Abstract
The current COVID-19 global pandemic poses immense challenges to global health, largely due to the difficulty to detect infection in the early stages of the disease, as well as the current lack of effective antiviral therapy. Research and understanding of the human immune system can provide important theoretical and technical support for the clinical diagnosis and treatment of COVID-19, the clinical implementations of which include immunoassays and immunotherapy, which play a crucial role in the fight against the pandemic. This review consolidates the current scientific evidence for immunoassay, which includes multiple methods of detecting antigen and antibody against SARS-CoV-2. We compared the characteristics, advantages and disadvantages, and clinical applications of these three detection techniques. In addition to detecting viral infections, knowledge on the body’s immunity against the virus is desirable; thus, the immunotherapy-based neutralizing antibody (nAb) detection methods were discussed. We also gave a brief introduction to the new immunoassay technology such as biosensing. This was followed by an in-depth and extensive review on a variety of immunotherapy methods. It includes convalescent plasma therapy, neutralizing antibody–based treatments targeting different regions of SARS-CoV-2, immunotherapy targeted on the host cell including inhibiting the host cell receptor and cytokine storm, as well as cocktail antibodies, cross-neutralizing antibodies, and immunotherapy based on cross-reactivity between viral epitopes and autoepitopes and autoantibody. Despite the development of various immunological testing methods and antibody therapies, the current global situation of COVID-19 is still tense. We need more efficient detection methods and more reliable antibody therapies. The up-to-date knowledge on therapeutic strategies will likely help clinicians worldwide to protect patients from life-threatening viral infections.
Keywords: SARS-CoV-2, COVID-19, Antibody, Immunity, Immunoassay, Immunotherapy
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus that causes coronavirus disease 2019 (COVID-19), the contagious disease responsible for the COVID-19 pandemic [1]. SARS-CoV-2 has four structural proteins, known as the S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins. It enters human cells by binding the S protein on its surface to the ACE2 receptor of human cells and thus proliferates in large numbers [2]. SARS-CoV-2 infection stimulates antigen-specific antibody responses.
Immunoassays and immunotherapy are the two broad areas of clinical application for antibody immunity. The primary goal of immunoassay is to detect the presence of antibodies, which can be used to monitor immunity and infection in the population, as well as to aid in the development of vaccines. As a result, developing a faster and more accurate antibody detection method is an important way to aid in the fight against the COVID-19 pandemic. Immunotherapy, on the other hand, could reduce mortality by offering a new way to treat patients without targeted drugs.
Therefore, the purpose of this review is to summarize the current scientific evidence for immunoassay and immunotherapy to further provide important theoretical and technical support for the clinical diagnosis and treatment of COVID-19.
Immunoassays
The term immunoassay refers to any assay that, at its core, depends on the binding of antigen and antibody. There are three types of tests available for COVID-19: molecular, antigen, and antibody (serology) testing. Molecular tests like the polymerase chain reaction (PCR) are considered to be the gold standard for diagnosis by amplifying and detecting its genetic material. However, this process can take hours and requires sophisticated lab equipment and technicians, and its detection sensitivity can vary depending on the sample collection handling method [3]. As an alternative, highly sensitive immunological methods that directly detect viral antigens or antibodies (separately or simultaneously) in clinical samples without sample preparation steps are necessary for the accurate diagnosis of COVID-19. These immune assays are fast (even within 15 min) and do not rely on professional technicians, equipment, and facilities.
Antigen assays are immunoassays that detect the specific viral proteins (antigens) using specific antibodies, allowing the capture of the entire virus or its fragments. Antigen tests have been developed for use in the laboratory or at the point of care, the latter being known as rapid antigen detection (RAD). SARS-CoV-2 RAD can detect the virus directly in respiratory samples. Furthermore, antigens are far more stable than RNA, making them less prone to degradation during transport and storage. Therefore, it may be a useful tool for the early diagnosis of the asymptomatic population. Both molecular and antigen tests detect whether a person is currently infected, while antibody (serology) mainly detects whether a person had an infection in the past (IgM can also detect recent or current infection), even if they had no symptoms of the illness. Antigen-specific antibody binding is used in the antibody detection assay to detect IgA, IgM, and IgG in blood, plasma, and serum samples. In a follow-up study of 173 patients [4], the antibody presence rate was < 40% in the first week after onset but quickly increased to 100.0% (IgA), 94.3% (IgM), and 79.8% (IgG) on the 15th day after onset. Therefore, antibody levels lag behind the infection, typically rising significantly in the second and third weeks after initial onset. This feature prevents antibody testing from being used for the timely diagnosis of COVID-19; however, the detection of IgG and IgM is beneficial to the study of the disease course. Thus, combining PCR and antibody detection can make up for their respective shortcomings. At the stage of large-scale vaccination, the significance of antibody detection mainly lies in monitoring the production of antibodies after a vaccine injection, in order to monitor the number of people immunized with the virus in the population and identify asymptomatic infections [5].
Among the four structural proteins of SARS-Cov-2, the most commonly used biomarkers for the detection of COVID-19 are the S and N protein antigens. As an evolutionary conserved and highly immunogenic phosphoprotein, N protein is the most frequently present protein in the SARS-CoV-2 structure. Thus, the SARS-CoV-2 antigen detection kits reported thus far primarily detect N protein [6]. The S protein is also very suitable as a diagnostic antigen because it is the main transmembrane protein of the virus and is highly immunogenic [7]. In addition, the spike protein exhibits amino acid sequence diversity among coronaviruses [8], enabling the specific detection of SARS-CoV-2. The SARS-CoV-2 spike detection enzyme-linked immunosorbent assay (ELISA) test has been developed for the quantitative detection of SARS-CoV-2 spike protein and is based on the solid-phase sandwich enzyme immunoassay. This test contains antibodies specific for the recombinant spike protein and can recognize both the recombinant as well as wild-type spike protein with 67.02 pg/ml sensitivity [9]. Both N and S proteins are highly antigenic and play an important role in inducing a host immune response and pathogenesis. Antibodies against N proteins are longer-lived and occur in greater abundance than antibodies against other viral components [4]. Several studies have been conducted to compare the sensitivity and specificity of the S and N proteins [4, 10, 11]. When Li et al. compared SARS-CoV-2 antigen detection from 20 studies [4], they concluded that the specificities of these two antigens were generally comparable, while antigen detection using the N protein antibodies was more sensitive than that using the S protein antibodies for early infections.
Two common technologies used in antigen detection are lateral flow immunoassay (LFIA) and chemiluminescence immunoassay (CLIA) (Table 1), while antibody detection methods fall into four categories: laboratory tests for ELISA [12], CLIA [13], LFIA [14], and neutralization test [15–17]. The LFIA method detects antigen or antibodies by using colloidal gold test paper, which is simple in design, portable, rapid, and easy to interpret, but is limited to qualitative detection and lacks sensitivity [18]. Both ELISA and CLIA are quantitative methods that involve labeling antibodies or antigens with enzymatic or chemical luminescent agents. In a meta-analysis using 24 articles, Mekonnen et al. [19] evaluated the diagnostic accuracy of serological antibody detection and found that the summary sensitivity/specificity of CLIA, ELISA, and LFIA were 92% (95% CI: 86–95%)/99% (CI: 97–99%), 86% (CI: 82–89%)/99% (CI: 98–100%), and 78% (CI: 71–83%)/98% (95% CI: 96–99%), respectively.
Table 1.
Summary of pros and cons of immunoassays
| Immunoassay | Main properties | Pros | Cons | Reference |
|---|---|---|---|---|
| LFIA | Based on the movement of a liquid sample through a polymeric strip with attached molecules that interact with the analyte |
• Visually recognizable signal; • Fast, low cost, portable, and easy-to-use; • Long shelf life, no need to refrigerate |
• Low sensitivity; • Qualitative or semi-quantitative result; • Good antibody preparation is obligatory; |
[14, 102] |
| CLIA | Labeling antibodies or antigens with chemical luminescent agents, and the luminescent markers were quantitatively or qualitatively detected after the reflection |
• High sensitivity and specificity; • Wide range of detection |
• Require professional laboratories and large instruments | [13, 103] |
| ELISA | An enzyme-labeled method of labeling primary or secondary antibody-specific antigens and antibodies with enzymes |
• High sensitivity; • High-throughput; |
• Time-consuming | [12, 104] |
| Neutralization test | Virus and antibody are mixed and incubated under appropriate conditions before testing the infection of the virus on the cell | • Can determine whether the antibody can neutralize the virus |
• Need sophisticated skills and biosafety laboratory to manipulate the live pathogen; • Time-consuming |
[15–17] |
| Biosensor assays | Combines a biological component with a physicochemical detector |
• Fast and sensitive response; • High-throughput; |
• Electrode not easy to maintain; • Need advanced skills; • Expensive labels |
[104] |
However, none of these three methods can determine whether the antibody has the ability to neutralize the virus. The presence of neutralizing antibodies (nAbs) is considered a functional correlate of immunity because it provides at least partial resistance to subsequent infections by virus–antigen binding to prevent interaction with host cells [20, 21]. Therefore, comparing new serological assays to virus-neutralizing tests is critical as part of the validation process. The current gold standard method to measure nAb is the conventional virus neutralization test. In the test, virus and nAbs are mixed and incubated under appropriate conditions before testing the infection of the virus on the cell [15–17]. The FDA stated in one of the templates [22] that plaque reduction neutralization test (PRNT) [23] is currently considered to be the gold standard for detecting and measuring nAb titers. Microneutralization assays and focus reduction neutralization testing are also accepted as comparable neutralization comparator methods. At the moment, the neutralization antibody assay is the gold standard for the diagnosis of antibodies and evaluation of the effectiveness of antibodies against COVID-19; however, the neutralization test requires a biosafety level 3 laboratory to manipulate the live pathogen [24] and a longer detection time; thus, it is far less popular than the methods listed above [15, 17] and cannot be used for large-scale screening. Therefore, neutralization assays using pseudovirus [25] and surrogate virus [26] were developed. Results of pseudovirus PRNT prepared by Yang et al. [27] were highly correlated with those obtained using live viruses (R2 = 0.6931, P < 0.005). Studies of surrogate viruses by Tan et al. [28] achieved a specificity of 99.93% and sensitivity of 95–100%, and neither pseudoviruses nor surrogate viruses required a biosafety level 3 laboratory [29]. They not only ensure the certainty of antibody detection but also reduce the limitation of test conditions.
In addition to the abovementioned traditional immunoassays, a wide range of technologies has also been developed to overcome the shortages in the testing area. Biosensing refers to the detection of biomolecules using an analytical device (i.e., biosensor) that combines a biological component with a physicochemical detector. Researchers have recently reported a variety of biosensing strategies such as electrochemical, optical, electrical, mechanical, and piezoelectric biosensors for the detection of pathogens [30, 31]. Among them, field-effect transistors have attracted the attention of scientists due to their miniaturized size, fast and sensitive response, and the potential for parallel sensing. Seo et al. [7] modified a graphene-based FET biosensor device (COVID-19 FET sensor) to detect spike protein on SARS-CoV-2. This device used probe linkers like 1-pyrenebutyric acid N-hydroxy succinimide ester (PBASE) to immobilize the specific antibody and can detect viral antigens with a sensitivity of 1 fg/ml. Furthermore, Cady et al. [32] introduced a multiplexed grating-coupled fluorescent plasmonics biosensor platform for the measurement of antibodies against SARS-CoV-2 in human blood serum and dried blood spot samples.
Immunotherapy
Immunotherapy has been shown to reliably enhance immune protection against cancer and viral infections, by modifying the patient’s own immune system or using antibodies provided from external sources. As the aggravation of physical damage and the increase in mortality of COVID-19 patients are all related to insufficient production of anti-SARS-CoV-2 antibodies, the use of antibody immunotherapy in improving the course of the disease and reducing mortality is of great research value. To that end, this section collects and describes the effects of several existing immunotherapies that use antibodies, with the hope of pointing out the future direction of immunotherapy.
Specific nAb-based therapies are being used to treat people who have been infected with the virus, and these therapies show great promise in reducing hospitalizations and deaths. Table 2 shows the summary of nAb-based therapy studies. A summarized diagram showing the current evidence for immunoassay and immunotherapy as well as the targeting sites is presented in Fig. 1.
Table 2.
Summary of studies on nAb-based therapies
| Publication | Ab type | Ab ID | Target | IC50 against pseudotyped SARS-CoV-2 (ug/ml) | IC50 against authentic SARS-CoV-2 (ug/ml) | Affinity (nmol/l) | Source | Registration number | Type of trial |
|---|---|---|---|---|---|---|---|---|---|
| Wu et al. [105] |
mAb mAb |
B38 H4 |
RBD RBD |
NR NR |
0.177 0.896 |
70.1 4.48 |
COVID-19 convalescent |
NR |
Clinical trial; phase 1 Initial isolation of nAbs |
| Cao et al. [106] | mAb | BD-368–2 | RBD | 0.0012 (HIV) | 0.015 | 0.82 | COVID-19 convalescent | NR | Animal model study |
| Shi et al. [42] | mAb | CB6 | RBD | 0.23 ~ 0.41 (HIV) | 0.036 | 2.49 | COVID-19 convalescent | NCT04780321 | Clinical trial; phase 2 |
| Ju et al. [41] | mAb | P2C-1F11 | RBD | 0.03 (HIV) | 0.03 | 2.21 | COVID-19 convalescent | NR | Initial isolation of nAbs |
| Hansen et al. [107] |
Cocktail Cocktail |
REGN10987 REGN10933 |
RBD RBD |
0.008 (HIV) 0.008 (HIV) |
0.007 0.009 |
NR NR |
Genetically humanized VI mice as well as COVID-19 convalescent |
Clinical trial; phase 2 Clinical trial; phase 3 |
|
| Rogers et al. [39] | mAb | CC6.29 | RBD | 0.002 (MLV) | 0.007 | 1.2 | COVID-19 convalescent | NR | Animal model study |
| Brouwer et al. [40] |
mAb mAb |
COVA1-18 COVA2-25 |
RBD RBD |
0.008 (HIV) 0.008 (HIV) |
0.007 0.009 |
NR NR |
COVID-19 convalescent |
NR NR |
Animal model study |
| Seydoux et al. [108] | mAb | CV30 | RBD | 0.03 (HIV) | NR | 3.6 | COVID-19 convalescent | NR | Initial isolation of nAbs |
| Liu et al. [43] |
mAb mAb mAb mAb |
2–15 2–17 5–24 4–8 |
RBD NTD NTD NTD |
0.005 (VSV) 0.168 (VSV) 0.013 (VSV) 0.032 (VSV) |
0.0007 0.007 0.008 0.009 |
NR NR NR NR |
COVID-19 convalescent |
NR NR NR NR |
Initial isolation of nAbs |
| Wu et al. [109] |
mAb mAb |
n3088 n3130 |
RBD RBD |
3.3 (HIV) 3.7 (HIV) |
2.6 4 |
3.7 (S1) 55.39 (S1) |
Phage-displayed single-domain antibody |
NR NR |
Initial isolation of nAbs |
| Sun et al. [110] | mAb | ab6 | RBD | NR | 0.35 | 11 | Phage-displayed single-domain antibody | NR | Initial isolation of nAbs |
| Chi et al. [50] |
mAb mAb |
4A8 0304-3H3 |
Non-RBD (S1, S-ECD) Non-RBD (S2, S-ECD) |
49 (HIV) NR |
0.61 0.04 |
92.7 (S1) 4.52 (S2) |
COVID-19 convalescent |
NR NR |
Initial isolation of nAbs |
| Jiang et al. [111] | Cross-neutralizing nAb | 47D11 | RBD | 0.061 (VSV) | 0.57 | 9.6 (RBD) | hACE2 transgenic mouse | NCT04644120 | Clinical trial; phase 1 |
| Sun et al. [112] | Cross-neutralizing nAb | S309 | RBD | 0.24 (MLV) | 0.079 | < 0.001 (RBD) | SARS convalescent | NR | Initial isolation of nAbs |
| Sia et al. [113] | Cross-neutralizing nAb | ADI-55688,55,689,55,690,55,951,55,993,56,000,56,010,56,032,56,046 | RBD | 0.05 ~ 1.40 (MLV) | 0.05 ~ 1.40 | < 10 | SARS convalescent | NR | Animal model study |
| Chi et al. [50] | Cross-neutralizing nAb |
COVA1-16 COVA2-02 |
RBD RBD |
0.131 (HIV) NR |
0.745 < 10 |
NR NR |
COVID-19 convalescent |
NR NR |
Initial isolation of nAbs |
| Tai et al. [70] | Cross-neutralizing nAb | 18F3,7B11 |
18F3: SARS-CoV and SARS-CoV-2 RBDs; 7B11: SARS-CoV RBD and not fully conserved in SARS-CoV-2 RBD |
< 10 (HIV) | NR | NR | SARS-CoV RBD-immunized mouse | NR | Initial isolation of nAbs |
| Baum et al. [60] | mAb | RGEN10933,REGN10987 | RBD | NR | NR | NR | Genetically humanized mice as well as COVID-19 convalescent |
Clinical trial; phase 3 Clinical trial; phase 2 Clinical trial; phase 3 |
|
| Lv et al. [114] | mAb | H014 | RBD | 3 nM | 38 nM | 27.8 nM | The phage antibody library | NR | Initial isolation of nAbs |
| Tychan [115] | mAb | TY027 | RBD | NR | NR | NR | Phage display | NCT04429529 | Clinical trial; phase 1 |
| Chen et al. [116] | mAb | LY3819253/Ly-CoV555 | RBD | NR | NR | NR | COVID-19 convalescent |
Clinical trial; phase 1 Clinical trial; phase 1 |
|
| Ju et al. [41] | mAb | JS106 (CB6) | RBD | NR | NR | NR | COVID-19 convalescent | NCT04441918 | Clinical trial; phase 1 |
| SAb [117] | pAb | SAB-185 | RBD | NR | NR | NR | Transgenic cattle |
Clinical trial; phase 1 Clinical trial; phase 1 |
|
| Sorrento Therapeutics Inc. [118] | mAb | STI-1499 (COVI-GUARD) | RBD | NR | NR | NR | COVID-19 convalescent | NCT04454398 | Clinical trial; phase 1 |
| Haschke et al. [119] | hrsACE2 | APN01 | RBD | NR | NR | NR | Recombinant human ACE2 protein |
Clinical trial; phase 1 Clinical trial; phase 2 |
|
| Gaborit et al. [120] | Cocktail | XAV-19 | RBD | NR | 2.5 | NR | Humanized animals | NCT04453384 | Clinical trial; phase 2 |
| Brii Biosciences Ltd [121] | mAb | BRII-196, BRII-198 | RBD | NR | NR | NR | COVID-19 convalescent |
Clinical trial; phase 1 Clinical trial; phase 1 Clinical trial; phase 2 |
|
| Sinocelltech Ltd. [122] | mAb | SCTA01 | RBD | NR | NR | NR | Mice | NCT04483375 | Clinical trial; phase 1 |
IC50 half-maximum inhibitory concentrations, Ab antibody, mAb monoclonal antibodies, pAb polyclonal antibody, nAb neutralizing antibody, hrsACE2 human recombinant soluble ACE2, NTD N-terminal domain, RBD receptor-binding domain, HIV human immunodeficiency virus, VSV vesicular stomatitis virus, MLV Moloney murine leukemia virus, NR not reported
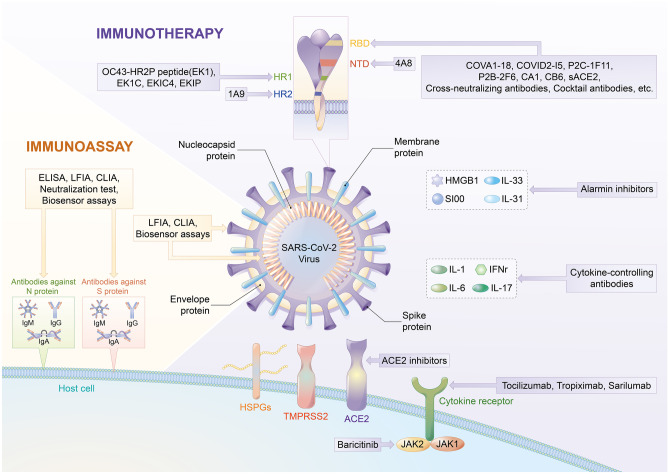
Fig. 1.
Schematic summary of immunoassay and immunotherapy for COVID-19. Among the four structural proteins of SARS-Cov-2, S and N proteins are commonly used as biomarkers for immunoassay. Two common technologies used in rapid antigen detection are LFIA and CLIA. As for antibody detection, ELISA, LFIA, CLIA, and neutralization tests are common assays. In addition, biosensor devices are developed for antigen and antibody detection. Immunotherapy includes neutralizing-antibody–based treatments targeting different regions of SARS-CoV-2, which include RBD and NTD on the S1 protein, as well as HR1 and HR2 on the S2 protein. Immunotherapy targeted on host cell ACE2 receptor, HSPGs, and IL-6 receptor can effectively inhibit the host cell receptor and cytokine storm. Antibodies that control the production of cytokines and alarmin inhibitors also play a role in inhibiting cytokine storms. S protein spike protein, N protein nucleocapsid protein, LFIA lateral flow immunoassay, CLIA chemiluminescence immunoassay, ELISA enzyme-linked immunosorbent assay, RBD receptor-binding domain, NTD N-terminal domain, HR heptad repeat, sACE2 soluble ACE2, HSPGs heparan sulfate proteoglycans
Convalescent Plasma Therapy
Convalescent plasma (CP) has been reported to be used in treating COVID-19 patients in the early stages of the SARS-CoV-2 pandemic, in the absence of approved specific antiviral agents. In a recent study [33], CP was used to treat 10 patients with severe COVID-19. Three days after the infusion of 200 ml CP, their clinical symptoms and blood oxygen saturation improved significantly, the level of viral loads and C-reactive protein decreased, and blood lymphocytes increased. After 7 days of treatment, CT scans revealed that all patients’ lung lesions had been absorbed. Similarly, Zeng et al. [34] reported that CP therapy improved clinical symptoms rapidly and was well tolerated in patients with severe COVID-19. Their research also demonstrated that CP could increase and maintain high nAb levels, as well as cause virus RNA to vanish within 7 days. The US FDA has approved CP to treat patients with severe COVID-19, but it is conditional on doctor approval [35]. It is worth noting that all patients received standard care, including antiviral and hormonal therapy, in addition to CP transfusion. Although CP can be obtained easily by removing blood cells of the donated blood, leaving behind plasma and antibodies, the large-scale application of CP treatment still faces significant challenges. These difficulties include a large number of CP deficiencies and donor-dependent variations in antibody specificities and titers [36]. Before CP therapy can be used widely, it must first be thoroughly evaluated in large-scale randomized controlled clinical trials. In addition, for CP therapy, a minimum effective nAb titer and optimal treatment time point must be determined.
RBD-Targeting nAb
Among the four structural proteins in SARS-COv-2, the S protein plays a key role in viral infection and pathogenesis [37]. It comprises subunits S1 and S2: S1 harbors the N-terminal domain (NTD) and the receptor-binding domain (RBD), whereas S2 harbors heptad repeat 1 (HR1) and HR2 [38]. The blood serum of COVID-19 patients contains a variety of SARS-CoV-2 antibodies, and RBD-binding antibodies have a higher neutralizing activity than non-RBD–binding antibodies. Rogers et al. [39] found that although viral infection induces a strong immune response against the non-RBD regions of the S protein, only a small part of this response is neutralizing. Brower et al. [40] isolated 19 nAbs, which target multiple antigenic sites on the S protein, and discovered that 74% of them targeted the RBD. Among them, COVA1-18 and COVID2-I5 were found to have the highest neutralizing activity against authentic SARS-CoV-2 virus, with half-maximum inhibitory concentrations (IC50) of 0.007 and 0.009 ug/ml, respectively. Ju et al. [41] isolated and characterized 206 monoclonal antibodies (mAbs) specific for the RBD from single B cells from eight individuals infected with SARS-CoV-2. P2C-1F11 and P2B-2F6 were two kinds of nAbs with IC50 values of 0.03 and 0.05 ug/ml, respectively.
Shi et al. [42] isolated two specific human mAbs (designated CA1 and CB6) targeting the RBD site from a patient recovering from COVID-19 infection. CB6 exhibited potent neutralization activity against SARS-CoV-2 in vitro, with an IC50 of 0.23–0.41 ug/ml for pseudovirus and 0.036 ug/ml for euvirus. In prophylactic settings, CB6 inhibited SARS-CoV-2 infection in rhesus monkeys, with a peak viral load of less than 103 RNA copies/ml. In treatment settings, CB6 inhibited the pathological lung damage and reduced the viral load by 3 lg. The RBD-binding antibodies that compete directly with ACE2 are the preferred prophylactic and therapeutic applications, and as reagents for defining antibody epitopes for vaccine design. Liu et al. [43] described the isolation of 61 SARS-CoV-2-neutralizing mAbs from five COVID-19 patients. Cryo-electron microscopy reconstructions revealed that the antibodies recognize the spike in its closed, ‘all RBD-down’ state. A 1.5-mg/kg mAb2-15 reduced both the infectious virus titers and RNA copy numbers in lung tissue by 4 logs in a golden hamster prevention experiment.
However, the problem of the continuous variation of RBD exists. Mutations in the mutant strain’s spike gene cause adaptations to the nAbs, making the vaccine less effective. Existing vaccines and therapeutic antibodies have been reported to be less effective in treating the newly discovered SA variant (B.1.351), cocktail therapy, or convalescent plasma as well. Targeting a relatively stable conserved peptide could be a response strategy to the high RBD variation of SARS-CoV-2. According to research, the antibody induced by the cross-reaction of SARS-CoV and MERS-CoV is targeted at the conserved peptide of coronavirus [44]. 3Clpro is related to virus replication, and the corresponding gene may be the coronavirus conserved sequence [45]. On the other hand, approaches to develop modified molecules with higher RBD-competing activity might be an option. Higuchi et al. [46] discovered that soluble ACE2 (sACE2), an engineered highly compatible extracellular domain of ACE2, effectively neutralizes the N501Y mutant strain and the escaped virus from convalescent plasma in a hamster model, and is currently undergoing phase II clinical trials in Europe. Lei et al. [47] created an ACE2-human IgG1 fragment crystallizable (Fc) fusion by connecting the extracellular domain of human ACE2 to the Fc region of the human immunoglobulin IgG1. This recombinant protein can neutralize pseudoviruses expressing S proteins of SARS-CoV-2 or SARS-CoV. It has also been demonstrated that the soluble recombinant AociCE2 receptor has a high affinity for the SARS S protein, with an affinity of 1.70 nM, which is comparable to mAb affinities. One potential limitation of the recombinant ACE2 strategy is that the increase in extracellular ACE2 levels may have unanticipated effects on the body, disrupting the normal combination of ACE2 in tissues [48].
Non-RBD Regions Targeting nAb
nAbs directed against non-RBD regions were detected in the plasma of COVID-19 convalescent patients [49], including nAbs against the N-terminal domain (NTD) and the S2 protein. S2 and NTD are generally not competitive inhibitors of RBD binding to ACE2 due to their distance from the binding site. Antibodies against S2 and NTD may inhibit virus binding to ACE2 or fusion with the cell membrane in an indirect manner by preventing the conformational change of S protein [50]. Chi et al. [50] isolated an antibody (4A8) from convalescent patients with COVID-19, which proved that the binding on the NTD of the SARS-CoV-2 S protein had a high neutralizing effect. Liu et al. [43] isolated three NTD-directed antibodies from five SARS-CoV-2-infected patients. In this study, all three antibodies showed high potency, with an IC50 value of 0.007 to 0.109 μg/ml against authentic SARS-CoV-2.
Previous studies have isolated several neutralizing mAbs to the S2 subunit, especially heptad repeat (HR) loops including HR1 and HR2 domains [51–53]. The HR region in the S2 subunit is conserved among various coronaviruses that can infect humans (HCoVs), and plays a central role in HCoV infections by forming the six-helix bundle (6-HB) core structure that mediates viral fusion [54]. 1A9 antibody is the only monoclonal antibody known to bind to the HR2 domain of the SARS-CoV-2 coronavirus S2 subunit and could neutralize in vitro infection of severe acute respiratory syndrome coronavirus [51]. Xia et al. [55] reported a modified OC43-HR2P peptide (EK1) that can bind HR1s of multiple HCoVs and form stable complexes, thereby inhibiting S protein-mediated fusion. In vivo studies demonstrated that intranasal administration of EK1 revealed highly protective effects and safety profiles, underscoring its clinical potential. Structural studies of EK1 highlighted its broad compatibility in accommodating HR1s from different HCoVs, thus consolidating its broad-spectrum inhibitory effect against pan-CoVs. As the potent stability of the SARS-CoV-2 6-HB structure might reduce the antiviral efficacy of EK1, Xia et al. [56] improved the inhibitory activity of EK1 by constructing lipopeptides EK1C and EK1P. Among a series of cholesteryl EK1 with multiple linkers, lipopeptide EK1C4 exhibited the most potent inhibitory activity against entry of pseudotyped coronaviruses and against in vitro infection by live coronaviruses. Intranasal application of EK1C4 showed strong protection of mice against HCoV-OC43 infection, suggesting that EK1C4 could be used to prevent and treat current and future SARS-related coronavirus infections. Moreover, Hoffmann et al. [57] found that the entry inhibitors EK1 and EK1C4 would still be active against the recently emerged SARS-CoV-2 variants B.1.1.7, B.1.351, and P.1 that harbor mutations in the S protein, indicating their strong inhibitory effects. In addition to the well-known protein targets, glycan targets are also worth studying because the S protein of SARS-CoV-2 is highly glycosylated. The isolated s309 nAb recognized glycan epitope on RBD indicated that the glycosylation of S protein might affect the development of nAbs against SARS-CoV-2 [58].
Cocktail Antibodies
The effect of convalescent plasma has been demonstrated in many trials and early clinical applications, but its application has been limited. The S protein of SARS-CoV-2 has undergone mutations that may make it resistant to single-targeting nAbs [40]. Mutations in the S protein’s E484, F490, Q493, and S494 have been shown to cause SARS-CoV-2 complete or partial resistance to some potential therapeutic antibodies [59]. To address this issue, preclinical studies show that combining two or more nAbs (cocktail antibodies) targeting distinct epitopes is a more effective and superior antibody therapy. The REGN10933 + REN10987 antibody cocktail, which binds to two nonoverlapping RBD epitopes, was able to neutralize all identified mutants [60–62].
Weinreich et al. [63] found that an antibody cocktail against SARS-CoV-2 can improve the prognosis by increasing the virus clearance rate, especially for patients with a slow start of virus immune response, and become an effective antiviral therapy. They showed that the REGN-COV2 antibody cocktail reduced the viral load and had a greater impact on patients who had not yet started the immune response or had a higher viral load at baseline. The safety results of the combined dose group of REGN-COV2 and placebo group were similar. Currently, the antibody cocktail is primarily used to treat confirmed COVID-19 patients who do not need supplemental oxygen and who are at high risk of progressing to severe COVID-19 [64]. It has been approved for use in the US and Brazil and is undergoing clinical trials in the UK.
Cocktail therapy has been used in other fields to varying degrees. Cocktail antibody therapy outperforms single antibody therapy in the treatment of SARS-CoV-2, particularly in terms of producing resistance to mutated viruses. It is necessary to develop better cocktail antibody combinations for different virus variants so that the treatment can be more targeted.
Cross-Neutralizing Antibodies
The primary amino acid sequences of S proteins in SARS-CoV and SARS-CoV-2 share 77.2% amino acid sequence identity, with 79.59% similarity and 74% identity in RBD domains [65, 66]. The high homology of two viral proteins allows for the isolation of SARS-CoV and SARS-CoV-2 cross-reactive antibodies. However, it is reported that SARS-CoV and SARS-CoV-2 cross-reactive antibodies can only neutralize one of the two viruses, but not the other, such as CR3022 [67]. Another antibody 515–5 isolated from patients with COVID-19 can effectively neutralize the SARS-CoV-2 virus and has only a weak but detectable neutralization effect on SARS-CoV [68]. Wan et al. [66] showed that the humanized antibody H014 could effectively neutralize SARS-CoV and SARS-CoV-2 by binding to a novel RBD conformational epitope. The cross-neutralizing antibody s309 isolated by Pinto et al. [69] from the memory B cells of SARS convalescent patients was shown to have high neutralizing activity against SARS-CoV and SARS-CoV-2 pseudovirus. Brouwer et al. [40] identified COVA1-16 and COVA2-02 from three COVID-19 patients, with IC50s of 2.5 and 0.61 μg/ml against pseudotyped SARS-CoV-2, respectively. Tai et al. [70] identified six SARS-CoV RBD-specific neutralizing mAbs that cross-reacted with SARS-CoV-2 RBD. Two of them, 18F3 and 7B11, can neutralize SARS-CoV-2 infection. It is worth noting that SARS-CoV and SARS-CoV-2 cross-neutralizing mAbs have recently been proved not to compete with hACE2 (e.g., nAbs ADI-55689/ADI-56046, 47D11, and S309), which indicates that there may be conserved epitopes between SARS-CoV-2 and SARS-CoV in addition to hACE2 binding sites [41, 66, 71, 72].
Immunotherapy Based on Inhibiting Host Cell–Virus Binding
Creating an antibody-like molecule that binds to host cells is another potentially promising strategy. The viral entry protein ACE2 and heparan sulfate proteoglycans (HSPGs) are all potential targets. Kruse et al. [48] proposed an antibody or single-chain antibody fragment (scFv) that could be engineered to deliver the SARS-CoV-2 S protein’s RBD by binding ACE2 and saturating available sites in host cells. ACE2 inhibitors are now commonly used as therapeutic drugs in addition to therapeutic antibodies, but studies have shown that inhibitor-blocked ACE2 can still bind to SARS-CoV-2 RBD in the closed conformation [46], implying that ACE2 inhibitors are ineffective as antiviral infection drugs.
Viral entry requires not only binding to the ACE2 receptor, but also priming of the virus’s spike (S) protein by the transmembrane protease serine 2 (TMPRSS2) by cleavage of the S proteins. The TMPRSS2 and ACE2 dual inhibitors in COVID-19 would be a novel antiviral class of drugs called “entry inhibitors.” For this purpose, Baby et al. [73] analyzed approximately 2800 US FDA-approved drugs by a virtual docking tool and found that lopinavir and valrubicin have the potential of dual-target inhibition whereby preventing SARS-CoV-2 entry to the host. In addition, due to the essential metabolic roles of ACE2, Baughn et al. [74] believed that TMPRSS2 may be a better candidate for targeted therapies. Leng et al. [75] have reported that intravenous transplantation of ACE2- and TMPRSS2- mesenchymal stem cells (MSCs) that are free from COVID-19 infection, improved the outcome of 7 enrolled patients with COVID-19 pneumonia 2–4 days after MSC transplantation. HSPGs are the primary sites for direct contact between the virus and cell surface [76]. Clausen et al. [77] demonstrated that the SARS-CoV-2 S protein interacts with heparan sulfate and ACE2 on the cell surface via RBD. Independently, ACE2 or heparan sulfate can bind to RBD of S protein, and the binding sites are adjacent. Heparan sulfate functions as a scaffold in the formation of a ternary complex with S protein and ACE2. Heparan sulfate increases the openness of RBD and promotes its binding to ACE2. Similarly, Clausen et al. [77] demonstrated that heparan sulfate promoted the entry of spike-dependent viral. Altogether, these findings point to HSPGs as a co-receptor for SARS-CoV-2 entry. The development of antibodies that target HSPGs could be an effective therapeutic strategy.
Immunotherapy Based on Inhibiting Cytokine Storm
The pro-inflammatory cytokine IL-6 may initiate an inflammatory cascade known as cytokine storm during the COVID-19 immune responses, resulting in increased alveolar–capillary blood–gas exchange dysfunction [66, 78]. Cytokine release syndrome is a significant contributor to acute respiratory distress syndrome and multiple organ failure. Many studies have found that cytokine storm is one of the important causes of the death of MERS, influenza, and SARS [79, 80]. IL-6 can be used as a target cytokine in the treatment of COVID-19-related acute respiratory distress syndrome. Clinical data show that serum IL-6 levels are significantly higher in ICU patients [81]. Michot et al. [82] reported a patient with respiratory failure associated with COVID-19. He had a rapid favorable outcome after two infusions of the anti-IL-6 receptor inhibitor tocilizumab, which indicated that anti-IL-6 receptor inhibitor therapy could reduce the risk of progression to SARS by mitigating the cytokine storm in the lungs with COVID-19. REMAP-CAP researchers [83] have shown that treatment with interleukin-6 receptor antagonists tropiximab and sarilumab improves the prognosis of critically ill patients, including survival, who receive organ support in the intensive care unit.
There are also some other cytokine antagonists, such as Baricitinib and fluvoxamine, that have been used in clinical trials with promising results. Baricitinib is a highly selective, effective, and safe JAK1 and JAK2 inhibitor that can inhibit IL-2, IL-6, IL-10, IFN-γ, and others. Baricitinib also inhibits endocytosis and virus aggregation through AP2-associated protein kinase 1 (AAK1) and cyclin G-associated kinase (GAK) [84].
Several studies have begun to investigate the feasibility of other cytokine therapies, in addition to treating patients with cytokine inhibitors. Tjan et al. recommended neutralizing antibody therapy in the early stages of infection and steroid therapy in severe cases after virus clearance was confirmed [85]. Other research has suggested that there may be existing monoclonal antibodies or specific antagonists that control the production of cytokines like IL-6, IL-1, IL-17, and IFN-γ [86]. The use of such cytokine-controlling antibodies can significantly improve the efficacy of neutralizing antibody therapy. Alarmins, such as HMGB1, IL-31, IL-33, and S100, have been shown in some studies on the mechanism of cytokines to aggravate cytokine storm and disease [87]. Alfarouk et al. described the specific mechanism of cytokine storms from the perspective of mitochondria: mitochondria are the focal point of cytokine storms. In the elderly population, the rational use of alarmins inhibitors or pharmacological or non-pharmacological modulators of mitochondrial function in neutralizing antibody therapy may improve efficacy and patient life quality [87, 88].
Autoantibodies Produced by Cross-Reaction Between Viral Epitopes and Autoepitopes
Autoantibodies are antibodies that mistakenly target and react with a person’s own tissues, organs, cells, and cell components. Usually, the immune system is able to discriminate between foreign substances (“non-self”) and the body’s own cells (“self”). When the immune system fails to distinguish between “self” and “non-self”, a large number of autoantibodies are produced, resulting in the development of autoimmune diseases. Autoimmune diseases activate complement; damage tissue; promote systemic inflammation [89]; cause damage to the skin, mucous membranes, and organs; as well as vascular inflammation such as thrombotic microangiopathy [90].
A variety of autoantibodies may be produced by COVID-19 patients, causing a prolonged period of immunodeficiency (T-cell and B-cell dysfunction [91], increased risk of new events of respiratory, diabetes, and cardiovascular diseases [92]), and a significant decline in the body function in some patients after recovery, a condition known as “post COVID-19 syndrome” [93].
Anti-NETs (neutrophil extracellular traps) antibodies have been discovered in COVID-19 patients’ serum [94]. NETs are bactericidal networks that form after the death of neutrophils and play a significant role in the pathogenesis of thrombovasculitis. However, the precise mechanism by which these antibodies inhibit NET clearance in the serum remains unknown [95]. Another research explored the development of autoantibodies in COVID-19 patients who had no prior autoimmune disease, and found that COVID-19 is linked to human autoantibody production [96].
On the other hand, autoantibodies may be closely related to cytokine storms, exacerbating the body’s inflammatory response and leading to acute respiratory distress (ARD) or acute renal failure (ARF). In other words, COVID-19 patients with positive autoantibodies (antinuclear and anti-neutrophilic cytoplasmic antibodies (ANCA)) had the worst clinical outcomes [96]. A study of the clinical and laboratory characteristics of 21 severe COVID-19 cases in China found that the prevalence of anti-52 kDa SSA/Ro antibody, anti-60 kDa SSA/Ro antibody, and antinuclear antibodies were 20%, 25%, and 50%, respectively [97].
The potential mechanism of autoantibody production has not been fully explained. One of the current hypotheses is the molecular simulation of infectious pathogens. Autoantigen is an endogenous antigen that is recognized as non-self by the immune system. Some SARS-CoV-2 epitopes showed cross-reactivity with autoantigen. By exposing the epitope to produce cross-reacting antibodies, the virus induces an autoimmune response [98]. Vojdani and Kharrazian [99] discovered that 21 out of 50 human tissue antigens had moderate to high strength responses to SARS-CoV-2 antibody, suggesting an antigenic cross-reaction between the SARS-CoV-2 epitope and various human tissue antigens. This cross-reaction can result in high titers of autoantibodies, inducing Guillain–Barre syndrome [100], lupus, antiphospholipid syndrome, and ANCA-associated vasculitis [98]. Franke et al. [101] found that high levels of antibodies were detected in patients’ cerebrospinal fluid. Indirect immunofluorescence of unfixed mouse brain sections showed the strongest IgG binding in most cases, implying that some human SARS-CoV-2 monoclonal antibodies may bind to brain tissue and induce autoimmune responses.
Therefore, when developing a neutralizing antibody against SARS-CoV-2 for clinical use, its cross-reaction should be taken into account to avoid autoimmune damage. Because autoantibodies are common in severe COVID-19 cases, and various cross-reaction mechanisms are still unknown, it is difficult to consider the overall effect of neutralizing antibody therapy on the body in a comprehensive manner. Therefore, it is critical to choose appropriate neutralizing antibodies against virus epitopes.
Conclusions
In recent years, understanding the immune response, especially the antibody immunity response, has led to the development of various clinical strategies for the detection and treatment of diseases such as COVID-19. Immunology against SARS-CoV-2 still represents several clinical and translational challenges, and more research is required to tailor immunoassays and immunotherapies to further improve detection and treatment efficacy.
Authors’ Opinion
The emergence of the COVID-19 pandemic resulting from the spread of the SARS-CoV-2 has inspired intensive efforts to develop immunoassays and immunotherapy to support clinical diagnosis and treatment. Lateral flow immunoassay (LFIA) that directly detect viral antigens or antibodies in clinical samples without any sample preparation step is necessary for the fast diagnosis of COVID-19. Highly sensitive immunological methods such as CLIA and ELISA can be used for high-throughput and accurate diagnosis. Virus-neutralizing tests, on the other hand, are crucial as they determine whether antibodies have the ability to neutralize the virus. A variety of biosensing strategies have also been developed.
The use of specific antibody immunotherapy is now one of the most effective therapeutic methods for COVID-19. In the early stages of the SARS-CoV-2 pandemic, convalescent plasma (CP) has been used in treating COVID-19 patients. Subsequently, monoclonal neutralizing antibodies (nAb) that interfere with different steps of viral infection were developed, with some showing considerable improvements in clinical results. The immunology against SARS-CoV-2 viruses still represents several clinical and translational challenges, and more research is required to tailor immunoassays and immunotherapies to further improve detection and treatment efficacy.
Abbreviations
- ANCA
Anti-neutrophilic cytoplasmic antibodies
- CLIA
Chemiluminescence immunoassay
- COVID-19
Coronavirus disease 2019
- CP
Convalescent plasma
- ELISA
Enzyme-linked immunosorbent assay
- Fc
Fragment crystallizable
- PRNT
Plaque reduction neutralization test
- HR
Heptad repeat
- hrsACE2
Human recombinant soluble ACE2
- HSPGs
Heparan sulfate proteoglycans
- Ig
Immunoglobin
- LFIA
Lateral flow immunochromatographic assays
- mAb
Monoclonal antibody
- N protein
Nucleocapsid protein
- nAb
Neutralizing antibody
- NET
Neutrophil extracellular traps
- NR
Not reported
- NTD
N-terminal domain
- pAb
Polyclonal antibody
- RBD
Receptor-binding domain
- S protein
Spike protein
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
Author Contribution
All authors, ZJC, BL, ZZhan, ZZhao, MX, PZ, JL, CH, JH, RC, and BS, made substantial contributions to the writing of this review, revised it critically for important intellectual content, approved the version to be published, and agreed to be accountable for all aspects of this review. All authors agreed with the content of this review and gave explicit consent to submit.
Funding
This study was supported by Guangzhou Institute of Respiratory Health Open Project (Funds provided by China Evergrande Group) (2020GIRHHMS04), Zhongnanshan Medical Foundation of Guangdong Province (ZNSA-2021005, ZNSA-2020001), State Key Laboratory of Respiratory Disease, Guangdong-Hong Kong-Macao Joint Laboratory of Respiratory Infectious Disease (GHMJLRID-Z-202102), Emergency Key Project of Guangzhou Laboratory (EKPG21-30–2), and Cultivation Project of the First Affiliated Hospital of Guangzhou Medical University (ZH202105).
Declarations
Conflict of Interest
The authors declare no conflict of interest related to this review.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhangkai J. Cheng, Bizhou Li, Zhiqing Zhan and Zifan Zhao contributed equally.
Contributor Information
Jianxing He, Email: hejx@vip.163.com.
Ruchong Chen, Email: chen_rch@163.com.
Baoqing Sun, Email: sunbaoqing@vip.163.com.
References
- 1.Srinivasan S, Cui H, Ga Z, Liu M, Lu S, Mkandawire W, Narykov O, Sun M, Korkin D (2020) Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses 12(4) [DOI] [PMC free article] [PubMed]
- 2.Malik YA. Properties of coronavirus and SARS-CoV-2. Malays J Pathol. 2020;42(1):3–11. [PubMed] [Google Scholar]
- 3.Li Y, Yao L, Li J, Chen L, Song Y, Cai Z, Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020;92(7):903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis. 2020;71(16):2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong DSY, Fragkou PC, Schweitzer VA, Chemaly RF, Moschopoulos CD, Skevaki C (2021) European Society of Clinical, M, Infectious Diseases Study Group for Respiratory, V., How to interpret and use COVID-19 serology and immunology tests. Clin Microbiol Infect [DOI] [PMC free article] [PubMed]
- 6.Black MA, Shen G, Feng X, Garcia-Beltran WF, Feng Y, Vasudevaraja V, Allison D, Lin LH, Gindin T, Astudillo M, Yang D, Murali M, Iafrate AJ, Jour G, Cotzia P, Snuderl M (2021) Analytical performance of lateral flow immunoassay for SARS-CoV-2 exposure screening on venous and capillary blood samples. J Immunol Methods 489:112909 [DOI] [PMC free article] [PubMed]
- 7.Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, Lee CS, Jun S, Park D, Kim HG, Kim SJ, Lee JO, Kim BT, Park EC, Kim SI. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 8.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayon Y, Croset M, Chirouze V, Tayot JL, Lagarde M. Phospholipid molecular species from human placenta lipids. Lipids. 1993;28(7):631–636. doi: 10.1007/BF02536058. [DOI] [PubMed] [Google Scholar]
- 10.Woo PC, Lau SK, Wong BH, Tsoi HW, Fung AM, Kao RY, Chan KH, Peiris JS, Yuen KY. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J Clin Microbiol. 2005;43(7):3054–3058. doi: 10.1128/JCM.43.7.3054-3058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burbelo PD, Riedo FX, Morishima C, Rawlings S, Smith D, Das S, Strich JR, Chertow DS, Davey RT, Cohen JI. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis. 2020;222(2):206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alhajj M, Farhana A (2021) Enzyme linked immunosorbent assay. In StatPearls, Treasure Island (FL) [PubMed]
- 13.Espejo AP, Akgun Y, Al Mana AF, Tjendra Y, Millan NC, Gomez-Fernandez C, Cray C. Review of current advances in serologic testing for COVID-19. Am J Clin Pathol. 2020;154(3):293–304. doi: 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, Zhang Y, Wang J, Huang B, Lin Y, Yang J, Cai W, Wang X, Cheng J, Chen Z, Sun K, Pan W, Zhan Z, Chen L, Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matusali G, Colavita F, Lapa D, Meschi S, Bordi L, Piselli P, Gagliardini R, Corpolongo A, Nicastri E, Antinori A, Ippolito G, Capobianchi MR, Castilletti C (2021) Inmi Covid-Laboratory, T., SARS-CoV-2 serum neutralization assay: a traditional tool for a brand-new virus. Viruses 13(4) [DOI] [PMC free article] [PubMed]
- 16.Garcia-Beltran WF, Lam EC, Astudillo MG, Yang D, Miller TE, Feldman J, Hauser BM, Caradonna TM, Clayton KL, Nitido AD, Murali MR, Alter G, Charles RC, Dighe A, Branda JA, Lennerz JK, Lingwood D, Schmidt AG, Iafrate AJ, Balazs AB (2021) COVID-19-neutralizing antibodies predict disease severity and survival. Cell 184(2):476–488e11 [DOI] [PMC free article] [PubMed]
- 17.Tian L, Elsheikh EB, Patrone PN, Kearsley AJ, Gaigalas AK, Inwood S, Lin-Gibson S, Esposito D, Wang L (2021) Towards quantitative and standardized serological and neutralization assays for COVID-19. Int J Mol Sci 22(5) [DOI] [PMC free article] [PubMed]
- 18.Shen B, Zheng Y, Zhang X, Zhang W, Wang D, Jin J, Lin R, Zhang Y, Zhu G, Zhu H, Li J, Xu J, Ding X, Chen S, Lu R, He Z, Zhao H, Ying L, Zhang C, Lv D, Chen B, Chen J, Zhu J, Hu B, Hong C, Xu X, Chen J, Liu C, Zhou K, Li J, Zhao G, Shen W, Chen C, Shao C, Shen X, Song J, Wang Z, Meng Y, Wang C, Han J, Chen A, Lu D, Qian B, Chen H, Gao H. Clinical evaluation of a rapid colloidal gold immunochromatography assay for SARS-Cov-2 IgM/IgG. Am J Transl Res. 2020;12(4):1348–1354. [PMC free article] [PubMed] [Google Scholar]
- 19.Mekonnen D, Mengist HM, Derbie A, Nibret E, Munshea A, He H, Li B, Jin T (2021) Diagnostic accuracy of serological tests and kinetics of severe acute respiratory syndrome coronavirus 2 antibody: a systematic review and meta-analysis. Rev Med Virol 31(3):e2181 [DOI] [PubMed]
- 20.Poland GA, Ovsyannikova IG, Kennedy RB. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396(10262):1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baumgarth N, Nikolich-Zugich J, Lee FE, Bhattacharya D. Antibody responses to SARS-CoV-2: let's stick to known knowns. J Immunol. 2020;205(9):2342–2350. doi: 10.4049/jimmunol.2000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.In vitro diagnostics EUAs. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas (Accessed 18 May, 2021 )
- 23.Abe KT, Li Z, Samson R, Samavarchi-Tehrani P, Valcourt EJ, Wood H, Budylowski P, Dupuis AP, 2nd Girardin RC, Rathod B, Wang JH, Barrios-Rodiles M, Colwill K, McGeer AJ, Mubareka S, Gommerman JL, Durocher Y, Ostrowski M, McDonough KA, Drebot MA, Drews SJ, Rini JM, Gingras AC (2020) A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight 5(19) [DOI] [PMC free article] [PubMed]
- 24.Favresse J, Gillot C, Di Chiaro L, Eucher C, Elsen M, Van Eeckhoudt S, David C, Morimont L, Dogné JM, Douxfils J. Neutralizing antibodies in COVID-19 patients and vaccine recipients after two doses of BNT162b2. Viruses. 2021;13(7):1364. doi: 10.3390/v13071364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nie J, Li Q, Wu J, Zhao C, Hao H, Liu H, Zhang L, Nie L, Qin H, Wang M, Lu Q, Li X, Sun Q, Liu J, Fan C, Huang W, Xu M, Wang Y. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valcourt EJ, Manguiat K, Robinson A, Chen JC, Dimitrova K, Philipson C, Lamoureux L, McLachlan E, Schiffman Z, Drebot MA, Wood H (2021) Evaluation of a commercially-available surrogate virus neutralization test for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Diagn Microbiol Infect Dis 99(4):115294 [DOI] [PMC free article] [PubMed]
- 27.Yang, Ren, Baoying Huang, A. Ruhan, Wenhui Li, Wenling Wang, Yao Deng, and Wenjie Tan. "Development and effectiveness of pseudotyped SARS-CoV-2 system as determined by neutralizing efficiency and entry inhibition test in vitro." Biosafety and health 2, no. 4 (2020): 226231 [DOI] [PMC free article] [PubMed]
- 28.Tan CW, Chia WN, Qin X, Liu P, Chen MI, Tiu C, Hu Z, Chen VC, Young BE, Sia WR, Tan YJ, Foo R, Yi Y, Lye DC, Anderson DE, Wang LF. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol. 2020;38(9):1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 29.Qiu X, Weng J, Jiang Z, Yan C, Gu H. SINS model in the management of biosafety level 2 laboratories: exploration and practice. Biosaf Health. 2019;1(3):129–133. doi: 10.1016/j.bsheal.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L, Ding L, Zhou J, Chen S, Chen F, Zhao C, Xu J, Hu W, Ji J, Xu H, Liu GL (2021) One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens Bioelectron 171:112685 [DOI] [PMC free article] [PubMed]
- 31.Shaffaf T, Ghafar-Zadeh E (2021) COVID-19 diagnostic strategies part II: protein-based technologies. Bioengineering (Basel) 8(5) [DOI] [PMC free article] [PubMed]
- 32.Cady NC, Tokranova N, Minor A, Nikvand N, Strle K, Lee WT, Page W, Guignon E, Pilar A, Gibson GN (2021) Multiplexed detection and quantification of human antibody response to COVID-19 infection using a plasmon enhanced biosensor platform. Biosens Bioelectron 171:112679 [DOI] [PMC free article] [PubMed]
- 33.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, Zhou M, Chen L, Meng S, Hu Y, Peng C, Yuan M, Huang J, Wang Z, Yu J, Gao X, Wang D, Yu X, Li L, Zhang J, Wu X, Li B, Xu Y, Chen W, Peng Y, Hu Y, Lin L, Liu X, Huang S, Zhou Z, Zhang L, Wang Y, Zhang Z, Deng K, Xia Z, Gong Q, Zhang W, Zheng X, Liu Y, Yang H, Zhou D, Yu D, Hou J, Shi Z, Chen S, Chen Z, Zhang X, Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A. 2020;117(17):9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng H, Wang D, Nie J, Liang H, Gu J, Zhao A, Xu L, Lang C, Cui X, Guo X, Zhou C, Li H, Guo B, Zhang J, Wang Q, Fang L, Liu W, Huang Y, Mao W, Chen Y, Zou Q. The efficacy assessment of convalescent plasma therapy for COVID-19 patients: a multi-center case series. Signal Transduct Target Ther. 2020;5(1):219. doi: 10.1038/s41392-020-00329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanne JH (2020) COVID-19: FDA approves use of convalescent plasma to treat critically ill patients. BMJ 368:m1256 [DOI] [PubMed]
- 36.Ku Z, Xie X, Davidson E, Ye X, Su H, Menachery VD, Li Y, Yuan Z, Zhang X, Muruato AE, i Escuer AG, Tyrell B, Doolan K, Doranz BJ, Wrapp D, Bates PF, McLellan JS, Weiss SR, Zhang, N, Shi PY, An Z, Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nat Commun. 2021;12(1):469. doi: 10.1038/s41467-020-20789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang N, Shang J, Jiang S, Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol. 2020;11:298. doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Du L. SARS-CoV-2 spike protein: a key target for eliciting persistent neutralizing antibodies. Signal Transduct Target Ther. 2021;6(1):95. doi: 10.1038/s41392-021-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He WT, Limbo O, Smith C, Song G, Woehl J, Yang L, Abbott RK, Callaghan S, Garcia E, Hurtado J, Parren M, Peng L, Ramirez S, Ricketts J, Ricciardi MJ, Rawlings SA, Wu NC, Yuan M, Smith DM, Nemazee D, Teijaro JR, Voss JE, Wilson IA, Andrabi R, Briney B, Landais E, Sok D, Jardine JG, Burton DR. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369(6506):956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, Bentlage AEH, van Haaren MM, Guerra D, Burger JA, Schermer EE, Verheul KD, van der Velde N, van der Kooi A, van Schooten J, van Breemen MJ, Bijl TPL, Sliepen K, Aartse A, Derking R, Bontjer I, Kootstra NA, Wiersinga WJ, Vidarsson G, Haagmans BL, Ward AB, de Bree GJ, Sanders RW, van Gils MJ. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J, Lan J, Yuan J, Wang H, Zhao J, Zhang S, Wang Y, Shi X, Liu L, Zhao J, Wang X, Zhang Z, Zhang L. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584(7819):115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 42.Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, Gao G, Hu X, Zhang Y, Tong Z, Huang W, Liu WJ, Wu G, Zhang B, Wang L, Qi J, Feng H, Wang FS, Wang Q, Gao GF, YuanZ YJ. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 43.Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, Luo Y, Chan JF, Sahi V, Figueroa A, Guo XV, Cerutti G, Bimela J, Gorman J, Zhou T, Chen Z, Yuen KY, Kwong PD, Sodroski JG, Yin MT, Sheng Z, Huang Y, Shapiro L, Ho DD. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584(7821):450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J, Zhao J, Mangalam AK, Channappanavar R, Fett C, Meyerholz DK, Agnihothram S, Baric RS, David CS, Perlman S. Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44(6):1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alturaiki W, Mubarak A, Al Jurayyan A, Hemida MG (2021) The pivotal roles of the host immune response in the fine-tuning the infection and the development of the vaccines for SARS-CoV-2. Hum Vaccin Immunother 1–13 [DOI] [PMC free article] [PubMed]
- 46.Higuchi Y, Suzuki T, Arimori T, Ikemura N, Mihara E, Kirita Y, Ohgitani E, Mazda O, Motooka D, Nakamura S, Sakai Y, Itoh Y, Sugihara F, Matsuura Y, Matoba S, Okamoto T, Takagi J, Hoshino A. Engineered ACE2 receptor therapy overcomes mutational escape of SARS-CoV-2. Nat Commun. 2021;12(1):3802. doi: 10.1038/s41467-021-24013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lei C, Qian K, Li T, Zhang S, Fu W, Ding M, Hu S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat Commun. 2020;11(1):2070. doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kruse RL (2020) Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res 9:72 [DOI] [PMC free article] [PubMed]
- 49.Wu F, Wang A, Liu M, Wang Q, Chen J, Xia S, Ling Y, Zhang Y, Xun J, Lu L, Jiang S, Lu H, Wen Y, Huang J (2020) Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. MedRxiv
- 50.Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, Zhang Z, Fan P, Dong Y, Yang Y, Chen Z, Guo Y, Zhang J, Li Y, Song X, Chen Y, Xia L, Fu L, Hou L, Xu J, Yu C, Li J, Zhou Q, Chen W. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lip KM, Shen S, Yang X, Keng CT, Zhang A, Oh HL, Li ZH, Hwang LA, Chou CF, Fielding BC, Tan TH, Mayrhofer J, Falkner FG, Fu J, Lim SG, Hong W, Tan YJ. Monoclonal antibodies targeting the HR2 domain and the region immediately upstream of the HR2 of the S protein neutralize in vitro infection of severe acute respiratory syndrome coronavirus. J Virol. 2006;80(2):941–950. doi: 10.1128/JVI.80.2.941-950.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duan J, Yan X, Guo X, Cao W, Han W, Qi C, Feng J, Yang D, Gao G, Jin G. A human SARS-CoV neutralizing antibody against epitope on S2 protein. Biochem Biophys Res Commun. 2005;333(1):186–193. doi: 10.1016/j.bbrc.2005.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elshabrawy HA, Coughlin MM, Baker SC, Prabhakar BS (2012) Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PLoS One 7(11):e50366 [DOI] [PMC free article] [PubMed]
- 54.Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, Ying T, Liu S, Shi Z, Jiang S, Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17(7):765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia S, Yan L, Xu W, Agrawal AS, Algaissi A, Tseng CTK, Wang Q, Du L, Tan W, Wilson IA, Jiang S, Yang B, Lu L (2019) A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv 5(4):eaav4580-eaav4580 [DOI] [PMC free article] [PubMed]
- 56.Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, Qi F, Bao L, Du L, Liu S, Qin C, Sun F, Shi Z, Zhu Y, Jiang S, Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, Krüger N, Graichen L, Hofmann-Winkler H, Kempf A, Winkler MS, Schulz S, Jäck HM, Jahrsdörfer B, Schrezenmeier H, Müller M, Kleger A, Münch J, Pöhlmann S (2021) SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 184(9):2384–2393.e12 [DOI] [PMC free article] [PubMed]
- 58.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 59.Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, Agudelo M, Barnes CO, Gazumyan A, Finkin S, Hagglof T, Oliveira TY, Viant C, Hurley A, Hoffmann HH, Millard KG, Kost RG, Cipolla M, Gordon K, Bianchini F, Chen ST, Ramos V, Patel R, Dizon J, Shimeliovich I, Mendoza P, Hartweger H, Nogueira L, Pack M, Horowitz J, Schmidt F, Weisblum Y, Michailidis E, Ashbrook AW, Waltari E, Pak JE, Huey-Tubman KE, Koranda N, Hoffman PR, West AP Jr, Rice CM, Hatziioannou T, Bjorkman PJ, Bieniasz PD, Caskey M, Nussenzweig MC (2020) Convergent antibody responses to SARS-CoV-2 infection in convalescent individuals. bioRxiv
- 60.Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M, Wei Y, Atwal GS, Murphy AJ, Stahl N, Yancopoulos GD, Kyratsous CA. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506):1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao H, Sun Y, Deng YQ, Wang N, Tan Y, Zhang NN, Li XF, Kong C, Xu YP, Chen Q, Cao TS, Zhao H, Yan X, Cao L, Lv Z, Zhu D, Feng R, Wu N, Zhang W, Hu Y, Chen K, Zhang RR, Lv Q, Sun S, Zhou Y, Yan R, Yang G, Sun X, Liu C, Lu X, Cheng L, Qiu H, Huang XY, Weng T, Shi D, Jiang W, Shao J, Wang L, Zhang J, Jiang T, Lang G, Qin CF, Li L, Wang X. Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection. Cell Res. 2021;31(1):25–36. doi: 10.1038/s41422-020-00444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang N, Sun Y, Feng R, Wang Y, Guo Y, Zhang L, Deng YQ, Wang L, Cui Z, Cao L, Zhang YJ, Li W, Zhu FC, Qin CF, Wang X. Structure-based development of human antibody cocktails against SARS-CoV-2. Cell Res. 2021;31(1):101–103. doi: 10.1038/s41422-020-00446-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, Perry C, Pan C, Hosain R, Mahmood A, Davis JD, Turner KC, Hooper AT, Hamilton JD, Baum A, Kyratsous CA, Kim Y, Cook A, Kampman W, Kohli A, Sachdeva Y, Graber X, Kowal B, DiCioccio T, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD, Trial I. REGN-COV2, a neutralizing antibody cocktail, in outpatients with COVID-19. N Engl J Med. 2021;384(3):238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.EMA reviewing data on monoclonal antibody use for COVID-19. European medicines agency, https://www.ema.europa.eu/en/news/ema-reviewing-data-monoclonal-antibody-use-covid-19, updated 04/02/2021
- 65.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wan J, Xing S, Ding L, Wang Y, Gu C, Wu Y, Rong B, Li C, Wang S, Chen K, He C, Zhu D, Yuan S, Qiu C, Zhao C, Nie L, Gao Z, Jiao J, Zhang X, Wang X, Ying T, Wang H, Xie Y, Lu Y, Xu J, Lan F (2020) Human-IgG-neutralizing monoclonal antibodies block the SARS-CoV-2 infection. Cell Rep 32(3):107918 [DOI] [PMC free article] [PubMed]
- 67.Zhou D, Duyvesteyn HME, Chen CP, Huang CG, Chen TH, Shih SR, Lin YC, Cheng CY, Cheng SH, Huang YC, Lin TY, Ma C, Huo J, Carrique L, Malinauskas T, Ruza RR, Shah PNM, Tan TK, Rijal P, Donat RF, Godwin K, Buttigieg KR, Tree JA, Radecke J, Paterson NG, Supasa P, Mongkolsapaya J, Screaton GR, Carroll MW, Gilbert-Jaramillo J, Knight ML, James W, Owens RJ, Naismith JH, Townsend AR, Fry EE, Zhao Y, Ren J, Stuart DI, Huang KA. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat Struct Mol Biol. 2020;27(10):950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 68.Wec AZ, Wrapp D, Herbert AS, Maurer DP, Haslwanter D, Sakharkar M, Jangra RK, Dieterle ME, Lilov A, Huang D, Tse LV, Johnson NV, Hsieh CL, Wang N, Nett JH, Champney E, Burnina I, Brown M, Lin S, Sinclair M, Johnson C, Pudi S, Bortz R, 3rd, Wirchnianski AS, Laudermilch E, Florez C, Fels JM, O'Brien CM, Graham BS, Nemazee D, Burton DR, Baric RS, Voss JE, Chandran K, Dye JM, McLellan JS, Walker LM. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369(6504):731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D (2020) Structural and functional analysis of a potent sarbecovirus neutralizing antibody. bioRxiv
- 70.Tai W, Zhang X, He Y, Jiang S, Du L (2020) Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antiviral Res 179:104820 [DOI] [PMC free article] [PubMed]
- 71.Wang C, Li W, Drabek D, Okba NMA, van Haperen R, Osterhaus A, van Kuppeveld FJM, Haagmans BL, Grosveld F, Bosch BJ. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11(1):2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baby K, Maity S, Mehta CH, Suresh A, Nayak UY, Nayak Y (2021) SARS-CoV-2 entry inhibitors by dual targeting TMPRSS2 and ACE2: an in silico drug repurposing study. Eur J Pharmacol 896:173922 [DOI] [PMC free article] [PubMed]
- 74.Baughn LB, Sharma N, Elhaik E, Sekulic A, Bryce AH, Fonseca R. Targeting TMPRSS2 in SARS-CoV-2 infection. Mayo Clin Proc. 2020;95(9):1989–1999. doi: 10.1016/j.mayocp.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leng Z, Zhu R, Hou W, Feng Y, Yang Y, Han Q, Shan G, Meng F, Du D, Wang S, Fan J, Wang W, Deng L, Shi H, Li H, Hu Z, Zhang F, Gao J, Liu H, Li X, Zhao Y, Yin K, He X, Gao Z, Wang Y, Yang B, Jin R, Stambler I, Lim LW, Su H, Moskalev A, Cano A, Chakrabarti S, Min KJ, Ellison-Hughes G, Caruso C, Jin K, Zhao RC. Transplantation of ACE2(-) mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging Dis. 2020;11(2):216–228. doi: 10.14336/AD.2020.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lang J, Yang N, Deng J, Liu K, Yang P, Zhang G, Jiang C (2011) Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS One 6(8):e23710 [DOI] [PMC free article] [PubMed]
- 77.Clausen, Thomas Mandel, Daniel Sandoval , Charlotte Spliid B, Jessica Pihl, Hailee Perrett R, Chelsea Painter D, Anoop Narayanan (2020) "SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2." Cell 183, no. 4 1043-1057 [DOI] [PMC free article] [PubMed]
- 78.Conti P, Ronconi G, Caraffa A, Gallenga CE, Ross R, Frydas I, Kritas SK. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by coronavirus-19 (COVID-19 or SARS-CoV-2): anti-inflammatory strategies. J Biol Regul Homeost Agents. 2020;34(2):327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 79.Hu B, Huang S, Yin L. The cytokine storm and COVID-19. J Med Virol. 2021;93(1):250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ryabkova VA, Churilov LP, Shoenfeld Y. Influenza infection, SARS, MERS and COVID-19: cytokine storm - the common denominator and the lessons to be learned. Clin Immunol. 2021;223:108652–108652. doi: 10.1016/j.clim.2020.108652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Broman N, Rantasarkka K, Feuth T, Valtonen M, Waris M, Hohenthal U, Rintala E, Karlsson A, Marttila H, Peltola V, Vuorinen T, Oksi J. IL-6 and other biomarkers as predictors of severity in COVID-19. Ann Med. 2021;53(1):410–412. doi: 10.1080/07853890.2020.1840621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michot JM, Albiges L, Chaput N, Saada V, Pommeret F, Griscelli F, Balleyguier C, Besse B, Marabelle A, Netzer F, Merad M, Robert C, Barlesi F, Gachot B, Stoclin A. Tocilizumab, an anti-IL-6 receptor antibody, to treat COVID-19-related respiratory failure: a case report. Ann Oncol. 2020;31(7):961–964. doi: 10.1016/j.annonc.2020.03.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Investigators RC, Gordon AC, Mouncey PR, Al-Beidh F, Rowan KM, Nichol AD, Arabi YM, Annane D, Beane A, van Bentum-Puijk W, Berry LR, Bhimani Z, Bonten MJM, Bradbury CA, Brunkhorst FM, Buzgau A, Cheng AC, Detry MA, Duffy EJ, Estcourt LJ, Fitzgerald M, Goossens H, Haniffa R, Higgins AM, Hills TE, Horvat CM, Lamontagne F, Lawler PR, Leavis HL, Linstrum KM, Litton E, Lorenzi E, Marshall JC, Mayr FB, McAuley DF, McGlothlin A, McGuinness SP, McVerry BJ, Montgomery SK, Morpeth SC, Murthy S, Orr K, Parke RL, Parker JC, Patanwala AE, Pettila V, Rademaker E, Santos MS, Saunders CT, Seymour CW, Shankar-Hari M, Sligl WI, Turgeon AF, Turner AM, van de Veerdonk FL, Zarychanski R, Green C, Lewis RJ, Angus DC, McArthur CJ, Berry S, Webb SA, Derde LPG. Interleukin-6 receptor antagonists in critically ill patients with COVID-19. N Engl J Med. 2021;384(16):1491–1502. doi: 10.1056/NEJMoa2100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Janik E, Niemcewicz M, Podogrocki M, Saluk-Bijak J, Bijak M (2021) Existing drugs considered as promising in COVID-19 therapy. Int J Mol Sci 22(11) [DOI] [PMC free article] [PubMed]
- 85.Tjan LH, Nagano T, Furukawa K, Nishimura M, Arii J, Fujinaka S, Iwata S, Sano S, Tohma Y, Nishimura Y, Mori Y. The neutralizing antibody response against severe acute respiratory syndrome coronavirus 2 and the cytokine/chemokine release in patients with different levels of coronavirus diseases 2019 severity: cytokine storm still persists despite viral disappearance in critical patients. JMA J. 2021;4(1):1–7. doi: 10.31662/jmaj.2020-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tabll AA, Shahein YE, Omran MM, Elnakib MM, Ragheb AA, Amer KE (2021) A review on monoclonal antibodies in COVID-19: role in immunotherapy, vaccine development and viral detection. Hum Antibodies [DOI] [PubMed]
- 87.Di Salvo E, Di Gioacchino M, Tonacci A, Casciaro M, Gangemi S. Alarmins, COVID-19 and comorbidities. Ann Med. 2021;53(1):777–785. doi: 10.1080/07853890.2021.1921252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alfarouk KO, Alhoufie STS, Hifny A, Schwartz L, Alqahtani AS, Ahmed SBM, Alqahtani AM, Alqahtani SS, Muddathir AK, Ali H, Bashir AHH, Ibrahim ME, Greco MR, Cardone RA, Harguindey S, Reshkin SJ. Of mitochondrion and COVID-19. J Enzyme Inhib Med Chem. 2021;36(1):1258–1267. doi: 10.1080/14756366.2021.1937144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karagianni P, Tzioufas AG (2019) Epigenetic perspectives on systemic autoimmune disease. J Autoimmun 104:102315 [DOI] [PubMed]
- 90.Xiao ZX, Miller JS, Zheng SG (2021) An updated advance of autoantibodies in autoimmune diseases. Autoimmun Rev 20(2):102743 [DOI] [PubMed]
- 91.Zuo Y, Estes SK, Ali RA, Gandhi AA, Yalavarthi S, Shi H, Sule G, Gockman K, Madison JA, Zuo M, Yadav V, Wang J, Woodard W, Lezak SP, Lugogo NL, Smith SA, Morrissey JH, Kanthi Y, Knight JS (2020) Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci Transl Med 12(570) [DOI] [PMC free article] [PubMed]
- 92.Ayoubkhani D, Khunti K, Nafilyan V, Maddox T, Humberstone B, Diamond I, Banerjee A (2021) Post-COVID syndrome in individuals admitted to hospital with COVID-19: retrospective cohort study. BMJ 372:n693 [DOI] [PMC free article] [PubMed]
- 93.Yong SJ. Long COVID or post-COVID-19 syndrome: putative pathophysiology, risk factors, and treatments. Infect Dis (Lond) 2021;53(10):737–754. doi: 10.1080/23744235.2021.1924397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ackermann M, Anders HJ, Bilyy R, Bowlin GL, Daniel C, De Lorenzo R, Egeblad M, Henneck T, Hidalgo A, Hoffmann M, Hohberger B, Kanthi Y, Kaplan MJ, Knight JS, Knopf J, Kolaczkowska E, Kubes P, Leppkes M, Mahajan A, Manfredi AA, Maueröder C, Maugeri N, Mitroulis I, Muñoz LE, Narasaraju T, Naschberger E, Neeli I, Ng LG, Radic MZ, Ritis K, Rovere-Querini P, Schapher M, Schauer C, Simon HU, Singh J, Skendros P, Stark K, Stürzl M, van der Vlag J, Vandenabeele P, Vitkov L, von Köckritz-Blickwede M, Yanginlar C, Yousefi S, Zarbock A, Schett G, Herrmann M (2021) Patients with COVID-19: in the dark-NETs of neutrophils. Cell Death Differ [DOI] [PMC free article] [PubMed]
- 95.Zuo Y, Yalavarthi S, Navaz SA, Hoy CK, Harbaugh A, Gockman K, Zuo M, Madison JA, Shi H, Kanthi Y, Knight JS (2021) Autoantibodies stabilize neutrophil extracellular traps in COVID-19. JCI Insight [DOI] [PMC free article] [PubMed]
- 96.Sacchi MC, Tamiazzo S, Stobbione P, Agatea L, De Gaspari P, Stecca A, Lauritano EC, Roveta A, Tozzoli R, Guaschino R, Bonometti R. SARS-CoV-2 infection as a trigger of autoimmune response. Clin Transl Sci. 2021;14(3):898–907. doi: 10.1111/cts.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou Y, Han T, Chen J, Hou C, Hua L, He S, Guo Y, Zhang S, Wang Y, Yuan J, Zhao C, Zhang J, Jia Q, Zuo X, Li J, Wang L, Cao Q, Jia E. Clinical and autoimmune characteristics of severe and critical cases of COVID-19. Clin Transl Sci. 2020;13(6):1077–1086. doi: 10.1111/cts.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu Y, Sawalha AH, Lu Q. COVID-19 and autoimmune diseases. Curr Opin Rheumatol. 2021;33(2):155–162. doi: 10.1097/BOR.0000000000000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vojdani A, Kharrazian D (2020) Potential antigenic cross-reactivity between SARS-CoV-2 and human tissue with a possible link to an increase in autoimmune diseases. Clin Immunol 217:108480 [DOI] [PMC free article] [PubMed]
- 100.Mantefardo B, Gube AA, Awlachew E, Sisay G. Novel coronavirus (COVID-19)-associated Guillain-Barre' syndrome: case report. Int Med Case Rep J. 2021;14:251–253. doi: 10.2147/IMCRJ.S305693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Franke C, Ferse C, Kreye J, Reincke SM, Sanchez-Sendin E, Rocco A, Steinbrenner M, Angermair S, Treskatsch S, Zickler D, Eckardt KU, Dersch R, Hosp J, Audebert HJ, Endres M, Ploner JC, Pruss H. High frequency of cerebrospinal fluid autoantibodies in COVID-19 patients with neurological symptoms. Brain Behav Immun. 2021;93:415–419. doi: 10.1016/j.bbi.2020.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Koczula KM, Gallotta A. Lateral flow assays. Essays Biochem. 2016;60(1):111–120. doi: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cinquanta L, Fontana DE, Bizzaro N. Chemiluminescent immunoassay technology: what does it change in autoantibody detection? Auto Immun Highlights. 2017;8(1):9. doi: 10.1007/s13317-017-0097-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Matabaro E, Ishimwe N, Uwimbabazi E, Lee BH. Current immunoassay methods for the rapid detection of aflatoxin in milk and dairy products. Compr Rev Food Sci Food Saf. 2017;16(5):808–820. doi: 10.1111/1541-4337.12287. [DOI] [PubMed] [Google Scholar]
- 105.Wu Y, Wang F, Shen C, Peng W, Li D, Zhao C, Li Z, Li S, Bi Y, Yang Y, Gong Y, Xiao H, Fan Z, Tan S, Wu G, Tan W, Lu X, Fan C, Wang Q, Liu Y, Zhang C, Qi J, Gao GF, Gao F, Liu L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368(6496):1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, Zhu Q, Zhang X, Zheng Y, Geng C, Chai X, He R, Li X, Lv Q, Zhu H, Deng W, Xu Y, Wang Y, Qiao L, Tan Y, Song L, Wang G, Du X, Gao N, Liu J, Xiao J, Su XD, Du Z, Feng Y, Qin C, Qin C, Jin R, Xie XS (2020) Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell 182(1):73–84e16 [DOI] [PMC free article] [PubMed]
- 107.Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, Fulton BO, Yan Y, Koon K, Patel K, Chung KM, Hermann A, Ullman E, Cruz J, Rafique A, Huang T, Fairhurst J, Libertiny C, Malbec M, Lee WY, Welsh R, Farr G, Pennington S, Deshpande D, Cheng J, Watty A, Bouffard P, Babb R, Levenkova N, Chen C, Zhang B, Romero Hernandez A, Saotome K, Zhou Y, Franklin M, Sivapalasingam S, Lye DC, Weston S, Logue J, Haupt R, Frieman M, Chen G, Olson W, Murphy AJ, Stahl N, Yancopoulos GD, Kyratsous CA. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369(6506):1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Seydoux E, Homad LJ, MacCamy AJ, Parks KR, Hurlburt NK, Jennewein MF, Akins NR, Stuart AB, Wan YH, Feng J, Whaley RE, Singh S, Boeckh M, Cohen KW, McElrath MJ, Englund JA, Chu HY, Pancera M, McGuire AT, Stamatatos L (2020) Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity 53(1):98–105e5 [DOI] [PMC free article] [PubMed]
- 109.Wu Y, Li C, Xia S, Tian X, Kong Y, Wang Z, Gu C, Zhang R, Tu C, Xie Y, Yang Z, Lu L, Jiang S, Ying T (2020) Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe 27(6):891–898e5 [DOI] [PMC free article] [PubMed]
- 110.Sun Z, Chen C, Li W, Martinez DR, Drelich A, Baek DS, Liu X, Mellors JW, Tseng CT, Baric RS, Dimitrov DS. Potent neutralization of SARS-CoV-2 by human antibody heavy-chain variable domains isolated from a large library with a new stable scaffold. MAbs. 2020;12(1):1778435. doi: 10.1080/19420862.2020.1778435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR, Li Q, Zhang L, Zhu Y, Si HR, Wang Q, Min J, Wang X, Zhang W, Li B, Zhang HJ, Baric RS, Zhou P, Yang XL, Shi ZL (2020) Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182(1):50–58 e8 [DOI] [PMC free article] [PubMed]
- 112.Sun SH, Chen Q, Gu HJ, Yang G, Wang YX, Huang XY, Liu SS, Zhang NN, Li XF, Xiong R, Guo Y, Deng YQ, Huang WJ, Liu Q, Liu QM, Shen YL, Zhou Y, Yang X, Zhao TY, Fan CF, Zhou YS, Qin CF, Wang YC (2020) A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28(1):124–133 e4 [DOI] [PMC free article] [PubMed]
- 113.Sia SF, Yan LM, Chin AWH, Fung K, Choy KT, Wong AYL, Kaewpreedee P, Perera R, Poon LLM, Nicholls JM, Peiris M, Yen HL. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583(7818):834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lv Z, Deng YQ, Ye Q, Cao L, Sun CY, Fan C, Huang W, Sun S, Sun Y, Zhu L, Chen Q, Wang N, Nie J, Cui Z, Zhu D, Shaw N, Li XF, Li Q, Xie L, Wang Y, Rao Z, Qin CF, Wang X. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369(6510):1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tychan Pte Ltd. Safety of TY027, a Treatment for COVID-19, in Humans, https://clinicaltrials.gov/ct2/show/NCT04429529, updated April 8, 2021
- 116.Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V, Shawa I, Adams AC, Van Naarden J, Custer KL, Shen L, Durante M, Oakley G, Schade AE, Sabo J, Patel DR, Klekotka P, Skovronsky DM, Investigators B. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with COVID-19. N Engl J Med. 2021;384(3):229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.SAb Biotherapeutics, Inc. Safety, Tolerability, and Pharmacokinetics of SAB-185 in Ambulatory Participants With COVID-19, https://clinicaltrials.gov/ct2/show/NCT04469179, updated October 26, 2021
- 118.Study to Evaluate STI-1499 (COVI-GUARD) in Patients With Moderate COVID19. https://clinicaltrials.gov/ct2/show/NCT04454398, updated January 8, 2021
- 119.Haschke M, Schuster M, Poglitsch M, Loibner H, Salzberg M, Bruggisser M, Penninger J, Krahenbuhl S. Pharmacokinetics and pharmacodynamics of recombinant human angiotensin-converting enzyme 2 in healthy human subjects. Clin Pharmacokinet. 2013;52(9):783–792. doi: 10.1007/s40262-013-0072-7. [DOI] [PubMed] [Google Scholar]
- 120.Gaborit B, Vanhove B, Vibet MA, Le Thuaut A, Lacombe K, Dubee V, Ader F, Ferre V, Vicaut E, Orain J, Le Bras M, Omnes A, Berly L, Jobert A, Morineau-Le Houssine P, Botturi K, Josien R, Flet L, Degauque N, Brouard S, Duvaux O, Poinas A, Raffi F, Group PS Evaluation of the safety and efficacy of XAV-19 in patients with COVID-19-induced moderate pneumonia: study protocol for a randomized, double-blinded, placebo-controlled phase 2 (2a and 2b) trial. Trials. 2021;22(1):199. doi: 10.1186/s13063-021-05132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.A Study of Human Monoclonal Antibodies, BRII-196 and BRII-198, https://clinicaltrials.gov/ct2/show/NCT04787211, updated August 30, 2021
- 122.Safety, Tolerability and Pharmacokinetics of SCTA01, an Anti-SARS-CoV-2 Monoclonal Antibody, in Healthy Chinese Subjects, https://clinicaltrials.gov/ct2/show/NCT04483375, updated March 23, 2021