Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has had severe consequences for health and the global economy. To control the transmission, there is an urgent demand for early diagnosis and treatment in the general population. In the present study, an automatic system for SARS-CoV-2 diagnosis is designed and built to deliver high specification, high sensitivity, and high throughput with minimal workforce involvement. The system, set up with cross-priming amplification (CPA) rather than conventional reverse transcription-polymerase chain reaction (RT-PCR), was evaluated using more than 1000 real-world samples for direct comparison. This fully automated robotic system performed SARS‐CoV‐2 nucleic acid-based diagnosis with 192 samples in under 180 min at 100 copies per reaction in a “specimen in data out” manner. This throughput translates to a daily screening capacity of 800–1000 in an assembly-line manner with limited workforce involvement. The sensitivity of this device could be further improved using a CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-based assay, which opens the door to mixed samples, potentially include SARS-CoV-2 variants screening in extensively scaled testing for fighting COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00449-021-02674-9.
Keywords: SARS-CoV-2 diagnosis, Robotic system, High throughput
Introduction
The COVID-19 pandemic, caused by the novel respiratory coronavirus 2 (SARS-CoV-2), has swept the world. Recently, statistical analysis of the number of daily cases in the European Union suggest an oncoming second wave of coronavirus [1, 2]. As of 20th July 2021, the virus had spread to 213 countries, causing more than 184 million confirmed infections and at least 4 million deaths (World Health Organization). The recent confirmed infections and deaths were dominated by the Delta variant of SARS-CoV-2 virus (B.1.427). This highly pathogenic mutant demonstrated a high transmission rate, a much shorter incubation period and a high death rate, which spreads in a sudden manner word wide. Without rigorous measures to test every possible case, the current drastic restrictions have limited ability in suppression over the transmission of the virus. While many measures to mitigate the multifactorial impact of COVID‐19 have been implemented, “diagnosis early and treat early” are essential for providing meaningful epidemiological data and for saving lives.
Epidemiological analysis can provide a strategy for social restriction policies; the identification of individuals currently or previously infected by SARS-CoV-2 is a critical component. Clinically, chest computed tomography (CT) imaging can be used to diagnose COVID-19 [3]. For large-scale screening, there are two standard logics for SARS‐CoV‐2 diagnostic modalities: testing for the existence of viral particles for current infection status, or using serum antibody tests to indicate the response to infection [4, 5]. Recently, there have been reports of people having a different strain of the coronavirus infection than the one that existed several months ago, despite the vaccinated population [6]. Unlike measles or smallpox vaccines, inactivated SARS-CoV-2 viral vaccines may not provide lifelong immunity [7, 8]. Hence, in this case, serology screening is indirect and cannot identify individuals who may transmit the virus. Laboratory-based diagnosis of active SARS‐CoV‐2 infection relies on the direct detection of virus‐specific nucleic acid regions (N, ORF1a gene, or E gene genes), most commonly obtained from the nasopharynx using throat swabs or sputum specimens [9, 10]. Robust detection technology is usually based on real-time reverse transcription polymerase chain reaction (RT-PCR). However, this assay is considered to have high specificity; its sensitivity is reportedly only 60–70% consistent with clinical status compared with CT examination [11–13]. Even when the initial RT-PCR test was negative, repeated RT-PCR tests have increased the cumulative positive rate of COVID-19, and in-hospital repeat runs of the test are recommended [13, 14]. The bottleneck of the sensitivity in nucleic acid tests is due to the preparation and purification of the low-load DNA/RNA from the initial clinical specimen and the insensitivity inherent in the detection method [15]. It is related to laboratory training level and experience in handling potentially highly infectious samples.
There is a significant demand for SARS-CoV-2 testing power to rapidly screen large numbers of individuals with high sensitivity to identify low infection loads at the asymptomatic stage. Setting up an RT-PCR test requires infrastructure, skilled personnel, and at least 4 h for the assay and analysis. It may take 12 h or even days to obtain the test report. The point-of-care test (POCT) system used in the US provides a test solution that takes less than 45 min, but the sensitivity has been reported to be as low as 5000 copies/ml, one sample per run involves manual loading, and the efficiency is not suitable for large-scale screening [16, 17]. Apart from POCT, other research teams have developed electrochemical sensing and nanoplasmonic sensing, which leverage electrical, optical, and electrochemical transducer systems. Indeed, there is a trade-off between high throughput and high sensitivity [18]. The growing realisation is that technologies that are ideal in population-wide screening for highly infectious samples require (i) minimal human intervention, (ii) minimal infrastructural setup, (iii) robust automation, (iv) high throughput, (v) high sensitivity/specificity, and (vi) reliable and automated data processing.
Despite the lag relative to manufacturing and service industries, the use of automation has gradually been implemented across the pharmaceutical, medical device, and biotechnology industries, from drug development to cell culturing processes [19]. The training level and labour costs in diagnostic laboratories are extremely high. The adoption of robotics and cognitive automation (R&CA) in a laboratory is not limited to enhancing productivity and reducing costs, but also increases reproducibility and accuracy. The biggest hurdle to industrialising the laboratory process is that the lab process was initially not designed for automation. R&CA for the laboratory cannot be limited to following entirely manual processes, such as linking individual laboratory devices using an automated guided vehicle (VGA) [20]. Transforming large-scale SARS‐CoV‐2 nucleic acid diagnosis screening into an R&CA assembly line requires the redesign and development of hardware to convert complicated molecular biological processes to a method suitable for R&CA.
To address the requirements for testing power in SARS-CoV-2 diagnoses, we have been interested in devising automated, rapid, and high-throughput diagnostic and profiling technologies. Here, we report the development and validation of a “specimens in data out” fully automated robotic system that can manage 192 samples in a batch and data reporting in less than 180 min (< 60 s/sample), with 50–100 copies/reaction viral titre sensitivity, which provides > 1000 specimens of testing power involving 2 people in two 8-hour shifts. We applied a system that implemented cross-priming amplification (CPA) for viral nucleic acid detection to a validation cohort of clinical specimens, which represented > 1000 samples. The system performance was directly compared with the standard RT-PCR approach under CDC-approved clinical conditions [21, 22].
Method
System overview
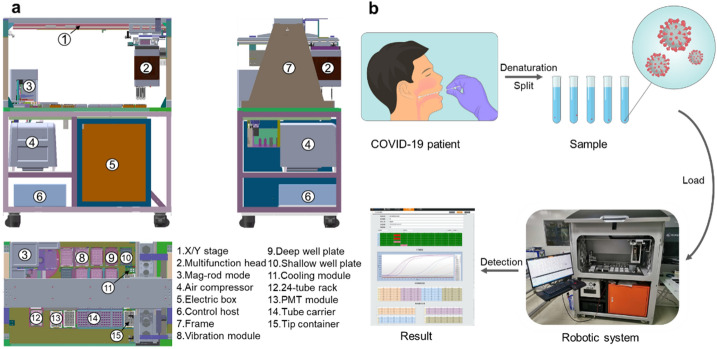
We described the construction of a modularized that is tailored to an automated experimental procedure using robotic instruments developed in-house (Fig. 1a). Swabs or sputum are placed into a lysis solution vial and heat-inactivated; later, they can be stored or transported before being loaded into the robotic system. The prepacked buffer and consumables are loaded into the system prior to the initiation of the automation process. The system contains 8-channel pipetting with a capacity of 8 × 24 specimens, 2-ml tube racks, and 96 formats of magnetic rod-based nucleic acid purification processors. The refrigeration module is prepared for the resuspended solution of CPA reaction mix with purified nucleic acid samples, and the CPA reaction is quantified using two fluorescent channels. The assay plate is transferred between individual functional units using a gripper located in the multifunctional head driven by a linear motor with < 50 µm accuracy at the X/Y-axis, and < 200 µm accuracy at the Z-axis. The entire system is commanded using an in-house developed controlling board that uses multi-core processors and FPGA communication to synchronise the 25 axes.
Fig. 1.
Automated robotic system for SARS-COV-2 diagnosis. a The mechanical structure and layout of the system shown from different orientations (front, rear, and top). The key functional modules of this system, including mag-rod module, multifunctional head with robot arm and single-photon counting-based PMT detector, are labelled with numbers. b A brief workflow of SARS-COV-2 detection from specimen preparation to nucleic acid extraction with enrichment, followed by fluorescent-based detection. A final report is generated after the raw data is generated from the PMT detection
System mechanical design
An isometric view of the computer-assisted draw is illustrated in Fig. 1. This in-house robotic system was designed with two compartments, including the working stack and the controlling section (Fig. 1a). The entire workflow was operated via a linear motor (TL09/12N + magnetplate) constructed series robotic arm (Fig. 1a-1) with an optical encoder (RELAIN30U1A), which facilitated the movement of the multifunction head (Fig. 1a-2) as the coordination of the assay plates process during the entire automation workflow. Samples from a bio-safety P2 lab were loaded into racks at 24 tubes per rack (Fig. 1a-14), then transferred into deep well plates (Fig. 1a-9) using disposable pipette from a tip container (Fig. 1a-15). They were further processed for lysis via a vibration module (Fig. 1a-8). The lysate was then prepared using a magnetic-rod module coupled to a DC motor (Fig. 1a-3), and the sample was loaded into a shallow plate. Quantification was performed using PMT (photomultiplier tubes) as the detection sensor. Other modules included an air compressor (Fig. 1a-4) that supported liquid pump movement. The air compressor (Fig. 1a-4), was located with an electric box (Fig. 1a-5), and the motor control host (Fig. 1a-6) was on the lower deck of the system. The entire sample loading and automation processing was divided into six steps, as illustrated using video recording (Supplement Video I–VI).
Electronic instrumentation
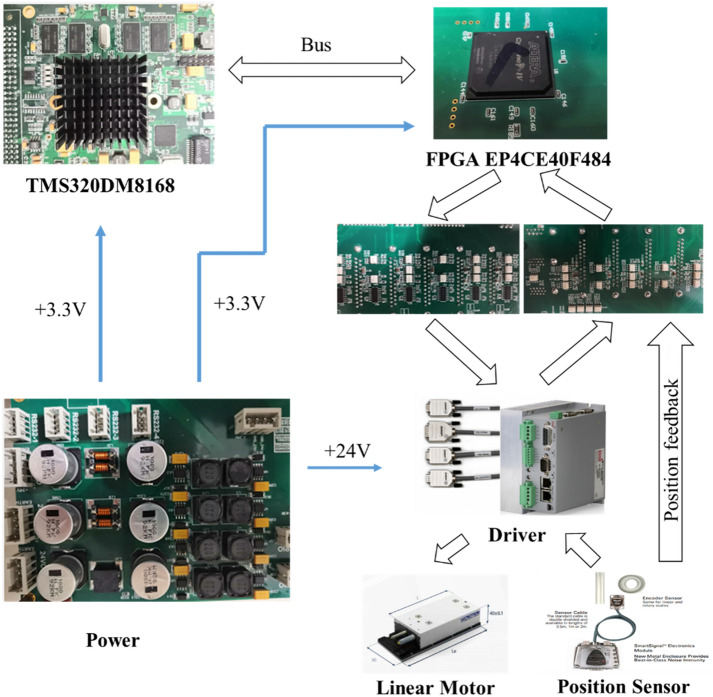
The instrumentation of the electrical and electronic elements that the device implemented needed to be highly synchronised for multi-module coordination. Therefore, the controlling subsystem was divided into eight modules. A general scheme shows the interaction between each module in Fig. 2. In detail, the multi-axis synchronous motion control system implements a high-speed and multi-coordinate system as the central control platform, which contains TMS320DM8168 as the main control chip, integrating ARM (1 GHz Cortex A8 RISC CPU) and floating-point DSP (Ti C674x VLIW 800 MHz). The chip with GPMC extends the FPGA to control 23 axes distributed over five CNC machine coordinate systems, synchronised motion, multiple external interruptions to trigger flashlight and camera shutter, and multiple input and output general control. The hardware implements an ARM + DSP + FPGA integrated multi-core integrated for high-speed control. The software core algorithm achieves 23-axis synchronous interpolation control and various complex computations within the 0.5 ms main loop. Computationally intensive tasks, especially in image preprocessing, are GPU-accelerated mainly using the open-source library OpenCV. This system uses a workstation with 192 GB of random-access memory (RAM) to buffer and efficiently process vast amounts of incoming imaging data.
Fig. 2.
Hardware of the automated robotic system
The control system includes the master computer and the slave embedded control board, which realises communication through an Ethernet bus; the embedded control system is used for data processing, and the processing results are uploaded to the master computer. The embedded control board includes a central control processing module, an FPGA module, and a peripheral control module; the peripheral control module includes a multi-axis linear motor control output module, multi-axis output for server motor control, a magnetic rod control module, a pipette liquid control module, a vibration heating module, and a PMT module.
The multi-axis output module includes multiple linear motors, drives, and grating scales, which are used to control the running speed, direction, and positioning of the motors. In addition, the state feedback module is implemented to feedback the running status of each motor. The grating ruler module is used to feedback each motor position. Finally, the universal input module is executed to read other signals, such as restricted travel positions, and the output module performs control outputs, such as DC motor, cylinders, and liquid pumps.
Controlling system movement
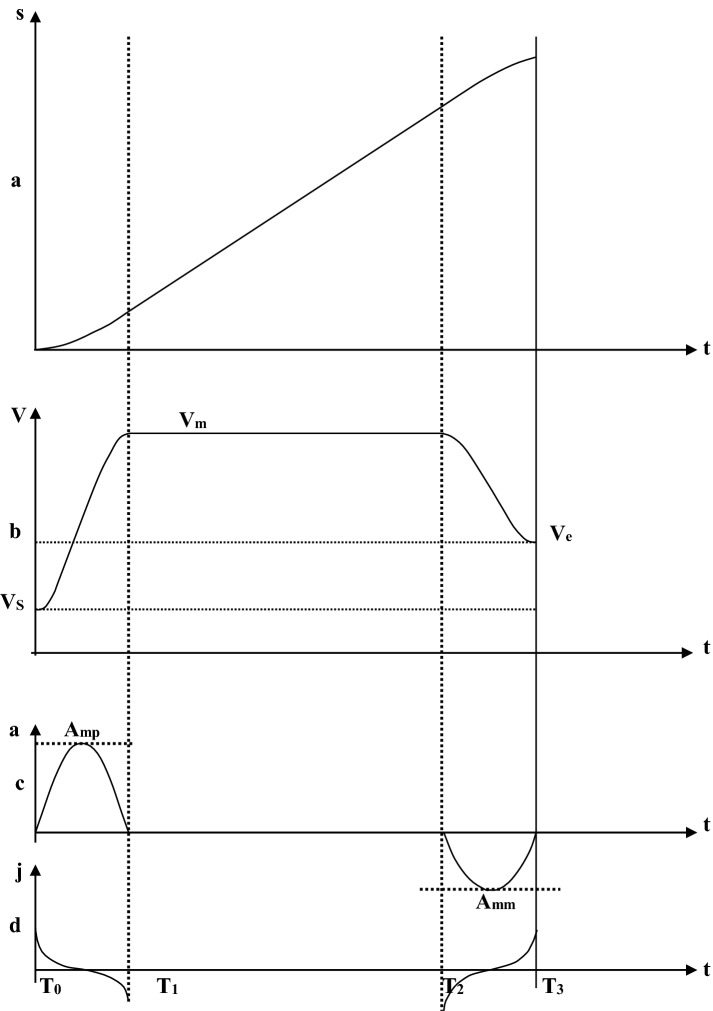
The entire system is regulated via a lower embedded control board using a high-performance dual-core processor via the main control chip. The ARM core of the dual-core processor, the main control processor, is used to communicate with the master system with control codes. In addition, the DSP core performs multi-axis motion control trajectory planning, motion control algorithm processing, and interpolation calculation with functional module control. The ARM core and DSP core of the dual-core processor exchange data through shared memory; the dual-core processor is connected to the FPGA processor through a 32-bit parallel bus for data exchange. The FPGA triggers the dual-core processor to perform interpolation output through interrupts; the FPGA receives the pulse value and direction value of each axis transmitted from the interpolation buffer, performs the synchronous interpolation of each axis, and realises the robot space movement. Movement control is accomplished according to the acceleration curve of the fifth-order polynomial acceleration, and the deceleration algorithm is a quadratic curve, which is a parabola with an opening. Figure 2 shows that the opening faces downward during the acceleration phase, and the opening faces upward during the deceleration phase.
The motion control curve is divided into three sections: the acceleration segment , the constant velocity section , and the deceleration section . is the maximum positive acceleration and is the maximum negative acceleration; is the starting speed; is the end speed, and it is a constant speed.
The jerk calculation formula is:
| 1 |
The acceleration calculation formula is:
| 2 |
When , there is maximum acceleration, and when , the acceleration is 0.
The speed calculation formula is:
| 3 |
When t = 0, the initial speed is ; when t = , the speed is ; when , the speed is ; when , the speed is .
Displacement calculation formula:
| 4 |
When t = 0, the initial displacement is 0; when , the displacement is ; when , the displacement is ; when , the displacement is .
Substituting into (3)
| 5 |
| 6 |
Similarly, substituting into Eq. (3) yields:
| 7 |
When the maximum negative acceleration is obtained, the time required from deceleration to T3–T2 (deceleration period time) is,
| 8 |
When , we enter formula Eq. (4) and obtain the displacement for the acceleration section:
| 9 |
| 10 |
Setting the total displacement as , the displacement of the constant speed section can be obtained as follows:
| 11 |
The time of the constant speed section is:
| 12 |
| 13 |
Hence, the FPGA controls the robot motor movement through the fifth-order polynomial S-curve, which can effectively reduce the vibration of frequent switching of machine speed and back and forth reversing, improving positioning accuracy, increasing operating speed, and shortening the run time to ensure the rapid completion of the experiment.
Samples preparations
Sample pretreatment
In most situations, the samples had all been inactivated by incubation at 56 °C for 30 min when they were received from the hospital. Then, 200 μl liquid was moved from a total 3–5 ml volume to sample tubes that contained 225 μl lysis buffer. Four 24-well plastic carriers were used to arrange all sample tubes in a certain order. All the processes were carried out in the BSL-2 laboratory and biosafety cabinet.
Nucleoid acid preparation
Both human (DNA) and SARS-CoV-2 (RNA) nucleoid acids were prepared using the TraceMag-Virus RNA/DNA Extraction Kit (KOALSON), which was adapted to the SARS-CoV-2 Integrated Nucleic Acid Preparation and Detection System. This kit consists of lysis buffer (225 μl for each sample, shown in the same way for the following components), binding buffer (405 μl), wash buffer I (500 μl), wash buffer II (500 μl), elution buffer (50 μl), magnetic beads (15 μl), and proteinase K (20 μl). All the reagents and relative consumables are prepackaged, so that the operator needs to remove the seal and put them in the right position according to the guidelines before the machine starts up.
Cross priming amplification (CPA) reaction
One-step: The amplification step was performed using the Diagnostic Kit for SARS-CoV-2 RNA (Isothermal Amplification Real-Time Fluorescence Assay) (UStar). This kit consisted of CPA buffer, CPA enzyme mix, positive control, and negative control. The CPA reaction system consisted of 28 μl CPA buffer, 2 μl CPA enzyme mix, and 20 μl template. These were mixed gently, and then sealed with 30 μl paraffin oil and incubated at 58 °C for 60 min. The detection channel used was FAM for the ORF and N genes of SARS-CoV-2.
Two-step: To improve the detection sensitivity, CRISPR technology was combined with the CPA reaction to become a two-step diagnostic kit for SARS-CoV-2 RNA. Both the CRISPR enzyme mix, and the CPA enzyme mix were made into glassy solids in different PCR tubes or 96-well plates. In step one, 30 μl buffer A was added to the CPA enzyme mix and stored at 4 ℃, and then incubated at 58 ℃ for 60 min after 20 μl template and 30 μl paraffin oil were added. In step two, 20 µl buffer B was added to the CRISPR enzyme mix and stored at 4 ℃ before it was added to the step one product and incubated at 37 ℃ for 30 min. The detection channels were FAM for the ORF gene of SARS-CoV-2, and CY5 for a human housekeeping gene as an internal control.
PMT detection
As the system designed, the fluorescent signal was collected using FAM: ET485/20 × ; ET535/30 m. CY5: ET640/30 × ; ET700/50 m. The single-point scanning time for each sample was 100 ms. Two channel signals were collected per minute.
RT-PCR
The control RT-PCR kit is a diagnostic kit for SARS-CoV-2 nucleic acid (real-time fluorescence assay) (BioPerfectus Technologies). The total 20 μl reaction system included: 7.5 μl reaction buffer; 5 μl enzyme mix; 4 μl SARS-CoV-2 reaction buffer; 3.5 μl RNase-free water. The PCR program consisted of 50 ℃, 30 min, 1 cycle; 95 ℃, 30 min, 1 cycle; 95 ℃, 10 s/55 ℃, 40 s, 45 cycles. The detection channels were FAM for the ORF gene, VIC for the N gene of SARS-CoV-2, and CY5 for a human housekeeping gene as an internal control. Positive result: as indicated for both FAM and VIC channel Ct ≤ 37 with an S-type curve. Negative result: as indicated for both FAM and VIC channel Ct > 40 or not detected. Suspected: as indicated FAM or VIC channel Ct ≤ 37, one another 37 < Ct ≤ 40, or both channels 37 < Ct ≤ 40.
Algorithm for distinguishing between positive and negative samples
To distinguish between positive and negative samples, one algorithm should be used to judge samples as positive or negative. Using weighted mean gradients (WMGs) is an effective tool for data analysis and has many merits, such as simplicity and noise reduction, WMGs were used for our judgments.
For the control positive and negative samples, WMGs that reduce noise can be calculated as:
| 14 |
where and are the magnitude and weight, respectively.
For control positive and negative samples, the average and variance of the WMGs can be described as follows:
| 15 |
| 16 |
Assuming an intermediate variable defined by:
For the samples, the positive and negative criteria are as follows:
| 17 |
where the subscripts P, N, and S indicate control, positive, and negative samples, respectively, and C is the constant coefficient. Each of our samples has 35 points, and there are 4 each of control, positive, and negative samples, respectively. In our algorithm, takes 0.90–0.95 (Fig. 3).
Fig. 3.
Control curves of a distance, b velocity, c acceleration, and d jerk
Results
System workflow and essential elements
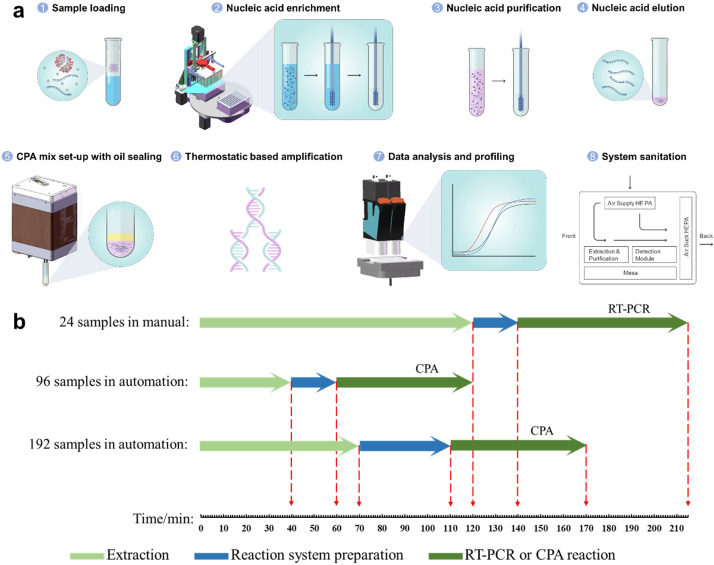
Having capacity for SARS‐CoV‐2 molecular detection with high sensitivity is an essential element in fighting multiple waves of pandemic globally. A centralised testing strategy has been implemented in many countries to enhance diagnostic throughput and quality assurance. This approach may be optimal for a direct connection between specimens and end-user scientists who perform large-scale batch assays. The system workflow is initiated after the inactivated samples are loaded into the system (Fig. 4a). Prior to the operation, all consumables and reagents are deposited into a cabinet with established airflow. First, after inactivation in the BSL-2 condition, up to 400 µl mix (specimen with lysis buffer) is transferred robotically into a binding solution, which contains magnetic beads for nucleic acid capture, and the total volume cannot exceed 1000 µl. Subsequently, in the combination linear motion module and gripper, two loading positions holding 96 deep well plates are rotated below the 96 magnetic rod matrix, which performs a 2-phase up-down movement at the Z-axis. The plastic cover around the magnetic rod can be ejected and disposed at the end of each run. The plastic rod cover works in tandem with the magnetic rod movement along the Z-axis, and magnetic beads can create associated/disassociated synchronised with the on/off association of the rod cover. This motion is not adopted from the manual procedure, and control of the speed, frequency, and workflow were optimised to harvest the maximum yield of nucleic acid in parallel with 96 throughput manner. The nucleic acid binding procedure takes 10 min, and three cycles of the magnetic rod module process take approximately 30 min. Hence, two batches of 96 samples (192 in total) can be processed in a coordinated manner to reduce the off-machine time. The purified nucleic acid product was dissolved in 30–50 µl elution buffer. Half of the product is prepared with CPA reaction for viral detection, and the other half is stored for reference. Some of the products are cross-examined using RT-PCR.
Fig. 4.
a Process of SARS-COV-2 detection. The samples were aliquoted and loaded into the system after an inactivation step which was performed at 56 ℃ for at least 30 min. Then, both SARS-COV-2 RNA and human DNA (overall nucleic acid) were captured by magnetic beads and were washed three times with different wash buffers. A heat-dry step was needed to evaporate the redundant ethanol in wash buffer III. The nucleic acid template gained from the heating elution step was mixed with the CPA reaction buffer. The reaction system was finally sealed with paraffin oil. The targets were then amplified using the thermostatic module at 58 ℃, and the raw data were processed by the system automatically. Finally, the equipment was cleaned by HEPA and UV lamp. b Comparison of time consumption of the manual and automatic system. The elapsed time for each sample of both the automated 96 and 192 throughputs were much shorter than the manual operation
The multifunctional head does not execute only the liquid handling and assay plate gripping; the PMT-based detection module is also used for CPA-detected viral copy quantification. In detail, two channels with an excitation of 485 nm/emission of 535 nm, and excitation of 640 nm/emission of 700 nm, respectively, for single or dual detection using the CPA reaction were designed. The excitation light path where the LED light source was located was physically separated at an angle of < 30 with a single-photon counter PMT attached, in conjunction with the aperture diaphragm and filters, reduces any large-angle scattering, which improves the signal-to-noise ratio, and enhances viral detection sensitivity. Finally, the time lapse of the CAP detection data was analysed in an automated manner. The total time consumption differed based on sample throughput (Fig. 4b). The single batch run of 96 samples consumed 120 min, as the run of 192 samples could be reduced due to the parallel robotic process, and the detection speed of the single-photon counter PMT was superior to that of the CAP reaction; hence, a speed of less than 60 s per reaction was achieved. In contrast, the manual RT-PCR with a 24-sample throughput consumed more time for nucleic acid preparation and detection. Therefore, with the entire robotic operation and the key functional components, throughput can be enhanced dramatically with minimal manual involvement.
Comparison of diagnostic performance and determination of detection sensitivity
The clinical sample was implemented for direct comparison between the gold standard CDC-recommended conventional RT-PCR assay and this fully automated process. As detailed in the method, the same targeted viral sequence as recommended by the CDC (N and ORF1a/b gene) was performed with 1129 specimens for neck-to-neck comparison. First, we cross-evaluated the automated yield rate of nucleoid acid extraction between the manual process and the conventional approach. The evaluation was performed using the human housekeeping gene (GAPDH) as an internal control; consistently, a 50–60% yield rate was obtained with both approaches. Noticeably, the number of minimum sample processes using automation could not be less than four, which means that it is less flexible than the manual method. In a total of 1129 specimens that used CAP combined with the automation process, 11/1129 samples were detected as SARS-CoV-2 positive, 4 samples showed statistical ambiguity, and the remaining 1114 samples were detected as SARS-CoV-2 negative. In comparison, manual RT-PCR showed that 15/1129 samples were reported as SARS-CoV-2 positive, while the others were negative (Fig. 5). Interestingly, apart from the three samples which fell between 3-sigma and 5-sigma statistically, the other data points were matched and were in 100% agreement. Unlike manual involvement in the data analysis of RT-PCR, the threshold cycle (Ct) quantified the titre number of viral particles, which could be manually adjusted based on operator experience under the circumstances; in contrast, the WMGs were automatically implemented in CPA quantification. There was no non-specific amplification of the negative control by any of the assays (data not shown), which is consistent with the result for 1114 samples as SARS-CoV-2 negative using RT-PCR as the comparison. Meanwhile, the data with statistical ambiguity as specious samples were retested for confirmation.
Fig. 5.
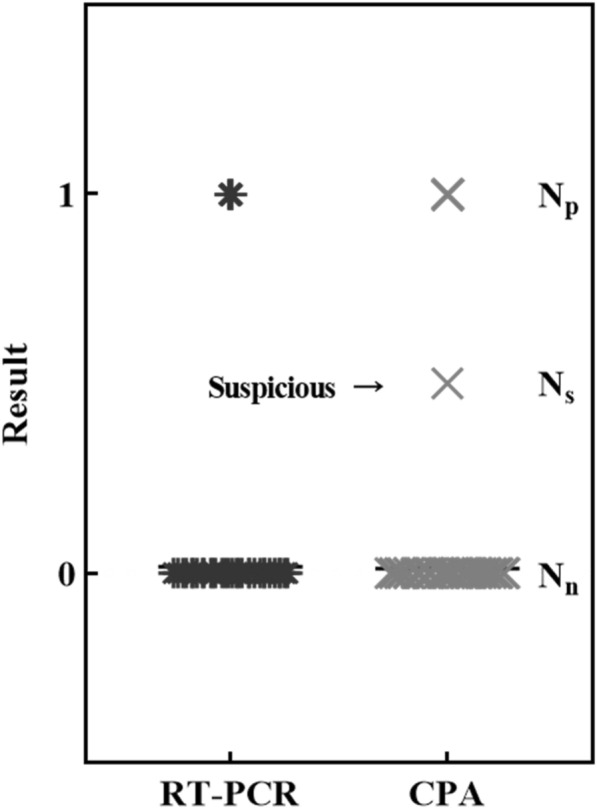
Direct comparison of CPA and RT-PCR for accuracy. More than 1000 specimens were used to perform an accuracy assessment with real-world samples. The data are shown in brief: negative samples of both RT-PCR and CPA, Nn = 1114; positive samples of RT-PCR, Np = 15; positive samples of CPA, Np = 11; suspicious samples of CPA, Ns = 4; total N = 1129; KAPPA test: κ = 0.73, P < 0.001 (substantial)
To assess the detection sensitivity of this automated approach, we conducted assays using serially diluted positive controls (25, 50, 100, 200, 500, 1000, and 2000 copies) as the template (Table 1). With more than 100 copies per reaction (not including 100) of the positive control, amplification and detection were observed in all assays (n = 3). Within 100 copies, amplification signals failed in one of 3 repeats. Hence, the limitation of CPA sensitivity is approximately Ct = 35, which is consistent with the viral titre of these ambiguous samples. Furthermore, the sensitivity was improved using a CRISPR-based detection probe as the second step after the CPA cycles (detailed in the Methods section). This sensitivity improvement can detect 50 copies per reaction with a reaction time of 30 min. These results suggested that the detection rate of this fully automated setup was comparable with that of the conventional CDC-recommended approach, but with superior throughput and limited manual involvement.
Table 1.
Sensitivity comparison of one-step and two-step detection strategy
| Copies/reaction | One-step 60 min | Two-step 90 min | Copies/ml | Ct value |
|---|---|---|---|---|
| 2000 | + + + | + + + | 40,000 | 30 ~ 31 |
| 1000 | + + + | + + + | 20,000 | 31 ~ 32 |
| 500 | + + + | + + + | 10,000 | 32 ~ 33 |
| 200 | + + + | + + + | 4000 | 33 ~ 34 |
| 100 | +−+ | + + + | 2000 | 34 ~ 35 |
| 50 | − | − + + | 1000 | 36 ~ 37 |
| 25 | – | − | 500 | > 37 |
| PC | + + + | + + + | – | – |
| NC | − | − | – | – |
The calculation criteria between different approaches were based on the following restricted conditions: The sample uptake volume from the original specimen for nucleic acid preparation was 200 µl or no less than 100 µl. The recovery rate of the extraction kit was estimated to be 50%. The volume of purified nucleic acid was 40 µl. CPA was used to quantify the volume of the amplification template to 20 µl. The absolute relevance between copies/ml and Ct value was verified using digital RT-PCR for the nucleic acid count as the reference obtained for this study from the CDC
Discussion
In the current development, we present a fully automated robotic system that can perform SARS‐CoV‐2 nucleic acid-based diagnosis in a “specimen in data out” manner. The aim of this development was to assess the combined system achievement for high specification, high sensitivity, and high throughput with minimum workforce involvement, and to evaluate the system using real-world evidence with more than a thousand clinical samples as the comparison study. We converted the biological process to suit the automation process and designed the system to extract nucleic acid from swabs or sputum. The robotic system implemented a CPA reaction to potentiate the throughput and sensitivity, rather than using RT-PCR, for SARS‐CoV‐2 nucleic acid detection in ORF1a/b and N genes. Moreover, an automated WMG data analysis approach with custom-developed algorithms was implemented for fast and accurate result profiling without user input. Our findings emphasise that understanding the specific sensitivity kinetics of the system should facilitate the screening strategy for large-scale testing in fighting COVID-19.
The sensitivity, specification, and throughput are the three most essential criteria for molecular diagnosis, which even serves as conflict metrics to achieve all three at the high-end. The design of the probes determined the specifications for SARS-CoV-2 diagnosis, as recommended by WHO in early 2020. Upton now, CDC from multiple regions have certified the CPA mix (in China, US and EU), as verified using large-scaled clinical data concerning sensitivity [23]. The single-photon counter can further enhance sensitivity under the current biological setup. Although we only used one optical channel with two viral genes for this clinical verification, further development may facilitate dual channels for detection of up to four genes, which may be applied to the influenza virus and SARS-CoV-2 double screening as two genes for each identification with high sensitivity. The throughput and walkway time are two independent measurements in SARS-CoV-2 diagnosis at present. Although the POCT setup on the machine is easy to use, in a real-world clinical lab, a regular workforce is required for sample confirmation, sample loading, and sample ejection. The workload for a clinical workforce, which requires an automation process, provides maximal no-need for man attended period as walkaway time and scalable parallel procedures. Hence, the deployment of a POCT testing device for extensively scaled screening is less optimal.
Screening for many infectious specimens in a centralised manner may be the next step for improving working efficiency. The concept of robotics and cognitive automation (R&CA) was implemented as total laboratory automation (TLA), which may be a solution for infectious samples [24, 25]. Indeed, the automated approach we developed and presented in this study, is consistent with the requirements and recommendations in the literature for reducing people’s involvement, minimising infrastructural setup, achieving high throughput with high sensitivity and specificity, and delivering reliable and automated data with high QC standards [24, 26]. However, the automation process is not limited to mimicking the entire manual process in a robotic manner. Biological reagents are critical for any molecular diagnosis; however, they were originally developed and designed for manual operation rather than for automation. Hence, adapting the biological process to automation, or redesigning the hardware to comply with a biological parallel process is crucial; consequently, it further enhances system efficiency and robustness.
To perform background screening for monitoring COVID-19 infection levels in a large population, the guideline for a 5-in-1 mixed sample SARS-CoV-2 diagnosis was discussed. The critical statistical assumption is that the overall COVID-19 infection rate in the target area is lower than 0.1%; indeed, this does not apply to clinical applications. Hence, using this robotic setup with 50 copies per reaction sensitivity in the 2-step CPA approach can quickly expand testing capacity and further improve throughput. In the post COVID-19 situation, background screening may be the most effective way to monitor the infection rate to provide meaningful epidemiological data. Furthermore, during the manuscript publication, the SARS-CoV-2 Delta variant becomes the mainstream of the infectious mutation, which is highly contagious. The advantage of our system is capable of operating the updated reagents, which use CRISPR for detecting the variable region in two-step automation approaches. As demonstrated in Supplement Fig S3, using one PMT fluorescent channel for ORF-gene detection, and the spare channel for detection of CRISPR bound variants sequence.
To sum up, our development converted the CAP reaction to suitable for automation with a tailored optical design and combined this with an online data analysis approach. Our system processed more than 1000 clinical samples for validation. The application-tailored process-specific automatic setup dramatically enhanced diagnostic screening power, and it achieved 50–100 viral copies per reaction and approximately 700–1000 reactions daily in an assembly-line manner. Although people have made a significant achievement regarding oral medicines for COVID-19, the 3–5 days treatment window is in the early phase of the infection. Hence, this diagnostic power is a vital capability in the current COVID-19 pandemic as in conventional clinical practice, which is also capable of variants detection.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors sincerely thank those who are on the front lines battling the SARS‐CoV‐2 virus. We also thank the strong technical support from, and the extraction kit provided by Guangzhou Koalson Bio-Technique Co. Ltd., in addition to the consociation with Hangzhou Ustar Biotechnologies Ltd. for the CPA relative optimisation test produced significant results. Groups interested in this SARS-CoV-2 integrated nucleic acid preparation and detection system can request information by emailing zhang_xiao@gibh.ac.cn.
Author contributions
JL, WF and ZH contributed equally to this work. ZH, WF and XZ were involved in designing the study and preparing the manuscript. XZ, JL, ZH, GS, JD, JQ and JL performed most of the experiments. KF, JL and ZZ analyzed the data. CD, KP, WG, KF and XZ contributed to the critical revision of the manuscript.
Funding
This work was partially funded by Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory) (No.198F021812), and the Instrument Developing Project of the Chinese Academy of Sciences (No. ZDZBGCH2018005, No. YZ201620).
Data availability
Supplementary materials.
Code availability
Not applicable.
Declarations
Conflict of interest
The manuscript has not been published before and is not being considered for publication elsewhere. All authors have contributed to the creation of this manuscript for important intellectual content and read and approved the final manuscript. We declare there is no conflict of interest.
Ethical approval
The clinical samples obtained were approved by the ethical board of Sun Yat-Sen Memorial Hospital.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Paiva HM, Afonso RJM, de Oliveira IL, Garcia GF. A data-driven model to describe and forecast the dynamics of COVID-19 transmission. PLoS ONE. 2020 doi: 10.1371/journal.pone.0236386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glass DH. European and US lockdowns and second waves during the COVID-19 pandemic. Math Biosci. 2020 doi: 10.1016/j.mbs.2020.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296(2):E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman T, Nissen K, Krambrich J, Ronnberg B, Akaberi D, Esmaeilzadeh M, Salaneck E, Lindahl J, Lundkvist A. Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS-CoV-2. Infect Ecol Epidemiol. 2020;10(1):1754538. doi: 10.1080/20008686.2020.1754538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qing-Bin M, Jing-Jing P, Xin W, Jia-Yao Y, Peng-Cheng L, Zi-Wei Q, Yong-Fen X, GuangJiang W, Zhi-Min H, Jian-Chun Y, Wen S. Clinical application of combined detection of SARS-CoV-2-specific antibody and nucleic acid. World J Clin Cases. 2020;8(19):4360–4369. doi: 10.12998/wjcc.v8.i19.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, Adriano A, Beese S, Dretzke J, di Ferrante Ruffano L, Harris IM, Price MJ, Dittrich S, Emperador D, Hooft L, Leeflang MM, Van den Bruel A, Cochrane C-DTAG. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. 2020 doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uddin M, Mustafa F, Rizvi TA, Loney T, Suwaidi HA, Al-Marzouqi AHH, Eldin AK, Alsabeeha N, Adrian TE, Stefanini C, Nowotny N, Alsheikh-Ali A, Senok AC. SARS-CoV-2/COVID-19: viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses. 2020;12(5):526. doi: 10.3390/v12050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basso D, Aita A, Navaglia F, Franchin E, Fioretto P, Moz S, Bozzato D, Zambon CF, Martin B, Dal Pra C, Crisanti A, Plebani M. SARS-CoV-2 RNA identification in nasopharyngeal swabs: issues in pre-analytics. Clin Chem Lab Med. 2020;58(9):1579–1586. doi: 10.1515/cclm-2020-0749. [DOI] [PubMed] [Google Scholar]
- 10.Torretta S, Zuccotti G, Cristofaro V, Ettori J, Solimeno L, Battilocchi L, D'Onghia A, Bonsembiante A, Pignataro L, Marchisio P, Capaccio P. Diagnosis of SARS-CoV-2 by RT-PCR using different sample sources: review of the literature. Ear Nose Throat J. 2021;100:131S–138S. doi: 10.1177/0145561320953231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanne J, Little B, Chung J, Elicker BM, Ketai LH. Essentials for radiologists on COVID-19: an update-radiology scientific expert panel. Radiology. 2020;296(2):E113–E114. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Tawfiq JA, Memish ZA. Diagnosis of SARS-CoV-2 infection based on CT scan vs RT-PCR: reflecting on experience from MERS-CoV. J Hosp Infect. 2020;105(2):154–155. doi: 10.1016/j.jhin.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirotsu Y, Mochizuki H, Omata M. Double-quencher probes improve detection sensitivity toward severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a reverse-transcription polymerase chain reaction (RT-PCR) assay. J Virol Methods. 2020 doi: 10.1016/j.jviromet.2020.113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Augustine R, Das S, Hasan A, Abhilash S, AbdulSalam S, Augustine P, Dalvi YB, Varghese R, Primavera R, Yassine HM, Thakor AS, Kevadiya BD. Rapid antibody-based COVID-19 mass surveillance: relevance, challenges, and prospects in a pandemic and post-pandemic world. J Clin Med. 2020 doi: 10.3390/jcm9103372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolters F, van de Bovenkamp J, van den Bosch B, van den Brink S, Broeders M, Chung NH, Favié B, Goderski G, Kuijpers J, Overdevest I, Rahamat-Langedoen J, Wijsman L, Melchers WJG, Meijer A. Multi-center evaluation of cepheid xpert® xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J Clin Virol. 2020 doi: 10.1016/j.jcv.2020.104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen S, Zhang XA, et al. High-coverage SARS-CoV-2 genome sequence acquired by target capture sequencing. J Med Virol. 2020;92(10):2221–2226. doi: 10.1002/jmv.26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan K, Zhang S, Zhang Y, Lu J, Holcombe M, Zhang X. A machine learning assisted, label-free, non-invasive approach for somatic reprogramming in induced pluripotent stem cell colony formation detection and prediction. Sci Rep. 2017;7(1):13496. doi: 10.1038/s41598-017-13680-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burger B, Maffettone PM, Gusev VV, Aitchison CM, Bai Y, Wang X, Li X, Alston BM, Li B, Clowes R, Rankin N, Harris B, Sprick RS, Cooper AI. A mobile robotic chemist. Nature. 2020;583(7815):237–241. doi: 10.1038/s41586-020-2442-2. [DOI] [PubMed] [Google Scholar]
- 21.Fang R, Li X, Hu L, You Q, Li J, Wu J, Xu P, Zhong H, Luo Y, Mei J, Gao Q. Cross-priming amplification for rapid detection of mycobacterium tuberculosis in sputum specimens. J Clin Microbiol. 2008;47(3):845–847. doi: 10.1128/JCM.01528-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu G, Hu L, Zhong H, Wang H, Yusa S, Weiss TC, Romaniuk PJ, Pickerill S, You Q. Cross priming amplification: mechanism and optimization for isothermal DNA amplification. Sci Rep. 2012;2:246. doi: 10.1038/srep00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung Y, Park GS, Moon JH, Ku K, Beak SH, Lee CS, Kim S, Park EC, Park D, Lee JH, Byeon CW, Lee JJ, Maeng JS, Kim SJ, Kim SI, Kim BT, Lee MJ, Kim HG. Comparative analysis of primer-probe sets for RT-qPCR of COVID-19 causative virus (SARS-CoV-2) ACS Infect Dis. 2020;6(9):2513–2523. doi: 10.1021/acsinfecdis.0c00464. [DOI] [PubMed] [Google Scholar]
- 24.Nasuto SJ, Hayashi Y. Anticipation: beyond synthetic biology and cognitive robotics. Biosystems. 2016;148:22–31. doi: 10.1016/j.biosystems.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 25.Santello M, Bianchi M, Gabiccini M, Ricciardi E, Salvietti G, Prattichizzo D, Ernst M, Moscatelli A, Jorntell H, Kappers AM, Kyriakopoulos K, Albu-Schaffer A, Castellini C, Bicchi A. Hand synergies: integration of robotics and neuroscience for understanding the control of biological and artificial hands. Phys Life Rev. 2016;17:1–23. doi: 10.1016/j.plrev.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cangelosi A, Stramandinoli F. A review of abstract concept learning in embodied agents and robots. Philos Trans R Soc Lond B Biol Sci. 2018;373(1752):20170131. doi: 10.1098/rstb.2017.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary materials.
Not applicable.