Abstract
Lysine acetylation (LysAc) is a conserved and important post-translational modification (PTM) that plays a key role in plant physiological and metabolic processes. Based on advances in Lys-acetylated protein immunoenrichment and mass-spectrometric technology, LysAc proteomics studies have been performed in many species. Such studies have made substantial contributions to our understanding of plant LysAc, revealing that Lys-acetylated histones and nonhistones are involved in a broad spectrum of plant cellular processes. Here, we present an extensive overview of recent research on plant Lys-acetylproteomes. We provide in-depth insights into the characteristics of plant LysAc modifications and the mechanisms by which LysAc participates in cellular processes and regulates metabolism and physiology during plant growth and development. First, we summarize the characteristics of LysAc, including the properties of Lys-acetylated sites, the motifs that flank Lys-acetylated lysines, and the dynamic alterations in LysAc among different tissues and developmental stages. We also outline a map of Lys-acetylated proteins in the Calvin–Benson cycle and central carbon metabolism–related pathways. We then introduce some examples of the regulation of plant growth, development, and biotic and abiotic stress responses by LysAc. We discuss the interaction between LysAc and Nα-terminal acetylation and the crosstalk between LysAc and other PTMs, including phosphorylation and succinylation. Finally, we propose recommendations for future studies in the field. We conclude that LysAc of proteins plays an important role in the regulation of the plant life cycle.
Keywords: lysine acetylproteomes, modified characteristics, plant growth and development, stress responses, PTM crosstalk
Lys-acetylproteomes have shed light on global lysine acetylation in plants and have demonstrated its extensive cellular functions. Here, the characteristics of lysine-acetylated proteins, the functions of lysine acetylation in plant growth, development, and stress response, and the crosstalk between lysine acetylation and other post-translational modifications are discussed. This review deepens our understanding of how lysine acetylation-dependent cellular processes help to regulate the plant life cycle.
Introduction
Post-translational modifications (PTMs) are complex processes that modulate proteins covalently by introducing new functional groups and modifying or removing the original functional groups; these modifications occur frequently after the proteins have been fully translated (Verdin and Ott, 2015; Millar et al., 2019). Lysine acetylation (LysAc) is a highly conserved, reversible PTM of both histones and nonhistones in prokaryotes and eukaryotes (Zhang et al., 2009; Rao et al., 2014). Allfrey et al. (1964) first reported that histones could be Lys-acetylated, and nonhistone proteins, high-mobility group (HMG) proteins, and tumor suppressor p53 were subsequently found to also be Lys-acetylated (Sterner et al., 1979; Gu and Roeder, 1997). Acetyl-coenzyme A (acetyl-CoA) serves as the source of the acetyl group for LysAc in addition to its function as an important intermediate precursor for the biosynthesis of various phytochemicals (Fatland et al., 2002; Chen et al., 2017). LysAc is performed by lysine acetyltransferases (KATs) and involves the deposition of acetyl groups from acetyl-CoA onto lysine, whereas deacetylation (LysDeAc) is catalyzed by lysine deacetylases (KDACs) and involves the removal of acetyl groups from lysine (Choudhary et al., 2014; Narita et al., 2019). The first KAT and KDAC were identified in the late 1990s (Brownell et al., 1996; Taunton et al., 1996). KATs can be grouped into three major families: the GNAT, the MYST, and p300/CBP (CREB-binding protein) families (Drazic et al., 2016). KDACs can also be grouped into three families (the RPD3/HDA1-like, Sir2, and HDT families), although the HDT type occurs only in plants (De Ruijter et al., 2003). KATs and KDACs seldom operate alone but instead combine with various subunits that define their substrate specificities and catalytic activities, thus forming multiprotein complexes (Shahbazian and Grunstein, 2007; Drazic et al., 2016). In general, LysAc masks positively charged lysine residues on proteins, disturbs ionic and hydrogen bonding, and increases protein hydrophobicity, thereby affecting the structures, functions, and activities of the target proteins, as well as their interactions with other biomolecules, including DNA and proteins (Choudhary et al., 2009; Wang et al., 2010; Zhao et al., 2010; Lehtimaki et al., 2015).
The best-known effects of LysAc are those that affect chromatin structure and gene expression through histone modification (Eberharter and Becker, 2002). LysAc decreases the affinity of histones, which generates a loose chromatin structure and promotes transcriptional activation, whereas LysDeAc leads to chromatin contraction and transcriptional inhibition (Grunstein, 1997; Struhl, 1998). In addition to nuclear substrates (e.g., histones, transcription factors [TFs], transcriptional coregulators), nonnuclear proteins/enzymes that participate in various biological processes, especially cellular metabolic processes, are also Lys-acetylated/deacetylated. This extends the functions of LysAc/LysDeAc from epigenetic control of chromatin dynamics and gene transcription to the regulation of cellular metabolism (Hentchel and Escalante-Semerena, 2015; Verdin and Ott, 2015; Chen et al., 2018). Hence, LysAc exerts key effects on various biological processes.
Early LysAc investigations focused mainly on histones (Law and Suttle, 2004; Shahbazian and Grunstein, 2007; Hollender and Liu, 2008). In 2006, Kim et al. first performed LysAc proteomics to analyze the LysAc regulatory network of HeLa cells and liver mitochondria of Mus musculus. Subsequently, numerous nonhistone proteins, including TFs, RNA splicing factors, chaperones, signal proteins, and cytoplasmic metabolic enzymes, were found to be Lys-acetylated. As a system-wide approach, LysAc proteomics enables the detection of Lys-acetylated proteins and sites and reveals that LysAc events occur extensively in nonhistones (Choudhary et al., 2009; Wang et al., 2010; Zhao et al., 2010). In 2011, the establishment of the Compendium of Protein Lysine Acetylation provided valuable information for elucidating the mechanism of LysAc regulation (Liu et al., 2011).
The development of LysAc proteomics occurred later in plants than in animals or microorganisms. Wu et al. (2011) and Finkemeier et al. (2011) first performed plant Lys-acetylproteome analyses in Arabidopsis thaliana. To date, plant Lys-acetylproteome analyses have focused mainly on higher plants, including A. thaliana (Finkemeier et al., 2011; Wu et al., 2011; Koenig et al., 2014; Hartl et al., 2017; Uhrig et al., 2017; Liu et al., 2018; Koskela et al., 2018; Bienvenut et al., 2020), Vitis vinifera (Melo-Braga et al., 2012; Liu et al., 2019), Pisum sativum (Smith-Hammond et al., 2014a), Glycine max (Smith-Hammond et al., 2014b; Li et al., 2021a), Oryza sativa (Nallamilli et al., 2014; He et al., 2016; Xiong et al., 2016; Wang et al., 2017; Li et al., 2018a, 2018b; Meng et al., 2018; Xue et al., 2018; Zhou et al., 2018), Fragaria ananassa (Fang et al., 2015), Medicago truncatula (Marx et al., 2016), Triticum aestivum (Zhang et al., 2016; Zhu et al., 2018; Guo et al., 2020), Brachypodium distachyon (Zhen et al., 2016), Picea asperata (Xia et al., 2016), Camellia sinensis (Xu et al., 2017; Jiang et al., 2018), Zea mays (Walley et al., 2018; Yan et al., 2020), Kandelia candel (Pan et al., 2018), Gossypium hirsutum (Singh et al., 2020), Hibiscus cannabinus (Chen et al., 2019), Paulownia tomentosa (Cao et al., 2019), Petunia hybrida (Zhao et al., 2020), Nicotiana benthamiana (Yuan et al., 2021), Populus tremula × Populus alba (Liao et al., 2021), Broussonetia papyrifera (Li et al., 2021b), and Phoebe zhennan (Zhao et al., 2021) (Table 1). By contrast, the Lys-acetylproteomes of lower plants are poorly studied and have been documented only in Phaeodactylum tricornutum (Chen et al., 2018) and Physcomitrium patens (Balparda et al., 2021). The Lys-acetylated proteins and sites detected by qualitative or quantitative LysAc proteomics techniques provide an overview of LysAc events in plants and serve as a foundation for further functional analysis.
Table 1.
Summary of Lys-acetylproteomes in plant species and Lys-acetylated sites.
| Species | Tissues/organs | Biotic stress | Abiotic stress | Number of Lys-acetylated proteins/sites/average sites | Reference |
|---|---|---|---|---|---|
| A. thaliana | Leaves | — | — | 55/64/1.16 | Wu et al. (2011) |
| A. thaliana | Leaves | — | — | 74/91/1.23 | Finkemeier et al. (2011) |
| A. thaliana | Mitochondria | — | — | 120/243/2.03 | Koenig et al. (2014) |
| A. thaliana | Leaves | — | — | 1022/2152/2.11 | Hartl et al. (2017) |
| A. thaliana | Seedlings | — | — | 909/1365/1.50 | Uhrig et al. (2017) |
| A. thaliana | Seedlings | — | — | 2638/5233/1.98 | Liu et al. (2018) |
| A. thaliana | Chloroplast | — | — | –/–/– | Koskela et al. (2018) |
| B. distachyon | Leaves | — | — | 353/6361.80 | Zhen et al. (2016) |
| B. papyrifera | Leaves | — | — | 3179/7130/2.24 | Li et al. (2021b) |
| C. sinensis | Leaves | — | — | 1752/3161/1.80 | Xu et al. (2017) |
| C. sinensis | Leaves | — | N starvation | 1286/2229/1.73 | Jiang et al. (2018) |
| F. ananassa | Leaves | — | — | 684/1392/2.04 | Fang et al. (2015) |
| G. hirsutum | Ovule | — | — | 1696/2754/1.62 | Singh et al. (2020) |
| G. max | Seeds | — | — | 245/400/1.63 | Smith-Hammond et al. (2014b) |
| G. max | Leaves | — | — | 1538/3148/2.05 | Li et al. (2021a) |
| H. cannabinus | Anther | — | — | 672/1204/1.79 | Chen et al. (2019) |
| K. candel | Leaves | — | Flooding | 617/1041/1.69 | Pan et al. (2018) |
| M. truncatula | Nodules | — | — | 734/–/– | Marx et al. (2016) |
| N. benthamiana | Leaves | Chinese wheat mosaic virus | — | 1964/4803/2.45 | Yuan et al. (2021) |
| O. sativa | Suspension cells | — | — | 44/60/1.36 | Nallamilli et al. (2014) |
| O. sativa | Seeds | — | — | 389/699/1.80 | He et al. (2016) |
| O. sativa | Leaves, stems, roots | — | — | 716/1337/1.88 | Xiong et al. (2016) |
| O. sativa | Seeds | — | — | 972/1817/1.87 | Wang et al. (2017) |
| O. sativa | Anther | — | — | 676/1354/2.00 | Li et al. (2018a) |
| O. sativa | Seeds | — | — | 692/1003/1.45 | Meng et al. (2018) |
| O. sativa | Callus, root, leaves, panicle | — | — | 890/1536/1.73 | Li et al. (2018b) |
| O. sativa | Leaves | — | Cold | 866/1353/1.56 | Xue et al. (2018) |
| O. sativa | Leaves | — | Oxidation | 1024/1669/1.63 | Zhou et al. (2018) |
| P. asperata | Embryo | — | — | 556/1079/1.94 | Xia et al. (2016) |
| P. hybrida | Corollas | — | — | 1148/2210/1.93 | Zhao et al. (2020) |
| P. patens | Gametophores | — | — | 638/–/– | Balparda et al. (2021) |
| P. tomentosa | Seedlings | Phytoplasma | — | 2893/5558/1.92 | Cao et al. (2019) |
| P. tremula × P. alba | Dormant buds | — | — | 3281/7594/2.31 | Liao et al. (2021) |
| P. tricornutum | Cells | — | — | 1220/2324/1.90 | Chen et al. (2018) |
| P. zhennan | Leaves | — | Drought | –/–/– | Zhao et al. (2021) |
| T. aestivum | Leaves | — | — | 277/416/1.50 | Zhang et al. (2016) |
| T. aestivum | Seeds | — | Drought | 442/716/1.62 | Zhu et al. (2018) |
| T. aestivum | Seeds | — | — | 722/1301/1.80 | Guo et al. (2020) |
| V. vinifera | Mesocarp and exocarp | Lobesia botrana | — | 97/138/1.42 | Melo-Braga et al. (2012) |
| V. vinifera | Leaves | — | Heat | 510/1135/2.23 | Liu et al. (2019) |
| Z. mays | Leaves | Cochliobolus carbonum | — | 912/2791/3.06 | Walley et al. (2018) |
| Z. mays | Leaves | — | — | 462/814/1.76 | Yan et al. (2020) |
In this paper, we review advances in plant Lys-acetylproteomes, focusing on the following three aspects: characteristics of Lys-acetylated proteins; functions of LysAc in plant growth, development, and stress response; and crosstalk between LysAc and other PTMs. We aim to present references for elucidating plant LysAc regulatory mechanisms and to provide perspectives for future research.
Characteristics of Lys-acetylated proteins
Distribution of Lys-acetylated sites
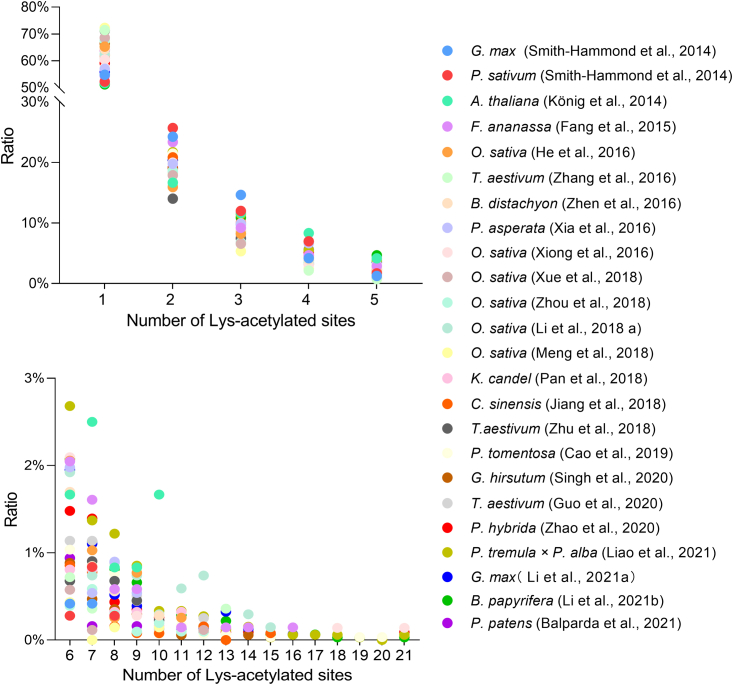
Relatively low numbers of plant Lys-acetylated proteins and sites were identified in early studies because of limitations associated with mass spectrometry and protein fractionation techniques, specificity of the anti-acetyl-lysine antibody, effects of the cell wall on protein extraction, and interference of plant secondary metabolites during protein affinity purification (Schilling et al., 2012; Nallamilli et al., 2014). Early work therefore identified fewer than 100 Lys-acetylated proteins, and the average number of Lys-acetylated sites per protein was 1.16–1.36 (Finkemeier et al., 2011; Wu et al., 2011; Nallamilli et al., 2014) (Table 1). With tremendous innovation and optimization of LysAc proteomics technologies, the numbers of identified Lys-acetylated sites are increasing, and the average number of Lys-acetylated sites detected per plant protein has increased to 1.50–3.06 (Table 1). A single Lys-acetylated protein typically contains 1–9 Lys-acetylated sites; proteins with 1–5 modified sites account for 92.35%–99.28% of all Lys-acetylated proteins, and proteins with only one Lys-acetylated site account for the largest proportion of Lys-acetylated proteins (49.83%–72.25%) (Figure 1 and Supplemental Table 1). A light-harvesting complex II (LHCII) protein identified in A. thaliana leaves contained the highest number of Lys-acetylated sites (29 sites) reported in any plant Lys-acetylproteome (Hartl et al., 2017).
Figure 1.
Distribution of Lys-acetylated sites in a single protein identified in plant Lys-acetylproteomes.
Detailed information can be found in Supplemental Table 1.
Lys-acetylated proteins with multiple modified sites
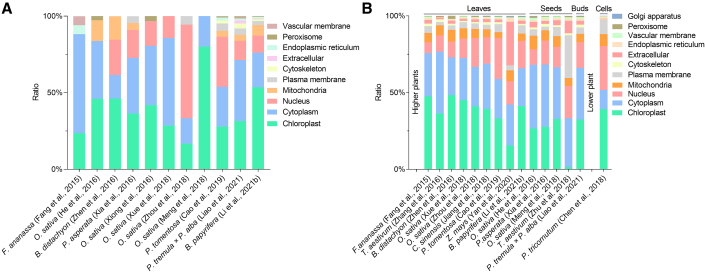
Because most Lys-acetylated proteins contain 1–5 Lys-acetylated sites (Figure 1 and Supplemental Table 1), it is interesting to obtain an overview of Lys-acetylated proteins with multiple modified sites. Here, we summarize some plant Lys-acetylproteomes to characterize the Lys-acetylated proteins with six or more modified sites. We find that these proteins include histones and nonhistone proteins and are distributed mainly in the chloroplast, nucleus, and cytoplasm (Figure 2A).
Figure 2.
Distribution of Lys-acetylated proteins across subcellular compartments.
(A) Distribution of proteins with multiple Lys-acetylated sites across subcellular compartments.
(B) Distribution of Lys-acetylated proteins identified in whole Lys-acetylproteomes across subcellular compartments. Leaves, seeds, buds, and cells indicate the materials analyzed to produce the plant Lys-acetylproteomes.
Very few Lys-acetylated sites have been identified in histone H1 and no common site has been reported in the literature. Numerous Lys-acetylated sites have been identified in H2A and H2B. However, because of the diversity of H2A and H2B tail sequences (Kawashima et al., 2015), the Lys-acetylated sites of the two histones are species- or tissue-specific. LysAc of H3 and H4 histones is a euchromatin modification (Jeon et al., 2014), and relatively fewer Lys-acetylated sites have been identified in these histones. Lys-acetylated sites in H3 and H4 show high conservation in different plant species and tissues. Although the Lys-acetylated sites detected in histones H2A and H2B are less conserved, continuous Lys-acetylated sites have been identified in these two histones, such as K10–K22 and K124–K155 in histone H2A and K7–K89 in histone H2B (Xia et al., 2016; Zhen et al., 2016; Singh et al., 2020). In brief, LysAc of histones H3 and H4 is conserved, whereas LysAc of histones H2A and H2B varies among plant species and developmental stages. Similar patterns have also been detected in animals and microorganisms (Zhang et al., 2013; Kwon et al., 2016).
Ribosomal proteins (RPs), elongation factors (EFs), and heat-shock proteins (HSPs) are the most commonly Lys-acetylated nonhistone proteins, with more than six modified sites. The 60S large ribosomal subunits and 40S small ribosomal subunits are the major proteins within the RP group and are distributed mainly in the chloroplast and cytoplasm, respectively. Both RPs and EFs contain many conserved Lys-acetylated sites. For example, K120, K204, K348, K359, and K368 of 60S RP L3 are highly conserved in O. sativa (Wang et al., 2017; Meng et al., 2018), G. hirsutum (Singh et al., 2020), P. asperata (Xia et al., 2016), and F. ananassa (Fang et al., 2015), whereas K232, K291, K427, and K482 of EF2 are highly conserved in O. sativa (He et al., 2016; Wang et al., 2017; Meng et al., 2018; Xue et al., 2018; Zhou et al., 2018), G. hirsutum (Singh et al., 2020), F. ananassa (Fang et al., 2015), and C. sinensis (Jiang et al., 2018). This suggests that LysAc is probably necessary for the regulation of protein synthesis and assembly. A KDAC (HDA714) has been shown to target RPs for LysDeAc, which is likely to affect the stability of the ribosome and its translational efficiency (Xu et al., 2021). However, rare homologous Lys-acetylated HSPs or conserved Lys-acetylated sites have been found in the current study.
Numerous Lys-acetylated sites have also been detected in chloroplast proteins, e.g., structural proteins corresponding to photosystems I and II (Xiong et al., 2016), ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) (Fang et al., 2015; Zhen et al., 2016; Wang et al., 2017; Xue et al., 2018), chlorophyll a/b-binding proteins (Xiong et al., 2016), chloroplast stem-loop-binding proteins (Fang et al., 2015; Jiang et al., 2018), oxygen-evolving enhancer proteins (Fang et al., 2015; Xiong et al., 2016), and enzymes involved in carbon assimilation, such as phosphoglycerate kinase (PGK) (He et al., 2016; Xia et al., 2016; Xiong et al., 2016; Zhen et al., 2016; Meng et al., 2018), fructose-bisphosphate aldolase (FBA) (Fang et al., 2015; Xia et al., 2016; Xiong et al., 2016; Wang et al., 2017; Li et al., 2018a), and sedoheptulose-bisphosphatase (SBP) (Fang et al., 2015). Stress-responsive proteins such as 14-3-3 protein (Li et al., 2018a), catalase (CAT) (Xiong et al., 2016), glutathione peroxidase (GPX) (Fang et al., 2015), and modified enzymes associated with other PTMs, e.g., phosphorylase (Meng et al., 2018) and methylase (Xiong et al., 2016; Liu et al., 2019), also possess multiple Lys-acetylated sites. However, whether LysAc has an effect on the functions of target proteins requires further verification.
Motif characterization of Lys-acetylated peptides
LysAc is usually distributed along the whole protein sequence and occurs around preferred amino acid residues. The protein sequence motifs of Lys-acetylated lysine residues are conserved in various plant species, tissues, or organs. Analyses of the motif model and the preference for amino acid residues surrounding Lys-acetylated sites can deepen our understanding of LysAc patterns. To date, analyses of LysAc motifs have mainly targeted all the identified LysAc peptides in Lys-acetylproteomes. KacH, KacY, KacF, KacK, KacR, KacT, KacS, F∗Kac, and KacN motifs (Kac denotes a Lys-acetylated lysine residue, an asterisk [∗] indicates a random amino acid residue, and the number of asterisks indicates the number of random amino acids in the motif) are highly conserved in different plants (Table 2). Most of the conserved residues are located at the −2 to +1 positions when the Lys-acetylated site is considered to occupy the 0 position. Significant enrichment has been detected for Y and H at +1 (He et al., 2016; Zhang et al., 2016; Zhen et al., 2016; Wang et al., 2017), L at −1 (Zhang et al., 2016), F at −2 to +2 (He et al., 2016; Xiong et al., 2016), V at −2 (Xia et al., 2016), and R from −8 to −4 and +2 to +8 (Meng et al., 2018), whereas K is generally excluded from −1.
Table 2.
Summary of the LysAc motifs in plant Lys-acetylproteomes.
| Species | Motif sequence | Reference |
|---|---|---|
| A. thaliana | KacY, KacF, F∗Kac, D∗Kac | Uhrig et al. (2017) |
| B. distachyon | KacH, KacY, KacF, Kac∗∗∗K, Kac∗I∗K | Zhen et al. (2016) |
| B. papyrifera | F∗Kac, Kac∗K, Kac∗H, Kac∗F, Kac∗R, Kac∗Y, Kac∗S, Kac∗T, Kac∗N, Kac∗D, Kac∗V, Kac∗W, Y∗Kac, T∗Kac, D∗KacR, Y∗KacS | Li et al. (2021b) |
| C. sinensis | KacH, KacF, KacR, KacK, KacT, KacS, KacN, Kac∗K, Kac∗∗K, K∗∗∗∗∗∗KacK, K∗∗∗∗∗∗∗∗KacK, Kac∗R, R∗∗∗∗∗∗KacK, Kac∗D, Kac∗E | Xu et al. (2017) |
| C. sinensis | KacH, KacK, KacR, KacT, KacS, KacN, Kac∗K, Kac∗∗K, Kac∗∗∗∗K, Kac∗∗∗∗∗∗K, Kac∗R, E∗∗KacK, EKac∗K, KacH∗K, Kac∗E, Kac∗D | Jiang et al. (2018) |
| F. ananassa | KacH, KacY, KacF, F∗Kac, L∗Kac | Fang et al. (2015) |
| G. hirsutum | KacH, KacF, KacK, KacR, KacT, KacS, KacN, KacV, RKacS, KacTE, KacVD, Kac∗E, Kac∗D, KacS∗∗∗∗∗K, A∗KacK, P∗KacK, C∗∗∗KacT | Singh et al. (2020) |
| G. max | KacH, KacF, KacK, KacR, KacT, KacS, KacN, KacKA, KacAK, KacRL, Kac∗K, Kac∗∗K, Kac∗D, Kac∗E, Kac∗R, Kac∗∗R, KacT∗∗∗∗∗∗∗∗K | Li et al. (2021a) |
| H. cannabinus | KacK, KacR, KKac, K∗∗Kac, K∗∗∗∗∗∗Kac, K∗∗∗Kac, K∗∗∗∗Kac, K∗∗∗∗∗Kac, Kac∗∗K, Kac∗∗∗K, Kac∗∗∗∗K, Kac∗∗∗∗∗K, Kac∗∗∗∗∗∗K, Kac∗∗A | Chen et al. (2019) |
| K. candel | KacK, K∗∗∗∗Kac, Kac∗∗∗∗K, KacR, KKac, EKac | Pan et al. (2018) |
| N. benthamiana | F∗Kac, D∗Kac, Kac∗K, Kac∗H, Kac∗F, Kac∗C, Kac∗A, Kac∗R, Kac∗Y, H∗Kac, C∗Kac, A∗Kac, V∗Kac∗K | Yuan et al. (2021) |
| O. sativa | KacH, KacY, KacF, F∗Kac, Kac∗∗∗∗R, Kac∗F | He et al. (2016) |
| O. sativa | KacH, KacY, KacF, F∗Kac, L∗Kac, Kac∗ F, FKac | Xiong et al. (2016) |
| O. sativa | KacH, KacY, KacT, F∗Kac, YKac, D∗KacK | Wang et al. (2017) |
| O. sativa | KacH, KacY, KacF, Kac∗∗∗K, K∗∗∗∗∗∗∗∗Kac, Kac∗∗∗R, Kac∗∗∗∗R | Meng et al. (2018) |
| O. sativa | KacH, KacY, KacT, KacS, F∗Kac, Kac∗∗∗∗∗∗K, Kac∗R, YKac, D∗KacK | Li et al. (2018b) |
| O. sativa | KacH, KacY, KacF, Kac∗∗∗K, K∗∗∗∗∗∗∗∗Kac, FKac, Kac∗I∗R, D∗∗Kac, Kac∗L∗R, KacF∗R, KacF∗∗R | Xue et al. (2018) |
| O. sativa | KacH, KacK, KacR, KacT, KacS, KacN, Kac∗K, Kac∗∗K, Kac∗∗∗∗∗∗K, K∗∗∗∗∗∗KacK | Zhou et al. (2018) |
| P. asperata | KacH, KacY, F∗Kac, K∗∗∗∗∗∗∗∗∗Kac, Kac∗F, YKac, V∗Kac | Xia et al. (2016) |
| P. hybrida | KacH, KacF, KacK, KacR, KacT, KacS, KacN, FKac, DKac, AKacK, Kac∗E, Kac∗D | Zhao et al. (2020) |
| P. tomentosa | KacH, KacK, KacR, KacT, KacS, KacN, K∗∗∗∗∗∗∗∗KacK, K∗∗∗∗∗∗∗KacK, K∗∗∗∗∗∗KacK, Kac∗K, Kac∗∗K, KacAK, Kac∗R, Kac∗D, Kac∗E, AKacK | Cao et al. (2019) |
| T. aestivum | KacH, KacY, KacF, LKac, FKac | Zhang et al. (2016) |
| P. tricornutum | KacH, KacY, KacF, FKac, LKac, YKac, LKacY, KacW, Kac∗F, Kac∗Y, Kac∗L, K∗∗∗Kac, K∗∗∗∗Kac, K∗∗∗∗∗Kac, I∗Kac∗L, I∗Kac, F∗∗Kac | Chen et al. (2018) |
| P. zhennan | GKacS、VKacS、LKacN、TKacV、NKacV、SKacV、D∗∗KacR、 YKacV、VKacK、NKacA | Zhao et al. (2021) |
| T. aestivum | KacK, K∗∗∗∗∗∗∗∗KacK, K∗∗∗∗∗∗∗∗∗KacK, Kac∗H | Zhu et al. (2018) |
| T. aestivum | KacH, KacF, KacK, KacR, KacT, KacS, KacN, FKac, Kac∗E, Kac∗D | Guo et al. (2020) |
| V. vinifera | KacY | Melo-Braga et al. (2012) |
| Z. mays | KacY, KacF, KacK, KacR, Kac∗∗R, AEKac, GKacK, EKac, KacL, DKac | Yan et al. (2020) |
Kac denotes Lys-acetylated lysine residue, asterisk (∗) indicates a random amino acid residue, and the number of asterisks indicates the number of random amino acids in the motif.
In histones, K is enriched across all LysAc motifs and is significantly enriched at +1, in contrast to Y, H, and F, which are enriched at +1 in the global Lys-acetylproteomes described above (Li et al., 2018a). E is enriched at the −1 and −3 positions in mitochondrial Lys-acetylated proteins of A. thaliana and P. sativum (Koenig et al., 2014; Smith-Hammond et al., 2014a). In the developing anthers of O. sativa, both T and D are significantly enriched at −1 in Lys-acetylated cytoplasmic proteins and nuclear proteins (Li et al., 2018a). There is a difference in LysAc motifs between histone and nonhistone proteins, and the LysAc motifs in certain subcellular Lys-acetylated proteins differ from those in the complete Lys-acetylproteome. Therefore, it is necessary to perform a compartment-specific LysAc motif analysis. In addition, residues such as Y, F, and H and some LysAc motifs are also enriched in human cells (Choudhary et al., 2014), Escherichia coli (Zhang et al., 2009), and plant pathogens such as Phytophthora sojae (Li et al., 2016). This indicates that plants and other organisms share commonly conserved LysAc motifs and LysAc events.
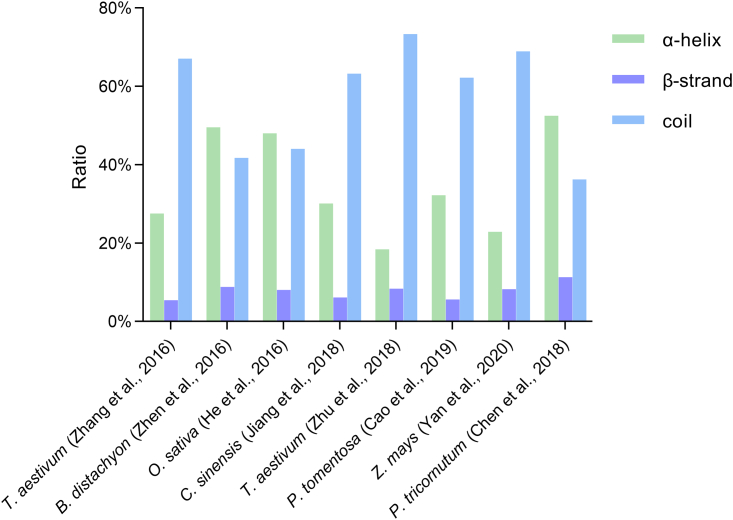
Predictions of the protein secondary structures that surround Lys-acetylated lysines reveal distinct distribution patterns in different plant species. More than 60% of Lys-acetylated sites are found in coils followed by α-helix and β-strand regions (Zhang et al., 2016; Jiang et al., 2018; Zhu et al., 2018; Cao et al., 2019; Yan et al., 2020), and more Lys-acetylated sites are found in the α helix than in the coil region in the Lys-acetylproteomes of O. sativa seeds (He et al., 2016) and B. distachyon leaves (Zhen et al., 2016) (Figure 3).
Figure 3.
Distribution of protein secondary structures surrounding Lys-acetylated sites (α helix, β strand, and coil).
Dynamic alterations of LysAc in different tissues, developmental stages, and conditions
In A. thaliana, the molecular weights and abundances of Lys-acetylated proteins differ among shoots, leaves, flowers, seeds, and roots (Wu et al., 2011), and the LysAc levels of roots and seedlings change dramatically during the diurnal cycle (Uhrig et al., 2017). In O. sativa, LysAc levels were higher in callus, leaves, and panicles than in roots (Li et al., 2018b), and post-anthesis seeds exhibited higher LysAc levels than flowers and pollen (Meng et al., 2018). In dormant buds of P. tremula × P. alba, there was a slight decrease in LysAc during dormancy release (Liao et al., 2021). Within 0–48 h after imbibition of O. sativa seeds, LysAc reached a higher level at 24 h (He et al., 2016). LysAc levels in radicles of P. asperata were higher at 14 days after partial desiccation treatment than at 0, 7, and 21 days (Xia et al., 2016). The LysAc levels in G. hirsutum ovules also changed from −1 to 0 days post anthesis (Singh et al., 2020).
Under drought stress, T. aestivum seeds showed enhanced LysAc levels at 20 days after flowering compared with 10, 15, 25, and 30 days, and the LysAc signal under drought stress was stronger than that under sufficient water conditions (Zhu et al., 2018). Increased LysAc levels were also detected in drought-stressed leaves of P. zhennan (Zhao et al., 2021) and virus-infected N. benthamiana (Yuan et al., 2021). Distinct LysAc was detected under nitrogen-, phosphorus-, and iron-deficient conditions in P. tricornutum (Chen et al., 2018). These results demonstrate that dynamic changes in LysAc differ among different tissues, developmental stages, and stresses, suggesting that LysAc is likely to play an important role in the regulation of plant growth.
Subcellular locations of Lys-acetylated proteins
Wu et al. (2011) localized Lys-acetylated proteins to the nucleus, plasma membrane, and chloroplast, whereas the chromocenter was hypo-Lys-acetylated. Subcellular localization predictions reveal that the number of Lys-acetylated proteins varies across subcellular compartments among plant species, tissues, and developmental stages (Fang et al., 2015; He et al., 2016; Xia et al., 2016; Xiong et al., 2016; Zhang et al., 2016; Zhen et al., 2016; Chen et al., 2018; Jiang et al., 2018; Li et al., 2018b, 2021b; Meng et al., 2018; Xue et al., 2018; Zhou et al., 2018; Zhu et al., 2018; Cao et al., 2019; Yan et al., 2020; Liao et al., 2021) (Figure 2B). Almost 90% of Lys-acetylated proteins are located in the chloroplast, cytoplasm, nucleus, and mitochondria of plant cells. However, in the lower plant P. tricornutum, more Lys-acetylated proteins are located in the nucleus than in the cytoplasm, and a relatively larger number of Lys-acetylated proteins are located in the plasma membrane compared with higher plants (Chen et al., 2018). Hence, the pattern of subcellular localization of Lys-acetylated proteins varies throughout plant species. The subcellular localization of Lys-acetylated proteins is also closely associated with transient plant growth or metabolic status (Jiang et al., 2018). For instance, an increased ratio of Lys-acetylated proteins located in the cell membrane and extracellular space was observed in T. aestivum seeds under drought conditions (Zhu et al., 2018).
Numerous proteins related to photosystem assembly, chlorophyll biosynthesis, and carbon assimilation are Lys-acetylated, suggesting that LysAc has a marked effect on chloroplast structure and photosynthetic processes (Fang et al., 2015; Zhen et al., 2016; Jiang et al., 2018). Koskela et al. (2018) identified the first chloroplast stroma-localized KAT, NUCLEAR SHUTTLE INTERACTING (NSI), in A. thaliana and determined that NSI was essential for dynamically reorganizing the photosynthetic state transitions of thylakoid protein complexes. Analysis of the chloroplast Lys-acetylome demonstrated that several specific photosynthetic proteins (e.g., PSBP-1, PSAH-1/2, LHCB1.4, KEA1, and KEA2) had decreased LysAc levels in the nsi mutant compared with the wild type. The LysAc level of K88 in PSBP-1 decreased more than 12-fold compared with the wild type (Koskela et al., 2018). In addition, some thylakoid proteins, such as LHCB6 and the ATPase β-subunit, had increased LysAc levels in the nsi mutant, suggesting that there is interplay between the LysAc of different proteins in the chloroplast (Koskela et al., 2018). Schmidt et al. (2017) extracted chloroplast ATP synthase from spinach chloroplasts and found that nine protein subunits, with the exception of membrane-embedded subunit III, were Lys-acetylated. However, systematic analyses of chloroplast Lys-acetylproteomes are still largely lacking in plants.
Plant mitochondria participate in biological processes and play key roles in the regulation of acetyl-CoA metabolism (Hartl and Finkemeier, 2012; Schwarzlaender et al., 2012; Xing and Poirier, 2012). Salvato et al. (2014) first reported the plant mitochondrial Lys-acetylproteome of the Solanum tuberosum tuber; however, only 3% of the mitochondrial proteins were Lys-acetylated. Later, 120 and 93 Lys-acetylated proteins were detected in A. thaliana and P. sativum mitochondria, respectively (Koenig et al., 2014; Smith-Hammond et al., 2014a). Approximately half of the Lys-acetylated proteins in P. sativum mitochondria were involved in primary metabolism (Smith-Hammond et al., 2014a). Proteins in complex V of the respiratory chain were strikingly Lys-acetylated compared with those of the other complexes in A. thaliana (Koenig et al., 2014). Comparative gene ontology (GO) term analysis of mitochondrial Lys-acetylated proteins from A. thaliana, O. sativa, M. musculus, and Homo sapiens indicated that 138 GO terms overlapped in these species, especially proteins in the tricarboxylic acid (TCA) cycle, mitochondrial electron transport chain, and ATP synthase (Hosp et al., 2017). This indicates a possible evolutionarily conserved role of LysAc in regulating the functions and activities of mitochondrial proteins and maintaining the operation of the TCA cycle. Balparda et al. (2021) also pointed out that more protein LysAc events occurred in TCA cycle enzymes and pyruvate decarboxylase (PDC) in both P. patens and A. thaliana. In addition, because of the relatively unique alkaline environment and elevated acetyl-CoA levels of mitochondria (Wagner and Payne, 2013), nonenzymatic LysAc occurs in mitochondrial proteins of A. thaliana in vitro; it is independent of KAT and occurs even when the mitochondria are denatured (Koenig et al., 2014). Hence, enzymatic and nonenzymatic patterns of LysAc are present in plant mitochondria.
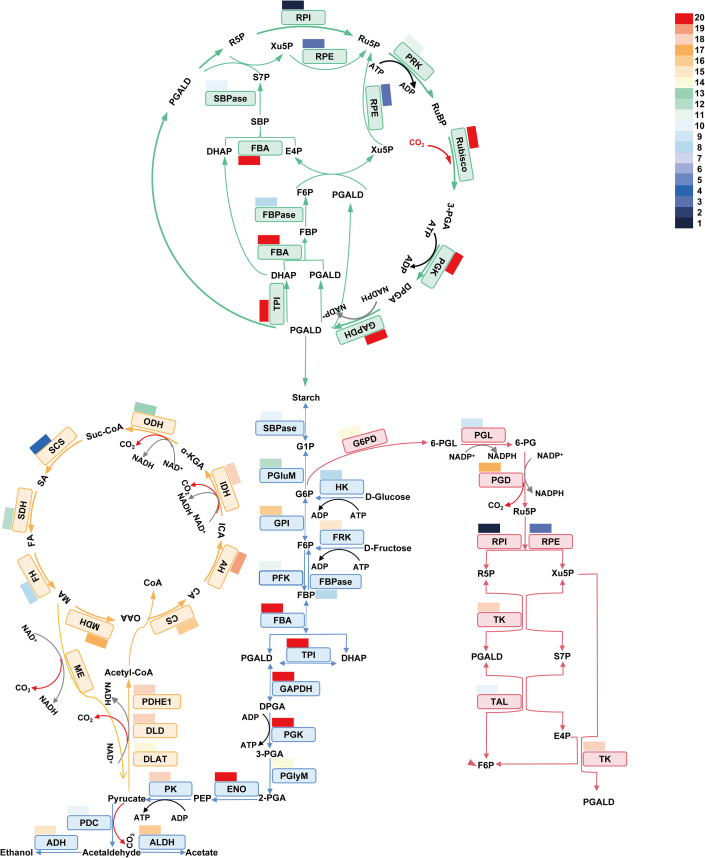
Characteristics of Lys-acetylated proteins in the Calvin–Benson cycle and central carbon metabolism
Increasing evidence has shown that proteins relevant to photosynthesis and carbon metabolism are extensively Lys-acetylated (Finkemeier et al., 2011; Wu et al., 2011; Fang et al., 2015; Xiong et al., 2016). Acetyl-CoA and NAD+ are key factors in cellular metabolic processes and are required for the catalysis of LysAc/LysDeAc (Choudhary et al., 2014; Baeza et al., 2016). To probe the LysAc landscape of enzymes that participate in the Calvin–Benson cycle and plant central carbon metabolism, we summarized the profiles of related enzymes from 20 reported plant Lys-acetylproteomes. As shown in Figure 4, enzymes of the Calvin–Benson cycle, glycolysis, and the TCA cycle are more strongly modified than those of the pentose phosphate pathway. Almost all of the enzymes involved in glycolysis and the TCA cycle undergo LysAc, consistent with reports in humans, animals, and microorganisms.
Figure 4.
Lys-acetylated model of proteins involved in the Calvin–Benson cycle and central carbon metabolism.
The Lys-acetylated enzymes relevant to the Calvin–Benson cycle and central carbon metabolism summarized from 20 Lys-acetylproteomes are noted with boxes of different colors to reflect their frequency of modification by LysAc. PRK, phosphoribulokinase; Rubisco, ribulose bisphosphate carboxylase/oxygenase; PGK, phosphoglycerate kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; TPI, triosephosphate isomerase; FBA, fructose-bisphosphate aldolase; FBPase, fructose-1,6-bisphosphatase; SBPase, sedoheptulose-1,7-bisphosphatase; RPE, ribulose phosphate epimerase; RPI, ribose-5-phosphate isomerase; PGluM, phosphoglucomutase; HK, hexokinase; GPI, glucose-6-phosphate isomerase; FRK, fructokinase; PFK, phosphofructokinase; PGlyM, 2,3-bisphosphoglycerate-independent phosphoglycerate mutase; ENO, enolase; PK, pyruvate kinase; PDC, pyruvate decarboxylase; ALDH, acetaldehyde dehydrogenase; ADH, alcohol dehydrogenase; PDHE1, pyruvate dehydrogenase complex E1 subunit; DLD, dihydrolipoyl dehydrogenase; DLAT, dihydrolipoamide acetyltransferase; CS, citrate synthase; AH, aconitate hydratase; IDH, isocitrate dehydrogenase; ODH, oxoglutarate dehydrogenase; SCS, succinyl-CoA synthetase; SDH, succinate dehydrogenase; FH, fumarate hydratase; ME, malic enzyme; MDH, malate dehydrogenase; G6PD, glucose-6-phosphate 1-dehydrogenase; PGL, 6-phosphogluconolactonase; PGD, 6-phosphogluconate dehydrogenase; TK, transketolase; TAL, transaldolase; R5P, ribose-5-phosphate; Ru5P, ribulose-5-phosphate; RuBP, ribulose-1,5-bisphosphate; 3-PGA, 3-phosphoglycerate; DPGA, 1,3-disphosphoglycerate; PGALD, 3-phosphoglyceraldehyde; DHAP, dihydroxyacetone phosphate; FBP, fructose-1,6-bisphosphate; F6P, fructose-6-phosphate; E4P, erythrose-4-phosphate; SBP, sedoheptulose-1,7-bisphosphate; S7P, sedoheptulose-7-phosphate; Xu5P, xylulose-5-phosphate; G1P, glucose-1-phosphate; G6P, glucose-6-phosphate; 2-PGA, 2-phosphoglycerate; PEP, phosphoenolpyruvate; OAA, oxaloacetate; CA, cis-aconitate; ICA, isocitrate; α-KGA, 2-oxoglutarate; SA, succinate; FA, fumarate; MA, malate; 6-PGL, 6-phosphoglucono-1,5-lactone; 6-PG, 6-phosphogluconate.
In the Calvin–Benson cycle, enzymes involved in the carboxylation and reduction of CO2 are strongly Lys-acetylated, whereas those involved in the regeneration of ribulose-1,5-bisphosphate (RuBP) show less modification (Figure 4). In glycolysis, enzymes associated with the phases of hexose phosphate cleavage and ATP and pyruvate production show greater LysAc levels than enzymes in the hexose phosphorylation phase. Isozymes (e.g., PGK, FBA, glyceraldehyde-3-phosphate dehydrogenase [GAPDH], and triosephosphate isomerase [TPI]) with chloroplast or cytosolic subcellular localization that participate in the Calvin–Benson cycle and glycolysis may be Lys-acetylated in both compartments simultaneously or in only one compartment. In the TCA cycle, LysAc is more common in proteins relevant to citric acid synthesis and oxidative decarboxylation than in proteins that participate in oxaloacetic acid regeneration. Enzymes of plant alcohol fermentation, such as PDC, acetaldehyde dehydrogenase, and alcohol dehydrogenase, are also Lys-acetylated. The pyruvate dehydrogenase complex is considered to be involved in the oxidative decarboxylation of pyruvate and the generation of acetyl-CoA, thereby linking the pathways of β-oxidation, glycolysis, and the TCA cycle (Milne et al., 2002). Three subunits of PDC (pyruvate dehydrogenase, dihydrolipoyl dehydrogenase, and dihydrolipoamide acetyltransferase) are strongly Lys-acetylated.
An increasing number of Lys-acetylated sites have been identified in numerous metabolic proteins; however, analyses of the biological effects of LysAc on target enzymes are still lacking (Lindahl et al., 2019). Here, we introduce some examples to show the effects of LysAc on the activities and functions of key metabolic enzymes. Rubisco catalyzes the carboxylation of RuBP and enables net CO2 assimilation into organic compounds, which is a rate-limiting step in photosynthesis (Carmo-Silva et al., 2015). A large number of Lys-acetylated sites have been identified in both the Rubisco large subunit (RBCL) and small subunit (RBCS). Previous studies have reported that LysAc can negatively regulate Rubisco activity (Finkemeier et al., 2011; Gao et al., 2016). However, recent studies have shown conflicting results. Under low-light conditions, the LysAc levels of Rubisco activase (RCA) and RBCL increased markedly, leading to significant increases in the activity and activation of Rubisco in the A. thaliana hda14 mutant (Hartl et al., 2017). Interestingly, another study reported that increased LysAc of Rubisco had no effect on its maximal activity (O'Leary et al., 2020). Therefore, specific Lys-acetylated sites appear to contribute to the different effects of LysAc on Rubisco activity.
Malate dehydrogenase (MDH) catalyzes malate oxidation and oxaloacetate reduction using NAD+/NADH as a co-substrate, and LysAc of MDH is conserved in plants (Sweetlove et al., 2010). The enzymatic activity of MDH in the direction of oxaloacetate reduction is negatively regulated by LysAc (Finkemeier et al., 2011). In P. patens, LysAc at K172 of mitochondrial MDH1 (mMDH1) doubles its catalytic rate in the direction of oxaloacetate reduction compared with the unmodified protein and is considered to be a requirement under conditions of high NAD+ demand. By contrast, LysAc of K172 has no effects on the enzymatic parameters of the malate oxidation reaction (Balparda et al., 2021). In A. thaliana, LysAc of K169 in mMDH1 (corresponding to K172 in P. patens) has no significant effects on kinetic parameters in the malate oxidation direction compared with the unmodified protein, whereas it can decrease the enzyme’s affinity for oxaloacetate and its catalytic efficiency in the oxaloacetate reduction direction. In addition, LysAc of K170 can decrease catalytic efficiency in both directions. LysAc of a C-terminal lysine (K334) of mMDH1 increases the catalytic efficiency of malate oxidation and decreases that of oxaloacetate reduction (Balparda et al., 2021). Similarly, LysAc of K99 and K140 in E. coli MDH and LysAc of K307 in human mMDH2 can enhance the catalytic efficiency of malate oxidation (Venkat et al., 2017). However, in E. coli and human, changes in catalytic efficiency are due to increased enzymatic activity caused by LysAc, whereas in A. thaliana, they are due to increased affinity for malate (Venkat et al., 2017; Balparda et al., 2021). Therefore, the effects of LysAc on MDH vary across different organisms.
In addition to its effect on enzyme activities, LysAc can also regulate the epigenetic characteristics of target proteins. GAPDH participates in the Calvin–Benson cycle and glycolysis by catalyzing the reversible conversion of glyceraldehyde-3-phosphate to 1,3-bisphosphoglyceric acid (Zaffagnini et al., 2013). GAPDH is also a transcriptional activator and can activate the expression of glycolytic genes, and LysAc of GAPDH1 in rice can stimulate the transcription of glycolytic genes (Zhang et al., 2017). In addition, LysDeAc of GAPDH1 can increase its enzymatic activity in the generation of 1,3-bisphosphoglyceric acid, similar to results observed in A. thaliana (Finkemeier et al., 2011). The increased LysAc level of GAPDH is thought to promote flux through glycolysis and inhibit flux through gluconeogenesis (Wang et al., 2010).
LysAc of metabolic enzymes is sufficient to affect their activities and appears to act as an effective feedback response to fine-tune metabolic flux and help plants acclimatize to a changing environment. However, more work should be devoted to exploring the functions of LysAc in the regulation of cellular metabolism in the future.
Low yield and stoichiometry of LysAc
Thousands of Lys-acetylated sites have been identified in plants. Nonetheless, not all Lys-acetylated proteins can be detected, especially those present at low abundance (Yan et al., 2020). Few overlapping Lys-acetylated sites exist in plant Lys-acetylproteomes, even within the same plant species. Accordingly, identified Lys-acetylated sites are likely to represent only a small fraction of all LysAc (Hosp et al., 2017). The numbers of Lys-acetylated proteins or sites detected in plants still lag behind those identified in mammals (Svinkina et al., 2015; Weinert et al., 2015). In addition to the effects of the cell wall on protein extraction and of secondary compounds that interfere with affinity purification, low yields of plant LysAc may also be due to: (1) anti-acetyl-lysine antibodies whose coverage of global plant Lys-acetylproteomes is less than optimal because they were originally developed in other organisms; and (2) differences in metabolic fluxes and patterns of acetyl-CoA consumption and production between animals and plants (Graham and Eastmond, 2002; Rothbart et al., 2012).
Compared with the relative percentage or fold change of Lys-acetylated peptides, stoichiometry analysis of LysAc can quantify the prevalence and reflect the physiological dynamics of LysAc modifications (Chen and Li, 2019). The stoichiometry of LysAc was reported to be very low in Saccharomyces cerevisiae (0.02%) (Weinert et al., 2014), E. coli (0.04%) (Weinert et al., 2017), and human (0.02%) (Hansen et al., 2019). O'Leary et al. (2020) found that the stoichiometry of four Lys-acetylated sites in A. thaliana RBCL was less than 1%, and that of one Lys-acetylated site in RBCS was 0.26%, suggesting that LysAc stoichiometry in plants is very low. However, a low LysAc stoichiometry seems to be sufficient to produce functional effects on proteins and affect cellular metabolism (Baeza et al., 2020). More efforts are needed in this field to obtain a more detailed view of the effects of low LysAc yield and stoichiometry.
Regulation of plant development and growth by LysAc
Bud dormancy release and seedling de-etiolation
LysAc of histones, signal effectors, and key metabolic enzymes is likely to play important roles in germination signaling pathways. Histones H2A and H2B and TFs are highly Lys-acetylated during bud dormancy release in P. tremula × P. alba (Liao et al., 2021). Numerous enzymes that participate in the degradation of lipids and amino acids to produce energy for bud breakage are differentially Lys-acetylated during bud break (Liao et al., 2021). Therefore, LysAc of histones, signal effectors, and key metabolic enzymes plays important roles in germination signaling pathways and reconfiguration of the metabolic system.
MYB, CONSTANS, and GRF are essential TFs for cell differentiation, photoperiod signal transduction, and shoot elongation (Putterill et al., 1995; Choi et al., 2004; Kornet and Scheres, 2009). These three TFs are Lys-acetylated in etiolated Z. mays seedlings during continuous white light illumination (Yan et al., 2020), suggesting that LysAc of these TFs is necessary for the regulation of chromatin organization and gene transcription during seedling de-etiolation. LysAc abundance shows differential alteration in various photosystem I and II proteins, and enzymes that participate in chlorophyll synthesis show increased LysAc levels under prolonged illumination (Yan et al., 2020). The LysAc levels of Rubisco in the Calvin–Benson cycle and pyruvate phosphate dikinase and phosphoenolpyruvate carboxylase in the C4 pathway increase with illumination time, and the enzyme activities are negatively regulated by LysAc. Furthermore, the majority of enzymes involved in energy metabolism are Lys-acetylated, suggesting a potential role for LysAc in the switch from skotomorphogenesis to photomorphogenesis in etiolated seedlings through its effects on transcriptional regulation, photosystem assembly, and metabolic enzyme activities (Yan et al., 2020).
Meiosis and pollen development
A large number of proteins related to the biological processes of meiosis and anther development are Lys-acetylated in developing O. sativa anthers before meiosis; these processes include chromatin silencing, pollen development, sporopollenin biosynthesis, callose deposition, fatty acid biosynthesis, and production of secretory proteins (Li et al., 2018a). More than half of the identified Lys-acetylated proteins are meiocyte proteins in O. sativa, and they are mainly enriched in the molecular processes of DNA synthesis, chromatin structure, RNA processing and transcriptional regulation, cell organization, vesicle transport, and protein degradation and folding (Li et al., 2018a).
Cytoplasmic male sterility (CMS) is a maternally inherited trait that causes plants to fail to generate functional pollen (Fujii et al., 2009). Approximately 92% of the differentially Lys-acetylated proteins (DAPs) in wild H. cannabinus and CMS lines are located in the cytoplasm, and 77% of the DAPs show increased LysAc in the wild-type line compared with the CMS line (Chen et al., 2019). The DAPs are mainly involved in the TCA cycle and energy metabolism, glycolysis, signal transduction, protein metabolism, fatty acid metabolism, and auxin transport, and most of them show decreased LysAc levels in the CMS line (Chen et al., 2019). Protein disulfide isomerase (PDIL) is essential for embryo maturation and pollen tube development (Wang et al., 2008). In the CMS line, the proteomic level of PDIL showed a 1.75-fold decrease and the LysAc level exhibited an 11.66-fold decrease compared with those in the wild type (Chen et al., 2019). Therefore, abnormal LysAc in the cytoplasm can mediate plant CMS by affecting energy synthesis and pollen development.
Leaf periodic albinism and fiber development
Compared with the number of differentially Lys-acetylated sites (DASs) between the prealbinization and albinotic stages of C. sinensis cv. ‘Anji Baicha’, more DASs were detected between the regreening and prealbinotic stages, consistent with changes in chlorophyll and carotene contents (Xu et al., 2017). Several LHC proteins (e.g., LHCA1, LHCA3, and LHCB1–5) showed different LysAc levels across the three stages (Xu et al., 2017). Carotenoid isomerase (CrtISO) plays a crucial role in the synthesis of the carotenoid precursors of abscisic acid (ABA) (Fang et al., 2008). The abundance of CrtISO was not altered at the protein accumulation level, but its LysAc changed significantly between the regreening and albinotic stages. In addition, the LysAc of enzymes mapped upstream of flavonoid biosynthesis was dramatically altered across the three stages (Xu et al., 2017). Therefore, LysAc can coordinately modulate periodic albinism in tea.
DAPs were located mainly in the cytoplasm, chloroplast, and mitochondria of wild-type G. hirsutum ovules before anthesis, but they were located mainly in the nucleus in fuzzless-lintless mutants (Singh et al., 2020). The DAPs in the wild type were significantly enriched in fatty acid metabolism, ribosome, TCA cycle, and oxidative phosphorylation, but the DAPs in the mutant line were mainly enriched in lipid metabolism, synthesis and degradation of ketone bodies, and folate biosynthesis (Singh et al., 2020). These results suggest that LysAc may be important for providing intermediates for the biosynthesis of macromolecules and metabolites and for meeting the energy needs associated with plant fiber development.
Germination and maturation of seeds
Compared with 0 h imbibition, most DAPs identified in T. aestivum seeds after 12 h and 24 h of imbibition exhibited increased LysAc levels (Guo et al., 2020). DAPs between 24 h and 0 h, as well as between 12 h and 0 h, were simultaneously enriched in the ribosome, glycolysis/gluconeogenesis, and carbon fixation pathways. DAPs between 12 h and 0 h showed greater enrichment in glyoxylate and dicarboxylate metabolism and proteasome pathways, whereas DAPs between 24 h and 0 h showed greater enrichment in biosynthesis of amino acids, the TCA cycle, and carbon metabolism (Guo et al., 2020). ABA 8′-hydroxylase and protein phosphatase 2C (PP2C) can promote seed germination via gibberellic acid biosynthesis and negative regulation of ABA signaling (Nambara and Marion-Poll, 2005; Cheng et al., 2017). Sorting nexin 1 (SNX1) and vacuolar protein sorting protein 72 (VPS72) participate in auxin transport (Shimada et al., 2006). All four of these hormone signaling proteins showed increased LysAc levels after seed imbibition (Guo et al., 2020).
Most of the Lys-acetylated proteins in P. asperata desiccated embryos are TFs or enzymes, whereas stress-responsive proteins and proteins with catalytic and oxidoreductase activities are more strongly Lys-acetylated in desiccated than in nondesiccated embryos (Xia et al., 2016). The reducing power generated by the pentose phosphate pathway is essential for maintaining the activity of antioxidant enzymes and preventing oxidative stress, and fatty acid metabolism is thought to enhance plant drought resistance by changing the lipid fluidity of cell membranes (Pandolfi et al., 1995; Rylott et al., 2006). Interestingly, Lys-acetylated proteins are significantly enriched in these two pathways in desiccated embryos relative to nondesiccated embryos. Hence, LysAc can be an active event and has various functions in the regulation of hormone signaling, energy supply, and protection of the embryo against oxidative damage during seed germination.
In O. sativa seeds collected 0, 3, and 7 days after pollination, LysAc preferentially occurred in non-TF proteins. DAPs showed higher LysAc levels at 3 and 7 days than at day 0 and were predominantly enriched in carbon metabolism, glycolysis, carbon fixation, and starch and sucrose metabolic pathways (Wang et al., 2017). Most of the DAPs related to glycolysis and starch and sucrose metabolism showed increased LysAc levels and a positive correlation with seed maturation. However, the LysAc levels of TCA-related DAPs increased in seeds from 0 to 3 days after pollination and remained unchanged from 3 to 7 days, suggesting that LysAc was likely to be involved in the inactivation of the TCA cycle to reduce cell damage caused by a shortage of internal oxygen and to divert carbon flux from energy production to storage deposits (Wang et al., 2017). Moreover, ADP-glucose pyrophosphorylase 2 and PDIL, which can promote seed maturation (Yamagata et al., 1982; Lee et al., 2007), showed increased LysAc levels after pollination.
For G. max seeds at the developmental stage of rapid storage oil and protein accumulation, the LysAc signal was highest in central cotyledonary mesophyll cells but lower on adaxial and abaxial surfaces and absent in the testa (Smith-Hammond et al., 2014b). In O. sativa seeds collected at 15 days post anthesis, starch synthesis-associated and metabolism-associated proteins and storage proteins were heavily Lys-acetylated (Meng et al., 2018). Therefore, LysAc potentially functions in the promotion of seed maturation by regulating starch metabolism, storage nutrient deposition, and carbon flux conversion during seed maturation.
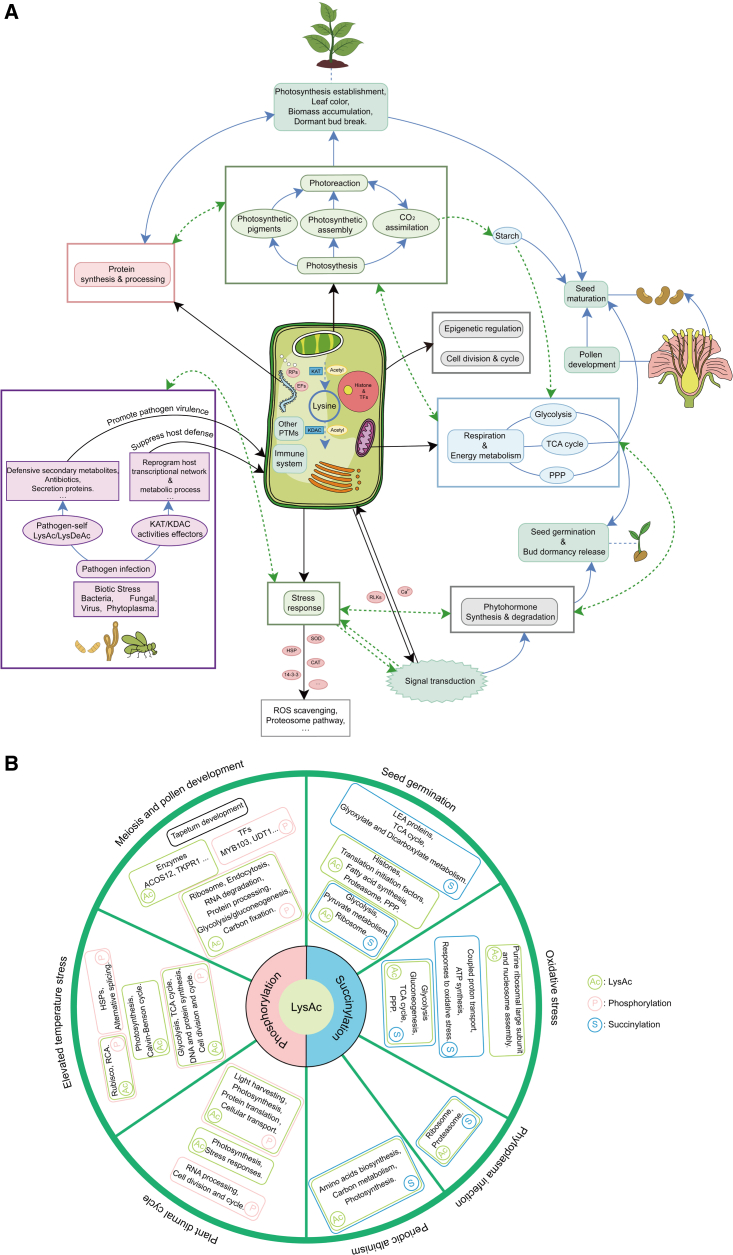
In summary, LysAc is an active event that occurs during plant development and in response to environmental signals. LysAc of histones, TFs, KATs, and KDACs can mediate plant physiological processes through epigenetic regulation, and LysAc of RPs and EFs can affect protein synthesis. Lys-acetylated proteins are enriched in diverse metabolic pathways, indicating that LysAc can regulate metabolic processes by affecting the activities and functions of associated proteins, thereby regulating plant growth and development. First, LysAc of proteins relevant to photosynthesis (e.g., structural proteins of the photosystems, LHCs, and proteins associated with photosynthetic pigment biosynthesis) influences photosystem assembly and affects the generation of ATP and NADPH. LysAc of enzymes in the Calvin–Benson cycle (e.g., Rubisco) supports the potential functions of LysAc in carbon fixation, further regulating the transport and deposition of starch, as well as photomorphogenesis, seedling de-etiolation, and leaf periodic albinism. Second, Lys-acetylated proteins are greatly enriched in carbon and energy metabolism pathways, especially glycolysis, the TCA cycle, and the pentose phosphate pathway. LysAc thus affects carbohydrate metabolism and switches of carbon and energy flux, further regulating dormant buds, germination, seed maturation, and fiber and pollen development. Third, LysAc of proteins relevant to meiosis suggests that LysAc is likely to regulate pollen development. Finally, LysAc of proteins related to the biosynthesis and degradation of phytohormones can affect phytohormone signaling and regulate seed germination and other physiological processes, as well as plant adaptation to stress conditions (Figure 5A).
Figure 5.
Putative LysAc networks in the plant life cycle.
(A) LysAc is involved in plant growth, development, and stress responses.
(B) Crosstalk between LysAc and other PTMs.
LysAc in plant stress responses
Plants have evolved a series of complex molecular mechanisms to withstand environmental stresses during long-term adaptation (Zhu, 2016). LysAc of histones can mediate plant stress responses by activating or inhibiting gene expression (Hu et al., 2019; Kumar et al., 2021). Lys-acetylproteomes provide a global analysis of LysAc networks under stressed conditions, especially for Lys-acetylated nonhistones.
LysAc regulates plant biotic stress responses
Expression of the HAT and HDAC genes in plants can be induced by biotic stresses, directly regulating cell LysAc and affecting host plant immunity and defense (Zhou et al., 2005; Ding et al., 2012; Xu et al., 2015; Song and Walley, 2016). To date, analyses of plant Lys-acetylproteomes related to biotic stresses such as pests (Melo-Braga et al., 2012), fungi (Walley et al., 2018), phytoplasma (Cao et al., 2019), and viruses (Yuan et al., 2021) have been reported. Most studies have suggested that LysAc participates in plant responses to biotic stresses by remodeling the transcriptional network and regulating metabolic processes and immune response.
The first report of a plant Lys-acetylproteome after pest infection concerned the exocarp and mesocarp of V. vinifera during Lobesia botrana infection (Melo-Braga et al., 2012) and identified 138 Lys-acetylated sites. However, the number of Lys-acetylated proteins in the mesocarp was much smaller than the numbers of phosphorylated and N-glycosylated proteins. The calcium-binding protein CaMCML and the aquaporin PIP1;3 have pivotal functions in the plant immune system (Ma et al., 2008). The two protein abundances changed differentially, but the LysAc levels of both proteins increased (Melo-Braga et al., 2012).
HC toxin (HCT) is an effector protein secreted by Cochliobolus carbonum that acts as a histone deacetylase inhibitor to induce hyperacetylation in both histones and nonhistones (Brosch et al., 1995; Walley et al., 2018). In Z. mays, 92.8% of the DAPs induced by HCT showed increased LysAc levels, and 52% of the DAPs were essential for plant defense, including histones, TFs, chromatin remodeling enzymes, and HAT. These results indicated that the hyperacetylation induced by HCT could promote pathogen virulence and reduce host plant immunity by reprogramming and breaking transcriptional networks (Walley et al., 2018).
Paulownia witches’ broom, the most widespread disease responsible for P. tomentosa mortality, is caused by a phytoplasma (Weintraub and Beanland, 2006). The DAPs in phytoplasma-infected P. tomentosa were enriched mainly in the peroxisome, fatty acid metabolism, glyoxylate and dicarboxylate metabolism, carbon fixation, and pentose phosphate pathways (Cao et al., 2019). RBCL and protochlorophyllide reductase (POR) play key roles in the regulation of starch and chlorophyll biosynthesis. The abundances of these two enzymes were not altered after infection, but their LysAc levels were increased. LysAc of K55 on RBCL1, K764 on RBCL2, and K78 and K339 on POR negatively regulated their enzyme activities (Cao et al., 2019).
Most of the DAPs in N. benthamiana leaves infected by Chinese wheat mosaic virus showed enhanced LysAc levels compared with uninfected plants, and various Lys-acetylated sites were located in conserved domains. In infected plants, DAPs involved in photosynthesis, carbon fixation, glyoxylate and dicarboxylate metabolism, and nitrogen metabolism showed increased LysAc levels, whereas DAPs that participate in fatty acid degradation, terpenoid backbone biosynthesis, and protein processing showed decreased LysAc levels (Yuan et al., 2021). Moreover, 51% of the Lys-acetylated proteins were located in the chloroplasts. The LysAc levels of proteins related to the photosystem II complex, photosystem I subunits, and parts of chloroplast ATP synthesis were dramatically increased after viral infection. Hence, LysAc has a key function in the regulation of photosynthesis during viral infection.
LysAc regulates plant abiotic stress responses
Numerous studies have reported that KATs and KDACs can activate stress-responsive genes and enhance abiotic stress tolerance by modulating histones and TFs (Kim et al., 2017; Shen et al., 2019; Zheng et al., 2020). Various stress-responsive proteins, including 14-3-3 protein, superoxide dismutase, CAT, and GPX, undergo LysAc. To date, plant Lys-acetylproteome studies have been performed for plant responses to water, nutrient, and temperature stresses. These studies have revealed that LysAc is likely to have a role in the regulation of stress adaptation by mediating signal transduction, respiration and energy metabolism, protein synthesis, and amino acid metabolism.
In T. aestivum under drought stress, the receptor-like kinase (RLK)-mediated signaling pathway and stress-responsive genes were activated (Zhu et al., 2018). Various enzymes involved in the glyoxylate module, the Calvin–Benson cycle, glycolysis/gluconeogenesis, the pentose phosphate pathway, and the TCA cycle showed altered LysAc levels. Stress-responsive proteins, starch biosynthetic proteins, and metabolic proteins also had significantly altered LysAc levels. Moreover, increased LysAc of ubiquitin-conjugating enzymes promoted the elimination of misfolded proteins and maintained protein stability by inhibiting proteasomal degradation (Zhu et al., 2018). In K. candel subjected to flooding stress, the transcript level of KAT increased while the content of acetyl-CoA decreased, consistent with the alteration of LysAc levels in DAPs (Pan et al., 2018). In addition, both Rubisco and RCA showed increased LysAc levels, but their activities were inhibited under flooding stress. Thus, an increase in RCA1 transcription is probably required to compensate for the decreased activity and maintain relatively high Rubisco activity.
Nutrient deficiency is an important limitation on plant growth. Compared with LysAc under nitrogen (N) starvation, the number of Lys-acetylated proteins in C. sinensis increased after N resupply (Jiang et al., 2018). The Lys-acetylated proteins were significantly enriched in the photosystem and thylakoid membrane after N resupply, whereas the DAPs were enriched in photosynthesis. The LysAc levels of electron transfer proteins, chlorophyll a/b-binding proteins, photosystem I and II subunits, cytochrome b6/f, and ATP synthases increased following N resupply in the short term (3 h) but decreased in the long term (3 days). Therefore, changes in plant LysAc levels caused by N limitation varied over time.
With regard to temperature stress, the LysAc levels of histones H3K27 and H3K36 in O. sativa increased significantly under cold stress (4°C). A protein–protein interaction network revealed that Lys-acetylated proteins were enriched mainly in ribosome, proteasome, glutathione metabolism, protein processing, and secondary metabolic biosynthesis (Xue et al., 2018). However, under high-temperature stress (35°C, 40°C, and 45°C), the LysAc levels of histones H2A.6 and H2A.4 increased. The DAPs that exhibited increased LysAc levels at 40°C were mainly related to photosynthesis, respiration, and DNA synthesis, whereas at 45°C they were mainly related to DNA synthesis (Liu et al., 2019).
In summary, under biotic stresses, the gene expression and enzyme activities of HATs and HDACs can change the host’s original LysAc patterns, especially for metabolism-associated enzymes. Pathogens can also deliver effectors with HAT and HDAC activities that may reprogram the host transcription and LysAc network, triggering perturbation of the host immune system. Moreover, LysAc of proteins related to pathogen virulence can promote infection and suppress host defenses. Under abiotic stresses, alteration of the external environment induces LysAc of intracellular signal transduction molecules, and activation of stress-responsive genes and proteins can enhance plant stress tolerance. Furthermore, numerous proteins associated with key metabolic processes, especially photosynthesis, energy metabolism, and protein degradation, show altered LysAc levels in response to stress exposure. Hence, LysAc plays a non-negligible role in biotic and abiotic stress responses (Figure 5A).
LysAc studies in lower plants
Because there are fewer studies of LysAc in lower plants than in higher plants, we focus here on P. tricornutum and P. patens. P. tricornutum is a model diatom species with a sequenced genome (Alipanah et al., 2015); it contains 24 and 15 genes encoding KATs and KDACs, respectively (Chen et al., 2018). Veluchamy et al. (2015) performed a genome-wide characterization of histone LysAc by describing the genome-wide distribution of K9/K14 in histone H3. Chen et al. (2018) analyzed the Lys-acetylproteome of P. tricornutum exposed to nitrogen, iron, and phosphate deficiencies and identified 2324 Lys-acetylated sites on 1220 proteins, including at least 39 histone Lys-acetylated sites. Over sixty-one percent of these Lys-acetylated proteins were also detected in higher plants (A. thaliana, V. vinifera, S. tuberosum, O. sativa, G. max, and F. ananassa), fungi (S. cerevisiae), and bacteria (Synechocystis sp. PCC 6803), revealing potentially conserved functions of LysAc among species. There were 17 Lys-acetylated motifs in P. tricornutum, and all except for LKacY, I∗Kac∗L, I∗Kac, KacW, and F∗∗Kac can be found in at least one species of higher plant, according to the 27 Lys-acetylproteomes summarized previously (Table 2). Consistent with the amino acid preferences of LysAc in higher plants, Y at the −1 and +1 positions, F at the −1/+1/+2 positions, and L at the −1/+2 positions are conserved in P. tricornutum. In terms of the protein secondary structures surrounding the Lys-acetylated lysine, the smallest number of Lys-acetylated sites are located in β-strand regions in both higher plants and P. tricornutum (Chen et al., 2018). More Lys-acetylated sites are found in α helices than in coil regions in P. tricornutum, similar to reports on O. sativa seeds (He et al., 2016) and B. distachyon leaves (Zhen et al., 2016), although other studies have reported opposite results (Zhang et al., 2016; Jiang et al., 2018; Zhu et al., 2018; Cao et al., 2019; Yan et al., 2020) (Figure 3). Almost all enzymes of fatty acid synthesis were found to be Lys-acetylated. Long-chain acyl-CoA synthetase catalyzes the esterification of free fatty acids (Shockey et al., 2002). LysAc of K407 and K425 in ptACSL1 and of K321 and K325 in ptACSL4 were confirmed in vivo. However, they exhibit distinct LysAc patterns under normal and nutrient-deficient conditions (Chen et al., 2018). These findings suggest that LysAc can regulate various cellular processes of P. tricornutum, especially fatty acid metabolism, and that there are both conserved and contrasting LysAc events in lower and higher plants.
P. patens is a premier model species for investigating questions in evolutionary, developmental, and cell biology (Rensing et al., 2020). In P. patens gametophores, 638 proteins were identified as acetylated at 1–9 lysine sites. Only eight proteins contained more than five Lys-acetylated sites, indicating that a relatively small number of LysAc events occur on a single protein compared with the number in higher plants (Balparda et al., 2021). Approximately fifty percent of the identified Lys-acetylated proteins were located in the mitochondria and plastids, and most TCA cycle enzymes were identified as Lys-acetylated, consistent with mitochondrial Lys-acetylproteomes in higher plants. Numerous orthologs of P. patens TCA cycle proteins, including MDH, and other mitochondrial proteins were Lys-acetylated in A. thaliana mitochondria also, whereas few acetylated lysines were common to both species (Balparda et al., 2021).
Interplay between LysAc and Nα-terminal acetylation
In addition to LysAc, proteins can also be modified by other types of acetylation, such as Nα-terminal acetylation (NTA), which is considered to be the closest cousin of KAC (Mukherjee et al., 2006; Giglione and Meinnel, 2021). NTA is a ubiquitous co-translational or post-translational modification that is catalyzed by Nα-terminal acetyltransferases (NATs) and involves the transfer of an acetyl group from acetyl-CoA to the protein α-amino group (Zhang et al., 2017; Aksnes et al., 2019). Distinctive features are observed in the two acetylations. First, LysAc is a reversible modification that occurs after translation, whereas NTA is irreversible and mainly occurs co-translationally (Bienvenut et al., 2020). Second, all known NATs belong to the GNAT superfamily, whereas KATs contain more families beyond the GNAT (Drazic et al., 2016). In addition, each member of the NAT family has complex preferences for different substrates in N-terminal residues, and protein NTA occurs independent of a single residue type or simple consensus motifs. By contrast, KATs target only lysine, and LysAc occurs at conserved motifs around preferred amino acid residues (Polevoda and Sherman, 2003; Bienvenut et al., 2012).
In multicellular organisms, KAT occurs more frequently than NTA (Linster and Wirtz, 2018). Regarding the interaction between LysAc and NTA, numerous identified NATs possess KAT activity as well. Bienvenut et al. (2020) identified eight plastid-associated NATs in A. thaliana, six of which possessed dual NAT and KAT activities and most of which could trigger hyper-LysAc in E. coli when GNATs were overexpressed. A. thaliana AtNAA60 is the first identified plasma membrane-anchored NAT; it also possesses KAT activity, although its KAT activity is much weaker than its NAT activity (Linster et al., 2020). On the other hand, NATs can show auto-LysAc activity. Dinh et al. (2015) identified the first chloroplast NAT (AtNAA70) in A. thaliana and demonstrated that it could auto-acetylate at K217, K254, and K265. AtNAA50 is located in the cytosol, endoplasmic reticulum (ER), and nuclei in A. thaliana, and it can be auto-Lys-acetylated in vitro as well (Armbruster et al., 2020).
Because both NATs and KATs require acetyl-CoA, a functional redundancy probably exists between them when targeting the same substrates. Accordingly, a possible interplay between NTA and LysAc may involve their competition for acetyl-CoA and substrates, even on the same lysine residue. However, studies of interactions between LysAc and NTA are still largely lacking.
Crosstalk between LysAc and other PTMs
Lysine can be modified by phosphorylation, succinylation, ubiquitylation, methylation, and other PTMs, and different PTMs can occur on the same or adjacent sites within one protein (Fischle et al., 2003; Soufi et al., 2012). Hence, there is a complex interaction of cooperation and competition between LysAc and other PTMs to regulate plant growth and development. Here, integrative analyses of the crosstalk between LysAc and other PTMs focus on phosphorylation and succinylation.
Crosstalk between LysAc and phosphorylation
Phosphorylation is one of the most important PTMs in both prokaryotes and eukaryotes and is mediated by kinases and phosphatases (Cohen, 2002; Ardito et al., 2017). Integrative analyses of crosstalk between LysAc and phosphorylation have revealed that the two PTMs regulate plant pollen development, abiotic stress, and the diurnal cycle through modification of numerous cellular pathways (Figure 5B).
During pollen development in developing anthers of O. sativa, near the time of meiosis, 189 proteins were found to be Lys-acetylated and phosphorylated simultaneously, and 103 dual-modified proteins overlapped in the meiocyte proteome (Li et al., 2018a). Analysis of the interaction network of the dual-modified proteins indicated that over half contained more phosphorylated sites than Lys-acetylated sites, although proteins involved in sugar metabolism and protein processing in the ER contained more Lys-acetylated sites. For proteins relevant to tapetum and pollen development, phosphorylation was more likely to modify TFs, whereas LysAc preferentially occurred on enzymes (Li et al., 2018a). In the H. cannabinus anther, 22 proteins were modified by both PTMs, and 63.6% (14/22) of these proteins showed opposite trends in the two PTMs (Chen et al., 2019).
In terms of abiotic stress, LysAc in V. vinifera had stronger modification effects on photosynthesis and Calvin–Benson cycle proteins under high temperature, whereas phosphorylation was observed more frequently in HSPs and proteins associated with alternative splicing (Liu et al., 2019). LysAc can decrease Rubisco activity, whereas phosphorylation has the opposite effect; however, both PTMs can decrease RCA activity (Boex-Fontvieille et al., 2014). The phosphorylation and LysAc levels of RCA and Rubisco increase simultaneously in response to heat, and the phosphorylation of Rubisco can compensate for decreased RCA activity (Liu et al., 2019). In T. aestivum seeds under drought stress, LysAc has a stronger effect on domains of the dual-modified proteins than does phosphorylation (Zhu et al., 2018).
LysAc and phosphorylation can also coordinately regulate the diurnal cycle in A. thaliana (Uhrig et al., 2017). The two PTMs occur preferentially on proteins of plant primary metabolism rather than secondary metabolism. LysAc predominantly modulates proteins that participate in metabolic processes and stress responses, whereas phosphorylation modulates more proteins involved in RNA processing, cell division, and the cell cycle. The abundance of the two PTMs in different tissues changes significantly during the diurnal cycle, and striking diurnal changes in LysAc and phosphorylation occur on enzymes associated with glycolysis and the Calvin–Benson cycle, respectively (Uhrig et al., 2017).
Crosstalk between LysAc and succinylation
Lysine succinylation is catalyzed by succinyltransferase and involves the addition of the succinyl group from succinyl-CoA to the lysine residue; desuccinylation is catalyzed by desuccinylase and is associated with the removal of succinyl groups from lysine (Zhang et al., 2011). Enzymes of succinyl-CoA metabolism can be highly Lys-acetylated. Among the proteins modified by both LysAc and succinylation, numerous common modified sites in polar/basic amino acids and nonpolar hydrophobic regions have been detected, and K∗E, K∗D, KK, and KR are enriched in both succinylation and LysAc (He et al., 2016; Xu et al., 2017), suggesting that LysAc has a closer interaction with succinylation than with other PTMs (Weinert et al., 2013). Crosstalk analyses of the two PTMs indicate that LysAc and succinylation enable the regulation of seed germination, leaf development, and biotic and abiotic stress responses through their effects on photosynthesis, carbon metabolism, amino acid metabolism, and protein synthesis and degradation (Figure 5B).
In terms of seed germination, 142 proteins exhibited LysAc and succinylation in O. sativa seeds collected 24 h after imbibition. In particular, 1-cysteine peroxiredoxin had nine Lys-acetylated sites, 13 succinylated sites, and seven overlapping sites. In addition, 133 commonly modified sites on 78 proteins were preferentially exposed on the protein surface (He et al., 2016). LysAc exerts stronger modification effects than succinylation on histones, translation initiation factors, and enzymes related to fatty acid synthesis, whereas succinylation can dramatically modify late embryogenesis abundant proteins and enzymes involved in the TCA cycle. The proteasome and pentose phosphate pathways are also enriched in Lys-acetylproteomes, whereas proteins related to glyoxylate and dicarboxylate metabolism are enriched only in succinylproteomes (He et al., 2016).
Regarding biotic stress, in seedlings of P. tomentosa infected by phytoplasma, more proteins were observed to undergo LysAc than succinylation, and there was a more striking change in LysAc than in succinylation (Cao et al., 2019). The dual-modified proteins were mainly enriched in ribosome and proteasome functions, revealing potential roles of LysAc and succinylation in protein synthesis, repair, and degradation after infection (Cao et al., 2019). Under oxidative stress, 723 proteins of O. sativa exhibited LysAc and succinylation. Specifically, 11 sites in RubL underwent LysAc and succinylation simultaneously. However, only 12%–32% of the Lys-acetylated or succinylated sites showed differences in abundance after H2O2 treatment, and most were decreased (Zhou et al., 2018). Almost all of the enzymes associated with glycolysis/gluconeogenesis, the TCA cycle, and the pentose phosphate pathway are modified by both PTMs. However, LysAc modulates more proteins related to nucleosome and purine ribosomal large subunit assembly, whereas succinylation occurs more frequently in proteins associated with coupled proton transport, ATP synthesis, and cellular responses to oxidative stress. Thus, succinylation and LysAc commonly participate in the regulation of plant physiological processes (Zhou et al., 2018).
Overall, numerous proteins are modified by more than one PTM, and these proteins are mainly involved in metabolic processes, especially primary metabolism, and thereby affect plant growth and adaptation to biotic and abiotic stresses. These results suggest that complex interactions among PTMs may coordinately regulate the plant life cycle (Figure 5B).
Perspectives
The enrichment and identification of Lys-acetylated proteins are prerequisites for the investigation of LysAc. LysAc proteomics has been performed in various plants and have revealed numerous Lys-acetylated histone and nonhistone proteins. However, recent plant Lys-acetylproteomics research has focused mainly on annual crops, less on perennial woody plants, and rarely on lower plants. The LysAc regulatory networks that act at specific plant developmental stages and the distribution of Lys-acetylated proteins among subcellular structures have also rarely been characterized. Hence, global LysAc proteomic analyses in more plant species, tissues, and subcellular structures are needed. There have also been few analyses of plant Lys-acetylproteomes in response to biotic and abiotic stresses. For biotic stresses, Lys-acetylproteomes have been evaluated in plants infected by only a few pests, fungi, and viruses, and responses to bacteria have rarely been described. Therefore, integration of the molecular mechanism by which LysAc affects host susceptibility and the effects of pathogenicity will provide new insights into the prevention of plant diseases. Investigations of the effects of abiotic stresses using Lys-acetylproteomes are also necessary.
Until recently, our understanding of KATs and KDACs was focused mainly on histones rather than nonhistone proteins. LysAc proteomics can only detect whether and where proteins are acetylated; it cannot assess the effects of LysAc on their activities (Santos-Rosa et al., 2003; Berndsen et al., 2007; Bienvenut et al., 2020). Furthermore, LysAc can occur nonenzymatically in mitochondria in vitro, even after heat inactivation (Koenig et al., 2014). To obtain a deeper understanding of the complex enzymatic mechanisms of LysAc and LysDeAc in the future, we can construct mutants that encode HATs or HDAs and use a combination of LysAc proteomics, transcriptomics, and immunoprecipitation to explore the functions and the target substrates of plant KATs and KDACs.
Numerous Lys-acetylated proteins identified by LysAc proteomics are enriched in various metabolic processes, especially in the central carbon metabolic pathway. Many Lys-acetylated proteins show altered LysAc levels that are independent of protein abundance. However, the effects of LysAc on protein folding, structure, and activity are still largely unknown. Moreover, system-wide analyses of LysAc mechanisms in specific pathways are remarkably limited. In the future, we can apply and incorporate site-specific direct LysAc into recombinant proteins and express them in E. coli or mutate Lys-acetylated K to R or Q to hinder the LysAc status and help to define the effects of LysAc in target proteins.
A competitive interplay probably exists between NTA and LysAc. Increasing evidence indicates that there are complex and extensive interactions between LysAc and other PTMs (Fischle et al., 2003; Soufi et al., 2012). However, there have been few plant studies of the interaction between LysAc and NTA, and crosstalk analyses of different PTMs have focused mainly on the crosstalk among LysAc, phosphorylation, and succinylation. Therefore, more investigations are needed on the effects of LysAc, NAT, and other PTMs on target proteins, the regulatory mechanisms of LysAc on key enzymes, and the means by which KATs and KDACs are modified by other PTMs.
Funding
This work was supported by the Second Tibetan Plateau Scientific Expedition and Research Program (2019QZKK0404), the National Natural Science Foundation of China (31770650), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA20020401), and the Fundamental Research Funds for the Central Universities.
Author contributions
L.X. collected and analyzed the data and wrote the manuscript. X.K., H.S., and Q.H. collected the data and provided advice. S.Z. designed the framework and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgments
No conflict of interest declared.
Published: November 24, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information can be found online at Plant Communications Online.
Supplemental information
Ratio indicates the proportion of Lys-acetylated proteins with a certain number of Lys-acetylated sites relative to all identified Lys-acetylated proteins in plant Lys-acetylproteomes.
References
- Aksnes H., Ree R., Arnesen T. Co-translational, post-translational, and non-catalytic roles of N-terminal acetyltransferases. Mol. Cell. 2019;73:1097–1114. doi: 10.1016/j.molcel.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipanah L., Rohloff J., Winge P., Bones A.M., Brembu T. Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Exp. Bot. 2015;66:6281–6296. doi: 10.1093/jxb/erv340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey V.G., Faulkner R., Mirsky A.E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardito F., Giuliani M., Perrone D., Troiano G., Lo Muzio L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int. J. Mol. Med. 2017;40:271–280. doi: 10.3892/ijmm.2017.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster L., Linster E., Boyer J.B., Brunje A., Eirich J., Stephan I., Bienvenut W.V., Weidenhausen J., Meinnel T., Hell R., et al. NAA50 is an enzymatically active Nα-acetyltransferase that is crucial for development and regulation of stress responses. Plant Physiol. 2020;183:1502–1516. doi: 10.1104/pp.20.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J., Smallegan M.J., Denu J.M. Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem. Sci. 2016;41:231–244. doi: 10.1016/j.tibs.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J., Lawton A.J., Fan J., Smallegan M.J., Lienert I., Gandhi T., Bernhardt O.M., Reiter L., Denu J.M. Revealing dynamic protein acetylation across subcellular compartments. J. Proteome Res. 2020;19:2404–2418. doi: 10.1021/acs.jproteome.0c00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balparda M., Elsasser M., Badia M.B., Giese J., Bovdilova A., Hudig M., Reinmuth L., Eirich J., Schwarzlander M., Finkemeier I., et al. Acetylation of conserved lysines fine-tunes mitochondrial malate dehydrogenase activity in land plants. Plant J. 2021 doi: 10.1111/tpj.15556. [DOI] [PubMed] [Google Scholar]
- Berndsen C.E., Albaugh B.N., Tan S., Denu J.M. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry. 2007;46:623–629. doi: 10.1021/bi602513x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienvenut W.V., Sumpton D., Martinez A., Lilla S., Espagne C., Meinnel T., Giglione C. Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-α-acetylation features. Mol. Cell. Proteomics. 2012;11:6. doi: 10.1074/mcp.M111.015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienvenut W.V., Bruenje A., Boyer J.-B., Muehlenbeck J.S., Bernal G., Lassowskat I., Dian C., Linster E., Dinh T.V., Koskela M.M., et al. Dual lysine and N-terminal acetyltransferases reveal the complexity underpinning protein acetylation. Mol. Syst. Biol. 2020;16:e9464. doi: 10.15252/msb.20209464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boex-Fontvieille E., Daventure M., Jossier M., Hodges M., Zivy M., Tcherkez G. Phosphorylation pattern of Rubisco activase in Arabidopsis leaves. Plant Biol. 2014;16:550–557. doi: 10.1111/plb.12100. [DOI] [PubMed] [Google Scholar]
- Brosch G., Ransom R., Lechner T., Walton J.D., Loidl P. Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell. 1995;7:1941–1950. doi: 10.1105/tpc.7.11.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell J.E., Zhou J.X., Ranalli T., Kobayashi R., Edmondson D.G., Roth S.Y., Allis C.D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Cao Y., Fan G., Wang Z., Gu Z. Phytoplasma-induced changes in the acetylome and succinylome of Paulownia tomentosa provide evidence for involvement of acetylated proteins in Witches' Broom disease. Mol. Cell. Proteomics. 2019;18:1703. doi: 10.1074/mcp.AAC119.001627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Silva E., Scales J.C., Madgwick P.J., Parry M.A.J. Optimizing Rubisco and its regulation for greater resource use efficiency. Plant Cell Environ. 2015;38:1817–1832. doi: 10.1111/pce.12425. [DOI] [PubMed] [Google Scholar]
- Chen C., Li C., Wang Y., Renaud J., Tian G., Kambhampati S., Saatian B., Vi N., Hannoufa A., Marsolais F., et al. Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat. Plants. 2017;3:814–824. doi: 10.1038/s41477-017-0023-7. [DOI] [PubMed] [Google Scholar]
- Chen P., Wei F., Li R., Li Z.Q., Kashif M.H., Zhou R.Y. Comparative acetylomic analysis reveals differentially acetylated proteins regulating anther and pollen development in kenaf cytoplasmic male sterility line. Physiol. Plant. 2019;166:960–978. doi: 10.1111/ppl.12850. [DOI] [PubMed] [Google Scholar]
- Chen Y., Li Y. Site-specific determination of lysine acetylation stoichiometries on the proteome-scale. Methods Enzymol. 2019;626:115–132. doi: 10.1016/bs.mie.2019.06.021. [DOI] [PubMed] [Google Scholar]
- Chen Z., Luo L., Chen R., Hu H., Pan Y., Jiang H., Wan X., Jin H., Gong Y. Acetylome profiling reveals extensive lysine acetylation of the fatty acid metabolism pathway in the diatom Phaeodactylum tricornutum. Mol. Cell. Proteomics. 2018;17:399–412. doi: 10.1074/mcp.RA117.000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Wang Z., Ren Z., Zhi L., Yao B., Su C., Liu L., Li X. SCFAtPP2-B11 modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana. Plos Genet. 2017;13:e1006947. doi: 10.1371/journal.pgen.1006947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D., Kim J.H., Kende H. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.) Plant Cell Physiol. 2004;45:897–904. doi: 10.1093/pcp/pch098. [DOI] [PubMed] [Google Scholar]
- Choudhary C., Weinert B.T., Nishida Y., Verdin E., Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cohen P. The origins of protein phosphorylation. Nat. Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- De Ruijter A.J.M., Van Gennip A.H., Caron H.N., Kemp S., Van Kuilenburg A.B.P. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Bellizzi M.d.R., Ning Y., Meyers B.C., Wang G.L. HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell. 2012;24:3783–3794. doi: 10.1105/tpc.112.101972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh T.V., Bienvenut W.V., Linster E., Feldman-Salit A., Jung V.A., Meinnel T., Hell R., Giglione C., Wirtz M. Molecular identification and functional characterization of the first N-acetyltransferase in plastids by global acetylome profiling. Proteomics. 2015;15:2426–2435. doi: 10.1002/pmic.201500025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A., Myklebust L.M., Ree R., Arnesen T. The world of protein acetylation. Biochim. Biophys. Acta. 2016;1864:1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Eberharter A., Becker P.B. Histone acetylation: a switch between repressive and permissive chromatin-second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Chai C., Qian Q., Li C., Tang J., Sun L., Huang Z., Guo X., Sun C., Liu M., et al. Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 2008;54:177–189. doi: 10.1111/j.1365-313X.2008.03411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Chen W., Zhao Y., Ruan S., Zhang H., Yan C., Jin L., Cao L., Zhu J., Ma H., et al. Global analysis of lysine acetylation in strawberry leaves. Front. Plant Sci. 2015;6:739. doi: 10.3389/fpls.2015.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatland B.L., Ke J.S., Anderson M.D., Mentzen W.I., Cui L.W., Allred C.C., Johnston J.L., Nikolau B.J., Wurtele E.S. Molecular characterization of a heteromeric ATP-citrate lyase that generates cytosolic acetyl-coenzyme A in Arabidopsis. Plant Physiol. 2002;130:740–756. doi: 10.1104/pp.008110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I., Laxa M., Miguet L., Howden A.J.M., Sweetlove L.J. Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 2011;155:1779–1790. doi: 10.1104/pp.110.171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Wang Y.M., Allis C.D. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Fujii S., Yamada M., Toriyama K. Cytoplasmic male sterility-related protein kinase, OsNek3, is regulated downstream of mitochondrial protein phosphatase 2C, DCW11. Plant Cell Physiol. 2009;50:828–837. doi: 10.1093/pcp/pcp026. [DOI] [PubMed] [Google Scholar]
- Gao X., Hong H., Li W.C., Yang L., Huang J., Xiao Y.L., Chen X.Y., Chen G.Y. Downregulation of Rubisco activity by non-enzymatic acetylation of RbcL. Mol. Plant. 2016;9:1018–1027. doi: 10.1016/j.molp.2016.03.012. [DOI] [PubMed] [Google Scholar]
- Giglione C., Meinnel T. Evolution-driven versatility of N terminal acetylation in photoautotrophs. Trends Plant Sci. 2021;26:375–391. doi: 10.1016/j.tplants.2020.11.012. [DOI] [PubMed] [Google Scholar]
- Graham I.A., Eastmond P.J. Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog. Lipid Res. 2002;41:156–181. doi: 10.1016/s0163-7827(01)00022-4. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Gu W., Roeder R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Guo W., Han L., Li X., Wang H., Mu P., Lin Q., Liu Q., Zhang Y. Proteome and lysine acetylome analysis reveals insights into the molecular mechanism of seed germination in wheat. Sci. Rep. 2020;10:13454. doi: 10.1038/s41598-020-70230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen B.K., Gupta R., Baldus L., Lyon D., Narita T., Lammers M., Choudhary C., Weinert B.T. Analysis of human acetylation stoichiometry defines mechanistic constraints on protein regulation. Nat. Commun. 2019;10:1055. doi: 10.1038/s41467-019-09024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl M., Finkemeier I. Plant mitochondrial retrograde signaling: post-translational modifications enter the stage. Front. Plant Sci. 2012;3:253. doi: 10.3389/fpls.2012.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl M., Fuessl M., Boersema P.J., Jost J.O., Kramer K., Bakirbas A., Sindlinger J., Ploechinger M., Leister D., Uhrig G., et al. Lysine acetylome profiling uncovers novel histone deacetylase substrate proteins in Arabidopsis. Mol. Syst. Biol. 2017;13:949. doi: 10.15252/msb.20177819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Wang Q., Li M., Damaris R.N., Yi X., Cheng Z., Yang P. Global proteome analyses of lysine acetylation and succinylation reveal the widespread involvement of both modification in metabolism in the embryo of germinating rice seed. J. Proteome Res. 2016;15:879–890. doi: 10.1021/acs.jproteome.5b00805. [DOI] [PubMed] [Google Scholar]
- Hentchel K.L., Escalante-Semerena J.C. Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol. Mol. Biol. Rev. 2015;79:321–346. doi: 10.1128/MMBR.00020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender C., Liu Z. Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 2008;50:875–885. doi: 10.1111/j.1744-7909.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- Hosp F., Lassowskat I., Santoro V., De Vleesschauwer D., Fliegner D., Redestig H., Mann M., Christian S., Hannah M.A., Finkemeier I. Lysine acetylation in mitochondria: from inventory to function. Mitochondrion. 2017;33:58–71. doi: 10.1016/j.mito.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Hu Y., Lu Y., Zhao Y., Zhou D.X. Histone acetylation dynamics integrates metabolic activity to regulate plant response to stress. Front. Plant Sci. 2019;10:1236. doi: 10.3389/fpls.2019.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., Kwon S., Lee Y.H. Histone acetylation in fungal pathogens of plants. Plant Pathol. J. 2014;30:1–9. doi: 10.5423/PPJ.RW.01.2014.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Gai Z., Wang Y., Fan K., Sun L., Wang H., Ding Z. Comprehensive proteome analyses of lysine acetylation in tea leaves by sensing nitrogen nutrition. BMC Genomics. 2018;19:840. doi: 10.1186/s12864-018-5250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T., Lorkovic Z.J., Nishihama R., Ishizaki K., Axelsson E., Yelagandula R., Kohchi T., Berger F. Diversification of histone H2A variants during plant evolution. Trends Plant Sci. 2015;20:419–425. doi: 10.1016/j.tplants.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Kim J.M., To T.K., Matsui A., Tanoi K., Kobayashi N.I., Matsuda F., Habu Y., Ogawa D., Sakamoto T., Matsunaga S., et al. Acetate-mediated novel survival strategy against drought in plants. Nat. Plants. 2017;3:17097. doi: 10.1038/nplants.2017.97. [DOI] [PubMed] [Google Scholar]
- Kim S.C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Koenig A.C., Hartl M., Boersema P.J., Mann M., Finkemeier I. The mitochondrial lysine acetylome of Arabidopsis. Mitochondrion. 2014;19:252–260. doi: 10.1016/j.mito.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Kornet N., Scheres B. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell. 2009;21:1070–1079. doi: 10.1105/tpc.108.065300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela M.M., Bruenje A., Ivanauskaite A., Grabsztunowicz M., Lassowskat I., Neumann U., Dinh T.V., Sindlinger J., Schwarzer D., Wirtz M., et al. Chloroplast acetyltransferase NSI is required for state transitions in Arabidopsis thaliana. Plant Cell. 2018;30:1695–1709. doi: 10.1105/tpc.18.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Thakur J.K., Prasad M. Histone acetylation dynamics regulating plant development and stress responses. Cell. Mol. Life Sci. 2021;78:4467–4486. doi: 10.1007/s00018-021-03794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O.K., Kim S., Lee S. Global proteomic analysis of lysine acetylation in zebrafish (Danio rerio) embryos. Electrophoresis. 2016;37:3137–3145. doi: 10.1002/elps.201600210. [DOI] [PubMed] [Google Scholar]
- Law R.D., Suttle J.C. Changes in histone H3 and H4 multi-acetylation during natural and forced dormancy break in potato tubers. Physiol. Plant. 2004;120:642–649. doi: 10.1111/j.0031-9317.2004.0273.x. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Hwang S.K., Han M., Eom J.S., Kang H.G., Han Y., Choi S.B., Cho M.H., Bhoo S.H., An G., et al. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.) Plant Mol. Biol. 2007;65:531–546. doi: 10.1007/s11103-007-9153-z. [DOI] [PubMed] [Google Scholar]
- Lehtimaki N., Koskela M.M., Mulo P. Posttranslational modifications of chloroplast proteins: an emerging field. Plant Physiol. 2015;168:768–775. doi: 10.1104/pp.15.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Lv B., Tan L., Yang Q., Liang W. Acetylome analysis reveals the involvement of lysine acetylation in diverse biological processes in Phytophthora sojae. Sci. Rep. 2016;6:29897. doi: 10.1038/srep29897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Zheng B., Zhao W., Ren T., Zhang X., Ning T., Liu P. Global analysis of lysine acetylation in soybean leaves. Sci. Rep. 2021;11:17858. doi: 10.1038/s41598-021-97338-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Chen C., Dong Y. A comprehensive examination of the lysine acetylation targets in paper mulberry based on proteomics analyses. PLoS One. 2021;16:e0240947. doi: 10.1371/journal.pone.0240947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ye J., Ma H., Lu P. Proteomic analysis of lysine acetylation provides strong evidence for involvement of acetylated proteins in plant meiosis and tapetum function. Plant J. 2018;93:142–154. doi: 10.1111/tpj.13766. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang Y., Bello B.K., Ajadi A.A., Tong X., Chang Y., Zhang J. Construction of a quantitative acetylomic tissue atlas in rice (Oryza sativa L.) Molecules. 2018;23:2843. doi: 10.3390/molecules23112843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X., Li Y., Hu Z., Lin Y., Zheng B., Ding J. Poplar acetylome profiling reveals lysine acetylation dynamics in seasonal bud dormancy release. Plant Cell Environ. 2021;44:1830–1845. doi: 10.1111/pce.14040. [DOI] [PubMed] [Google Scholar]
- Lindahl A.J., Lawton A.J., Baeza J., Dowell J.A., Denu J.M. Site-specific lysine acetylation stoichiometry across subcellular compartments. Methods Mol. Biol. 2019;1983:79–106. doi: 10.1007/978-1-4939-9434-2_6. [DOI] [PubMed] [Google Scholar]
- Linster E., Wirtz M. N-terminal acetylation: an essential protein modification emerges as an important regulator of stress responses. J. Exp. Bot. 2018;69:4555–4568. doi: 10.1093/jxb/ery241. [DOI] [PubMed] [Google Scholar]
- Linster E., Layer D., Bienvenut W.V., Dinh T.V., Weyer F.A., Leemhuis W., Bruenje A., Hoffrichter M., Miklankova P., Kopp J., et al. The Arabidopsis Nα-acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response. New Phytol. 2020;228:554–569. doi: 10.1111/nph.16747. [DOI] [PubMed] [Google Scholar]
- Liu G.T., Jiang J.F., Liu X.N., Jiang J.Z., Sun L., Duan W., Li R.M., Wang Y., Lecourieux D., Liu C.H., et al. New insights into the heat responses of grape leaves via combined phosphoproteomic and acetylproteomic analyses. Hortic. Res. 2019;6:100. doi: 10.1038/s41438-019-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Yu F., Yang Z., Wang T., Xiong H., Chang C., Yu W., Li N. Establishment of dimethyl labeling-based quantitative acetylproteomics in Arabidopsis. Mol. Cell. Proteomics. 2018;17:1010–1027. doi: 10.1074/mcp.RA117.000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Cao J., Gao X., Zhou Y., Wen L., Yang X., Yao X., Ren J., Xue Y. Cpla 1.0: an integrated database of protein lysine acetylation. Nucleic Acids Res. 2011;39:D1029–D1034. doi: 10.1093/nar/gkq939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Smigel A., Tsai Y.C., Braam J., Berkowitz G.A. Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol. 2008;148:818–828. doi: 10.1104/pp.108.125104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx H., Minogue C.E., Jayaraman D., Richards A.L., Kwiecien N.W., Siahpirani A.F., Rajasekar S., Maeda J., Garcia K., Del Valle-Echevarria A.R., et al. A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat. Biotechnol. 2016;34:1198–1205. doi: 10.1038/nbt.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Braga M.N., Verano-Braga T., Leon I.R., Antonacci D., Nogueira F.C.S., Thelen J.J., Larsen M.R., Palmisano G. Modulation of protein phosphorylation, N-glycosylation and lys-acetylation in grape (Vitis vinifera) mesocarp and exocarp owing to Lobesia botrana infection. Mol. Cell. Proteomics. 2012;11:945–956. doi: 10.1074/mcp.M112.020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Lv Y., Mujahid H., Edelmann M.J., Zhao H., Peng X., Peng Z. Proteome-wide lysine acetylation identification in developing rice (Oryza sativa) seeds and protein co-modification by acetylation, succinylation, ubiquitination, and phosphorylation. Biochim. Biophys. Acta Proteins Proteom. 2018;1866:451–463. doi: 10.1016/j.bbapap.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Millar A.H., Heazlewood J.L., Giglione C., Holdsworth M.J., Bachmair A., Schulze W.X. The scope, functions, and dynamics of posttranslational protein modifications. Annu. Rev. Plant Biol. 2019;70:119–151. doi: 10.1146/annurev-arplant-050718-100211. [DOI] [PubMed] [Google Scholar]
- Milne J.L.S., Shi D., Rosenthal P.B., Sunshine J.S., Domingo G.J., Wu X.W., Brooks B.R., Perham R.N., Henderson R., Subramaniam S. Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: a multifunctional catalytic machine. EMBO J. 2002;21:5587–5598. doi: 10.1093/emboj/cdf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Keitany G., Li Y., Wang Y., Ball H.L., Goldsmith E.J., Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- Nallamilli B.R.R., Edelmann M.J., Zhong X., Tan F., Mujahid H., Zhang J., Nanduri B., Peng Z. Global analysis of lysine acetylation suggests the involvement of protein acetylation in diverse biological processes in rice (Oryza sativa) PLoS One. 2014;9:e89283. doi: 10.1371/journal.pone.0089283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Narita T., Weinert B.T., Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019;20:156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- O'Leary B.M., Scafaro A.P., Fenske R., Duncan O., Stroher E., Petereit J., Millar A.H. Rubisco lysine acetylation occurs at very low stoichiometry in mature Arabidopsis leaves: implications for regulation of enzyme function. Biochem. J. 2020;477:3885–3896. doi: 10.1042/BCJ20200413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Wang L., Chen S., Lv X., Lu S., Cheng C.L., Tan F., Chen W. Protein acetylation as a mechanism for Kandelia candel's adaption to daily flooding. Tree Physiol. 2018;38:895–910. doi: 10.1093/treephys/tpx162. [DOI] [PubMed] [Google Scholar]
- Pandolfi P.P., Sonati F., Rivi R., Mason P., Grosveld F., Luzzatto L. Targeted disruption of the housekeeping gene encoding glucose-6-phosphate-dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J. 1995;14:5209–5215. doi: 10.1002/j.1460-2075.1995.tb00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B., Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Rao R.S.P., Thelen J.J., Miernyk J.A. Is Lys-Nε-acetylation the next big thing in post-translational modifications? Trends Plant Sci. 2014;19:550–553. doi: 10.1016/j.tplants.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Rensing S.A., Goffinet B., Meyberg R., Wu S.Z., Bezanilla M. The moss Physcomitrium (Physcomitrella) patens: a model organism for non-seed plants. Plant Cell. 2020;32:1361–1376. doi: 10.1105/tpc.19.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart S.B., Lin S., Britton L.M., Krajewski K., Keogh M.C., Garcia B.A., Strahl B.D. Poly-acetylated chromatin signatures are preferred epitopes for site-specific histone H4 acetyl antibodies. Sci. Rep. 2012;2:489. doi: 10.1038/srep00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylott E.L., Eastmond P.J., Gilday A.D., Slocombe S.P., Larson T.R., Baker A., Graham I.A. The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal beta-oxidation is essential for seedling establishment. Plant J. 2006;45:930–941. doi: 10.1111/j.1365-313X.2005.02650.x. [DOI] [PubMed] [Google Scholar]
- Salvato F., Havelund J.F., Chen M., Rao R.S.P., Rogowska-Wrzesinska A., Jensen O.N., Gang D.R., Thelen J.J., Moller I.M. The potato tuber mitochondrial proteome. Plant Physiol. 2014;164:637–653. doi: 10.1104/pp.113.229054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Valls E., Kouzarides T., Martinez-Balbas M. Mechanisms of P/CAF auto-acetylation. Nucleic Acids Res. 2003;31:4285–4292. doi: 10.1093/nar/gkg655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B., Rardin M.J., MacLean B.X., Zawadzka A.M., Frewen B.E., Cusack M.P., Sorensen D.J., Bereman M.S., Jing E., Wu C.C., et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline application to protein acetylation and phosphorylation. Mol. Cell. Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C., Beilsten-Edmands V., Mohammed S., Robinson C.V. Acetylation and phosphorylation control both local and global stability of the chloroplast F-1 ATP synthase. Sci. Rep. 2017;7:44068. doi: 10.1038/srep44068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzlaender M., Koenig A.C., Sweetlove L.J., Finkemeier I. The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J. Exp. Bot. 2012;63:1735–1750. doi: 10.1093/jxb/err374. [DOI] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Shen Y., Lei T., Cui X., Liu X., Zhou S., Zheng Y., Guerard F., Issakidis-Bourguet E., Zhou D.X. Arabidopsis histone deacetylase HDA15 directly represses plant response to elevated ambient temperature. Plant J. 2019;100:991–1006. doi: 10.1111/tpj.14492. [DOI] [PubMed] [Google Scholar]
- Shimada T., Koumoto Y., Li L., Yamazaki M., Kondo M., Nishimura M., Hara-Nishimura I. AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 2006;47:1187–1194. doi: 10.1093/pcp/pcj103. [DOI] [PubMed] [Google Scholar]
- Shockey J.M., Fulda M.S., Browse J.A. Arabidopsis contains nine long-chain acyl-coenzyme A synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 2002;129:1710–1722. doi: 10.1104/pp.003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P.K., Gao W., Liao P., Li Y., Xu F.C., Ma X.N., Long L., Song C.P. Comparative acetylome analysis of wild-type and fuzzless-lintless mutant ovules of upland cotton (Gossypium hirsutum Cv. Xu142) unveils differential protein acetylation may regulate fiber development. Plant Physiol. Biochem. 2020;150:56–70. doi: 10.1016/j.plaphy.2020.02.031. [DOI] [PubMed] [Google Scholar]
- Smith-Hammond C.L., Hoyos E., Miernyk J.A. The pea seedling mitochondrial Nε-lysine acetylome. Mitochondrion. 2014;19:154–165. doi: 10.1016/j.mito.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Smith-Hammond C.L., Swatek K.N., Johnston M.L., Thelen J.J., Miernyk J.A. Initial description of the developing soybean seed protein Lys-Nε-acetylome. J. Proteomics. 2014;96:56–66. doi: 10.1016/j.jprot.2013.10.038. [DOI] [PubMed] [Google Scholar]
- Song G., Walley J.W. Dynamic protein acetylation in plant-pathogen interactions. Front. Plant Sci. 2016;7:421. doi: 10.3389/fpls.2016.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi B., Soares N.C., Ravikumar V., Macek B. Proteomics reveals evidence of cross-talk between protein modifications in bacteria: focus on acetylation and phosphorylation. Curr. Opin. Microbiol. 2012;15:357–363. doi: 10.1016/j.mib.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Sterner R., Vidali G., Allfrey V.G. Studies of acetylation and deacetylation in high mobility group proteins: identification of the sites of acetylation in HMG-1. J. Biol. Chem. 1979;254:1577–1583. [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Gene Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Svinkina T., Gu H., Silva J.C., Mertins P., Qiao J., Fereshetian S., Jaffe J.D., Kuhn E., Udeshi N.D., Carr S.A. Deep, quantitative coverage of the lysine acetylome using novel anti-acetyl-lysine antibodies and an optimized proteomic workflow. Mol. Cell. Proteomics. 2015;14:2429–2440. doi: 10.1074/mcp.O114.047555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L.J., Beard K.F.M., Nunes-Nesi A., Fernie A.R., Ratcliffe R.G. Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci. 2010;15:462–470. doi: 10.1016/j.tplants.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Taunton J., Hassig C.A., Schreiber S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Uhrig R.G., Schlapfer P., Mehta D., Hirsch-Hoffmann M., Gruissem W. Genome-scale analysis of regulatory protein acetylation enzymes from photosynthetic eukaryotes. BMC Genomics. 2017;18:514. doi: 10.1186/s12864-017-3894-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluchamy A., Rastogi A., Lin X., Lombard B., Murik O., Thomas Y., Dingli F., Rivarola M., Ott S., Liu X., et al. An integrative analysis of post-translational histone modifications in the marine diatom Phaeodactylum tricornutum. Genome Biol. 2015;16:102. doi: 10.1186/s13059-015-0671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat S., Gregory C., Sturges J., Gan Q., Fan C. Studying the lysine acetylation of malate dehydrogenase. J. Mol. Biol. 2017;429:1396–1405. doi: 10.1016/j.jmb.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E., Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- Wagner G.R., Payne R.M. Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley J.W., Shen Z., McReynolds M.R., Schmelz E.A., Briggs S.P. Fungal-induced protein hyperacetylation in maize identified by acetylome profiling. Proc. Natl. Acad. Sci. U S A. 2018;115:210–215. doi: 10.1073/pnas.1717519115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Boavida L.C., Ron M., McCormick S. Truncation of a protein disulfide isomerase, PDIL2-1, delays embryo sac maturation and disrupts pollen tube guidance in Arabidopsis thaliana. Plant Cell. 2008;20:3300–3311. doi: 10.1105/tpc.108.062919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hou Y., Qiu J., Li Z., Zhao J., Tong X., Zhang J. A quantitative acetylomic analysis of early seed development in rice (Oryza sativa L.) Int. J. Mol. Sci. 2017;18:1376. doi: 10.3390/ijms18071376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert B.T., Moustafa T., Iesmantavicius V., Zechner R., Choudhary C. Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 2015;34:2620–2632. doi: 10.15252/embj.201591271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert B.T., Satpathy S., Hansen B.K., Lyon D., Jensen L.J., Choudhary C. Accurate quantification of site-specific acetylation stoichiometry reveals the impact of sirtuin deacetylase CobB on the E. coli acetylome. Mol. Cell. Proteomics. 2017;16:759–769. doi: 10.1074/mcp.M117.067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert B.T., Schoelz C., Wagner S.A., Iesmantavicius V., Su D., Daniel J.A., Choudhary C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Weinert B.T., Iesmantavicius V., Moustafa T., Scholz C., Wagner S.A., Magnes C., Zechner R., Choudhary C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 2014;10:833. doi: 10.15252/msb.156513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub P.G., Beanland L. Insect vectors of phytoplasmas. Annu. Rev. Entomol. 2006;51:91–111. doi: 10.1146/annurev.ento.51.110104.151039. [DOI] [PubMed] [Google Scholar]
- Wu X., Oh M.H., Schwarz E.M., Larue C.T., Sivaguru M., Imai B.S., Yau P.M., Ort D.R., Huber S.C. Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 2011;155:1769–1778. doi: 10.1104/pp.110.165852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Jing D., Kong L., Zhang J., OuYang F., Zhang H., Wang J., Zhang S. Global lysine acetylome analysis of desiccated somatic embryos of Picea asperata. Front. Plant Sci. 2016;7:1927. doi: 10.3389/fpls.2016.01927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Poirier Y. The protein acetylome and the regulation of metabolism. Trends Plant Sci. 2012;17:423–430. doi: 10.1016/j.tplants.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Peng X., Cheng Z., Liu W., Wang G.L. A comprehensive catalog of the lysine-acetylation targets in rice (Oryza sativa) based on proteomic analyses. J. Proteomics. 2016;138:20–29. doi: 10.1016/j.jprot.2016.01.019. [DOI] [PubMed] [Google Scholar]
- Xu J., Xu H., Liu Y., Wang X., Xu Q., Deng X. Genome-wide identification of sweet orange (Citrus sinensis) histone modification gene families and their expression analysis during the fruit development and fruit-blue mold infection process. Front. Plant Sci. 2015;6:607. doi: 10.3389/fpls.2015.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Liu Q., Chen Z., Yue Y., Liu Y., Zhao Y., Zhou D.X. Histone deacetylases control lysine acetylation of ribosomal proteins in rice. Nucleic Acids Res. 2021;49:4613–4628. doi: 10.1093/nar/gkab244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.X., Chen W., Ma C.L., Shen S.Y., Zhou Y.Y., Zhou L.Q., Chen L. Proteome and acetyl-proteome profiling of Camellia sinensis cv. ‘Anjin Baicha’ during periodic albinism reveals alterations in photosynthetic and secondary metabolite biosynthetic pathways. Front. Plant Sci. 2017;8:2104. doi: 10.3389/fpls.2017.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C., Liu S., Chen C., Zhu J., Yang X., Zhou Y., Guo R., Liu X., Gong Z. Global proteome analysis links Lysine acetylation to diverse functions in Oryza Sativa. Proteomics. 2018;18:1700036. doi: 10.1002/pmic.201700036. [DOI] [PubMed] [Google Scholar]
- Yamagata H., Sugimoto T., Tanaka K., Kasai Z. Biosynthesis of storage proteins in developing rice seeds. Plant Physiol. 1982;70:1094–1100. doi: 10.1104/pp.70.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Shen Z., Gao Z.F., Chao Q., Qian C.R., Zheng H., Wang B.C. A comprehensive analysis of the lysine acetylome reveals diverse functions of acetylated proteins during de-etiolation in Zea mays. J. Plant Physiol. 2020;248:153158. doi: 10.1016/j.jplph.2020.153158. [DOI] [PubMed] [Google Scholar]
- Yuan B., Liu T., Cheng Y., Gao S., Li L., Cai L., Yang J., Chen J., Zhong K. Comprehensive proteomic analysis of lysine acetylation in Nicotiana benthamiana after sensing CWMV infection. Front. Microbiol. 2021;12:672559. doi: 10.3389/fmicb.2021.672559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M., Fermani S., Costa A., Lemaire S.D., Trost P. Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front. Plant Sci. 2013;4:450. doi: 10.3389/fpls.2013.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhao Y., Zhou D.X. Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Res. 2017;45:12241–12255. doi: 10.1093/nar/gkx825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C.F., Grishin N.V., Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Zheng S., Yang J.S., Chen Y., Cheng Z. Comprehensive profiling of protein lysine acetylation in Escherichia coli. J. Proteome Res. 2013;12:844–851. doi: 10.1021/pr300912q. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Song L., Liang W., Mu P., Wang S., Lin Q. Comprehensive profiling of lysine acetylproteome analysis reveals diverse functions of lysine acetylation in common wheat. Sci. Rep. 2016;6:21069. doi: 10.1038/srep21069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Xia N., Zhao D.G. Analysis of acetylated proteomics in response to drought stress of Phoebe zhennan leaves. Plant Physiol. J. 2021;57:1371–1381. [Google Scholar]
- Zhao H., Zhong S., Sang L., Zhang X., Chen Z., Wei's Q., Chen G., Liu J., Yu Y. PaACL silencing accelerates flower senescence and changes the proteome to maintain metabolic homeostasis in Petunia hybrida. J. Exp. Bot. 2020;71:4858–4876. doi: 10.1093/jxb/eraa208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen S., Deng X., Wang J., Zhu G., Cao H., Yuan L., Yan Y. First comprehensive proteome analyses of lysine acetylation and succinylation in seedling leaves of Brachypodium distachyon L. Sci. Rep. 2016;6:31576. doi: 10.1038/srep31576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Ge J., Bao C., Chang W., Liu J., Shao J., Liu X., Su L., Pan L., Zhou D.X. Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Mol. Plant. 2020;13:598–611. doi: 10.1016/j.molp.2019.12.011. [DOI] [PubMed] [Google Scholar]
- Zhou C.H., Zhang L., Duan J., Miki B., Wu K.Q. Histone deacetylase19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell. 2005;17:1196–1204. doi: 10.1105/tpc.104.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Finkemeier I., Guan W., Tossounian M.A., Wei B., Young D., Huang J., Messens J., Yang X., Zhu J., et al. Oxidative stress-triggered interactions between the succinyl- and acetyl-proteomes of rice leaves. Plant Cell Environ. 2018;41:1139–1153. doi: 10.1111/pce.13100. [DOI] [PubMed] [Google Scholar]
- Zhu G.R., Yan X., Zhu D., Deng X., Wu J.S., Xia J., Yan Y.M. Lysine acetylproteome profiling under water deficit reveals key acetylated proteins involved in wheat grain development and starch biosynthesis. J. Proteomics. 2018;185:8–24. doi: 10.1016/j.jprot.2018.06.019. [DOI] [PubMed] [Google Scholar]
- Zhu J.K. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ratio indicates the proportion of Lys-acetylated proteins with a certain number of Lys-acetylated sites relative to all identified Lys-acetylated proteins in plant Lys-acetylproteomes.