Abstract
Structure-based drug discovery (SBDD) is an indispensable approach for the design and optimization of new therapeutic agents. Here we highlight the rapid progress that has turned cryo-electron microscopy (cryoEM) into an exceptional SBDD tool, and the wealth of new structural information it is providing for high-value pharmacologic targets. We review key advantages of a technique that directly images vitrified biomolecules without the need for crystallization, both in terms of a broader array of systems that can be studied and the different forms of information it can provide, including heterogeneity and dynamics. Further, we discuss near- and far-future developments, working in concert to achieving the resolution and throughput necessary for cryoEM to make a widespread impact on the SBDD pipeline.
Keywords: cryo-electron microscopy, single-particle analysis, pharmacology, small molecule, biologics, structure-based drug discovery
Structure-based drug discovery in the era of cryoEM
Structure-based drug discovery (SBDD) leverages atomic-level information of a therapeutic target to rationally design and ascertain in silico chemical species effective against that target. SBDD began in the 1980s with the development of enzyme-targeting drugs such as captopril and dorzolamide [1–3], and is an answer to the often costly, time consuming, and low-yielding high-throughput screening (HTS) performed in the wet lab. The creation of these FDA-approved drugs and others was enabled by newly available crystallographic structural models in combination with advances in computer-aided molecular modeling. Since then, SBDD has rapidly accelerated with both the use of more sophisticated computational techniques for drug discovery and the rapidly expanded compendium of crystallographic structures of therapeutic targets. From 1999 to 2013, the majority (78) of 113 approved first-in-class drugs were discovered through structure-based approaches [4].
Despite the rapid growth of SBDD, the motivation to further expand its use is greater than ever. SBDD can be critical for providing creative approaches to drugging well-validated targets previously considered undruggable, as demonstrated with K-Ras(G12C), which required crystallographic structures to identify a previously unknown binding pocket that avoids competition with picomolar-affinity GDP/GTP [5]. Since target validation is one of the primary challenges in optimal resource allocation for discovery and development efforts, first-in-class drug-like molecules can provide new insights into target validity and disease application, like in the case of the small molecule bromodomain inhibitors (+)-JQ-1 and I-BET762. These compounds were widely shared and successfully used to characterize and validate the importance of bromodomains in a variety of diseases, leading to a large number of clinical candidates [6]. Even for targets with existing FDA-approved drugs, there is often a clinical need for further SBDD. For example, higher selectivity may be desired like in the case of erdafitinib, which was designed to be more selective than its predecessors towards the fibroblast growth factor receptor over closely related off-target receptors [7]. There may be a need for modified potency or efficacy, or to provide distinct receptor subtype selectivity, such as the sphingosine 1-phosphate (S1P) inhibitor siponimod, whose selection for S1P1 over S1P3 is critical for its improved efficacy and safety profile compared to prior non-selective S1P inhibitors [8]. Furthermore, as in the case of numerous antiviral, antibacterial, and anticancer agents, there may be a need to rationally target a mutated viral protease, bacterial ribosomal subunit, or oncogenic kinase that has developed resistance to existing drugs [9–11].
The most significant bottleneck for successful SBDD efforts has been a lack of high-resolution structural information of the biological target. While small and well-ordered biological molecules are amenable to crystallography, there has been a shortage of high-resolution structures of difficult-to-crystallize proteins, such as transmembrane receptors or dynamic complexes, which comprise a large proportion of validated targets. Crystallographic structures often involve significant modifications to the target protein such as truncations, thermostabilizing mutations, or the insertion of entire orthotopic domains for thermal stability, which may place limitations on the interpretation of structural information for SBDD. Most critically, an enormous number of targets have simply been so far “uncrystallizable.”
The need for accurate structures is increasingly being met by cryo-electron microscopy (cryoEM) at a sufficiently high resolution for computationally-aided drug design, which is the topic of this review. In contrast to X-ray crystallography, cryoEM does not require that the target of interest form a crystal lattice. Instead, the purified biological macromolecule of interest is flash-frozen in a thin layer of vitreous (non-crystalline) ice and imaged directly with transmission electron microscopy. The recorded cryoEM projections of individual particles—hundreds of thousands to millions in any given dataset—are then used to reconstruct a 3-dimensional electrostatic potential map (see Glossary) into which the macromolecule is modeled (see [12] for a review of cryoEM). This technique thus lends itself well to the structural determination of protein complexes, and proteins with lower thermal stability and higher dynamic motion, as well as transmembrane proteins in lipid micelles. With continued improvement in resolution, cryoEM has emerged as a remarkably powerful new tool for drug design.
CryoEM in drug discovery
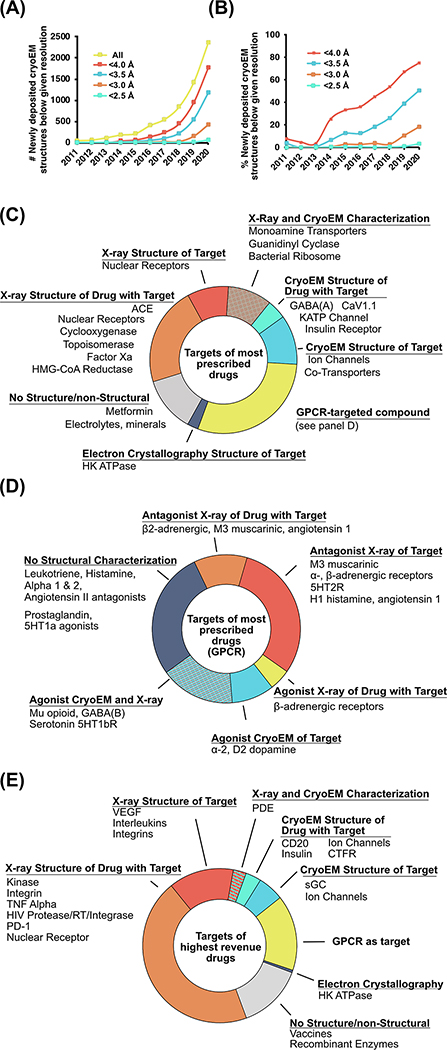
Prior to 2014, cryoEM rarely provided the sub-4.0 Å resolution often required for SBDD. However, the explosive progress of the methodology in the last few years has brought an outpouring of high-resolution structural data that was previously inaccessible. The “quantum jump” of cryoEM owes itself to many advances such as direct electron detectors for recording images, improved computational methods, and hardware parallelization for processing large datasets, a timeline of which is reviewed in detail elsewhere [13]. Furthermore, the nature of cryoEM as a direct-visualization technique has allowed for the rapid diagnosis of biochemical problems such as sample aggregation and instability, leading to rapid improvement of specimen quality through genetic and biochemical modifications, binders to stabilize mobile elements, or improved detergents to extract membrane proteins from their native environment. As a result, the number of cryoEM structures deposited in the Protein Data Bank (PDB)i with a resolution of 4.0 Å or better has grown from 16 prior to 2014 to 1,753 new structures deposited in 2020 alone (Figure 1A). The proportion of newly deposited structures with resolution better than 4.0Å and 3.5Å has increased from 36% and 12% in 2015 to 75% and 50% in 2020, respectively. Perhaps most impressively, the proportion of deposited structures in 2020 with resolutions higher than 3.0Å and 2.5Å, previously almost nonexistent, are now a non-negligible 18% and 3% respectively (Figure 1B).
Figure 1: The improvement in resolution of cryoEM and its contribution to the structural characterization of protein drug targets.
A) The absolute number of newly deposited cryoEM structures in the PDB below given resolutions and B) percentage of newly deposited cryoEM structures in the PDB below given resolutions [61]. C) Chart of the targets of the 200 most-prescribed drugs of 2018 broken down by the target’s structural characterization, D) chart of the targets of the 44 most-prescribed drugs targeting GPCRs broken down by structural characterization, and E) chart of the targets of the 200 highest-sales drugs (as a proxy for newer drugs) of 2018 broken down by the target’s structural characterization. Data manually curated in 2020 by identification of the protein target, if applicable, of the 200 most prescribed and 200 highest sales drugs as curated by the Njardarson lab for 2019 [14], followed by the identification of related structures in the PDB.
To assess the impact of cryoEM on the field of SBDD, we surveyed the structural data relating to targets of the 200 most frequently prescribed drugs in the US in 2018 [14]. Structural information is available in the PDB for 72% of these targets, which was determined by X-ray crystallography (42%), cryoEM (15%), or by both methods (15%) (Figure 1C). Many of the targets characterized by cryoEM include transmembrane proteins such as ion channels (GABAA, CaV, NaV, KATP), active states of G protein-coupled receptors (GPCRs), and transporters (serotonin transporter, NaCl transporter). GPCRs represent the targets of 44 out of the 200 most prescribed drugs, which include agonists, antagonists, and inverse agonists (Figure 1D; note that while antagonists and inverse agonists are pharmacologically distinct, here we group them together as “antagonists” because the experimental data required for differentiation is often unavailable). Thirty-two of these GPCRs (73%) have had some form of structural characterization, which includes crystal structures with antagonists (44%) or agonists (7%), cryoEM structures with agonists (9%), or characterization with both X-ray crystallography and cryoEM (20%). Of note, the vast majority of GPCR crystal structures have been solved in complex with antagonists, owing primarily to the challenge of obtaining high-quality crystals of very dynamic active-state GPCRs.
While cryoEM has made an impressive impact on the list of most prescribed drugs, this list is biased towards those that have had many years on the market. To improve our understanding of the role cryoEM can play in future drug discovery, we also considered the 200 highest revenue drugs of 2018 as a proxy for the products of relatively recent discovery programs [14], hereafter referred to as “newer drugs” (Figure 1E). Noticeable differences exist between the top-prescribed and newer drugs. In particular, a substantial fraction of the newer drugs has been characterized with crystallography. This reflects the importance of structural data in modern times: even when a discovery effort is not ‘structure driven’, it is rare that structures are not pursued, as they can provide critical data for lead optimization and further discovery. Additionally, the fraction characterized by cryoEM on this list is smaller but still appreciable considering the long drug development timelines and the short period for which high-resolution cryoEM has been possible. These drugs and targets include biologics, ion channels, and GPCRs, a list that is similarly enriched in highly dynamic macromolecules that are less amenable to crystallization.
What cryoEM brings to SBDD: Accessing new types of structures
While there are many targets of FDA-approved drugs that have proven fully tractable for X-ray crystallography, cryoEM has opened the door to many additional targets that have not been easy to crystallize—generally, larger and more dynamic proteins and protein complexes. Access to challenging intracellular complexes such as ribosomes of pathogens, chromatin-modifying complexes, and transcriptional machinery has been improved by cryoEM [15–17]. Notably, a first-in-class inhibitor of mitochondrial transcription was recently described in complex with human mitochondrial RNA polymerase using cryoEM [18].
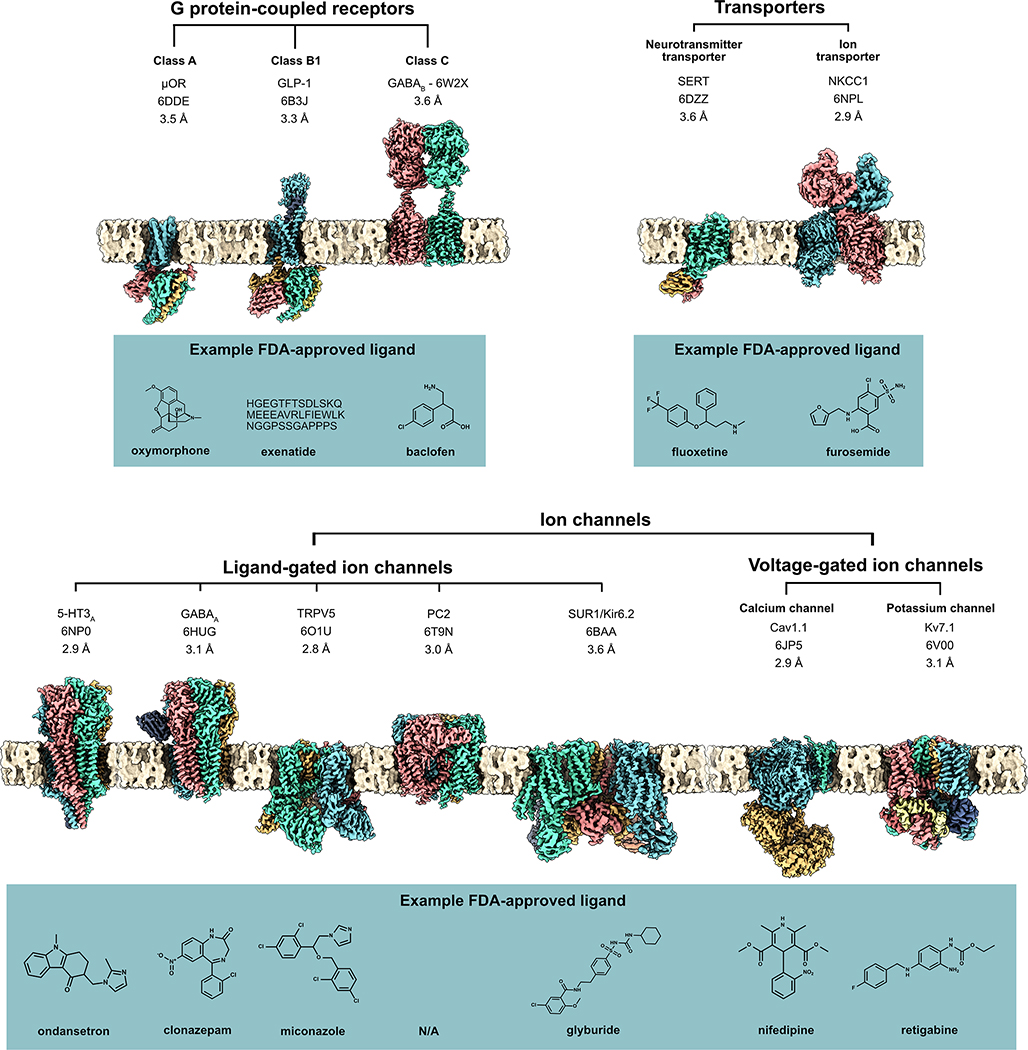
However, there is no space where cryoEM has had a larger impact than that of membrane proteins. GPCRs, ion channels, and transporters are well-represented in both the most prescribed and the newer drugs, yet these targets are notoriously difficult for X-ray crystallography. Although lipidic cubic phase crystallization has provided some headway in the GPCR field, the proteins generally required heavy modification with thermostabilizing mutations and fusion domains to promote crystal formation, requiring an extensive screening of constructs and conditions to produce somewhat engineered stabilized conformations (for examples, see [19,20]). In contrast, cryoEM has the ability to directly image detergent- or nanodisc-solubilized membrane proteins, often as native or near-native constructs, requiring only biochemical stability. The tractability of this method is reflected in the wide range of membrane protein structures that have now been determined, often to high resolution. The majority of these membrane proteins have long been the targets of approved drugs, with their structures only recently obtained (Figure 2, Key Figure).
Figure 2, Key Figure: High-resolution cryoEM maps of a wide variety of membrane protein drug targets.
This includes G protein-coupled receptors (GPCRs) and transporters (top row) and ion channels (bottom row), with an example FDA-approved ligand for each receptor (blue boxes). PDB IDs and resolution provided below protein name. Membrane generated with CHARMM-GUIviii [62] and figure rendered in ChimeraXix [63]. GABAA (PDB ID 6HUG) and TRPV5 (PDB ID 6O1U) were determined in lipid nanodiscs; all others displayed were obtained in detergent micelles.
Some of the most dramatic advances in the use of cryoEM for transmembrane proteins have come from the design of new reagents that preserve transmembrane protein structure, protect the protein during cryoEM grid preparation, and provide homogeneous samples for high-resolution analysis. For example, the use of synthetic detergents such as n-dodecyl β-D-maltoside (DDM) and lauryl maltose neopentyl glycol (LMNG) allows for the efficient extraction and solubilization of transmembrane proteins from the cell membrane while preserving in most cases a near-native protein structure. It should be noted that detergent micelles may pose their own challenges, including the persistence of empty or protein-bound micelles that lead to heterogeneity and compromise data processing and alignment, as well as surfactant effects on sample blotting and vitrification that may be hard to predict (detergents are reviewed in [21]). One important alternative to detergent micelles is the lipid nanodisc, which can in principle provide a near-native lipid bilayer for structural and biophysical studies. The use of lipid nanodiscs for membrane protein drug targets has been pivotal and extensive: Nanodiscs in combination with cryoEM have elucidated the functional mechanisms and high-resolution structures of the TRPV1 and TRPV5 ion channels (in the case of TRPV1, lipid was essential for inhibitor binding), the GABAA ligand-gated ion channel, human P-glycoprotein, and recently GPCR-β-arrestin complexes, to name a few [22–26]. For further reading on nanodiscs, see [27,28]. Notably, cryoEM can even determine structures of proteins embedded in liposomes, allowing the visualization of ion channels in more native-like electrochemical gradients, as well as the study of pore-forming proteins [29,30].
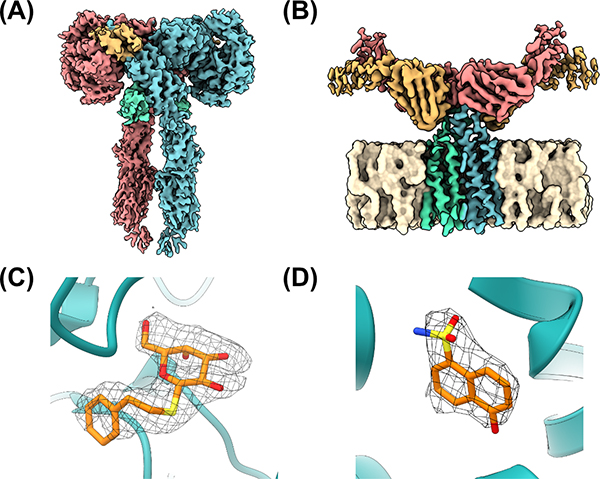
Another key area in which cryoEM has made an enormous impact in the last few years is in the realm of biologic drugs. Even though the list of most prescribed drugs possesses relatively few biological agents outside of insulin and its derivatives, the newer drugs are replete with them. Crystallization techniques for biologics have indeed improved by focusing only on the salient domains for drug-target recognition; however, cryoEM has provided unprecedented levels of structural detail for several key biologic drugs engaging near full-length proteins. One particularly impactful example is the insulin receptor, a dimeric receptor tyrosine kinase that plays key roles in the modulation of glucose homeostasis. Disruption of insulin receptor signaling is involved in several diseases such as type II diabetes, which affects an estimated 9.3% (463 million people) worldwide [31]. Two groups have made substantial progress in this area using cryoEM, the first resolving the structure of the insulin receptor ectodomain bound to either one or two molecules of insulin at resolutions of 4.3 and 7.2 Å, with the second group obtaining the structure of the insulin receptor ectodomain bound to four molecules of insulin at a nominal resolution of 3.1 Å (Figure 3A). These structures have highlighted distinct binding sites for insulin, as well as the conformational changes required for the activation of this key pharmacologic target [32,33].
Figure 3: Demonstration of the utility of cryoEM for both small-molecule and biologics discovery.
CryoEM maps of (A) the insulin receptor bound to insulin (PDB ID 6PXV) and (B) CD20 in complex with rituximab Fabs (PDB ID 6VJA). (C) Accurate modeling of PETG into a cryoEM map of β-galactosidase using GemSpot (PDB ID 6CVM). (D) Fragment-based discovery for PKM2 with cryoEM density allowing for correct identification and placement of discovery fragment (PDB ID 6TTF).
Other examples abound: cryoEM has shed new light on both old and new targets of biological therapeutics, from the HER2-trastuzamab-pertuzumab complex to a neutralizing monoclonal antibody to SARS-CoV-2, paving the way for further discovery and development of both biosimilars and first-in-class biologics [34,35]. Another notable example is B-lymphocyte antigen CD20, a well-established therapeutic target in leukemia and autoimmune diseases, although its functional role remains unclear. Despite a molecular weight of ~35 kDa, well-resolved cryoEM structures were obtained for CD20 in complex with the Fab portion of the therapeutic monoclonal antibodies rituximab, ofatumumab, and obinutuzumab (Figure 3B) [36,37]. Additionally, negative stain EM of the full rituximab-CD20 complex revealed that rituximab induces cyclic higher-order structures [36], yielding unexpected insight into activation of the complement system of innate immunity. This level of structural characterization would most likely prove impossible for crystallization given the presence of both highly flexible and membrane-spanning elements in the complex.
Notwithstanding these rapid successes, there are still areas of membrane protein structure determination in which cryoEM has yet to make an impact. One such area is that of monomeric membrane proteins smaller than 50–70 kDa that do not display appreciable intra- or extra-cellular domains. Due to little or no features extending beyond the noisy detergent micelle or lipid nanodisc, the alignment of projections from such particles are challenging for high resolution work, such as an inactive-state GPCR in the absence of a signaling partner. Given the large number of proteins that fall in this category, this is an active area of research with several strategies under investigation. These strategies have seen some level of success such as with CD20 as noted above, and more progress is soon expected with the increased use of asymmetry-conferring synthetic binding partners such as large fusion proteins, antibody fragments, nanobodies and nanobody derivatives, and scaffolding proteins.
The power of computational approaches combined with cryoEM
Single-particle cryoEM leverages projections from millions of directly visualized particles to reproduce an electrostatic potential map, often involving tens of terabytes of raw data. As such, the methodology has benefitted enormously from rapidly evolving computational approaches that simultaneously address the need for ever-higher resolution and also a more nuanced understanding of particle dynamics. However, there are several intrinsic challenges to obtaining high-confidence models of ligand-target complexes with cryoEM compared to X-ray crystallography. One challenge is the relative paucity of cryoEM structures with resolution higher than 2.5Å, which is often necessary for a human modeler to be able to unambiguously place the ligand and also begin to resolve water molecules in the binding site. Additionally, modeling into cryoEM maps is a fundamentally different process from that of crystallography in which there is bidirectional feedback between modeling and map refinement. Briefly, in X-ray crystallography, the map and model are refined together such that one informs and refines the other in reciprocal space, with a rigorous and well-defined set of statistical measurements that provide key information regarding model accuracy and, crucially, generation of difference maps between the experimental data and the model. In contrast, cryoEM map refinement is an entirely independent process that only relies on the EM projections collected, and the refined map is then used to separately build and refine the model in real space. The independence of these two processes make the true statistical correlation between map quality and model accuracy a somewhat arbitrary process in which the rigorous calculation of a difference map is not straightforward, although recent progress has been made [38]. There are also physical differences, such as the fact that X-rays produce an electron density map whereas cryoEM produces an electrostatic potential map. Such differences, taken together, make the application of crystallographic model validation tools problematic for cryoEM, which needs to develop improved metrics for the accuracy of maps and models.
One way, at least in part, to address these challenges and new approaches to structure determination is powerful computational techniques and accurate molecular force fields for modeling macromolecules and their interactions with ligands in cryoEM maps. The combination of real-space and reciprocal-space refinement with the OPLS3e molecular mechanics force field, implemented in the workhorse PHENIX software suiteii, has generated substantially improved geometry statistics for both biomolecular and small molecule components of refined structures [39]. OPLS3e refinement has also been incorporated into GemSpot, our own pipeline which integrates a variety of computational methodsiii into a single workflow to improve the accuracy of ligand poses in cryoEM maps (Figure 3C) [40]. New computational tools have also allowed for the use of cryoEM in fragment-based drug discovery, in which high-solubility, low molecular weight ‘fragment’ compounds are soaked, often in a cocktail of more than one structurally distinct compound, into a biomolecular target. Solving the resulting structure can then elucidate key high-affinity interactions in multiple different sites of a ligand-binding pocket, which can then be combined into a lead compound. However, this method requires a high-quality, high-resolution ligand density in order to assign the correct pose and/or chemical entity, which has historically been a challenge for cryoEM. Saur et al. recently succeeded in using cryoEM for fragment-based drug discovery, using both the highly tractable case of beta galactosidase as well as the more therapeutically relevant and challenging case of the kinase PKM2 [41]. Although it was necessary to use in-house software in order to place the ligands in a combined map- and chemical interaction-driven approach, they were able to convincingly model a ~150 molecular weight fragment bound to beta galactosidase. Perhaps even more impressively, they were able to identify which fragments bound to PKM2 out of a four-compound cocktail (Figure 3D). Thus, the continuous development and interplay of computational approaches with cryoEM density maps provides a powerful platform with which to model macromolecular complexes at high resolution.
The quickly evolving landscape of cryoEM
Improving accessibility and throughput
High-resolution cryoEM of macromolecules requires substantial costs for the establishment, maintenance, and daily operations of sophisticated instrumentation. As a result, this has been largely restricted to resource-rich institutions—posing a significant challenge for the democratization of this tool. However, this barrier is slowly being lowered. National facilities now provide a resource to train scientists and collect high resolution data in a way that is similar to synchrotrons for X-ray radiationiv–vii. Many major pharmaceutical companies have also recently invested in in-house state-of-the-art cryoEM facilities for both data collection and processing. Moreover, a fundamental attribute of cryoEM that holds future promise is that the facilities required are far more replicable than the ultra-costly synchrotrons and linear accelerators for crystallography. With developments in progress to harness lower-energy 100 keV electron beams [42], the future may involve relatively affordable in-house scopes that could allow for the use of cryoEM wherever drug discovery takes place. Given that 49% of the drugs approved by the FDA in 2018 were owned by small and mid-sized companies, such advancements will allow for more widescale leveraging of this technology.
The new rapid pace at which cryoEM can be performed is demonstrated by the recent structural characterization of proteins involved in the SARS-CoV-2 virus responsible for the 2019–2020 global coronavirus pandemic. Within months of the viral outbreak, structures were obtained by cryoEM for several conformations of the viral spike protein alone and its complexes with complexes with human angiotensin converting enzyme or numerous neutralizing human antibody fragments [35,43–46]. The structure of the repurposed drug remdesivir, recently granted breakthrough FDA approval for treating COVID-19, bound to the SARS-CoV-2 RNA polymerase has also been determined by cryoEM [47]. Given that X-ray crystallography has been the traditional method for viral RNA polymerase structural determination, this is a somewhat surprising coup that emphasizes the utility of cryoEM in rapid-response research.
Moreover, resolution continues to advance dramatically, with a recent report of apoferritin, a model complex for cryoEM, at 1.25 Å—essentially, allowing for unambiguous placement of individual atoms and in some cases even resolving hydrogens and protonation states [48,49]. There is little doubt that further improvements in instrumentation will continue to break records for the practically achievable resolution for well-behaved samples.
The use of cryoEM for drug discovery and development is expected to further benefit from increased automation in nearly all aspects of the technique. In terms of grid preparation, automated tools are emerging to address the challenges of reproducibility and sample waste [50–52]. These efforts will not only improve automation and accessibility but begin to address other challenges in cryoEM grid preparation such as decreasing particle exposure to the air-water interface [53]. Efforts are also underway to address certain user-involved and partially subjective aspects of cryoEM such as particle picking, using machine learning approaches and deep neural networks to improve speed and accuracy [54,55]. Such automation and machine learning approaches are expected to enmesh into even the core of single-particle classification and improve the entire cryoEM pipeline.
Major hardware and also software improvements are also expected to increase the accessibility of cryoEM for SBDD. The development of larger, higher-speed cameras continues unabated. A standard data collection with an older direct detector camera and 1 movie collected per stage shift produced on the order of 50 movies per hour, whereas newer detectors combined with 9 or 12 movies collected per stage shift can produce upwards of 200 movies per hour, translated to millions of particle projections per 24h of data collection. Further, while many of the highest resolution structures today are obtained with a 300 keV microscope, these machines are large and expensive, both in terms of upfront cost and maintenance contracts. In many cases for the thin samples used in single-particle analysis, a 200 keV microscope may be sufficient, and even a 100 keV microscope could be used to obtain structures of up to 3.4 Å resolution [42].
New windows into molecular dynamics
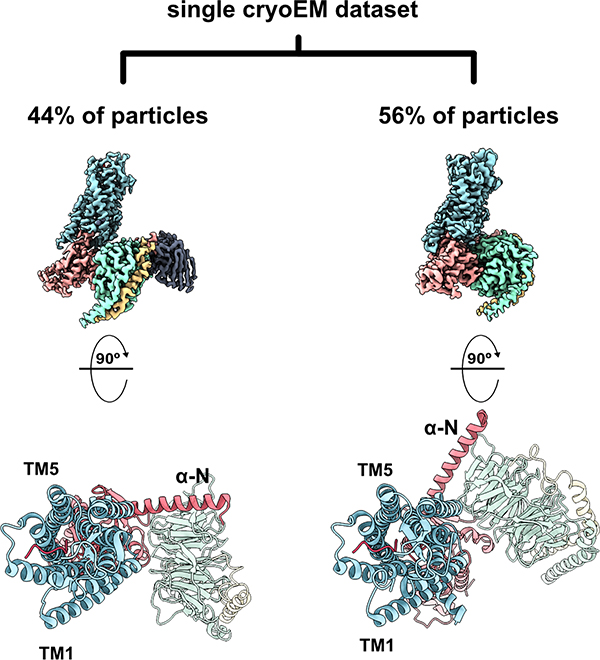
Combining these improvements in both hardware and data processing will continue to unlock one of the most promising features of cryoEM. While X-ray structures are restricted into a crystal lattice, cryoEM visualizes a vitrified sample with an ensemble of states – a conformational continuum or series of distinct energy minimum states that can collectively provide a powerful window into macromolecular dynamics. Computational tools such 2D- and 3D-classification and focused refinement of subregions are able to harness the heterogeneity of particles within a data set to model the motions of large, flexible components of a macromolecule [56]. In one recent example from our group, single-particle analysis of the Neurotensin 1 receptor from a single set of conditions revealed both a previously identified G protein, agonist bound state, and a novel intermediate state along the G protein coupling pathway (Figure 4) [57]. More recently, the application of a deep learning framework to a cryoEM dataset revealed the conformational dynamics of the 26S proteasome to an unprecedented level of atomic detail [58]. As both the resolution and classification tools continue to improve, we will be able to characterize even finer conformational variability. These developments, coupled with computational techniques like molecular dynamics simulations and machine learning approaches, will allow for more accurate modeling of the complex process of ligand binding and expose new—and druggable—intermediate states.
Figure 4: From a single cryoEM dataset, 3-dimensional classification of projections revealed two distinct conformers representing two distinct GPCR-G protein interaction states representing two thermodynamically comparable conformers.
In the canonical state (left, PDB ID 6OS9), the receptor engages the G protein in a prototypical fashion in which the nucleotide binding pocket is primed for GTP binding. In the non-canonical state (right, PDB ID 6OSA), the G protein heterotrimer is rotated by 45° compared to the canonical state, representing an intermediate ligand-bound receptor state along the G protein coupling pathway. TM: transmembrane helix; α-N: N-terminal alpha helix of G protein.
Concluding remarks
Despite the leaps and strides that have brought cryoEM to relevance in SBDD, it remains to be fully leveraged for the valuable information that it will provide in the discovery and optimization of all classes of therapeutics. Building on advances such as 3D variability analysis and deep learning algorithms that reconstruct conformational continuums, in combination with larger and higher quality data sets, it is possible that smaller magnitude, higher-resolution dynamic motions of proteins and even their ligands can be characterized through further development of these types of computational tools. To this end, we also expect that the development of time resolved cryoEM approaches will give unprecedented insights into the critical association and dissociation processes underlying the dynamic character of macromolecular complexes, providing further opportunities for drug targeting [59,60]. Given that crystallographic and most cryoEM models provide but a snapshot of an energy minimum, and that a nuanced understanding of mechanisms and intermediate states is critical for developing novel modes of action, such information on conformational dynamics could prove groundbreaking for rational drug design and optimization. Between these types of advancements in resolution, processing, exploration of dynamics, and accessibility, single particle cryoEM is poised to make enormous contributions to drug discovery and human health.
Outstanding Questions.
Can we increase the throughput of cryo-electron microscopy (cryoEM) techniques to allow for a rapid iterative approach to structurally informed drug optimization?
How can we improve automation to waste less sample and enhance reproducibility?
How do we leverage deep learning algorithms to improve the data processing pipeline through automation of tasks such as grid screening and particle picking?
How can we improve the capabilities of cryoEM to interrogate smaller, symmetric proteins?
Can cryoEM data processing advancements provide insight into the effects of ligand binding on target conformational dynamics?
Highlights.
Cryo-electron microscopy (cryoEM) has recently obtained resolutions sufficient for informing structure-based drug design (SBDD).
The use of cryoEM allows access to new types of structures for SBDD, such as transmembrane proteins and difficult-to-crystalize complexes.
New computational tools combined with cryoEM data provide key data for understanding ligand-protein interactions.
Advances in cryoEM are rapidly improving accessibility and throughput, including increased automation in both hardware and software, and technical advances to improve signal to noise ratios and speed.
New computational tools are unharnessing the ability of cryoEM to capture intermediate and equilibrium states of ligand-protein complexes.
Acknowledgements:
The authors acknowledge the following funding sources: R01 NS092695 (G.S.), T32 GM089626 (J.G.M.).
Glossary
- Agonist
a substance that increases the activation of a receptor to produce a biological response.
- Antagonist (also neutral antagonist)
a substance that has no activity in the absence of agonist or inverse agonist but can block the activity of either.
- Biologic drug
a drug that is manufactured in a living system, which can include recombinant proteins, sugars, gene therapies, or nucleic acids. Also known as biopharmaceuticals.
- Electrostatic potential map (also coulomb potential map)
a 2-dimensional or 3-dimensional representation of the charge distribution in a given sample. In EM, an electrostatic potential map is generated through the scattering of electrons due to coulombic forces as they interact with a sample.
- Electron density map
a plot of electron density versus position within the repeating units of a crystal, represented in 2D- or 3D-space, that is derived from an X-ray diffraction pattern.
- Fragment-based drug discovery
a drug screening approach in which low molecular weight chemical fragments are assessed for their ability to bind the target of interest, with the goal of using these fragments as initial entry points for further medicinal chemistry.
- Inverse agonist
a substance that decreases the basal activation of a receptor to produce a biological response.
- Lipidic cubic phase crystallization
a technique in which a membrane-mimetic matrix is used for the stabilization and crystallization of membrane proteins in a lipid environment, typically for X-ray crystallographic studies.
- Lipid nanodisc
a hockey puck-shaped composite of a phospholipid bilayer encircled by an amphipathic helical belt protein called membrane scaffold protein.
- Negative stain EM
a technique in which a heavy metal salt stain embeds and fixes a biological specimen for EM visualization at room temperature. The approach results in EM images with enhanced inverse contrast between the specimen and its surroundings, and thus the term “negative”. Although the dried metal salt allows visualization of only the shape of the specimen at low resolution (~2nm), this technique is valuable for simple and quick assessment of sample quality.
- Time resolved cryoEM
an approach in which biological samples are flash frozen at specific intervals along a dynamic conformational transition and visualized using cryoEM.
Footnotes
Protein Data Bank (PDB): https://www.rcsb.org/
PHENIX software: http://phenix-online.org/download/
Schrödinger software stack: https://www.schrodinger.com/downloads/releases
Stanford-SLAC Cryo-EM Center: https://cryoem.slac.stanford.edu/s2c2/
Pacific Northwest Cryo-EM Center: https://pncc.labworks.org
National Center for CryoEM Access and Training: https://nccat.nysbc.org
Diamond Light Source https://www.diamond.ac.uk
ChimeraX molecular visualization program: https://www.cgl.ucsf.edu/chimerax/
CHARMM-GUI molecular system builder: https://www.charmm-gui.org
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Byers LD and Wolfenden R (1972) A potent reversible inhibitor of carboxypeptidase A. J. Biol. Chem. 247, 606–608 [PubMed] [Google Scholar]
- 2.Patlak M (2004) From viper’s venom to drug design: treating hypertension. FASEB J. 18, 421. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin JJ et al. (1989) Thienothiopyran-2-sulfonamides: novel topically active carbonic anhydrase inhibitors for the treatment of glaucoma. J. Med. Chem. 32, 2510–2513 [DOI] [PubMed] [Google Scholar]
- 4.Eder J et al. (2014) The discovery of first-in-class drugs: origins and evolution. Nat. Rev. Drug Discov. 13, 577–587 [DOI] [PubMed] [Google Scholar]
- 5.Ostrem JM et al. (2013) K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang G et al. (2015) Discovery of Chemical Inhibitors of Human Bromodomains. Chem. Rev. 115, 11625–11668 [DOI] [PubMed] [Google Scholar]
- 7.Perera TPS et al. (2017) Discovery and Pharmacological Characterization of JNJ-42756493 (Erdafitinib), a Functionally Selective Small-Molecule FGFR Family Inhibitor. Mol. Cancer Ther. 16, 1010–1020 [DOI] [PubMed] [Google Scholar]
- 8.Pan S et al. (2013) Discovery of BAF312 (Siponimod), a Potent and Selective S1P Receptor Modulator. ACS Med. Chem. Lett. 4, 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ali A et al. (2010) Structure-based design, synthesis, and structure-activity relationship studies of HIV-1 protease inhibitors incorporating phenyloxazolidinones. J. Med. Chem. 53, 7699–7708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srivastava A et al. (2011) New target for inhibition of bacterial RNA polymerase: “switch region.” Curr. Opin. Microbiol. 14, 532–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia Y et al. (2016) Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 534, 129–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danev R et al. (2019) Cryo-Electron Microscopy Methodology: Current Aspects and Future Directions. Trends Biochem. Sci. 44, 837–848 [DOI] [PubMed] [Google Scholar]
- 13.Renaud J-P et al. (2018) Cryo-EM in drug discovery: achievements, limitations and prospects. Nat. Rev. Drug Discov. 17, 471–492 [DOI] [PubMed] [Google Scholar]
- 14.McGrath NA et al. (2010) A Graphical Journey of Innovative Organic Architectures That Have Improved Our Lives. J. Chem. Educ. 87, 1348–1349 [Google Scholar]
- 15.Shalev-Benami M et al. (2017) Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin. Nat. Commun. 8, 1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu Q et al. (2018) Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex. Cell 174, 1117–1126.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H et al. (2020) Structure of the transcription coactivator SAGA. Nature 577, 717–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonekamp NA et al. (2020) Small-molecule inhibitors of human mitochondrial DNA transcription. Nature 588, 712–716 [DOI] [PubMed] [Google Scholar]
- 19.Kruse AC et al. (2013) Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waltenspühl Y et al. (2020) Crystal structure of the human oxytocin receptor. Sci. Adv. 6, eabb5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thonghin N et al. (2018) Cryo-electron microscopy of membrane proteins. Methods 147, 176–186 [DOI] [PubMed] [Google Scholar]
- 22.Gao Y et al. (2016) TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang S et al. (2019) Structural insight into TRPV5 channel function and modulation. Proc. Natl. Acad. Sci. U. S. A. 116, 8869–8878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masiulis S et al. (2019) GABAA receptor signalling mechanisms revealed by structural pharmacology. Nature 565, 454–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alam A et al. (2019) Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363, 753–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staus DP et al. (2020) Structure of the M2 muscarinic receptor-β-arrestin complex in a lipid nanodisc. Nature 579, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denisov IG and Sligar SG (2017) Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev. 117, 4669–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Efremov RG et al. (2017) Lipid Nanodiscs as a Tool for High-Resolution Structure Determination of Membrane Proteins by Single-Particle Cryo-EM. Methods Enzymol. 594, 1–30 [DOI] [PubMed] [Google Scholar]
- 29.Yao X et al. (2020) Cryo-EM analysis of a membrane protein embedded in the liposome. Proc. Natl. Acad. Sci. U. S. A. 117, 18497–18503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang SS et al. (2019) The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1. Nat. Commun. 10, 4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeedi P et al. (2019) Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 157, 107843. [DOI] [PubMed] [Google Scholar]
- 32.Scapin G et al. (2018) Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature 556, 122–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchikawa E et al. (2019) Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. eLife 8, e48630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao Y et al. (2019) Cryo-EM Structure of HER2-trastuzumab-pertuzumab complex. PloS One 14, e0216095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinto D et al. (2020) Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 [DOI] [PubMed] [Google Scholar]
- 36.Rougé L et al. (2020) Structure of CD20 in complex with the therapeutic monoclonal antibody rituximab. Science 367, 1224–1230 [DOI] [PubMed] [Google Scholar]
- 37.Kumar A et al. (2020) Binding mechanisms of therapeutic antibodies to human CD20. Science 369, 793–799 [DOI] [PubMed] [Google Scholar]
- 38.Yamashita K et al. (2021) Cryo-EM single particle structure refinement and map calculation using Servalcat. bioRxiv DOI: 10.1101/2021.05.04.442493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Zundert GCP et al. (2020) Macromolecular refinement of X-ray and cryo-electron microscopy structures with Phenix / OPLS3e for improved structure and ligand quality. bioRxiv DOI: 10.1101/2020.07.10.198093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson MJ et al. (2020) GemSpot: A Pipeline for Robust Modeling of Ligands into Cryo-EM Maps. Structure 28, 707–716.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saur M et al. (2020) Fragment-based drug discovery using cryo-EM. Drug Discov. Today 25, 485–490 [DOI] [PubMed] [Google Scholar]
- 42.Naydenova K et al. (2019) CryoEM at 100 keV: a demonstration and prospects. IUCrJ 6, 1086–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chi X et al. (2020) A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369, 650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L et al. (2020) Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 [DOI] [PubMed] [Google Scholar]
- 45.Walls AC et al. (2020) Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181, 281–292.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan R et al. (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kokic G et al. (2021) Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 12, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakane T et al. (2020) Single-particle cryo-EM at atomic resolution. Nature 587, 152–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yip KM et al. (2020) Atomic-resolution protein structure determination by cryo-EM. Nature 587, 157–161 [DOI] [PubMed] [Google Scholar]
- 50.Ravelli RBG et al. (2020) Cryo-EM structures from sub-nl volumes using pin-printing and jet vitrification. Nat. Commun. 11, 2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klebl DP et al. (2020) Need for Speed: Examining Protein Behavior during CryoEM Grid Preparation at Different Timescales. Structure 28, 1238–1248.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rubinstein JL et al. (2019) Shake-it-off: a simple ultrasonic cryo-EM specimen-preparation device. Acta Crystallogr. 75, 1063–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noble AJ et al. (2018) Reducing effects of particle adsorption to the air-water interface in cryo-EM. Nat. Methods 15, 793–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-Azzawi A et al. (2020) DeepCryoPicker: fully automated deep neural network for single protein particle picking in cryo-EM. BMC Bioinformatics 21, 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner T et al. (2019) SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakane T et al. (2018) Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, e36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato HE et al. (2019) Conformational transitions of a neurotensin receptor 1-Gi1 complex. Nature 572, 80–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Z et al. (2020) Deep manifold learning reveals hidden dynamics of proteasome autoregulation. bioRxiv DOI: http://arxiv.org/abs/2012.12854 [Google Scholar]
- 59.Mäeots M-E et al. (2020) Modular microfluidics enables kinetic insight from time-resolved cryo-EM. Nat. Commun. 11, 3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dandey VP et al. (2020) Time-resolved cryo-EM using Spotiton. Nat. Methods 17, 897–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berman HM (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jo S et al. (2008) CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 [DOI] [PubMed] [Google Scholar]
- 63.Pettersen EF et al. (2021) UCSF CHIMERAX : Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]