Abstract
Investigation of plant-bacteria interactions requires quantification of in planta bacterial titers by means of cumbersome and time-consuming colony-counting assays. Here, we devised a broadly applicable tool for bioluminescence-based quantitative and spatial detection of bacteria in plants. We developed vectors that enable Tn7 transposon-mediated integration of the luxCDABE luciferase operon into a specific genomic location found ubiquitously across bacterial phyla. These vectors allowed for the generation of bioluminescent transformants of various plant pathogenic bacteria from the genera Pseudomonas, Rhizobium (Agrobacterium), and Ralstonia. Direct luminescence measurements of plant tissues inoculated with bioluminescent Pseudomonas syringae pv. tomato DC3000 (Pto-lux) reported bacterial titers as accurately as conventional colony-counting assays in Arabidopsis thaliana, Solanum lycopersicum, Nicotiana benthamiana, and Marchantia polymorpha. We further showed the usefulness of our vectors in converting previously generated Pto derivatives to isogenic bioluminescent strains. Importantly, quantitative bioluminescence assays using these Pto-lux strains accurately reported the effects of plant immunity and bacterial effectors on bacterial growth, with a dynamic range of four orders of magnitude. Moreover, macroscopic bioluminescence imaging illuminated the spatial patterns of Pto-lux growth in/on inoculated plant tissues. In conclusion, our vectors offer untapped opportunities to develop bioluminescence-based assays for a variety of plant-bacteria interactions.
Keywords: plant immunity, bacterial virulence, bioluminescence imaging, Pseudomonas syringae, Arabidopsis thaliana, Marchantia polymorpha
Quantification of in planta bacterial growth by colony-counting assays is cumbersome and time consuming. This study reports the design of ready-to-use vectors that enable site-specific genomic integration of luciferase genes for bioluminescence-based quantitative and spatial analysis of bacteria in plants.
Introduction
Plants are colonized by a multitude of microbes, such as bacteria, fungi, and oomycetes. Plant-colonizing microbes can be classified as pathogens, mutualists, and commensals, which can have negative, positive, and neutral consequences for plant health, respectively. Bacterial pathogens instigate plant immunity through the activation of cell-surface receptors, but they are able to suppress the resulting defense responses by deploying a repertoire of effectors (Dou and Zhou, 2012), enabling them to proliferate in plants and cause disease. Plants can restrict pathogen growth by inducing another layer of plant immunity, known as effector-triggered immunity (ETI), through the recognition of specific effectors by intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) (Cui et al., 2015).
Traditionally, in planta growth of bacteria is quantified by plating serial dilutions of tissue extracts onto plates containing selective medium and counting the number of bacterial colonies that appear. This so-called colony-counting assay has been instrumental for studies of interactions between plants and bacterial pathogens, providing molecular insights into the mechanisms of plant immunity and bacterial virulence. However, colony-counting assays are cumbersome and time consuming and demand skilled hands to obtain reproducible results; they can be a bottleneck, especially when high-throughput assays or large-scale quantitative data are required. Moreover, colony-counting assays do not allow for the collection of spatial information on bacterial growth in planta because of tissue disruption during the sample preparation step.
Quantification of light emission from genetically engineered bioluminescent bacteria has been used as an alternative approach to estimating the growth of pathogenic bacteria in infected animal and plant hosts (Fan et al., 2008; Howe et al., 2010; Cruz et al., 2014). For this purpose, bacterial luciferase operons, such as luxCDABE of Photorhabdus luminescens and Vibrio fischeri, are particularly useful. The luxCDABE operon expresses a heterodimeric luciferase encoded by luxA and luxB that catalyzes the oxidation of reduced flavin mononucleotide (FMNH2) and a long-chain fatty aldehyde, resulting in light emission at a wavelength of 490 nm (Meighen, 1993). FMNH2 is continuously supplied in aerobically growing bacterial cells, and the long-chain fatty aldehyde is continuously regenerated by a fatty acid reductase complex encoded by luxC, luxD, and luxE (Meighen, 1993). Thus, metabolically active bacteria expressing luxCDABE spontaneously emit light without an external supply of luciferase substrates.
The introduction of a plasmid vector carrying luxCDABE is an easy way to generate bioluminescent bacteria (Hikichi et al., 1998; 1999). However, bioluminescence emitted by such bacteria is influenced by plasmid copy numbers and is unstable without antibiotic selection. To circumvent these problems, stable chromosomal integration of luxCDABE has been employed (Cruz et al., 2014; Fan et al., 2008; Howe et al., 2010; Tsuge et al., 1999). For instance, Fan et al. (2008) used an artificial transposon that induces random transposition of luxCDABE to generate bioluminescent Pseudomonas syringae pv. tomato DC3000 (Pto) and P. syringae pv. maculicola. They demonstrated that the bioluminescence emitted by these bacteria can be used as a proxy to quantify bacterial growth in Arabidopsis thaliana leaves. However, the use of these bioluminescent P. syringae strains has been limited by lack of evidence that the random insertion of luxCDABE does not cause undesired gene disruption. Determining the genomic location of randomly inserted luxCDABE takes time but is essential if further genetic analysis of bacterial genes is planned.
Among transposable elements, the Tn7 transposon is characterized by its integration mode. Transposition of Tn7 occurs at a specific and neutral site, known as the Tn7 attachment (attTn7), in a specific orientation at high frequency. AttTn7 is located downstream of the glmS gene, which encodes an essential glucosamine-fructose-6-phosphate aminotransferase (Peters and Craig, 2001) and is found across bacterial phyla, including Proteobacteria, Firmicutes, and Bacteroidetes (Wiles et al., 2018). Previous reports have shown that Tn7 is active in the genera Pseudomonas, Xanthomonas, Rhizobium, Ralstonia, Burkholderia, Erwinia, Pectobacterium, and Pantoea to which many, if not all, phytopathogenic bacteria belong (Choi et al., 2006; Choi and Schweizer, 2006; Jittawuttipoka et al., 2009; Kernell; Kovács et al., 2009; Kernell Burke et al., 2015; Romero-Jimenez et al., 2015; Zhang et al., 2021; Riccardo et al., 2021). Thus, Tn7-based genetic manipulation tools should be broadly applicable to studies on plant-bacterial pathogen interactions.
Here, we developed Tn7-based vectors for site-specific chromosomal integration of Photorhabdus luminescens luxCDABE. The broad applicability of these vectors was demonstrated by the successful bioluminescence tagging of various plant pathogenic bacteria from the genera Pseudomonas, Rhizobium (Agrobacterium), and Ralstonia. We then provided evidence that our bioluminescence assays with bioluminescent Pto (Pto-lux) and its derivatives allow for the quantitative evaluation of the effects of plant immunity and bacterial virulence on bacterial growth in a wide range of plant species. Moreover, using Pto-lux, we were able to visualize spatially and temporally variable patterns of Pto growth in/on inoculated tissues of several plant species.
Results
Construction of pBJ vectors
Pto can infect crops such as tomato and model plants such as A. thaliana and the liverwort Marchantia polymorpha, and it thus serves as a model bacterial pathogen for studying molecular and evolutionary plant-bacterial pathogen interactions (Xin et al., 2018; Gimenez-Ibanez et al., 2019). Previously, a bioluminescent Pto was generated by transposon-mediated random insertion of luxCDABE into the genome and was used to quantify bacterial growth in A. thaliana (Fan et al., 2008). However, the use of this bioluminescent Pto strain has been limited, as random insertion of luxCDABE may have caused undesired gene disruption. To circumvent this problem, we thought to employ pBEN276, which has been successfully used to induce Tn7-mediated integration of luxCDABE into the genome of Salmonella enterica (Howe et al., 2010). However, our attempt to transform Pto with pBEN276 was not successful, most likely because the replicon of the plasmid is not compatible with Pto.
To enable Tn7-mediated chromosomal integration of luxCDABE into Pto and other plant-colonizing bacteria, we took advantage of the broad-host-range vectors pBBR1-MCS2, pBBR1-MCS5, and pLAFR3, which replicate in a wide range of Proteobacteria (Kovach et al., 1995; Staskawicz et al., 1987). We constructed seven vectors, pBJ1 to pBJ7, with two different constitutive promoters for luxCDABE expression and three different antibiotic resistance genes (Figure 1 and Table 1). The mobilization functions of the parental plasmids of the pBJ vectors, pBBR1-MCS2, pBBR1-MCS5, and pLAFR3, have been well defined (Staskawicz et al., 1987; Szpirer et al., 2001). We then introduced the pBJ vectors into bacteria, a process that we demonstrated in this study can be performed by electroporation. After the introduction of pBJ vectors into bacteria, Tn7-mediated transposition of luxCDABE was induced by arabinose. All pBJ vectors with their nucleotide sequences and features are available at Addgene (https://www.addgene.org/Akira_Mine/).
Figure 1.
Schematic representation of pBJ vectors.
tnsABCD are the genes required for transposition at the attTn7 site and are controlled by an arabinose-inducible promoter (PBAD). The luxCDABE operon encoding luciferase is flanked by Tn7 transposon arms Tn7L and Tn7R (indicated by vertical lines) and is controlled by either of the constitutive promoters Pfrr or Pkan. Detailed information on the features of pBJ vectors is available in Table 1.
Table 1.
Features of pBJ vectors.
| Vector name | Promoter | Replicon | Resistance | Vector backbone | Other features |
|---|---|---|---|---|---|
| pBJ1 | Pfrr | pBBR1 | gentamicin | pBBRMCS-5 | mob |
| pBJ2 | Pkan | pBBR1 | gentamicin | pBBRMCS-5 | mob |
| pBJ3 | Pfrr | pBBR1 | kanamycin | pBBRMCS-2 | mob |
| pBJ4 | Pkan | pBBR1 | kanamycin | pBBRMCS-2 | mob |
| pBJ5 | Pfrr | pBBR1, RK2 | gentamicin | pBBRMCS-5 | mob |
| pBJ6 | Pkan | pBBR1, RK2 | gentamicin | pBBRMCS-5 | mob |
| pBJ7 | Pkan | RK2 | tetracycline | pLAFR3 | mob |
pBJ vectors possess either of the constitutive promoters Pfrr or Pkan for luxCDABE expression and have different broad-host-range replicons and antibiotic resistance genes to broaden their applicability. All pBJ vectors were constructed from mobilizable plasmids and can therefore be transferred by conjugation.
pBJ vectors enabled site-specific chromosomal integration of luxCDABE into a variety of plant pathogenic bacteria
Using pBJ vectors, we managed to successfully generate bioluminescent transformants of Pto, the disarmed Agrobacterium tumefaciens strain GV3101, and Japanese isolates of pathogenic bacteria, including P. syringae strains SUPP1331 and SUPP1141, A. tumefaciens strain CH3, and Ralstonia solanacearum strain OE1-1 (Figure 2A and Supplemental Figure 1). Sequencing analysis confirmed that integration of luxCDABE occurred consistently at attTn7, located 25 bp downstream of glmS in all transformed bacterial strains/species (Supplemental Table 1). To investigate whether insertion of luxCDABE at attTn7 causes a growth penalty, we compared the growth of luxCDABE-tagged strains of Pto, P. syringae SUPP1331, A. tumefaciens CH3, and R. solanacearum OE1-1 with that of their parental strains in liquid medium. The data indicated that all luxCDABE-tagged strains grew as well as their parental strains (Supplemental Figure 2). This is consistent with the generally accepted idea that attTn7 is a neutral genomic site for bacterial fitness (Peters and Craig, 2001).
Figure 2.
Characterization of Pseudomonas syringae pv. tomato DC3000 (Pto) with insertion of luxCDABE at attTn7.
(A)Pto and Pto tagged with Pkan- or Pfrr-driven luxCDABE (Pto Pkan:lux or Pto Pfrr:lux) were observed under white light (left panels), and their bioluminescence was detected in the dark (right panels).
(B)Pto Pkan:lux or Pto Pfrr:lux were cultured in King's B medium without antibiotic selection for 4 days. Aliquots of overnight cultures were transferred to new medium every day. Bioluminescence was quantified every day and normalized to bacterial density (OD600). Dots and bars indicate normalized bioluminescence of three independent cultures and their means, respectively.
(C) Growth of Pto, Pto Pkan:lux, and Pto Pfrr:lux (infiltrated at OD600 = 0.001) in leaves of A. thaliana Col-0 was measured at 3 days post inoculation (dpi) by standard colony-counting assays. For each treatment, eight biological replicates collected from different leaves were analyzed. No statistically significant differences were found (adjusted P < 0.05, pairwise t-test with the Benjamini-Hochberg correction for multiple hypothesis testing; n = 8).
(D) Disease symptoms at 3 dpi of A. thaliana Col-0 leaves infiltrated with mock, Pto, Pto Pkan:lux, or Pto Pfrr:lux at OD600 = 0.01.
We compared the activities of two different constitutive promoters, Pfrr and Pkan, used for luxCDABE expression. Pfrr is derived from a constitutively expressed gene encoding the ribosome-recycling factor in Escherichia coli (Liu et al., 2005). Pkan, also known as neomycin phosphotransferase II (nptII) promoter, has been used for constitutive gene expression in a wide range of Proteobacteria, including the genera Pseudomonas (Fan et al., 2008), Xanthomonas (Song and Yang, 2010), Rhizobium (Xi et al., 1999), Ralstonia (Sichwart et al., 2011), Burkholderia (Park et al., 2010), and Erwinia (Peng et al., 2021). Pto with Pkan-driven luxCDABE (Pto Pkan:lux) showed stronger luminescence than Pto with Pfrr-driven luxCDABE (Pto Pfrr:lux) in vitro (Figure 2A and 2B). Both Pto derivatives maintained light emission for 4 days in non-selective in vitro cultures (Figure 2B), indicating that luxCDABE is stably maintained in the Pto genome. We then inoculated A. thaliana accession Col-0 with Pto Pkan:lux and Pto Pfrr:lux, and we measured bacterial titers at 3 days post inoculation (dpi) using conventional colony-counting assays. Pto Pkan:lux and Pto Pfrr:lux growth was similar to that of their parental wild-type Pto (Figure 2C). Disease symptoms caused by Pto, Pto Pkan:lux, and Pto Pfrr:lux were also comparable (Figure 2D). Taken together, these data allowed us to conclude that the stable integration of luxCDABE at attTn7 does not affect Pto growth and virulence in A. thaliana.
Bioluminescence accurately reported bacterial titers in various plant-Pto interactions
We next tested whether bioluminescence emitted by Pto Pkan:lux and Pto Pfrr:lux could be used as a proxy to estimate bacterial titers in planta. Pto Pkan:lux and Pto Pfrr:lux were syringe infiltrated into leaves of A. thaliana Col-0 at varying doses. Leaf discs were excised from the bacteria-infiltrated leaves and placed in a light-reflecting, white 96-well plate, followed by direct measurement of bioluminescence intensities (relative light units [RLUs]) (Supplemental Figure 3A). After bioluminescence measurement, the same leaf discs were subjected to conventional colony-counting assays to determine bacterial titers (colony-forming units [CFUs]) (Supplemental Figure 3B). We found that RLUs and CFUs showed a strong positive correlation, with a regression slope close to 1 (Figure 3A and Supplemental Figure 4A). Similar results were obtained with P. syringae SUPP1331 tagged with Pfrr-driven luxCDABE (Supplemental Figure 4B) and A. tumefaciens strain CH3 tagged with Pkan-driven luxCDABE (Supplemental Figure 4C). We noticed that bioluminescence emitted by Pto Pkan:lux in infected leaves was approximately 10 times greater than that emitted by Pto Pfrr:lux (Figure 3A and Supplemental Figure 4A). Because higher bioluminescence emission increases the lower detection limit, we decided to employ Pto Pkan:lux (hereafter referred to as Pto-lux) for further bioluminescence assays in this study.
Figure 3.
Bioluminescence accurately reports bacterial titers in various plant-Pto interactions.
(A–D)A. thaliana(A), Nicotiana benthamiana(B), Solanum lycopersicum(C), and Marchantia polymorpha(D) were inoculated with Pto Pkan:lux (Pto-lux hereafter) at varying doses (from low to high: OD600 = 0.0002, 0.001, and 0.01 for A. thaliana; 0.00001, 0.00005, and 0.0005 for N. benthamiana; 0.000002, 0.00001, and 0.0001 for S. lycopersicum; 0.001, 0.01, and 0.1 for M. polymorpha). Log10-transformed CFUs and RLUs at 2 dpi were calculated and plotted along the y and x axes, respectively. Regression equations and Pearson's correlation coefficients (R) are shown above the plots.
(E) Similar correlation analysis was performed using the data from all tested plants. At, A. thaliana; Nb, N. benthamiana; Sl, S. lycopersicum; Mp, M. polymorpha.
N. benthamiana, S. lycopersicum, and M. polymorpha have also been used in combination with Pto to explore molecular and evolutionary aspects of plant-bacterial pathogen interactions. N. benthamiana elicits ETI against Pto through recognition of the HopQ1-1 effector (Wei et al., 2007). S. lycopersicum cv. M82 and M. polymorpha Takaragaike accessions, Tak-1 and Tak-2, are susceptible to Pto (Du et al., 2014; Gimenez-Ibanez et al., 2019). We tested whether RLU of Pto-lux can also be used as a proxy to measure Pto growth in these plant species. Leaves of N. benthamiana and S. lycopersicum cv. M82 were syringe infiltrated with Pto-lux, and thalli of M. polymorpha Tak-1 were vacuum infiltrated. The bacterial inoculation of N. benthamiana and S. lycopersicum and that of M. polymorpha were performed independently in two different laboratories. As shown in Figure 3, we found strong positive correlations between RLUs and CFUs, with regression slopes close to 1, in all tested plant species. To our surprise, a strong positive correlation and a regression slope close to 1 were still evident when we incorporated all the RLUs and CFUs obtained from different plant species into one plot (Figure 3E), even though the data were obtained in two laboratories using different experimental conditions. These results indicate that our quantitative bioluminescence assay is a highly reliable and robust method for the assessment of bacterial titers in planta.
Development of the Pto-lux system for evaluation of the effects of plant immunity and bacterial effectors on bacterial growth in A. thaliana
We sought to make use of Pto-lux to recapitulate various aspects of the A. thaliana-Pto pathosystem. A. thaliana Col-0 is susceptible to Pto but becomes resistant when the AvrRpt2 or AvrRpm1 effector is expressed in Pto. This occurs because ETI is activated through the recognition of AvrRpt2 and AvrRpm1 by the corresponding NLRs, RPS2 and RPM1, respectively (Axtell and Staskawicz, 2003; Mackey et al., 2002, 2003). To test whether the ETI-mediated reduction in bacterial growth could be monitored using Pto-lux, we transformed Pto-lux with pLAFR3 carrying avrRpt2 (Pto-lux avrRpt2), avrRpm1 (Pto-lux avrRpm1), or empty pLAFR3 (Pto-lux EV). The resulting Pto-lux derivatives were used to inoculate A. thaliana Col-0 and rpm1 rps2 plants. Both conventional colony-counting and bioluminescence assays detected the reduction in growth of Pto-lux avrRpt2 and Pto-lux avrRpm1 compared with Pto-lux EV in Col-0 (Figure 4A and 4B). In rpm1 rps2, the growth of Pto-lux avrRpt2 and Pto-lux avrRpm1 was slightly higher than that of Pto-lux EV (Figure 4A and 4B). This probably reflects the virulence effects of the AvrRpt2 and AvrRpm1 effectors in the absence of their corresponding receptors (Tsuda et al., 2009). Importantly, we observed a strong positive correlation between RLUs and CFUs, with a regression slope close to 1 (Figure 4C). These results demonstrated that our bioluminescence assays can quantitatively evaluate the effects of ETI and bacterial effectors on bacterial growth.
Figure 4.
Bioluminescence assays accurately evaluate the effects of plant immunity on Pto growth in A. thaliana.
(A–C) Leaves of Col-0 and rpm1 rps2 plants were infiltrated with Pto Pkan:lux (Pto-lux hereafter) carrying the empty vector (Pto-lux EV), avrRpt2 (Pto-lux avrRpt2), or avrRpm1 (Pto-lux avrRpm1) at OD600 = 0.001. Log10-transformed CFUs (A) and RLUs (B) at 2 dpi were calculated from eight biological replicates collected from different leaves and were plotted along the y and x axes, respectively (C).
(D–F) Leaves of Col-0 and dde2 ein2 pad4 sid2 (deps) were infiltrated with Pto-lux EV and Pto-lux avrRpt2 at OD600 = 0.001. Log10-transformed CFUs (D) and RLUs (E) at 2 dpi were calculated from eight biological replicates collected from different leaves and were plotted along the y and x axes, respectively (F). (A, B, D, and E) Statistically significant differences are indicated by different letters (adjusted P < 0.05, pairwise t-test with the Benjamini-Hochberg correction for multiple hypothesis testing; n = 8).
To investigate whether Pto-lux could be used to detect increased bacterial growth in a susceptible genotype compared with its corresponding wild-type control, we inoculated Col-0 and dde2 ein2 pad4 sid2 (deps) plants with Pto-lux EV and Pto-lux avrRpt2. The deps quadruple mutant is defective in defense signaling mediated by the phytohormones ethylene, jasmonic acid, and salicylic acid, and it is therefore highly susceptible to Pto and Pto avrRpt2 (Tsuda et al., 2009). Both colony-counting and bioluminescence assays detected an approximately 100-fold increase in the growth of Pto-lux EV and Pto-lux avrRpt2 in deps compared with Col-0 (Figure 4D and 4E). Measured CFUs and RLUs showed a strong positive correlation and a regression slope close to 1 (Figure 4F). The results obtained using virulent and avirulent Pto-lux derivatives in combination with A. thaliana Col-0 and deps plants collectively indicated that our bioluminescence assays have a dynamic range of 103–107 CFU/cm2, which can mostly cover the range of Pto growth in resistant and susceptible genotypes of A. thaliana.
Previous studies have reported a number of hypovirulent Pto mutants, such as Pto hrcC− and Pto ΔavrPtoΔavrPtoB. Pto hrcC− is deficient in the type III secretion system required for effector delivery (Yuan and He, 1996), whereas Pto ΔavrPtoΔavrPtoB lacks two functionally redundant effectors, AvrPto and AvrPtoB (Yuan and He, 1996; Lin and Martin, 2005; He et al., 2006). To test whether Pto-lux can be used to evaluate attenuation in bacterial growth caused by the lack of these virulence factors, we introduced luxCDABE to Pto hrcC− and Pto ΔavrPto ΔavrPtoB (Pto-lux hrcC− and Pto-lux ΔavrPto ΔavrPtoB) at the attTn7 site using pBJ2 (Supplemental Figure 5). It is worth noting that the resulting bioluminescent strains can be considered isogenic to Pto-lux, as all strains carry luxCDABE at the same genomic location (Supplemental Table 1). Colony-counting and bioluminescence assays both showed that Pto-lux hrcC− and Pto-lux ΔavrPto ΔavrPtoB grew less than Pto-lux in Col-0 (Figure 5A and 5B), which was further supported by a strong positive correlation between CFUs and RLUs and a regression slope close to 1 (Figure 5C). These results emphasize that pBJ vectors make it possible to generate isogenic bioluminescent strains of previously generated Pto mutants for comparative bioluminescence-based growth assays.
Figure 5.
Bioluminescence assays accurately evaluate the effects of bacterial virulence factors on Pto growth in A. thaliana.
(A–C) Leaves of Col-0 were infiltrated with Pto-lux, Pto-lux hrcC−, or Pto-lux ΔAvrPto ΔAvrPtoB at OD600 = 0.0005. Log10-transformed CFUs (A) and RLUs (B) at 2 dpi were calculated from eight biological replicates collected from different leaves of three plants and were plotted along the y and x axes, respectively (C).
(D–F)A. thaliana Col-0 plants were spray-inoculated with Pto-lux, Pto-lux COR−, and Pto-lux ΔcmaA at OD600 = 0.1. Log10-transformed CFUs (D) and RLUs (E) at 3 dpi were calculated from eight biological replicates, each of which was collected from three different leaves of a single plant, and were plotted along the y and x axes, respectively (F).
(A, B, C, and E) Statistically significant differences are indicated by different letters (adjusted P < 0.05, pairwise t-test with the Benjamini-Hochberg correction for multiple hypothesis testing; n = 8).
Pto produces the phytotoxin coronatine (COR) to re-open closed stomata, thereby gaining access to the apoplast for multiplication (Melotto et al., 2006). Hence, COR-deficient (COR−) Pto strains show reduced growth compared with their wild-type counterpart. To investigate whether our bioluminescence assays could differentiate between growth of Pto and its COR− strain, we used pBJ2 to generate a bioluminescent transformant of the Tn5 transposon-mutagenized COR− strain Pto DC3118 (Ma et al., 1991) (Pto-lux COR−) (Supplemental Figure 5). Because the exact position of transposon insertion has not been determined in this COR− strain (Ma et al., 1991), we also used a suicide vector-based targeted gene deletion to generate a Pto-lux mutant that lacked the cmaA gene required for COR production (Brooks et al., 2004) (Pto-lux ΔcmaA) (Supplemental Figure 6). These bioluminescent strains were used to spray-inoculate Col-0 plants. The growth of Pto-lux COR− and Pto-lux ΔcmaA was reduced compared with that of Pto-lux in both colony-counting and bioluminescence assays, and CFUs and RFUs showed a strong positive correlation and a regression slope close to 1 (Figure 5D–5F). Thus, in addition to transformation of existing Pto mutants by pBJ vectors, it is also possible to use the gene deletion approach with Pto-lux to evaluate the virulence functions of bacterial genes based on bioluminescence assays.
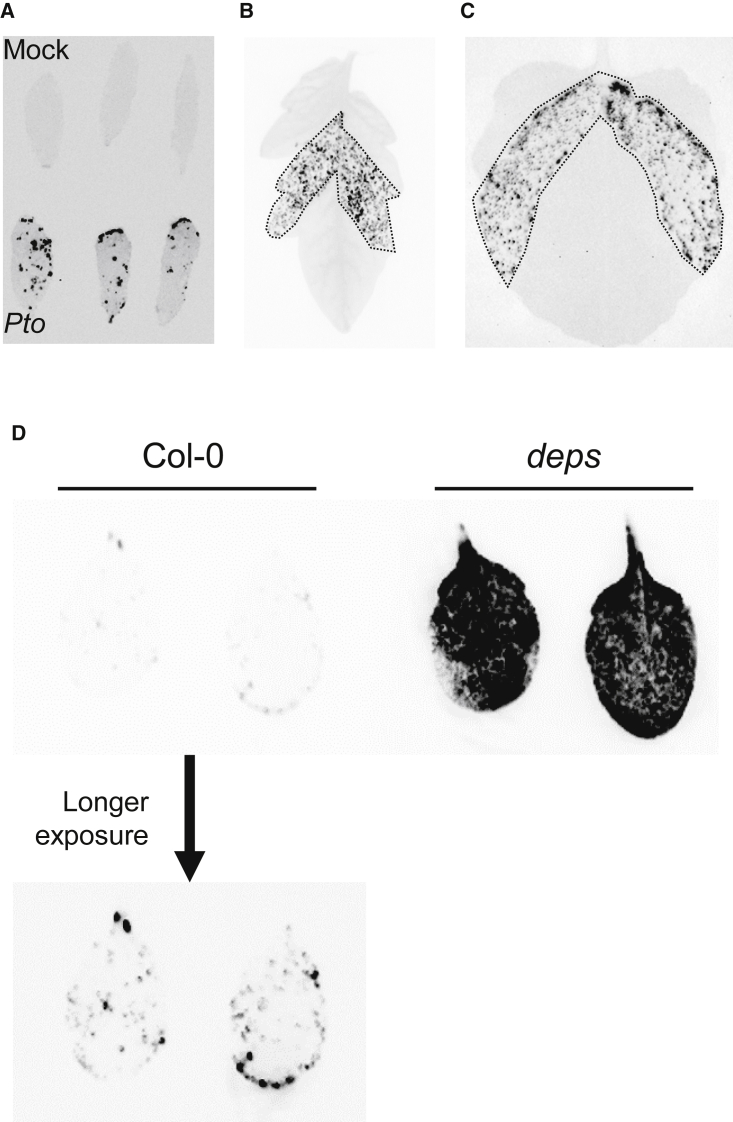
Macroscopic bioluminescence imaging illuminated heterogenous patterns of Pto-lux growth in infected leaves
Colony-counting assays include a cell/tissue disruption step and therefore cannot be used to collect information on the spatial growth of bacteria in infected tissues. We took advantage of Pto-lux to visualize the sites of bacterial multiplication at a macroscopic level using a luminescence imager. Despite the fact that suspensions of Pto-lux were syringe infiltrated into whole areas of A. thaliana Col-0 leaves, spotty bioluminescence signals were observed (Figure 6A). Similar results were obtained in S. lycopersicum and N. benthamiana (Figure 6B and 6C). These results suggested that Pto multiplication is spatially variable in infected leaves. By contrast, bioluminescence signals of A. tumefaciens CH3-lux in infiltrated areas of N. benthamiana leaves were not spotty, but rather uniform (Supplemental Figure 7). This suggests that Pseudomonas and Agrobacterium bacteria may have distinct spatial multiplication patterns. Intriguingly, we observed Pto-lux bioluminescence signals in almost the entire leaves of immunocompromised deps A. thaliana plants (Figure 6D), suggesting that plant immunity affects spatial multiplication patterns of this bacterial pathogen. Collectively, these results emphasize the usefulness of our bioluminescence imaging approach for spatial analysis of plant-bacteria interactions.
Figure 6.
Bioluminescence illuminates heterogenous multiplication of Pto in inoculated leaves.
(A) Bioluminescence of A. thaliana Col-0 leaves was detected 2 days after infiltration with Pto-lux at OD600 = 0.001.
(B and C) Bioluminescence of S. lycopersicum cv. M82 (B) and N. benthamiana(C) leaves was detected 2 days after infiltration with Pto-lux at OD600 = 0.0001. The dotted lines surround the area where bacterial suspensions were infiltrated.
(D) Bioluminescence of Col-0 and deps leaves was detected 2 days after infiltration with Pto-lux at OD600 = 0.001.
The growth of Pto-lux on M. polymorpha thalli
We further explored the value of our Pto-lux system by investigating the interaction between Pto and the liverwort M. polymorpha, an emerging and attractive pathosystem for elucidating the evolution of molecular mechanisms that underlie plant-bacteria interactions (Gimenez-Ibanez et al., 2019). As spatiotemporal patterns of Pto growth in M. polymorpha have not yet been properly described, we monitored the growth of Pto-lux in M. polymorpha Tak-1 over time. When we inoculated with a bacterial suspension at OD600 = 0.01, Pto-lux growth at the basal regions of thalli was saturated at 2 dpi, and the bacterial titer was maintained up to at least 4 dpi under our experimental conditions (Figure 7A). Bioluminescence imaging revealed that Pto-lux preferentially colonized and grew at the basal region of the thalli at early time points, whereas at later time points, they grew at the side edges of the thalli (Figure 7B). The bioluminescence signals derived from the inoculum were observed outside the thalli at 0 dpi but diminished after 1 dpi (Figure 7B). Disease symptoms associated with chlorosis were observed at later time points, mainly at the basal region (Figure 7B). These temporal colonization patterns fit well with the bacterial titers that were measured at the basal region (Figure 7A). Further time-course assays of Pto-lux growth in M. polymorpha demonstrated that CFUs and RLUs correlated well, with a regression slope close to 1, over 7 days (Supplemental Figure 8). Thus, bioluminescence imaging with Pto-lux revealed the temporal and spatial dynamics of Pto-M. polymorpha interactions.
Figure 7.
Pto-lux serves as a new tool for genetic dissection of M. polymorpha-Pto interactions.
(A and B) Fourteen-day-old Tak-1 thalli were vacuum infiltrated with Pto-lux at OD600 = 0.01 and sampled at 0, 1, 2, 3, and 7 dpi. Log10-transformed RLUs were calculated from eight biological replicates collected from the thalli of different plants (A). Bright field images were merged with pseudocolored bioluminescence images (pink) (B).
(C) Fourteen-day-old Tak-1 thalli were vacuum infiltrated with Pto-lux, Pto-lux hrcC−, Pto-lux COR−, or Pto-lux ΔavrPto ΔavrPtoB at OD600 = 0.01 and sampled at 2 dpi. Log10-transformed RLUs were calculated from eight biological replicates collected from the thalli of different plants.
(D) Fourteen-day-old Tak-1, Tak-2, BC3-38, BoGa L2, and BoGa L5 thalli were vacuum infiltrated with Pto-lux at OD600 = 0.01 and sampled at 0 and 2 dpi. Log10-transformed RLUs were calculated from eight biological replicates collected from the thalli of different plants.
(A, C, and D) Statistically significant differences are indicated by different letters (adjusted P < 0.05, pairwise t-test with the Benjamini-Hochberg correction for multiple hypothesis testing; n = 8).
Pto virulence and infection of M. polymorpha depend on bacterial effectors, as shown by the use of the Pto mutants Pto hrcC−, Pto COR−, and Pto ΔavrPto ΔavrPtoB (Gimenez-Ibanez et al., 2019). Consequently, we tested whether these reported observations could be recapitulated using our Pto-lux system. Consistent with the previous study, our bioluminescence assay showed that a functional type III secretion system and the effectors AvrPto and AvrPtoB, but not COR, were required for full virulence of Pto against M. polymorpha (Figure 7C). This result confirms that our bioluminescence assay is robust and compatible for evaluating the functions of candidate bacterial effectors in different plant models.
Currently, M. polymorpha accessions Tak-1 (male) and Tak-2 (female) and Botanical Garden Osnabrück (BoGa) accessions (male and female) are broadly utilized as wild-type strains in molecular genetic studies (Okada et al., 2000; Ishizaki et al., 2008; Althoff et al., 2014; Buschmann et al., 2016). Spores produced by crossing Takaragaike accessions (Tak-1 and Tak-2) or BoGa accessions have been used to generate mutant or transgenic plants and to perform forward genetic screening (Ishizaki et al., 2008, 2013a, 2013b; Sugano et al., 2014; Honkanen et al., 2016). However, these male and female lines very often display distinct phenotypes. It is therefore important to test whether these accessions differ in their levels of bacterial resistance. Tak-1 and Tak-2 and their backcrossed progeny BC3-38 displayed the same level of resistance against Pto-lux (Figure 7D). BoGa-L5 (male) and BoGa-L2 (female) were comparably resistant (Figure 7D). Thus, these results suggested that comparing transgenic plants generated from spores produced by crossing Tak-1 and Tak-2 plants or BoGa-L5 and BoGa-L2 plants with their parental plants is fairly straightforward in terms of bacterial resistance. Moreover, our bioluminescence assays revealed that the BoGa-L5 and BoGa-L2 plants were slightly more susceptible than the Takaragaike accessions (Figure 7D).
Finally, we used Pto-lux to compare different methods of bacterial inoculation in M. polymorpha. We first tested whether Pto could colonize M. polymorpha by simply dipping the thalli into the bacterial suspension without applying a vacuum. Significant Pto-lux growth was observed after dip inoculation, although the bacterial titer at 2 dpi was lower than that achieved with vacuum infiltration (Supplemental Figure 9A). The use of a filter paper on top of solid medium for plant growth prior to inoculation can sometimes affect thalli growth in an uncontrolled way, an effect that is not observed for thalli grown directly on agar. Therefore, we tested whether thalli on agar plates without filter paper could be used for the Pto infection assay (Supplemental Figure 10A). We poured the bacterial suspension onto thalli grown on agar plates and then performed vacuum infiltration. The bacterial suspension was then poured off, and the thalli were cultured further on the same plates (Supplemental Figure 10A). We found that the agar-based method was as feasible as the established filter paper method for Pto-lux growth measurement in M. polymorpha thalli (Supplemental Figure 9B).
Discussion
Several studies have reported the generation of bioluminescent bacteria chromosomally tagged with the luxCDABE luciferase operon and described their use in the study of plant-bacteria interactions (Tsuge et al., 1999; Fan et al., 2008; Cruz et al., 2014). However, these studies employed genetic tools designed for the specific bacterial strains used or that could cause undesired gene disruption by random insertion of the transgene. Thus, an efficient and broadly applicable tool for bioluminescence tagging is needed for researchers looking to manipulate their own bacterial isolates. In this study, we have developed pBJ vectors that can be used to readily generate bioluminescent bacteria by inducing Tn7-mediated chromosomal integration of luxCDABE into a specific and neutral site known as attTn7. It has been shown that attTn7 is conserved across bacterial phyla ranging from Proteobacteria and Firmicutes to Bacteroidetes (Wiles et al., 2018). pBJ vectors collectively confer resistance to three different antibiotics, contain two different constitutive promoters for luxCDABE expression (Table 1), and were constructed from broad-host-range vectors that are known to replicate in a wide range of Proteobacteria. Owing to these features of pBJ vectors, we were able to generate bioluminescent transformants of various phytopathogenic bacteria from different genera, including Pseudomonas, Rhizobium (Agrobacterium), and Ralstonia. The promoter region of luxCDABE in pBJ1, pBJ3, and pBJ5 can be easily replaced after digestion with the XhoI restriction enzyme, which can be helpful for users who intend to use different promoters for luxCDABE expression in their bacterial isolates. Taken together, the characteristics of pBJ vectors enable them to serve as versatile tools for bioluminescence tagging of a wide range of Proteobacteria. A future challenge will be to evaluate and/or modify pBJ vectors for the manipulation of Tn7-compatible bacteria other than Proteobacteria.
Using pBJ vectors, we generated Pto-lux and demonstrated its use for bioluminescence-based quantification of bacterial titers in a rapid and high-throughput manner. Unlike conventional colony-counting assays, our bioluminescence assays do not require sample grinding, preparation and plating of serial dilutions, and manual colony counting, thereby saving substantial time, reducing labor costs, and minimizing human error (Supplemental Figure 3). In addition, because our bioluminescence assays do not require tissue disruption, the release of plant metabolites, which is inevitable in colony-counting assays and may cause undesired effects on bacterial growth on plates, can be avoided. These effects should not be taken lightly, as increasing evidence suggests that various plant species accumulate defense metabolites in specialized cells such as trichomes and/or in intracellular vesicles and organelles (e.g., oil bodies and vacuoles) (Hatsugai et al., 2009; Huchelmann et al., 2017; Romani et al., 2020). A potential limitation of our bioluminescence assay is the possibility that bioluminescence may not accurately reflect bacterial numbers at infection stages when bacterial metabolism decreases, such as biofilm formation.
Our Pto-lux system has solved the limitations that are inherent to a similar system reported previously (Fan et al., 2008). Unlike the random insertion of luxCDABE used by Fan et al., Tn7-mediated site-specific transposition in our system allowed for comparative analysis of isogenic bioluminescent transformants of wild-type Pto and previously generated Pto derivatives with altered virulence, such as Pto avrRpt2, Pto avrRpm1, Pto hrcC−, Pto COR−, and Pto ΔavrPto ΔavrPtoB (Figures 4 and 5). These Pto derivatives have been used extensively to study molecular mechanisms of plant innate immunity and bacterial virulence (He et al., 2006; Kim et al., 2009; Cui et al., 2013; Geng et al., 2016; Mine et al., 2017, 2018). In addition, because the genomic location of luxCDABE in Pto-lux is clearly defined, it is straightforward to use homologous recombination techniques to generate targeted gene knockins and knockouts in Pto-lux. This is exemplified by the genetic analysis of COR function using Pto-lux ΔcmaA (Figure 5D and 5E).
Macroscopic bioluminescence imaging revealed spatially variable growth of Pto-lux in various plant species. For instance, we showed that even when bacterial suspensions were infiltrated into entire areas of A. thaliana leaves, the bioluminescence signals of Pto-lux were spotty (Figure 6A). Although an undetectable bioluminescence signal does not necessarily mean absence of bacterial growth, this result suggests that Pto multiplication is not uniform but rather heterogenous in infected tissues. By contrast, bioluminescence signals of Pto-lux were observed in almost the entire areas of leaves in immunocompromised deps plants (Figure 6C), suggesting that plant immunity affects the spatial patterns of bacterial multiplication. We were also able to visualize spatiotemporal patterns of Pto-lux growth in M. polymorpha Tak-1 thalli over time (Figure 7B), which generally supported the reported observation that Pto growth is lower at meristematic apical notches than at the basal regions of thalli (Gimenez-Ibanez et al., 2019). It is reasonable to speculate that reproductive organs, such as apical notches and gemma cups, are resistant to Pto infection in order to ensure the survival of M. polymorpha. It would be an interesting future challenge to explore plant and bacterial factors that affect spatially variable bacterial growth in plants. Our system would be similarly applicable to the visualization of bacteria inside roots, as was the case with luxCDABE-tagged R. solanacearum (Planas-Marques et al., 2020). However, visualization of bioluminescent bacteria inside woody, lignified tissues may require physical tissue dissection before observation.
Bioluminescence-based quantitative growth assays are very efficient and can be used to optimize experimental conditions for plant-bacteria interactions. As an example, we established experimental conditions in which Tak-1, Tak-2, and their progeny BC3-38, displayed the same level of bacterial resistance (Figure 7D), ensuring fair genetic comparisons of transgenic or mutant plants to the Tak-1 and Tak-2 parents. Our findings are in sharp contrast to those of Gimenez-Ibanez et al. (2019), who reported differences in Pto growth on Tak-1 and Tak-2. Major differences between the two studies are plant age and growth conditions. We used 14-day-old thalli grown under continuous light, whereas Gimenez-Ibanez et al. used 2- to 4-week-old thalli grown under long day conditions (16 h light/8 h dark cycle). Because developmental differences between Tak-1 and Tak-2 become more prominent as plants grow older, we speculate that the use of relatively young thalli and sampling from the defined basal region, where Pto preferentially colonize and grow, are key for fair comparisons between the genotypes. Moreover, we found that the male and female lines of BoGa accessions shared a similar level of resistance but were slightly more susceptible to Pto-lux than Takaragaike accessions (Figure 7D). Thus, we argue that our experimental system with Pto-lux will facilitate genetic analysis of M. polymorph-Pto interactions.
Bacteria are causative agents of plant diseases and are, at the same time, major constituents of the collective assemblage of plant-colonizing microbes called the plant microbiota, which affects plant health (Pfeilmeier et al., 2016; Hacquard et al., 2017). Thus, in addition to advancing our understanding of plant-bacterial pathogen interactions, it is becoming increasingly important to elucidate the mechanisms by which plants and bacterial members of the plant microbiota establish mutualistic relationships. We anticipate that the bioluminescence-based quantitative and spatial detection of bacteria enabled by pBJ vectors will serve as a new tool to facilitate these rapidly growing research fields of plant-bacteria interactions.
Methods
Plant materials and growth conditions
A. thaliana accession Col-0 and its mutants rpm1-3 rps2-101C (Mackey et al., 2003) and dde2-2 ein2-1 pad4-1 sid2-2 (Tsuda et al., 2009), N. benthamiana, S. lycopersicum cv. M82, and M. polymorpha accessions Tak-1 (male), Tak-2 (female), BC3-38 (female), BoGa-L5 (male), and Boga-L2 (female) (Althoff et al., 2014; Bowman et al., 2017; Yamaoka et al., 2018) were used in this study. A, thaliana plants were grown in a climate chamber at 22°C with a 10-h light period and 60% relative humidity. N. benthamiana plants were grown in an air-conditioned room at 22°C with a 16-h light period. S. lycopersicum plants were grown in a climate chamber with a 16 h light/8 h dark cycle at 24°C during the day and 22°C at night with 60% relative humidity. M. polymorpha gemmae or thalli were grown in a walk-in growth chamber at 22°C under 50–60 μmol photons m−2 s−1 continuous white LED light.
Construction of pBJ vectors
The pBBR1-MCS5 derivative pJN105 was cut with XhoI and NotI. The larger fragment was treated with T4 DNA polymerase followed by circularization with T4 DNA ligase, yielding pJN105ΔpBAD. Two DNA fragments encoding tnsABCD or luxCDABE were amplified by PCR from pBEN276 (Howe et al., 2010) using the primer sets Pfrr_inside_F plus New_Tn7L_pJN_M13F or Pfrr_inside_R plus pBEN_Pamp_pJN_M13R, respectively. These DNA fragments were assembled using the NEBuilder HiFi DNA Assembly Cloning Kit (NEB, E2621) into ApaI-digested pJN105ΔpBAD, yielding pBJ1. The kanamycin promoter (Pkan) was amplified from pBBR1-MCS2 using Pkan_pBJ_F2 and Pkan_pBJ_R2 and assembled into XhoI-digested pBJ1 to replace the frr promoter (Pfrr), resulting in pBJ2.
pBJ3 and pBJ4 were designed to confer kanamycin resistance instead of the gentamicin resistance conferred by pBJ1/2. A DNA fragment was amplified from pBEN276 using Pfrr_inside_R plus pBEN_Pamp_pJN_M13R, and another DNA fragment was amplified from pBJ1 using Pfrr_inside_F plus pBJ_dXhoI_Tn7R. These two fragments were then assembled into ApaI-digested pBBR1-MCS2, yielding pBJ3. pBJ4 was constructed by replacing Pfrr in pBJ3 with Pkan as described above.
To broaden the host range of the pBJ vectors, a broad-host-range RK2 replicon was amplified from pFREE-RK2 using OriV_RK2_Fwd plus OriV_RK2_Rev and assembled into NaeI-digested pBJ1 and pBJ2 to produce pBJ5 and pBJ6, respectively. For a similar purpose, the broad-host-range cosmid vector pLAFR3 was cut with EcoRI and treated with T4 DNA polymerase, followed by further digestion with PstI. This linearized vector was used as a recipient for a NaeI-SbfI restriction fragment of pBJ2 containing the tnsABCD-Pkan-luxCDABE cassette, yielding pBJ7.
Primers used for the construction of pBJ vectors are listed in Supplemental Table 2.
Generation of bioluminescent bacteria
pBJ vectors were used to generate chromosomally luxCDABE-tagged P. syringae, A. tumefaciens, and R. solanacearum. A step-by-step protocol for generation of luxCDABE-tagged Pto by pBJ2 is given in the supplemental experimental procedures. Pto, P. syringae SUPP1331, and SUPP1141 are resistant to rifampicin. A. tumefaciens GV3101 and CH3 are resistant to rifampicin/gentamicin and rifampicin/kanamycin, respectively. Antibiotics were used at the following concentrations: rifampin, 50 μg/ml; kanamycin, 50 μg/ml; gentamicin, 10 μg/ml; and tetracycline, 20 μg/ml. Single colonies of Pto, Pto hrcC−, Pto COR−, Pto ΔavrPto ΔavrPtoB, and P. syringae SUPP1331 and SUPP1141 were inoculated into King’s B (KB) medium with the appropriate antibiotics. The overnight cultures were washed twice with 300 mM sucrose at room temperature and suspended in 300 mM sucrose. The competent cells were transformed with pBJ1 (gentamicin resistance), pBJ2 (gentamicin resistance), pBJ4 (kanamycin resistance), pBJ5 (gentamicin resistance), pBJ6 (gentamicin resistance), or pBJ7 (tetracycline resistance) by electroporation using a Gene Pulser Xcell PC system (Bio-Rad). The electroporation conditions were 2.5 kV, 25 μF, and 200 Ω. Bacteria were recovered for 2–4 h at 28°C in KB media, then streaked onto KB plates with appropriate antibiotics and incubated at 28°C for 4–5 days. Bioluminescent colonies were detected using a FUSION Solo S system (Vilber), inoculated into KB medium containing 0.1% L-arabinose, and cultured overnight at 28°C to induce transposition. The cultures were streaked onto KB plates with rifampicin and placed in an incubator at 28°C for 2–3 days until single colonies appeared. Bioluminescent colonies were patched onto KB plates with appropriate antibiotics to select gentamicin-, kanamycin-, or tetracycline-sensitive colonies, confirming the absence of pBJ vectors. Insertion of luxCDABE into the attTn7 sites was determined by sequencing. pLAFR3 carrying avrRpt2 or avrRpm1 was introduced by electroporation into Pto chromosomally tagged with Pkan-driven luxCDABE (Pto-lux), which had been generated with pBJ2.
A single colony of A. tumefaciens GV3101 was inoculated into LB medium containing rifampicin and gentamicin, grown at 28°C to OD600 = 0.8, collected by centrifugation, and suspended in ice-chilled 20 mM CaCl2. The competent cells were mixed with pBJ3 or pBJ4 and flash-frozen in liquid nitrogen, followed by incubation at 37°C for 30 min. Bacteria were recovered in SOC medium at 28°C for 2 h and then streaked onto LB plates with rifampicin, gentamicin, and kanamycin. A single colony of A. tumefaciens CH3 was inoculated into LB medium containing rifampicin, grown overnight, collected by centrifugation, and washed with and suspended in ice-chilled 10% glycerol. The competent cells were transformed with pBJ7 by electroporation (25 kV, 25 μF, and 200 Ω). Bacteria were recovered in SOC medium at 28°C for 2 h and then streaked onto LB plates with rifampicin and tetracycline. After 2–3 days of incubation at 28°C, single bioluminescent colonies of these A. tumefaciens strains were inoculated into LB medium containing 0.1% L-arabinose and cultured overnight at 28°C to induce transposition. The culture was streaked onto LB plates with the appropriate antibiotics and placed in an incubator at 28°C for 2 days until single colonies appeared. Bioluminescent colonies were patched onto LB plates with the appropriate antibiotics to select kanamycin- or tetracycline-sensitive colonies, confirming the absence of pBJ vectors. Chromosomal integration of luxCDABE was determined by sequencing.
A single colony of R. solanacearum OE1-1 (Kanda et al., 2003) was inoculated into PY medium, grown at 30°C to OD600 = 0.8, collected by centrifugation, and suspended in ice-chilled 10% glycerol. The competent cells were transformed with pBJ7 by electroporation (3 kV, 25 μF, and 300 Ω). Bacteria were recovered in SOC medium at 30°C for 2 h and then streaked onto PY plates containing tetracycline, 0.0005% crystal violet, and 0.02% 2,3,5-triphenyltetrazolium chloride. A single bioluminescent colony was then inoculated into PY medium containing 0.1% L-arabinose and cultured overnight at 28°C to induce transposition. The culture was streaked onto PY plates with crystal violet and 2,3,5-triphenyltetrazolium chloride and placed in an incubator at 30°C for 2 days until single colonies appeared. Bioluminescent colonies were patched onto PY plates with or without tetracycline to select tetracycline-sensitive colonies, confirming the absence of pBJ7. Chromosomal integration of luxCDABE was determined by sequencing.
Deletion of cmaA from Pto-lux
Two DNA fragments containing the 1-kb upstream or downstream region of cmaA were PCR-amplified from genomic DNA of Pto using the primer sets cmaA_up1000_F plus cmaA_del_Rev or cmaA_del_Fwd plus cmaA_down_1000_Rev. These fragments were assembled into XbaI-digested pK18mobsacB. This construct was introduced into Pto-lux by electroporation. Bacteria were recovered in KB medium at 28°C for 2 h and spotted onto a KB plate with rifampicin and kanamycin. The deletion of cmaA was induced by counter-selection on a KB plate containing rifampicin and 10% sucrose and was confirmed by size reduction of a PCR fragment amplified by cmaA_conf_Fwd plus cmaA_conf_Rev. Primers used for the generation of Pto-lux ΔcmaA are listed in Supplemental Table 2.
Inoculation of plants with P. syringae
Bacterial suspensions in sterile water were directly infiltrated into leaves of 4- to 5-week-old A. thaliana, 5- to 6-week-old N. benthamiana, or 3- to 4-week-old S. lycopersicum using a needleless syringe. In each independent experiment, one biological replicate consisted of four leaf discs (4 mm diameter), and six to eight biological replicates were collected for each treatment. Alternatively, 4- to 5-week-old A. thaliana plants were spray-inoculated with bacterial suspensions in 10 mM MgCl2 with 0.04% Silwet L-77. After inoculation, plants were kept under covers for 1 h, and then the covers were removed. In each independent experiment, one biological replicate consisted of six leaf discs (4 mm diameter) excised from three different leaves of the same plant, and eight biological replicates were collected for each treatment.
M. polymorpha gemmae were placed on half-strength Gamborg's B5 agar covered with a Whatman filter paper (cat. no. 1001-085) and grown for 14 days at 22°C under continuous white LED light. The 14-day-old plants were transferred to fresh empty Petri dishes, submerged in the bacterial suspensions, and incubated with or without vacuum for 5 min. After the incubations, plants were placed onto a Whatman filter paper that had been soaked with Milli-Q water in fresh empty Petri dishes. Inoculated plants were kept in a climate chamber at 22°C under a 16 h light/8 h dark cycle. Alternatively, M. polymorpha gemmae were grown on half-strength Gamborg's B5 agar without the filter paper, and the bacterial suspension was poured into the cultured agar plate and vacuum infiltrated (Supplemental Figure 10A). The bacterial suspension was decanted, and plants were further cultured in a climate chamber at 22°C under a 16 h light/8 h dark cycle. In the time-course experiment shown in Supplemental Figure 8, vacuum-inoculated plants were placed on a water-soaked filter paper in a Petri dish and incubated with the lid sealed with surgical tape (ambient humidity) for sampling at 2 and 3 dpi and with the lid sealed with Parafilm (high humidity) for sampling at 3 and 7 dpi. In each independent experiment, one biological replicate consisted of one thallus disc (5 mm diameter) excised from an individual plant, and eight biological replicates were collected for each treatment.
Quantification of P. syringae growth
Bioluminescence measurements were performed with the GloMax Navigator Microplate Luminometer (Promega) or the FLUOstar Omega (BMG Labtech) plate reader. For A. thaliana, N. benthamiana, and S. lycopersicum, leaf discs (4 mm diameter) were punched using a biopsy punch and placed in a white, light-reflecting 96-well plate (Corning, no. 3912). The plate containing leaf discs in each well was placed in the sample drawer of the GloMax Navigator and kept in the dark by closing the lid for 10 min to reduce background signals. Then, the luminescence of each sample was measured for 10 s. After measurement, leaf discs were pulverized in 400 μl of 5 mM MgSO4, and serial dilutions were made. From each dilution, 10 μl was streaked on KB plates containing rifampicin. Log10-transformed RLUs and CFUs per cm2 leaf surface area were calculated. For M. polymorpha, a thallus disc (5 mm diameter) was punched from the basal region of the thallus using a biopsy punch (Figure S10B). The thallus discs were transferred to wells of a white-reflecting 96-well plate (VWR, no. 738-0016). The plate was placed in the FLUOstar Omega (BMG Labtech) plate reader and kept in the dark for 10 min before measurement to reduce background signals. Then, luminescence of each sample was measured for 5 s. After the measurement, the thallus discs were ground in 400 μl of 10 mM MgSO4 using a mixing mill (MM400, Retsch) at 30 Hz for 5 min, and serial dilutions were made. From each dilution, 10 μl was streaked on KB plates containing rifampicin. The plates were then incubated at 22°C for 3 days until the colonies became sufficiently visible for counting. Log10-transformed RLUs and CFUs per cm2 thallus surface area were calculated. Pairwise comparison was performed using the R function pairwise.t.test with pooled SD, and the Benjamini-Hochberg method was used to correct for multiple hypothesis testing. Linear regression and calculation of correlation coefficients were performed using the R functions lm and cor, respectively.
Bioluminescence imaging
The FUSION Solo S (Vilber) or ChemiDoc MP Imaging System (Bio-Rad) was used to visualize the luminescence of plant tissues inoculated with Pto-lux or A. tumefaciens CH3-lux. Detached leaves of A. thaliana or S. lycopersicum were placed on a wet paper and then placed into the sample drawer of the FUSION Solo S instrument. Before imaging, samples were kept inside the instrument with the lid closed to reduce background signals. M. polymorpha thalli on a Whatman filter paper were imaged using the ChemiDoc MP Imaging System. After 10 min of dark adaptation, images were obtained using signal accumulation mode. The images were reduced to the signal-showing pixels, hue shifted, and merged with microscopy images of the infected thalli using the GNU image manipulation program.
Funding
This work was supported by JST PRESTO (JPMJPR17Q6) and Grant-in-Aid for Scientific Research (B) (19H02960) to A. Mine, by the Ritsumeikan Global Innovation Research Organization to A.T., and by funds from the Max Planck Society and the “Priority Programme 2237 MAdLand” funded by the Deutsche Forschungsgemeinschaft (NA 946/1-1) to H.N.
Author contributions
A. Mine designed the research. A. Mine and H.N. conceived the experiments. A. Mine, A. Matsumoto, T.S., K.M., and H.N. performed the experiments and analyzed the data. A. Mine, H.N., and A.T. wrote the paper.
Acknowledgment
We thank Yuichi Takikawa (Shizuoka University) for providing P. syringae SUPP1331 and SUPP1141 and A. tumefaciens CH3, Yasufumi Hikichi (Kochi University) for providing R. solanacearum OE1-1, and Noriko Shimizu (Ritsumeikan University) for technical assistance. We also thank Kenichi Tsuda (Huazhong Agricultural University) and Tatsuya Nobori (Salk Institute) for critical reading of the manuscript and Rozina Kardakaris (MPIPZ) for editing the manuscript. pBEN276, pBBR1MCS-2, and pFREE-RK2 were gifts from Pierre Germon (Addgene plasmid no. 85168), Kenneth Peterson (Addgene plasmid no. 85168), and Morten Norholm (Addgene plasmid no. 92054), respectively. pK18mobsacB was obtained from the National BioResource Project (NIG, Japan): E. coli. The tomato resource used in this study was provided by the National BioResource Project (NBRP), MEXT, JAPAN. Supplemental Figures 3 and 10 were created using BioRender.com. No conflict of interest declared.
Published: July 20, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Accession numbers
The accession numbers for the genes discussed in this article are as follows: RPM1 (AT3G07040), RPS2 (AT4G26090), DDE2 (AT5G42650), EIN2 (AT5G03280), PAD4 (AT3G52430), SID2 (At1g74710), avrPto (avrPto1; PSPTO_4001), avrPtoB (hopAB2; PSPTO_3087), and cmaA (PSPTO_4709).
Supplemental information
References
- Althoff F., Kopischke S., Zobell O., Ide K., Ishizaki K., Kohchi T., Zachgo S. Comparison of the MpEF1alpha and CaMV35 promoters for application in Marchantia polymorpha overexpression studies. Transgenic Res. 2014;23:235–244. doi: 10.1007/s11248-013-9746-z. [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Staskawicz B.J. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Kohchi T., Yamato K.T., Jenkins J., Shu S., Ishizaki K., Yamaoka S., Nishihama R., Nakamura Y., Berger F., et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017;171:287–304 e215. doi: 10.1016/j.cell.2017.09.030. [DOI] [PubMed] [Google Scholar]
- Brooks D.M., Hernandez-Guzman G., Kloek A.P., Alarcon-Chaidez F., Sreedharan A., Rangaswamy V., Penaloza-Vazquez A., Bender C.L., Kunkel B.N. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 2004;17:162–174. doi: 10.1094/MPMI.2004.17.2.162. [DOI] [PubMed] [Google Scholar]
- Buschmann H., Holtmannspotter M., Borchers A., O'Donoghue M.T., Zachgo S. Microtubule dynamics of the centrosome-like polar organizers from the basal land plant Marchantia polymorpha. New Phytol. 2016;209:999–1013. doi: 10.1111/nph.13691. [DOI] [PubMed] [Google Scholar]
- Choi K.H., Schweizer H.P. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 2006;1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- Choi K.H., DeShazer D., Schweizer H.P. mini-Tn7 insertion in bacteria with multiple glmS-linked attTn7 sites: example Burkholderia mallei ATCC 23344. Nat. Protoc. 2006;1:162–169. doi: 10.1038/nprot.2006.25. [DOI] [PubMed] [Google Scholar]
- Cruz A.P., Ferreira V., Pianzzola M.J., Siri M.I., Coll N.S., Valls M. A novel, sensitive method to evaluate potato germplasm for bacterial wilt resistance using a luminescent Ralstonia solanacearum reporter strain. Mol. Plant Microbe Interact. 2014;27:277–285. doi: 10.1094/MPMI-10-13-0303-FI. [DOI] [PubMed] [Google Scholar]
- Cui F., Wu S., Sun W., Coaker G., Kunkel B., He P., Shan L. The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol. 2013;162:1018–1029. doi: 10.1104/pp.113.219659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J.E. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- Dou D., Zhou J.M. Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe. 2012;12:484–495. doi: 10.1016/j.chom.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Du M., Zhai Q., Deng L., Li S., Li H., Yan L., Huang Z., Wang B., Jiang H., Huang T., et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell. 2014;26:3167–3184. doi: 10.1105/tpc.114.128272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Crooks C., Lamb C. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 2008;53:393–399. doi: 10.1111/j.1365-313X.2007.03303.x. [DOI] [PubMed] [Google Scholar]
- Geng X., Shen M., Kim J.H., Mackey D. The Pseudomonas syringae type III effectors AvrRpm1 and AvrRpt2 promote virulence dependent on the F-box protein COI1. Plant Cell Rep. 2016;35:921–932. doi: 10.1007/s00299-016-1932-z. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Zamarreno A.M., Garcia-Mina J.M., Solano R. An evolutionarily ancient immune system governs the interactions between Pseudomonas syringae and an early-diverging land plant lineage. Curr. Biol. 2019;29:2270–2281.e4. doi: 10.1016/j.cub.2019.05.079. [DOI] [PubMed] [Google Scholar]
- Hacquard S., Spaepen S., Garrido-Oter R., Schulze-Lefert P. Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 2017;55:565–589. doi: 10.1146/annurev-phyto-080516-035623. [DOI] [PubMed] [Google Scholar]
- Hatsugai N., Iwasaki S., Tamura K., Kondo M., Fuji K., Ogasawara K., Nishimura M., Hara-Nishimura I. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 2009;23:2496–2506. doi: 10.1101/gad.1825209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P., Shan L., Lin N.C., Martin G.B., Kemmerling B., Nurnberger T., Sheen J. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- Hikichi Y., Nasu-Nakazawa Y., Kitanosono S., Suzuki K., Okuno T. The behavior of genetically lux-marked Ralstonia solanacearum in grafted tomato cultivars resistant or susceptible to bacterial wilt. Ann. Phytopathol. Soc. Jpn. 1999;65:597–603. doi: 10.3186/jjphytopath.65.597. [DOI] [Google Scholar]
- Hikichi Y., Suzuki K., Toyoda K., Horikoshi M., Hirooka T., Okuno T. Successive observation of growth and movement of genetically lux-marked Pseudomonas cichorii and the response of host tissues in the same lettuce leaf. Ann. Phytopathol. Soc. Jpn. 1998;64:519–525. doi: 10.3186/jjphytopath.64.519. [DOI] [Google Scholar]
- Honkanen S., Jones V.A.S., Morieri G., Champion C., Hetherington A.J., Kelly S., Proust H., Saint-Marcoux D., Prescott H., Dolan L. The mechanism forming the cell surface of tip-growing rooting cells is conserved among land plants. Curr. Biol. 2016;26:3238–3244. doi: 10.1016/j.cub.2016.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K., Karsi A., Germon P., Wills R.W., Lawrence M.L., Bailey R.H. Development of stable reporter system cloning luxCDABE genes into chromosome of Salmonella enterica serotypes using Tn7 transposon. BMC Microbiol. 2010;10:197. doi: 10.1186/1471-2180-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huchelmann A., Boutry M., Hachez C. Plant glandular trichomes: natural cell factories of high biotechnological interest. Plant Physiol. 2017;175:6–22. doi: 10.1104/pp.17.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K., Chiyoda S., Yamato K.T., Kohchi T. Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 2008;49:1084–1091. doi: 10.1093/pcp/pcn085. [DOI] [PubMed] [Google Scholar]
- Ishizaki K., Johzuka-Hisatomi Y., Ishida S., Iida S., Kohchi T. Homologous recombination-mediated gene targeting in the liverwort Marchantia polymorpha L. Sci. Rep. 2013;3:1532. doi: 10.1038/srep01532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K., Mizutani M., Shimamura M., Masuda A., Nishihama R., Kohchi T. Essential role of the E3 ubiquitin ligase nopperabo1 in schizogenous intercellular space formation in the liverwort Marchantia polymorpha. Plant Cell. 2013;25:4075–4084. doi: 10.1105/tpc.113.117051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jittawuttipoka T., Buranajitpakorn S., Fuangthong M., Schweizer H.P., Vattanaviboon P., Mongkolsuk S. Mini-Tn7 vectors as genetic tools for gene cloning at a single copy number in an industrially important and phytopathogenic bacteria, Xanthomonas spp. FEMS Microbiol. Lett. 2009;298:111–117. doi: 10.1111/j.1574-6968.2009.01707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda A., Yasukohchi M., Ohnishi K., Kiba A., Okuno T., Hikichi Y. Ectopic expression of Ralstonia solanacearum effector protein PopA early in invasion results in loss of virulence. Mol. Plant Microbe Interact. 2003;16:447–455. doi: 10.1094/MPMI.2003.16.5.447. [DOI] [PubMed] [Google Scholar]
- Kernell Burke A., Duong D.A., Jensen R.V., Stevens A.M. Analyzing the transcriptomes of two quorum-sensing controlled transcription factors, RcsA and LrhA, important for Pantoea stewartii virulence. PLoS One. 2015;10:e0145358. doi: 10.1371/journal.pone.0145358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.G., Geng X., Lee S.Y., Mackey D. The Pseudomonas syringae type III effector AvrRpm1 induces significant defenses by activating the Arabidopsis nucleotide-binding leucine-rich repeat protein RPS2. Plant J. 2009;57:645–653. doi: 10.1111/j.1365-313X.2008.03716.x. [DOI] [PubMed] [Google Scholar]
- Kovach M.E., Elzer P.H., Hill D.S., Robertson G.T., Farris M.A., Roop R.M., 2nd, Peterson K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Kovács K., Hill P.J., Grierson D., Dodd C.E.R., Pamfil D., Fray R.G. Development of a novel inducible bioluminescent and antibiotic resistance tagging system and its use to investigate the role of antibiotic production by Pectobacterium carotovorum ssp. carotovorum during potato tuber infection. Eur. J. Plant Pathol. 2009;125:655–664. doi: 10.1007/s10658-009-9513-4. [DOI] [Google Scholar]
- Lin N.C., Martin G.B. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol. Plant Microbe Interact. 2005;18:43–51. doi: 10.1094/MPMI-18-0043. [DOI] [PubMed] [Google Scholar]
- Liu M., Durfee T., Cabrera J.E., Zhao K., Jin D.J., Blattner F.R. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J. Biol. Chem. 2005;280:15921–15927. doi: 10.1074/jbc.M414050200. [DOI] [PubMed] [Google Scholar]
- Ma S.-W., Morris V.L., Cuppels D.A. Characterization of a DNA region required for production of the phytotoxin coronatine by Pseudomonas syringae pv. tomato. Mol. Plant Microbe Interact. 1991;4:69–74. [Google Scholar]
- Mackey D., Holt B.F., 3rd, Wiig A., Dangl J.L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Mackey D., Belkhadir Y., Alonso J.M., Ecker J.R., Dangl J.L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Meighen E.A. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 1993;7:1016–1022. doi: 10.1096/fasebj.7.11.8370470. [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Mine A., Seyfferth C., Kracher B., Berens M.L., Becker D., Tsuda K. The defense phytohormone signaling network enables rapid, high-amplitude transcriptional reprogramming during effector-triggered immunity. Plant Cell. 2018;30:1199–1219. doi: 10.1105/tpc.17.00970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine A., Berens M.L., Nobori T., Anver S., Fukumoto K., Winkelmuller T.M., Takeda A., Becker D., Tsuda K. Pathogen exploitation of an abscisic acid- and jasmonate-inducible MAPK phosphatase and its interception by Arabidopsis immunity. Proc. Natl. Acad. Sci. U S A. 2017;114:7456–7461. doi: 10.1073/pnas.1702613114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S., Fujisawa M., Sone T., Nakayama S., Nishiyama R., Takenaka M., Yamaoka S., Sakaida M., Kono K., Takahama M., et al. Construction of male and female PAC genomic libraries suitable for identification of Y-chromosome-specific clones from the liverwort, Marchantia polymorpha. Plant J. 2000;24:421–428. doi: 10.1046/j.1365-313x.2000.00882.x. [DOI] [PubMed] [Google Scholar]
- Park J.Y., Lee Y.H., Yang K.Y., Kim Y.C. AiiA-mediated quorum quenching does not affect virulence or toxoflavin expression in Burkholderia glumae SL2376. Lett. Appl. Microbiol. 2010;51:619–624. doi: 10.1111/j.1472-765X.2010.02940.x. [DOI] [PubMed] [Google Scholar]
- Peng J., Schachterle J.K., Sundin G.W. Orchestration of virulence factor expression and modulation of biofilm dispersal in Erwinia amylovora through activation of the Hfq-dependent small RNA RprA. Mol. Plant Pathol. 2021;22:255–270. doi: 10.1111/mpp.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.E., Craig N.L. Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol. 2001;2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- Pfeilmeier S., Caly D.L., Malone J.G. Bacterial pathogenesis of plants: future challenges from a microbial perspective: challenges in bacterial molecular plant pathology. Mol. Plant Pathol. 2016;17:1298–1313. doi: 10.1111/mpp.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Marques M., Kressin J.P., Kashyap A., Panthee D.R., Louws F.J., Coll N.S., Valls M. Four bottlenecks restrict colonization and invasion by the pathogen Ralstonia solanacearum in resistant tomato. J. Exp. Bot. 2020;71:2157–2171. doi: 10.1093/jxb/erz562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldan R., Nattapong S., Marcel B.-P., Indra B., Huang Wei E., Preston Gail M. From macro to micro: a combined bioluminescence-fluorescence approach to monitor bacterial localization. Environmental Microbiology. 2021;23(4):2070–2085. doi: 10.1111/1462-2920.15296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani F., Banic E., Florent S.N., Kanazawa T., Goodger J.Q.D., Mentink R.A., Dierschke T., Zachgo S., Ueda T., Bowman J.L., et al. Oil body formation in Marchantia polymorpha is controlled by MpC1HDZ and serves as a defense against arthropod herbivores. Curr. Biol. 2020;30:2815–2828.e8. doi: 10.1016/j.cub.2020.05.081. [DOI] [PubMed] [Google Scholar]
- Romero-Jimenez L., Rodriguez-Carbonell D., Gallegos M.T., Sanjuan J., Perez-Mendoza D. Mini-Tn7 vectors for stable expression of diguanylate cyclase PleD∗ in Gram-negative bacteria. BMC Microbiol. 2015;15:190. doi: 10.1186/s12866-015-0521-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichwart S., Hetzler S., Broker D., Steinbuchel A. Extension of the substrate utilization range of Ralstonia eutropha strain H16 by metabolic engineering to include mannose and glucose. Appl. Environ. Microbiol. 2011;77:1325–1334. doi: 10.1128/AEM.01977-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Yang B. Mutagenesis of 18 type III effectors reveals virulence function of XopZ(PXO99) in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 2010;23:893–902. doi: 10.1094/MPMI-23-7-0893. [DOI] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S.S., Shirakawa M., Takagi J., Matsuda Y., Shimada T., Hara-Nishimura I., Kohchi T. CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 2014;55:475–481. doi: 10.1093/pcp/pcu014. [DOI] [PubMed] [Google Scholar]
- Szpirer C.Y., Faelen M., Couturier M. Mobilization function of the pBHR1 plasmid, a derivative of the broad-host-range plasmid pBBR1. J. Bacteriol. 2001;183:2101–2110. doi: 10.1128/JB.183.6.2101-2110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Stoddard T., Glazebrook J., Katagiri F. Network properties of robust immunity in plants. PLoS Genet. 2009;5:e1000772. doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge S., Ikawa Y., Hikichi Y., Nakazawa-Nasu Y., Suzuki K., Kubo Y., Horino O. Behavior of bioluminescent transconjugants of Xanthomonas oryzae pv. oryzae in compatible and incompatible rice leaves. Ann. Phytopathol. Soc. Jpn. 1999;65:93–99. doi: 10.3186/jjphytopath.65.470. [DOI] [Google Scholar]
- Wei C.-F., Kvitko B.H., Shimizu R., Crabill E., Alfano J.R., Lin N.-C., Martin G.B., Huang H.-C., Collmer A. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 2007;51:32–46. doi: 10.1111/j.1365-313X.2007.03126.x. [DOI] [PubMed] [Google Scholar]
- Wiles T.J., Wall E.S., Schlomann B.H., Hay E.A., Parthasarathy R., Guillemin K. Modernized tools for streamlined genetic manipulation and comparative study of wild and diverse proteobacterial lineages. mBio. 2018;9 doi: 10.1128/mBio.01877-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi C., Lambrecht M., Vanderleyden J., Michiels J. Bi-functional gfp- and gusA-containing mini-Tn5 transposon derivatives for combined gene expression and bacterial localization studies. J. Microbiol. Methods. 1999;35:85–92. doi: 10.1016/s0167-7012(98)00103-1. [DOI] [PubMed] [Google Scholar]
- Xin X.F., Kvitko B., He S.Y. Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 2018;16:316–328. doi: 10.1038/nrmicro.2018.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka S., Nishihama R., Yoshitake Y., Ishida S., Inoue K., Saito M., Okahashi K., Bao H., Nishida H., Yamaguchi K., et al. Generative cell specification requires transcription factors evolutionarily conserved in land plants. Curr. Biol. 2018;28:479–486.e5. doi: 10.1016/j.cub.2017.12.053. [DOI] [PubMed] [Google Scholar]
- Yuan J., He S.Y. The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J. Bacteriol. 1996;178:6399–6402. doi: 10.1128/jb.178.21.6399-6402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cao Y., Zhang L., Ohnishi K., Hikichi Y., Li J. The Tn7-based genomic integration is dependent on an attTn7 box in the glms gene and is site-specific with monocopy in Ralstonia solanacearum species complex. Mol. Plant Microbe Interact. 2021 doi: 10.1094/MPMI-11-20-0325-SC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.