Abstract
Objective
We assessed the clinical performance of novel Roche Elecsys SARS-CoV-2 Antigen fully automated electrochemiluminescence immunoassay (ECLIA).
Design and methods
We tested 160 subjects, 110 (68.8%), with positive molecular test for SARS-CoV-2 infection in nasopharyngeal samples, with Altona Diagnostics RealStar SARS-CoV-2 RT-PCR Kit and Roche Elecsys SARS-CoV-2 Antigen.
Results
Highly significant correlation was found between Elecsys SARS-CoV-2 Antigen ECLIA and cycle threshold (Ct) values of SARS-CoV-2 S and E genes (both r = −0.91; p < 0.001). The area under the curve (AUC), sensitivity and specificity of Elecsys SARS-CoV-2 Antigen ECLIA were 0.83, 0.43 and 1.00 in all samples, 0.99, 0.87 and 0.99 in those with both Ct values < 30, as well as 1.00, 1.00 and 0.89 in samples with both Ct values < 25.
Conclusion
Roche Elecsys SARS-CoV-2 Antigen ECLIA may be a surrogate of molecular testing for identification of super-spreaders.
Keywords: COVID-19, SARS-CoV-2, Laboratory medicine, Diagnosis, Immunoassay
1. Introduction
Although the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA remains the gold standard in coronavirus disease 2019 (COVID-19) diagnostics, widespread usage of molecular tests is plagued by bottlenecks such as relatively limited throughput and long turnaround time. The poor availability of reagents for performing molecular assays in many countries is another of such drawbacks. According to a recent worldwide survey disseminated by the American Association of Clinical Chemistry (AACC), the large majority of testing facilities (i.e., up to 80%) are engaged in strenuous challenges to perform routine or urgent SARS-CoV-2 diagnostic testing due to staff shortage and difficulties in reagents supply [1], thus leaving many laboratories with large backlogs of untested samples. Owing to the paramount volume of diagnostics tests still needed to contrast the ongoing COVID-19 pandemic, rapid antigen tests are increasingly proposed as feasible alternatives for high-throughput and fast diagnosis of SARS-CoV-2 infections, especially in subjects carrying high viral load [2,3]. Therefore, the purpose of this study was to assess the clinical performance of the novel Elecsys SARS-CoV-2 Antigen immunoassay.
2. Materials and methods
The study population consisted of all consecutive subjects referred with suspected COVID-19 (either symptomatic or for being close contacts of SARS-CoV-2 positive subjects) to the Laboratory Medicine Service of Pederzoli Hospital (Peschiera del Garda, Verona, Italy), between August 16 and September 15, 2021. A nasopharyngeal swab (Virus swab UTM Copan, Brescia, Italy) was taken at admission and immediately conveyed to the laboratory. SARS-CoV-2 molecular testing was performed with Altona Diagnostics RealStar SARS-CoV-2 RT-PCR Kit (Altona Diagnostics GmbH, Hamburg, Germany), a real-time reverse transcription polymerase chain reaction (rRT-PCR) based on double amplification and detection of SARS-CoV-2 E and S genes. The assay was performed with Bio-Rad CFX96™ Deep Well Dx Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA), whilst results were classified as positive when cycle threshold (Ct) values of both SARS-CoV-2 S and E genes were <45.
SARS-CoV-2 antigen testing was performed with the novel electrochemiluminescence immunoassay (ECLIA) Roche Elecsys SARS-CoV-2 Antigen on Roche Cobas 6000 (Roche Diagnostics GmbH, Mannheim, Germany). This technique is based on a one-step double antibody sandwich assay for detecting SARS-CoV-2 nucleocapsid antigen in nasopharyngeal and oropharyngeal swabs. The test sample is reactive when the cut-off index (COI) is ≥ 1. According to manufacturer's declaration the limit of detection is 22 Median Tissue Culture Infectious Dose (TCID50)/mL, whilst the imprecision ranges between 1.9 and 3.5%.
The clinical performance of Elecsys SARS-CoV-2 Antigen ECLIA was compared with results of molecular testing by Spearman's correlation, receiver operating characteristic (ROC) curve analysis, and calculation of diagnostic sensitivity and specificity. Results were expressed as median and interquartile range (IQR). The statistical analysis was carried out with Analyse-it software (Analyse-it Software Ltd, Leeds, UK). This study was part of routine clinical laboratory operations for SARS-CoV-2 screening and diagnosis at the local facility, so that patient informed consent and Ethical Committee approval were unnecessary. The study was conducted in accordance with the Declaration of Helsinki, under the terms of relevant local legislation.
3. Results
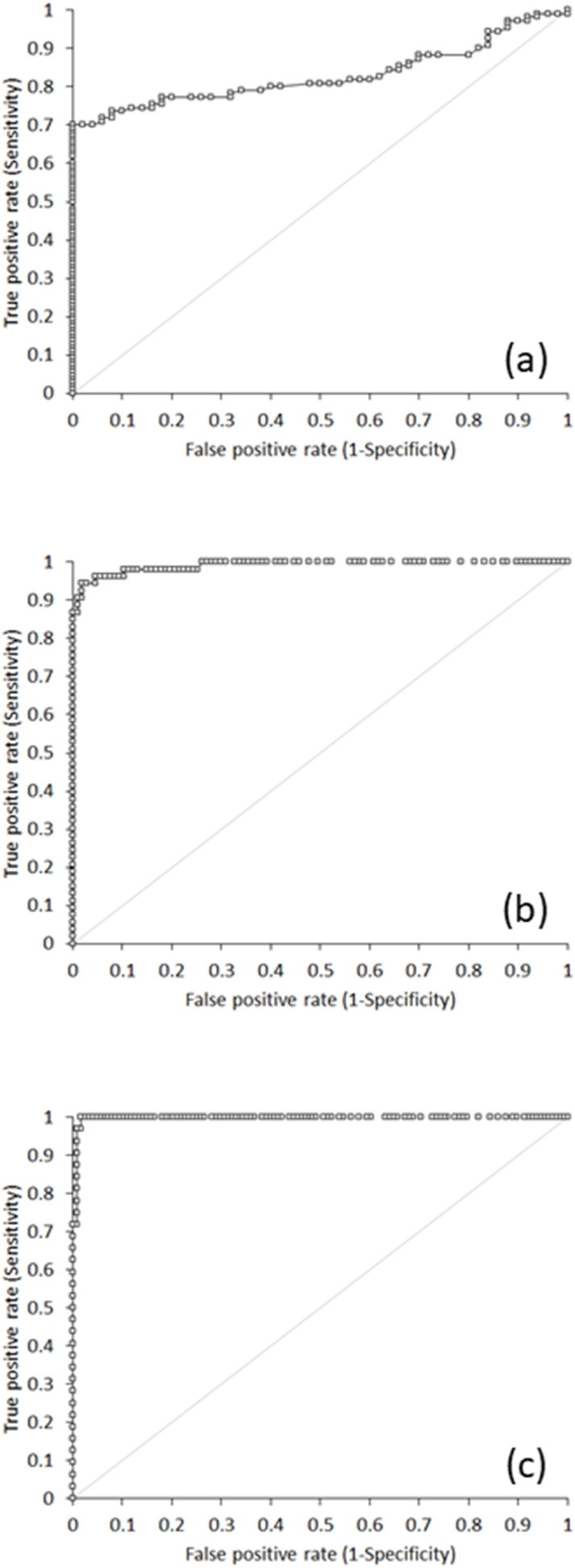
The final study population consisted of 160 patients (median age 38 years, IQR 24–58 years; 69 women), 110 of whom (68.8%) positive at molecular testing (i.e., Ct values of both SARS-CoV-2 S and E genes <45). A highly significant correlation was observed between values of Elecsys SARS-CoV-2 Antigen ECLIA and measurable Ct values of SARS-CoV-2 S and E genes (both r = −0.91; 95% CI, −0.94 to −0.87; p < 0.001). The diagnostic performance of Elecsys SARS-CoV-2 Antigen ECLIA is summarized in Table 1, either cumulative or stratified for the Ct values obtained in nasopharyngeal samples. The overall area under the curve (AUC) for detecting SARS-CoV-2 positive samples was 0.83, with 0.43 sensitivity and 1.00 specificity at manufacturer's recommended COI ≥1 (Fig. 1a). Such performance considerably increased in detecting samples with higher viral load (i.e., Ct values of both genes <30), displaying an AUC of 0.99, with 0.87 sensitivity and 0.99 specificity at COI ≥1 (Fig. 1b). In samples with even higher viral loads (i.e., Ct values of both genes <25), Elecsys SARS-CoV-2 Antigen ECLIA exhibited further improved diagnostic performance, with 1.00 AUC, 1.00 sensitivity and 0.89 specificity (Fig. 1c).
Table 1.
Clinical performance of Roche Elecsys SARS-CoV-2 Antigen fully automated electrochemiluminescence immunoassay (ECLIA) stratified according to cycle threshold (Ct) values.
| Ct values | AUC | Sensitivity | Specificity |
|---|---|---|---|
| All samples | 0.83 (95%CI, 0.77–0.89; p < 0.001 | 0.43 (95%CI, 0.333–0.525) | 1.00 (95%CI, 0.93–1.00) |
| <30 | 0.99 (95%CI, 0.98–1.00; p < 0.0001) | 0.87 (95%CI, 0.75–0.945) | 0.99 (95%CI, 0.95–1.00) |
| <25 | 1.00 (95%CI, 0.99–1.00; p < 0.001) | 1.00 (95%CI, 0.89–1.00) | 0.89 (95%CI, 0.82–0.94) |
AUC, area under the curve; Ct, cycle threshold.
Fig. 1.
Receiver Operating Characteristics (ROC) curves of Roche Elecsys SARS-CoV-2 Antigen fully automated electrochemiluminescence immunoassay (ECLIA) against molecular testing in (a) all nasopharyngeal samples, (b) nasopharyngeal samples with cycle threshold (Ct) values < 30 and (c) nasopharyngeal samples with cycle threshold (Ct) values < 25.
4. Discussion
More than 100 years after the dramatic Spanish flu pandemic which caused nearly 50 million casualties in 1918/19 [4], humanities is now challenged by COVID-19, a new life-threatening viral disease caused by the beta coronavirus SARS-CoV-2, which has already infected over 320 million people and has caused over 5 million deaths according to the Johns Hopkins University of Medicine Coronavirus Resource Center [5]. Such a paramount number of infections and deaths is placing an unprecedented pressure on clinical laboratories all around the world, which now struggle to provide timely test results for early and accurate diagnosis of SARS-CoV-2 infections to all those who need them.
Recent evidence convincingly suggests that widespread testing, also encompassing efficient screening of high-risk contacts of SARS-CoV-2 positive cases and their household members, represents an essential step toward effective control of local COVID-19 outbreaks [6,7]. Therefore, the use of SARS-CoV-2 antigen immunoassays represent a valuable perspective for largely enhancing the current testing capacity, provided that the assay has been clinically validated before its introduction into routine practice [3].
The results of our investigation, which entailed the clinical assessment of the novel, fully automated Elecsys SARS-CoV-2 Antigen ECLIA, attests that this technique provides remarkable performance not only for detecting subjects with high SARS-CoV-2 viral load (i.e., AUC of 1.00 for Ct values of both S and E genes <25), but also as a potentially useful surrogate test for molecular diagnostics, since its displayed acceptable accuracy also as population screening test (AUC of 0.83 on all samples, with 1.00 specificity). Based on its remarkably high specificity, a positive test results would hence not necessarily require to confirm sample positivity with rRT-PCR, whilst the excellent sensitivity for detecting nasopharyngeal samples with SARS-CoV-2 Ct values < 25 will enable to efficiently and timely identify positive subjects with high viral load and increased risk of generating large outbreaks (i.e., the so-called super-spreading events). Notably, Gniazdowski et al. reported that over 97% of SARS-CoV-2 viral cultures are negative in nasopharyngeal samples displaying Altona Ct values < 26.2 [7]. At a similar Ct threshold, Elecsys SARS-CoV-2 Antigen ECLIA displayed 1.00 AUC, 1.00 sensitivity and 0.89 specificity, which would make it a potentially valuable surrogate of molecular testing for mass SARS-CoV-2 screening and for specific identification of super-spreaders [8].
Ethical approval
All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments.
Funding
None.
Author contributions
All authors were involved in designing study, analysing the data and writing the manuscript.
Declaration of competing interest
The authors declare that they have no competing interest.
References
- 1.American Association of Clinical Chemistry COVID-19 survey results. https://www.aacc.org/science-and-research/covid-19-resources/aacc-covid-19-testing-survey/full-survey-results Available at:
- 2.Mattiuzzi C., Henry B.M., Lippi G. Making sense of rapid antigen testing in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) diagnostics. Diagnosis (Berl) 2020 Nov 26 doi: 10.1515/dx-2020-0131. dx-2020-0131. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 3.Bohn M.K., Lippi G., Horvath A.R., Erasmus R., Grimmler M., Gramegna M., et al. IFCC interim guidelines on rapid point-of-care antigen testing for SARS-CoV-2 detection in asymptomatic and symptomatic individuals. Clin. Chem. Lab. Med. 2021 doi: 10.1515/cclm-2021.0455. [DOI] [PubMed] [Google Scholar]
- 4.Taubenberger J.K., Morens D.M. The 1918 influenza pandemic and its legacy. Cold Spring Harb. Perspect. Med. 2020;10(10) doi: 10.1101/cshperspect.a038695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johns Hopkins University of Medicine Coronavirus Resource Center COVID-19 dashboard. https://coronavirus.jhu.edu/map.html Available at:
- 6.Chas J., Nadal M., Siguier M., Fajac A., Denis M., Morand-Joubert L., Pialoux G. Broad-based SARS-CoV-2 testing program for healthcare workers in a primary care hospital in France. Infect. Dis. News. 2021;51:556–559. doi: 10.1016/j.idnow.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gniazdowski V., Morris C.P., Wohl S., Mehoke T., Ramakrishnan S., Thielen P., et al. Repeat COVID-19 molecular testing: correlation of SARS-CoV-2 culture with molecular assays and cycle thresholds. Clin. Infect. Dis. 2020 Oct 27 doi: 10.1093/cid/ciaa1616. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goyal A., Reeves D.B., Cardozo-Ojeda E.F., Schiffer J.T., Mayer B.T. Viral load and contact heterogeneity predict SARS-CoV-2 transmission and super-spreading events. Elife. 2021;10 doi: 10.7554/eLife.63537. [DOI] [PMC free article] [PubMed] [Google Scholar]