Abstract
The usefulness of randomly amplified polymorphic DNA method (RAPD) to identify each species of genus Erysipelothrix and for epidemiological analysis of this genus was studied. Eighty-one strains and 18 random primers were tested. Among the tested primers, the primers NK51 (GGTGGTGGTATC) and NK6 (CCCGCGCCCC) produced noticeable results. The primer NK51 revealed four species-specific RAPD patterns. Of the 66 strains of E. rhusiopathiae, 64 had the same unique band of 884 bp. Of the 12 strains of E. tonsillarum, 11 produced a 1,265-bp band. In addition, two strains, previously thought to be E. rhusiopathiae, produced the 1,265-bp band, suggesting that they had been misclassified. One strain of E. tonsillarum produced the 884-bp band, suggesting that it too was E. rhusiopathiae. The E. rhusiopathiae strain of serovar 13 produced a 650-bp band, and the strain of serovar 18 produced a clear 420-bp band as well as three weak bands of 1,265, 918, and 444 bp. The primer NK6 revealed 14 RAPD patterns that were not serovar specific. However, different patterns were produced among strains of the same serovar showing that the RAPD method is able to identify the genetic variations of strains of this genus and can rapidly and easily differentiate strains of the same serovar. Based on these results, we concluded that the RAPD method with primers NK51 and NK6 is a rapid and reliable method to identify the species of this genus; we also concluded that this method might be a useful tool for the epidemiological analysis of the Erysipelothrix species.
Erysipelothrix rhusiopathiae, a gram-positive, slender, straight or slightly curved rod, is known to be the causative agent of erysipelas in swine and erysipeloid in humans. This bacterium has been isolated from many species of wild and domestic animals in most parts of the world (31).
Until recently, the genus Erysipelothrix was thought to be comprised of only one species, E. rhusiopathiae. However, Takahashi et al. (19, 20) showed that this genus comprises at least two distinct species by the DNA-DNA hybridization: E. rhusiopathiae—comprising serovars 1a, 1b, 2, 4, 5, 6, 8, 9, 11, 12, 15, 16, 17, 19, and 21, and type N—and E. tonsillarum—comprising serovars 3, 7, 10, 14, 20, 22, and 23. Moreover, they inferred that serovars 13 and 18 might be members of a new and separate species (20). Current bacteriological culture methods require at least 3 days to isolate this bacterium and about 10 days to determine its serovars (21). To replace these time-consuming methods, primer pair MO101-MO102, primer pair ER1-ER2 and a four primer pair set (ER1F-ER1R, ER2F-ER2R, ER3F-ER3R, and ER4F-ER4R) were developed by Makino et al. (9), Shimoji et al. (17), and Takeshi et al. (22), respectively, for rapid and direct detection using PCR method.
However, each primer has some drawbacks. The primers MO101-MO102 is genus specific and is unable to differentiate species. The primers ER1-ER2 is able to identify E. rhusiopathiae strains but is unable to identify E. tonsillarum strains and the E. rhusiopathiae strains of serovars 13 and 18 due to the lack of amplification products with DNAs extracted from these strains. The primers ER1F-ER1R, ER2F-ER2R, ER3F-ER3R, and ER4F-ER4R are considered sufficient to identify each species of this genus. However, due to a difference of size as small as 3 bp among the amplification products, it is difficult to differentiate them. Moreover, four amplification tubes per sample are necessary for identification of the species.
On the other hand, recently the randomly amplified polymorphic DNA (RAPD) method with one random primer has shown the ability to differentiate bacteria and strains of one genus at species level, and even some effectiveness as a tool for epidemiological and taxonomic studies (5–7, 10, 11, 14–16, 24–28, 30). In the study described in this paper we examined the possibility of identifying species and differentiating strains using the RAPD method and the usefulness of this method in epidemiological analysis of genus Erysipelothrix.
MATERIALS AND METHODS
Bacterial strains and biochemical tests.
The details of the 81 Erysipelothrix strains used in this study are shown in Table 1. These comprise 54 field isolates (8, 12, 13, 18), reference strains for 23 serovars (including subserovars 1a and 1b) and type N, and type strains of E. rhusiopathiae and E. tonsillarum (20). Biochemical characterization of the strains were made on the basis of the carbohydrate fermentation patterns, test-tube growth in gelatin medium, production of H2S in triple sugar iron agar (Difco Laboratories) slants, and catalase and oxidase production, as described previously (18, 20). The carbohydrate fermentation test was carried out using nutrient broth supplemented with 1% Andrade's indicator and 10% horse serum (20, 29). The serovars were identified by the agar gel double-diffusion precipitation technique using the heat-stable antigen extracted from the cell wall of each strain and rabbit antisera representing serovars 1 through 23 of the Erysipelothrix species (20).
TABLE 1.
Erysipelothrix sp. strains used in this study and RAPD patterns produced by each primer
| Species and serovar | Strain | Source | RAPD pattern with primer:
|
PCR results with primers:
|
||
|---|---|---|---|---|---|---|
| NK51 | NK6 | MO101-MO102d | ER1-ER2e | |||
| E. rhusiopathiae | ||||||
| 1a | ME-7a | Unknown | A | a | + | + |
| E176 | Fish | A | b | + | + | |
| E157 | Fish | A | a | + | + | |
| 1b | 422/1E1a | Porcine spleen | A | a | + | + |
| E019 | Fish | A | g | + | + | |
| E038 | Fish | A | a | + | + | |
| K040 | Wild boar meat | A | a | + | + | |
| K075 | Wild boar meat | A | a | + | + | |
| 2 | ATCC 19414b | Pig with endocarditis | A | c | + | + |
| R32E11a | Unknown | A | d | + | + | |
| NF4E1 | Porcine spleen | A | a | + | + | |
| 115 | Chicken | A | g | + | + | |
| 17.2a | Chicken | A | a | + | + | |
| 10.2a | Chicken | A | a | + | + | |
| E037 | Fish | A | a | + | + | |
| K003 | Wild boar meat | A | e | + | + | |
| N026 | Chicken | A | d | + | + | |
| 4 | Doggerscharbea | Fish | A | a | + | + |
| 212 | Chicken meat | A | g | + | + | |
| 213 | Chicken meat | A | g | + | + | |
| E127 | Fish | A | f | + | + | |
| 5 | Pécs 67a | Porcine tonsil | A | d | + | + |
| AKO | Chicken | A | a | + | + | |
| 148 | Chicken | A | g | + | + | |
| 2.2a | Chicken | A | g | + | + | |
| 2.4a | Chicken | A | g | + | + | |
| K059 | Wild boar meat | A | g | + | + | |
| 64 | Chicken | A | g | + | + | |
| 6 | Tuzoka | Bustard | A | g | + | + |
| 36.4a | Chicken | A | g | + | + | |
| 136 | Chicken | A | f | + | + | |
| 20.4a | Chicken | A | f | + | + | |
| N009 | Chicken | A | g | + | + | |
| K002 | Wild boar meat | A | g | + | + | |
| N025 | Chicken | A | f | + | + | |
| 8 | Godaa | Godwit | A | d | + | + |
| 47 | Chicken | A | g | + | + | |
| E024 | Fish | A | g | + | + | |
| N008 | Chicken | A | g | + | + | |
| r4.1a | Chicken meat | A | g | + | + | |
| r4.2a | Chicken meat | A | g | + | + | |
| r6.1a | Chicken meat | A | g | + | + | |
| r6.3a | Chicken meat | A | g | + | + | |
| 9 | Kapareka | Fish | A | a | + | + |
| E077 | Fish | A | f | + | + | |
| E112 | Fish | A | h | + | + | |
| 280 | Chicken | A | g | + | + | |
| K052 | Wild boar meat | A | f | + | + | |
| 11 | IV12/8a | Porcine tonsil | A | g | + | + |
| K021 | Wild boar meat | A | a | + | + | |
| 12 | Pécs 9a | Porcine tonsil | A | a | + | + |
| 88 | Chicken meat | A | g | + | + | |
| 97 | Chicken meat | A | g | + | + | |
| 96 | Chicken meat | A | g | + | + | |
| E146 | Fish | A | g | + | + | |
| 15 | Pécs 3597a | Porcine tonsil | A | d | + | + |
| E073 | Fish | B | i | + | − | |
| 16 | Tanzaniaa | Parrot | A | d | + | + |
| K037 | Wild boar meat | B | j | + | − | |
| 17 | 545a | Porcine spleen | A | d | + | + |
| 19 | 2017a | Porcine spleen | A | g | + | + |
| E053 | Fish | A | d | + | + | |
| E051 | Fish | A | g | + | + | |
| K031 | Deer meat | A | d | + | + | |
| 21 | Bãno 36a | Sheep dip | A | a | + | + |
| N | MEW22a | Unknown | A | a | + | + |
| E. tonsillarum | ||||||
| 3 | Witllinga | Fish | B | j | + | − |
| 7 | ATCC 43339c | Porcine tonsil | B | j | + | − |
| ATCC 43338 | Porcine tonsil | B | j | + | − | |
| P-43 | Fish | B | j | + | − | |
| K015 | Wild boar meat | B | j | + | − | |
| K004 | Wild boar meat | B | j | + | − | |
| 10 | Lengyel-Pa | Squirrel | B | j | + | − |
| K024 | Wild boar meat | A | g | + | + | |
| 14 | Iszap-4a | Mud of zoo pond | B | j | + | − |
| 20 | 2553a | Porcine spleen | B | k | + | − |
| 22 | Bãno 107a | Sheep dip | B | l | + | − |
| 23 | KS20Aa | Pig slurry | B | j | + | − |
| Erysipelothrix spp. | ||||||
| 13 | Pécs 56a | Porcine tonsil | C | m | + | − |
| 13 | Shiribeshi17 | Pigpen litter | C | m | + | − |
| 18 | 715a | Porcine spleen | D | n | + | − |
DNA preparation.
Total DNAs from the strains listed in Table 1 were prepared using the method described by Makino et al. (9). Briefly, bacterial cells of a 24-h culture were suspended in 200 μl of TES buffer (50 mM Tris-HCl, 5 mM EDTA, 50 mM NaCl [pH 8.0]), containing 10 μl of lysozyme (10 mg/ml) and 10 μl of N-acetylmuramidase SG (1 mg/ml). They were then incubated for 30 min at 37°C before the addition of 10 μl of 10% sodium dodecyl sulfate and 10 μl of proteinase K (20 mg/ml). After further incubation at 55°C for 60 min, the crude DNA preparation was treated with RNase, extracted three times with phenol-chloroform, precipitated with ethanol, and dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). The DNA concentration was determined using the GeneQuanto pro RNA/DNA Calculator (Pharmacia Biotech ltd, Cambridge, England) and adjusted to the working concentration.
RAPD PCR and gel electrophoresis.
Random primers from the collection of primers available in our laboratory were randomly chosen, and a total of 18 primers were tested to amplify the DNAs. These were 7 10-mer primers (guanine-plus-citosine contents of 60 to 100%) and 11 12-mer primers (guanine-plus-citosine contents of 41.7 to 58.3%). The PCR amplification was performed with the Program Temperature Control System PC-700 (Astec Co., Ltd., Tokyo, Japan). The amplification was carried out in a 50-μl reaction mixture containing 5 μl of 10× reaction buffer (Sawady Technology Co., Ltd., Tokyo, Japan); 200 μM (each) dATP, dTTP, dCTP, and dGTP; 200 nM random primer; 2.5 U of Taq DNA polymerase (Sawady Technology Co.), and 500 ng of template DNA under a drop of mineral oil. The cycling program was 4 cycles at 94°C for 5 min, 34°C for 5 min, and 72°C for 5 min; 30 cycles at 94°C for 1 min, 34°C for 1 min, and 72°C for 2 min; and a final incubation at 72°C for 10 min. An aliquot of 10 μl of the amplified products was subjected to electrophoresis in 2% agarose gel (Iwai Chemicals Co., Tokyo, Japan), stained with ethidium bromide, and photographed under UV light. All the DNA samples were amplified by using the primers MO101-MO102 (9) and ER1-ER2 (17).
RESULTS
Of the 18 primers tested, 15 produced one or more RAPD patterns; among these the primers designated NK51 (GGTGGTGGTATC) and NK6 (CCCGCGCCCC) produced noticeable results.
The primer NK51 produced four species-specific RAPD patterns. Three of these patterns were composed of a single band of 884, 1,265, 650 bp and were designated RAPD patterns A, B, and C, respectively. One was composed of a clear band of 420 bp and three weak bands of 1,265, 918, and 444 bp, and it was designated RAPD pattern D. The RAPD pattern A was produced with DNAs extracted from the type strain of E. rhusiopathiae and from the reference strains of serovars 1a, 1b, 2, 4, 5, 6, 8, 9, 11, 12, 15, 16, 17, 19, and 21 and type N, believed to be E. rhusiopathiae. The RAPD pattern B was produced with DNAs extracted from the type strain of E. tonsillarum and from the reference strains of serovars 3, 7, 10, 14, 20, 22, and 23, considered to be E. tonsillarum. The RAPD patterns C and D were produced, respectively, with DNAs extracted from E. rhusiopathiae strains of serovars 13 and 18 (Fig. 1). The same RAPD patterns, A, B, C, or D, were produced with DNAs extracted from the 54 field isolates; among them, RAPD patterns and taxonomic classification of 51 strains were in agreement with the classification based on the serovars, as the serovar reference strains mentioned above (Table 1). However, three strains showed RAPD patterns that were not in agreement with the classification based on the serovars. The strains E073 and K037 of serovars 15 and 16, classified as E. rhusiopathiae, showed the RAPD pattern B produced from the type strain of E. tonsillarum. The strain K024 of serovar 10, classified as E. tonsillarum, showed the RAPD pattern A produced from the type strain of E. rhusiopathiae. All 81 strains showed positive results with the primer MO101-MO102. Among the 66 E. rhusiopathiae strains, 64 showed positive results; 11 of 12 E. tonsillarum strains and the E. rhusiopathiae strains of serovars 13 and 18 showed negative results with the primers ER1-ER2. However, in a manner similar to the results obtained with the primer NK51, the three strains described above showed different results. The strains E073 and K037 had negative results, and the strain K024 had a positive result (Table 1). Of the biochemical tests, these three strains showed the ability to ferment saccharose (data not shown).
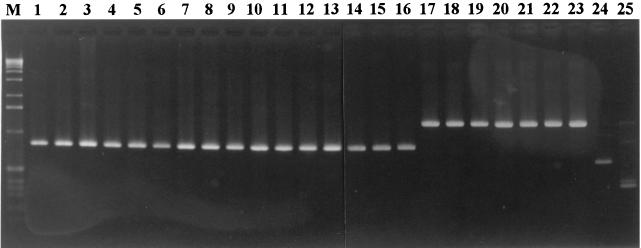
FIG. 1.
RAPD patterns of the Erysipelothrix sp. serovar reference strains produced with the primer NK51. Lanes 1 to 16 show RAPD pattern A (884 bp) produced from the E. rhusiopathiae strains: lane 1, ME-7 (serovar 1a); lane 2, 422/1E1 (serovar 1b); lane 3, ATCC 19414 (type strain, serovar 2); lane 4, Doggerscharbe (serovar 4); lane 5, Pécs67 (serovar 5); lane 6, Tuzok (serovar 6); lane 7, Goda (serovar 8); lane 8, Kaparek (serovar 9); lane 9, IV 12/8 (serovar 11); lane 10, Pécs9 (serovar 12); lane 11, Pécs3597 (serovar 15); lane 12, Tanzania (serovar 16); lane 13, 545 (serovar 17); lane 14, 2017 (serovar 19); lane 15, Bãno36 (serovar 21); lane 16, MEW22 (type N). Lanes 17 to 23 show RAPD pattern B (1,265 bp) produced from the E. tonsillarum strains: lane 17, Witlling (serovar 3); lane 18, ATCC 43339 (type strain, serovar 7); lane 19, Lengyel-P (serovar 10); lane 20, Iszap-4 (serovar 14); lane 21, 2553 (serovar 20); lane 22, Bãno107 (serovar 22); lane 23, KS20A (serovar 23). Lanes 24 and 25 show RAPD patterns C (650 bp) and D (420, 444, 918, and 1,265 bp, sizes from bottom to top), respectively, produced from Erysipelothrix spp. Pécs56 (serovar 13) and 715 (serovar 18). Lane M, 1-kb ladder (GIBCO-BRL).
The primer NK6 produced 14 RAPD patterns designated as RAPD patterns a through n (Fig. 2). No serovar-specific RAPD pattern was produced. However, different RAPD patterns were produced from strains of the same serovars (Table 1). Sixty-four E. rhusiopathiae strains showed RAPD patterns varying from a through h, among which the number of bands differed from one to eight, and 11 E. tonsillarum strains showed RAPD patterns j, k, or l, which differed by one band. The strain K024 of serovar 10 showed the RAPD pattern g. The E. rhusiopathiae strains of serovars 13 and 18 showed the RAPD patterns m and n. The strain K037 of serovar 16 showed the RAPD pattern j, and the strain E073 of serovar 15 showed a unique RAPD pattern, i (Table 1).
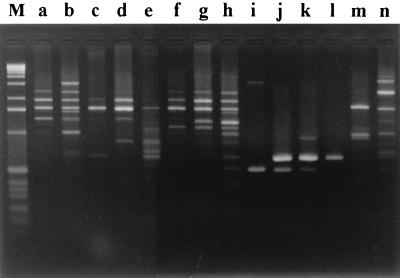
FIG. 2.
RAPD patterns obtained from the 81 Erysipelothrix strains with the primer NK6. The strains of lanes a to i are E. rhusiopathiae: lane a, ME-7 (serovar 1a); lane b, E176 (serovar 1a); lane c, ATCC 19414 (type strain, serovar 2); lane d, R32E11 (serovar 2); lane e, K003 (serovar 2); lane f, E127 (serovar 4); lane g, Tuzok (serovar 6); lane h, E112 (serovar 9); lane i, E073 (serovar 15). The strains of lanes j to l are E. tonsillarum: lane j, ATCC 43339 (type strain, serovar 7); lane k, 2553 (serovar 20); lane l, Bãno107 (serovar 22). The strains of lanes m and n are, respectively, Erysipelothrix spp. Pécs56 (serovar 13) and 715 (serovar 18). Lane M, 1-kb ladder (GIBCO-BRL).
DISCUSSION
In this study, we identified a random primer, NK51, able to produce species-specific RAPD patterns for E. rhusiopathiae (A), E. tonsillarum (B), and even for the E. rhusiopathiae strains of serovars 13 (C) and 18 (D), which shows that the RAPD method can be used to identify the species of genus Erysipelothrix. The results obtained with serovar reference strains and the 51 (98%) field isolates were in agreement with the classification based on the DNA-DNA hybridization, serotyping, and biochemical characteristics reported by Takahashi et al. (20). It should be noted that among the 81 strains used, the strains E073 and K037 showed atypical biochemical and molecular biological characteristics, and the strain K024 showed an atypical molecular biological characteristic. If the taxonomic classification of these strains is based only on the serovars, the strains E073 and K037 must be classified as E. rhusiopathiae and the strain K024 must be classified as E. tonsillarum. However, studies carried out by Chooromoney et al. (4) and Ahrné et al. (1) using the multilocus enzyme electrophoresis and restriction fragment length polymorphisms, respectively, reported that some strains of same serovars can be classified into different clusters, casting doubt on the classification based on serotyping. On the other hand, the RAPD performed with the primer NK51 was able to differentiate and classify these atypical strains, and the results were in agreement with the PCR using those previously developed primers, suggesting that they had been misclassified. Thus, strains E073 and K037 might be considered E. tonsillarum, and the strain K024 might be considered E. rhusiopathiae. In previous studies, because only strains of serovars 3, 7, 10, 14, 20, 22, and 23, classified as E. tonsillarum based on the DNA-DNA hybridization, were able to ferment saccharose, this fermentation ability has been considered a unique characteristic of these serovars and of E. tonsillarum species (20). However, we found the strains E073 and K037 of serovars 15 and 16 were also able to ferment saccharose and had the molecular biological characteristic of E. tonsillarum, and the strain K024 of serovar 10 was able to ferment saccharose but had the molecular biological characteristic of the E. rhusiopathiae species. These results demonstrated that the RAPD method can be used to classify Erysipelothrix strains into the genetically correlated species and also suggest that, similar to serotyping, the ability to ferment saccharose is somewhat questionable as a means of identifying the species of this genus. Moreover, as the primer NK51 produced bands from both of the two species, from the strains of serovars 13 and 18, and the characteristic bands of each species differ from each other by more than 200 bp, the identification of these species can be carried out using only one primer and one amplification tube per sample. Thus, this method might be useful in identifying the species of genus Erysipelothrix, and it might be easier than the method developed earlier (9, 17, 22).
Furthermore, we used the primer NK6 and found that RAPD patterns that differed by one to eight bands were produced with DNAs of E. rhusiopathiae strains of the same serovar, and RAPD patterns that differed by one band were produced with DNAs of E. tonsillarum strains. Moreover, when RAPD patterns of strains E073 and K037 (serovars 15 and 16) and strain K024 (serovar 10) were compared with those of strains of the same serovar, no similar band was found. These results proved that RAPD is not only able to differentiate the strains of the same serovar but is also able to identify the genetic diversity among strains of these species. Similarities and differences among strains of the same serovar have been described by comparing the cell protein composition using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (2, 3, 23); genetic diversities have been described by multilocus enzyme electrophoresis (4). Our study differs from earlier studies in identifying the strains of these species and determining the genetic diversity based directly on the DNA amplification.
Based on these results, we conclude that RAPD carried out using the primers NK51 and NK6 might be a rapid and reliable method of identifying the species and strains of this genus; it also might be a useful tool for epidemiological studies of the genus Erysipelothrix. We are now identifying the nucleotide sequences of the amplified products and attempting to design a specific primer able to identify bacteria of this genus and differentiate each species. Furthermore, in view of the genetic diversity, and the similarities of biochemical characteristics among the Erysipelothrix strains, further molecular biological studies with a large collection of isolates of each serovar would be needed to elucidate the taxonomic relationship of the serovars with species of the genus Erysipelothrix.
REFERENCES
- 1.Ahrné S, Stenströn I, Jensen N E, Petterson B, Uhlén M, Molin G. Classification of Erysipelothrix strains on the basis of restriction fragment length polymorphisms. Int J Syst Bacteriol. 1995;45:382–385. doi: 10.1099/00207713-45-2-382. [DOI] [PubMed] [Google Scholar]
- 2.Bernáth S, Kucsera G, Kádár I, Horváth G, Morovján G Y. Comparison of the protein patterns of Erysipelothrix rhusiopathiae strains by SDS-PAGE and autoradiography. Acta Vet Hung. 1997;45:417–425. [PubMed] [Google Scholar]
- 3.Bernáth S, Morovján G Y, Sztojkov V, Szita G. Comparison of the protein composition of Erysipelothrix rhusiopathiae strains of subtype 1a isolated from ducks and pigs. Acta Vet Hung. 1998;46:211–217. [PubMed] [Google Scholar]
- 4.Chooromoney K N, Hampson D J, Eamens G J, Turner M J. Analysis of Erysipelothrix rhusiopathiae and Erysipelothrix tonsillarum by multilocus enzyme electrophoresis. J Clin Microbiol. 1994;32:371–376. doi: 10.1128/jcm.32.2.371-376.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzsimons N A, Cogan T M, Condon S, Beresford T. Phenotypic and genotypic characterization of non-starter lactic acid bacteria in mature Cheddar cheese. Appl Environ Microbiol. 1999;65:3418–3426. doi: 10.1128/aem.65.8.3418-3426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grayson T H, Cooper L F, Atienzar F A, Knowles M R, Gilpin M L. Molecular differentiation of Renibacterium salmoninarum isolates from worldwide locations. Appl Environ Microbiol. 1999;65:961–968. doi: 10.1128/aem.65.3.961-968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayford A E, Petersen A, Vogensen F K, Jakobsen M. Use of conserved randomly amplified polymorphic DNA (RAPD) fragments and RAPD pattern for characterization of Lactobacillus fermentum in Ghanaian fermented maize dough. Appl Environ Microbiol. 1999;65:3213–3221. doi: 10.1128/aem.65.7.3213-3221.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanai Y, Hayashidani H, Kaneko K, Ogawa M, Takahashi T, Nakamura M. Occurrence of zoonotic bacteria in retail game meat in Japan with special reference to Erysipelothrix. J Food Prot. 1997;60:328–331. doi: 10.4315/0362-028X-60.3.328. [DOI] [PubMed] [Google Scholar]
- 9.Makino S, Okada Y, Maruyama T, Ishikawa K, Takahashi T, Nakamura M, Ezaki T, Morita H. Direct and rapid detection of Erysipelothrix rhusiopathiae DNA in animals by PCR. J Clin Microbiol. 1994;32:1526–1531. doi: 10.1128/jcm.32.6.1526-1531.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makino S, Okada Y, Maruyama T, Kaneko S, Sasakawa C. PCR-based random amplified polymorphic DNA fingerprinting of Yersinia pseudotuberculosis and its practical applications. J Clin Microbiol. 1994;32:65–69. doi: 10.1128/jcm.32.1.65-69.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitsuda T, Kuroki H, Ishikawa N, Imagawa T, Ito S, Miyamae T, Mori M, Uehara S, Yokota S. Molecular epidemiological study of Haemophilus influenzae serotype b strains obtained from children with meningitis in Japan. J Clin Microbiol. 1999;37:2548–2552. doi: 10.1128/jcm.37.8.2548-2552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakazawa H, Hayashidani H, Higashi J, Kaneko K, Takahashi T, Ogawa M. Occurrence of Erysipelothrix spp. in broiler chickens at an abattoir. J Food Prot. 1998;61:907–909. doi: 10.4315/0362-028x-61.7.907. [DOI] [PubMed] [Google Scholar]
- 13.Nakazawa H, Hayashidani H, Higashi J, Kaneko K, Takahashi T, Ogawa M. Occurrence of Erysipelothrix spp. in chicken meat parts from a processing plant. J Food Prot. 1998;61:1207–1209. doi: 10.4315/0362-028x-61.9.1207. [DOI] [PubMed] [Google Scholar]
- 14.Okazaki M, Watanabe T, Morita K, Higurashi Y, Araki K, Shukuya N, Baba S, Watanabe N, Egami T, Furuya N, Kanamori M, Shimazaki S, Uchimura H. Molecular epidemiological investigation using a randomly amplified polymorphic DNA assay of Burkholderia cepacia isolates from nosocomial outbreaks. J Clin Microbiol. 1999;37:3809–3814. doi: 10.1128/jcm.37.12.3809-3814.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popovic T, Kim C, Reiss J, Reeves M, Nakao H, Golaz A. Use of molecular subtyping to document long-term persistence of Corynebacterium diphtheriae in South Dakota. J Clin Microbiol. 1999;37:1092–1099. doi: 10.1128/jcm.37.4.1092-1099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen H N, Olsen J E, Rasmussen O F. RAPD analysis of Yersinia enterocolitica. Lett Appl Microbiol. 1994;19:359–362. doi: 10.1111/j.1472-765x.1994.tb00475.x. [DOI] [PubMed] [Google Scholar]
- 17.Shimoji Y, Mori Y, Hyakutake K, Sekizaki T, Yokomizo Y. Use of an enrichment broth cultivation-PCR combination assay for rapid diagnosis of swine erysipelas. J Clin Microbiol. 1998;36:86–89. doi: 10.1128/jcm.36.1.86-89.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiono H, Hayashidani H, Kaneko K, Ogawa M, Muramatsu M. Occurrence of Erysipelothrix rhusiopathiae in retail raw pork. J Food Prot. 1990;53:856–858. doi: 10.4315/0362-028X-53.10.856. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi T, Fujisawa T, Benno Y, Tamura Y, Sawada T, Suzuki S, Muramatsu M, Mitsuoka T. Erysipelothrix tonsillarum sp. nov. isolated from tonsils of apparently healthy pigs. Int J Syst Bacteriol. 1987;37:166–168. [Google Scholar]
- 20.Takahashi T, Fujisawa T, Tamura Y, Suzuki S, Muramatsu M, Sawada T, Benno Y, Mitsuoka T. DNA relatedness among Erysipelothrix rhusiopathiae strains representing all twenty-three serovars and Erysipelothrix tonsillarum. Int J Syst Bacteriol. 1992;42:469–473. doi: 10.1099/00207713-42-3-469. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi T, Zarkasie K, Mariana S, Sumadi, Ogata M. Serological and pathogenic characterization of Erysipelothrix rhusiopathiae isolates from tonsils of slaughter pigs in Indonesia. Vet Microbiol. 1989;21:165–175. doi: 10.1016/0378-1135(89)90029-1. [DOI] [PubMed] [Google Scholar]
- 22.Takeshi K, Makino S, Ikeda T, Takada N, Nakashiro A, Nakanishi K, Oguma K, Katoh Y, Sunagawa H, Ohyama T. Direct and rapid detection by PCR of Erysipelothrix sp. DNAs prepared from bacterial strains and animal tissues. J Clin Microbiol. 1999;37:4093–4098. doi: 10.1128/jcm.37.12.4093-4098.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura Y, Takahashi T, Zarkasie K, Nakamura M, Yoshimura H. Differentiation of Erysipelothrix rhusiopathiae and Erysipelothrix tonsillarum by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of cell proteins. Int J Syst Bacteriol. 1993;43:111–114. doi: 10.1099/00207713-43-1-111. [DOI] [PubMed] [Google Scholar]
- 24.Tilsala-Timisjärvi A, Alatossava T. Strain-specific identification of probiotic Lactobacillus rhamnosus with randomly amplified polymorphic DNA-derived PCR primers. Appl Environ Microbiol. 1998;64:4816–4819. doi: 10.1128/aem.64.12.4816-4819.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tynkkynen S, Satokari R, Saarela M, Mattila-Sandholm T, Saxelin M. Comparison of ribotyping, randomly amplified polymorphic DNA analysis, and pulsed-field gel electrophoresis in typing of Lactobacillus rhamnosus and L. casei strains. Appl Environ Microbiol. 1999;65:3908–3914. doi: 10.1128/aem.65.9.3908-3914.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warner J M, Oliver J D. Randomly amplified polymorphic DNA analysis of starved and viable but nonculturable Vibrio vulnificus cells. Appl Environ Microbiol. 1998;64:3025–3028. doi: 10.1128/aem.64.8.3025-3028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warner J M, Oliver J D. Randomly amplified polymorphic DNA analysis of clinical and environmental isolates of Vibrio vulnificus and other Vibrio species. Appl Environ Microbiol. 1999;65:1141–1144. doi: 10.1128/aem.65.3.1141-1144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White T G, Shuman R D. Fermentation reactions of Erysipelothrix rhusiopathiae. J Bacteriol. 1961;82:595–599. doi: 10.1128/jb.82.4.595-599.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams J G K, Kubelik A R, Livak K J, Rasfalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood R L. Erysipelas. In: Straw B E, D'Allaire S, Mengeling W L, Taylor D J, editors. Diseases of swine. 8th ed. Ames: Iowa State University Press; 1999. pp. 419–430. [Google Scholar]