Abstract
A total of 1,210 clinical isolates of Escherichia coli collected from a university hospital in southern Taiwan were screened for production of extended-spectrum β-lactamases (ESBLs). Expression of classical ESBLs (resistant to extended-spectrum β-lactam agents and susceptible to β-lactam inhibitors) was inferred in 18 isolates by the phenotypic confirmatory test. These included 10 isolates producing CTX-M-3, 2 strains carrying SHV-12, 1 strain harboring SHV-5, 1 strain expressing TEM-10, and 4 strains producing unidentifiable ESBLs with a pI of 8.05, 8.0, or 7.4. Eighteen isolates that showed decreased susceptibilities to ceftazidime and/or cefotaxime, negative results for the confirmatory test, and high-level resistance to cefoxitin (MICs of ≥128 μg/ml) were also investigated. Five isolates were found to produce CMY-2 AmpC enzymes, one isolate carried both CTX-M-3 and CMY-2, and the remaining three and nine isolates expressed putative AmpC β-lactamases with pIs of >9.0 and 8.9, respectively. Thus, together with the isolate producing CTX-M-3 and CMY-2, 19 (1.6%) isolates produced classical ESBLs. Pulsed-field gel electrophoresis revealed that all isolates carrying CTX-M-3 and/or CMY-2 were genetically unrelated, indicating that dissemination of resistance plasmids was responsible for the spread of these two enzymes among E. coli in this area. Among the 16 isolates expressing CTX-M-3 and/or CMY-2, 5 might have colonized outside the hospital environment. Our data indicate that CTX-M-3 and CMY-2, two β-lactamases initially identified in Europe, have been disseminated to and are prevalent in Taiwan.
The emergence of plasmid-mediated extended-spectrum β-lactamases (ESBLs) among members of the family Enterobacteriaceae has become a growing worldwide problem (17, 19–23, 29, 31, 32, 34, 41). The Bush group 2 (or Ambler molecular class A) ESBLs possess an extended hydrolysis spectrum directed toward oxyimino-β-lactams and aztreonam but remain susceptible to inhibition by β-lactamase inhibitors (6). Most of the ESBLs in Escherichia coli and Klebsiella pneumoniae are derived from TEM- or SHV-type β-lactamases by one or more amino acid substitutions that confer resistance to extended-spectrum cephalosporins (17, 19, 29, 31, 34, 41). Recently, more and more non-TEM- and non-SHV-derived ESBLs have been identified over an extremely wide geographic area, such as CTX-M-related enzymes found in Europe, South America, and Mediterranean countries (2, 11, 13, 29, 32) and Toho-1 and Toho-2 found in Japan (16, 25). Unlike TEM and SHV producers, reports of the outbreaks caused by non-TEM- and non-SHV-ESBL-producing organisms and knowledge of clinical impacts of these enzymes are still limited (29, 32).
Along with ESBLs, the emergence of plasmid-mediated Ambler class C cephalosporinases (or Bush group 1 cephalosporinases) has occurred among clinical isolates of the Enterobacteriaceae recently (3, 4, 12, 15, 33, 40, 41). It is believed that these enzymes are derived from AmpC chromosomally located cephalosporinases (6, 29). The plasmid-mediated cephalosporinases that are related most closely to AmpC cephalosporinase from Pseudomonas aeruginosa are represented by MOX-1 identified in Japan and CMY-1 in Korea (3, 15); those related most closely to AmpC cephalosporinase from Citrobacter freundii are represented by CMY-2 and LAT-1 found in Greece (4, 40), and those related most closely to AmpC enzyme from Enterobacter cloacae are represented by MIR-1 identified in the United States (33). These enzymes can produce resistance to cephamycins, extended-spectrum cephalosporins, and aztreonam and, unlike class A ESBLs, they are not inhibited by β-lactamase inhibitors (6).
SHV-derived enzymes have been identified as the most common ESBLs among clinical isolates of K. pneumoniae in Taiwan (22, 41); however, little is known about the prevalence and characteristics of ESBLs among E. coli isolates in this country. The present study was conducted to determine the prevalence and genotypes of classical ESBLs (resistant to extended-spectrum cephalosporins and susceptible to inhibition by β-lactam inhibitors) among clinical isolates of E. coli in southern Taiwan. We present here the first description of the presence of CTX-M-3 in the Far East. The first identification of the CMY-2 AmpC enzyme in this area is also described.
MATERIALS AND METHODS
Selection of clinical isolates and patients.
Between January and September 1999, 2,047 clinical isolates of E. coli were consecutively collected in the Department of Pathology, National Cheng Kung University Hospital, a 900-bed university hospital in southern Taiwan. A total of 1,210 isolates, including those from different patients or those from the same patient but with different antimicrobial susceptibilities, were selected in this study. These isolates were identified by using the conventional techniques (10) and/or the API 20E system (bioMérieux, Marcy l'Etoile, France). The medical records of patients harboring ESBL-producing isolates or AmpC hyperproducers were reviewed.
Susceptibility tests and confirmation of ESBL production.
MICs of the antibiotics were determined by means of the agar dilution method according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS) (28). The antimicrobial agents and their sources were as follows: ampicillin and cofoxitin (Sigma Chemical Company, St. Louis, Mo.); aztreonam (Bristol-Myers Squibb, New Brunswick, N.J.); ceftazidime (Glaxo Group Research, Ltd., Greenford, United Kingdom); cefotaxime and cefuroxime (Hoechst-Roussel Pharmaceuticals, Inc., Somerville, N.J.); ceftriaxome (Hoffmann-La Roche, Inc., Utley, N.J.); and imipenem (Merck Sharp & Dohme, West Point, Pa.). E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as quality reference strains.
EBSL production was detected by means of phenotypic confirmatory tests as recommended by the NCCLS (28). E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as negative and positive controls, respectively.
IEF and enzyme inhibition assay.
Crude preparations of β-lactamases from clinical isolates and their transconjugants were obtained by sonication as described previously (5). Isoelectric focusing (IEF) analysis was performed by the method of Matthew et al. (27) with an LKB Multiphor apparatus on prepared PAGplate gels (pH 3.5 to 9.5; Pharmacia Biotech Asia Pacific, Hong Kong, China). Enzyme activities of β-lactamases were detected by overlaying the gel with 0.5 mM nitrocefin in 0.1 M phosphate buffer (pH 7.0). Extracts from TEM-1-, SHV-1-, CMY-1-, SHV-5-, and CTX-M-3-producing strains were used as standards for pIs of 5.4, 7.6, 8.0, 8.2, and 8.4, respectively. In addition, the pI was also calculated by reference to a calibration curve constructed from the relative mobility of the isoelectric focusing markers (Pharmacia Biotech Asia Pacific). Inhibition assay was carried out by overlaying the gels with 0.5 mM nitrocefin with or without 0.3 mM cloxacillin or 0.3 mM clavulanic acid in 0.1 M phosphate buffer (pH 7.0) (9).
Conjugation experiments and plasmid analysis.
Conjugation experiments were performed as described previously (36) with streptomycin-resistant E. coli C600 as the recipient (1). Transconjugants were selected on tryptic soy agar plates supplemented with 500 μg of streptomycin (Sigma) per ml and 10 μg of ceftazidime, 10 μg of cefotaxime, or 64 μg of cefoxitin per ml.
Plasmids from clinical isolates and transconjugants were extracted by a rapid alkaline lysis procedure (38). For the restriction enzyme analysis of transconjugant plasmids, EcoRI and PstI (Roche Molecular Biochemicals, Mannheim, Germany) were used. Digested and nondigested DNA samples were analyzed by electrophoresis on 0.8% agarose gels. The gels were stained with ethidium bromide (Sigma), and plasmid bands were visualized under UV light. The plasmid sizes of transconjugants were estimated by adding up restriction fragments.
PCR amplification and DNA sequencing.
Plasmid preparations from clinical isolates and their transconjugants were used as templates in PCR reactions. To amplify the entire sequences of blaTEM-, blaSHV-, blaCMY-1-, and blaCTX-M-related genes, oligonucleotide primers specific for these genes were used as described previously (13, 26, 30, 41). The β-lactamases that can be amplified with the primers for blaCMY-1 are CMY-1 and CMY-8 (41). LAT-type AmpC β-lactamases and CMY-2-related β-lactamases were genetically closely related, while both of them shared only approximately 42% amino acid identities to CMY-1 (3, 4, 12, 40). Thus, oligonucleotide primers AmpC-1C (5′CTGCTGCTGACAGCCTCTTT) and AmpC-1B (5′-TTTTCAAGAATGCGCCAGGC-3′), corresponding to nucleotides 28 to 47 and 1136 to 1117, respectively, of the blaCMY-2 structural gene (4), were used to amplify an internal fragment of about 95% of blaCMY-2- and blaLAT-related genes. CMY-1-related β-lactamases were not amplified with the primer pair. PCR reactions for blaTEM, blaSHV, blaCMY-1, and blaCTX-M genes were run under conditions as described previously (13, 26, 30, 41). The PCR conditions for the blaCMY-2-related genes were as follows: 3 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C; and finally 7 min at 72°C. The amplicons were purified with PCR Clean Up Kits (Roche Molecular Biochemicals) and were sequenced on an ABI Prism 377 Sequencer Analyzer (Applied Biosystems, Foster City, Calif.).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was carried out with a contour-clamped homogeneous electric field system (Pulsaphor Plus; Pharmacia LKB Biotechnology, Uppsala, Sweden) as described previously (8). The genomic DNAs were prepared as described by Piggot et al. (35) and were digested overnight with 10 U of SfiI (New England Biolabs, Beverly, Mass.). DNA was electrophoresed through a 1% agarose gel in Tris-borate-EDTA buffer at 150 V for 30 h, with pulse times ranging from 5 to 35 s. The DNA bands were visualized by staining of the gel with ethidium bromide and were photographed. Bacteriophage lambda DNA concatemers (Gibco-BRL, Gaithersburg, Md.) were used as size standards.
RESULTS
NCCLS confirmatory tests for ESBLs.
Of the 1,210 nonrepetitive clinical isolates of E. coli screened for susceptibilities to ceftazidime and cefotaxime, the MICs of ceftazidime and/or cefotaxime for 50 isolates were ≥2 μg/ml. These isolates were tested with the NCCLS phenotypic confirmatory test for ESBLs. Eighteen isolates gave positive results, indicating production of classical ESBLs by these isolates. Among the 32 isolates negative for the NCCLS confirmatory test, 18 showed high-level resistance to cefoxitin (MICs of ≥128 μg/ml), and 14 showed only slightly decreased susceptibilities to ceftazidime (MICs of 2 to 8 μg/ml), cefotaxime (MICs of 2 to 8 μg/ml), and cefoxitin (MICs of 16 to 64 μg/ml). The 18 isolates producing classical ESBLs and the 18 isolates negative for the NCCLS confirmatory test but with high-level resistance to cefoxitin (MICs of ≥128 μg/ml) were included for further study.
IEF and enzyme inhibition assay.
The results of IEF are summarized in Table 1. On IEF gels, all β-lactamases produced by the 18 classical ESBL-producing isolates were inhibited by 0.3 mM clavulanic acid but not by 0.3 mM cloxacillin. The enzymes with pIs of >9.0, 9.0, and 8.9 detected in the 18 isolates with high-level resistance to cefoxitin were inhibited by cloxacillin but not by clavulanic acid and thus were tentatively classified as AmpC enzymes. The other two enzymes, with pIs of 5.4 and 8.4, were inhibited by clavulanic acid but not by cloxacillin. Of the 18 AmpC producers, 1 also possessed the β-lactamases with pIs of 8.4 and 5.4, which matched the enzymes produced by 10 of the 18 classical ESBL producers, indicating expression of a classical ESBL by this isolate. Thus, a total 19 isolates were considered to carry classical ESBLs.
TABLE 1.
Antimicrobial susceptibilities, IEF of β-lactamases, and identified β-lactamases of ESBL or AmpC producers
| Isoelectric point(s) | No. of isolates | MIC (μg/ml) ofa:
|
Genotype(s) of β-lactamase(s) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CXM | CAZ | CTX | CRO | ATM | FOX | IPM | |||
| ESBL producers | ||||||||||
| Clinical isolates (pI) | ||||||||||
| 8.4, 5.4 | 10 | >256 | >256 | 4–16 | ≥256 | >256 | 4–64 | 4–16 | ≤0.25 | CTX-M-3, TEM-1 |
| 8.2, 5.4 | 2 | >256 | 64–128 | 128–256 | 16–32 | 8–16 | 128–256 | 8–16 | ≤0.25 | SHV-12, TEM-1 |
| 8.2, 5.4 | 1 | >256 | 64 | 128 | 32 | 8 | 128 | 2 | 0.25 | SHV-5, TEM-1 |
| 8.05, 5.4 | 1 | >256 | >256 | 0.5 | 8 | 8 | 1 | 8 | 0.25 | Unidentified, TEM-1 |
| 8.0, 5.4 | 2 | >256 | >256 | 2–4 | ≥128 | ≥256 | 4–8 | 4–16 | 0.25 | Unidentified, TEM-1 |
| 7.4, 5.4 | 1 | >256 | 64 | 0.5 | 8 | 1 | 0.5 | 8 | 0.25 | Unidentified, TEM-1 |
| 5.6 | 1 | >256 | 32 | >256 | 8 | 16 | >256 | 8 | 0.5 | TEM-10 |
| Transconjugants (plasmid size [kb]) | ||||||||||
| 8.4 (65) | 9 | >256 | >256 | 4–16 | ≥128 | ≥256 | 4–32 | 4–16 | ≤0.25 | CTX-M-3 |
| 8.2 (60) | 2 | >256 | 64 | 128 | 32 | 16 | 128 | 4–8 | 0.25 | SHV-12 |
| 8.2 (60) | 1 | >256 | 64 | 128 | 32 | 16 | 128 | 8 | 0.25 | SHV-5 |
| 8.05 (55) | 1 | >256 | >256 | 0.5 | 16 | 8 | 1 | 16 | 0.13 | Unidentified |
| 8.0 (70) | 2 | >256 | >256 | 2 | ≥256 | >256 | 4 | 4 | 0.25 | Unidentified |
| ESBL and AmpC producers | ||||||||||
| Clinical isolate (pI) | ||||||||||
| >9.0, 8.4, 5.4 | 1 | >256 | >256 | 32 | >256 | >256 | 32 | >256 | 0.25 | CMY-2, CTX-M3, TEM-1 |
| AmpC hyperproducers | ||||||||||
| Clinical isolates (pI) | ||||||||||
| >9.0, 9.0, 5.4 | 1 | >256 | >256 | 256 | 128 | 256 | 32 | >256 | 0.25 | Unidentified, CMY-2, TEM-1 |
| >9.0, 5.4 | 3 | >256 | >256 | ≥128 | 8–32 | 8–32 | 16–32 | >256 | ≤0.25 | Unidentified, TEM-1 |
| 9.0, 5.4 | 4 | >256 | 32–64 | 32–64 | 8–16 | 16–32 | 8–16 | ≥128 | ≤0.25 | CMY-2, TEM-1 |
| 8.9, 5.4 | 9 | >256 | 32–64 | 2–16 | 2–16 | 2–16 | 4–32 | ≤256 | ≥0.25 | Unidentified, TEM-1 |
| Transconjugants (plasmid size [kb]) | ||||||||||
| 9.0 (90) | 4 | >256 | 32–64 | 32–64 | 8–16 | 16–32 | 8–16 | ≥128 | ≤0.25 | CMY-2 |
| 8.9 (75) | 5 | >256 | 32–64 | 2–8 | 2–8 | 2–8 | 8–32 | ≥256 | ≤0.25 | Unidentified |
MICs were determined by the agar dilution test. Antimicrobial agents: AMP, ampicillin; CXM, cefuroxime; CAZ, ceftazidime; CTX, cefotaxime; CRO, ceftriaxone; ATM, aztreonam; FOX, cefoxitin; IPM, imipenem.
Transfer of resistance.
For the 18 classic ESBL-producing isolates, cefotaxime resistance was transferred from 9 of 10 isolates producing β-lactamases with a pI of 8.4, from the isolate producing β-lactamases with a pI of 8.05, and from 2 of 2 isolates producing β-lactamases with a pI of 8.0. Ceftazidime resistance was transferred from three of three isolates that carried the β-lactamase with a pI of 8.2. Transfer of the β-lactamases with pIs of 7.4 and 5.6 was not successful. Transfer of the cefoxitin resistance was achieved for four of five isolates possessing the β-lactamase with a pI of 9.0 and for five of nine isolates producing the β-lactamase with a pI of 8.9. Transfer of resistance from the isolate containing the β-lactamases with pIs of >9.0 and 8.4 was not achieved. The sizes of plasmids transferred to E. coli are shown in Table 1.
Susceptibility tests.
The susceptibilities of the studied organisms and their transconjugants to various β-lactams are summarized in Table 1. For the 18 classic ESBL-producing isolates, the β-lactamases with pIs of 8.4 and 8.0 conferred high-level resistance to cefotaxime (MICs of ≥128 μg/ml), whereas the β-lactamases with pIs of 5.6 and 8.2 conferred high-level resistance to ceftazidime (MICs of ≥128 μg/ml). Two isolates producing β-lactamases with pIs of 8.05 and 7.4, respectively, showed slightly decreased susceptibilities to cefotaxime (MICs of 8 μg/ml) and ceftazidime (MICs of 0.5 μg/ml). For the 18 putative AmpC hyperproducers, the β-lactamases with pIs of >9.0, 9.0, and 8.9 were associated with the resistance to cefoxitin. The β-lactamases with a pI of >9.0 seemed to confer high-level resistance to ceftazidime (MICs of ≥128 μg/ml) as well.
PCR and sequence analysis.
The results of PCR and sequence analysis are summarized in Table 1. The blaTEM genes were amplified from all studied isolates of E. coli. The isolate harboring the enzyme with a pI of 5.6 was found to carry TEM-10 by nucleotide sequencing. All the other isolates contained TEM-1. The blaCTX-M-related genes were amplified for all 11 isolates expressing pI 8.4 β-lactamases. The pI 8.4 β-lactamases were confirmed as CTX-M-3 enzymes by sequence analysis. The blaSHV-related genes were amplified for all three isolates producing the pI 8.2 β-lactamases. Two isolates were shown to carry SHV-12, and one was shown to harbor SHV-5 by sequence analysis. The blaCMY-2-related genes were amplified from four isolates producing β-lactamases with pIs of 9.0 and 5.4, and the isolate containing pI > 9.0, 9.0, and 5.4 β-lactamases and sequence analysis confirmed that all PCR products were blaCMY-2. The blaCMY-1-related genes were not amplified in any studied isolates. The genotypes of all transconjugants were consistent with those of their donors.
PFGE.
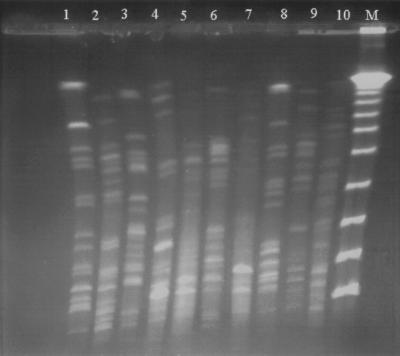
PFGE was performed to determine whether clonal spreading was responsible for dissemination of CTX-M-3 and CMY-2. The results of PFGE analyses are summarized in Table 2 and partially shown in Fig. 1. All these isolates revealed different PFGE patterns, indicating that they were from different clones.
TABLE 2.
Selected clinical data and PFGE profiles of 16 E. coli isolates producing CTX-M-3 or CMY-2
| Patient data (no., sex/age [yr]) | pIs of β-lactamases | Underlying diseases | Type of infectiona | Specimen | Ward/possible reservoir of isolatesb | PFGE pattern |
|---|---|---|---|---|---|---|
| 1, F/52 | 8.4, 5.4 | Breast cancer | UTI | Urine | ER/community hospital | I |
| 2, M/85 | 8.4, 5.4 | Cerebrovascular disease | Suspected UTI | Urine | ER/nursing home | II |
| 3, M/65 | 8.4, 5.4 | Gouty arthritis | Wound infection | Blood | SICU-1/SICU-1 | III |
| 4, M/84 | 8.4, 5.4 | Chronic hepatitis | Pneumonia | Blood | ER/ward-6B | IV |
| 5, M/19 | 8.4, 5.4 | Traumatic intracranial hemorrhage | Wound infection | Wound | SICU-1/SICU-1 | V |
| 6, F/78 | 8.4, 5.4 | Cerebrovascular disease | Aspiration pneumonia | Sputum | ER/ward-10B | VI |
| 7, M/85 | 8.4, 5.4 | Gastric cancer | Suspected pneumonia | Sputum | SICU-2/SICU-2 | VII |
| 8, M/76 | 8.4, 5.4 | Normal pressure hydrocephalus | Wound infection | Wound | OPD/community | VIII |
| 9, F/58 | 8.4, 5.4 | Liver cirrhosis | Pressure sore | Wound | Ward-6B/ward-6B | IX |
| 10, F/32 | 8.4, 5.4 | Multiple fractures | Wound infection | Wound | Ward-7A/ward-7A | X |
| 11, M/2 | >9.0, 8.4, 5.4 | Ureterovesicular stricture | Suspected UTI | Urine | OPD/community | XI |
| 12, F/84 | >9.0, 9.0, 5.4 | Cholelithiasis | Suspected UTI | Urine | Ward-8B/ward-8B | XII |
| 13, F/80 | 9.0, 5.4 | Liver cyst | Pneumonia | Blood | Ward-6B/ward-6B | XIII |
| 14, F/33 | 9.0, 5.4 | Acute pyelonephritis | Acute pyelonephritis | Urine | OPD/community | XIV |
| 15, F/52 | 9.0, 5.4 | Cholelithiasis | Cholecystitis | Bile | OR/community | XV |
| 16, F/84 | 9.0, 5.4 | Sublaxation of cervical spine | UTI | Urine | Ward-7C/ward-7C | XVI |
UTI, urinary tract infection.
ER, emergency room; OPD, outpatient department; OR, operation room; SICU, surgical intensive care unit.
FIG. 1.
PFGE profiles of SfiI-digested genomic DNAs from 10 E. coli isolates producing CTX-M-3 and/or CMY-2. Lanes 1 to 3, 8, and 9, CTX-M-3-producing isolates recovered from patients 1, 3, 4, 9, and 5, respectively (Table 2); lanes 4 to 6 and 10, CMY-2-producing isolates from patients 15, 14, 13, and 16, respectively; lane 7, isolate producing CTX-M-3 and CMY-2 from patient 12; lane M, a lambda ladder (Gibco-BRL) which served as molecular size marker.
Patient data.
The sources from which the 16 isolates producing CTX-M-3 or CMY-2 were recovered, as well as selected clinical features of the patients carrying these organisms, are summarized in Table 2. Positive cultures of CTX-M-3 or CMY-2 producers were obtained from eight patients after 72 h of hospitalization. CTX-M-3 producers were isolated from four patients in the emergency room: patients 4 and 6 were just discharged from this hospital in less than 1 week before isolation of the organisms; patient 1 had been hospitalized at a community hospital for 1 month before she was referred to our hospital; and patient 2 was a nursing home resident. Patients 8, 11, and 14 had been hospitalized 6 months to 2 years before clinical presentations of their infections. Patient 15 had no history of hospitalization before she underwent emergency cholecystectomy at our hospital. Two CTX-M-3 producers and one CMY-2 producer were isolated from blood samples. Since it was not known that they were infected with ESBL producers, patient 3 was treated with gentamicin and piperacillin, and patient 4 was treated with cefotaxime, cefuroxime, ciprofloxacin, and gentamicin. Both of them died of sepsis within 1 week of blood culturing. Patient 13 was successfully treated with an imipenem-containing regimen. All other patients have done well with or without administration of imipenem or else died of causes unrelated to their infections.
DISCUSSION
Production of classical ESBLs was detected in 18 of 1,210 clinical isolates of E. coli by the NCCLS confirmatory test. Together with one isolate carrying an ESBL and an AmpC enzyme, the prevalence of ESBL producers among E. coli in this study was 1.6%. The rate of prevalence was lower than those reported in other Far-Eastern countries, such as Korea (4.8%) (31) and Japan (2.9 to 8.1%) (20, 21) and was also lower than the rate of 8.5% among K. pneumoniae isolates in this area (41).
SHV-derived ESBLs have been found to be prevalent among ESBL-producing K. pneumonia isolates (75%) in Taiwan (22, 41). In this study, it was interesting to find that, unlike K. pneumoniae, only 3 (15.8%) of 19 E. coli isolates that produced classical ESBLs carried SHV-derived enzymes, whereas 15 isolates (78.9%) harbored non-TEM and non-SHV ESBLs. CTX-M-3, a novel class A ESBL recently identified in Poland (13), was responsible for 57.9% of the ESBLs in this study. This enzyme has been found to be prevalent among clinical Enterobacteriaceae isolates in a Warsaw hospital (32) and has rarely been described as occurring elsewhere in the world. Although the presence of CTX-M-related β-lactamases, such as CTX-M-2 and Toho-1, have been reported in Japan (16, 20), The presence of CTX-M-3 has never been reported in that country. Therefore, this is the first report of the persistence and prevalence of CTX-M-3 outside of Europe. It is interesting but unclear how CTX-M-3 was imported into Taiwan. All of our plasmids encoding CTX-M-3 were of the same size and revealed the same restriction pattern (data not shown). Compared with the plasmids from the Warsaw hospital on the basis of the restriction patterns shown in the literature (13, 32), our plasmids seemed to be related to those with the PstI restriction pattern A2+ (32). An international collaborative study is needed to elucidate how this enzyme spread across countries and continents.
Resistance to cefoxitin in E. coli is considered uncommon and is usually attributed to overexpression of the species-specific chromosomal cephalosporinase (24). In our initial survey, 32 isolates were resistant to cefoxitin, a level which outnumbered classic ESBL producers. The putative AmpC enzymes were transferable in half of the 18 isolates with high-level resistance to cefoxitin. The data suggested the emergence of plasmid-mediated class C β-lactamases among clinical isolates of E. coli in Taiwan. Recently, we have identified a CMY-l-related AmpC enzyme, designated CMY-8, from clinical isolates of K. pneumoniae (41). None of the isolates of E. coli in this study carried CMY-1 or CMY-8, but rather five isolates were found to carry CMY-2. CMY-2 was first identified in a clinical isolate of K. pneumoniae from Greece (4) and has rarely been observed in other parts of the world. Therefore, the present study is also the first report of the spread of CMY-2 to the Far East and, together with the findings of our previous study, indicates the presence of both CMY-1-related and CMY-2 β-lactamases in Taiwan.
Of the 36 isolates that were studied completely, 16 carried unrecognized ESBLs or AmpC enzymes (Table 1). The enzyme with a high pI of >9.0 was not transferable, suggesting that it belongs to chromosome-encoded AmpC β-lactamases. Overexpression, mutations of the AmpC structural genes, or both might be responsible for their decreased susceptibilities to extended-spectrum cephalosporinases (7, 14, 18). Sequencing their AmpC structural genes followed by cloning of these genes if necessary should be the first step toward elucidating the resistance mechanisms of these isolates. The enzyme with a high pI of 8.9 revealed high-level resistance to cefoxitin. In addition, this enzyme was transferable and was not susceptible to inhibition by clavulanic acid. All of these findings suggested that the β-lactamase should be a plasmid-mediated class C cephalosporinase. Of 18 AmpC hyperproducers, 9 expressed this enzyme, indicating that it was also an important β-lactamase prevalent among E. coli isolates in Taiwan. Of 18 ESBL producers confirmed by the NCCLS method, 4 did not yield any amplicons for SHV or CTX-M genes. They all carried TEM-1, a restricted-spectrum β-lactamase (6, 24). The resistance phenotypes could be transferred to E. coli recipients by conjugation experiments in three of them. These data suggested that they harbored non-TEM and non-SHV ESBLs. PCRs with more primers specific for other known ESBLs or AmpC enzymes and/or cloning experiments will be performed to determine if they are novel β-lactamases.
Previous studies from Poland showed that both plasmid dissemination and clonal spread contributed to the concurrent outbreaks of CTX-M-3-producing organisms of the family Enterobacteriaceae in the Warsaw hospital (13, 32). Seven CTX-M-3-producing C. freundii isolates collected at that hospital over a 4-month period were genetically related, and those researchers found that the nosocomial strain could be maintained for a relatively long time in the hospital environment. All isolates harboring CTX-M-3 or CMY-2 in the present study were genetically unrelated, suggesting that the dissemination of resistance plasmids was responsible for the prevalence of these two enzymes among E. coli isolates in this area. Ten of these isolates obviously were nosocomial strains of this hospital because their hosts had been hospitalized for more than 72 h or were just discharged from this hospital (Table 2). Two isolates were considered to be from a community hospital and a nursing home, respectively. Three patients had been hospitalized 6 months to 2 years before presentations of their infections. It is very difficult to determine whether the isolates from the latter three patients were community strains or colonized on these patients at the time of their previous hospitalization. However, even if these isolates were originally from this hospital, it is very likely that CTX-M-3 and CMY-2 have been disseminated to the community environment in this area because these patients had been discharged for months to years. Isolation of a CMY-2-producing strain from a patient with no history of hospitalization further supports the speculation. Although no evidence of nosocomial outbreak caused by CTX-M-3- or CMY2-producing E. coli strains was found over a 9-month period in this hospital, the fact that the genes encoding these two β-lactamases may disseminate to other members of the family Enterobacteriaceae and the possibility that CTX-M-3 producers could be maintained in hospitals for a long time necessitate close monitoring of these two enzymes among clinical isolates of Enterobacteriaceae in this hospital to prevent such an occurrence (13, 32).
With the NCCLS confirmatory test, the isolate producing both CTX-M-3 and CMY-2 showed a <5-mm increase in a zone diameter for either ceftazidime or cefotaxime in combination with clavulanic acid versus its zone when tested alone. According to the NCCLS criteria (28), the isolate initially was not regarded as an ESBL producer. CMY-2, which was not susceptible to the inhibition of clavulanic acid (4), was possibly responsible to the negative result. The targets of the NCCLS confirmatory test are ESBLs that are susceptible to inhibition by β-lactam inhibitors. There is no mention of testing or reporting results for AmpC hyperproducers or the isolates that produce both ESBLs and AmpC enzymes. The presentation of our case suggests that classical ESBLs, when they are coexistent with AmpC enzymes, are difficult to detect with the susceptibility testing methods used by routine clinical microbiology laboratories. However, since AmpC enzymes could also confer resistance to oxyiminocephalosporins, the detection of classical ESBLs from AmpC hyperproducers might not provide more help than susceptibility results for clinicians in the selection of effective antimicrobial agents. Thus, rather than reporting mechanisms of resistance, modification of cephalosporin results on laboratory reports as recommended by Tenover et al. (39) should increase the accuracy of the susceptibility test reporting in this case.
Timely identification of ESBL producers and early use of appropriate antibiotic regimens are important in the successful treatment of bloodstream infections with ESBL producers (37). Imipenem-containing regimens have been shown to yield the most favorable results (37). Without appropriate antibiotic treatment, both patients with bacteremia caused by CTX-M-3 producers died of sepsis. On the other hand, the patient with bacteremia caused by a CMY-2 producer survived after treatment with an imipenem-containing regimen. These findings emphasize the importance of early detection of ESBLs and appropriate antibiotic treatment in such patients. Further case control studies that include more patients are needed in order to determine the clinical impacts of these two enzymes and appropriate treatment regimens.
In conclusion, CTX-M-3 and CYM-2, two β-lactamases initially found in Europe, have been disseminated to and are prevalent among clinical isolates of E. coli in Taiwan. Dissemination of resistance plasmids is responsible for the spread of these two enzymes in southern Taiwan. More importantly, these two enzymes might have spread to the community environment in this area.
ACKNOWLEDGMENTS
We kindly thank J. Kim, Dankook University, Korea, for provision of a K. pneumoniae strain carrying CMY-1.
This work was partially supported by grants NCKUH89-054 from National Cheng Kung University Hospital and NSC89-2314-B-006-031 from the National Science Council, Republic of China.
REFERENCES
- 1.Bachmann B J, Low K B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980;44:1451–1456. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas J M. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Stemplinger I, Jungwirth R, Wilhelm R, Chong Y. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other β-lactamase genes. Antimicrob Agents Chemother. 1996;40:1926–1930. doi: 10.1128/aac.40.8.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauernfeind A, Stemplinger I, Jungwirth R, Giamarellou H. Characterization of the plasmidic β-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob Agents Chemother. 1996;40:221–224. doi: 10.1128/aac.40.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauernfeind A, Grimm H, Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 1990;18:294–298. doi: 10.1007/BF01647010. [DOI] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caroff N, Espaze E, Berard I, Richet H, Reynaud A. Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum β-lactamase production. FEMS Microbiol Lett. 1999;173:459–465. doi: 10.1111/j.1574-6968.1999.tb13539.x. [DOI] [PubMed] [Google Scholar]
- 8.Chu G, Vollrath D, Davis R W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986;234:1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- 9.Danel F, Hall L M C, Gur D, Livermore D M. OXA-15, an extended-spectrum variant of OXA-2 β-lactamase, isolated from a Pseudomonas aeruginosa strain. Antimicrob Agents Chemother. 1997;41:785–790. doi: 10.1128/aac.41.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farmer J J., III . Enterobacteriaceae: introduction and identification. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 438–449. [Google Scholar]
- 11.Gazouli M, Tzelepi E, Sidorenko S V, Tzouvelekis L S. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A β-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob Agents Chemother. 1998;42:1259–1262. doi: 10.1128/aac.42.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazouli M, Tzouvelekis L S, Vatopoulos A C, Tzelepi E. Transferable class C β-lactamases in Escherichia coli strains isolated in Greek hospitals and characterization of two enzyme variants (LAT-3 and LAT-4) closely related to Citrobacter freundii AmpC β-lactamase. J Antimicrob Chemother. 1998;42:419–425. doi: 10.1093/jac/42.4.419. [DOI] [PubMed] [Google Scholar]
- 13.Gniadkowski M, Schneider I, Palucha A, Jungwirth R, Mikiewicz B, Bauernfeind A. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamases that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob Agents Chemother. 1998;42:827–832. doi: 10.1128/aac.42.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haruta S, Nukaga M, Taniguchi K, Sawai T. Resistance to oxyimino β-lactams due to a mutation of chromosomal β-lactamase in Citrobacter freundii. Microbiol Immunol. 1998;42:165–169. doi: 10.1111/j.1348-0421.1998.tb02267.x. [DOI] [PubMed] [Google Scholar]
- 15.Horii T, Arakawa Y, Ohta M, Sugiyama T, Wacharotayankun R, Ito H, Kato N. Characterization of a plasmid-borne and constitutively expressed blaMOX-1 gene encoding AmpC-type β-lactamase. Gene. 1994;139:93–98. doi: 10.1016/0378-1119(94)90529-0. [DOI] [PubMed] [Google Scholar]
- 16.Ishii Y, Ohno A, Taguchi H, Imajo S, Ishiguro M, Matsuzawa H. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob Agents Chemother. 1995;39:2269–2275. doi: 10.1128/aac.39.10.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacoby G A, Han P. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996;34:908–911. doi: 10.1128/jcm.34.4.908-911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaffe A, Chabbert Y A, Derlot E. Selection and characterization of β-lactam-resistant Escherichia coli K-12 mutants. Antimicrob Agents Chemother. 1983;23:622–625. doi: 10.1128/aac.23.4.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarlier V, Nicolas M-H, Fournier G, Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 20.Kawakami S, Ono Y, Yamamoto M, Matumura M, Okamoto R, Inoue M, Miyazawa Y. Extended-spectrum β-lactamase (ESBL) produced by Escherichia coli and Klebsiella pneumoniae isolated from Teikyo University Hospital-the second report. Kansenshogaku Zasshi. 2000;74:24–29. doi: 10.11150/kansenshogakuzasshi1970.74.24. [DOI] [PubMed] [Google Scholar]
- 21.Lewis M T, Yamaguchi K, Biedenbach D J, Jones R N. In vitro evaluation of cefepime and other broad-spectrum β-lactams in 22 medical centers in Japan: a phase II trial comparing two annual organism samples. The Japan Antimicrobial Resistance Study Group. Diagn Microbiol Infect Dis. 1999;35:307–315. doi: 10.1016/s0732-8893(99)00120-0. [DOI] [PubMed] [Google Scholar]
- 22.Liu P Y F, Tung J C, Ke S C, Chen S L. Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a district hospital in Taiwan. J Clin Microbiol. 1998;36:2759–2762. doi: 10.1128/jcm.36.9.2759-2762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livermore D M, Yuan M. Antibiotic resistance and production of extended-spectrum β-lactamases amongst Klebsiella spp. from intensive care units in Europe. J Antimicrob Chemother. 1996;38:409–424. doi: 10.1093/jac/38.3.409. [DOI] [PubMed] [Google Scholar]
- 24.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;34:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma L, Ishii Y, Ishiguro M, Matsuzawa H, Yamaguchi K. Cloning and sequencing of the gene encoding Toho-2, a class A β-lactamase preferentially inhibited by tazobactam. Antimicrob Agents Chemother. 1998;42:1181–1186. doi: 10.1128/aac.42.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mabilat C, Goussard S. PCR detection and identification of genes for extended-spectrum β-lactamases. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 553–559. [Google Scholar]
- 27.Matthew M, Harris M, Marshall M J, Rose G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Ninth informational supplement. M100–S9. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 29.Nordmann P. Trends in β-lactam resistance among Enterobacteriaceae. Clin Infect Dis. 1998;27(Suppl. 1):S100–S106. doi: 10.1086/514905. [DOI] [PubMed] [Google Scholar]
- 30.Nüesch-Inderbinen M T, Hächler H, Kayser F H. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur J Clin Microbiol Infect Dis. 1996;15:398–402. doi: 10.1007/BF01690097. [DOI] [PubMed] [Google Scholar]
- 31.Pai H, Lyu S, Lee J H, Kim J, Kwon Y, Kim J, Choe K W. Survey of extended-spectrum β-lactamases in clinical isolates of Escherichia coli and Klebsiella pneumoniae: prevalence of TEM-52 in Korea. J Clin Microbiol. 1999;37:1758–1763. doi: 10.1128/jcm.37.6.1758-1763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palucha A, Mikiewicz B, Hryniewicz W, Gniadkowski M. Concurrent outbreaks of extended-spectrum β-lactamase-producing organisms of the family Enterobacteriaceae in a Warsaw hospital. J Antimicrob Chemother. 1999;44:489–499. doi: 10.1093/jac/44.4.489. [DOI] [PubMed] [Google Scholar]
- 33.Papanicolaou G A, Medeiros A A, Jacoby G A. Novel plasmid-mediated β-lactamase (MIR-1) conferring resistance to oxyimino- and α-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990;34:2200–2209. doi: 10.1128/aac.34.11.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philippon A, Arlet G, Lagrange P H. Origin and impact of plasmid-mediated extended-spectrum beta-lactamases. Eur J Clin Microbiol Infect Dis. 1994;13(Suppl. 1):S17–S29. doi: 10.1007/BF02390681. [DOI] [PubMed] [Google Scholar]
- 35.Piggot P J, Amjad M, Wu J J, Sandoval H, Castro J. Genetic and physical maps of Bacillus subtilis 168. In: Harwood C R, Cutting S M, editors. Molecular biology methods for bacillus. West Sussex, England: John Wiley & Sons, Ltd.; 1990. pp. 493–543. [Google Scholar]
- 36.Provence D L, Curtiss R., III . Gene transfer in gram-negative bacteria. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 319–347. [Google Scholar]
- 37.Schiappa D A, Hayden M K, Matushek M G, Hashemi F N, Sullivan J, Smith K Y, Miyashiro D, Quinn J P, Weinstein R A, Trenholme G M. Ceftazidime-resistant Klebsiella pneumoniae and Escherichia coli bloodstream infection: a case-control and molecular epidemiologic investigation. J Infect Dis. 1996;174:529–536. doi: 10.1093/infdis/174.3.529. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi S, Nagano Y. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J Clin Microbiol. 1984;20:608–613. doi: 10.1128/jcm.20.4.608-613.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenover F C, Mohammed M J, Gorton T S, Dembek Z F. Detection and reporting of organisms producing extended-spectrum β-lactamases: survey of laboratories in Connecticut. J Clin Microbiol. 1999;37:4065–4070. doi: 10.1128/jcm.37.12.4065-4070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzouvelekis L S, Tzelepi E, Mentis A F. Nucleotide sequence of a plasmid-mediated cephalosporinase gene (blaLAT-1) found in Klebsiella pneumoniae. Antimicrob Agents Chemother. 1994;38:2207–2209. doi: 10.1128/aac.38.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan J J, Wu S M, Tsai S H, Wu J J, Su I J. Prevalence of SHV-12 among clinical isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamase and identification of a novel AmpC enzyme (CMY-8) in southern Taiwan. Antimicrob Agents Chemother. 2000;44:1438–1442. doi: 10.1128/aac.44.6.1438-1442.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]