqcount package 68. Data normalization and differential expression analysis were conducted using DESeq2 package 69. Gene set enrichment analysis (GSEA) on gene ontology biological processes were done using GSEA java package (gsea2–2.2.1.jar) from the Broad Institute 70,71.
ChIP sequencing and analysis.
Cells were fixed and cross-linked with 1.42% formaldehyde. The cross-linking was quenched with 125 mM glycine, and cells were washed and collected with ice-cold PBS containing 1x protease & phosphatase inhibitor (Pierce). The nuclei were isolated by lysing the cell pellets in RIPA buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1x protease and phosphatase inhibitor cocktail (Pierce)). The chromatin was then sheared by sonication using Bioruptor Pico (Diagenode). The lysates were cleared by centrifugation.
For chromatin immunoprecipitation, the lysates were pre-cleared by incubation with Protein A/G Plus Agarose (Pierce). Clear lysates were incubated overnight at 4 °C with appropriate primary antibodies. The antibody complexes were obtained with ChIP-Grade Protein A/G Plus Agarose (Pierce) and washed five times with RIPA buffer. The DNA was recovered as described 72. Control immunoprecipitation was done in parallel without antibodies. Raw fastq reads for all ChIP-seq experiments were processed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), low-quality reads were removed with trimmomatic (DOI: 10.1093/bioinformatics/btu170) v0.33 with SLIDINGWINDOW:4:30 and quality reads were aligned to the mm9 (mouse) and hg19 (human) reference genome using Bowtie version 1.2.2 73 with the following criteria: --best --chunkmbs 320. To directly compare ChIP-seq between samples, uniquely mapped reads for each mark were normalized by total reads per condition, sorted, and indexed using SAMtools version 1.9 74. Model-based analysis of ChIP-seq (MACS version 1.4.2) 75 was used to identify antibody-IP enrichment over input background. MACS2 was used to identify the differential binding of each protein in different conditions with the following criteria: bdgdiff –g 60 –l 120. To visualize ChIP-seq libraries on the IGV browser, we used deepTools version 2.7.15 to generate bigwig files by scaling the bam files to reads per kilobase per million (RPKM) using the following criteria: bamCoverage –b–normalizeUsing RPKM–smoothLength 300–binSize 30–extendReads 200 –o.
Quantitative real-time PCR.
Total RNA was isolated using TRIzol Reagent (Invitrogen), reverse-transcribed with SuperScript III First-Strand Synthesis System (Invitrogen), and amplified with SYBR Green PCR Master Mix (Invitrogen), according to the manufacturers’ protocols. The expression levels were normalized with mouse Hprt1 or human HPRT1 mRNA in each sample. The primer sequences are provided in Supplementary Table 2.
siRNA knockdown.
siRNAs targeting human G9A (siGENOME SMARTpool M-006937–01), human SETDB1 (siGENOME SMARTpool M-020070–00) and non-targeting control siRNA pool (siGENOME non-targeting D-001210–01) were purchased from GE Dharmacon/Horizon and used according to the manufacturer’s protocol.
Generation of lentiviral constructs.
The lentiviral construct encoding 3xFlag-TERT was prepared using human TERT construct from Dr. Steven Artandi (Stanford University) 76. The cDNAs encoding 3xFlag-tagged full-length and truncated forms of TERT were amplified using PCR and subcloned into TOPO TA cloning vector, and sequentially into pHAGE-EF1a lentiviral vector. The catalytically inactive (CI) TERT construct was generated by substituting the aspartic acid at position 712 residue with the alanine 77 by site-directed mutagenesis. The pHAGE-EF1a-IRES-EGFP empty vector was used as the control. The sequences were confirmed by sequencing.
Immunoprecipitation and immunoblot.
Immunoprecipitation and immunoblotting were carried out as previously described 78 with a minor modification. For coimmunoprecipitation, the differentiated neuronal cells were lysed on ice in a modified RIPA buffer. The cell supernatant was incubated at 4 °C overnight with appropriate primary antibodies, and ChIP-Grade Protein A/G Plus Agarose (Pierce) was then added and incubated for an additional hour at 4 °C. Immunoprecipitates were washed five times with RIPA buffer, recovered with SDS sample buffer, and subjected to immunoblot analysis. Protein samples were subjected to NuPAGE (Invitrogen), transferred to PVDF membrane (Bio-Rad), and probed with primary antibodies. Immunoreactivity was visualized with appropriate HRP-conjugated secondary antibodies (Cell Signaling Tech) followed by SuperSignal Chemiluminescent Substrate (Pierce). Quantitative densitometric analysis of select gel band intensities was carried out using ImageJ software.
Proteomic analysis.
Immunoprecipitates were separated on SDS-PAGE. Gel bands were digested overnight with trypsin (Pierce) following destaining, reduction with DTT and alkylation with iodoacetamide (Sigma). The samples then underwent solid-phase extraction cleanup with an Oasis HLB plate (Waters) and the resulting samples were injected onto an Orbitrap Fusion Lumos mass spectrometer coupled to an Ultimate 3000 RSLC-Nano liquid chromatography system. Samples were injected onto a 75 um i.d., 75-cm long EasySpray column (Thermo) and eluted with a gradient from 0–28% buffer B over 90 min. Buffer A contained 2% (v/v) ACN and 0.1% formic acid in water, and buffer B contained 80% (v/v) ACN, 10% (v/v) trifluoroethanol, and 0.1% formic acid in water. The mass spectrometer was operated in positive ion mode with a source voltage of 1.5 kV and an ion transfer tube temperature of 275 °C. MS scans were acquired at 120,000 resolution in the Orbitrap and up to 10 MS/MS spectra were obtained in the ion trap for each full spectrum acquired using higher-energy collisional dissociation (HCD) for ions with charges 2–7. Dynamic exclusion was set for 25 s after an ion was selected for fragmentation.
Raw MS data files were analyzed using Proteome Discoverer v2.2 (Thermo) with peptide identification performed using Sequest HT searching against the human protein database from UniProt. Fragment and precursor tolerances of 10 ppm and 0.6 Da were specified, and three missed cleavages were allowed. Carbamidomethylation of Cys was set as a fixed modification, with oxidation of Met set as a variable modification. The false-discovery rate (FDR) cutoff was 1% for all peptides.
ELISA analysis.
Terminally differentiated human neurons were infected with EGFP or TERT lentiviral constructs and cultured for the indicated time periods. Amyloid-β levels were quantified with sandwich enzyme-linked immunosorbent assay (ELISA) kit (Invitrogen) according to the manufacturer’s instructions and normalized to total protein levels determined by BCA assay (Pierce).
Antibodies.
Anti-BDNF (ab108319, Abcam, 1:1,000), anti-Aβ (Clone 6E10, SIG-39320, Covance, 1:1,000), anti-TERT (ab32020, Abcam, 1:500), anti-H3K9me3 (ab8898, Abcam, 1:30), anti-KDM1A (ab17721, Abcam, 1:300), anti-GFAP (Z-0334, Dako, 1:4,000), anti-IBA-1 (019–19741, Wako, 1:1,000), anti-APOE (NB110–60531, Novus, 1:2,000), anti-SIRT1 (ab32441, Abcam, 1:1,000), anti-LRP1 (ab92544, Abcam, 1:3,000), anti-HSP90 (4877, Cell Signaling Tech, 1:1,000), anti-HSP70 (4872, Cell Signaling Tech, 1:1,000), anti-FOXO3A (ab12162, Abcam, 1:1,000), anti-CREBBP (7389, Cell Signaling Tech, 1:1,000), anti-non-phospho (active) β-Catenin (8814, Cell Signaling Tech, 1:500), anti-β-Catenin (total) (8480, Cell Signaling Tech, 1:500), anti-POLR2A (MA1–46093, Thermo Scientific, 1:1,000), anti-TCF7 (2206, Cell Signaling Tech, 1:30), anti-Flag (Clone M2, F1804, Sigma, 1:3,000), anti-Tubulin (T5168, Sigma, 1:10,000), anti-mouse IgG, HRP-linked secondary antibody (7076, Cell Signaling Tech, 1:10,000) and anti-rabbit IgG, HRP-linked secondary antibody (7074, Cell Signaling Tech, 1:10,000) were obtained from commercial sources.
Perfusion and immunohistochemistry.
Mice under anesthesia were transcardially perfused with saline and then with 4% paraformaldehyde in PBS (pH 7.4). The brains were post-fixed in the same fixative and embedded in paraffin. Subsequently, the brain sections were deparaffinized, hydrated, and blocked with Biocare blocking reagent (Biocare Medical) according to standard procedures. After the epitope unmasking, the sections were incubated with primary antibodies, followed by secondary Envision plus antibodies with HRP-labeled polymer (Dako). The antigen was visualized using DAB chromogen system (Dako). Quantitative analysis of immunohistochemical staining intensities was carried out using ImageJ software.
Golgi-Cox staining of mouse brains.
The whole mouse brains were freshly isolated and stained with FD Rapid GolgiStain kit (FD NeuroTechnologies Inc), according to manufacturer’s procedures. The impregnated mouse brains were processed for Golgi-Cox staining with the kit. Serial cryostat sections of 100 μm thickness were cut coronally and mounted on gelatin-coated slides (FD NeuroTechnologies Inc). The slides were stained with staining solution, dehydrated, cleared, and coverslipped with Eukitt Quick-hardening mounting medium (Sigma). The morphology of dendritic spines of neurons were examined using the confocal reflection microscopy (Leica Microsystems) to take advantage of the stain strong reflectance. The epi-illumination was scanned with the 488 nm laser through a 63x NA 1.4 objective in a Leica SP8 laser scanning confocal system equipped with the acousto-optical tunable filter (AOBS) and 5-channel spectral detector. The spectral range of one detector was set to a narrow range (~8 nm) centered at 488 nm with the reflectance suppression option turned off and the pinhole size set to 1 Airy unit. Additional signals were collected during the same scans by other detectors set to longer wavelengths. The imaging was performed in 3-D by collecting z-stack images processed to form the maximum intensity projection (MIP) image using Leica LAS software.
Telomerase activity assay.
Telomerase activity assay was measured using the telomeric repeat amplification protocol (TRAP), as previously described 79. The cells were harvested with 1x RIPA buffer. The sample lysates were centrifuged to remove cellular debris and the protein concentration was determined by BCA protein assay (Pierce). The detection of telomerase activity was analyzed based on real-time quantitative PCR with TRAP master mix (1x SYBR Green PCR Master Mix, 100 ng HPLC-purified ACX and TS primers, 1 mM EDTA) with control samples (heat-inactivated samples, positive controls, and lysis buffer-only controls). The standard curve was created by serial dilutions of telomerase-positive samples (HEK293 cells) (R2 > 0.90). The relative telomerase activity (RTA) of samples was calculated based on the following equation: RTA = 10[(Ct sample-Yint)/slope)].
In vitro binding assay.
For the preparation of TERT and β-Catenin proteins, the constructs expressing 3xFlag-TERT and Flag-β-CATENNIN were transfected separately in HEK293 cells. The cells were lysed with a denaturing lysis buffer (62.5 mM Tris, 2% SDS, 5 mM EDTA) to avoid possible contamination by any interacting proteins. After centrifugation at 21,000 × g for 10 min, each cell extract (the supernatant) was diluted with modified RIPA buffer and Flag-tagged proteins were purified with anti-FLAG M2 Affinity agarose slurry (Sigma) at 4 °C for 3 hr. After four washes, each precipitated Flag-tagged protein was competitively eluted using 3×FLAG peptide from the agarose resin by incubation with the elution buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 0.5% NP-40, 3 mM imidazole, 750 ng/μl 3×FLAG peptide (APExBIO)) at 4 °C for 1 hr. For in vitro binding assay, the purified 3xFlag-TERT and Flag-β-CATENNIN were then incubated together in HGMC binding buffer (50 mM HEPES [pH 7.4], 100 mM NaCl, 20% glycerol, 0.1% NP-40, 5 mM MgCl2, 5 mM CaCl2, 1 mM EDTA, 1x phosphatase and phosphatase inhibitor cocktail; Pierce) for 15 min at 30 °C, followed by co-immunoprecipitation with anti-TERT antibody in a modified RIPA buffer. The precipitated products were washed, resolved by SDS-PAGE and analyzed by immunoblots.
tPA activity assay.
tPA proteolytic activity was assessed using SensoLyte AMC tPA Activity Assay kit (AS-72160, AnaSpec) according to the manufacturer’s protocol. Measurements were performed in mouse cortical and hippocampal homogenates prepared in a modified RIPA buffer without protease inhibitors. tPA activity was presented as relative fluorescence unit (RFU) after normalization for total protein content.
Mouse behavioral test.
The evaluation of learning and memory in mice was carried out using modified Barnes maze, similarly to that previously described 80,81. The apparatus consisted of a clear grey circular open disk (92 cm diameter) with 20 circular holes (5 cm diameter) (Panlab Harvard Apparatus). The holes were equally spaced around the perimeter and located 2 cm from the edge of the maze. A black escape box (20 cm x 9 cm x 9 cm) was positioned beneath one of these holes, and distinct visual cues were placed at three points around the maze. The overhead lights and a buzzer (3 kHz, 85 dB) were used as aversive stimuli to encourage the mice to seek out the escape hole. The mice were given 180 s for each trial to identify and enter the escape hole. The mice received three trials per day with a 15-min inter-trial interval on four consecutive days during the acquisition and retention phases. The latency to enter the escape hole was recorded. Between each trial, the maze and escape hole were thoroughly cleaned with 70% ethanol to remove any other clues that might affect performance in subsequent trials.
Statistical analysis
No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications 16,82,83. All data distributions were assumed to be normal, but this was not formally tested. The mice and cells were randomly assigned to control or experimental groups, except in experiments that required specific genotypes and ages. The investigators were generally not blinded to allocation during experiments and outcome assessment. No animal or sample was excluded from the analysis. All statistical analyses were performed using GraphPad Prism 8; the statistic tests and p values are described in the indicated figure legends.
Data availability
The RNA-Seq data have been deposited in Gene Expression Omnibus (GEO) under accession numbers GSE163523, GSE163524 and GSE163525, and the ChIP-seq data in Sequence Read Archive (SRA) under accession numbers PRJNA633993 and PRJNA633994. The mass spectrometry data have been deposited in MassIVE under accession numbers MSV000088190. All other data are available from the corresponding author upon reasonable request.
Extended Data
Extended Data Fig. 1: Mouse primary cortical and hippocampal neuronal culture and gene expression profile of histone methyltransferases and demethylases in 5xFAD neurons.
a, Brightfield images of primary hippocampal and cortical neurons isolated from 5xFAD last-stage embryos (E18.5). b, Immunoblots for full-length APP and oligomeric amyloid-β in primary cortical and hippocampal neurons from 5xFAD and non-transgenic control mice at 1, 8, 14 and 21 DIV. A tubulin was used as a loading control. Experiments in a-b were repeated three times independently with similar results. c, d, mRNA expression levels of each histone methyltransferase (c) or demethylase (d) in cortical and hippocampal neurons isolated from 5xFAD and non-transgenic control mice at 2~3-month-old. Transcript levels were normalized to Hprt1 mRNA (n = 3 per group). e, Quantification of KDM1A staining intensity in the CA1 hippocampal subfield of 5xFAD and wildtype littermate control mice (n = 4 per group, p = 0.0003). Data are mean ± s.e.m. ***P < 0.001; ns, not significant (two-tailed unpaired t-test).
Extended Data Fig. 2: Generation of Cre-inducible Tert knock-in mouse (R26-CAG-LSL-mTert).
a, Genotyping results of the original ES targeted lines carrying the R26-CAG-LSL-mTert-IRES-eGFP-pA alleles. b, Representative photographs of chimeric mice obtained from targeted ES cells. c, Aβ immunostaining in the hippocampus of adult (7-month-old) control and Tert-activated R26-CAG-LSL-mTert; 5xFAD; Camk2a-CreERT2 mice. Experiments were repeated three times independently with similar results. Scale bar, 300 μm.
Extended Data Fig. 3: The effects of TERT induction on neuroinflammation associated with activation of astrocytes and microglia.
a, Immunohistochemical staining for the astrocytic marker GFAP in the CA1 hippocampal subfield of adult control and Tert-activated R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice. Scale bar, 100 μm. b, Quantitative comparison of GFAP-positive astrocytes in the hippocampus (n = 4 per group, 8-month-old, p = 0.0177). c, IBA-1 immunostaining in the CA1 hippocampal subfield of adult control and Tert-activated R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice. Scale bar, 100 μm. d, Quantification of IBA1-positive activated microglia in the mouse hippocampus (n = 4 per group, p = 0.0015). Data are mean ± s.e.m. *P < 0.05, **P < 0.01 (two-tailed unpaired t-test).
Extended Data Fig. 4: Significantly up- or downregulated genes identified in RNA-Seq of Tert-activated R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mouse neurons.
a, mRNA levels of Tert and Terc in control and Tert-activated neurons isolated from R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mouse brains (n = 4 per group; p = 0.0391, p > 0.9999, respectively). b, mRNA levels of significantly downregulated genes in Tert-activated neurons compared to control (n = 4 per group; p = 0.0089, p = 0.0001, p = 0.0031, p = 0.0002, p = 0.0375, p = 0.0462, p = 0.0714, p = 0.0011, p = 0.0084, p = 0.0002, p = 0.0498, p = 0.0015, p = 0.002, p = 0.0438, respectively). c, mRNA levels of significantly upregulated genes in Tert-activated neurons compared to control (n = 4 per group; p = 0.0029, p = 0.0026, p = 0.0048, p = 0.0019, p = 0.0024, p = 0.0129, p = 0.0154, p = 0.0036, p = 0.0032, p = 0.0044, p = 0.0005, p = 0.0311, p = 0.01, p = 0.0063, p = 0.0063, p = 0.0225, p = 0.0005, p = 0.0403, p = 0.0129, respectively). d,e, Validation of App and ApoE mRNA (d) and protein (e) expression levels in the mouse brains of control (-TAM) and Tert-activated (+TAM) R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mice by quantitative RT-PCR (n = 4 per group; p = 0.0134, p = 0.0061, respectively) and immunoblotting. Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant (two-tailed unpaired t-test).
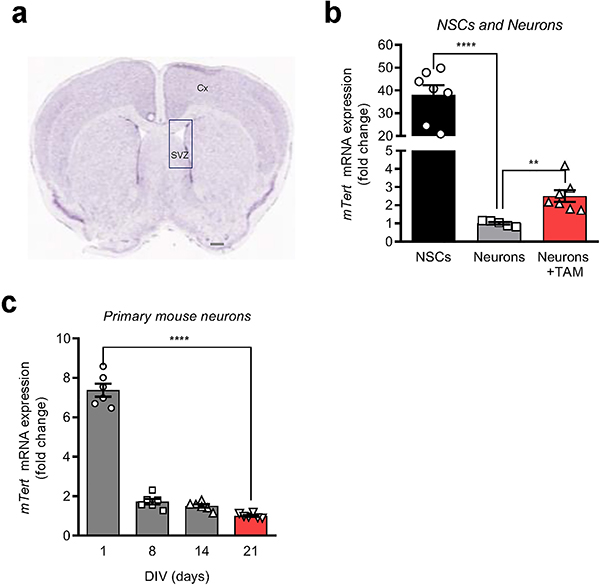
Extended Data Fig. 5: Tert expression in mouse adult neurons and neural stem cells as well as during neuronal maturation.
a, Mouse brain section showing the subventricular zone (SVZ) and cerebral cortex (Cx) for harvesting NSCs and neurons. Scale bar, 500 μm. b, mRNA levels of Tert gene in neural stem cells (NSCs) and neurons isolated from the brains of R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mouse with or without tamoxifen treatment (n = 7 (NSCs), 5 (neurons), 7 (neurons + TAM group); p < 0.0001, p = 0.0033, respectively). c, Tert mRNA levels during neuronal maturation of primary cortical and hippocampal neurons from 3xTg-AD mice at 1, 8, 14 and 21 DIV (n = 6 per group; day 1 vs. day 21: p < 0.0001). Data are mean ± s.e.m. **P < 0.01, ****P < 0.0001 (two-tailed unpaired t-test).
Extended Data Fig. 6: Sixty-four (64) pathways activated in both mouse cortical and hippocampal neurons isolated from TERT-AD mice upon Tert activation.
Boxplots showing the Tert-induced fold changes of all the upregulated coding genes in Tert-activated cortical (Mouse_C) and hippocampal (Mouse_H) neurons isolated from R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mouse brains compared to each untreated and matched control group. For all box plots, each dot represents the average value of differentially expressed gene found in the comparison; centre lines denote medians; box limits denote 25th–75th percentile (Q1-Q3); whiskers are drawn up to the smallest or largest observed value that is still within 1.5 times the interquartile range below the first quartile or above the third quartile, respectively; all other observed points are plotted as outliers. p values were calculated by two-tailed Student’s t test.
Extended Data Fig. 7: TERT levels in NDC- and APPDp-derived neurons, cloning of wild-type and catalytically inactive Flag-hTERT lentiviral vector and quantification of immunoblots shown in Fig.4.
a,b, TERT mRNA (a) and protein (b) levels in the neurons derived from NDC- and APPDp-derived iPSCs. c, Quantification of immunoblots in Fig. 4d. The values were normalized to respective control band intensity (n = 3; TERT: p = 0.0053, p = 0.0007, respectively, G9A: p < 0.0001, p < 0.0001, respectively, SETDB1: p < 0.0001, p < 0.0001, respectively). d, Schematic of wild-type Flag-tagged human TERT lentiviral expression construct. e, Immunoblots for the confirmation of 3xFlag-TERT expression in HEK293 cells. A tubulin was used as a loading control. Experiments were repeated three times independently with similar results. f, Quantification of immunoblots in Fig. 4g (n = 3 per group; APP: p = 0.0024, p < 0.0001, respectively, SIRT1: p = 0.0006, p = 0.0002, respectively, HSP70: p = 0.0228, p = 0.0037, respectively). Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant (two-tailed unpaired t-test (a) or two-way ANOVA with Tukey’s multiple comparisons test (c,f)). g, Schematic of catalytically inactive (CI) human TERT lentiviral expression construct. The white asterisk indicates the position of the single mutation D712A, which renders the protein catalytically inactive. h, Immunoblots for the confirmation of Flag-tagged catalytically inactive TERT expression in HEK293 cells. A tubulin was used as a loading control. Experiments were repeated three times independently with similar results. i, mRNA expression levels of each gene indicated in EGFP-, wildtype (WT) TERT- or catalytically inactive (CI) TERT-transduced APPDp neurons (n = 4; EGFP vs. WT and EGFP vs. CI: TERT: p = 0.0003, p = 0.0003, respectively, SIRT1: p = 0.0031, p = 0.0014, respectively, BDNF: p = 0.0001, p < 0.0001, respectively, PSD95: p = 0.0081, p = 0.0021, respectively, HSF1: p = 0.0014, p = 0.0005, respectively, HSP70–1: p = 0.0041, p = 0.0006, respectively, NRF2: p = 0.0053, p = 0.0052, respectively, HO1: p = 0.0024, p = 0.0005, respectively). Transcript levels were normalized to HPRT1 mRNA. Data are mean ± s.e.m. **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant (two-way ANOVA with Tukey’s multiple comparisons test)
Extended Data Fig. 8: Thirteen (13) pathways activated in mouse cortical and hippocampal AD neurons as well as in human iPSC-derived APPDp neurons upon TERT activation.
Violin plots showing the TERT-induced fold changes of all the upregulated coding genes in Tert-activated cortical neurons (Mouse_C) and hippocampal neurons (Mouse_H) isolated from R26-CAG-LSL-mTert; 3xTg-AD; Camk2a-CreERT2 mouse brains as well as TERT-activated human iPSC-derived APPDp neurons (Human) compared to each matched control group.
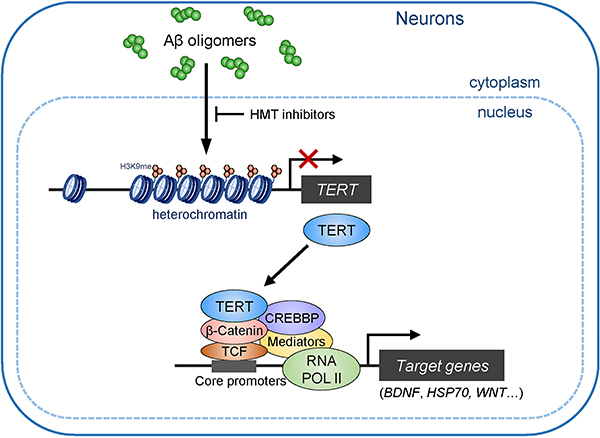
Extended Data Fig. 9: TERT contributes to β-Catenin/TCF-mediated transactivation in AD neurons.
At the early pathological stage of AD, amyloid-β (Aβ) oligomers induce the transcriptional repression of TERT gene via the propagation of heterochromatin in neurons. Genetic depletion and pharmacological inhibition of H3K9 methyltransferases (HMTs) can de-repress TERT gene suppression. TERT protein is able to interact with RNA pol II core transactivation machinery through β-Catenin and triggers the transcriptional induction of specific genes associated with neuronal survival and synaptic function in AD neurons, enabling to alleviate cognitive deficits.
Supplementary Material
Acknowledgements
The authors thank all the members of R.A.D laboratory for the discussion and constructive suggestions for this project; Dr. Steven Artandi for providing human TERT cDNA construct; Dr. Lawrence Goldstein for providing iPSC lines from NDC and APPDp patient. This work was supported by the National Institutes of Health (R01 CA084628), the Mathers Foundation (R.A.D), and a generous gift from Robert and Renee Belfer to the Neurodegeneration Consortium (R.A.D.). This study made use of the MD Anderson’s Advanced Technology Genomics Core (NCI CA016672), Research Histology, Pathology, and Imaging Core (DHHS/NCI P30 CA16672), and MDACC Advanced Microscopy Core (NIH 1S10 RR029552), and UT Southwestern Proteomics Core.
Competing interests
The authors declare no competing interests specifically related to this work. R.A.D. is a founder, advisor and/or director of Tvardi Therapeutics, Inc., Nirogy Therapeutics, Inc., Stellanova Therapeutics, Inc., Sporos Bioventures, LLC and Asylia Therapeutics, Inc. which are focused on therapies for cancer, fibrosis and/or inflammation.
References
- 1.Huang YD & Mucke L Alzheimer Mechanisms and Therapeutic Strategies. Cell 148, 1204–1222, doi: 10.1016/j.cell.2012.02.040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selkoe DJ & Hardy J The amyloid hypothesis of Alzheimer’s disease at 25years. Embo Mol Med 8, 595–608, doi:DOI 10.15252/emmm.201606210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iaccarino HF et al. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature 540, 230–+, doi: 10.1038/nature20587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naslund J et al. Correlation between elevated levels of amyloid beta-peptide in the brain and cognitive decline. Jama-J Am Med Assoc 283, 1571–1577, doi:DOI 10.1001/jama.283.12.1571 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Tarasoff-Conway JM et al. Clearance systems in the brain-implications for Alzheimer disease. Nat Rev Neurol 11, 457–470, doi: 10.1038/nrneurol.2015.119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haass C & Selkoe DJ Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Bio 8, 101–112, doi: 10.1038/nrm2101 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Schindowski K, Belarbi K & Buee L Neurotrophic factors in Alzheimer’s disease: role of axonal transport. Genes Brain Behav 7 Suppl 1, 43–56, doi: 10.1111/j.1601-183X.2007.00378.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miranda M, Morici JF, Zanoni MB & Bekinschtein P Brain-Derived Neurotrophic Factor: A Key Molecule for Memory in the Healthy and the Pathological Brain. Front Cell Neurosci 13, 363, doi: 10.3389/fncel.2019.00363 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sampaio TB, Savall AS, Gutierrez MEZ & Pinton S Neurotrophic factors in Alzheimer’s and Parkinson’s diseases: implications for pathogenesis and therapy. Neural Regen Res 12, 549–557, doi: 10.4103/1673-5374.205084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagahara AH & Tuszynski MH Potential therapeutic uses of BDNF in neurological and psychiatric disorders. Nat Rev Drug Discov 10, 209–219, doi: 10.1038/nrd3366 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Chiodi I & Mondello C Telomere-independent functions of telomerase in nuclei, cytoplasm, and mitochondria. Front Oncol 2, doi:ARTN 133 10.3389/fonc.2012.00133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunes C & Rudolph KL The Role of Telomeres in Stem Cells and Cancer. Cell 152, 390–393, doi: 10.1016/j.cell.2013.01.010 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Blasco MA Telomeres and human disease: Ageing, cancer and beyond. Nat Rev Genet 6, 611–622, doi: 10.1038/nrg1656 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Wong KK et al. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature 421, 643–648, doi: 10.1038/nature01385 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Yankner BA, Lu T & Loerch P The aging brain. Annu Rev Pathol-Mech 3, 41–66, doi: 10.1146/annurev.pathmechdis.2.010506.092044 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Jaskelioff M et al. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 469, 102–106, doi: 10.1038/nature09603 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spilsbury A, Miwa S, Attems J & Saretzki G The Role of Telomerase Protein TERT in Alzheimer’s Disease and in Tau-Related Pathology In Vitro. J Neurosci 35, 1659–1674, doi: 10.1523/Jneurosci.2925-14.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eitan E et al. Excitotoxic and Radiation Stress Increase TERT Levels in the Mitochondria and Cytosol of Cerebellar Purkinje Neurons. Cerebellum 15, 509–517, doi: 10.1007/s12311-015-0720-6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang HJ et al. Ectopic expression of the catalytic subunit of telomerase protects against brain injury resulting from ischemia and NMDA-induced neurotoxicity. J Neurosci 24, 1280–1287, doi: 10.1523/JNEUROSCI.4082-03.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Brien RJ & Wong PC Amyloid precursor protein processing and Alzheimer’s disease. Annu Rev Neurosci 34, 185–204, doi: 10.1146/annurev-neuro-061010-113613 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang S et al. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet 36, 877–882, doi: 10.1038/ng1389 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Pang PT et al. Cleavage of proBDNF by tPA/plasmin is essential for long-term hippocampal plasticity. Science 306, 487–491, doi: 10.1126/science.1100135 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Oddo S et al. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: Intracellular A beta and synaptic dysfunction. Neuron 39, 409–421, doi:Doi 10.1016/S0896-6273(03)00434-3 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Oakley H et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J Neurosci 26, 10129–10140, doi: 10.1523/Jneurosci.1202-06.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dixit D, Ghildiyal R, Anto NP & Sen E Chaetocin-induced ROS-mediated apoptosis involves ATM-YAP1 axis and JNK-dependent inhibition of glucose metabolism. Cell Death Dis 5, e1212, doi: 10.1038/cddis.2014.179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chase KA et al. Examining the effects of the histone methyltransferase inhibitor BIX-01294 on histone modifications and gene expression in both a clinical population and mouse models. PLoS One 14, e0216463, doi: 10.1371/journal.pone.0216463 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J et al. Chromatin Landscape Defined by Repressive Histone Methylation during Oligodendrocyte Differentiation. J Neurosci 35, 352–365, doi: 10.1523/Jneurosci.2606-14.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madisen L et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–U311, doi: 10.1038/nn.2467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamagata Y et al. Kinase-Dead Knock-In Mouse Reveals an Essential Role of Kinase Activity of Ca2+/Calmodulin-Dependent Protein Kinase II alpha in Dendritic Spine Enlargement, Long-Term Potentiation, and Learning. J Neurosci 29, 7607–7618, doi: 10.1523/Jneurosci.0707-09.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fakhoury M Microglia and Astrocytes in Alzheimer’s Disease: Implications for Therapy. Curr Neuropharmacol 16, 508–518, doi: 10.2174/1570159X15666170720095240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lackie RE et al. The Hsp70/Hsp90 Chaperone Machinery in Neurodegenerative Diseases. Front Neurosci-Switz 11, doi:ARTN 254 10.3389/fnins.2017.00254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenzweig R, Nillegoda NB, Mayer MP & Bukau B The Hsp70 chaperone network. Nat Rev Mol Cell Biol, doi: 10.1038/s41580-019-0133-3 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Perera ON et al. Telomerase promotes formation of a telomere protective complex in cancer cells. Sci Adv 5, eaav4409, doi: 10.1126/sciadv.aav4409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong S et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716, doi: 10.1126/science.aad8373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palop JJ & Mucke L Amyloid-beta-induced neuronal dysfunction in Alzheimer’s disease: from synapses toward neural networks. Nat Neurosci 13, 812–818, doi: 10.1038/nn.2583 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Israel MA et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature 482, 216–U107, doi: 10.1038/nature10821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin WP et al. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J Biol Chem 281, 21745–21754, doi: 10.1074/jbc.M602909200 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Herskovits AZ & Guarente L SIRT1 in Neurodevelopment and Brain Senescence. Neuron 81, 471–483, doi: 10.1016/j.neuron.2014.01.028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans CG, Wisen S & Gestwicki JE Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1–42) aggregation in vitro. J Biol Chem 281, 33182–33191, doi: 10.1074/jbc.M606192200 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Vallee A & Lecarpentier Y Alzheimer Disease: Crosstalk between the Canonical Wnt/Beta-Catenin Pathway and PPARs Alpha and Gamma. Front Neurosci-Switz 10, doi:ARTN 459 10.3389/fnins.2016.00459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park JI et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 460, 66–72, doi: 10.1038/nature08137 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu H, Fu W & Mattson MP The catalytic subunit of telomerase protects neurons against amyloid beta-peptide-induced apoptosis. J Neurochem 75, 117–124, doi: 10.1046/j.1471-4159.2000.0750117.x (2000). [DOI] [PubMed] [Google Scholar]
- 43.Dulac C Brain function and chromatin plasticity. Nature 465, 728–735, doi: 10.1038/nature09231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonasio R, Tu S & Reinberg D Molecular signals of epigenetic states. Science 330, 612–616, doi: 10.1126/science.1191078 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidler C, Kovalchuk O & Kovalchuk I Epigenetic Regulation of Cellular Senescence and Aging. Front Genet 8, doi:ARTN 138 10.3389/fgene.2017.00138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snigdha S et al. H3K9me3 Inhibition Improves Memory, Promotes Spine Formation, and Increases BDNF Levels in the Aged Hippocampus. J Neurosci 36, 3611–3622, doi: 10.1523/Jneurosci.2693-15.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christopher MA et al. LSD1 protects against hippocampal and cortical neurodegeneration. Nat Commun 8, 805, doi: 10.1038/s41467-017-00922-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang ZH et al. Destabilization of beta-catenin by mutations in presenilin-1 potentiates neuronal apoptosis. Nature 395, 698–702, doi:Doi 10.1038/27208 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Kitazawa M et al. Blocking IL-1 Signaling Rescues Cognition, Attenuates Tau Pathology, and Restores Neuronal beta-Catenin Pathway Function in an Alzheimer’s Disease Model. J Immunol 187, 6539–6549, doi: 10.4049/jimmunol.1100620 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Ferrari GV & Moon RT The ups and downs of Wnt signaling in prevalent neurological disorders. Oncogene 25, 7545–7553, doi: 10.1038/sj.onc.1210064 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Kim KC et al. Overexpression of Telomerase Reverse Transcriptase Induces Autism-like Excitatory Phenotypes in Mice. Mol Neurobiol 53, 7312–7328, doi: 10.1007/s12035-015-9630-3 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Rao S, Ye N, Hu H, Shen Y & Xu Q Variants in TERT influencing telomere length are associated with paranoid schizophrenia risk. Am J Med Genet B Neuropsychiatr Genet 171B, 317–324, doi: 10.1002/ajmg.b.32403 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Lee J et al. TERT promotes cellular and organismal survival independently of telomerase activity. Oncogene 27, 3754–3760, doi: 10.1038/sj.onc.1211037 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Miwa S et al. Decreased mTOR signalling reduces mitochondrial ROS in brain via accumulation of the telomerase protein TERT within mitochondria. Aging-Us 8, 2551–+, doi: 10.18632/aging.101089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomas-Loba A et al. Telomerase Reverse Transcriptase Delays Aging in Cancer-Resistant Mice. Cell 135, 609–622, doi: 10.1016/j.cell.2008.09.034 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Eitan E et al. Novel telomerase-increasing compound in mouse brain delays the onset of amyotrophic lateral sclerosis. Embo Mol Med 4, 313–329, doi: 10.1002/emmm.201200212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu HY, Fu WM & Mattson MP The catalytic subunit of telomerase protects neurons against amyloid beta-peptide-induced apoptosis. J Neurochem 75, 117–124, doi:DOI 10.1046/j.1471-4159.2000.0750117.x (2000). [DOI] [PubMed] [Google Scholar]
- 58.Blackburn EH, Epel ES & Lin J Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198, doi: 10.1126/science.aab3389 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Chakravarti D et al. Telomere dysfunction activates YAP1 to drive tissue inflammation. Nat Commun 11, 4766, doi: 10.1038/s41467-020-18420-w (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thai TH et al. Regulation of the germinal center response by microRNA-155. Science 316, 604–608, doi: 10.1126/science.1141229 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Kaech S & Banker G Culturing hippocampal neurons. Nat Protoc 1, 2406–2415, doi: 10.1038/nprot.2006.356 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Beaudoin GM 3rd et al. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat Protoc 7, 1741–1754, doi: 10.1038/nprot.2012.099 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Raja WK et al. Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer’s Disease Phenotypes. PLoS One 11, e0161969, doi: 10.1371/journal.pone.0161969 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brennand KJ et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225, doi: 10.1038/nature09915 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu BY & Zhang SC Differentiation of spinal motor neurons from pluripotent human stem cells. Nat Protoc 4, 1295–1304, doi: 10.1038/nprot.2009.127 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brewer GJ & Torricelli JR Isolation and culture of adult neurons and neurospheres. Nat Protoc 2, 1490–1498, doi: 10.1038/nprot.2007.207 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Guo W, Patzlaff NE, Jobe EM & Zhao X Isolation of multipotent neural stem or progenitor cells from both the dentate gyrus and subventricular zone of a single adult mouse. Nat Protoc 7, 2005–2012, doi: 10.1038/nprot.2012.123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, doi: 10.1093/bioinformatics/btu638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mootha VK et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273, doi: 10.1038/ng1180 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Terranova C et al. An Integrated Platform for Genome-wide Mapping of Chromatin States Using High-throughput ChIP-sequencing in Tumor Tissues. J Vis Exp, doi: 10.3791/56972 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25, doi: 10.1186/gb-2009-10-3-r25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079, doi: 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137, doi: 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong FL et al. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150, 481–494, doi: 10.1016/j.cell.2012.07.012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weinrich SL et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet 17, 498–502, doi: 10.1038/ng1297-498 (1997). [DOI] [PubMed] [Google Scholar]
- 78.Shim HS, Wei M, Brandhorst S & Longo VD Starvation Promotes REV1 SUMOylation and p53-Dependent Sensitization of Melanoma and Breast Cancer Cells. Cancer Res 75, 1056–1067, doi: 10.1158/0008-5472.Can-14-2249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Herbert BS, Hochreiter AE, Wright WE & Shay JW Nonradioactive detection of telomerase activity using the telomeric repeat amplification protocol. Nat Protoc 1, 1583–1590, doi: 10.1038/nprot.2006.239 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Castellano JM et al. Human umbilical cord plasma proteins revitalize hippocampal function in aged mice. Nature 544, 488–492, doi: 10.1038/nature22067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patil SS, Sunyer B, Hoger H & Lubec G Evaluation of spatial memory of C57BL/6J and CD1 mice in the Barnes maze, the Multiple T-maze and in the Morris water maze. Behav Brain Res 198, 58–68, doi: 10.1016/j.bbr.2008.10.029 (2009). [DOI] [PubMed] [Google Scholar]
- 82.Sahin E et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470, 359–365, doi: 10.1038/nature09787 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iaccarino HF et al. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature 540, 230–235, doi: 10.1038/nature20587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data have been deposited in Gene Expression Omnibus (GEO) under accession numbers GSE163523, GSE163524 and GSE163525, and the ChIP-seq data in Sequence Read Archive (SRA) under accession numbers PRJNA633993 and PRJNA633994. The mass spectrometry data have been deposited in MassIVE under accession numbers MSV000088190. All other data are available from the corresponding author upon reasonable request.