Highlights
-
•
The Legionella genus contains nine core effectors.
-
•
Three Legionella pneumophila core effectors are required for intracellular growth.
-
•
The Legionella genus core effectors display functional conservation among orthologs.
-
•
One Legionella core effector requires an accessory protein to perform its function.
Keywords: Legionella, Icm; Dot; T4SS; Effectors; Legionella, Icm/Dot
Abstract
The intracellular pathogen Legionella pneumophila, as well as other Legionella species, utilize the Icm/Dot type-IV secretion system to translocate an exceptionally large and diverse repertoire of effectors into their host cells. However, only nine core effectors were found to be present in all analyzed Legionella species. In this study, we investigated the core effectors, and used intracellular growth complementation to determine whether orthologs of core effectors perform the same function in different Legionella species. We found that three out of the nine L. pneumophila core effectors are required for maximal intracellular growth. Examination of orthologous core effectors from four Legionella species spread over the Legionella phylogenetic tree revealed that most of them perform the same function. Nevertheless, some of the orthologs of the core effector LegA3 did not complement the L. pneumophila legA3 deletion mutant for intracellular growth. LegA3 is encoded as part of an operon together with another gene, which we named legA3C, encoding a non-translocated protein. We found that LegA3 and LegA3C physically interact with each other, are both required for maximal intracellular growth, and the LegA3-LegA3C orthologous pairs from all the Legionella species examined fully complement the L. pneumophila legA3 deletion mutant for intracellular growth. Our results indicate that the Legionella core effectors orthologs generally perform the same function and establish that LegA3 requires LegA3C to fulfill its conserved function.
Graphical abstract
1. Introduction
Legionella pneumophila and other Legionella species grow intracellularly in human macrophages and amoebae (Moliner et al., 2010; Boamah et al., 2017; Burillo et al., 2017). To establish a replicative niche inside eukaryotic cells, Legionella modulate host-cell functions using the Icm/Dot type-IV secretion system (Segal et al., 1998; Vogel et al., 1998). The Icm/Dot system is essential for L. pneumophila pathogenesis, since it delivers effector proteins into the host cells and these effectors directly modulate various host cell processes for the benefit of the bacteria (reviewed in (Isberg et al., 2009; Hubber and Roy, 2010; Gomez-Valero et al., 2011; Xu and Luo, 2013; Qiu and Luo, 2017)). Previously, we and others have determined the complete genomic sequence of numerous Legionella species (Burstein et al., 2016; Gomez-Valero et al., 2019). These analyses indicated that the Icm/Dot components, encoded by 25 genes organized in two separate genomic regions, are highly conserved across the Legionella genus (Burstein et al., 2016; Gomez-Valero et al., 2019). The majority of the Icm/Dot proteins constitute the membrane complex through which the effectors are translocated into host cells (Ghosal et al., 2017; Chetrit et al., 2018, 2019; Park et al., 2020a) and several cytoplasmic components were also identified (Coers et al., 2000; Feldman et al., 2005).
In contrast to the high conservation of the Icm/Dot secretion system components, genomic sequencing studies have demonstrated a high degree of plasticity in the effector repertoires encoded by different Legionella species (Zusman et al., 2008; Ninio et al., 2009; Burstein et al., 2016; Gomez-Valero et al., 2019). Studies indicate that thousands of effectors are present in the Legionella genus, and to date, over 300 Icm/Dot effectors have been validated in the most pathogenic and the best studied Legionella species, L. pneumophila (Burstein et al., 2009; Huang et al., 2011; Zhu et al., 2011; Lifshitz et al., 2013). The predicted effector repertoires of different Legionella species were found to be largely non-overlapping and only seven validated and two putative core effectors were found to be shared by all species studied (Burstein et al., 2016; Gomez-Valero et al., 2019). In the light of the tremendous number of effectors found in the Legionella genus, the identification of so few core effectors was surprising. Analysis of this group of effectors indicated that their phylogenetic tree is in good agreement with the species tree, they are highly similar at the protein level and their GC content is similar to the average genomic GC content of each species (Burstein et al., 2016; Gomez-Valero et al., 2019). Together, these findings suggest that the core effectors evolved as part of the Legionella genus for an extended period of time.
To date, the function of two of the core effectors was uncovered. The L. pneumophila LegA3/AnkH - lpg2300 is the only core effector that was found to be present in all the bacteria known to harbor an Icm/Dot secretion system which include all the Legionella species examined, Coxiella burnetii (a human pathogen, and a potential bioterrorism agent (Azad, 2007)), Rickettsiella grylli and Diplorickettsia massiliensis (both are pathogens of arthropods (Leclerque and Kleespies, 2008; Mathew et al., 2012)). The LegA3 effector contains a single ankyrin repeat at its N-terminus and mutants lacking the legA3 gene are impaired for intracellular growth in several hosts (O'Connor et al., 2012; Burstein et al., 2016; Shames et al., 2017; Park et al., 2020b). More recently, the LegA3 core effector was shown to interact with the host LARP7 component of the 7SK snRNP complex leading to host global transcriptional reprogramming (Von Dwingelo et al., 2019). Another L. pneumophila core effector, MavN/IroT - lpg2815, was found in all the Legionella species examined and in R. grylli. Mutants lacking the mavN gene are impaired for intracellular growth in several hosts (Isaac et al., 2015; Portier et al., 2015; Burstein et al., 2016; Shames et al., 2017; Park et al., 2020b). MavN is localized to the Legionella-containing vacuole (LCV), its transcription is strongly induced under iron-restricted conditions and it is required for efficient iron acquisition during intracellular growth, as well as for the acquisition of other transition-metal-ions (Isaac et al., 2015; Portier et al., 2015; Christenson et al., 2019).
The information regarding the five other validated core effectors (VipF - lpg0103, RavC - lpg0107, CetLp1 - lpg0140, lpg2832 and lpg3000) and the two putative core effectors (lpg0086 and lpg1356) is limited. Most of these core effectors are not required for intracellular growth in host cells (Burstein et al., 2016; Shames et al., 2017; Park et al., 2020b). Lpg3000 is required for intracellular growth in Hartmannella vermiformis, and lpg0086 is required for intracellular growth in several host cells (Shames et al., 2017; Park et al., 2020b). In addition, most of these effectors contain no known protein sequence domains, except for lpg1356, which harbors multiple Sel1 repeats and VipF, which contains an acetyltransferase domain and might be involved in membrane trafficking (Shohdy et al., 2005; Young et al., 2016).
The goal of this study was to investigate the functional conservation of core effectors and bacterial proteins they might interact with. To this end, we examined core effectors orthologs from four Legionella species spread over the Legionella phylogenic tree for complementation of the corresponding L. pneumophila deletion mutants. We examined intracellular growth complementation mediated by core effectors orthologs. Our results reveal that most of the core effectors orthologs investigated function similarly as their L. pneumophila ortholog. Interestingly, we also found that the widely conserved core effector, LegA3, requires an adjacent non-translocated protein in order to mediate its conserved function during infection.
2. Results
2.1. The Legionella genus contains nine core effectors
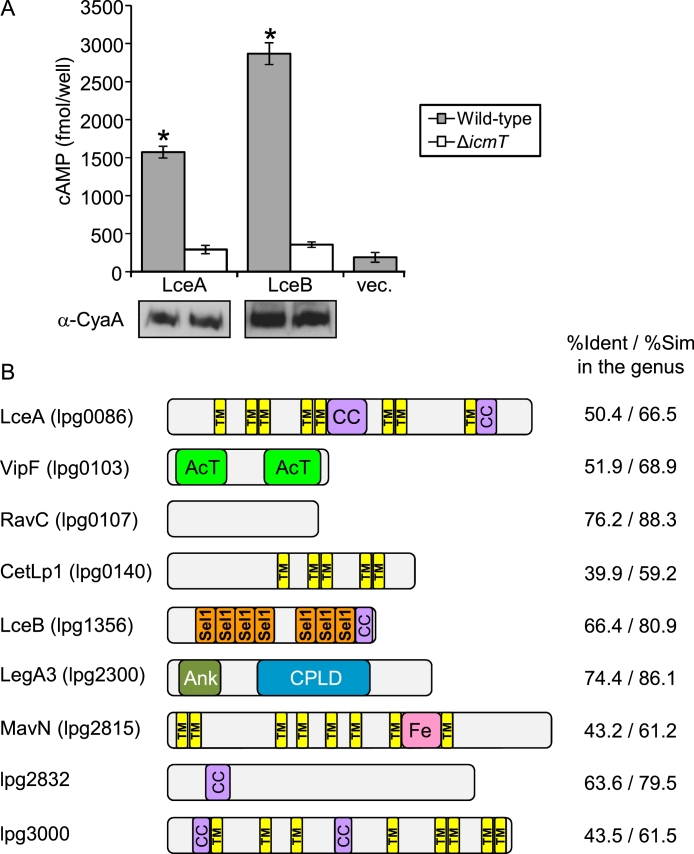
Previous analysis of more than 300 validated L. pneumophila effectors in 41 Legionella species revealed that only seven of these effectors are present in all the Legionella species examined and these effectors were designated “core effectors” (Burstein et al., 2016). Later, examination in 18 additional Legionella species indicated that these seven core effectors are present in all 59 Legionella species examined (Gomez-Valero et al., 2019). Beside these seven validated core effectors, two additional putative core effectors are also present in all 59 Legionella species for which the genomic sequence is available (see Dataset S1) and these putative core effectors harbor features similar to the ones found in validated effector proteins (Burstein et al., 2009). Examination of these two putative core effectors for translocation into host cells revealed that they are translocated into host cells in an Icm/Dot dependent manner (Fig. 1A), thus increasing the number of validated core effectors in the Legionella genus to nine. We named the two core effectors newly validated in this study LceA (lpg0086) and LceB (lpg1356) for Legionellaconserved effectors.
Fig. 1.
The Legionella genus harbors nine core effectors. A. The L. pneumophila wild-type strain JR32 (gray bars) and the icmT deletion mutant GS3011 (white bars) harboring the CyaA fusion proteins (indicated below each bar) were used to infect HL-60-derived human macrophages, and the cAMP levels of the infected cells were determined. Vector control is indicated as “vec.”. The bar heights represent the mean amounts of cAMP per well obtained in at least three independent experiments; error bars indicate standard deviations. The cAMP levels of each fusion were found to be significantly different (*, P < 0.01, Student's t-test) between the wild-type strain and the icmT deletion mutant. The effectors were examined by Western blot analysis for their expression in the wild-type strain (left) and the icmT deletion mutant (right) using an anti-CyaA antibody. B. Domain architecture of L. pneumophila core effectors orthologs. The known domains of each effector are shown. The domains presented are as follows: AcT - N-acetyltransferase; Ank, ankyrin repeat; Sel1 – Sel1 repeat, which represents a subfamily of tetratricopeptide repeat (TPR); Fe – iron binding domain (Isaac et al., 2015); CPLD - cysteine protease-like domain (Von Dwingelo et al., 2019); CC – coiled-coil region (determined using Paircoil (Berger et al., 1995)); TM – transmembrane domains (determined using TOPCONS (Tsirigos et al., 2015)). The average percent identity and similarity (%Ident /%Sim) were determined among the orthologs in all the 59 Legionella species using BLAST all-against-all.
2.2. Analysis of the Legionella genus core effectors
To gain some insights regarding these nine core effectors, we examined their degree of conservation in the genus and their domains organization (Fig. 1B). Four of these core effectors (LceA, CetLp1, MavN and lpg3000) contain multiple transmembrane domains and are the least conserved proteins among the core effectors (Fig. 1B, Dataset S2). The other five core effectors (VipF, RavC, LceB, LegA3 and lpg2832), which harbor no transmembrane domains, are more conserved (Dataset S2) and three of them contain known domains (VipF contains an acetyltransferase domain, LceB harbors multiple Sel1 repeats and LegA3 contains an ankyrin domain and a cysteine protease-like domain, Fig. 1B). The most conserved core effector (RavC) contains no transmembrane domains or any other known domains.
As indicated above, four out of the nine core effectors harbor multiple (more than four) transmembrane domains (Fig. 1B). This observation led us to examine all the L. pneumophila validated effectors (Dataset S3) for the presence of multiple transmembrane domains using the TOPCONS program (Tsirigos et al., 2015) and the result of this analysis is presented in Table 1. Altogether, 16 validated L. pneumophila effectors harbor multiple predicted transmembrane domains (Table 1). This observation suggests that effectors containing multiple transmembrane domains are enriched among core effectors since 4 out of 9 core effectors harbor these domains, whereas only 12 out of all the other effectors (300 effectors) are predicted to harbor multiple transmembrane domains. The enrichment of effectors harboring multiple predicted transmembrane domains among core effectors might suggest that these effectors perform important functions during infection and once acquired (or evolved de novo) they are kept in the bacteria.
Table 1.
Legionella pneumophila effectors harboring multiple transmembrane domains.
| Lpg# | Name | Size (AA) | TMsa | Sp.b | Domainsc | Int.d | Known functione. |
|---|---|---|---|---|---|---|---|
| lpg1661 | – | 372 | 10 | 54 | AcT | ||
| lpg0086 | LceA | 648 | 8 | 59 | CC | + | |
| lpg2552 | LecE | 555 | 8 | 27 | Activates Pah1 | ||
| lpg2815 | MavN | 683 | 8 | 59 | + | Iron acquisition | |
| lpg2888 | – | 637 | 8 | 54 | CC | + | |
| lpg3000 | – | 612 | 8 | 59 | CC | + | |
| lpg1822 | CetLp4 | 339 | 6 | 3 | CC | + | |
| lpg0059 | Ceg2 | 368 | 5 | 2 | |||
| lpg0140 | CetLp1 | 444 | 5 | 59 | |||
| lpg0716 | LciE | 337 | 5 | 6 | CC | Activated by Cu | |
| lpg2806 | CetLp7 | 460 | 5 | 22 | CC | ||
| lpg2884 | MavP | 245 | 5 | 27 | |||
| lpg0621 | SidA | 474 | 4 | 21 | CC | ||
| lpg2164 | LecC | 154 | 4 | 21 | |||
| lpg2804 | Lem29 | 468 | 4 | 34 | CC | ||
| lpg2885 | – | 184 | 4 | 9 | VGCC |
TMs – transmembrane domains, as was determined using TOPCONS (Tsirigos et al., 2015).
Number of Legionella species in which the effectors were found, the maximal number of Legionella species for which the genomic sequence was available was 59, four of the effectors listed are core effectors present in all 59 species (underlined).
The domains identified were: CC - Coiled-Coil regions; AcT – Acyltransferase; VGCC - Voltage gated chloride channel.
Requirement for intracellular growth was determined in this study and in (Burstein et al., 2016; Shames et al., 2017; Park et al., 2020b).
The function of LecE was determined by (Viner et al., 2012), the function of MavN was determined by (Isaac et al., 2015; Portier et al., 2015) and the copper activation of LciE was determined by (Linsky et al., 2020).
In line with previous analyses indicating that effectors are often located in hypervariable genomic regions (Zusman et al., 2008; Ninio et al., 2009), we were interested to examine whether the nine core effectors are located in conserved genomic regions having a similar synteny between Legionella species. Examination of the genomic regions encoding the core effectors revealed that their genomic regions are conserved in the Legionella genus (Fig. S1). This observation suggests that besides being conserved in the whole Legionella genus, the genomic regions of the genes encoding the Legionella core effectors are also conserved. These genomic regions might be less prone to change and thus contributing to the fact that the core effectors were not lost during evolution. However, not all the effectors located in these conserved genomic regions are core effectors (for example ylfA and legP, Fig. S1), suggesting the core effectors were kept in all the Legionella species due to the importance of their functions and not only due to their location in relatively conserved genomic regions.
2.3. Three L. pneumophila core effectors are required for maximal intracellular growth in Acanthamoeba castellanii
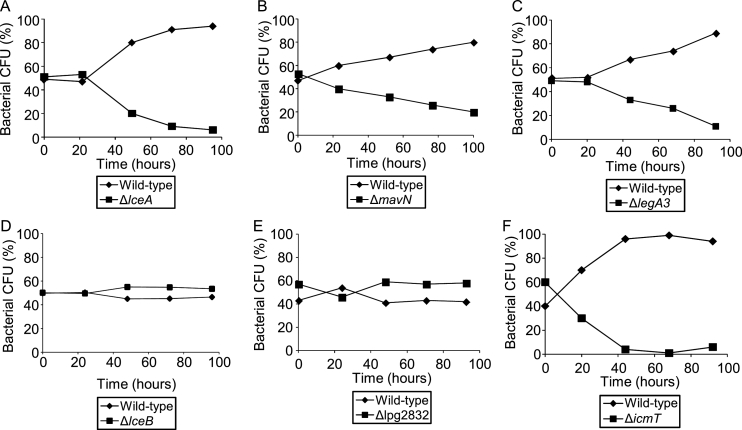
We aimed to determine whether core effectors orthologs from different Legionella species perform the same function as the L. pneumophila core effectors orthologs. To that end, we studied the intracellular growth of deletion mutants in all the nine core effectors using a competition assay with A. castellanii as a host. Effector deletion mutants usually show weak intracellular growth phenotypes (O'Connor et al., 2012; Park et al., 2020b), and the same was found with deletion mutants of core effectors (Burstein et al., 2016). Therefore, we used a well-established intracellular competition assay, which is more sensitive than regular intracellular growth assays (Kessler et al., 2013; Dolinsky et al., 2014; Feldheim et al., 2016; Goodwin et al., 2016), to analyze the core effectors deletion mutants. Three of the L. pneumophila core effectors (LceA, MavN and LegA3) were found to have a clear intracellular growth phenotype (Fig. 2) and the other six core-effectors grew similarly to the wild-type strain (Fig. 2 and Fig. S2). The three L. pneumophila core effectors orthologs with an intracellular growth phenotype were examined for functional conservation of their orthologs in the Legionella genus, as described below.
Fig. 2.
Three L. pneumophila core effectors are required for maximal intracellular growth in the environmental host A. castellanii. Intracellular competition assay in A. castellanii between L. pneumophila wild type strain JR32 and the lceA deletion mutant (A); the mavN deletion mutant (B); the legA3 deletion mutant (C) the lceB deletion mutant (D); the lpg2832 deletion mutant (E) and the icmT deletion mutant as a negative control (F). CFU – colony forming units. The data shown are representative of three independent experiments.
2.4. Most of the LceA and MavN orthologs perform the same function
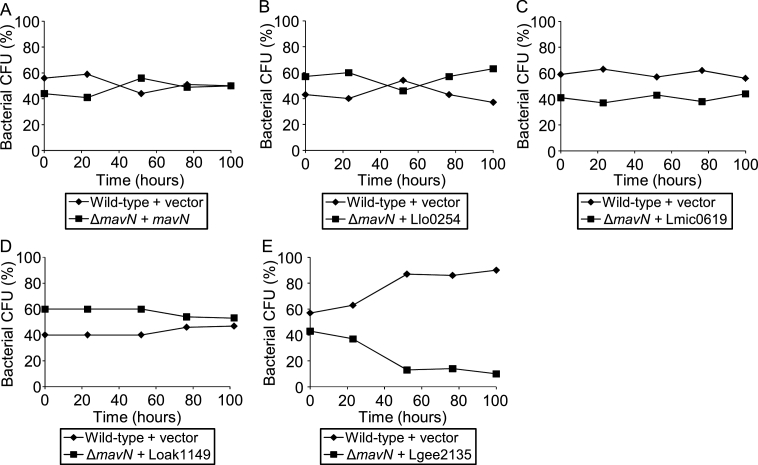
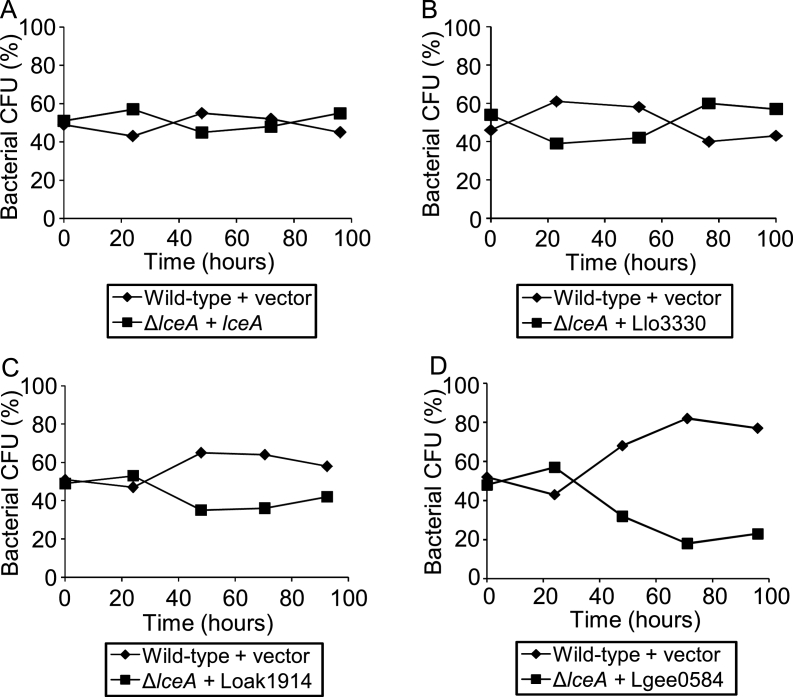
To determine whether the function of the core effectors required for intracellular growth is conserved in the Legionella genus, the LceA and MavN orthologs from four Legionella species were chosen as representatives of the major clades of the Legionella phylogenetic tree (Burstein et al., 2016): L. longbeachae from the clade which includes L. pneumophila; L. micdadei from the second major clade; L. oakridgensis from the deep-branching clade and L. geestiana as a deep-rooting Legionella species. The lceA and mavN genes from L. pneumophila, as well as their orthologs from the four Legionella species indicated above, were cloned under Ptac control (induced by IPTG) and introduced into the L. pneumophila lceA and mavN deletion mutants. Complementation analysis using competition assays clearly indicated that the L. longbeachae and L. oakridgensis orthologs of LceA and MavN, as well as the L. micdadei ortholog of MavN (we were unable to clone the L. micdadei lceA ortholog), complemented the intracellular growth phenotype of the L. pneumophila corresponding deletion mutants (Fig. 3A-D and Fig. 4A-C). However, for both effectors, no complementation was obtained with the orthologs from the deep-rooting, evolutionarily distant, species L. geestiana (Fig. 3E and Fig. 4D), this result might occur due to expression problem of these proteins in L. pneumophila or due to change in their function (see Discussion).
Fig. 3.
Most of the MavN orthologs complemented the L. pneumophila mavN deletion mutant for intracellular growth. Intracellular competition assay in A. castellanii between L. pneumophila wild type strain JR32 containing a vector and the mavN deletion mutant complemented with: the L. pneumophila MavN (A); the L. longbeachae MavN ortholog Llo0254 (B); the L. micdadeii MavN ortholog Lmic0619 (C); the L. oakridgensis MavN ortholog Loak1149 (D) and the L. geestiana MavN ortholog Lgee2135 (E). All MavN orthologs were introduced on a plasmid under Ptac control in the same way as the L. pneumophila mavN gene. CFU – colony forming units. The data shown are representative of three independent experiments.
Fig. 4.
Most of the LceA orthologs complemented the L. pneumophila lceA deletion mutant for intracellular growth. Intracellular competition assay in A. castellanii between L. pneumophila wild type strain JR32 containing a vector and the lceA deletion mutant complemented with: the L. pneumophila LceA (A); the L. longbeachae LceA ortholog Llo3330 (B); the L. oakridgensis LceA ortholog Loak1914 (C) and the L. geestiana LceA ortholog Lgee0584 (D). All LceA orthologs were introduced on a plasmid under Ptac control in the same way like the L. pneumophila lceA gene. CFU – colony forming units. The data shown are representative of three independent experiments.
Collectively, these results indicate that excluding the L. geestiana orthologs, the LceA and MavN orthologs examined perform the same function during infection as their L. pneumophila orthologs.
2.5. Some LegA3 orthologs do not complement the L. pneumophila legA3 deletion mutant
The LegA3 core effector is unique since it is present in all sequenced bacteria harboring an Icm/Dot type-IV secretion system, including C. burnetii, R. grylli and D. massiliensis. The L. pneumophila LegA3 is 467 amino acids long and it contains a single ankyrin domain at its N-terminus and a cysteine protease-like domain (Fig. 1B).
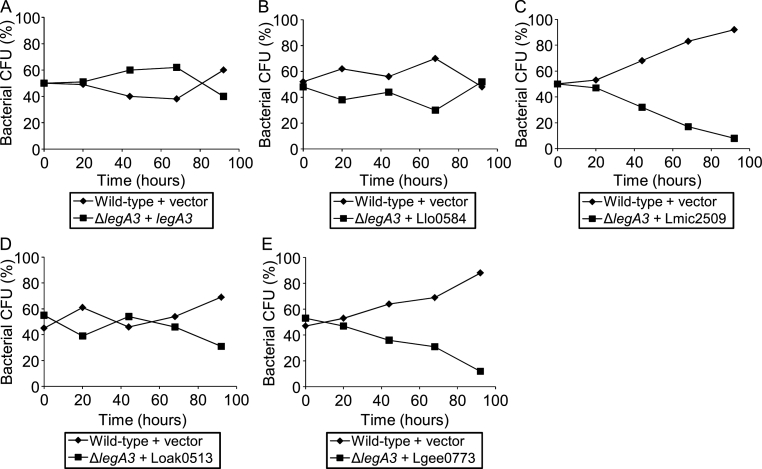
To examine whether LegA3 orthologs perform the same function, the legA3 orthologs from the four Legionella species indicated above were cloned under Ptac control and introduced into the L. pneumophila legA3 deletion mutant. The results obtained from this analysis were unexpected (Fig. 5): the L. longbeachae and L. oakridgensis orthologs of LegA3 complemented the intracellular growth phenotype of the L. pneumophila legA3 deletion mutant (Fig. 5B and D). However, no complementation was obtained with the L. micdadei and L. geestiana orthologs of LegA3 (Fig. 5C and E). This result was surprising since the L. longbeachae, L. micdadei and L. oakridgensis orthologs of LegA3 are highly homologous to the L. pneumophila LegA3 (Dataset S2), yet the L. micdadei ortholog did not complement the intracellular growth phenotype of the L. pneumophila legA3 deletion mutant. This might indicate that either some of the orthologs of the LegA3 core effector have changed their function during evolution, or that another factor is required for the LegA3 orthologs in order to function in L. pneumophila.
Fig. 5.
Intracellular growth complementation of the L. pneumophila legA3 deletion mutant by different LegA3 orthologs. Intracellular competition assay in A. castellanii between L. pneumophila wild type strain JR32 containing a vector and the legA3 deletion mutant complemented with: the L. pneumophila LegA3 (A); the L. longbeachae LegA3 ortholog Llo0584 (B); the L. micdadeii LegA3 ortholog Lmic2509 (C); the L. oakridgensis LegA3 ortholog Loak0513 (D) and the L. geestiana LegA3 ortholog Lgee0773 (E). All LegA3 orthologs were introduced on a plasmid under Ptac control in the same way like the L. pneumophila legA3 gene. CFU – colony forming units. The data shown are representative of three independent experiments.
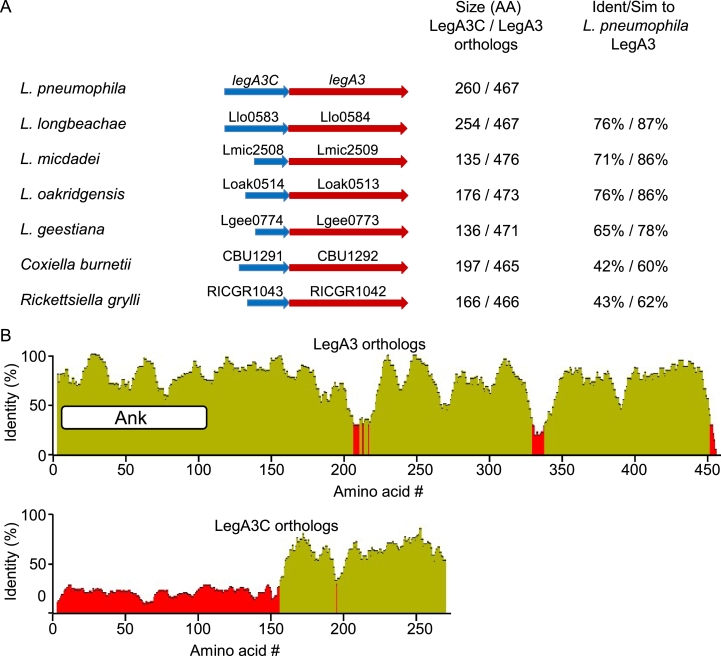
2.6. LegA3C is encoded from the same transcriptional unit as LegA3 and is conserved only at its C-terminus
The rather surprising results described above led us to examine the genomic region of legA3. We found that lpg2301 is located immediately upstream to legA3 (lpg2300), with no intergenic region between them. Additionally, it was previously shown that lpg2301 and legA3 form an operon containing a single transcription start site located upstream to lpg2301 (Sahr et al., 2012). A similar genomic organization of legA3 and lpg2301 orthologs like the one found in L. pneumophila was also found in all the Legionella species examined, as well as in C. burnetii, R. grylli and D. massiliensis (Fig. 6A). Therefore, we named lpg2301 LegA3C for LegA3companion. Unlike the LegA3 orthologs, which have a similar size in all of the species examined, the size of the LegA3C orthologs differs considerably: ranging from 135 to 260 amino acids (Fig. 6A). Examination of the protein sequences of the LegA3 and LegA3C revealed that LegA3 is highly conserved throughout its entire length, whereas LegA3C is conserved only at its C-terminus (Fig. 6B). In addition, the LegA3C C-terminus is less conserved in comparison to LegA3 (Fig. 6B). LegA3C was previously examined for translocation into host cells using the BlaM reporter system and it did not translocate (Zhu et al., 2011). We examined LegA3C for translocation using the CyaA reporter system and obtained the same result (Fig. S3). These results indicate that even though LegA3 is translocated into host cells and LegA3C is not, they are encoded from the same transcriptional unit and their function might be related.
Fig. 6.
Genomic organization of the legA3C-legA3 operon and similarity of the proteins it encodes. A. legA3C-legA3 operon organization in representative bacteria harboring the Icm/Dot secretion system. The size of the LegA3C and LegA3 orthologs and the degree of identity and similarity of the LegA3 orthologs to the L. pneumophila LegA3 are indicated on the right. B. Protein identity plot of Legionella LegA3 and LegA3C orthologs. The regions colored in red indicate low degree of identity. Ank indicates the location of the predicted ankyrin protein-protein binding domain. The sequence similarity analysis was performed using the Geneious software (https://www.geneious.com).
2.7. LegA3C, similarly to LegA3, is required for intracellular growth in amoeba
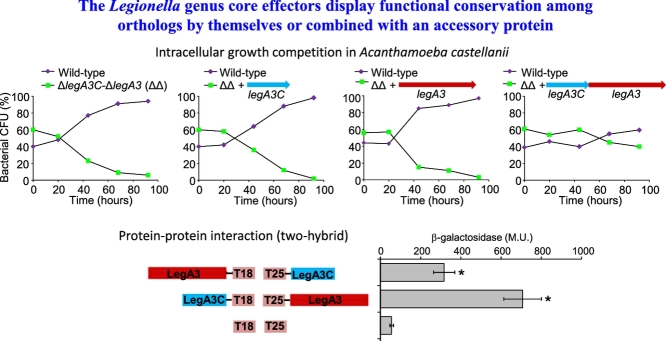
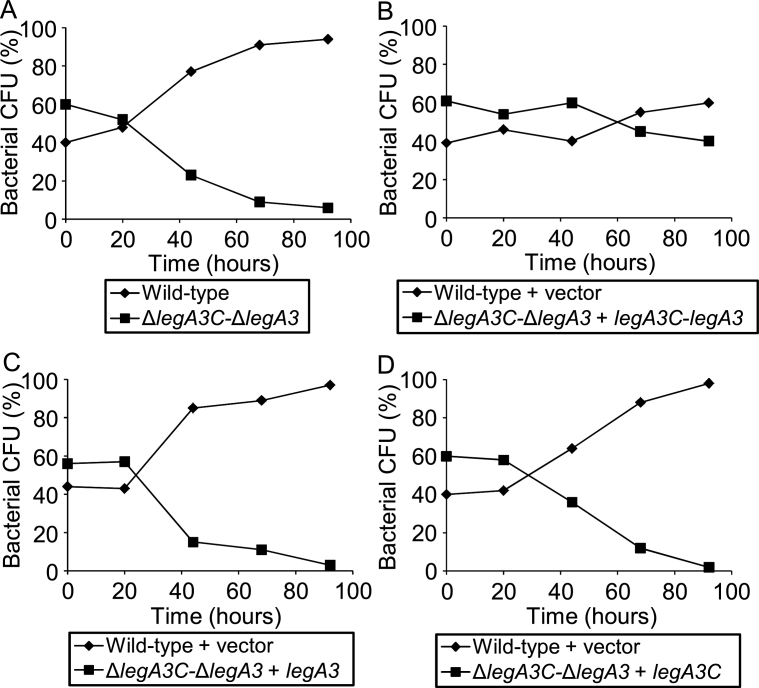
To determine if legA3C is required for intracellular growth, we generated a double deletion mutant of legA3C and legA3. This double deletion mutant was examined in a competition assay versus the wild type strain and was outcompeted after four days (Fig. 7A). The intracellular growth defect of the L. pneumophila legA3C-legA3 double deletion mutant was complemented when both legA3C and legA3 genes were introduced on a plasmid (Fig. 7B). However, the intracellular growth defect of the L. pneumophila legA3C-legA3 double deletion mutant was not complemented when each of the two genes (either legA3C or legA3) was introduced separately (Fig. 7CD), indicating that both legA3C and legA3 are required for intracellular growth in amoeba. This result suggests that LegA3C, even though not translocated into host cells, is required for L. pneumophila intracellular growth.
Fig. 7.
Both LegA3 and LegA3C are required for maximal intracellular growth of L. pneumophila. Intracellular competition assay in A. castellanii between L. pneumophila wild type strain JR32 (or the wild-type strain containing a vector in the complementation experiments) and the legA3C-legA3 double deletion mutant (A); the legA3C-legA3 double deletion mutant containing the legA3C-legA3 genes (B); the legA3C-legA3 double deletion mutant containing the legA3 gene by itself (C); and the legA3C-legA3 double deletion mutant containing the legA3C gene by itself (D). The legA3C-legA3 operon, the legA3C and legA3 genes were all introduced on a plasmid under Ptac control. CFU – colony forming units. The data shown are representative of three independent experiments.
2.8. The L. pneumophila LegA3 directly interacts with LegA3C
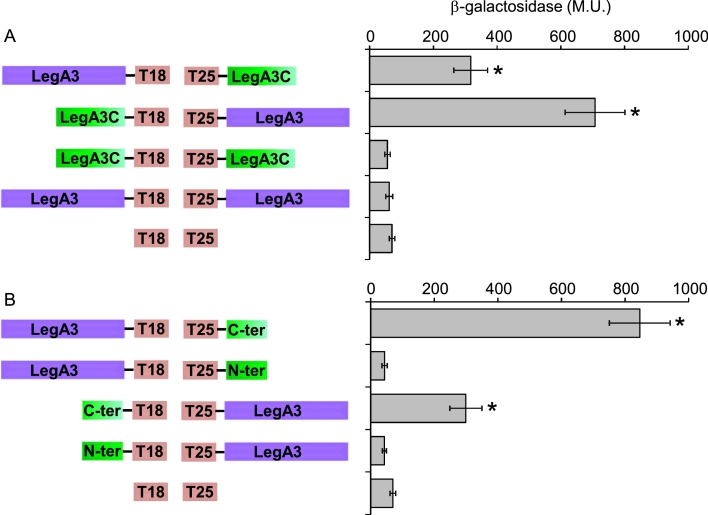
To further explore the connection between LegA3 and LegA3C, we examined whether LegA3C directly interacts with the LegA3 core effector using a bacterial two-hybrid system (Karimova et al., 1998). The results indicated that the non-translocated protein LegA3C and the LegA3 core effector directly interact with one another (Fig. 8A). Examination of self-interaction indicated that these two proteins do not self-interact (Fig. 8A) and no interaction was observed for any of the constructs with the vector controls (Fig. S4). As elaborated above, LegA3C can be divided into two regions, a non-conserved N-terminal domain and a relatively conserved C-terminal domain (Fig. 6). Examination of these two domains of LegA3C using the two-hybrid system indicates that the conserved C-terminal part of LegA3C is required and sufficient for interaction with LegA3, while the N-terminal part does not interact with LegA3 (Fig. 8B). These results indicate that the relatively conserved part of LegA3C interacts with LegA3 in the bacterial cell.
Fig. 8.
LegA3 and LegA3C interact with one another, via the C-terminal domain of LegA3C. Bacterial two-hybrid analysis of the L. pneumophila LegA3 and LegA3C interaction. The fusions of legA3 (purple) and legA3C (green) to the CyaA domains (T18 and T25) of the two-hybrid system are indicated on the left. Full-length fusions of LegA3 and LegA3C (A) and full-length fusions of LegA3 with LegA3C C-terminal and N-terminal fusions (C-ter and N-ter, respectively) (B) were examined. Data (expressed in Miller units [M.U.]) are the average ± standard deviations (error bars) of the results of at least three different experiments. The levels of interaction obtained between the different constructs were found to be significantly higher (*, P < 10−4, Student's t-test), when comparing to the background interaction of the two vectors control (T18 and T25). In addition, all the constructs when examined with a vector controls showed no interaction (Fig. S4).
2.9. The LegA3-LegA3C orthologous pairs complement the L. pneumophila legA3 deletion mutant
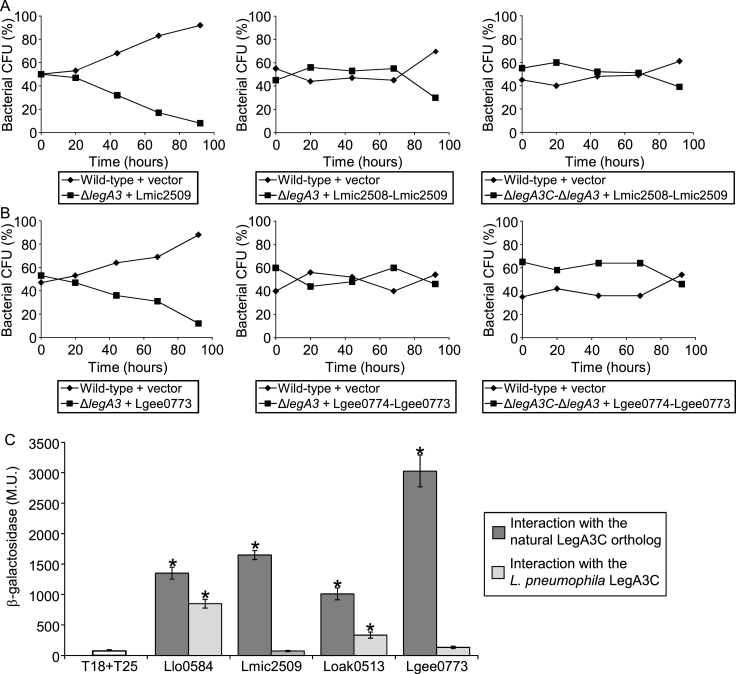
To further determine whether the LegA3C and LegA3 orthologs form functional pairs, we used the single legA3 deletion mutant and the legA3C-legA3 double deletion mutant, to examine the L. micdadei and L. geestiana legA3C-legA3 orthologous operons for complementation using the amoeba competition assay. These species were chosen since their LegA3 ortholog did not complement the L. pneumophila legA3 deletion mutant when introduced by itself (Fig. 5). When the L. micdadei or L. geestiana legA3C-legA3 orthologous operons (Lmic2508-Lmic2509 and Lgee0774-Lgee0773) were examined in both the legA3 single deletion mutant and in the legA3C-legA3 double deletion mutant, clear complementation was obtained (Fig. 9AB) and the complemented mutants were not outcompeted by the wild-type strain. These results indicate that in the case of L. micdadei and L. geestiana, the reason for the lack of complementation observed with their legA3 orthologs was the absence of their natural partners (the LegA3C orthologs), which probably function together with their corresponding LegA3 core effector orthologs.
Fig. 9.
The L. micdadeii and L. geestiana legA3C-legA3 orthologous operons complement the L. pneumophila legA3C-legA3 deletion mutant for intracellular growth. A. Intracellular competition assay in A. castellanii between L. pneumophila wild type strain JR32 containing a vector and the legA3 deletion mutant complemented with (from left to right): the L. micdadeii LegA3 ortholog Lmic2509; the legA3 deletion mutant complemented with the L. micdadeii legA3C-legA3 orthologous operon Lmic2508-Lmic2509 and the legA3C-legA3 double deletion mutant complemented with the L. micdadeii legA3C-legA3 orthologous operon Lmic2508-Lmic2509. B. Same as panel A but with the L. geestiana LegA3 ortholog Lgee0773 or the L. geestiana legA3C-legA3 orthologous operon Lgee0774-Lgee0773. CFU – colony forming units. The data shown are representative of three independent experiments (to ease the comparison, the data presented in Figs. 5C and 5E is presented here as well). C. Intraspecies and interspecies interaction between LegA3 and LegA3C orthologs. Bacterial two-hybrid analysis of interaction between natural pairs of LegA3 and LegA3C (dark gray bars) and interspecies interaction between the LegA3 ortholog from different bacteria and the L. pneumophila LegA3C (light gray bars). Data (expressed in Miller units [M.U.]) are the average ± standard deviations (error bars) of the results of at least three different experiments. The levels of interaction obtained between the different constructs were found to be significantly higher (*, P < 10−4, Student's t-test), when comparing the interaction between each pair and the two vectors control (T18 and T25).
2.10. Interaction of LegA3 and LegA3C is required for their function
To further understand the relations between the LegA3 and LegA3C orthologs present in the other Legionella species, we cloned both the LegA3 and LegA3C orthologs from the four Legionella species described above into the two-hybrid system. First, we examined the natural pairs of LegA3 and LegA3C orthologs for interaction with each other and all four pairs were found to interact (Fig. 9C). We hypothesized that the lack of complementation observed with the L. micdadei and L. geestiana LegA3 orthologs was due to a lack of interaction between them and the L. pneumophila LegA3C ortholog. We hence examined interspecies interaction using the L. pneumophila LegA3C and the L. micdadei and L. geestiana LegA3 orthologs (Lmic2509 and Lgee0773) and no interaction was observed. However, when the same analysis was performed with the L. pneumophila LegA3C and the L. longbeachae and L. oakridgensis LegA3 orthologs (Llo0584 and Loak0513) significant levels of interaction were observed (Fig. 9C). These results support our hypothesis and perfectly coincide with the intracellular growth analysis that showed that the L. longbeachae and L. oakridgensis LegA3 complemented the L. pneumophila legA3 deletion mutant without the need for their natural LegA3C ortholog (Fig. 5B and D). Sequence comparison of the five LegA3C orthologs did not reveal any region in the LegA3C orthologs where several amino acids were similar in L. pneumophila, L. longbeachae, and L. oakridgensis, but not in L. micdadei and L. geestiana (Fig. S5). Collectively, these results show that as long as there is an interacting pair of Legionella LegA3 and LegA3C orthologs in the L. pneumophila legA3 deletion mutant, its intracellular phenotype is complemented to wild-type levels. This suggests that all the LegA3 orthologs examined perform the same conserved function during infection.
3. Discussion
The Legionella genus in general and L. pneumophila in particular contain the largest number of effectors known in a genus or in a specific species, respectively (Burstein et al., 2016; Gomez-Valero et al., 2019). It has been shown in L. pneumophila that in most cases a deletion mutant of a single effector or even a deletion of several effectors in the same strain does not result in an intracellular growth phenotype (O'Connor et al., 2011; Shames et al., 2017; Park et al., 2020b). This observation is explained by functional redundancy among effectors (O'Connor et al., 2011, 2012), which is emphasized by the diverse repertoire of effectors present in the Legionella genus (Burstein et al., 2016). Surprisingly, only three out of the nine core effectors were found to be required for optimal intracellular growth, suggesting that no other effector can perform their function and substitute for their absence. The other six core effectors, even though selected for throughout the course of evolution of the Legionella genus, were found not to be required for intracellular growth in A. castellanii. The lack of intracellular growth phenotype was unexpected since it suggests that these core effectors could have been lost during the course of evolution, as they might fulfill non-essential functions or that they might have redundant effectors that can substitute for their function. Both these options, if correct, could have led to the loss of these core effectors at least from some of the Legionella species. The high conservation on one hand and lack of an intracellular phenotype on the other can be explained in at least two ways. It is known that Legionella replicate in many amoebae hosts (Fields, 1996; Moliner et al., 2010; Boamah et al., 2017) and this ability probably shaped the composition of the Legionella genus core effectors. The core effectors that were found not to be required for intracellular growth in A. castellanii might be required for intracellular growth in other amoebae species that are prevalent in the environment. Such amoebae would be encountered by all the Legionella species and thus select for maintaining these core effectors in all of them. An example for this possibility was recently described for the lpg3000 core effector which was found to be required for maximal intracellular growth only in H. vermiformis (Park et al., 2020b). Another option is that these core effectors are required for intracellular growth under specific host conditions. For example, encystment and excystment are processes mediated by amoeba to survive harsh conditions and return to regular growth, respectively. These processes might require specific effectors in the bacteria to survive such conditions in the amoeba, but these effectors are not required during regular intracellular growth in amoeba. The fact that the nine core effectors were all selected for during Legionella evolution and therefore remained intact in all the species examined, suggests that all of them probably do not have functionally redundant effectors and that under certain conditions they are crucial for the survival of Legionella in one of their hosts, a process that constantly shapes the arsenal of effectors.
Out of the nine core effectors examined in this study, MavN was found to be required for intracellular growth (Fig. 2) and it was shown before to be required for iron acquisition during intracellular growth (Isaac et al., 2015; Portier et al., 2015). Examination of its orthologs indicated that most of them complement the corresponding L. pneumophila deletion mutant for intracellular growth (Fig. 3), suggesting that they perform the same function like their L. pneumophila ortholog. However, the L. geestiana ortholog of mavN did not complement the intracellular growth phenotype. The core effector MavN was previously studied and amino acids critical for their functions were identified (Isaac et al., 2015; Christenson et al., 2019). Examination of the conservation of these amino acids, located in the iron binding loop, indicated that two of them are conserved in the whole genus (Fig. S6). However, a third critical histidine residue, at position 445, is altered in L. geestiana (Fig. S6), in which loop number seven containing these three critical amino acids is dramatically different in comparison to the other species (Fig. S6). Since a change of this single amino acid from histidine to alanine in the L. pneumophila MavN ortholog abolished the activity of the protein (Christenson et al., 2019), it might very well be that the histidine to methionine alteration in L. geestiana led to the lack of intracellular growth complementation in amoeba (Fig. 3). The LceA core effector orthologs showed results similar to the MavN core effector orthologs (compare Figs. 3 and 4), however LceA was not studied before and there are no amino acids known to be critical for its function. Therefore, we looked for amino acids that are conserved in all the LceA orthologs but are altered in L. geestiana (Fig. S7). We identified two such amino acids located between transmembrane domains 3 and 4 and the difference in one or both of these amino acids might be responsible for the lack of complementation that was observed with the L. geestiana LceA ortholog. The lack of complementation by both the MavN and LceA orthologs from L. geestiana and the identification of changes in conserved amino acids in these proteins might indicate that the function of core effectors has changed in this evolutionary distant Legionella species.
One of the most interesting findings presented here was the discovery of LegA3C which interacts with LegA3, the most widely conserved effector found in the Legionella genus. We showed that LegA3C is required for intracellular growth and only interacting pairs of LegA3 - LegA3C orthologs complement the intracellular growth phenotype of the legA3 deletion mutant, indicating that LegA3C is critical for LegA3 in order to perform its function. Even though elucidating the specific role of LegA3C is beyond the scope of this paper, several possibilities for the function of LegA3C can come to mind and the most likely one is that LegA3C might function as a chaperone of LegA3 affecting its stability or translocation. Unlike the Icm/Dot secretion system that utilizes the IcmS-IcmW-LvgA chaperone complex to aid the translocation of multiple effectors (Ninio et al., 2005; Vincent and Vogel, 2006; Cambronne and Roy, 2007; Lifshitz et al., 2013; Meir et al., 2020), the type-III secretion system was shown to utilize specific effector-chaperone pairs (Page and Parsot, 2002; Ghosh, 2004). In type-III secretion systems the specific effector-chaperone pairs are usually encoded from genes located one next to the other, the effector and its dedicated chaperone specifically interact with one another and the chaperones were shown to affect the stability and/or translocation of their cognate effectors (Page and Parsot, 2002; Ghosh, 2004). In a similar manner, LegA3C might affect LegA3 stability or translocation. As far as translocation is concerned LegA3C might affect LegA3 translocation by directing it to the Icm/Dot secretion system (as was shown with type-III effectors chaperones), or LegA3C might affect the translocation of LegA3 in a different manner. It was previously shown, that the rate of LegA3 translocation is slow in comparison to other Legionella effectors (Charpentier et al., 2009), this can suggest the LegA3C might affect the hierarchy of translocation of LegA3 compared to other effectors.
Even though the effectors in the Legionella genus are highly diverse (Burstein et al., 2016), nine effectors were kept during the course of evolution in all the Legionella species examined, but the function of only two of them was uncovered thus far (Isaac et al., 2015; Portier et al., 2015; Christenson et al., 2019; Von Dwingelo et al., 2019). Uncovering the conserved function of the rest of the Legionella genus core effectors will bring to light genes that play key roles in the pathogenicity and survival of all Legionella species and therefore were critical to maintain in all the species of the Legionella genus.
4. Materials and methods
4.1. Bacterial strains and media
The L. pneumophila wild-type strain used in this work was JR32, a streptomycin-resistant, restriction-negative mutant of L. pneumophila Philadelphia-1, which is a wild-type strain in terms of intracellular growth (Sadosky et al., 1993). In addition, mutant strains derived from JR32, other Legionella species and E. coli strains that were used in this study are listed in Dataset S4. Bacterial media, plates, and antibiotic were as previously described (Segal and Shuman, 1997).
4.2. Plasmid construction
To construct isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible constructs, the L. pneumophila lceA and legA3C genes, the lceA, mavN and legA3 orthologs from L. longbeachae, L. micdadei, L. oakridgensis and L. geestiana, as well as the L. pneumophila legA3C-legA3 operon and the orthologous operons from L. micdadei and L. geestiana were amplified by PCR. The primers used for the PCR are listed in Dataset S6. The PCR products were then digested with the suitable restriction enzymes included in the primers and cloned into pUC-18. The inserts were sequenced to verify that no mutations were introduced during the PCR. The inserts were then digested with the same enzymes and cloned into pMMB207C downstream to the Ptac promoter (the genes were cloned without any tag in order not to affect their functionality) to generate the plasmids listed in Dataset S5.
To construct plasmids for the two-hybrid analysis, the L. pneumophila full-length legA3 and legA3C genes, the C-terminal and N-terminal truncations of the L. pneumophila legA3C gene and the L. longbeachae, L. micdadei, L. oakridgensis and L. geestiana legA3 and legA3C orthologous genes were cloned into the pT18 and/or pT25 vectors of the Bordetella pertussis cyaA two-hybrid system (Karimova et al., 1998). All these genes and gene-fragments were PCR amplified using primers designed to generate in-frame fusions with the T18 or T25 fragments of the cyaA gene (Dataset S6). The resulting plasmids are listed in Dataset S5. The inserts of all the plasmids were sequenced, introduced as pairs into E. coli IO7012D and lacZ expression levels were measured by using the β-galactosidase assay as previously described (Feldman et al., 2005).
To construct CyaA fusions for translocation analysis, the pMMB-cyaA-C (Zusman et al., 2007) vector, which contains a polylinker under Ptac control at the end of the cyaA gene, was used. The L. pneumophila lceA (lpg0086), lceB (lpg1356) and legA3C (lpg2301) genes were amplified by PCR using a pair of primers containing suitable restriction sites (Dataset S6). The PCR products were subsequently digested with the relevant restriction enzymes and cloned into pMMB-cyaA-C to generate the plasmids listed in Dataset S5. The plasmids were sequenced and introduced into L. pneumophila JR32 wild-type strain and the icmT deletion mutant. Cyclic AMP (cAMP) levels were measured as previously described (Lifshitz et al., 2014).
To construct deletion substitution mutants in the L. pneumophila lceA and lceB genes and a double deletion mutant of the L. pneumophila legA3C-legA3 genes, a 1-kb DNA fragment located on each side of the planned deletion was amplified by PCR using the primers listed in Dataset S6. The primers were designed to contain a SalI site at the location of the deletion. The two fragments that were amplified were cloned into pUC-18, digested with the suitable enzymes, and sequenced. The plasmids generated are listed in Dataset S5. The resulting plasmids were digested with the suitable enzymes and the inserts were used for a four-way ligation containing the Kanamycin (Km) resistance cassette (Pharmacia) digested with SalI and the pUC-18 vector digested with EcoRI and BamHI. The generated plasmids pZB-lpg0086–4way, pZB-lpg1356–4way and pED-pUC-2301up-2300down-Km (Dataset S5) were digested with PvuII, which cuts on both sides of the pUC-18 polylinker and the resulting fragment was cloned into the pLAW344 allelic exchange vector and digested with EcoRV to generate the plasmids pZB-lpg0086–4way-LAW, pZB-lpg1356–4way-LAW and pED-d2301up-2300down-Km (Dataset S5). The allelic exchange was performed as previously described (Segal and Shuman, 1997).
4.3. Intracellular growth assays
Intracellular competition assays of L. pneumophila strains in A. castellanii (ATCC 30,234) were performed as previously described (Kessler et al., 2013; Dolinsky et al., 2014; Feldheim et al., 2016; Goodwin et al., 2016). Briefly, L. pneumophila strains were grown for 72 h on CYE plates or CYE plates containing chloramphenicol and IPTG for the complementation assays. The bacteria were calibrated and mixed in a ratio of 1:1. The mixture of bacteria was plated on CYE plate and at least 100 colonies were examined on CYE plates with and without Km to ensure a ratio of 1:1 at the beginning of the competition assay. The same mixture (approximately 106 bacteria) was used to infect a confluent flask of A. castellanii (approximately 107 cells) in one-fourth of proteose peptone-yeast extract-glucose (PYG) medium (Moffat and Tompkins, 1992), in which L. pneumophila cannot grow (data not shown). The supernatant of each flask was sampled at intervals of 24 h, and a 1 ml sample was removed and centrifuged for 10 min at 810 g to precipitate the amoebae. Due to the lower plating efficiency of L. pneumophila on CYE plates containing Km after amoeba infection, the infection progeny was plated first on CYE plates and only then at least 100 colonies were examined on CYE plates containing Km. The ratio between the two strains of examined bacteria was determined at each time point. In addition, mutants showing an intracellular growth phenotype were also tested in a similar competition experiment in AYE media, to confirm that and they were not outcompeted by the wild-type strain, indicating that their phenotype was due to difference in intracellular replication (data not shown).
4.4. Two-hybrid analysis
The plasmids constructed for the two-hybrid analysis were introduced as pairs into E. coli IO7012D (DcyaA), and were grown on Luria-Bertani plates containing ampicillin and chloramphenicol for 18 h. Three individual colonies from each clone were selected and grown overnight at 30 °C in Luria-Bertani broth containing ampicillin and chloramphenicol. The assays were performed with 50 l of culture, and the substrate used for -galactosidase hydrolysis was o-nitrophenyl--d-galactopyranoside.
4.5. Western blot analysis
The formation of a CyaA fusion proteins in L. pneumophila was validated by Western blot analysis using the primary anti CyaA antibody 3D1 (Santa Cruz Biotechnology, Inc.) diluted 1:500 and goat anti-mouse IgG conjugated to horseradish peroxidase (Jackson Immunoresearch Laboratories, Inc.) diluted 1:10,000 as the secondary antibody.
Funding
This work was supported in part by grant 2013-240 from the United States–Israel Binational Science Foundation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Natalya Boufan, Elena Degtyar and Ram Viner for plasmids and strains construction. We thank David Burstein for his help with the bioinformatic analyses and for reading the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2022.100105.
Appendix. Supplementary materials
References
- Azad A.F. Pathogenic Rickettsiae as bioterrorism agents. Clin. Infect. Dis. 2007;45(Suppl 1):S52–S55. doi: 10.1086/518147. [DOI] [PubMed] [Google Scholar]
- Berger B., Wilson D.B., Wolf E., Tonchev T., Milla M., Kim P.S. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA. 1995;92(18):8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boamah D.K., Zhou G., Ensminger A.W., O'Connor T.J. From many hosts, one accidental pathogen: the diverse protozoan hosts of Legionella. Front. Cell Infect. Microbiol. 2017;7:477. doi: 10.3389/fcimb.2017.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burillo A., Pedro-Botet M.L., Bouza E. Microbiology and epidemiology of Legionnaire's disease. Infect. Dis. Clin. North Am. 2017;31(1):7–27. doi: 10.1016/j.idc.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Burstein D., Zusman T., Degtyar E., Viner R., Segal G., Pupko T. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5(7) doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D., Amaro F., Zusman T., Lifshitz Z., Cohen O., Gilbert A.J., Pupko T., Shuman H.A., Segal G. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat. Genet. 2016;48:167–175. doi: 10.1038/ng.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne E.D., Roy C.R. The Legionella pneumophila IcmSW complex interacts with multiple dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 2007;3(12):e188. doi: 10.1371/journal.ppat.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier X., Gabay J.E., Reyes M., Zhu J.W., Weiss A., Shuman H.A. Chemical genetics reveals bacterial and host cell functions critical for type IV effector translocation by Legionella pneumophila. PLoS Pathog. 2009;5(7) doi: 10.1371/journal.ppat.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetrit D., Hu B., Christie P.J., Roy C.R., Liu J. A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nat. Microbiol. 2018;3(6):678–686. doi: 10.1038/s41564-018-0165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenson E.T., Isaac D.T., Yoshida K., Lipo E., Kim J.S., Ghirlando R., Isberg R.R., Banerjee A. The iron-regulated vacuolar Legionella pneumophila MavN protein is a transition-metal transporter. Proc. Natl. Acad. Sci. USA. 2019;116(36):17775–17785. doi: 10.1073/pnas.1902806116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J., Kagan J.C., Matthews M., Nagai H., Zuckman D.M., Roy C.R. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 2000;38(4):719–736. doi: 10.1046/j.1365-2958.2000.02176.x. [DOI] [PubMed] [Google Scholar]
- Dolinsky S., Haneburger I., Cichy A., Hannemann M., Itzen A., Hilbi H. The Legionella longbeachae Icm/Dot substrate SidC selectively binds phosphatidylinositol 4-phosphate with nanomolar affinity and promotes pathogen vacuole-endoplasmic reticulum interactions. Infect. Immun. 2014;82(10):4021–4033. doi: 10.1128/IAI.01685-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim Y.S., Zusman T., Speiser Y., Segal G. The Legionella pneumophila CpxRA two-component regulatory system: new insights into CpxR's function as a dual regulator and its connection to the effectors regulatory network. Mol. Microbiol. 2016;99(6):1059–1079. doi: 10.1111/mmi.13290. [DOI] [PubMed] [Google Scholar]
- Feldman M., Zusman T., Hagag S., Segal G. Coevolution between nonhomologous but functionally similar proteins and their conserved partners in the Legionella pathogenesis system. Proc. Natl. Acad. Sci. USA. 2005;102(34):12206–12211. doi: 10.1073/pnas.0501850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B.S. The molecular ecology of Legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- Ghosal D., Chang Y.W., Jeong K.C., Vogel J.P., Jensen G.J. In situ structure of the Legionella Dot/Icm type IV secretion system by electron cryotomography. EMBO Rep. 2017;18(5):726–732. doi: 10.15252/embr.201643598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D., Jeong K.C., Chang Y.W., Gyore J., Teng L., Gardner A., Vogel J.P., Jensen G.J. Molecular architecture, polar targeting and biogenesis of the Legionella Dot/Icm T4SS. Nat. Microbiol. 2019;4(7):1173–1182. doi: 10.1038/s41564-019-0427-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 2004;68(4):771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L., Rusniok C., Cazalet C., Buchrieser C. Comparative and functional genomics of Legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front. Microbiol. 2011;2:208. doi: 10.3389/fmicb.2011.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L., Rusniok C., Carson D., Mondino S., Perez-Cobas A.E., Rolando M., Pasricha S., Reuter S., Demirtas J., Crumbach J., Descorps-Declere S., Hartland E.L., Jarraud S., Dougan G., Schroeder G.N., Frankel G., Buchrieser C. More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc. Natl. Acad. Sci. USA. 2019;116(6):2265–2273. doi: 10.1073/pnas.1808016116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin I.P., Kumova O.K., Ninio S. A conserved OmpA-like protein in Legionella pneumophila required for efficient intracellular replication. FEMS Microbiol. Lett. 2016;363(16) doi: 10.1093/femsle/fnw173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Boyd D., Amyot W.M., Hempstead A.D., Luo Z.Q., O'Connor T.J., Chen C., Machner M., Montminy T., Isberg R.R. The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol. 2011;13(2):227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber A., Roy C.R. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- Isaac D.T., Laguna R.K., Valtz N., Isberg R.R. MavN is a Legionella pneumophila vacuole-associated protein required for efficient iron acquisition during intracellular growth. Proc. Natl. Acad. Sci. USA. 2015;112(37):E5208–E5217. doi: 10.1073/pnas.1511389112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg R.R., O'Connor T.J., Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 2009;7(1):13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G., Pidoux J., Ullmann A., Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA. 1998;95(10):5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A., Schell U., Sahr T., Tiaden A., Harrison C., Buchrieser C., Hilbi H. The Legionella pneumophila orphan sensor kinase LqsT regulates competence and pathogen-host interactions as a component of the LAI-1 circuit. Environ. Microbiol. 2013;15(2):646–662. doi: 10.1111/j.1462-2920.2012.02889.x. [DOI] [PubMed] [Google Scholar]
- Leclerque A., Kleespies R.G. Type IV secretion system components as phylogenetic markers of entomopathogenic bacteria of the genus Rickettsiella. FEMS Microbiol. Lett. 2008;279(2):167–173. doi: 10.1111/j.1574-6968.2007.01025.x. [DOI] [PubMed] [Google Scholar]
- Lifshitz Z., Burstein D., Schwartz K., Shuman H.A., Pupko T., Segal G. Identification of novel Coxiella burnetii Icm/Dot effectors and genetic analysis of their involvement in modulating a mitogen-activated protein kinase pathway. Infect. Immun. 2014;82(9):3740–3752. doi: 10.1128/IAI.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifshitz Z., Burstein D., Peeri M., Zusman T., Schwartz K., Shuman H.A., Pupko T., Segal G. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl. Acad. Sci. USA. 2013;110(8):E707–E715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsky M., Vitkin Y., Segal G. A novel Legionella genomic island encodes a copper-responsive regulatory system and a single Icm/Dot effector protein transcriptionally activated by copper. MBio. 2020;11(1) doi: 10.1128/mBio.03232-19. e03232-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew M.J., Subramanian G., Nguyen T.T., Robert C., Mediannikov O., Fournier P.E., Raoult D. Genome sequence of Diplorickettsia massiliensis, an emerging Ixodes ricinus-associated human pathogen. J. Bacteriol. 2012;194(12):3287. doi: 10.1128/JB.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir A., Mace K., Lukoyanova N., Chetrit D., Hospenthal M.K., Redzej A., Roy C., Waksman G. Mechanism of effector capture and delivery by the type IV secretion system from Legionella pneumophila. Nat. Commun. 2020;11(1):2864. doi: 10.1038/s41467-020-16681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J.F., Tompkins L.S. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 1992;60(1):296–301. doi: 10.1128/iai.60.1.296-301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moliner C., Fournier P.E., Raoult D. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol. Rev. 2010;34(3):281–294. doi: 10.1111/j.1574-6976.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- Ninio S., Celli J., Roy C.R. A Legionella pneumophila effector protein encoded in a region of genomic plasticity binds to Dot/Icm-modified vacuoles. PLoS Pathog. 2009;5(1) doi: 10.1371/journal.ppat.1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio S., Zuckman-Cholon D.M., Cambronne E.D., Roy C.R. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 2005;55(3):912–926. doi: 10.1111/j.1365-2958.2004.04435.x. [DOI] [PubMed] [Google Scholar]
- O'Connor T.J., Adepoju Y., Boyd D., Isberg R.R. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Natl. Acad. Sci. USA. 2011;108(36):14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T.J., Boyd D., Dorer M.S., Isberg R.R. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science. 2012;338(6113):1440–1444. doi: 10.1126/science.1229556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A.L., Parsot C. Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 2002;46(1):1–11. doi: 10.1046/j.1365-2958.2002.03138.x. [DOI] [PubMed] [Google Scholar]
- Park D., Chetrit D., Hu B., Roy C.R., Liu J. Analysis of Dot/Icm Type IVB Secretion system subassemblies by cryoelectron tomography reveals conformational changes induced by DotB binding. MBio. 2020;11(1) doi: 10.1128/mBio.03328-19. e03328-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.M., Ghosh S., O'Connor T.J. Combinatorial selection in amoebal hosts drives the evolution of the human pathogen Legionella pneumophila. Nat. Microbiol. 2020;5(4):599–609. doi: 10.1038/s41564-019-0663-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier E., Zheng H., Sahr T., Burnside D.M., Mallama C., Buchrieser C., Cianciotto N.P., Hechard Y. IroT/mavN, a new iron-regulated gene involved in Legionella pneumophila virulence against amoebae and macrophages. Environ. Microbiol. 2015;17(4):1338–1350. doi: 10.1111/1462-2920.12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Luo Z.Q. Legionella and Coxiella effectors: strength in diversity and activity. Nat. Rev. Microbiol. 2017;15(10):591–605. doi: 10.1038/nrmicro.2017.67. [DOI] [PubMed] [Google Scholar]
- Sadosky A.B., Wiater L.A., Shuman H.A. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 1993;61(December):5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahr T., Rusniok C., Dervins-Ravault D., Sismeiro O., Coppee J.Y., Buchrieser C. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol. 2012;9(4):503–519. doi: 10.4161/rna.20270. [DOI] [PubMed] [Google Scholar]
- Segal G., Shuman H.A. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 1997;65(12):5057–5066. doi: 10.1128/iai.65.12.5057-5066.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G., Purcell M., Shuman H.A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA. 1998;95(4):1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shames S.R., Liu L., Havey J.C., Schofield W.B., Goodman A.L., Roy C.R. Multiple Legionella pneumophila effector virulence phenotypes revealed through high-throughput analysis of targeted mutant libraries. Proc. Natl. Acad. Sci. USA. 2017;114(48):E10446–E10454. doi: 10.1073/pnas.1708553114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohdy N., Efe J.A., Emr S.D., Shuman H.A. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc. Natl. Acad. Sci. USA. 2005;102(13):4866–4871. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirigos K.D., Peters C., Shu N., Kall L., Elofsson A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015;43(W1):W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent C.D., Vogel J.P. The Legionella pneumophila IcmS-LvgA protein complex is important for Dot/Icm-dependent intracellular growth. Mol. Microbiol. 2006;61(3):596–613. doi: 10.1111/j.1365-2958.2006.05243.x. [DOI] [PubMed] [Google Scholar]
- Viner R., Chetrit D., Ehrlich M., Segal G. Identification of two Legionella pneumophila effectors that manipulate host phospholipids biosynthesis. PLoS Pathog. 2012;8(11) doi: 10.1371/journal.ppat.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J.P., Andrews H.L., Wong S.K., Isberg R.R. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279(5352):873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- Von Dwingelo J., Chung I.Y.W., Price C.T., Li L., Jones S., Cygler M., Abu Kwaik Y. Interaction of the Ankyrin H core effector of Legionella with the host LARP7 component of the 7SK snRNP complex. MBio. 2019;10(4) doi: 10.1128/mBio.01942-19. e01942-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Luo Z.Q. Cell biology of infection by Legionella pneumophila. Microbes Infect. 2013;15(2):157–167. doi: 10.1016/j.micinf.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.H., Caldwell T.A., McKenzie A.M., Kokhan O., Berndsen C.E. Characterization of the structure and catalytic activity of Legionella pneumophila VipF. Proteins. 2016;84(10):1422–1430. doi: 10.1002/prot.25087. [DOI] [PubMed] [Google Scholar]
- Zhu W., Banga S., Tan Y., Zheng C., Stephenson R., Gately J., Luo Z.Q. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE. 2011;6(3):e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman T., Degtyar E., Segal G. Identification of a hypervariable region containing new Legionella pneumophila Icm/Dot translocated substrates by using the conserved icmQ regulatory signature. Infect. Immun. 2008;76(10):4581–4591. doi: 10.1128/IAI.00337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman T., Aloni G., Halperin E., Kotzer H., Degtyar E., Feldman M., Segal G. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 2007;63(5):1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.