Abstract
The endoderm, one of the three primary germ layers, gives rise to lung, liver, stomach, intestine, colon, pancreas, bladder, and thyroid. These endoderm-originated organs are subject to many life-threatening diseases. However, primary cells/tissues from endodermal organs are often difficult to grow in vitro. Human pluripotent stem cells (hPSCs), therefore, hold great promise for generating endodermal cells and their derivatives for the development of new therapeutics against these human diseases. Although a wealth of research has provided crucial information on the mechanisms underlying endoderm differentiation from hPSCs, increasing evidence has shown that metabolism, in connection with epigenetics, actively regulates endoderm differentiation in addition to the conventional endoderm inducing signals. Here we review recent advances in metabolic and epigenetic regulation of endoderm differentiation.
Keywords: metabolic switch, epigenetic remodeling, endodermal gene expression, histone crotonylation, endoderm differentiation
Efficient Endoderm Differentiation in vitro Has Important Implications in Regenerative Medicine
During gastrulation, the totipotent epiblasts in a developing embryo form three germ layers, namely ectoderm, endoderm and mesoderm, which are the foundations for future tissue and organ development. Endoderm is the tissue precursor to organs like pancreas, liver, stomach, intestines, lungs, bladder, and thyroid [1]. These endodermal organs are essential for survival, and their dysfunction has been associated with many diseases, including diabetes, gastroenteritis, colitis, fatty liver disease, acute liver failure, and various types of cancer. However, it is technically and ethically challenging to grow primary endodermal cells/tissues in vitro. Thus, efficient induction of pure endodermal cells from human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and induced human pluripotent stem cells (hiPSCs), is a prerequisite for successful differentiation of downstream endoderm-derived tissues/organs for regenerative medicine [2].
Recent research advances have uncovered several crucial signaling pathways and genetic factors that regulate endodermal differentiation of PSCs. However, emerging evidence is revealing an active role of metabolic reprogramming and epigenetic modification in chromatin remodeling required for key endodermal gene expression during endoderm differentiation. The metabolism-coupled epigenetic remodeling integrates nutritional cues with canonical developmental signals and is critical for a highly coordinated endoderm lineage commitment of PSCs. This metabolism-epigenetics link may further pave the way for novel therapeutic strategies against diseases associated with endodermal organ dysfunction. Here, we review the recent advances on metabolic and epigenetic regulation of endoderm differentiation. First, we introduce canonical signaling pathways and factors that regulate endoderm differentiation. We then describe the metabolic switch during endoderm differentiation from PSCs and the connection between metabolism and epigenetics. We further review the various epigenetic states of the regulatory elements of endodermal genes in PSCs and their remodeling during endoderm differentiation. Finally, we discuss the importance of metabolism-coupled epigenetics in regulating endoderm differentiation, raise outstanding questions, and envision future directions.
Signaling Transduction and Transcriptional Regulation of Endoderm Differentiation
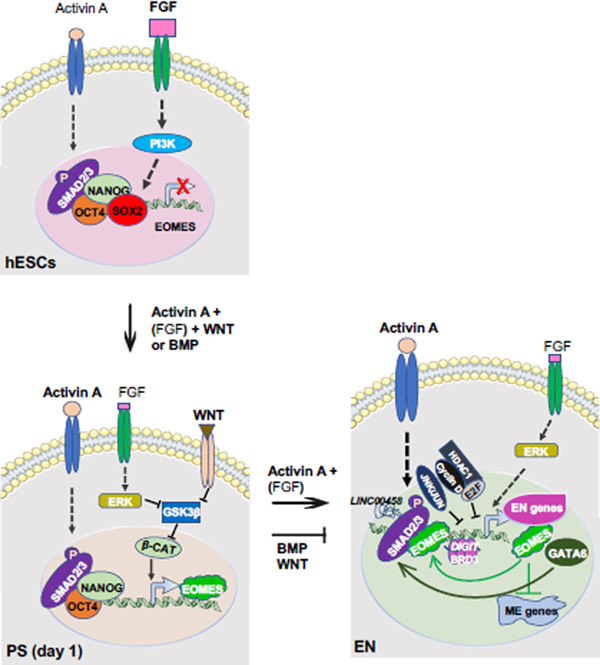
During in vivo mouse embryogenesis, the epiblast (Embryonic day 5.5 (E5.5)) differentiates into primitive streak (PS) around E6.5, which generates mesendoderm, a transient germ layer giving rise to mesoderm and endoderm, around E7.0-E7.5 [3–5]. When induced for endoderm differentiation in vitro, hPSCs differentiate into PS or mesendoderm around 24 hours after induction, then to endodermal cells after 3 – 5 days, depending on different protocols. Several developmental signals are required for differentiation of PS and endoderm, including WNT, BMP, FGF, and the Nodal/Activin A pathway [6–9] (Figure 1). WNT, BMP, or FGF signaling are essential for differentiation of PS or mesendoderm, as inhibition of any of the signals impairs PS formation on day 1 of endoderm differentiation from hESCs [8, 9]. After PS is generated, high Nodal/Activin A and low endogenous FGF signals effectively differentiate cells into endoderm in the following days, while BMP and WNT signaling promotes PS to mesoderm lineage and represses endoderm differentiation [8, 10, 11] (Figure 1). Nodal/Activin A binds and activates a heteromeric complex of TGF-β receptors, which phosphorylate SMAD2/3 [12]. Phosphorylated SMAD2/3 (p-SMAD2/3) translocate to the nucleus, bind to the regulatory elements of key endoderm lineage specifiers, such as FOXA2, SOX17, GSC and GATA6, and promote their expression [13, 14] (Figure 1). Endoderm differentiation of mouse ESCs (mESCs) can also be induced by similar signals or experimental conditions in vitro [15]. It is worth noting that the dosage of these signals dictates their impacts on cell fate determination, as low levels of Nodal/Activin A and high levels of FGF are required for pluripotency maintenance of hPSCs [16–18].
Figure 1. Canonical developmental signals and factors important for differentiation of PS and endoderm.
In primed hESCs, low Activin A and high FGF signaling promote the expression of pluripotency genes via their downstream effectors like SMAD2/3 and PI3K, respectively. OCT4, SOX2, and NANOG bind to the regulatory elements of EOMES and repress its expression in undifferentiated hESCs. After induction of endoderm differentiation by exogeneous Activin A with BMP or WNT, reduction of SOX2 allows NANOG to promote EOMES expression. Meanwhile, low level of FGF signaling activates ERK (MAPK), which inhibits GSK3β, an inhibitor of WNT/β-catenin signaling. Exogeneous WNT signals also inhibit GSK3β to activate β-catenin to promote EOMES expression. EOMES then interacts with phosphorylated SMAD2/3 to induce itself, GATA6, and genes characteristically expressed in the PS and endoderm. After PS is generated, exogeneous Nodal/Activin A and endogenous FGF [(FGF)] signaling effectively differentiate cells into endoderm in the following days, while BMP and WNT signaling promotes PS to mesoderm lineage and represses endoderm differentiation. Additionally, GATA6, LINC00458 which interacts with SMAD2/3, and BRD3 condensates with DIGIT also promote endoderm differentiation. Cyclin D which recruits E2F and HDAC1, and JNK/JUN family bind to endoderm loci to block endoderm differentiation. (FGF): endogenous FGF; β-CAT: β-catenin, EN: endoderm; ME: mesoderm.
Many additional factors that modulate endoderm differentiation have recently been identified. NANOG, a key pluripotency factor, is also important for induction of EOMES, a T-box gene important for embryonic development of PS and endoderm [19]. In undifferentiated hESCs, NANOG binds together with OCT4, SOX2, and SMAD2/3 to the regulatory elements of EOMES and represses EOMES expression. Upon endoderm differentiation, the reduction of SOX2 allows NANOG to promote the expression of EOMES, which then interacts with SMAD2/3, to induce the expression of itself and other PS and endodermal genes [19] (Figure 1). GATA6, a highly conserved zinc-finger transcription factor, is required for efficient endoderm formation in hPSCs by cooperating with EOMES/SMAD2/3 [20–23]. Factors that regulate cell cycles can also influence endoderm differentiation of hPSCs. For example, environmental WNT levels can control the G1 length distribution of a hPSC population, and hPSCs with a short G1 length tend to differentiate into mesendoderm versus neuroectoderm [24]. Cyclin D, a key cell cycle regulator, coordinates cell cycle progress with cell fate decisions in hPSCs. While cyclin D binds to endodermal loci together with E2F and HDAC1 to block endodermal gene expression, it also binds to neuroectodermal loci with SP1 and p300 to activate neuroectodermal gene expression [25]. As cyclin D expression is absent in early G1 and increased in later G1, hPSCs are biased to endoderm differentiation in early G1 and to neuroectoderm in later G1 [25]. Long noncoding RNAs (LncRNAs), including LINC00458 and DIGIT, also regulate endoderm differentiation of hPSCs. LINC00458 is upregulated on soft substrate for hPSC differentiation towards an early endodermal lineage through SMAD2/3 [26]. DIGIT interacts with the bromodomain and extra-terminal domain protein BRD3 to form phase-separated condensates, which bind enhancers of endodermal transcriptional factors (TFs) and promote endoderm differentiation of hPSCs [27, 28]. Additionally, five genes in the mitogen-activated protein kinase (MAPK) JNK/JUN pathway have recently been identified as key barriers of endoderm differentiation of hESCs. These genes co-occupy ESC enhancers with OCT4, NANOG and SMAD2/3 to inhibit the exit from the pluripotent state, and thereby endoderm differentiation [29].
Metabolic Switch during Endoderm Differentiation from PSCs
PSCs at different pluripotent states and differentiated endodermal cells possess distinct metabolic programs to maintain their growth and functions. hESCs, a type of primed PSCs, heavily rely on glycolysis and one-carbon catabolism of methionine for rapid growth and pluripotency maintenance [30–32]. Naïve mESCs, however, can dynamically switch from glycolysis to oxidative phosphorylation on demand without impacting their pluripotency [33]. Instead, they are critically dependent on threonine catabolism (another one-carbon metabolism pathway) [34, 35] and glutamine [36] for self-renewal and maintenance of pluripotency.
After exiting from the pluripotent state, cells begin to acquire cell fate-specific metabolic programs during differentiation into three germ layers. For instance, differentiation of hESCs into endoderm and mesoderm, but not ectoderm, leads to a metabolic switch from glycolysis to oxidative phosphorylation [37]. In endodermal cells, this metabolic switch is associated with a drastic change of mitochondria [38], including morphological change from punctate forms in hESCs into elongated and fused extensive network in endodermal cells. The contents of mitochondrial DNA (mtDNA) and mass are also significantly increased, accompanied with enhanced ATP production, and increased expression of genes involved in oxidative phosphorylation [38]. The high-methionine metabolic flux in hESCs is also shifted to a low-consumption state in endodermal cells, as endodermal cells only require a low amount of methionine for cell growth and are not affected by methionine deprivation [32]. Another metabolic pathway that is presumably induced during PS and meso-endoderm but not ectoderm differentiation is the mevalonate metabolic pathway. Inhibition of this pathway by statins suppresses PS differentiation while promoting ectoderm differentiation (neurons) from mESCs [39].
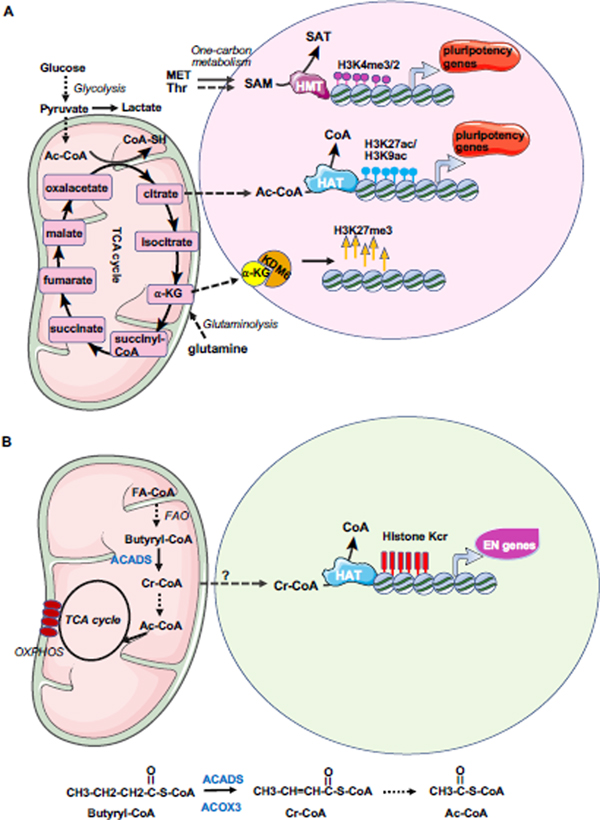
Emerging evidence shows that changes in metabolic states are active regulators of cell fate decisions, particularly via influencing epigenetics [40] (Figure 2). Metabolites, including S-adenosylmethionine (SAM), acetyl-CoA, and α-ketoglutarate (α-KG), are substrates or cofactors for epigenetic-modifying complexes [32, 35, 41, 42] [36]. For example, acetyl-CoA is the acetyl-donor for histone acetylation. The high rate of glycolysis in hESCs is essential for maintaining the acetyl-CoA level, and thus the histone acetylation level and pluripotency gene expression [31] (Figure 2A). SAM is the major cellular methyl donor for histone and DNA methylation, while α-KG, an intermediate of the TCA cycle, is a cofactor for histone and DNA demethylases (also called dioxygenase). Therefore, they are important for regulation of histone methylation (Figure 2A). In naïve mESCs, elevated α-KG promotes histone/DNA demethylation and maintains pluripotency [36]. SAM is regulated by one-carbon metabolism of threonine and methionine and its fluctuation is correlated with global H3K4me3, which is important for pluripotency maintenance [32, 35]. Crotonyl-CoA, a β-oxidation intermediate of butyryl-CoA, is an acyl-donor for histone crotonylation that specifically promotes endoderm differentiation [43] (Figure 2B). Farnesyl diphosphate, an intermediate metabolite from mevalonate metabolism, can be covalently attached to the cysteine residues within carboxyl-terminal CaaX motifs of Lamin B1 to anchor it to membranes, thus facilitating PS differentiation from mESCs [39]. In the following sections, we will review in detail the dynamic epigenetic regulation of endoderm differentiation.
Figure 2. Metabolic influence on ESC maintenance and endoderm differentiation.
(A) hESCs heavily rely on glycolysis and acetyl-CoA, and one-carbon metabolism of methionine and SAM for maintenance of epigenetics required for pluripotency. mESCs are dependent on glutamine metabolism and α-KG as well as one-carbon metabolism of threonine and SAM for maintaining pluripotency and the epigenetic status. Ac-CoA: acetyl-CoA; HMT: histone methyltransferase; HAT: histone acetyltransferase, SAM: S-adenosylmethionine, α-KG: α-ketoglutarate.
(B) Crotonyl-CoA from fatty acid oxidation (FAO) enhances histone crotonylation on endodermal genes and promotes endoderm differentiation. Upon endoderm differentiation, butyryl-CoA, a short-chain fatty acyl-CoA which may be produced from the longer-chain fatty-acyl-CoA via β-oxidation (or fatty acid oxidation), is catalyzed to produce crotonyl-CoA by ACADS. Crotonyl-CoA can be degraded to form 2 molecules of acetyl-CoA, which enters TCA cycle to reduce NAD+ to NADH for oxidative phosphorylation. Meanwhile, crotonyl-CoA can be present in the nucleus to enhance histone crotonylation on the regulatory elements of endodermal genes to promote endodermal gene expression. Kcr: lysine crotonylation; Cr-CoA: crotonyl-CoA, FAO: fatty acid oxidation; FA-CoA: fatty acyl-CoA; OXPHOS: oxidative phosphorylation; ACADS: acyl-CoA dehydrogenase short chain (in mitochondria); ACOX3: acyl-CoA oxidase (in peroxisome).
Profound Epigenetic Changes during Endoderm Differentiation Are Important for Efficient Induction of Endodermal Gene Expression
Genome-wide transcriptional and chromatin state mapping has revealed profound epigenetic changes coupled with the observed metabolic switch on both promoters and enhancers of endodermal genes during endoderm differentiation of ESCs [14, 44]. Particularly, promoters of many developmental genes are marked by bivalent chromatin marks H3K4me3 (an active mark) and H3K27me3 (a repressive mark) in ESCs that poise them for lineage-specific activation or repression [45–47]. Differentiation of ESCs into endoderm leads to rapid removal of H3K27me3 mark, resulting in monovalent formation and transcriptional activation of these genes [14, 44, 48].
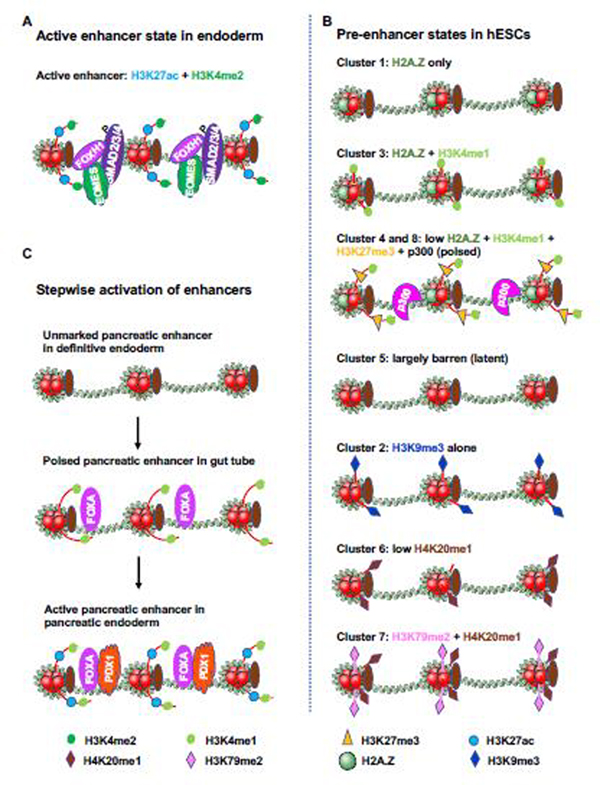
A recent transcriptional and epigenetic profiling of pure progenitor and endodermal cell populations showed that the enhancers of PS and endodermal genes in endodermal cells are highly enriched with active chromatin marks H3K27ac and H3K4me2 and are co-occupied with EOMES, SMAD2/3/4 and FOXH1 [8] (Figure 3A), indicating that these enhancers are active for gene expression. In the uncommitted hESCs, these endoderm enhancers are inactive for gene expression, but are marked by a multiplicity of distinct histone modifications and/or chromatin regulators for future activation,; therefore, they are referred to as pre-enhancer states [8]. Eight clusters of pre-enhancer states for endodermal enhancers in hESCs are identified based on their diverse chromatin marks (Figure 3B). Cluster 1 is occupied by H2A.Z only. Because H2A.Z-loaden nucleosomes are unstable and are readily displaced by transcription factors (TFs) [49, 50], these H2A.Z-marked endoderm pre-enhancers more readily attract EOMES, SMAD2/3/4, and FOXH1 upon differentiation, and become rapidly activated within 3 days of endoderm induction despite virtual absence of H3K4me1 [8], an epigenetic mark enriched at active and primed enhancers. Cluster 2 pre-enhancers are occupied by H3K9me3 alone, a repressive heterochromatic mark, and Cluster 5 is in a “latent” pre-enhancer state lacking any known histone modifications, suggesting these states are not appropriate for quick enhancer activation after differentiation [8]. Clusters 4 and 8 are marked by H3K27me3, together with H3K4me1 and p300, which place these pre-enhancers in a poised state for quick activation [8]. Given that endodermal genes are not only differentially induced during endoderm differentiation with distinct temporal dynamics and amplitudes, but also will be further differentially regulated during the subsequent differentiation into various endoderm-derived lineages, the presence of multiple pre-enhancer states would allow enhancers to be targeted by different complexes to form distinct chromatin structures for a highly coordinated, lineage-specific expression of different endodermal genes in response to various signals. Therefore, the establishment of a permissive chromatin landscape in ESCs is a prelude for efficient endoderm differentiation.
Figure 3. States of endodermal gene enhancers in hESCs and endodermal cells.
(A) Active enhancers are highly enriched for H3K27ac and H3K4me2, and are co-occupied by SMAD2/3/4, EOMES and FOXH1. The figure is adapted from [8]
(B) Chip-seq was performed in undifferentiated hESCs with chromatin marks and modifiers including H3K4me1, H3K4me2, H3K4me3, H2A.Z, H3K27me3, H3K9me3, H3K27ac, H3K9ac, H3K79me2, H4K20me1, CHD1, CHD7, EZH2, HDAC2, RBBP5, JARID1A and p300. The abundance of the marks/modifiers in the future endoderm enhancers were analyzed, organized by unbiased clustering, and illustrated. The active enhancer state in endodermal cells is highly enriched for H3K27ac and H3K4me2. Cluster 4 and 8 are characterized by low H2A.Z, H3K4me1, H3K27me3 and p300, representing the “poised” enhancer state for quick activation. H2A.Z renders nucleosome unstable and facilitates transcriptional factors to bind DNA. Cluster 2 is enriched by H2A.Z and H3K4me1, marking the enhancer for subsequent activation. Those pre-enhancers are devoid of any chromatin marks/modifiers as illustrated in cluster 5, are latent in activation. The presence of multiplicity of pre-enhancer states allows a precise control of enhancer activation under the influence of both lineage inducing signals and effectors. The figure is adapted from [8].
(C) Stepwise activation of pancreatic enhancers. Pancreatic enhancers are unmarked in definitive endoderm, are poised by H3K4me1 and FOXA proteins in gut tube, and are active in pancreatic endoderm after H3K27ac deposition and PDX1 binding. The figure is adapted from [51].
In support of this notion, a stepwise developmental transition at endoderm-derived cell lineage-specific enhancers from unmarked chromatin, H3K4me1 marked poised chromatin, to H3K27ac marked active chromatin has been discovered using purified cell populations from different defined stages of pancreatic and hepatic differentiation of hESCs [51] (Figure 3C). The acquisition of the poised enhancer state in the primitive gut tubes, which is first recognized by the pioneer TFs FOXA1 and FOXA2, predicts their ability to respond to inductive signals toward different lineages. Consistently, FOXA2 is required for pancreatic and foregut and the subsequent hepatic differentiation through enhancer priming and chromatin remodeling [52, 53]. Collectively, epigenetic priming of endodermal gene promoters and enhancers is important for the acquisition of a poised chromatin state for hESC-derived endodermal lineage intermediates to acquire developmental competence toward specific lineages.
Dynamic Regulation of H3K27 Trimethylation during Endoderm Differentiation
A critical element in epigenetic remodeling for differentiation is the establishment of the repressive H3K27me3 domain on differentiation genes in ESCs and the quick removal of this mark upon induction of differentiation [54, 55]. This process is dynamically regulated by the H3K27 trimethyltransferase Polycomb Group (PcG) protein complex and histone demethylases.
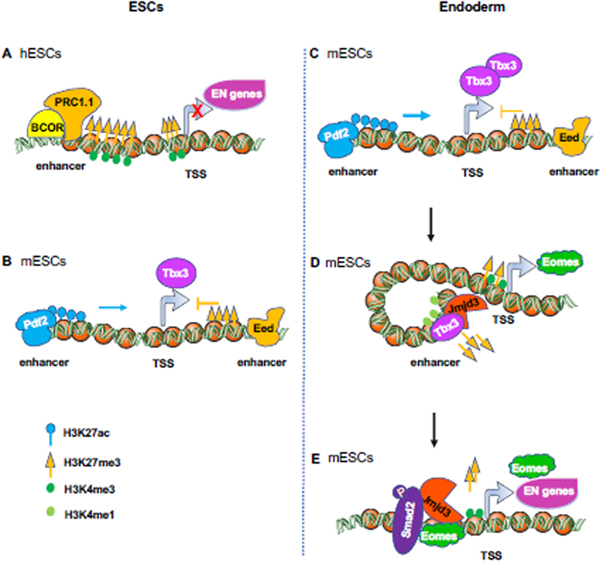
In hESCs, the expression of differentiation genes is actively repressed by PcG protein-mediated H3K27 trimethylation. Deletion of EZH2, the methyltransferase subunit of PRC2, leads to loss of promoter-localized H3K27me3, desuppression of key developmental genes, and defects of self-renewal, proliferation and differentiation [56]. BCOR, a BCL6 interacting corepressor [57] and a member of a non-canonical PRC1.1 complex, but neither PRC2 nor canonical PRC complexes, specifically mediates suppression of endodermal and mesodermal genes in hESCs [58] (Figure 4A). BCOR is first recruited to BCOR-responsive target promoters by a C-terminal linker region, then recruits PRC1.1 and canonical PRC complexes to maintain the H3K27me3 level. Its N-terminal repression domain also functions in part through facilitating H3K27me3 deposition at its target genes [58].
Figure 4. Dynamic regulation of H3K27 trimethylation on endodermal genes in ESCs and endodermal cells.
(A) BCOR, a member of PRC1.1 complex, specifically represses the expression of endodermal and mesodermal genes in hESCs through enhancement of H3K27me3.
(B-C) Eed and Dpf2 antagonize the expression of Tbx3 in mESCs. (B) Eed, a subunit of PRC2 complex binds an intragenic Tbx3 enhancer to oppose Dpf2 (a subunit of SWI-SNF complex)-dependent Tbx3 expression and mesendodermal differentiation in mESCs. (C) Dpf2 binds the distal enhancer of Tbx3, increases the H3K27ac level, and promotes the expression of Tbx3 during mesendodermal differentiation of mESCs.
(D-E) Jmjd3 and Eomes function in a positive feedback loop to promote endoderm differentiation from mESCs. (D) Jmjd3 physically associates with Tbx3, then binds to a bivalent enhancer (marked by H3K4me1 and H3K27me3) of Eomes, which in turn leads to a reduction of H3K27me3 deposition in the enhancer, and an interaction between enhancer and promoter, thereby promoting Eomes expression. (E) Jmjd3 and Smad2, recruited by Eomes, co-bind to the bivalent promoters (marked by H3K4me3 and H3K27me3) of core endoderm differentiation genes such as Eomes, Sox17, Gsc, Gata6, Mixl1 and Foxa2 to remove H3K27me3 for their promoter activation[48].
In mESCs, on the other hand, deletion of Ezh2 or Eed, a PRC2 subunit, has little effect on the deposition of promoter-localized H3K27me3 and their self-renewal or proliferation, partially due to compensation from Ezh1 [59–61]. Instead, the PRC complex suppresses endodermal gene expression through repression of Tbx3 (Figure 4B and 4C). Tbx3 is a TF required for mESC pluripotency maintenance [62], but it also paradoxically functions to specify mesendoderm cell fate by binding to key lineage determining factors like Eomes and enhancing paracrine Nodal signaling during mESC differentiation and in murine and Xenopus embryos [63] (Figure 4B). During mESC differentiation, the expression of Tbx3 is regulated by the antagonistical effects between the Brg or Brahma-associated factors (BAF) complex and PRC2 complex [64] (Figure 4B and 4C). Dpf2, a subunit of Baf45, binds the distal enhancer of Tbx3, increases the H3K27ac level, and promotes the expression of Tbx3 upon differentiation. In contrast, PRC2 subunit Eed binds an intragenic Tbx3 enhancer to oppose Dpf2-dependent Tbx3 expression and mesendodermal differentiation of mESCs [64].
Upon initiation of differentiation, the removal of H3K27me3 repressive mark present in poised pre-enhancers in ESCs is carried out by H3K27me3 demethylases, including the Jumonji domain-containing proteins, UTX (KDM6a) and JMJD3 (KDM6b) [48, 65–67]. UTX or JMJD3 deficiency does not affect the pluripotency of either human or mouse ESCs [48, 68, 69]. Endoderm differentiation of hESCs induces the expression of UTX or JMJD3, which is crucial for the expression of WNT3, a WNT signaling agonist that promotes PS formation before Nodal/Activin A in the early stage, and DKK1, a WNT signaling antagonist that inhibits mesoderm differentiation in the later stage [10, 68]. In mESCs, Jmjd3 physically associates with Tbx3 and binds to a bivalent enhancer (marked by H3K4me1 and H3K27me3) of Eomes, which reduces H3K27me3 deposition, induces an enhancer-promoter interaction, and promotes Eomes expression [48] (Figure 4D). Eomes further recruits Jmjd3 and Smad2 to co-bind to the bivalent promoters of core endoderm differentiation genes such as Eomes, Sox17, Gsc, Gata6, Mixl1, and Foxa2 to remove H3K27me3 for their promoter activation [48] (Figure 4E). Thus, Jmjd3 and Eomes function in a positive feedback loop to promote endoderm differentiation. Collectively, UTX or JMJD3-mediated H3K27me3 removal is important for the rapid induction of endoderm differentiation genes.
Histone Crotonylation Promotes Endoderm Differentiation of hESCs
Histone crotonylation, a newly identified histone acylation mark [70], activates gene expression just like histone acetylation but with an even stronger effect [70]. Histone crotonylation is also biochemically similar to histone acetylation, as both have a widespread distribution across genomes, are catalyzed by histone acetyltransferase p300 [70, 71], and are removed by histone deacetylases like SIRT1–3 and class I HDACs [72–75]. However, the physiological role of histone crotonylation is unclear. A recent study reported that a key function of this unique histone modification is to promote endoderm differentiation of hESCs [43] (Figure 2B). Histone crotonylation level is low in hESCs, but is increased in endodermal cells differentiated from hESCs and is highly enriched in endodermal cells of mouse embryos. Genome-wide transcriptional and chromatin profiling showed that histone crotonylation is highly enriched on regulatory elements of meso-endodermal genes just like H3K27ac, and that alterations of histone crotonylation levels are correlated with changes of gene expression. Furthermore, H4K77cr and H4K91cr, which reside on the nucleosome lateral surface and the histone-histone interface, respectively, are detected only in endodermal cells. Mutation of lysine at either site to arginine abolishes crotonylation and impairs endoderm differentiation, indicating that acylation on either site is important for endoderm differentiation [43].
Histone crotonylation is sensitive to cellular metabolism. Crotonyl-CoA, the donor for histone crotonylation, is a dehydrogenation product from butyryl-CoA catalyzed by mitochondrial acyl-CoA dehydrogenase short chain (ACADS) or peroxisomal acyl-CoA oxidase 3 (ACOX3). Crotonyl-CoA can also be generated by acyl-CoA synthetase short chain family members (ACSS2) directly from crotonate [71]. Upon endoderm differentiation, ACADS, ACOX3 and ACSS2 are induced to produce crotonyl-CoA that in turn enhances histone crotonylation deposition on regulatory elements of meso-endodermal genes. Knockout of ACADS or ACOX3 reduces intracellular crotonyl-CoA concentration, decreasing histone crotonylation deposition on promoters of endodermal genes and impairing endoderm differentiation while increasing ectoderm differentiation. Conversely, exogeneous addition of crotonate enhances intracellular crotonyl-CoA concentration, increasing histone crotonylation and promoting homogeneous endodermal gene expression [43, 76]. Therefore, intracellular crotonyl-CoA concentration is positively correlated with the abundance of histone crotonylation, which is important to specify meso-endoderm lineage commitment.
The metabolic switch-associated reduction of glycolytic metabolism that occurs during mesoderm and endoderm differentiation of hPSCs [37] is believed to be important to release stem cells from pluripotency thereby promoting differentiation [31, 77]. However, whether and how the increase of oxidative phosphorylation during this metabolic remodeling actively impacts this early-stage differentiation is unclear. Since crotonyl-CoA is produced during oxidation of fatty acids and amino acids, the discovery of histone crotonylation in promoting endoderm differentiation provides a direct link between endoderm differentiation-associated increase of oxidative phosphorylation, epigenetic reprogramming, and endodermal gene expression. In support of the notion that fatty acid oxidation, but not glucose oxidation, contributes to the increased oxidative phosphorylation in endoderm, addition of glucose does not increase oxygen consumption rates in endodermal and mesodermal cells [37] and hyperglycemia (high blood glucose) inhibits endoderm differentiation in vitro and in vivo [78].
DNA methylation Regulates Endoderm Differentiation
DNA methylation is stably maintained in somatic tissues. However, the mammalian genome undergoes two waves of global demethylation to reset the epigenome to a basic totipotent state during sexual reproduction, once in the primordial germ cells and the other in the preimplantation embryos [79]. The naïve mESCs, which are isolated from the preimplantation blastocysts, therefore, have a low global DNA methylation level and can maintain their self-renewal even in the absence of all three DNA methyltransferases (DNMTs) or all three Ten-eleven translocation (TET) family of dioxygenases, the DNA demethylases [80–82]. On the other hand, DNA methylation is required to silence pluripotency factors and to further pose a fundamental epigenetic barrier that guides and restricts differentiation [79, 83, 84]. An appropriate level of DNA methylation is, therefore, important for differentiation of mESCs. Consistently, loss of the TET enzymes leads to defects in mESC differentiation in vitro and PS patterning during mouse embryo gastrulation [81, 82].
In contrast to mESCs, the primed hESCs, which are at a later developmental stage, require DNMT1, but not DNMT3A nor DNMT3B, for survival [85]. Upon differentiation, the interplay between DNMT3B and TET proteins is required to maintain an appropriate amount of DNA methylation to ensure that lineage-specific transcription, which is silenced at hESCs, is robustly induced [86]. SALL3, a sal-like C2H2-type zinc-finger protein and an inhibitor of DNMT3B, regulates the differentiation propensity of hiPSCs through modulation of DNA methylation [87]. SALL3 expression in hiPSCs correlates positively with ectoderm differentiation capacity and negatively with meso-endoderm differentiation capacity. In hiPSCs, SALL3 binds to the CpG islands in gene bodies of WNT3A and WNT5A, where it inhibits the DNA methyltransferase activity of DNMT3B and decreases the gene body methylation. Knocking down SALL3 induces aberrant DNA hypermethylation and inhibits ectoderm differentiation while enhancing endoderm/mesoderm differentiation [87]. However, how SALL3 deficiency-induced DNMT3B-catalyzed CpG methylation leads to the enhanced meso-endodermal gene expression remains unclear.
Concluding Remarks
Increasing evidence from the past decade has demonstrated that metabolism, in connection with epigenetics, actively participates in regulating endoderm differentiation of PSCs. This metabolism-epigenetics coupling allows for a highly coordinated, rapid, and homogeneous endoderm lineage commitment and enhances the differentiation efficiency. This link opens the door for development of novel therapeutic manipulation of endoderm differentiation via supplementation of different key driving metabolites, which may help to advance PSC-based therapies for human diseases affecting endodermal organs. These findings also imply that altered epigenetic modifications may contribute to many other human diseases associated with metabolic dysregulation, including metabolic syndrome, developmental defects, cancer, and neurodegenerative diseases. Future studies along this line might have the potential for development of novel therapies against these human diseases.
Despite these advancements, much remains unknown. For instance, what signals trigger the metabolic switch during meso-endoderm differentiation and how these signals may interact with epigenetics are still unknown. What determines the epigenetic diversity of pre-enhancer configurations in ESCs, and how different chromatin marks are then deposited at the different intermediates along the differentiation process are also not clear. More importantly, how structurally similar epigenetic marks, such as acetylation and crotonylation, are specifically deposited on the regulatory elements of meso-endodermal genes at various differentiation stages remains elusive.
It is also worth noting that not all epigenetic marks are sensitive to metabolic alterations. For example, only a few histone methylation marks, including H3K4me3, are sensitive to methionine/threonine metabolism and the intracellular fluctuation of SAM [32, 35, 88]. Histone acetylation is also not as sensitive to metabolic supplementation as histone crotonylation [43]. One explanation for this phenomenon lies in the biochemistry of the epigenetic modification enzymes, which have different affinities (Km) towards different metabolite substrates. The Michaelis-Menten model of enzyme kinetics indicates that when the intracellular concentration of their small molecule substrates, such as SAM and acyl-CoA, is lower than the respective Km, the production of epigenetic marks will be sensitive to the metabolic availability of the substrate. Consequently, a low metabolite concentration and/or a high Km are associated with a high sensitivity of epigenetic marks to metabolism. Since the intracellular concentration of crotonyl-CoA in mel1 hESCs is about 600- to 1000-fold less than that of acetyl-CoA, it is reasonable that this low concentration is linked to histone crotonylation being highly sensitive to the change of intracellular crotonyl-CoA concentration upon endoderm differentiation as compared to histone acetylation [43]. Consistently, while histone acetylation is not sensitive to addition of exogeneous acetate in mel1 hESCs and HeLa cells where the intracellular acetyl-CoA concentration is saturate (e.g. 30 pmol/106 cells in mel1 hESCs) [43, 71], the H3K27ac and H3K9ac levels are enhanced by acetate supplementation in H1 hESCs with an intracellular acetyl-CoA concentration of 7 pmol/106 cells [31]. In the case of histone methylation, H3K4me3 is most sensitive to conditions that restrict SAM availability compared to many other histone methylation marks, possibly due to a comparatively high Km of its methyltransferase for SAM [32, 35, 88, 89]. Additionally, H3K4me3 is highly abundant in the euchromatin in mESCs, which may also contribute to its high sensitivity to the restriction of intracellular SAM [35, 90]. Therefore, the distinct sensitivity of different epigenetic marks to metabolism is an outcome of combined influences from the intrinsic kinetic characteristics of transferases and/or erasers, the intracellular concentration of metabolites, and the abundance of the histone epigenetic marks.
Another key question is how the global metabolic switch during endoderm differentiation specifically affects chromatin features at specific loci and causes specific gene expression alterations. One possibility is that DNA sequence-specific TFs recruit enzymes that produce or consume metabolites to influence local substrate concentrations at specific chromatin loci. The observation that crotonyl-CoA producing enzymes are induced and localized in the nucleus upon induction of endoderm differentiation of hESCs supports this notion [43]. Future investigations on how chromatin remodeling factors (e.g. writers, readers, and erasers) interact with metabolic enzymes (or their metabolites) and key differentiation TFs to coordinate endoderm differentiation will shed new light on the mechanisms underlying metabolic and epigenetic regulation of endoderm differentiation (see Outstanding questions box).
Outstanding Questions.
What signals trigger the metabolic switch during meso-endoderm differentiation and how these signals may interact with epigenetics?
What determines the epigenetic diversity of pre-enhancer configurations in ESCs, and how different chromatin marks are then stepwise deposited at the different intermediates along the differentiation process?
How are structurally similar epigenetic marks, such as acetylation and crotonylation, specifically deposited on the regulatory elements of meso-endodermal genes at different differentiation stages?
Why do different epigenetic marks have different sensitivities to alteration of metabolism? How do chromatin remodeling factors (e.g. writers, readers, and erasers) interact with metabolic enzymes (or their metabolites) and key differentiation TFs to coordinate endoderm differentiation?
How to manipulate epigenetics with key driving metabolites to advance PSC-based therapy for human diseases affecting endodermal organs?
Highlights.
Metabolic reprogramming upon endoderm differentiation of ESCs is coupled with profound epigenetic changes.
Epigenetic remodeling induces permissive chromatin state for endodermal gene expression.
Dynamics of H3K27 trimethylation during endoderm differentiation is modulated by PRC and demethylases.
Histone crotonylation promotes endoderm differentiation of hESCs in response to metabolic reprogramming.
Acknowledgements
We thank Drs. Paul Wade and Raja Jothi, and members of the Li laboratory for critical reading of the manuscript. We also thank Christine Caufield-Noll, NIH Library, for manuscript editing assistance. Images used to create figures were downloaded from Service Medical ART: SMART (https://smart.servier.com/image-set-download/). The work related to this article was supported by the Intramural Research Program of National Institute of Environmental Health Sciences of National Institutes of Health to X.L. (Z01 ES102205). We apologize to those colleagues whose work has not been cited due to the space limit.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nowotschin S et al. (2019) The endoderm: a divergent cell lineage with many commonalities. Development 146 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yiangou L et al. (2018) Human Pluripotent Stem Cell-Derived Endoderm for Modeling Development and Clinical Applications. Cell Stem Cell 22 (4), 485–499. [DOI] [PubMed] [Google Scholar]
- 3.Tam PP and Beddington RS (1987) The formation of mesodermal tissues in the mouse embryo during gastrulation and early organogenesis. Development 99 (1), 109–26. [DOI] [PubMed] [Google Scholar]
- 4.Lawson KA et al. (1991) Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113 (3), 891–911. [DOI] [PubMed] [Google Scholar]
- 5.Tada S et al. (2005) Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development 132 (19), 4363–74. [DOI] [PubMed] [Google Scholar]
- 6.McLean AB et al. (2007) Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells 25 (1), 29–38. [DOI] [PubMed] [Google Scholar]
- 7.Sumi T et al. (2008) Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development 135 (17), 2969–79. [DOI] [PubMed] [Google Scholar]
- 8.Loh KM et al. (2014) Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell 14 (2), 237–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sui L et al. (2013) Signaling pathways during maintenance and definitive endoderm differentiation of embryonic stem cells. Int J Dev Biol 57 (1), 1–12. [DOI] [PubMed] [Google Scholar]
- 10.Naujok O et al. (2014) The generation of definitive endoderm from human embryonic stem cells is initially independent from activin A but requires canonical Wnt-signaling. Stem Cell Rev Rep 10 (4), 480–93. [DOI] [PubMed] [Google Scholar]
- 11.Bernardo AS et al. (2011) BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9 (2), 144–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu MY and Hill CS (2009) Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell 16 (3), 329–43. [DOI] [PubMed] [Google Scholar]
- 13.Brown S et al. (2011) Activin/Nodal signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells 29 (8), 1176–85. [DOI] [PubMed] [Google Scholar]
- 14.Kim SW et al. (2011) Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol 357 (2), 492–504. [DOI] [PubMed] [Google Scholar]
- 15.Mfopou JK et al. (2014) Efficient definitive endoderm induction from mouse embryonic stem cell adherent cultures: a rapid screening model for differentiation studies. Stem Cell Res 12 (1), 166–77. [DOI] [PubMed] [Google Scholar]
- 16.Beattie GM et al. (2005) Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells 23 (4), 489–95. [DOI] [PubMed] [Google Scholar]
- 17.Singh AM et al. (2012) Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell 10 (3), 312–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding VM et al. (2010) FGF-2 modulates Wnt signaling in undifferentiated hESC and iPS cells through activated PI3-K/GSK3beta signaling. J Cell Physiol 225 (2), 417–28. [DOI] [PubMed] [Google Scholar]
- 19.Teo AK et al. (2011) Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev 25 (3), 238–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chia CY et al. (2019) GATA6 Cooperates with EOMES/SMAD2/3 to Deploy the Gene Regulatory Network Governing Human Definitive Endoderm and Pancreas Formation. Stem Cell Reports 12 (1), 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heslop JA et al. (2021) GATA6 defines endoderm fate by controlling chromatin accessibility during differentiation of human-induced pluripotent stem cells. Cell Rep 35 (7), 109145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi ZD et al. (2017) Genome Editing in hPSCs Reveals GATA6 Haploinsufficiency and a Genetic Interaction with GATA4 in Human Pancreatic Development. Cell Stem Cell 20 (5), 675–688 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiyaboonchai A et al. (2017) GATA6 Plays an Important Role in the Induction of Human Definitive Endoderm, Development of the Pancreas, and Functionality of Pancreatic beta Cells. Stem Cell Reports 8 (3), 589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang J et al. (2019) Control over single-cell distribution of G1 lengths by WNT governs pluripotency. PLoS Biol 17 (9), e3000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pauklin S et al. (2016) Initiation of stem cell differentiation involves cell cycle-dependent regulation of developmental genes by Cyclin D. Genes Dev 30 (4), 421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YF et al. (2020) Control of matrix stiffness promotes endodermal lineage specification by regulating SMAD2/3 via lncRNA LINC00458. Sci Adv 6 (6), eaay0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daneshvar K et al. (2020) lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nat Cell Biol 22 (10), 1211–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daneshvar K et al. (2016) DIGIT Is a Conserved Long Noncoding RNA that Regulates GSC Expression to Control Definitive Endoderm Differentiation of Embryonic Stem Cells. Cell Rep 17 (2), 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li QV et al. (2019) Genome-scale screens identify JNK-JUN signaling as a barrier for pluripotency exit and endoderm differentiation. Nat Genet 51 (6), 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J et al. (2011) UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J 30 (24), 4860–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moussaieff A et al. (2015) Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab 21 (3), 392–402. [DOI] [PubMed] [Google Scholar]
- 32.Shiraki N et al. (2014) Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab 19 (5), 780–94. [DOI] [PubMed] [Google Scholar]
- 33.Vlaski-Lafarge M et al. (2019) Bioenergetic Changes Underline Plasticity of Murine Embryonic Stem Cells. Stem Cells 37 (4), 463–475. [DOI] [PubMed] [Google Scholar]
- 34.Wang J et al. (2009) Dependence of mouse embryonic stem cells on threonine catabolism. Science 325 (5939), 435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shyh-Chang N et al. (2013) Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339 (6116), 222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carey BW et al. (2015) Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518 (7539), 413–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cliff TS et al. (2017) MYC Controls Human Pluripotent Stem Cell Fate Decisions through Regulation of Metabolic Flux. Cell Stem Cell 21 (4), 502–516 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S et al. (2020) TGFbeta-dependent mitochondrial biogenesis is activated during definitive endoderm differentiation. In Vitro Cell Dev Biol Anim 56 (5), 378–385. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto-Uchida Y et al. (2016) The mevalonate pathway regulates primitive streak formation via protein farnesylation. Sci Rep 6, 37697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chisolm DA and Weinmann AS (2018) Connections Between Metabolism and Epigenetics in Programming Cellular Differentiation. Annu Rev Immunol 36, 221–246. [DOI] [PubMed] [Google Scholar]
- 41.Cai L et al. (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42 (4), 426–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wellen KE et al. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324 (5930), 1076–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang Y et al. (2021) Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell 28 (4), 748–763 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie R et al. (2013) Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell 12 (2), 224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernstein BE et al. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125 (2), 315–26. [DOI] [PubMed] [Google Scholar]
- 46.Mikkelsen TS et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448 (7153), 553–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan G et al. (2007) Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1 (3), 299–312. [DOI] [PubMed] [Google Scholar]
- 48.Kartikasari AE et al. (2013) The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. EMBO J 32 (10), 1393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin C et al. (2009) H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41 (8), 941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z et al. (2012) Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 151 (7), 1608–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang A et al. (2015) Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 16 (4), 386–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Genga RMJ et al. (2019) Single-Cell RNA-Sequencing-Based CRISPRi Screening Resolves Molecular Drivers of Early Human Endoderm Development. Cell Rep 27 (3), 708–718 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee K et al. (2019) FOXA2 Is Required for Enhancer Priming during Pancreatic Differentiation. Cell Rep 28 (2), 382–393 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee TI et al. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125 (2), 301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boyer LA et al. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441 (7091), 349–53. [DOI] [PubMed] [Google Scholar]
- 56.Collinson A et al. (2016) Deletion of the Polycomb-Group Protein EZH2 Leads to Compromised Self-Renewal and Differentiation Defects in Human Embryonic Stem Cells. Cell Rep 17 (10), 2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huynh KD et al. (2000) BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev 14 (14), 1810–23. [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z et al. (2018) A Non-canonical BCOR-PRC1.1 Complex Represses Differentiation Programs in Human ESCs. Cell Stem Cell 22 (2), 235–251 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riising EM et al. (2014) Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell 55 (3), 347–60. [DOI] [PubMed] [Google Scholar]
- 60.Shen X et al. (2008) EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32 (4), 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chamberlain SJ et al. (2008) Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26 (6), 1496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niwa H et al. (2009) A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460 (7251), 118–22. [DOI] [PubMed] [Google Scholar]
- 63.Weidgang CE et al. (2013) TBX3 Directs Cell-Fate Decision toward Mesendoderm. Stem Cell Reports 1 (3), 248–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang W et al. (2019) The BAF and PRC2 Complex Subunits Dpf2 and Eed Antagonistically Converge on Tbx3 to Control ESC Differentiation. Cell Stem Cell 24 (1), 138–152 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agger K et al. (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449 (7163), 731–4. [DOI] [PubMed] [Google Scholar]
- 66.Hong S et al. (2007) Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A 104 (47), 18439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lan F et al. (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449 (7163), 689–94. [DOI] [PubMed] [Google Scholar]
- 68.Jiang W et al. (2013) Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res 23 (1), 122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee S et al. (2012) UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell 22 (1), 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan M et al. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146 (6), 1016–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sabari BR et al. (2015) Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell 58 (2), 203–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bao X et al. (2014) Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madsen AS and Olsen CA (2012) Profiling of substrates for zinc-dependent lysine deacylase enzymes: HDAC3 exhibits decrotonylase activity in vitro. Angew Chem Int Ed Engl 51 (36), 9083–7. [DOI] [PubMed] [Google Scholar]
- 74.Wei W et al. (2017) Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res 27 (7), 898–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sabari BR et al. (2017) Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol 18 (2), 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fang Y and Li X (2021) A simple, efficient, and reliable endoderm differentiation protocol for human embryonic stem cells using crotonate. STAR Protocols 2, 100659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gu W et al. (2016) Glycolytic Metabolism Plays a Functional Role in Regulating Human Pluripotent Stem Cell State. Cell Stem Cell 19 (4), 476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen ACH et al. (2017) Hyperglycemia impedes definitive endoderm differentiation of human embryonic stem cells by modulating histone methylation patterns. Cell Tissue Res 368 (3), 563–578. [DOI] [PubMed] [Google Scholar]
- 79.Messerschmidt DM et al. (2014) DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 28 (8), 812–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsumura A et al. (2006) Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11 (7), 805–14. [DOI] [PubMed] [Google Scholar]
- 81.Dai HQ et al. (2016) TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature 538 (7626), 528–532. [DOI] [PubMed] [Google Scholar]
- 82.Dawlaty MM et al. (2014) Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell 29 (1), 102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jackson M et al. (2004) Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol 24 (20), 8862–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Isagawa T et al. (2011) DNA methylation profiling of embryonic stem cell differentiation into the three germ layers. PLoS One 6 (10), e26052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liao J et al. (2015) Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet 47 (5), 469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verma N et al. (2018) TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nat Genet 50 (1), 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuroda T et al. (2019) SALL3 expression balance underlies lineage biases in human induced pluripotent stem cell differentiation. Nat Commun 10 (1), 2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tang S et al. (2017) Methionine metabolism is essential for SIRT1-regulated mouse embryonic stem cell maintenance and embryonic development. EMBO J 36 (21), 3175–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schvartzman JM et al. (2018) Metabolic regulation of chromatin modifications and gene expression. J Cell Biol 217 (7), 2247–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Azuara V et al. (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8 (5), 532–8. [DOI] [PubMed] [Google Scholar]