Abstract
Citrobacter rodentium (formerly Citrobacter freundii biotype 4280 and Citrobacter genomospecies 9) was described on the basis of biochemical characterization and DNA-DNA hybridization data and is the only Citrobacter species known to possess virulence factors homologous to those of the human pathogens enteropathogenic Escherichia coli and enterohemorrhagic E. coli. These virulence factors are encoded on the locus of enterocyte effacement (LEE), a pathogenicity island required for the characteristic attaching and effacing (AE) pathology seen in infection with these three pathogens. C. rodentium, which apparently infects only mice, provides a useful animal model for studying the molecular basis of AE pathology. No work has been done to assess differences in pathogenicity between C. rodentium isolates from diverse sources. Here, we report the examination of 15 C. rodentium isolates using a battery of genetic and biochemical approaches. No differences were observed between the isolates by repetitive-element sequence-based PCR analysis, biochemical analysis, and possession of LEE-specific virulence factors. These data suggest that members of the species are clonal. We further characterized an atypical E. coli strain from Japan called mouse-pathogenic E. coli (MPEC) that, in our hands, caused the same disease as C. rodentium. Applying the same battery of tests, we found that MPEC possesses LEE-encoded virulence factors and is indistinguishable from the previously characterized C. rodentium isolate DBS100. These results demonstrate that MPEC is a misclassified C. rodentium isolate and that members of this species are clonal and represent the only known attaching and effacing bacterial pathogen of mice.
Citrobacter rodentium, previously designated Citrobacter freundii biotype 4280 and Citrobacter genomospecies 9, is the etiologic agent of transmissible murine colonic hyperplasia (TMCH), a naturally occurring disease of laboratory mice characterized by epithelial cell hyperproliferation in the descending colon (1, 5, 33). C. rodentium infection in most adult mice is self-limiting, with little morbidity or mortality. In contrast, suckling mice are more susceptible, developing diarrhea, retarded growth, and rectal prolapse that is associated with colonic inflammation, and show significant mortality (2). Pathologic changes in TMCH include grossly detectable thickening and increased rigidity of the colon and microscopic evidence of epithelial cell hyperplasia, including marked crypt elongation, increased numbers of mitotic figures, and goblet cell loss. These histopathologic alterations are maximal 2 to 3 weeks postinfection and resolve by 2 months postinfection. Recent work by Higgins and coworkers (11) has demonstrated that C. rodentium infection results in a Th1-type immune response, characterized by elevated levels of interleukin-12, gamma interferon, and tumor necrosis factor alpha.
Ultrastructural examination of colonic tissue from mice infected with C. rodentium shows large numbers of bacteria intimately attached to the epithelial cell surface, effacement of the normal brush border, and pedestal-like extensions of the epithelial cells beneath the adherent bacteria (17, 31). These ultrastructural changes, called attaching and effacing (AE) lesions, are indistinguishable from those caused by enteropathogenic Escherichia coli (EPEC) (24, 30) and enterohemorrhagic E. coli (EHEC) (8). The genes necessary and sufficient for the AE pathology are located on a 35-kb pathogenicity island, the locus of enterocyte effacement (LEE) (22). Expression of the EPEC LEE in a laboratory strain of E. coli is sufficient to confer AE activity (23). C. rodentium has also been shown to possess homologs of LEE-located genes (28, 31) and is currently the only known murine AE pathogen.
In the late 1960s and early 1970s, outbreaks of rectal prolapse and diarrhea associated with moderate mortality were reported in several mouse colonies (1, 4, 7, 9). Both Brennan et al. (4), at the Argonne National Laboratory, and Ediger et al. (9), at the Frederick Cancer Research Center, isolated an atypical C. freundii strain from affected mice and demonstrated that the isolates were the etiologic agents of their respective outbreaks. C. freundii biotype 4280 was isolated by Barthold et al. (1) from a third outbreak and was shown to be essentially the same as the Argonne National Laboratory and Frederick isolates. Barthold et al. also went on to fulfill Koch's postulates with this strain (1). Subsequent studies on the microbial and molecular pathogenesis of these atypical C. freundii strains have been done with Barthold's C. freundii biotype 4280 isolate (28, 31, 32), which was later reclassified as C. rodentium (33).
In 1964, prior to these outbreaks in the United States, a closed colony of DDY mice at the National Institute of Health in Japan experienced an explosive outbreak of diarrhea accompanied by high mortality in suckling animals. Attempts to eradicate the etiologic agent were unsuccessful, and the colony was depopulated (25). A previously unidentified pathogen, later named mouse-pathogenic E. coli (MPEC), was isolated from infected animals and identified as the etiologic agent. A gram-negative, nonmotile rod, MPEC was classified as an atypical E. coli isolate based on its biochemical characteristics (26).
Clinically, MPEC disease, also known as infectious megaenteron of mice, is very similar to TMCH. It occurs most frequently and is most severe in suckling mice and is characterized clinically by diarrhea, coat ruffling, weight loss, and high rates of mortality (25, 26). Histopathological examination of the small intestine shows a replacement of the mature villus cells with immature crypt-type cells and elongated crypts as a result of MPEC-induced hyperplasia. There is a distinct lack of inflammatory reactions. Similar architectural changes are observed in the large intestine, with the addition of a marked depletion of goblet cells, most likely due to their hyperplasia-induced replacement by immature crypt-type cells (25, 26). The clinical signs and the histopathological changes seen in the large intestine closely resemble those reported for C. rodentium disease (1, 2). While small intestine involvement is not considered a hallmark of C. rodentium disease, Barthold and coworkers (2) did report similar alterations in their most severely affected mice.
Further work on MPEC, undertaken by Itoh and coworkers, has focused on the in vivo analysis of the role of host genetic background and intestinal microbiota in susceptibility to disease (12–15). Similarly, Barthold et al. (3) have examined the ability of diet and genetic background to modulate pathogenesis. To date, unlike C. rodentium, no work has been published on the virulence factors of MPEC. In addition, a comparative assessment of the diseases caused by these two bacteria has not been undertaken in the same strain of mouse.
Clinical disease progression, gross colonic lesions, and microscopic changes seen in both C. rodentium and MPEC infections suggested similar mechanisms of pathogenesis. Since LEE-located genes are required for virulence in C. rodentium (28, 32), we investigated the possibility that MPEC might also have a LEE and might be a murine AE E. coli strain. We now report that MPEC does possess a LEE, and, based on several lines of investigation, we conclude that MPEC actually is a C. rodentium strain. Isolation of C. rodentium from mouse colonies on two different continents provided an opportunity to compare the population genetics of the species. To do this, we compared our collection of C. rodentium isolates using several different approaches, and we conclude that the species is clonal.
MATERIALS AND METHODS
Media, bacterial strains, and growth conditions.
Bacteria were stored in Luria-Bertani (LB) broth (American Bioanalytical, Natick, Mass.) with 50% glycerol at −80°C. Bacteria were grown at 37°C in LB broth, on LB agar (American Bioanalytical), on MacConkey lactose agar, or on eosin-methylene blue (EMB) agar (Difco Laboratories, Detroit, Mich.). Where indicated, ampicillin was added at a final concentration of 100 μg/ml. The bacterial strains used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and oligonucleotides used in this study
| Strain or oligonucleotide | Descriptiona | Reference or source |
|---|---|---|
| C. rodentium (previously described) | ||
| DBS100 | Prototype TMCH isolate, ATCC 51459, original biotype 4280 | 1 |
| DBS125 | University of Missouri–Columbia isolate 1136-89, biotype 4280 | 33 |
| DBS126 | University of Missouri–Columbia isolate 2896-81, biotype 4280 | 33 |
| CDC1843-73T | ATCC 51116 (species 9 type strain) | 5 |
| CDC2643-76 | ATCC 51637 | 5 |
| CDC3311-75 | ATCC 51638 | 5 |
| C. rodentium (this study) | ||
| DBS425 | University of Washington 95-4234, from blood | 20 |
| DBS426 | University of Washington 95-4240, from liver | 20 |
| DBS427 | University of Washington 95-4241, from blood | 20 |
| DBS428 | University of Washington 95-4243, from liver | 20 |
| DBS494 | University of Washington 96-4212B | 20 |
| DBS495 | University of Washington 96-4212W | 20 |
| DBS496 | University of Washington FVO 10, from colon | 20 |
| DBS500 | University of Washington, 6-18-96 | 20 |
| DBS557 | Oak Ridge National Laboratory 3431e, from colon | This report |
| MPEC | E. coli O115a,c:K(B) strain Ex-33 | 15 |
| Citrobacter type strains | ||
| CDC621-64 | C. freundii, ATCC 8090 (species 1) | 5 |
| CDC3613-63 | C. koseri, ATCC 27156 (species 2) | 5 |
| CDC9020-77 | C. amalonaticus, ATCC 25407 (species 3) | 5 |
| CDC2991-81 | C. farmeri, ATCC 51112 (species 4) | 5 |
| CDC460-61 | C. youngae, ATCC 29935 (species 5) | 5 |
| CDC80-58 | C. braakii, ATCC 51113 (species 6) | 5 |
| CDC876-58 | C. werkmanii, ATCC 51114 (species 7) | 5 |
| CDC4696-86 | C. sedlakii, ATCC 51115 (species 8) | 5 |
| CDC4693-86 | C. gillenii, ATCC 51117 (species 10) | 5 |
| CDC2970-59 | C. murliniae, ATCC 51118 (species 11) | 5 |
| E. coli DH5α | F− φ80dlacZΔ endA recA hsdR supE thi gyrA Δ(lacZYA argF) | BRL |
| Oligonucleotides | ||
| CF2974 | 5′-CGGGATCCAATAAGACTGAAGCAAC-3′ | This study |
| SAL013 | 5′-GCTAAGCTTATATGATTAGAC-3′ | This study |
| ESPB1 | 5′-TGAGACAGTTGGCACATTGC-3′ | 28 |
| ESPB2 | 5′-TGGTGGTACAACTCTTCGAGC-3′ | 28 |
| BOXA1R | 5′-CTACGGCAAGGCGACGCTGACG-3′ | 34 |
| ERIC1R | 5′-ATGTAAGCTCCTGGGGATTCAC-3′ | 34 |
| ERIC2 | 5′-AAGTAAGTGACTGGGGTGAGCG-3′ | 34 |
| REP1R-I | 5′-IIIICGICGICATCIGGC-3′ | 34 |
| REP2-I | 5′-ICGICTTATCIGGCCTAC-3′ | 34 |
I, inosine.
Biotyping, serotyping, and DNA relatedness.
Biotyping was performed by the Microbiology Laboratory at Massachusetts General Hospital (Boston, Mass.) using API 20E strips and a Vitek system with GNI cards (bioMérieux Vitek, Hazelwood, Mo.). Serotyping was performed by the E. coli Reference Center at Pennsylvania State University (University Park, Pa.). Antibiotic disk diffusion tests were performed by the Microbiology Laboratory in the Division of Comparative Medicine at the Massachusetts Institute of Technology (MIT). DNA relatedness tests were performed as described previously (33) at the Centers for Disease Control and Prevention (Atlanta, Ga.). Briefly, bacteria were grown in brain heart infusion broth at 37°C with shaking until they reached the late logarithmic phase. DNA was extracted and purified as described previously (6). DNA samples were labeled enzymatically with [32P]dCTP using a nick translation reagent kit (Bethesda Research Laboratories, Inc., Gaithersburg, Md.) as directed by the manufacturer. Relatedness between pairs of bacterial DNAs was determined by the hydroxyapatite method. DNA samples to be hybridized were incubated at 60°C; thermal elution profiles were obtained as described previously (6).
PCR, cloning, and nucleotide sequence determination.
Using primers (Table 1) complementary to the 3′ end of the eaeA gene (CF2974 and SAL013) and an internal fragment from the espB gene (ESPB1 and ESPB2) (28), PCR was performed. Reaction mixtures contained 20 pmol of each primer, 5 nmol of each deoxynucleoside triphosphate, approximately 1 μg of genomic DNA template, and 0.6 U of Taq DNA polymerase (Promega Corp., Madison, Wis.) in a final concentration of 50 mM KCl–10 mM Tris (pH 8.3)–1.5 mM MgCl2. An initial denaturation step of 94°C for 2 min in a DNA engine (MJ Research Inc., Watertown, Mass.) was followed by 30 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1.5 min, with a final extension step of 72° for 5 min. The PCR products were examined in an ethidium bromide-stained 0.8% agarose gel. The eaeA amplicons from CDC1843-73T and MPEC were cloned using the pGEM-T Easy Vector system (Promega Corp.) and introduced into E. coli strain DH5α by high-voltage electroporation. Plasmid DNA was recovered using the QIAprep plasmid kit (Qiagen, Chatsworth, Calif.) and quantified by UV spectrophotometry.
Nucleotide sequences were determined for the 3′ end of the eaeA genes of CDC1843-73T and MPEC and for the tir genes of DBS100 and CDC1843-73T using an ABI Prism 310 genetic analyzer (Perkin-Elmer Corp., Norwalk, Conn.) as recommended by the manufacturer.
Rep-PCR.
Repetitive-element sequence-based PCR (rep-PCR) was performed as described previously (34). Briefly, genomic DNA was isolated using the DNeasy tissue kit (Qiagen) as recommended for bacteria and quantified by UV spectrophotometry. Each 25-μl PCR mix contained 20 pmol of each primer (previously reported in reference 34, Table 1), 150 ng of template DNA, and one Ready-To-Go PCR bead (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). An initial denaturation step of 95°C for 7 min in a DNA engine (MJ Research Inc.) was followed by 30 cycles of 95°C for 1 min, 52°C (for ERIC and BOX primers; Table 1) or 40°C (for REP primers; Table 1) for 1 min, and 65°C for 8 min, with a final extension step of 65°C for 16 min. The PCR products were examined in an ethidium bromide-stained 1.5% agarose gel.
FAS.
The fluorescent actin staining (FAS) assay of Knutton et al. (19), as modified for C. rodentium by Newman et al. (28), was used. Briefly, 5 × 104 HEp-2 cells (ATCC CCL-23) were seeded on glass coverslips in 24-well plates and grown overnight at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated calf serum and 2 mM l-glutamine. Bacteria were grown overnight in LB, washed with phosphate-buffered saline, and diluted 1:100 into DMEM supplemented with 0.1 M HEPES (pH 7.0). Bacteria were then incubated standing at 37°C and in 5% CO2 for 1 h and inoculated onto the HEp-2-coated coverslips at an approximate multiplicity of infection of 100. Midway through a 6-h incubation at 37°C and 5% CO2, the cell culture medium was replaced with fresh medium. At the conclusion of the 6-h incubation, coverslips were washed six times with phosphate-buffered saline, fixed with 4% paraformaldehyde, and permeabilized with 0.2% Triton X-100. Coverslips were then double fluorescently labeled for F-actin and bacteria and examined on a Nikon Labophot epifluorescence microscope. F-actin was labeled with Texas red-conjugated phalloidin (Molecular Probes, Eugene, Oreg.) and visualized with a 580-nm dichroic filter. C. rodentium cells were labeled with an anti-C. rodentium polyclonal rabbit antibody and visualized with a Cascade blue-conjugated goat anti-rabbit immunoglobulin G antibody (Molecular Probes) using a 400-nm dichroic filter. Coverslips labeled only with anti-C. rodentium primary antibody and the Cascade blue-conjugated secondary antibody showed no crossover when viewed with the 580-nm filter.
Infection study.
Three-week-old outbred Swiss Webster mice of both sexes (Taconic Laboratories, Germantown, N.Y.) were housed in microisolator cages within an Association for Assessment and Accreditation of Laboratory Animal Care–approved facility and maintained on pelleted rodent chow and water ad libitum. Water was supplemented with streptomycin (5 mg/ml) for the period between 48 and 24 h prior to bacterial inoculation. All experiments were approved by the MIT Animal Care and Use Committee. Mice were inoculated by oral gavage with 100 μl of a bacterial culture grown overnight in LB broth or with 100 μl of sterile LB broth. Each strain was inoculated at approximately 109 CFU/mouse, as determined by plate counts on MacConkey lactose agar. Similarly, colonization levels were determined by plating dilutions of feces from individual animals on MacConkey lactose agar on day 7 postinoculation. Animals were euthanized 10 days postinoculation. The distal colon of each mouse was collected aseptically. The colon was fixed in neutral-buffered formalin, processed for routine paraffin embedding, sectioned, and stained with hematoxylin and eosin (H&E) for routine histology. H&E-stained colonic sections were scored blindly for necrosis, hyperplasia, and inflammation by a veterinary pathologist using an increasing scale of severity from 0 (none) to 4 (severe), with 1 being minimal, 2 being mild, and 3 being moderate.
Transmission electron microscopy.
The distal 2 mm of the colon was processed for transmission electron microscopy. Briefly, tissue was fixed in 2% glutaraldehyde–3% paraformaldehyde–0.1 M sodium cacodylate (pH 7.4) buffer for 75 min at room temperature and then washed three times with 0.1 M sodium cacodylate (pH 7.4). Samples were postfixed with 1% OsO4 for 2.5 h in the dark on ice, washed three times with 0.1 M sodium cacodylate (pH 7.4), and stored at 4°C in the dark. After being rinsed once with 0.5% aqueous uranyl acetate, samples were stained for 2 h with 0.5% aqueous uranyl acetate at room temperature, rinsed with distilled water, dehydrated through a graded ethanol series, washed with propylene oxide for 1 min, incubated overnight in 50% propylene oxide–50% Epon, and embedded the following day in 100% Epon. Specimens were sectioned, stained with uranyl acetate and lead citrate, and examined on a JEOL 1200CX electron microscope.
Nucleotide sequence accession numbers.
The following nucleotide sequences have been submitted to GenBank and assigned the accession numbers shown in parentheses: the 3′ end of the eaeA gene from CDC1843-73T (AF301147), the 3′ end of the eaeA gene from MPEC (AF301146), the tir gene from DBS100 (AF301617), and the tir gene from CDC1843-73T (AF301618).
RESULTS
MPEC is a misclassified C. rodentium isolate.
Initial examination of MPEC on MacConkey lactose agar and EMB agar revealed a colony morphology not typical of E. coli but identical to that of C. rodentium. On MacConkey lactose agar, the colony morphology, a result of delayed lactose fermentation due to the absence of LacY, is a pale pink center surrounded by a clear rim, and on EMB agar, it is a colorless colony. Further testing by disk diffusion assay for ampicillin showed that MPEC, like many Citrobacter isolates (10) and all of the C. rodentium isolates in Table 1 (data not shown), possesses an inducible β-lactamase activity.
After these observations, we characterized MPEC further. Along with the six previously described C. rodentium strains (33) (Table 1), MPEC was examined for its serological and biochemical characteristics. Serotyping for E. coli O and H antigens showed an identical reactivity profile—O152 and O173:NM—for all strains tested except CDC1843-73T, which was typed as O56, O57, X13:NM. Biochemical analysis using the API 20E strip yielded an identical profile for all of the strains—atypical C. freundii (negative for indole production and unable to produce H2S). DBS100, CDC1843-73T, and MPEC were further characterized using the Vitek system. All three strains were biochemically identical and identified as Enterobacter amnigenus. The results from both biochemical testing methodologies are in agreement with the results reported by O'Hara et al. (29), in which the ability of several commercial biochemical identification tests to properly identify members of the 11 newly described Citrobacter species was examined.
Finally, genomic DNA from MPEC was tested by DNA-DNA hybridization to that of DBS100 and CDC1843-73T and was found to be more than 96% related to both strains, with a divergence of less than 0.2% within related DNA sequences at an annealing temperature of 60°C.
From these experiments, we concluded that MPEC is a misclassified C. rodentium isolate. This led us to investigate further the virulence factors present in MPEC.
Characterization of LEE-specific loci.
A key characteristic of C. rodentium as a species is the possession of virulence factors homologous to those found in the LEE pathogenicity island of the human pathogens EPEC and EHEC. We have demonstrated by Southern analysis that the six previously reported C. rodentium strains (Table 1) have genes homologous to both the eaeA and espB (formerly eaeB) genes of the EPEC LEE, while the type strains for the other 10 Citrobacter species do not (33).
To assess whether known C. rodentium isolates possess eaeA and espB genes, PCR was performed using the primers shown in Table 1. An amplicon of the expected size was generated for both primer sets in all C. rodentium isolates but not in any of the Citrobacter type strains (data not shown).
Comparison of the published sequences of eaeA genes from various AE species has shown that the 5′ two-thirds of the gene is highly conserved and encodes the membrane-spanning domain, while the 3′ third of the gene encodes the receptor-binding domain(s) and is more divergent between AE species. This divergence has been proposed to account for observed differences in host tropism and intestinal localization among AE species (27). The nucleotide sequences of the 3′ end of the eaeA gene from CDC1843-73T and MPEC were determined on both strands. Comparison to the published DBS100 C. rodentium eaeA sequence (31) showed that the three sequences are more than 99% identical on the nucleotide level. The complete sequence of the MPEC eaeA gene has recently been added to GenBank (accession no. AB040740) and is also more than 99% identical to that of DBS100 across the entire coding region.
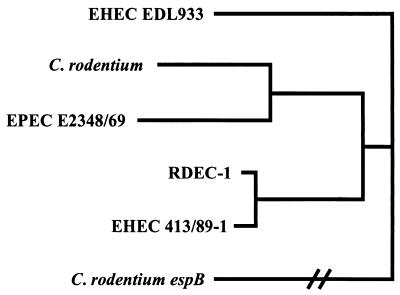
Furthermore, the nucleotide sequence of the MPEC tir gene has been also added to GenBank (accession no. AB026719). The tir gene is required to produce the AE phenotype both in vitro (18) and in vivo (21). Published tir sequences show high sequence divergence between AE species (Fig. 1, Table 2). The tir genes of DBS100 and CDC1843-73T were sequenced on both strands and found to be identical. Comparison to the published MPEC tir sequence showed a single amino acid difference at amino acid 149 (Thr to Asn) in the protein products and three conservative nucleotide substitutions.
FIG. 1.
Clustal alignment of full-length Tir proteins, showing that C. rodentium Tir is most similar to that of the human EPEC strain E2348/69. When carboxyl-terminal intimin peptides are similarly analyzed, C. rodentium clusters with the rabbit EPEC strain RDEC-1, demonstrating the difficulty in identifying a precursor organism for the C. rodentium LEE. The tree was arbitrarily anchored with the C. rodentium EspB protein, as Tir does not show appreciable homology to any protein previously reported. GenBank accession numbers are AF125993 (EHEC EDL933), AF022236 (EPEC E2348/69), AF045568 (RDEC-1), AJ223063 (EHEC 413/89-1), and AF177537 (C. rodentium EspB).
TABLE 2.
Tir protein homology to DBS100 Tira
| Strain | % Identity | % Similarity |
|---|---|---|
| EPEC strain E2348/69 (O127:H6) | 76 | 83 |
| EHEC strain EDL933 (O157:H7) | 58 | 70 |
| EPEC strain RDEC-1 (O15:H−) | 68 | 78 |
| EHEC strain 413/89-1 (O26:H−) | 68 | 78 |
Sequence accession numbers are the same as in Fig. 1. Percent identity and similarity were calculated using NCBI's BLASTP 2.0 program (http://www.ncbi.nlm.nih.gov/gorf/bl2.html).
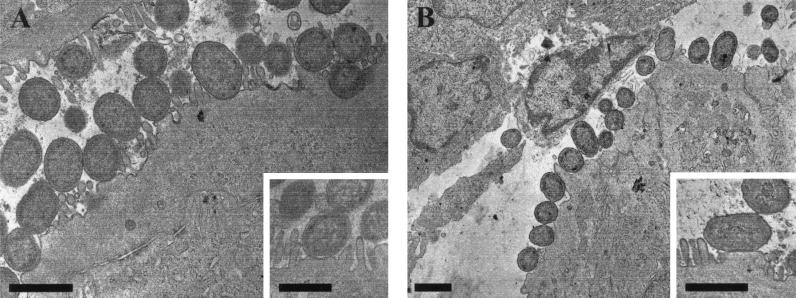
Possession of LEE-encoded genes does not necessarily imply a functional phenotype. To determine if both CDC1843-73T and MPEC are capable of producing AE lesions in vitro, both strains were tested in the FAS assay and found to be positive (data not shown). To confirm that the positive results in the FAS assay represented true AE pedestal formation, sections of colonic tissue from mice infected with CDC1843-73T and MPEC were examined by transmission electron microscopy. Both strains were able to intimately adhere to colonic epithelial cells, to efface the surrounding microvilli, and to mediate actin pedestal formation (Fig. 2).
FIG. 2.
Transmission electron micrographs showing AE pedestal induction on mouse colonic epithelial cells by (A) CDC1843-73T (original magnification, ×12,000; inset, ×20,000) and (B) MPEC (original magnification, ×6,000; inset, ×25,000). Bars, 1 μm.
From these experiments, we conclude that all C. rodentium isolates possess genes homologous to those found in the LEE. Comparisons of the sequences of two separate genes previously shown to exhibit divergence among other AE species demonstrate no appreciable divergence among C. rodentium isolates, even among those recovered from mice on separate continents. The nearly identical nucleotide sequences between C. rodentium strain DBS100 and MPEC further support our conclusion that MPEC is a misclassified C. rodentium isolate.
Whole-genome analysis of MPEC and C. rodentium isolates.
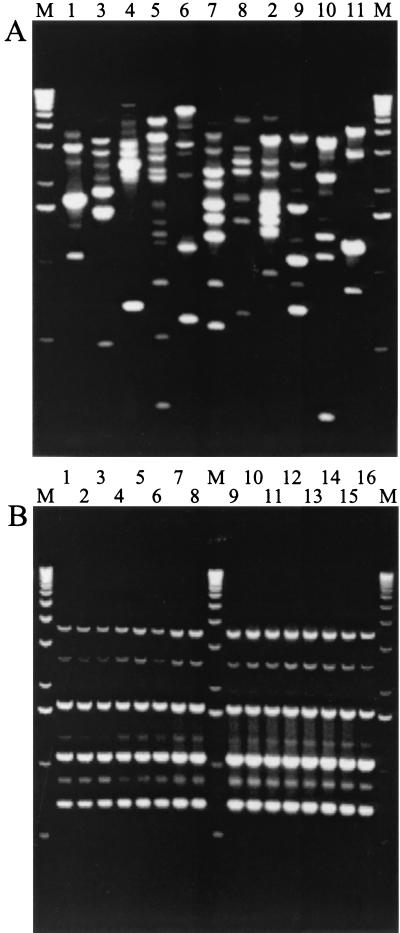
The lack of sequence divergence in LEE-located genes suggested that the species may be clonal. To assess this possibility, we further characterized the strains by rep-PCR, a high-resolution genotyping method that takes advantage of conserved repetitive DNA sequences that are dispersed throughout the bacterial genome (34). Previously, this approach has been applied to another member of the Citrobacter genus, Citrobacter koseri (formerly Citrobacter diversus) (35), with good results. Figure 3B shows the results obtained using the REP primers (Table 1) with the C. rodentium isolates. All strains produced an identical amplification pattern that differs from those produced by the type strains of the other 10 Citrobacter species (Fig. 3A). Similar results were obtained using two other sets of previously described primers, BOX and ERIC (Table 1; data not shown). From these experiments, we conclude that all C. rodentium strains isolated to date represent a clonal population.
FIG. 3.
Rep-PCR analysis of whole-genome DNA from Citrobacter species type strains (A) and C. rodentium isolates (B) using the REP primer pair. All C. rodentium isolates produced an identical banding pattern (B), which is in contrast to the Citrobacter species type strains, for which no two species exhibit an identical profile (A). Lane M, 1-kb ladder. (A) Lane numbers represent the Citrobacter species numbers given in Table 1. (B) Lane numbers represent the 16 C. rodentium isolates in the order listed in Table 1.
In vivo pathogenicity of selected C. rodentium strains.
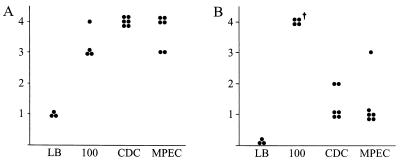
The ability to form AE lesions in vitro suggested that C. rodentium-induced TMCH and MPEC-induced infectious megaenteron of mice might be the same disease in vivo. To assess the comparative virulence of DBS100, MPEC, and CDC1843-73T, a mouse infection study was performed. CDC1843-73T was selected because it possesses a different O:H profile than either DBS100 or MPEC, it is the type strain for the species, and there is some confusion about the original source of this isolate (5, 33). Colonization was quantitatively measured on day 7 postinfection by bacterial fecal counts (Table 3). No differences were observed in colonization levels between DBS100 and CDC1843-73T, although animals challenged with MPEC were on average colonized at a level 10-fold lower than the other two groups. Mice were euthanized on day 10 postinfection. Two mice from the DBS100-infected group died on day 9 postinfection. Both animals had had clinical signs of TMCH and gross colonic hyperplasia that was indistinguishable from that of other members of the group. Both animals were excluded from further analysis. H&E-stained colonic sections were scored blindly for necrosis, hyperplasia, and inflammation by a veterinary pathologist, using an increasing scale of severity from 0 to 4. Figure 4 shows the scores for both hyperplasia and necrosis. Levels of colonic hyperplasia were indistinguishable between the inoculated strains. A representative comparison is shown in Fig. 5 for a mock-infected (sterile broth) mouse (Fig. 5A) and a mouse infected with MPEC (Fig. 5B). The changes are characteristic of TMCH, including crypt elongation, increased number of cells per crypt, and few goblet cells. Unexpectedly, a significant amount of necrosis was observed in mice infected with DBS100 but not in mice in the other infection groups (Fig. 5C). Furthermore, mice infected with DBS100 demonstrated multifocal erosion and ulceration, heavy bacterial colonization, intense suppuration, transmural inflammation, and invasive hyperplasia. Sections from mice infected with CDC1843-73T and MPEC were characterized by moderate to heavy bacterial colonization and submucosal edema. Occasionally more severe damage typical of the DBS100-infected mice, such as focal surface necrosis, crypt invasion, and abscess formation, was seen in these two infection groups.
TABLE 3.
Colonization levels 7 days postinfection
| Inoculum | Mean log CFU/g of feces ± SE | No. of mice |
|---|---|---|
| Sterile broth | 0.00 ± 0.00 | 3 |
| DBS100 | 9.18 ± 0.17 | 5a |
| CDC1843-73T | 9.01 ± 0.09 | 6 |
| MPEC | 8.03 ± 0.21b | 6 |
Feces were not obtained from the sixth animal in this group.
Statistically different from both DBS100 and CDC1843-73T (P < 0.005) using a two-tailed unpaired t test.
FIG. 4.
Graphic representation of histologic scores for hyperplasia (A) and necrosis (B) of H&E-stained colonic tissue, showing that all three strains were able to induce similar levels of hyperplasia while only DBS100 (labeled 100) induced necrosis. Each dot represents one animal. The y axis represents an increasing scale of severity from 0 to 4, as described in Materials and Methods. The Mann-Whitney U test was used to calculate P values. The dagger indicates a statistically significant difference (P < 0.05) between CDC1843-73T (CDC) and MPEC. All groups in both panels are statistically different from the applicable LB controls (P < 0.05).
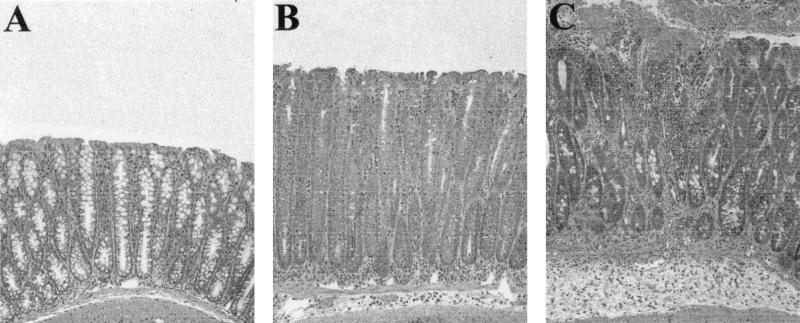
FIG. 5.
H&E-stained colon sections. (A) Section from a mock-infected (sterile broth) mouse showing normal colonic crypt architecture. (B) Section from an MPEC-infected mouse showing crypt hyperplasia, goblet cell depletion, and mild inflammation. (C) DBS100-induced necrosis showing epithelial damage, severe mucosal inflammation with an accompanying inflammatory exudate, and submucosal edema. All panels are at an identical level of magnification.
DISCUSSION
In this report, we characterized the genetic, biochemical, and murine virulence of C. rodentium strains isolated from several different animal facilities, including the previously reported and misclassified MPEC strain from Japan (26). Our objective in this study was to compare these isolates in order to better understand the population genetics and intrarelatedness of the species.
To date, AE E. coli strains have been described that are capable of infecting not only humans but also many types of animals (27), though apparently not mice. Recent work by Janda et al. has reclassified AE Hafnia alvei strains as E. coli isolates (16). This suggests that the LEE has been transferred to only one other species, C. rodentium, which coincidentally is the only known AE species pathogenic for mice. In the late 1960s, Nakagawa et al. (26) reported a strain of E. coli that caused a murine diarrheal disease, termed infectious megaenteron of mice, whose pathology closely resembled that of C. rodentium-induced TMCH. We hypothesized that MPEC might possess the LEE and represent a novel murine model for AE disease pathogenesis. Encouraged by the detection of an MPEC eaeA homolog by PCR, we further characterized MPEC and noted that it possesses an inducible β-lactamase gene and a delayed lactose fermentation phenotype on MacConkey lactose agar. Both observations are consistent with those seen with C. rodentium, and a review of the original MPEC report (26) showed that Nakagawa observed a similar C. rodentium-like colony morphology on MacConkey lactose agar. Biochemical profiling and DNA-DNA hybridization results clearly proved that MPEC is a strain of C. rodentium, not E. coli. This conclusion afforded the opportunity to investigate the clonality of the C. rodentium isolates in our collection, which now included a strain that was isolated on a separate continent contemporary to the earliest reports of C. rodentium in the literature.
We characterized the C. rodentium isolates using a variety of approaches. Initial characterization using PCR demonstrated that all isolates possessed both an eaeA and an espB gene, both of which were absent in other Citrobacter species. Rep-PCR was further applied to both the C. rodentium isolates and the type strains for the other Citrobacter species. Using three different primer sets, no differences were observed among the profiles generated by the C. rodentium isolates. The C. rodentium profile for each primer set was further different from the profiles generated by the other Citrobacter species. These results suggest that isolates of C. rodentium are clonal.
We were further interested in characterizing a subset of the isolates for their sequence divergence in LEE-specific genes. Two genes in particular, eaeA and tir, show sequence divergence between isolates of AE E. coli and have been shown to be necessary for AE disease pathogenesis. The eaeA gene encodes the intimin protein, the principal adhesin for AE bacteria. The carboxyl-terminal third of intimin contains the receptor-binding domain and is hypothesized to be involved in host specificity and tissue tropism. The tir gene encodes the intimin receptor Tir, which is translocated into host cells by a type III secretion system. The sequence of MPEC tir has recently been added to GenBank. Sequence analysis of the carboxyl-terminal third of eaeA and full-length tir from DBS100, CDC1843-73T, and MPEC shows that the proteins produced are nearly identical.
To assess whether these three isolates produced an identical disease in vivo, we performed a murine infection study. Colonic sections were assessed by a veterinary pathologist, who concluded that the disease induced by all three strains was the same, spanning a range of severity that correlated with the strain used for inoculation. DBS100-infected animals showed the most severe pathology, and MPEC-infected animals showed the least. MPEC colonized Swiss-Webster mice at a level 10-fold lower than the other two groups and produced a pathology similar to that previously reported for C. rodentium, colonic hyperplasia with little inflammatory response. CDC1843-73T colonized at a level similar to that of DBS100 but did not cause the high level of morbidity and mortality seen with DBS100. The ability of the three strains to elicit the AE phenotype in vivo was confirmed by transmission electron microscopy. We conclude that all three strains cause TMCH and induce the formation of AE pedestals beneath intimately adhered bacteria. Furthermore, a strain-dependent degree of severity is superimposed on the reproducible hyperplasia characteristic of TMCH, resulting in a continuum of pathology from minimal inflammation to necrosis and severe colitis.
Historically, TMCH was described by Barthold et al. (2) as a murine model for hyperplasia and tumor promotion in which the “basic lesion is mucosal hyperplasia with variable imposition of inflammatory changes” (1). More recently, Higgins et al. (11) have reported that C. rodentium causes a mucosal Th1 response and colitis similar to that seen in inflammatory bowel disease. Our observations obtained here with three different C. rodentium strains demonstrate a continuum of disease severity that ranges from hyperplasia to colitis. The strain differences observed in our in vivo study may be due to the accumulation of spontaneous mutations during in vitro passage. Alternatively, there may be allelic differences which would not be detected by our rep-PCR analysis that account for the attenuated phenotype. Serial in vivo passage of the strains could be done to clarify the source of these differences. Though necrosis was not seen with either CDC1843-73T or MPEC, typical TMCH was observed, confirming the ability of all three strains to induce the same basic disease.
In this study, DBS100 caused necrosis and severe colitis, a disease state more similar to that observed in suckling mice than the typical TMCH observed in adult mice. Prior to infection, mice received streptomycin in the water to normalize possible differences in the microflora. This may have influenced the severity of the disease. Alternatively, DBS100 could possess a putative virulence factor not found in either CDC1843-73T or MPEC. However, we favor the idea that an increased number of CFU at the time of inoculation, resulting in an increased rate of AE lesion formation early in infection, causes more severe disease. Work is ongoing to investigate the conditions under which C. rodentium can be utilized as a model system for studying hyperplasia, colitis, and the transition between the two responses.
In this study, we have examined the population genetics of the species C. rodentium. Our data suggest a clonal origin for the species, which is now distributed globally. Further studies are needed to define the mechanism by which C. rodentium causes epithelial hyperplasia with limited inflammation versus necrosis with severe colitis. We believe that studies with laboratory mice infected with C. rodentium will continue to provide insights into cytokinetics, cancer risk, and, potentially, inflammatory bowel disease.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant CA63112 from the National Cancer Institute to D.B.S. and National Institutes of Health fellowship F32 CA76716 to J.V.N. S.A.L. was supported by NIEHS grant ES07020. The EM Laboratory is supported by NIH BRS Shared Instrumentation grant 1S10 RR-5734-01 to the MIT Biology Department.
We thank Kikuji Itoh for generously providing the MPEC strain, Lillian Maggio-Price and Charmaine Foltz for providing C. rodentium strains, James Versalovic for discussions about rep-PCR, Patricia Reilly for assistance with all aspects of electron microscopy, and Sharda Jha for technical assistance.
REFERENCES
- 1.Barthold S W, Coleman G L, Bhatt P N, Osbaldiston G W, Jonas A M. The etiology of transmissible murine colonic hyperplasia. Lab Anim Sci. 1976;26:889–894. [PubMed] [Google Scholar]
- 2.Barthold S W, Coleman G L, Jacoby R O, Livstone E M, Jonas A M. Transmissible murine colonic hyperplasia. Vet Pathol. 1978;15:223–236. doi: 10.1177/030098587801500209. [DOI] [PubMed] [Google Scholar]
- 3.Barthold S W, Osbaldiston G W, Jonas A M. Dietary, bacterial, and host genetic interactions in the pathogenesis of transmissible murine colonic hyperplasia. Lab Anim Sci. 1977;27:938–945. [PubMed] [Google Scholar]
- 4.Brennan P C, Fritz T E, Flynn R J, Poole C M. Citrobacter freundii associated with diarrhea in laboratory mice. Lab Anim Care. 1965;15:266–275. [PubMed] [Google Scholar]
- 5.Brenner D J, Grimont P A D, Steigerwalt A G, Fanning G R, Ageron E, Riddle C F. Classification of citrobacteria by DNA hybridization: designation of Citrobacter farmeri sp. nov., Citrobacter youngae sp. nov., Citrobacter braakii sp. nov., and three unnamed genomospecies. Int J Syst Bacteriol. 1993;43:645–658. doi: 10.1099/00207713-43-4-645. [DOI] [PubMed] [Google Scholar]
- 6.Brenner D J, McWhorter A C, Knutson J K, Steigerwalt A G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982;15:1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brynjolfsson G, Lombard L S. Colitis cystica in mice. Cancer. 1969;23:225–229. doi: 10.1002/1097-0142(196901)23:1<225::aid-cncr2820230130>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 8.Donnenberg M S, Tzipori S, McKee M L, O'Brien A D, Alroy J, Kaper J B. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Investig. 1993;92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ediger R D, Kovatch R M, Rabstein M M. Colitis in mice with a high incidence of rectal prolapse. Lab Anim Sci. 1974;24:488–494. [PubMed] [Google Scholar]
- 10.Frederiksen W, Søgaard P. The genus Citrobacter. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 2744–2753. [Google Scholar]
- 11.Higgins L M, Frankel G, Douce G, Dougan G, MacDonald T T. Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect Immun. 1999;67:3031–3039. doi: 10.1128/iai.67.6.3031-3039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh K, Ueda K, Fujiwara K. Susceptibility of germ-free mice to infectious megaenteron. Microbiol Immunol. 1980;24:281–290. doi: 10.1111/j.1348-0421.1980.tb02831.x. [DOI] [PubMed] [Google Scholar]
- 13.Itoh K, Maejima K, Ueda K, Fujiwara K. Difference in susceptibility of mice raised under barrier-sustained (SPF) or conventional conditions to infectious megaenteron. Microbiol Immunol. 1979;23:909–913. doi: 10.1111/j.1348-0421.1979.tb02824.x. [DOI] [PubMed] [Google Scholar]
- 14.Itoh K, Maejima K, Ueda K, Fujiwara K. Effect of intestinal flora on megaenteron of mice. Microbiol Immunol. 1978;22:661–672. doi: 10.1111/j.1348-0421.1978.tb00419.x. [DOI] [PubMed] [Google Scholar]
- 15.Itoh K, Matsui T, Tsuji K, Mitsuoka T, Ueda K. Genetic control in the susceptibility of germfree inbred mice to infection by Escherichia coli O115a,c:K(B) Infect Immun. 1988;56:930–935. doi: 10.1128/iai.56.4.930-935.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janda J M, Abbott S L, Albert M J. Prototypal diarrheagenic strains of Hafnia alvei are actually members of the genus Escherichia. J Clin Microbiol. 1999;37:2399–2401. doi: 10.1128/jcm.37.8.2399-2401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson E, Barthold S W. The ultrastructure of transmissible murine colonic hyperplasia. Am J Pathol. 1979;97:291–314. [PMC free article] [PubMed] [Google Scholar]
- 18.Kenny B, DeVinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 19.Knutton S, Baldwin T, Williams P H, McNeish A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maggio-Price L, Nicholson K L, Kline K M, Birkebak T, Suzuki I, Wilson D L, Schauer D, Fink P J. Diminished reproduction, failure to thrive, and altered immunologic function in a colony of T-cell receptor transgenic mice: possible role of Citrobacter rodentium. Lab Anim Sci. 1998;48:145–155. [PubMed] [Google Scholar]
- 21.Marches O, Nougayrede J P, Boullier S, Mainil J, Charlier G, Raymond I, Pohl P, Boury M, De Rycke J, Milon A, Oswald E. Role of Tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect Immun. 2000;68:2171–2182. doi: 10.1128/iai.68.4.2171-2182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDaniel T K, Kaper J B. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 24.Moon H W, Whipp S C, Argenzio R A, Levine M M, Giannella R A. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun. 1983;41:1340–1351. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muto T, Nakagawa M, Isobe Y, Saito M, Nakano T, Imaizumi K. Infectious megaenteron of mice. I. Manifestation and pathological observation. Jpn J Med Sci Biol. 1969;22:363–374. doi: 10.7883/yoken1952.22.363. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa M, Sakazaki R, Muto T, Saito M, Hagiwara T, Imaizumi K. Infectious megaenteron in mice. II. Detection of coliform organisms of an unusual biotype as the primary cause. Jpn J Med Sci Biol. 1969;22:375–382. [PubMed] [Google Scholar]
- 27.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman J V, Zabel B A, Jha S S, Schauer D B. Citrobacter rodentium espB is necessary for signal transduction and for infection of laboratory mice. Infect Immun. 1999;67:6019–6025. doi: 10.1128/iai.67.11.6019-6025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Hara C M, Roman S B, Miller J M. Ability of commercial identification systems to identify newly recognized species of Citrobacter. J Clin Microbiol. 1995;33:242–245. doi: 10.1128/jcm.33.1.242-245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothbaum R J, Partin J C, Saalfield K, McAdams A J. An ultrastructural study of enteropathogenic Escherichia coli infection in human infants. Ultrastruct Pathol. 1983;4:291–304. doi: 10.3109/01913128309140582. [DOI] [PubMed] [Google Scholar]
- 31.Schauer D B, Falkow S. Attaching and effacing locus of a Citrobacter freundii biotype 4280 that causes transmissible murine colonic hyperplasia. Infect Immun. 1993;61:2486–2492. doi: 10.1128/iai.61.6.2486-2492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schauer D B, Falkow S. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect Immun. 1993;61:4654–4661. doi: 10.1128/iai.61.11.4654-4661.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schauer D B, Zabel B A, Pedraza I F, O'Hara C M, Steigerwalt A G, Brenner D J. Genetic and biochemical characterization of Citrobacter rodentium sp. nov. J Clin Microbiol. 1995;33:2064–2068. doi: 10.1128/jcm.33.8.2064-2068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Versalovic J, Schneider M, de Bruijn F J, Lupski J R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 35.Woods C R, Jr, Versalovic J, Koeuth T, Lupski J R. Analysis of relationships among isolates of Citrobacter diversus by using DNA fingerprints generated by repetitive sequence-based primers in the polymerase chain reaction. J Clin Microbiol. 1992;30:2921–2929. doi: 10.1128/jcm.30.11.2921-2929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]