Abstract
One of the best examples of sexual dimorphism is the development and function of the gonads, ovaries and testes, which produce sex-specific gametes, oocytes, and spermatids, respectively. The development of these specialized germ cells requires sex-matched somatic support cells. The sexual identity of somatic gonadal cells is specified during development and must be actively maintained during adulthood. We previously showed that the transcription factor Chinmo is required to ensure the male sexual identity of somatic support cells in the Drosophila melanogaster testis. Loss of chinmo from male somatic gonadal cells results in feminization: they transform from squamous to epithelial-like cells that resemble somatic cells in the female gonad but fail to properly ensheath the male germline, causing infertility. To identify potential target genes of Chinmo, we purified somatic cells deficient for chinmo from the adult Drosophila testis and performed next-generation sequencing to compare their transcriptome to that of control somatic cells. Bioinformatics revealed 304 and 1549 differentially upregulated and downregulated genes, respectively, upon loss of chinmo in early somatic cells. Using a combination of methods, we validated several differentially expressed genes. These data sets will be useful resources to the community.
Keywords: testis, ovary, Chinmo, sexual identity, sex transformation
Introduction
In all sexually reproducing species, distinct transcriptional programs instruct the development of male and female individuals. Resulting sex differences manifest in the anatomy, physiology, and behavior of adult populations. Importantly, inappropriate sexual specification underpins numerous developmental defects and pathologies across many species, including human disorders of sexual development (reviewed in Bashamboo and McElreavey 2015). Therefore, dissecting the underlying mechanisms for sex determination and maintenance are of critical importance.
The first mechanism of sex determination was characterized in the fruit fly Drosophila melanogaster and controls the expression of Doublesex (Dsx), a key effector of sexual differentiation. Dsx is the founding member of the doublesex/Mab-3-related transcription factor family and regulates many known sex differences in Drosophila morphology and behavior (reviewed in Camara et al. 2008). Importantly, Dsx plays a well-documented role in establishing sex-specific somatic gonads (DeFalco et al. 2003; Le Bras and Van Doren 2006; Camara et al. 2019). In somatic cells that require knowledge of their sex, an alternative splicing cascade yields female-specific expression of the RNA-binding proteins Sex-lethal (Sxl) and Transformer (Tra; Boggs et al. 1987; Inoue et al. 1990; Bell et al. 1991). In female somatic cells, Tra binds to dsx pre-mRNA to yield the female isoform dsxF, while male somatic cells lack Sxl and Tra and produce the default male dsxM mRNA (Inoue et al. 1992; Lynch and Maniatis 1996). The resulting sex-specific DsxM and DsxF proteins control transcriptional programs that instruct male and female sexual differentiation, respectively.
Sexual dimorphism is perhaps most apparent in the development of reproductive organs—ovaries and testes—which must produce sex-specific gametes to ensure continuation of the species. In many sexually reproducing species, including Drosophila melanogaster, the testis and ovary harbor germ cells that give rise to sperm and oocytes, respectively. These gametes depend on signals from somatic support cells housed in the same tissue throughout their development (Leatherman and Dinardo 2008, 2010; Waterbury et al. 2000; Wawersik et al. 2005).
Anchored to the apex of the testis, a tight cluster of postmitotic cells termed the hub contributes to the male stem cell niche, producing secreted factors that support both germline stem cells (GSCs) and somatic cyst stem cells (CySCs; Figure 1A, reviewed in Greenspan et al. 2015). GSCs undergo oriented mitotic divisions to produce daughter cells with asymmetric cell fates: the cell adjacent to the hub self-renews and remains a GSC, while the other daughter cell, now a gonialblast (Gb), is physically displaced from the stem cell niche and begins to differentiate (Hardy et al. 1979; Yamashita et al. 2003). Each Gb undergoes four rounds of mitosis with incomplete cytokinesis to produce 2-, 4-, 8-, and 16-cell spermatogonial cysts. Each 16-cell cyst undergoes meiosis to generate mature spermatids (Fuller 1998). GSCs are supported by CySCs, which act as an extended niche in concert with hub cells (Leatherman and Dinardo 2008, 2010). Each Gb is ensheathed by two differentiating cyst cells, which both exit the cell cycle and continue growing in size to support germ cells throughout spermatogenesis (Lenhart and DiNardo 2015; Shields et al. 2014).
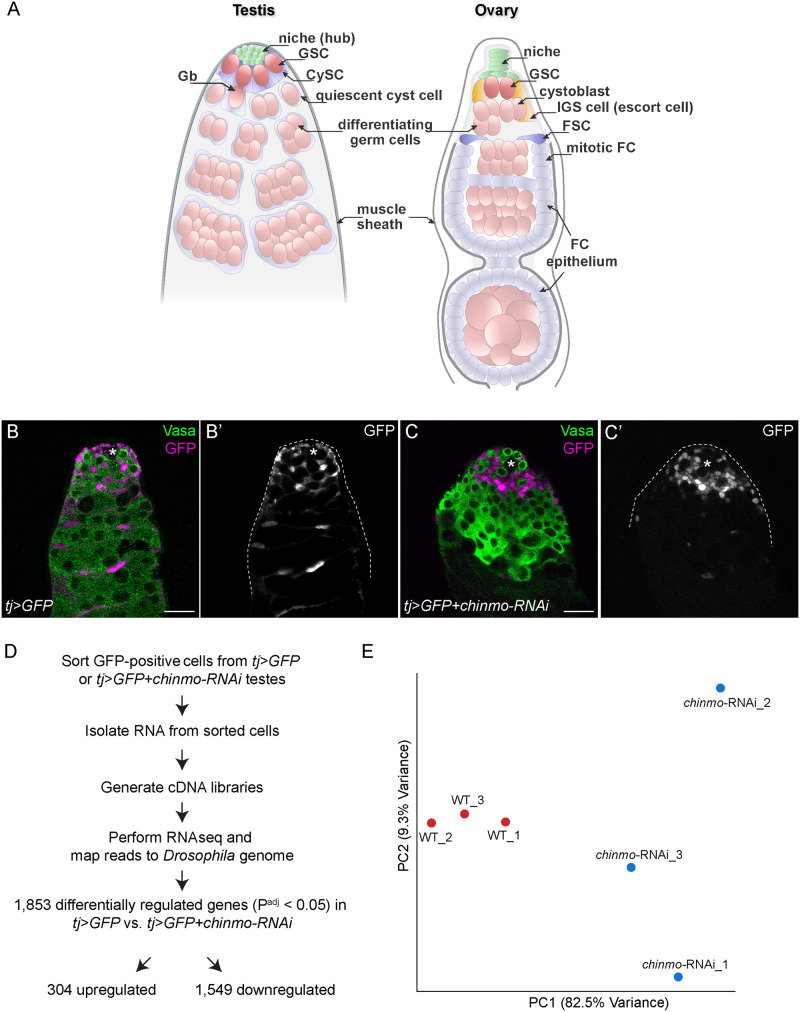
Figure 1.
Transcriptomic profiling of chinmo-deficient CySCs. (A) Diagram of the adult Drosophila testis (left) and ovary (right). (Left) The testis niche is composed of quiescent somatic cells termed “hub” cells (green). The niche supports ∼8–12 GSCs (dark pink) and ∼13 somatic CySCs (dark blue). The GSC divides to produce a Gb (light pink) that differentiates. The Gb is ensheathed by two quiescent cyst cells (light blue), daughter cells of the CySCs. These same two cyst cells continue to ensheath the Gb as it divides four more times with incomplete cytokinesis, giving rise to spermatogonia (light pink and as “differentiating germ cells”) that enter meiosis. (Right) The ovarian niche (green) supports approximately two to three GSCs (dark pink). The GSC divides to produce a cystoblast that begins differentiation. IGS cells (light orange, also called escort cells) wrap around the cystoblast as it differentiates. Approximately one to four FSCs (dark blue) reside in the middle of the germarium and produce mitotic follicle cells (FCs) which function as epithelium that envelops the 16-cell germ cyst. (B, C) Representative confocal image of a control tj>GFP testis (B) and a tj>GFP+chinmo-RNAi testis (C) at 2 days of adulthood. In the control testis, GFP (magenta) was expressed in the nuclei of tj-expressing somatic support cells (i.e., CySCs and cyst cells) that ensheathed early germ cells (B). In the tj>GFP +chinmo-RNAi testis, GFP (magenta) was still expressed in the nuclei of tj-expressing somatic support cells (C). However, these chinmo-deficient somatic support cells were in close contact with each other and no longer properly enveloping germ cells, demonstrating that they were becoming sex-transformed. Vasa (green) marks germ cells. An asterisk marks the niche. Scale bar = 20 μm (D) Work-flow of the RNA-seq. Briefly, we purified GFP-positive cells from tj>GFP or tj>GFP+chinmo-RNAi adult testes at 2–3 days after eclosion. We isolated total RNA from these cells and generated cDNA libraries, which were used for the RNA-seq. The reads were mapped to the Drosophila genome (dm6). 1853 genes were differentially expressed (i.e., FC ≥ 2 and Padj ≤ 0.05) in tj>GFP+chinmo-RNAi cells compared with tj>GFP cells, with 304 upregulated and 1549 downregulated. (E) Principal component analysis of tj>GFP (red circles labeled “WT”) and tj>GFP+chinmo-RNAi (blue circles labeled “chinmo-RNAi”). The WT samples clustered together and were distinct from the chinmo-RNAi samples. Genotypes (B) w/Y; tj-Gal4, UAS-GFPnls/UAS-GFPnls; +/+, (C) w/Y; tj-Gal4, UAS-GFPnls/+; UAS-chinmo-RNAi/UAS-Dcr-2, (E) w/Y; tj-Gal4, UAS-GFPnls/UAS-GFPnls; +/+ (labeled “WT”) and w/Y; tj-Gal4, UAS-GFPnls/+; UAS-chinmo-RNAi/UAS-Dcr-2 (labeled “chinmo-RNAi”)
Multiple somatic cell types contribute to female germline development in the ovary (Figure 1A, reviewed in (Drummond-Barbosa 2019; Rust and Nystul 2020)). In each ovary, 16–20 ovarioles each comprise a chain of developing egg chambers: mature oocytes reside at the base of the ovary, whereas the apical end of each ovariole houses two to three female GSCs. The most apical chamber in the ovariole, the germarium, contains a stack of six to seven niche cells. GSCs divide asymmetrically to produce a cystoblast that, aided by somatic inner germarial sheath (IGS) cells, undergoes four rounds of mitotic division similar to male GSC differentiation. The 16-cell germ cyst is enclosed in an egg chamber that goes through 14 stages of differentiation. Developing egg chambers are surrounded by a follicle epithelium that assembles around the germline in the germarium. These somatic follicle cells are descendants of approximately one to four follicle stem cells (FSCs) that reside at the 2a/2b boundary in the germarium (Fadiga and Nystul 2019). FSCs may be functionally analogous to CySCs as both require similar proliferative pathways and exhibit similar competitive behaviors (Amoyel et al. 2013, 2014; Cook et al. 2017; Issigonis et al. 2009; Leatherman and Dinardo 2008; Michel et al. 2012; Vied et al. 2012).
Although germ cells autonomously specify their sex identity, they also rely on cues from sex-specific somatic cell types. In the early developing testis, the male somatic gonad produces the IL-6-like ligand Unpaired to activate the sole Drosophila STAT family protein Stat92E in male germ cells (Sheng et al. 2009; Wawersik et al. 2005). This male-specific Stat92E activation is required for male germ cell development. In contrast, female germ cells lack Stat92E activity. Consistent with this, several lines of research have demonstrated that in order for gametogenesis to proceed successfully, the sex identity of somatic support cells in the gonad must match that of the neighboring germ cells (Grmai et al. 2018; Ma et al. 2014; Sheng et al. 2009; Waterbury et al. 2000; Wawersik et al. 2005).
We initially characterized a target of Stat92E, the Broad complex, Tramtrack, and Bric-à-brac (BTB)/Zinc finger (ZF)-containing transcription factor Chronologically inappropriate morphogenesis (Chinmo), for its role in adult CySC maintenance. CySC clones lacking chinmo are rapidly lost from the testis niche, indicating that Chinmo is required autonomously for CySC niche residency (Flaherty et al. 2010). Interestingly, Chinmo is also required to preserve the male sexual identity of adult CySCs. Although Chinmo is expressed in all cell types of the testis stem cell niche, including CySCs, it is absent from somatic cells in the ovary (Grmai et al. 2018; Ma et al. 2014). Loss of chinmo in adult CySCs, either by endogenous mutation or tissue-specific depletion, causes CySCs to transform into epithelial-like cells (Grmai et al. 2018; Ma et al. 2014). These sex-transformed somatic cells exhibit gene expression and morphology reminiscent of follicle cells in the adult ovary. The resulting germline-soma mismatch in sex identity upon somatic chinmo depletion causes male sterility (Grmai et al. 2018).
Despite the importance of somatic Chinmo in preserving the male germline, to date no direct Chinmo targets have been reported. Identifying factors that are regulated by Chinmo in adult CySCs will reveal important insights about Chinmo as a regulator of male stem cell identity. In pursuit of such factors, we performed transcriptomic profiling and bioinformatic analyses of purified control and chinmo-depleted CySCs. Additionally, we used a positional weight matrix (PWM) for DNA bound by the Chinmo zinc finger domains to survey the Drosophila genome for loci containing these sites (Enuameh et al. 2013). By comparing the list of genes with putative Chinmo binding sites to our RNA-seq, we narrowed down our list of differentially expressed genes to those that may be direct Chinmo targets. Some of these genes, such as β heavy spectrin (βH-spectrin; Flybase: karst (kst)), polychaetoid (pyd), and scribble (scrib), function in epithelial polarity and structure. We validated these targets as being enriched both in wild-type (WT) ovarian follicle cells and in sex-transformed testicular somatic cells. These data suggest that in WT testicular somatic cells, Chinmo might repress the expression of factors that promote epithelial morphogenesis.
Materials and methods
Fly stocks
We used traffic jam (tj)-Gal4 (Kyoto Stock Center-104055); UAS-GFPnls (Bloomington Drosophila Stock Center (BDSC-4775); UAS-chinmoHMS00036-RNA nterference (RNAi) (BDSC-33638); UAS-Dcr-2 (BDSC-24651); mirr-lacZcre2 (BDSC-10880); chinmoST (Ma et al. 2014); and the protein trap scrib-GFP (scribCA07683) (Buszczak et al. 2007; a gift from Ronald Davis, Scripps Research Florida, FL, USA). We maintained crosses at 25°C in a 12-h light/dark incubator.
Flies were reared on food made with these ingredients: 1800 mL Molasses (LabScientific, Catalog no. FLY-8008-16), 266 g Agar (Mooragar, Catalog no. 41004), 1800 g Cornmeal (LabScientific, Catalog no. FLY-8010-20), 744 g Yeast (LabScientific, Catalog no. FLY-8040-20F), 47 L Water, 56 g Tegosept (Sigma no. H3647-1KG), 560 mL Reagent Alcohol (Fisher no. A962P4), and 190 mL Propionic Acid (Fisher no. A258500).
Testis dissociation, flow cytometry, and RNA isolation for RNA-seq
We dissected testes from adult Drosophila tj>GFP (genotype: w/Y; tj-Gal4, UAS-GFPnls/UAS-GFPnls; +/+) or tj>GFP+chinmo-RNAi (genotype: w/Y; tj-Gal4, UAS-GFPnls/+; UAS-chinmo-RNAi/UAS-Dcr-2) males at 2 or 3 days post-eclosion. The testis dissociation protocol was generously provided by Margaret Fuller (Stanford University, Palo Alto, CA, USA) and modified slightly for this experiment. Testes were dissected into Schneider’s medium and transferred to prepared tubes with 500 µL of Schneider’s medium with L-glutamine. Once testes sunk to the bottom of the tubes, the media was carefully aspirated using a Pasteur pipet. To each tube was added 150 µL 0.25% Trypsin + EDTA; 150 µL collagenase (starting concentration 1 mg/mL) + 1% BSA. Samples were vortexed vigorously for 15 min at room temperature; after confirming that testes were no longer intact, the enzymatic reactions were terminated by adding 1 mL Schneider’s medium + 12.5% Fetal Bovine Serum (FBS), 0.1 mg/mL gentamicin, 2 mM EDTA. Cell suspensions were filtered through a 100 µm Falcon filter into an Eppendorf tube and centrifuged at 5000 rpm for 10 min at 4°C. Supernatants were removed carefully and cell pellets were resuspended in Schneider’s/FBS/gentamicin/EDTA solution. If necessary, samples of the same genotype were pooled at this point. GFP-expressing somatic cells were purified from the resulting filtrate by FACS using a Sony SY3200 highly automated parallel sorting cell sorter into 750 µL TRIzol LS (Thermo Fisher). RNA was isolated and precipitated according to manufacturer’s instructions. RNA extracts were cleaned using RNeasy Mini Kit (QIAGEN) according to manufacturer’s instructions. RNA quality and quantity were determined using the Agilent 1200 Bioanalyzer.
RNA-sequencing and analysis
The RNA-sequencing was performed by the Genome Technology Center at the NYU Langone Medical Center. One nanogram of total RNA was used for library prep, and cDNA was amplified by using Nugen Ovation RNA-Seq System V2 kit (Part No. 7102-32), 100 ng of Covaris-fragmented cDNA were used as input to prepare the libraries, using the Ovation Ultralow Library system (Nugen, Part 0330-32), and amplified by 10 cycles of PCR. The samples were mixed into two pools and run in two 50-nucleotide paired-end read rapid run flow cell lanes on the Illumina HiSeq 2500 sequencer.
Bioinformatics
Bioinformatic analysis was performed by the Applied Bioinformatics Laboratories at NYU Langone Medical Center. Sequencing results were demultiplexed and converted to FASTQ format using Illumina Bcl2FastQ software. Reads were aligned to the dm6 release of the Drosophila melanogaster genome using the splice-aware STAR aligner. PCR duplicates were removed using the Picard toolkit (http://broadinstitute.github.io/picard). The HTSeq package (Love et al. 2014) was utilized to generate counts for each gene based on how many aligned reads overlap its exons. These counts were then used to test for differential expression using negative binomial generalized linear models implemented by the DESeq2 R package (Anders et al. 2015). The adjusted P-value (Padj) was generated by using the False Discovery Rate with the Benjamini–Hochberg method. A scatter (Volcano) plot was generated using Stata 15.1 (StataCorp LLC, College Station, TX, USA). For Supplementary Tables S1 and S2, we chose a cutoff of fold change (FC) ≤ −2.0 or ≥ 2.0 and Padj ≤ 0.05.
Genome-wide analysis of Chinmo binding sites
We obtained a PWM for Chinmo from Fly Factor Survey (https://mccb.umassmed.edu/ffs/; Enuameh et al. 2013), the consensus being (G/A)ATGCAC(T/C)(T/N)NN (Supplementary Tables S3 and S4, READ ME Tab). We used a web-based program PWMScan (https://ccg.epfl.ch//pwmtools/pwmscan.php; Ambrosini et al. 2018) to search the Drosophila genome for Chinmo binding sites that matched that PWM with a stringent P value less than 1 × 10−5 (recommended by developers of the PWMScan website; Ambrosini et al. 2018). We then compared the list of locations of Chinmo binding sites with the list of genes and their locations (https://genome.ucsc.edu/cgi-bin/hgTables?command=start). We report in Supplementary Tables S3 and S4 differentially regulated genes in chinmo-deficient CySCs with at least one Chinmo binding site in noncoding regions, defined as 1500 bps upstream of the transcription start site, introns, and 1500 bps downstream of the termination sequence.
Antibody staining
Immunofluorescence was performed as described in Flaherty et al. (2010). The following primary antibodies were used: goat anti-Vasa (1:50, dC-13, Santa Cruz), rabbit anti-Vasa (1:3000; gift of Ruth Lehmann, Whitehead Institute, MA, USA), guinea pig anti-Tj (1:5000; gift of Dorothea Godt, University of Toronto, ON, Canada), mouse anti-Fasciclin (Fas) 3 (1:50; Developmental Studies Hybridoma Bank (DSHB), rat anti-DE-cad (1:25, DSHB), mouse anti-Pyd (clone Pyd2) (1:10) (DSHB); rabbit anti-βH-spectrin (1:250) (gift of Claire Thomas, Penn State University, PA, USA); rabbit anti-GFP (1:250) (Invitrogen), mouse anti-β-gal (1:50) (DSHB). We used fluorescent secondary antibodies at 1:250 (Jackson Laboratories). The samples were then mounted in Vectashield. We collected fluorescent images at 63× magnification using a Zeiss LSM 700 confocal microscope.
Results
Next-generation sequencing of FACS-purified, chinmo-depleted somatic cells
We isolated viable control tj>GFP cells (Figure 1B) or viable chinmo-depleted somatic cells (tj>chinmo-RNAi; Figure 1C) by flow cytometry based on their lack of propidium iodide uptake and their expression of GFP in the tj domain (i.e., CySCs and early cyst cells; Fairchild et al. 2016). We chose to isolate these cells at 2–3 days of adulthood because this is the time point when physical changes in the testis stem cell niche are observed (Figure 1C; Ma et al. 2014). The isolated RNA was processed for expression profiling, and the sequencing was performed using Illumina HiSeq2500 Paired-End 50 Cycle Flow Cell (Figure 1D). Principal Component Analysis revealed that control (labeled “WT”) and tj>chinmo-RNAi samples were distinct clusters (Figure 1E). The variability between the chinmo-RNAi samples is likely a result of the early time point we chose to maximize the chance of detecting transcriptional changes at the initiation of male-to-female sex transformation.
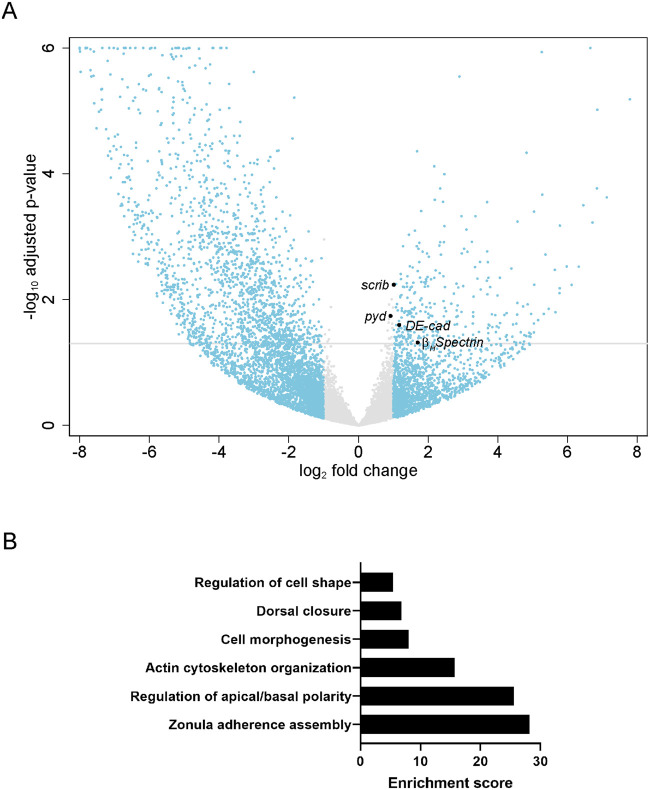
For the bioinformatics analyses, we chose an arbitrary cutoff of Padj ≤ 0.05 and FC ≥ 2 and found 304 upregulated genes and 1549 downregulated genes (Supplementary Tables S1, S2, and Figure 2A). We analyzed the differentially regulated genes in tj>chinmo-RNAi somatic cells for presence of a Chinmo binding site in noncoding regions. We found that 99 significantly upregulated genes (Padj ≤ 0.05) had at least 1 Chinmo binding site, including βH-spectrin (kst) with 3 sites, pyd and scrib, with 2 sites each (Supplementary Table S3). This analysis did not identify any Chinmo binding sites in DE-cadherin (DE-cad) (Flybase: shotgun). Of the differentially downregulated genes, 208 had at least one Chinmo binding site (Supplementary Table S4).
Figure 2.
Genotype ontology and gene expression in chinmo-depleted somatic cells. (A) Scatter (Volcano) plot for genes in chinmo-depleted somatic cells (tj>GFP+chinmo-RNAi) compared with controls (tj>GFP). The x-axis is the log2 of the fold change and the y-axis is the negative log10 of the adjusted P-value (Padj). Gray line indicates Padj = 0.05. Gray circles indicate genes with log2 (FC) between −1 and 1 (corresponding to FC between 0.5 and 2). Blue circles indicate genes with log2 (FC) greater than 1 (corresponding to FC >2). The larger black circles indicate the differentially upregulated genes βH-spectrin, pyd, scrib, and DE-cad. (B) Gene Ontology. DAVID Functional Annotation Clustering analysis depicting enriched biological processes for the 100 upregulated genes with putative Chinmo binding sites. The enrichment score plotted on the y-axis is based on the Fisher’s exact P-values of each gene in the group. The higher the score, the more enriched is the group. Genotype: (A) w/Y; tj-Gal4, UAS-GFPnls/+; UAS-chinmo-RNAi/UAS-Dcr-2.
To identify common biological functions among differentially expressed genes, we analyzed the 99 upregulated annotated genes with putative Chinmo binding sites using DAVID (Database for Annotation, Visualization and Integrated Discovery; Huang da et al. 2009a, b). Consistent with the epithelial morphogenesis triggered upon chinmo loss, we found that most significantly enriched gene ontology terms were related to actin organization, apical-basal polarity, cytoskeleton and cell junction (Figure 2B). We then selected four genes for further validation based on availability of resources, including antibodies, enhancer- or protein-trap lines, and known roles in the ovarian follicular epithelium: βH-spectrin, DE-cad, pyd, and scrib (Figure 2A). The expression patterns of these four factors were assessed in WT ovaries, WT testes, and in testes in which chinmo was somatically depleted, either by use of the chinmoST loss-of-function allele or by expression of UAS-chinmo-RNAi using tj-Gal4.
chinmo-depleted somatic cells upregulate βH-spectrin
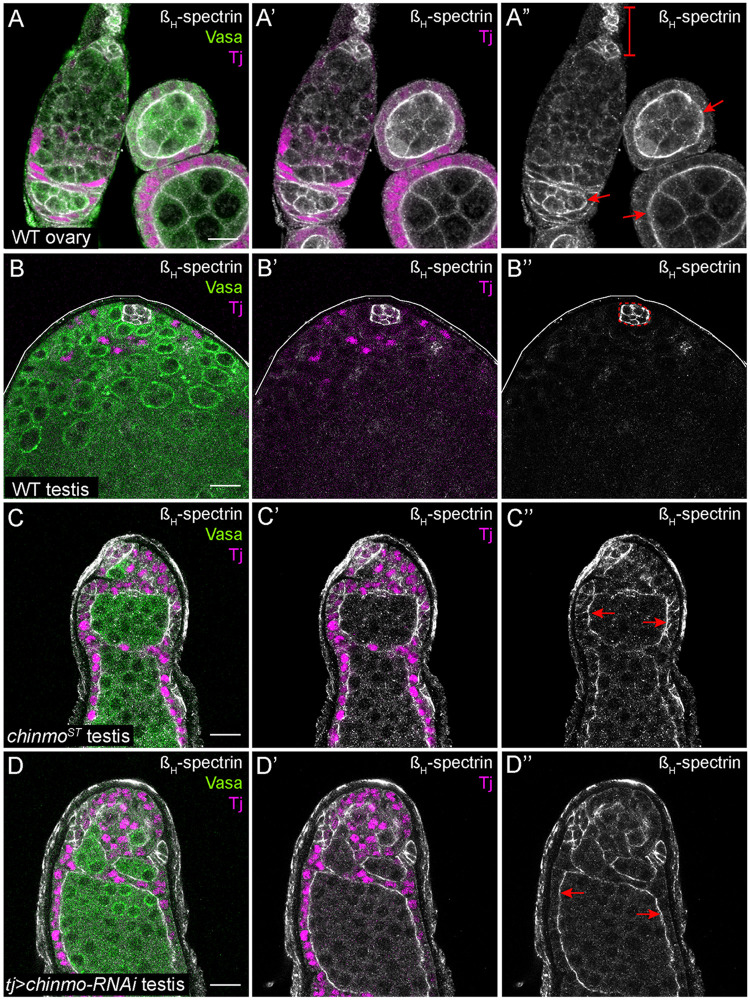
Spectrins serve structural roles in cells as they line the intracellular side of the plasma membrane (Liem 2016). Spectrin dimers are composed of α and β subunits that interact in an anti-parallel manner. Two αβ dimers then associate to form a tetramer (Byers et al. 1987; Dubreuil et al. 1996). In Drosophila, there is a second type of spectrin dimer composed of the same α subunit and a higher molecular weight protein called βH spectrin (Dubreuil et al. 1990). In Drosophila embryos and adult follicle cells, the spectrin cytoskeleton is polarized: αβH-spectrin dimers localize to the apical domain while αβ-spectrin dimers are found at the basolateral margin (Lee et al. 1997; Thomas and Williams 1999). Consistent with prior work, an xy-section of the adult germarium showed that βH-spectrin is expressed in niche cells (Figure 3A”, bracket; Zarnescu and Thomas 1999). Furthermore, βH-spectrin is enriched at the apical side of follicle cells in the germarium and in later egg chambers (Figure 3A”, arrows). In a WT testis, βH-spectrin was expressed exclusively in niche cells (outlined in Figure 3B”). In contrast, βH-spectrin was ectopically expressed in all somatic cells in a chinmoST mutant testis and in testes somatically depleted for chinmo, and this was particularly apparent in the apical domain of feminized, follicle-like cells that line the periphery of the testis adjacent to the muscle sheath (Figure 3, C” and D”, arrows).
Figure 3.
chinmo-deficient somatic cells upregulate βH-Spectrin. (A–A”) xy-section of a WT ovary shows that βH-spectrin (white) is present in somatic cells. Bracket in A” indicates the niche cells. Arrows in A” indicate apical enrichment of βH-spectrin in follicle cells. (B–B”) In a WT testis, βH-spectrin (white) is expressed strongly in niche cells (outlined by a dashed line in B”) and at very low levels in other somatic cells. (C–D”) In a chinmoST mutant testis (C–C”) or a testis somatically depleted for chinmo (D–D”), βH-spectrin (white) is ectopically expressed in feminized somatic cells. Arrows in C” and D” indicate apical enrichment of βH-spectrin in chinmo-depleted somatic cells. In (A–D), Vasa (green) marks the germline and Tj (magenta) marks cyst cells. Scale bar = 10 μm. Time point in (A–D) is 7–8 days post-eclosion. Genotypes: (A) +/+; +/+; +/+ (OregonR), (B) +/Y; +/+; +/+ (OregonR), (C) w/Y; chinmoST/chinmoST; +/+, and (D) w/Y; tj-Gal4/+; UAS-chinmo-RNAi/UAS-Dcr-2 (labeled “tj>chinmo-RNAi”)
chinmo-depleted somatic cells upregulate DE-cad
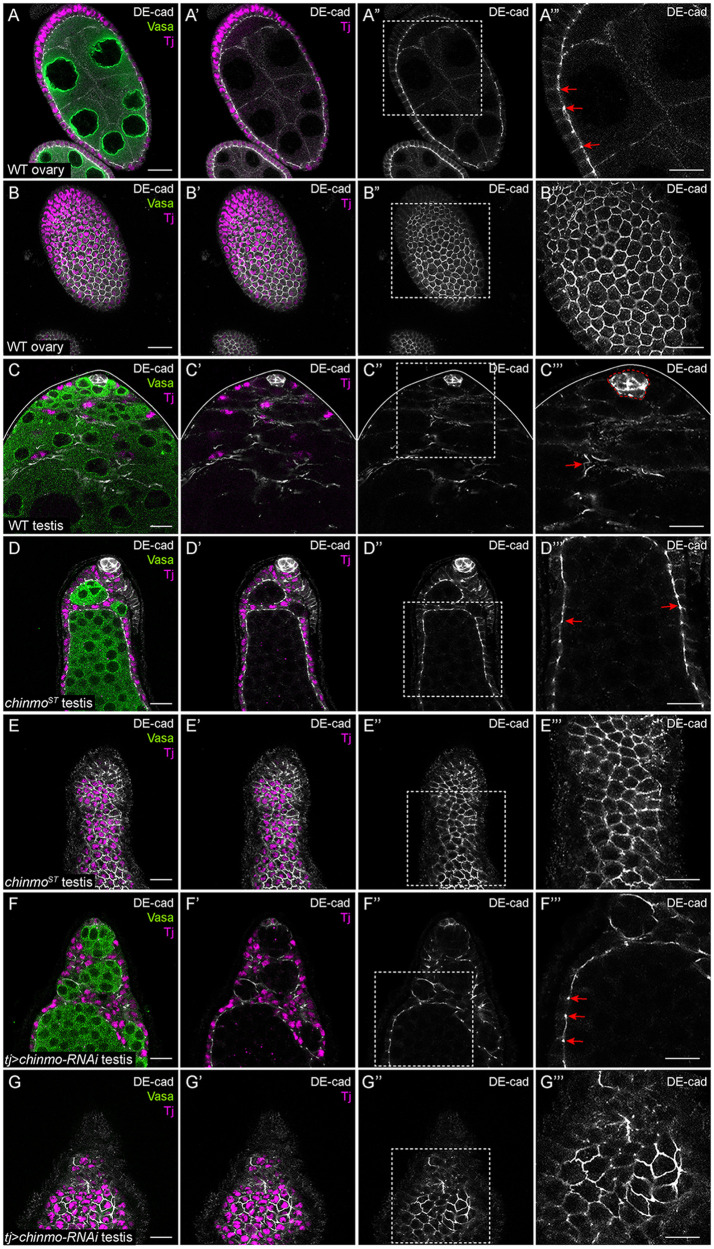
A homolog of classical vertebrate cadherins, Drosophila DE-cad mediates cell-cell adhesion through homophilic interactions (Tepass et al. 1996; Hill et al. 2001). DE-cad is a main component of adherens junctions (AJs), which maintain epithelial integrity (Harris and Tepass 2010). The DE-cad intracellular domain associates with cytoskeletal proteins like Armadillo/β-catenin, which in turn anchors actin filaments to the plasma membrane (Bulgakova et al. 2012). Consistent with prior work, xy-sections of a WT ovary showed that DE-cad was expressed in AJs in the somatic follicle cells (Figure 4, A–B’”, arrows; Franz and Riechmann 2010). In a WT testis, DE-cad was expressed strongly in somatic niche cells (outlined in Figure 4C”’) and at lower levels in somatic support cells (Figure 4C’”, arrow). DE-cad levels strongly increased in feminized, follicle-like somatic cells in chinmoST testes or in chinmo-depleted somatic cells (Figure 4, D–G’”, arrows). Middle xy-sections of feminized testes showed enrichment of DE-cad in AJs (Figure 4, D’” and 4F’”, arrows).
Figure 4.
chinmo-deficient somatic cells upregulate DE-cad. (A–A’”) Middle xy-section of a WT ovary shows that DE-cad (white) is expressed at AJs (arrows in A’”) in somatic follicle cells. (B–B’”) An apical xy-section of a WT ovary shows that DE-cad (white) is apically enriched in follicle cells. (C–C”’) In a WT testis, DE-cad (white) is expressed strongly in the niche (outlined by a dashed line in C’”) and at a modest level in the somatic cells (arrow, C’”). (D–D”’) Middle xy-section of a chinmoST mutant testis shows that DE-cad (white) is ectopically expressed at AJs (arrows, D’”) in feminized somatic cyst cells. (E–E’”) An apical xy-section of a chinmoST mutant testis reveals apical enrichment of DE-cad (white) in the feminized somatic cells. (F–F’”) Middle xy-section of a tj>chinmo-RNAi testis shows that DE-cad (white) is ectopically expressed at AJs (arrows, F’”) in feminized somatic cyst cells. (G–G’”) An apical xy-section of a tj>chinmo-RNAi testis shows apical enrichment of DE-cad (white) in the feminized follicle-like cells. In (A–G), Vasa (green) marks the germline and Tj (magenta) marks cyst cells. Scale bar = 10 μm. (A’”–G”’) is a magnification of the boxed regions in (A”–G”), respectively. Time point in (A–G) is 7–8 days post-eclosion. Genotypes: (A, B) +/+; +/+; +/+ (OregonR), (C) +/Y; +/+; +/+ (OregonR), (D, E) w/Y; chinmoST/chinmoST; +/+ (F, G) w/Y; tj-Gal4/+; UAS-chinmo-RNAi/UAS-Dcr-2 (labeled “tj>chinmo-RNAi”).
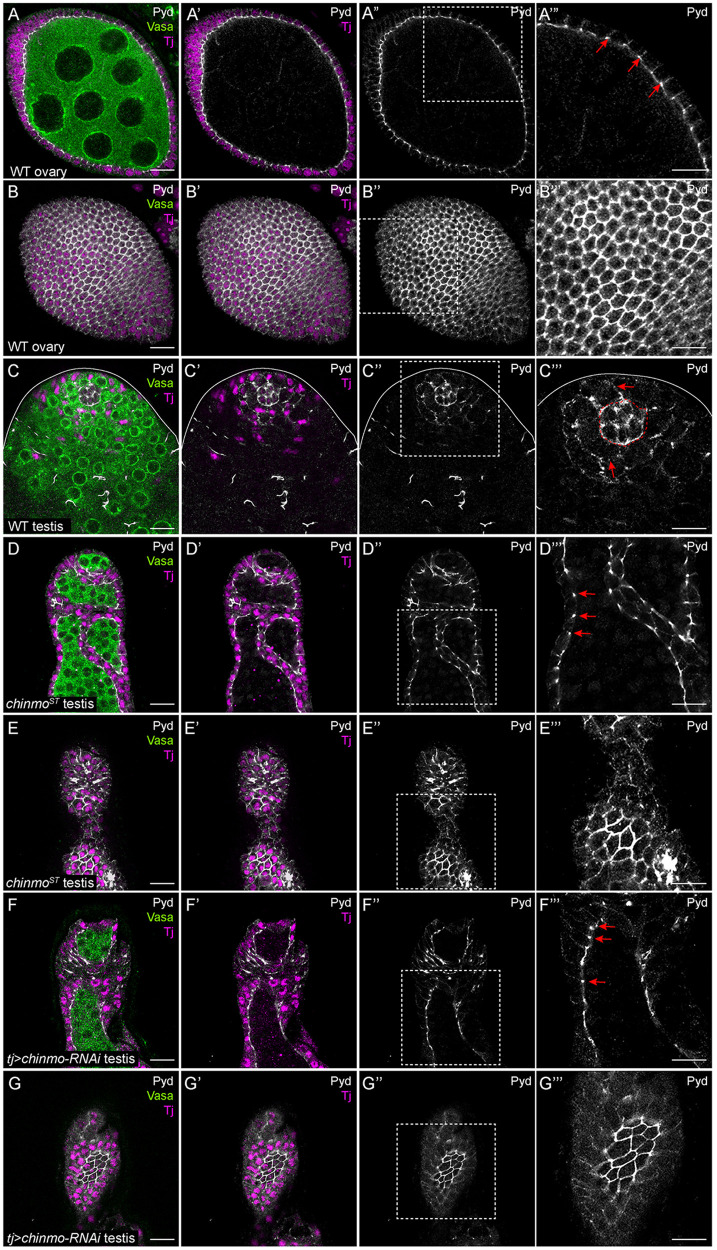
chinmo-depleted somatic cells upregulate Pyd
Pyd is the single zonula occludens (ZO) protein in Drosophila and the homolog of ZO-1 (Takahisa et al. 1996; Wei and Ellis 2001). A member of the membrane-associated guanylate kinase family of proteins, ZO-1 is a primary component of tight junctions, which maintain barriers between epithelial cells. (Takahisa et al. 1996; Nelson 2008; Fanning and Anderson 2009). In invertebrates, the barrier-forming junction between epithelial cells is called the septate junction, and it is localized more basally than AJs (Furuse and Tsukita 2006). Surprisingly, in several epithelia in Drosophila, Pyd localizes apically at AJs with DE-cad and not to the septate junction (Wei and Ellis 2001; Jung et al. 2006; Seppa et al. 2008; Choi et al. 2011). Consistent with prior work, a middle and apical xy-section of a WT ovary showed that Pyd was expressed exclusively in somatic follicle cells (Figure 5, A–B’”, arrows; Djiane et al. 2011). In a WT testis, Pyd was expressed strongly in niche cells (outlined in Figure 5C’”) and at lower levels in CySCs (Figure 5C’”, arrows). In testes somatically deficient in chinmo, Pyd expression increased substantially in feminized, follicle-like cells and localized to AJs (Figure 5, D–G’”, arrows).
Figure 5.
chinmo-deficient somatic cells upregulate Pyd. (A–A’”) Middle xy-section of a WT ovary shows that Pyd (white) is expressed strongly in somatic follicle cells. Arrows (A’”) indicate Pyd expression in AJs. (B–B’”) An apical xy-section of a WT ovary shows the apical enrichment of Pyd (white) in the follicle cells. (C–C’”) In a WT testis, Pyd (white) is expressed at moderate levels in the niche (outlined by a dashed line in C’”) and at low levels in other somatic cells (C”’, arrows). (D–D’”) Middle xy-section of a chinmoST mutant testis shows that Pyd (white) is ectopically expressed in AJs (arrows in D’”) in feminized somatic cyst cells. (E–E’”) An apical xy-section of a chinmoST mutant testis shows apical enrichment in the feminized cells. (F–F’”) Middle xy-section of a tj>chinmo-RNAi testis shows that Pyd (white) is ectopically expressed in AJs (arrows in F’”) in feminized somatic cyst cells. (G–G’”) An apical xy-section of a tj>chinmo-RNAi testis shows apical enrichment of Pyd (white) in the feminized follicle-like cells. In (A–G), Vasa (green) marks the germline and Tj (magenta) marks cyst cells. Scale bar = 10 μm. (A’”–G”’) is a magnification of the boxed regions in (A”–G”), respectively. Time point in (A–G) is 7–8 days post-eclosion. Genotypes: (A, B) +/+; +/+; +/+ (OregonR), (C) +/Y; +/+; +/+ (OregonR), (D, E) w/Y; chinmoST/chinmoST; +/+, (F, G) w/Y; tj-Gal4/+; UAS-chinmo-RNAi/UAS-Dcr-2 (labeled “tj>chinmo-RNAi”).
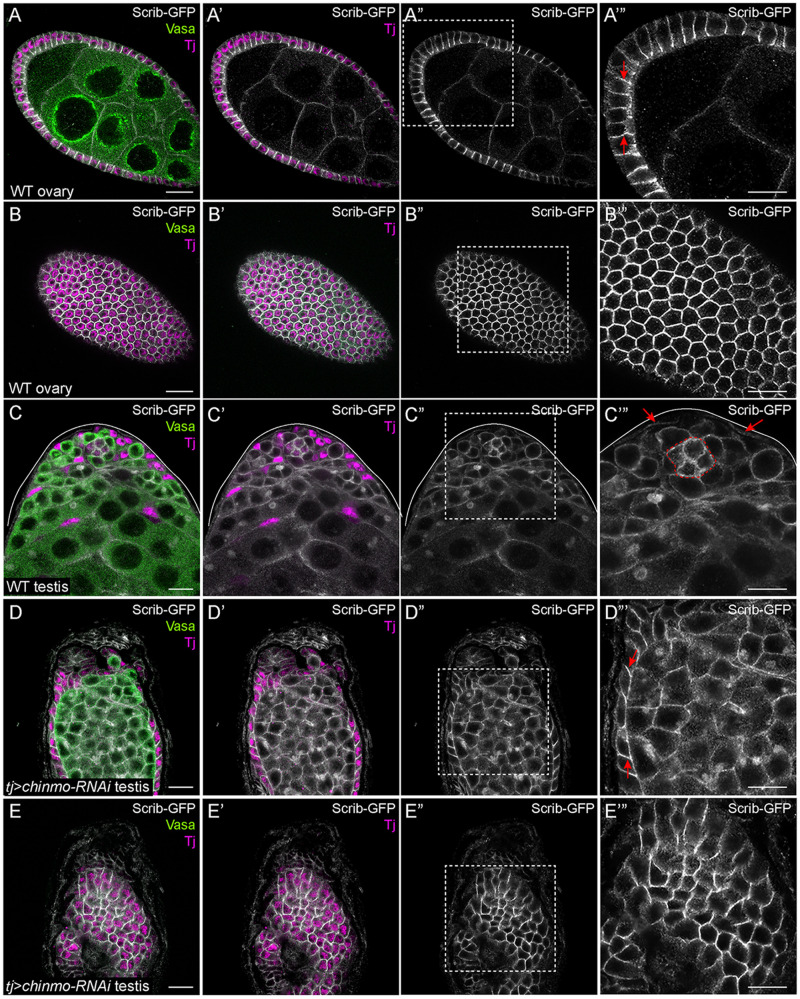
chinmo-depleted somatic cells upregulate Scrib
A scaffolding protein that localizes to the septate junction of epithelial cells, Scrib contains leucine-rich repeats and PDZ domains (Bilder and Perrimon 2000; Bryant and Huwe 2000; Santoni et al. 2002). Both types of domains are critical for Scrib localization and stabilization at the plasma membrane (Albertson et al. 2004; Zeitler et al. 2004). scrib mutant follicle cell clones display loss of apico-basal polarity, characterized by a disrupted actin cytoskeleton and multi-layered morphology (Bilder et al. 2000). In a WT ovary, Scrib is restricted to the lateral membranes of the follicle epithelium (Bilder et al. 2000; Kronen et al. 2014). We used a Scrib-GFP protein trap that recapitulates Scrib protein localization (Batz et al. 2014; Buszczak et al. 2007). In a middle xy-section of a WT ovary, Scrib-GFP was strongly expressed in the lateral domain of follicle cells (Figure 6, A–B’”, arrows; Bilder et al. 2000; Kronen et al. 2014). Consistent with prior report, in a WT testis, Scrib was expressed strongly in niche cells (outlined in Figure 6C’”) and at lower levels in somatic support cells (Figure 6C’”, arrows; Papagiannouli 2013). In testes in which chinmo was somatically depleted, Scrib-GFP expression increased in the lateral domain of feminized, follicle-like cells (Figure 6, D–E’”, arrows).
Figure 6.
chinmo-deficient somatic cells upregulate Scrib. (A–A’’’) Middle xy-section of a WT ovary shows that Scrib (white) is expressed strongly in the lateral domains (arrows in A’”) of follicle cells and at lower levels in the germline. (B–B’’’) An apical xy-section of a WT ovary shows the apical enrichment of Scrib (white) in the follicle cells. (C–C’’’) In a WT testis, Scrib (white) is expressed in the niche (outlined in C’”), at moderate levels in germ cells, and at low levels in somatic cyst cells (arrows in C’”). (D–D’’’) Middle xy-section of a tj>chinmo-RNAi testis shows that Scrib (white) is ectopically expressed in the lateral domain (arrows in D’”) of epithelial feminized somatic cells. (E–E’’’) An apical xy-section of a tj>chinmo-RNAi testis shows apical enrichment of Scrib (white) in the feminized follicle-like cells. In (A–E), Vasa (green) marks the germline and Tj (magenta) marks cyst cells. Scale bar = 10 μm. (A’”–E”’) is a magnification of the boxed regions in (A”–E”), respectively. Time point in (A–E) is 7–8 days post-eclosion. Genotypes: (A, B) w/w; +/+; scrib-GFP/scrib-GFP (C) w/Y; +/+; scrib-GFP/scrib-GFP, and (D, E) w/Y; tj-Gal4/UAS-Dcr-2; UAS-chinmo-RNAi/scrib-GFP (labeled “tj>chinmo-RNAi”).
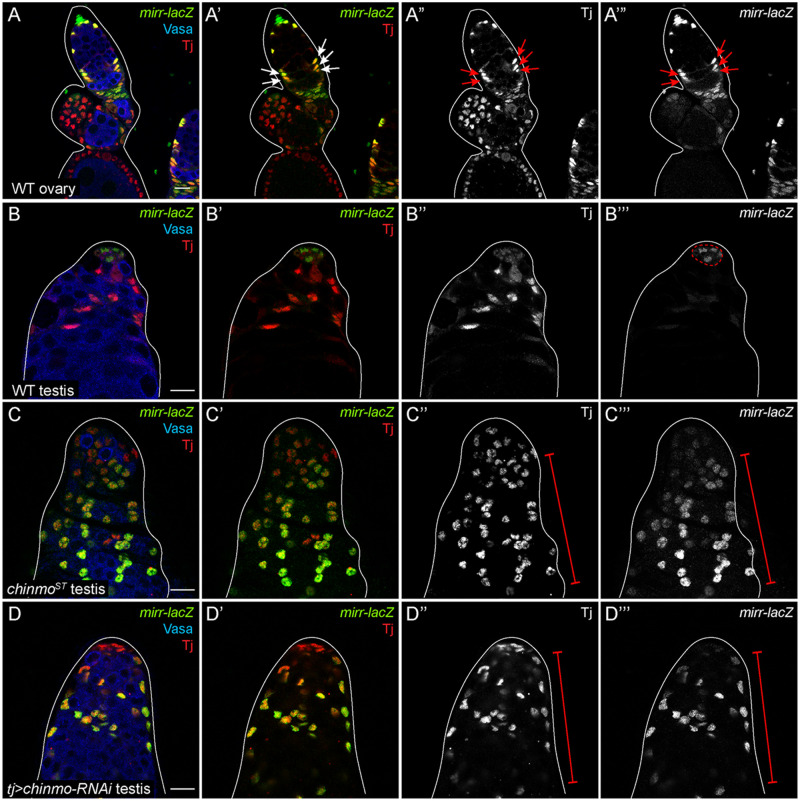
chinmo-depleted somatic cells upregulate Mirror
A recent report revealed that Mirror (Mirr), an Iroquois complex homeobox transcription factor, is expressed in IGS cells and early follicle cells in the germarium, in addition to its known expression in dorsal follicle cells at stage 10 of oogenesis (Jordan et al. 2000; Zhao et al. 2000; Cavodeassi et al. 2001; Xi et al. 2003; Tu et al. 2020). Importantly, mirr transcripts were increased 2.1-fold in chinmo-depleted somatic cells compared with controls, but this result was not statistically significant (P < 0.349). Nevertheless, we decided to validate these results. We confirmed that a mirr-LacZ enhancer trap was robustly expressed in tj-positive, early follicle cells in a WT germarium (Figure 7, A–A’”, arrows). In a WT testis, mirr was expressed strongly in niche cells (outlined in Figure 7B’”) and at much lower levels in the somatic CySCs (Figure 7B’”). Consistent with increased mirr transcripts in the RNA-seq, mirr-LacZ was strongly expressed in all somatic cells in a chinmoST mutant testis and in a testis somatically depleted for chinmo (Figure 7, C–D”’, brackets). These results indicate that during sex transformation, chinmo-deficient CySCs upregulate mirr.
Figure 7.
chinmo-deficient somatic cells upregulate mirr. (A–A’”) Middle xy-section of a WT germarium shows that mirr (green) is present in early FSCs (arrows in A’–A’”). (B–B’”) In a WT testis, mirr (green) is expressed strongly in niche cells (outlined in B””) and at a modest level in other somatic cells. (C–C’”) In a chinmoST mutant testis, mirr (green) is ectopically expressed in feminized somatic cells (bracket in C” and C’”). (D–D’”) mirr (green) is ectopically expressed in the feminized somatic cells (brackets in D” and D’”) in a tj>chinmo-RNAi testis. In (A–D), Vasa (blue) marks the germline and Tj (red) marks cyst cells. Scale bar = 10 μm. Time point in (A–D) is 7–8 days post-eclosion. Genotypes: (A) w/w; +/+; mirr-lacZ/TM6B, Tb, (B) w/Y; +/+; mirr-lacZ/TM6B, Tb, (C) w/Y; chinmoST/chinmoST; mirr-lacZ/TM6B, Tb, (D) w/Y; tj-Gal4/UAS-Dcr-2; UAS-chinmo-RNAi/mirr-lacZ (labeled “tj>chinmo-RNAi”).
Discussion
Here, we report the transcriptional profiling of purified WT somatic cyst cells or those depleted for the putative transcriptional repressor Chinmo. We demonstrate that their respective transcriptional profiles are distinct and that depletion of Chinmo triggers a dramatic transcriptional response within 2 days of its loss. Our interest lies in identifying genes and gene networks that are direct or indirect targets of Chinmo and the perturbations that regulate sex transformation of male cyst cells. Using a combination of immunofluorescence, enhancer traps and protein traps, we validated upregulated candidate genes broadly implicated in epithelial architecture. Our work demonstrates that chinmo-dependent feminization is accompanied with dramatic molecular changes in epithelial morphogenesis marked by ectopic expression of cell polarity regulators like DE-cad, Pyd, and Scrib, cytoskeleton regulators like βH-spectrin, and cell fate regulators like Mirr. Furthermore, some of these genes (pyd, scrib, and βH-spectrin) harbor multiple putative Chinmo binding sites in noncoding regions, suggesting that they might be direct Chinmo targets.
Despite the fact that Chinmo regulates a variety of processes, from neuronal temporal development to sex determination, very few Chinmo target genes are known. In fact, most publications concerning Chinmo have revealed upstream regulators of the chinmo gene and its transcripts and not factors acting downstream of Chinmo (Ostrin et al. 2006; Flaherty et al. 2010; Wu et al. 2012; Liu et al. 2015; Syed et al. 2017). We found only one other RNA-seq that reported Chinmo target genes in larval neural stem cells (Narbonne-Reveau et al. 2016). Although βH-spectrin is a common target of both RNA-seq data sets, it is regulated in opposite directions, positively by Chinmo in neural tumors (Narbonne-Reveau et al. 2016) and negatively in testicular somatic cells (this study). Future work will be needed to determine whether βH-spectrin and other genes are direct targets of Chinmo in vivo and whether there is any cell type specificity. Ideally, this would involve ChIP-seq experiments using an endogenously tagged Chinmo (currently lacking in the field) in purified CySCs and in larval stem cells. Such ChIP-seq experiments would be useful in validating the Chinmo PWM [(G/A)ATGCAC(T/C)(T/N)NN] identified through bacterial one-hybrid (B1H) approaches (Enuameh et al. 2013). We note that our whole-genome screen for sites matching the Chinmo B1H PWM identified large genes, suggesting that Chinmo binding to chromatin in vivo may be more complex.
Data availability
Strains are available upon request. A PWM for Chinmo was obtained by B1H assays (Enuameh et al. 2013). We used a web-based program PWMScan (https://ccg.epfl.ch//pwmtools/pwmscan.php; Ambrosini et al. 2018) to search the Drosophila genome for Chinmo binding sites. Supplementary Tables S1 and S2 contain the list of genes that are differentially up- or downregulated (Padj ≤ 0.05) in somatic cells depleted for chinmo, respectively. Supplementary Tables S3 and S4 contain the list of genes that are differentially up- and downregulated, respectively, in chinmo-depleted somatic cells that contain at least one Chinmo binding site. The RNA-seq data in this study have been deposited at NCBI’s Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number (GSE148230). Supplementary material is available at figshare: https://doi.org/10.6084/m9.figshare.13604270.
Acknowledgments
We are grateful to Marc Amoyel, Salva Herrera, Michael Burel, Alessandro Bailetti, Shally Margolis, and other Bach lab members for help with dissecting testes. We thank Dr Selase Enuameh, Dr Scot Wolf, and Dr Michael Brodsky (University of Massachusetts Medical School, MA, USA) for generously sharing the Chinmo binding site derived from their bacterial one-hybrid studies. We thank Minx Fuller for providing us with the FACS protocol. We thank Ronald Davis, Ruth Lehmann, Dorothea Godt, Claire Thomas, Jessica Treisman, the Bloomington Stock Center (BDSC) for fly stocks, and antibodies. We also thank the Cytometry and Cell Sorting laboratory and the Genome Technology Center at NYU Langone Medical Center for FACS-sorting GFP-positive cells from adult testes and performing the RNA-seq, respectively. We thank Igor Dolgalev for the PCA analysis.
Funding
This work was funded by the National Institute of General Medical Sciences (R01GM085075 to E.A.B.); the New York State Stem Cell Science (NYSTEM; C028132 and C32577GG to E.A.B.); United Negro College Fund/Merck Science (L.G.). The BDSC is supported by a grant from the Office of the Director of the National Institutes of Health under Award Number (P40OD018537). FlyBase is supported by a grant from the National Human Genome Research Institute at the U.S. National Institutes of Health (U41 HG000739). The GTC is partially supported by the Cancer Center Support Grant (P30CA016087) at the Laura and Isaac Perlmutter Cancer Center at NYU Langone Medical Center.
Conflicts of interest: We declare no conflicts of interest.
Literature cited
- Albertson R, Chabu C, Sheehan A, Doe CQ.. 2004. Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J Cell Sci. 117:6061–6070. [DOI] [PubMed] [Google Scholar]
- Ambrosini G, Groux R, Bucher P.. 2018. Pwmscan: A fast tool for scanning entire genomes with a position-specific weight matrix. Bioinformatics. 34:2483–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel M, Sanny J, Burel M, Bach EA.. 2013. Hedgehog is required for CySC self-renewal but does not contribute to the GSC niche in the Drosophila testis. Development. 140:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel M, Simons BD, Bach EA.. 2014. Neutral competition of stem cells is skewed by proliferative changes downstream of Hh and Hpo. EMBO J. 33:2295–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. Htseq–a python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashamboo A, McElreavey K.. 2015. Human sex-determination and disorders of sex-development (DSD). Semin Cell Dev Biol. 45:77–83. [DOI] [PubMed] [Google Scholar]
- Batz T, Forster D, Luschnig S.. 2014. The transmembrane protein Macroglobulin complement-related is essential for septate junction formation and epithelial barrier function in Drosophila. Development. 141:899–908. [DOI] [PubMed] [Google Scholar]
- Bell LR, Horabin JI, Schedl P, Cline TW.. 1991. Positive autoregulation of Sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 65:229–239. [DOI] [PubMed] [Google Scholar]
- Bilder D, Li M, Perrimon N.. 2000. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 289:113–116. [DOI] [PubMed] [Google Scholar]
- Bilder D, Perrimon N.. 2000. Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature. 403:676–680. [DOI] [PubMed] [Google Scholar]
- Boggs RT, Gregor P, Idriss S, Belote JM, McKeown M.. 1987. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell. 50:739–747. [DOI] [PubMed] [Google Scholar]
- Bryant PJ, Huwe A.. 2000. Lap proteins: what’s up with epithelia? Nat Cell Biol. 2:E141–143. [DOI] [PubMed] [Google Scholar]
- Bulgakova NA, Klapholz B, Brown NH.. 2012. Cell adhesion in Drosophila: versatility of cadherin and integrin complexes during development. Curr Opin Cell Biol. 24:702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, et al. 2007. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 175:1505–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers TJ, Dubreuil R, Branton D, Kiehart DP, Goldstein LS.. 1987. Drosophila spectrin. II. Conserved features of the alpha-subunit are revealed by analysis of cDNA clones and fusion proteins. J Cell Biol. 105:2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara N, Whitworth C, Dove A, Van Doren M.. 2019. Doublesex controls specification and maintenance of the gonad stem cell niches in Drosophila. Development. 146:dev170001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara N, Whitworth C, Van Doren M.. 2008. The creation of sexual dimorphism in the Drosophila soma. Curr Top Dev Biol. 83:65–107. [DOI] [PubMed] [Google Scholar]
- Cavodeassi F, Modolell J, Gomez-Skarmeta JL.. 2001. The iroquois family of genes: from body building to neural patterning. Development. 128:2847–2855. [DOI] [PubMed] [Google Scholar]
- Choi W, Jung KC, Nelson KS, Bhat MA, Beitel GJ, et al. 2011. The single Drosophila ZO-1 protein Polychaetoid regulates embryonic morphogenesis in coordination with Canoe/Afadin and Enabled. Mol Biol Cell. 22:2010–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook MS, Cazin C, Amoyel M, Yamamoto S, Bach E, et al. 2017. Neutral competition for Drosophila follicle and cyst stem cell niches requires vesicle trafficking genes. Genetics. 206:1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco TJ, Verney G, Jenkins AB, McCaffery JM, Russell S, et al. 2003. Sex-specific apoptosis regulates sexual dimorphism in the Drosophila embryonic gonad. Dev Cell. 5:205–216. [DOI] [PubMed] [Google Scholar]
- Djiane A, Shimizu H, Wilkin M, Mazleyrat S, Jennings MD, et al. 2011. Su(dx) e3 ubiquitin ligase-dependent and -independent functions of Polychaetoid, the Drosophila ZO-1 homologue. J Cell Biol. 192:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond-Barbosa D. 2019. Local and physiological control of germline stem cell lineages in Drosophila melanogaster. Genetics. 213:9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil RR, Byers TJ, Stewart CT, Kiehart DP.. 1990. A Beta-spectrin isoform from Drosophila (Beta H) is similar in size to vertebrate Dystrophin. J Cell Biol. 111:1849–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil RR, MacVicar G, Dissanayake S, Liu C, Homer D, et al. 1996. Neuroglian-mediated cell adhesion induces assembly of the membrane skeleton at cell contact sites. J Cell Biol. 133:647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE.. 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enuameh MS, Asriyan Y, Richards A, Christensen RG, Hall VL, et al. 2013. Global analysis of Drosophila Cys(2)-His(2) zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome Res. 23:928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadiga J, Nystul TG.. 2019. The follicle epithelium in the Drosophila ovary is maintained by a small number of stem cells. eLife. 8: e49050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild MJ, Yang L, Goodwin K, Tanentzapf G.. 2016. Occluding junctions maintain stem cell niche homeostasis in the fly testes. Curr Biol. 26:2492–2499. [DOI] [PubMed] [Google Scholar]
- Fanning AS, Anderson JM.. 2009. Zonula occludens-1 and -2 are cytosolic scaffolds that regulate the assembly of cellular junctions. Ann N Y Acad Sci. 1165:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, et al. 2010. Chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev Cell. 18:556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz A, Riechmann V.. 2010. Stepwise polarisation of the Drosophila follicular epithelium. Dev Biol. 338:136–147. [DOI] [PubMed] [Google Scholar]
- Fuller MT. 1998. Genetic control of cell proliferation and differentiation in Drosophila spermatogenesis. Semin Cell Dev Biol. 9:433–444. [DOI] [PubMed] [Google Scholar]
- Furuse M, Tsukita S.. 2006. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 16:181–188. [DOI] [PubMed] [Google Scholar]
- Greenspan LJ, de Cuevas M, Matunis E.. 2015. Genetics of gonadal stem cell renewal. Annu Rev Cell Dev Biol. 31:291–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grmai L, Hudry B, Miguel-Aliaga I, Bach EA.. 2018. Chinmo prevents transformer alternative splicing to maintain male sex identity. PLoS Genet. 14:e1007203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RW, Tokuyasu KT, Lindsley DL, Garavito M.. 1979. The germinal proliferation center in the testis of Drosophila melanogaster. J Ultrastruct Res. 69:180–190. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Tepass U.. 2010. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol. 11:502–514. [DOI] [PubMed] [Google Scholar]
- Hill E, Broadbent ID, Chothia C, Pettitt J.. 2001. Cadherin superfamily proteins in Caenorhabditis elegans and Drosophila melanogaster. J Mol Biol. 305:1011–1024. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA.. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA.. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Inoue K, Hoshijima K, Higuchi I, Sakamoto H, Shimura Y.. 1992. Binding of the Drosophila Transformer and Transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc Natl Acad Sci U S A. 89:8092–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Hoshijima K, Sakamoto H, Shimura Y.. 1990. Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature. 344:461–463. [DOI] [PubMed] [Google Scholar]
- Issigonis M, Tulina N, de Cuevas M, Brawley C, Sandler L, et al. 2009. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science. 326:153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan KC, Clegg NJ, Blasi JA, Morimoto AM, Sen J, Stein D, et al. 2000. The homeobox gene mirror links EGF signalling to embryonic dorso-ventral axis formation through notch activation. Nat Genet. 24:429–433. [DOI] [PubMed] [Google Scholar]
- Jung AC, Ribeiro C, Michaut L, Certa U, Affolter M.. 2006. Polychaetoid/ZO-1 is required for cell specification and rearrangement during Drosophila tracheal morphogenesis. Curr Biol. 16:1224–1231. [DOI] [PubMed] [Google Scholar]
- Kronen MR, Schoenfelder KP, Klein AM, Nystul TG.. 2014. Basolateral junction proteins regulate competition for the follicle stem cell niche in the Drosophila ovary. PLoS One. 9:e101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras S, Van Doren M.. 2006. Development of the male germline stem cell niche in Drosophila. Dev Biol. 294:92–103. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S.. 2008. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 3:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S.. 2010. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol. 12:806–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Brandin E, Branton D, Goldstein LS.. 1997. Alpha-spectrin is required for ovarian follicle monolayer integrity in Drosophila melanogaster. Development. 124:353–362. [DOI] [PubMed] [Google Scholar]
- Lenhart KF, DiNardo S.. 2015. Somatic cell encystment promotes abscission in germline stem cells following a regulated block in cytokinesis. Dev Cell. 34:192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem RK. 2016. Cytoskeletal integrators: the spectrin superfamily. Cold Spring Harb Perspect Biol. 8: a018259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yang CP, Sugino K, Fu CC, Liu LY, et al. 2015. Opposing intrinsic temporal gradients guide neural stem cell production of varied neuronal fates. Science. 350:317–320. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW, Maniatis T.. 1996. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 10:2089–2101. [DOI] [PubMed] [Google Scholar]
- Ma Q, Wawersik M, Matunis EL.. 2014. The JAK-STAT target Chinmo prevents sex transformation of adult stem cells in the Drosophila testis niche. Dev Cell. 31:474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel M, Kupinski AP, Raabe I, Bokel C.. 2012. Hh signalling is essential for somatic stem cell maintenance in the Drosophila testis niche. Development. 139:2663–2669. [DOI] [PubMed] [Google Scholar]
- Narbonne-Reveau K, Lanet E, Dillard C, Foppolo S, Chen CH, et al. 2016. Neural stem cell-encoded temporal patterning delineates an early window of malignant susceptibility in Drosophila. eLife. 5: e13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ. 2008. Regulation of cell-cell adhesion by the Cadherin-Catenin complex. Biochem Soc Trans. 36:149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, et al. 2006. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 16:466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannouli F. 2013. The internal structure of embryonic gonads and testis development in Drosophila melanogaster requires scrib, lgl and dlg activity in the soma. Int J Dev Biol. 57:25–34. [DOI] [PubMed] [Google Scholar]
- Rust K, Nystul T.. 2020. Signal transduction in the early Drosophila follicle stem cell lineage. Curr Opin Insect Sci. 37:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni MJ, Pontarotti P, Birnbaum D, Borg JP.. 2002. The lap family: a phylogenetic point of view. Trends Genet. 18:494–497. [DOI] [PubMed] [Google Scholar]
- Seppa MJ, Johnson RI, Bao S, Cagan RL.. 2008. Polychaetoid controls patterning by modulating adhesion in the Drosophila pupal retina. Dev Biol. 318:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng XR, Posenau T, Gumulak-Smith JJ, Matunis E, Van Doren M, et al. 2009. JAK-STAT regulation of male germline stem cell establishment during Drosophila embryogenesis. Dev Biol. 334:335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields AR, Spence AC, Yamashita YM, Davies EL, Fuller MT.. 2014. The actin-binding protein profilin is required for germline stem cell maintenance and germ cell enclosure by somatic cyst cells. Development. 141:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed MH, Mark B, Doe CQ.. 2017. Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. eLife. 6: e26287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahisa M, Togashi S, Suzuki T, Kobayashi M, Murayama A, et al. 1996. The Drosophila tamou gene, a component of the activating pathway of extramacrochaetae expression, encodes a protein homologous to mammalian cell-cell junction-associated protein ZO-1. Genes Dev. 10:1783–1795. [DOI] [PubMed] [Google Scholar]
- Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Torok T, et al. 1996. Shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 10:672–685. [DOI] [PubMed] [Google Scholar]
- Thomas GH, Williams JA.. 1999. Dynamic rearrangement of the spectrin membrane skeleton during the generation of epithelial polarity in Drosophila. J Cell Sci. 112:2843–2852. [DOI] [PubMed] [Google Scholar]
- Tu R, Duan B, Song X, Chen S, Scott A, et al. 2021. Multiple niche compartments orchestrate stepwise germline stem cell progeny differentiation. Curr Biol. 31:827-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vied C, Reilein A, Field NS, Kalderon D.. 2012. Regulation of stem cells by intersecting gradients of long-range niche signals. Dev Cell. 23:836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterbury JA, Horabin JI, Bopp D, Schedl P.. 2000. Sex determination in the Drosophila germline is dictated by the sexual identity of the surrounding soma. Genetics. 155:1741–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawersik M, Milutinovich A, Casper AL, Matunis E, Williams B, et al. 2005. Somatic control of germline sexual development is mediated by the JAK/STAT pathway. Nature. 436:563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ellis HM.. 2001. Localization of the Drosophila MAGUK protein Polychaetoid is controlled by alternative splicing. Mech Dev. 100:217–231. [DOI] [PubMed] [Google Scholar]
- Wu YC, Chen CH, Mercer A, Sokol NS.. 2012. let-7-Complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev Cell. 23:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi R, McGregor JR, Harrison DA.. 2003. A gradient of JAK pathway activity patterns the anterior-posterior axis of the follicular epithelium. Dev Cell. 4:167–177. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, Fuller MT.. 2003. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 301:1547–1550. [DOI] [PubMed] [Google Scholar]
- Zarnescu DC, Thomas GH.. 1999. Apical spectrin is essential for epithelial morphogenesis but not apicobasal polarity in Drosophila. J Cell Biol. 146:1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitler J, Hsu CP, Dionne H, Bilder D.. 2004. Domains controlling cell polarity and proliferation in the Drosophila tumor suppressor Scribble. J Cell Biol. 167:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Woolner S, Bownes M.. 2000. The Mirror transcription factor links signalling pathways in Drosophila oogenesis. Dev Genes Evol. 210:449–457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. A PWM for Chinmo was obtained by B1H assays (Enuameh et al. 2013). We used a web-based program PWMScan (https://ccg.epfl.ch//pwmtools/pwmscan.php; Ambrosini et al. 2018) to search the Drosophila genome for Chinmo binding sites. Supplementary Tables S1 and S2 contain the list of genes that are differentially up- or downregulated (Padj ≤ 0.05) in somatic cells depleted for chinmo, respectively. Supplementary Tables S3 and S4 contain the list of genes that are differentially up- and downregulated, respectively, in chinmo-depleted somatic cells that contain at least one Chinmo binding site. The RNA-seq data in this study have been deposited at NCBI’s Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number (GSE148230). Supplementary material is available at figshare: https://doi.org/10.6084/m9.figshare.13604270.