Abstract
Dead-End (DND1) is an RNA-binding protein involved in translational regulation. Defects in DND1 gene causes germ cell tumors and sterility in rodents. Experimental studies with human somatic cancer cells indicate that DND1 has anti-proliferative and pro-apoptotic function in some while oncogenic function in other cells. We examined The Cancer Genome Atlas data for gene alterations and gene expression changes in DND1 in a variety of human cancers. We found that DND1 is amplified, deleted or mutated in multiple human cancers. In different cancers, DND1 alteration correlates with increased diagnosis age of patients, shift in tumor spectrum or change of tumor sites and in some cases is significantly associated with worse survival for cancer patients. For 15 cancers, we retrieved expression data of thousands of genes that co-expressed with DND1. We found that these cancers contain different percentage of genes that are positively or negatively co-expressed with DND1. Ingenuity Pathway Analysis was performed to explore the biological implications of these genes. More than 10 canonical pathways were identified and each cancer type exhibits unique pathway profiles. Comparison analysis across all 15 cancer types showed that some cancers exhibit strikingly similar profiles of DND1-correlated signaling pathway activation or suppression. Our data reinforce the notion that the biological role of DND1 is cell-type specific and suggest that DND1 may play opposing role by exerting anti-proliferative effects in some cancer cells while being pro-proliferative in others. Our study provides valuable insights to direct experimental investigations of DND1 function in somatic cancers.
Keywords: DND1, Somatic cancers, TCGA, Co-expression, IPA
Highlights
-
•
DND1 is altered with different frequencies in multiple human cancers.
-
•
DND1 changes in cancers correlate with clinical outcomes including worse prognosis.
-
•
DND1 is co-expressed with a large number of genes across multiple cancer types.
-
•
DND1 correlates with activation or suppression of canonical biological pathways.
1. Introduction
The Dead-End 1 (DND1) is an RNA-binding protein containing two RNA recognition motifs (RRMs) in tandem, followed by a double-stranded RNA-binding motif at the carboxyl-terminus [1]. DND1 is a multifunctional protein and has been found to exhibit diverse molecular activities. Its role in translation regulation has been extensively studied [[2], [3], [4], [5]]. DND1 can bind specific mRNAs such as p27 [2], LATS2 [2], geminin [6] and trim36 [7] etc. and block microRNA (miRNA) access from the 3′-untranslated regions (3′-UTR) of target mRNAs and inhibit miRNA-mediated mRNA degradation, thus up-regulating translation. In contrast, DND1 has also been shown to function as a translation suppressor by recruiting the CCR4-NOT deadenylase complex [5]. Other functions of DND1 include activation of germline-specific translation of nanos1 through promoting initiation [8] and modifying the activity of target protein such as activator protein 1 [9].
DND1 is essential for primordial germ cell (PGC) survival [10]. The Dnd1Ter mutation, with an arginine residue at amino acid 190 converted to a premature stop codon, causes PGC loss in all mouse genetic backgrounds, leading to infertility in both genders [11]. In addition, male Dnd1Ter mice on the 129 strain background also develop testicular germ cell tumors (TGCTs) [[10], [11], [12], [13], [14], [15]]. Furthermore, the WKY/Ztm rat strain carrying homozygous mutation in Dnd1 was found to develop congenital testicular and ovarian teratomas, and exhibit infertility with complete penetrance in both genders [16].
Although DND1's role in germ cell development and tumorigenesis has been extensively studied, multiple research reports in recent years suggest that DND1 may also be involved in somatic cancers. DND1 has been shown to exert tumor suppressive effects in breast cancer cells [17], hepatocellular carcinoma cells [18], tongue squamous cell carcinoma (TSCC), skin cancer and acute myeloid leukemia [[19], [20], [21]]. In addition, Dnd1Ter was indicated to possess tumorigenic properties in the intestine as it significantly increased polyp number and mass in the Apc+/Min mouse model of intestinal polyposis [22]. On the contrary, silencing DND1 suppressed SW48 colorectal cancer cell proliferation [23]. Hence DND1 is an anti-proliferative, pro-apoptotic tumor suppressor in a variety of cancers, but may also exert oncogenic function in others. In this study we examined DND1 status in human somatic cancers by using megadata from the publicly accessible cBioPortal platform (cbioportal.org, last accessed on 20 Nov 2021) [24,25]. We examined DND1 gene alterations in a number of cancers and the association of DND1 alterations with several clinical outcomes. In addition, we analyzed genes significantly co-expressed with DND1 in 15 cancers developed from breast, bladder, colon, brain, liver, lung, ovary, pancreas, prostate, kidney, stomach, skin and testes. Our study provides valuable insights for future experimental investigations of the role of DND1 in somatic cancers.
2. Methods
2.1. cBioPortal database and Ingenuity Pathway Analysis
The analysis of DND1 gene status and its association with clinical outcomes in human cancers was performed on cBioPortal platform (www.cbioportal.org; last accessed on Nov 20th, 2021)[24,25]. Gene co-expression data were retrieved from cBioPortal, which contains gene name, cytoband, spearman's correlation, p-value and q-value (the p value adjusted for the False Discovery Rate) for each gene that is correlated with DND1 in mRNA expression. The data for genes that have a q-value less than 0.01 were uploaded into Qiagen's IPA system (www.ingenuity.com) for core analysis to determine canonical pathways in each cancer. For statistical and performance reasons, the number of analyzed genes were limited to 2000 when there are more than 2000 significant genes using q < 0.01 as a cutoff. Comparison analysis was also performed and unsupervised hierarchical clustering heatmap was used to demonstrate the patterns of the pathways impacted by DND1 in different cancers.
2.2. Venn diagram
The Venn diagrams were generated through the website www.interactivenn.net [26].
2.3. Statistics
Wilcoxon Test was used for mutation count and diagnosis age comparisons; Chi-squared Test was used for tumor sites and tumor subtype comparisons. The p value has been adjusted for the False Discovery Rate.
3. Results
3.1. DND1 is altered in multiple human cancers
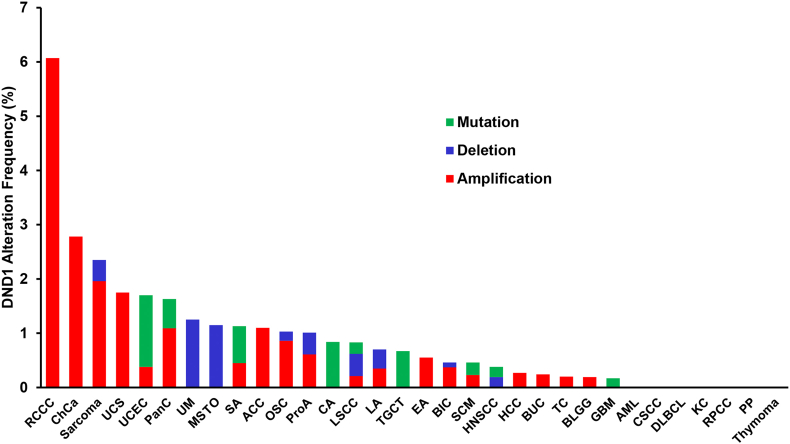
Emerging in vitro evidence suggests that DND1 may play a role in somatic cancers apart from its well-known function in mouse testicular germ cell tumor formation. The Cancer Genome Atlas (TCGA) database serves as a repository recording the mutational and gene expression changes in a large cohort of patients' tumor samples. We first investigated whether DND1 gene is altered in human cancers by querying DND1 in 32 TCGA PanCancer Atlas Studies on cBioPortal platform [24,25](Suppl. Table 1). DND1 shows various alteration frequency across different cancer types, with kidney renal clear cell carcinoma (RCCC) containing the highest rate of DND1 alteration (6.07%), followed by cholangiocarcinoma (ChCa) and sarcoma (Fig. 1, Suppl. Table 1). In studies other than TCGA, DND1 alteration has also been identified in human cancers with various frequencies (Suppl. Figure 1).
Fig. 1.
The alteration of DND1 gene in human cancers as identified by 32 TCGA PanCancer Atlas studies. RCCC, Kidney Renal Clear Cell Carcinoma; ChCa, Cholangiocarcinoma; UCS, Uterine Carcinosarcoma; UCEC, Uterine Corpus Endometrial Carcinoma; PanC, Pancreatic Adenocarcinoma; UM, Uveal Melanoma; MSTO, Mesothelioma; SA, Stomach Adenocarcinoma; ACC, Adrenocortical Carcinoma; OSC, Ovarian Serous Cystadenocarcinoma; ProA, Prostate Adenocarcinoma; CA, Colorectal Adenocarcinoma; LSCC, Lung Squamous Cell Carcinoma; LA, Lung Adenocarcinoma; TGCT, Testicular Germ Cell Tumors; EA, Esophageal Adenocarcinoma; BIC, Breast Invasive Carcinoma; SCM, Skin Cutaneous Melanoma; HNSCC, Head and Neck Squamous Cell Carcinoma; LHCC, Liver Hepatocellular Carcinoma; BUC, Bladder Urothelial Carcinoma; TC, Thyroid Carcinoma; BLGG, Brain Lower Grade Glioma; GBM, Glioblastoma Multiforme; AML, Acute Myeloid Leukemia; CSCC, Cervical Squamous Cell Carcinoma; DLBCL, Diffuse Large B-Cell Lymphoma; KC, Kidney Chromophobe; RPCC, Kidney Renal Papillary Cell Carcinoma; PP, Pheochromocytoma and Paraganglioma.
Interestingly, DND1 exhibits different patterns of alteration across cancers. Based on TCGA PanCancer Atlas studies, DND1 amplification prevails in RCCC, ChCa, sarcoma and uterine carcinosarcoma (UCS) etc. In contrast, mutation of DND1 is the major alteration in uterine corpus endometrial carcinoma (UCEC), colorectal adenocarcinoma (CA) and TGCT. DND1 deletion has been identified in sarcoma, uveal melanoma (UM), mesothelioma (MSTO), ovarian serous cystadenocarcinoma (OSC), prostate adenocarcinoma (ProA), lung squamous cell carcinoma (LSCC) and lung adenocarcinoma (LA) etc (Fig. 1, Suppl. Table 1).
3.2. DND1 alternation in cancers is associated with several clinical outcomes
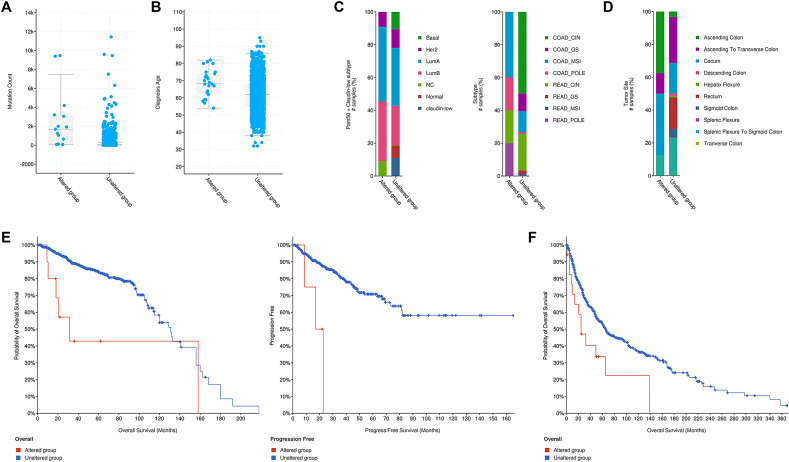
We next examined whether the above DND1 alterations in cancers correlates with any clinical outcomes. We found that enhanced mutation count was identified in colorectal cancers with DND1 alteration (Fig. 2A). In addition, DND1 alteration correlates with increased diagnosis age of prostate cancer patients (Fig. 2B), shift in tumor spectrum of breast (Fig. 2C, Left) and colon cancers (Fig. 2C, Right) and change of tumor sites for colon cancer (Fig. 2D). Furthermore, prostate cancer patients with DND1 gene alteration exhibit significantly poorer prognosis in terms of overall and progress free survivals compared to patients with normal DND1 (Fig. 2E). Similarly, DND1 gene alteration is also significantly associated with a worse overall survival for melanoma patients (Fig. 2F). Interestingly, although DND1 alterations occur with the highest frequency in RCCC among all cancers (Fig. 1), it is not significantly associated with any clinical outcomes of RCCC (data not shown), which could be due to either the sample size not being large enough to detect the impact of DND1 alteration on RCCC clinical outcomes, or the non-significant role of DND1 in determining RCCC clinical outcomes.
Fig. 2.
DND1 alteration is associated with clinical features of a variety of human cancers. (A) The comparison of mutation counts in colorectal cancers between DND1 altered and unaltered groups. q = 1.126e-5; (B) The comparison of the age at the diagnosis of prostate cancer between DND1 altered and unaltered groups. q = 4.521e-6; (C) The comparison of breast (Left; q = 1.663e-4) and colorectal cancer spectrum (Right; q = 1.838e-5) between DND1 altered and unaltered groups. (D) The comparison of colorectal tumor sites between DND1 altered and unaltered groups, q = 6.792e-3. (E) The comparison of overall (Left, Logrank test p-value: 2.711e-3) and progress free survivals (Right, Logrank test p-value: 1.341e-4) of prostate cancer patients between DND1 altered and unaltered groups. (F) The comparison of overall survivals of melanoma patients between DND1 altered and unaltered groups (Logrank test p-value: 4.410e-3).
3.3. DND1 is co-expressed with a large number of genes across different cancer types
Furthermore, we examined the potential molecular impact of DND1 on malignant somatic cells. Since DND1 regulates translation via increasing or decreasing target mRNA stability [1], we queried genes that are co-expressed with DND1 in 14 somatic cancer types: breast invasive carcinoma (BIC), bladder urothelial carcinoma (BUC), colorectal adenocarcinoma (CA), glioblastoma multiforme (GBM), liver hepatocellular carcinoma (HCC), lung adenocarcinoma (LA), lung squamous cell carcinoma (LSCC), ovarian serous cystadenocarcinoma (OSC), pancreatic adenocarcinoma (PanA), prostate adenocarcinoma (ProA), kidney renal clear cell carcinoma (RCCC), kidney renal papillary cell carcinoma (RPCC), stomach adenocarcinoma (SA), and skin cutaneous melanoma (SCM). In addition, as DND1 plays an important role in TGCT formation in 129 mouse strain [10,15], we also queried its co-expressed genes in human TGCT. The TCGA studies of these cancer types contain large sample numbers incorporating data from tissues of 149–1084 patients (Suppl. Table 1), which ensures accuracy and precision in detecting correlations.
All 15 cancer types contained genes that are significantly co-expressed with DND1 (adjusted p < 0.01, Suppl. Data 1). The top ten positively and negatively co-expressed genes for each cancer type are listed in Suppl. Table 2. Of note, except for the genes that are located near DND1 on chromosome 5 (e.g., WDR55, TRIM41, FAM193B, HARS2, APBB3 and FCHSD1 etc.), none of those top co-expressed genes are shared by more than three cancer types, indicating that the gene sets that are strongly impacted by DND1 may be cancer type-specific.
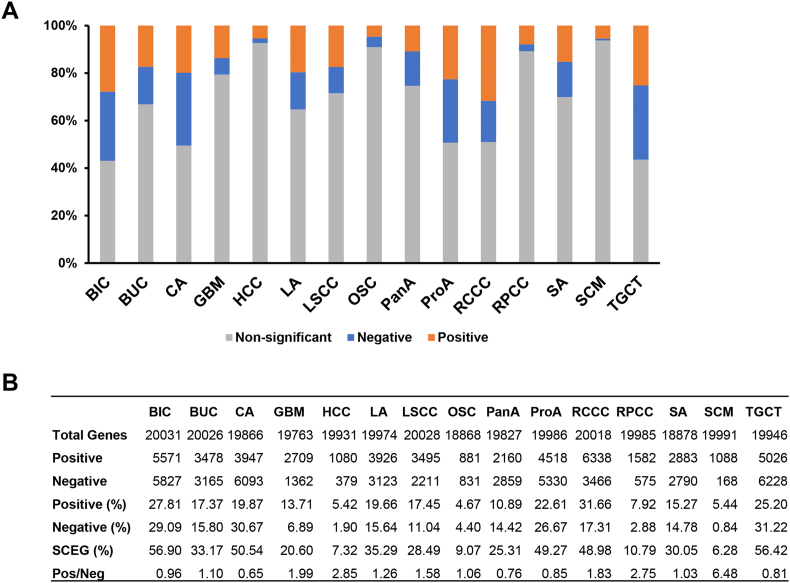
Genes significantly co-expressed with DND1 are present with different percentages in different cancer type (Fig. 3). HCC, OSC, RPCC and SCM have lower percentage of DND1-co-expressed genes (7.32%, 9.07%, 10.79% and 6.28%) compared to others, while close to or over half of BIC, CA, ProA, RCCC and TCGT genes are significantly co-expressed with DND1 (56.9%, 50.54%, 49.27%, 48.98 and 56.42%; Fig. 3A and B). Interestingly, the ratio between positively and negatively co-expressed genes also varies greatly. In BIC, BUC, ProA and SA, there is nearly equal amount of positively and negatively co-expressed genes (Fig. 3A and B). However, in HCC, RPCC and SCM, the positively co-expressed genes evidently outnumber those that are negatively co-expressed with DND1 (2.85:1, 2.75:1 and 6.48:1, respectively; Fig. 3A and B). In contrast, in CA and PanA, there are much less genes positively co-expressed with DND1 (Fig. 3A and B).
Fig. 3.
DND1 is significantly co-expressed with other genes in a variety of human cancers. (A) The percentage of genes that are positively (orange bars) or negatively (blue bars) co-expressed with DND1 in a variety of human cancers with q value less than 0.01. (B) Summary of the number of total genes that are expressed in a variety of human cancers, the number and percentage of genes that are significantly positively or negatively co-expressed with DND1, as well as the ratio between them (Pos/Neg). BIC, breast invasive carcinoma; BUC, bladder urothelial carcinoma; CA, colorectal adenocarcinoma; GBM, glioblastoma; HCC, hepatocellular carcinoma; LA, lung adenocarcinoma; LSCC, lung squamous cell carcinoma; OSC, ovarian serous cystadenocarcinoma; PanA, pancreatic adenocarcinoma; ProA, prostate adenocarcinoma; RCCC, renal clear cell carcinoma; RPCC, renal papillary cell carcinoma; SA, stomach adenocarcinoma; SCM, skin cutaneous melanoma; TGCT, testicular germ cell tumor. SCEG, significantly co-expressed genes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
It is interesting to note that even in different cancer types originating from the same organ, DND1 appears to function differently. For example, as mentioned above, in RCCC, 48.98% genes are significantly co-expressed with DND1, while in RPCC, another kidney cancer subtype, this number reduces to 10.79% (Fig. 3A and B). These data, together with various gene co-expression patterns in different cancers, as described above, indicate that the biological role of DND1 in somatic cells may be highly cell type-specific.
3.4. DND1 correlates with activation or suppression of canonical biological pathways
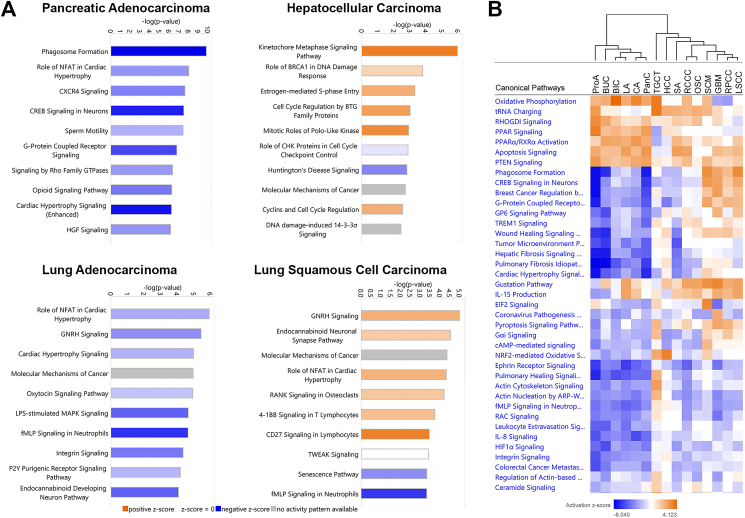
To explore the biological implication of genes that significantly co-express with DND1, we next performed Ingenuity Pathway Analysis. In each cancer type, more than 10 canonical pathways were identified and the top ten are shown in Fig. 4A and Suppl. Figure 1. Interestingly, each cancer type exhibits unique pathway profiles. In PanA, LA, BIC, BUC, CA and ProA, DND1 expression is mostly associated with pathway suppression, that is, DND1 expression conversely correlates with the expression of genes in the according pathways. In contrast, in HCC, LSCC, and RPCC etc., DND1 expression largely correlates with activation of pathways (Fig. 4A and Suppl. Figure 2).
Fig. 4.
DND1 expression is significantly associated with signaling activation or suppression in a variety of human cancers. (A) The top ten signaling pathways in four cancer types, of which the activation or suppression are significantly associated with DND1 expression. (B) Unsupervised clustering heatmap of signaling pathways across fifteen human cancers, whose activation or suppression are associated with DND1 expression. BIC, breast invasive carcinoma; BUC, bladder urothelial carcinoma; CA, colorectal adenocarcinoma; GBM, glioblastoma; HCC, hepatocellular carcinoma; LA, lung adenocarcinoma; LSCC, lung squamous cell carcinoma; OSC, ovarian serous cystadenocarcinoma; PanA, pancreatic adenocarcinoma; ProA, prostate adenocarcinoma; RCCC, renal clear cell carcinoma; RPCC, renal papillary cell carcinoma; SA, stomach adenocarcinoma; SCM, skin cutaneous melanoma; TGCT, testicular germ cell tumor.
Furthermore, comparison analysis was performed across all 15 cancer types and unsupervised hierarchical clustering heatmap was generated (Fig. 4B). Strikingly, PanA, CA, LA, BIC, BUC and ProA exhibit similar profiles of DND1-correlated signaling pathway activation and suppression (Fig. 4B). Furthermore, it is noteworthy that DND1 expression is associated with activation of a variety of well-known anti-cancerous pathways such as PPAR signaling [27], PPARα/RXRα activation [27], apoptosis and PTEN signalings [28] and with deactivation of pro-cancer pathways such as G-protein coupled receptor pathway [29], TREM1 signaling [30,31], and the tumor microenvironment signaling [[32], [33], [34], [35]] (Fig. 4A and B and Suppl. Figure 2). These results are consistent with the previously reported anti-proliferative and pro-apoptotic roles of DND1. On the other hand, it is intriguing that DND1 is also associated with the activation of pro-tumorigenesis pathways such as oxidative phosphorylation [36] and RHOGDI signaling [37], as well as the suppression of phagosome formation etc (Fig. 4B).
Because the DND1-assoicated pathway profiles of PanA, CA, LA, BIC, and BUC are closely clustered together (Fig. 4B), we next examined DND1-co-expressed genes that are shared among these five cancer types. Although among the aforementioned 15 cancers, the top ten genes that are positively or negatively associated with DND1 in mRNA expression in each cancer are highly cancer type-specific (Suppl. Table 2), by examining a larger gene list, we found 787 genes shared by PanA, CA, LA, BIC, and BUC, which are positively co-expressed with DND1 while there are 521 common genes that are conversely co-expressed with DND1 in these five cancer types (q < 0.01; Suppl. Figure 3).
4. Discussion
In this study, by taking advantage of the megadata available on cBioPortal, we examined the DND1 status in a variety of human cancers, its association with cancer clinical outcomes and genes significantly co-expressed with DND1. We also performed integrated analysis of genes that are significantly co-expressed with DND1 and identified biological pathways that are potentially impacted by DND1 in human cancer cells. To our best knowledge, this is the first study that systematically analyzes DND1 in a large number of human cancer patients across different cancer types. However, the association between DND1 alteration and clinical outcomes (Fig. 2) have to be interpreted with caution, as they can be confounded by many different variables that are not controlled for in the analyses. Nonetheless, overall, our results indicate that DND1 likely plays a part in human somatic cancer. This study will serve as a prelude for future experimental investigation of DND1 function in cancers.
Since DND1 can function in translational regulation by either stabilizing or degrading mRNA [1], identification of a large number of genes that significantly co-express with DND1 in human cancers (Fig. 3) is expected. However, the percentage of significantly co-expressed genes (SCEG) varies greatly among different cancer types, from 6.28% in SCM to 56.9% in BIC, and even between cancer subtypes that originate from the same organ (e.g, 48.98% in RCCC and 10.79% in RPCC). In addition, in many of the cancers we examined, the expression of DND1 seems to predominantly either positively or conversely related with others. Similarly, the IPA of these co-expressed genes also revealed that DND1 is mainly associated with pathway activation in some cancers such as HCC, LSCC and RPCC etc., but deactivation in others such as PanA, LA, BUC and CA etc. All these data indicate the context-dependence of DND1 function, which may result from the complexity of DND1 regulators and co-factors.
Further more, our IPA results showed that DND1 is associated with activation of a variety of well-known anti-cancerous pathways such as apoptosis and PTEN signaling, and with deactivation of pro-cancer pathways such as G-protein coupled receptor pathway, TREM1 signaling and the tumor microenvironment pathway (Fig. 4). These results are consistent with the previously reported anti-proliferative and pro-apoptotic roles of DND1. However, DND1 seems to also exhibit pro-cancerous features as it also associates with the activation of oxidative phosphorylation and RHOGDI signaling. These seemingly contradictory data suggest DND1 may play dual and opposing roles by exerting anti- and pro-proliferative effects in cancer cells. Of note, DND1 expression is associated with tRNA charging and phagosome formation signaling in multiple cancers (Fig. 4), which also supports the dual role of DND1, as both these signaling pathways have been indicated to possess both anti- and pro-tumorigenesis functions [[38], [39], [40]]. Furthermore, although several in vitro experiments have indicated the tumor suppressive function of DND1 in cell lines of breast cancer, liver cancer and TSCC etc., in SW48 colorectal cancer cells, silencing DND1 suppressed cell proliferation and DND1 overexpression reversed the tumor-suppressive roles of miR-24, indicating a tumor-promoting role of DND1 [23]. High DND1 level also indicates a poor prognosis in prostate cancer [41]. Further investigation will be needed to experimentally verify the effects of DND1 on the signaling pathways identified by the IPA. If the above proposed dual roles of DND1 indeed exist, it will be important to explore the molecular mechanisms that decide the alternate roles of DND1. Information from our analysis presented here is one step that could improve cancer diagnosis and treatment in the future.
Funding sources
Y. Z has been funded by NIGMS [1SC2GM135111-01], NIH RCMI [U54MD007605] and CPRIT [RP180748].
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101206.
Contributor Information
Yun Zhang, Email: yun.zhang@tsu.edu.
Angabin Matin, Email: matin_a@mercer.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Zhang Y., Godavarthi J.D., Williams-Villalobo A., Polk S., Matin A. The role of DND1 in cancers. Cancers. 2021;13 doi: 10.3390/cancers13153679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kedde M., Strasser M.J., Boldajipour B., Vrielink J.A., Slanchev K., le Sage C., Nagel R., Voorhoeve P.M., van Duijse J., Orom U.A., Lund A.H., Perrakis A., Raz E., Agami R. RNA-binding protein Dnd1 inhibits MicroRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 3.Aguero T., Jin Z., Owens D., Malhotra A., Newman K., Yang J., King M. Combined functions of two RRMs in Dead-end1 mimic helicase activity to promote nanos1 translation in the germline. Mol. Reprod. Dev. 2018;85:896–908. doi: 10.1002/mrd.23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki A., Niimi Y., Shinmyozu K., Zhou Z., Kiso M., Saga Y. Dead end1 is an essential partner of NANOS2 for selective binding of target RNAs in male germ cell development. EMBO Rep. 2016;17:37–46. doi: 10.15252/embr.201540828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaji M., Jishage M., Meyer C., Suryawanshi H., Der E., Yamaji M., Garzia A., Morozov P., Manickavel S., McFarland H.L., Roeder R.G., Hafner M., Tuschl T. DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs. Nature. 2017;543(7646):568–572. doi: 10.1038/nature21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S., Zeng M., Sun H., Deng W., Lu Y., Tao D., Liu Y., Zhang S., Ma Y. Zebrafish Dnd protein binds to 3'UTR of geminin mRNA and regulates its expression. BMB Rep. 2010;43(6):438–444. doi: 10.5483/bmbrep.2010.43.6.438. 410.5483/bmbrep.2010.5443.5486.5438. [DOI] [PubMed] [Google Scholar]
- 7.Mei W., Jin Z., Lai F., Schwend T., Houston D., King M., Yang J. Maternal Dead-End1 is required for vegetal cortical microtubule assembly during Xenopus axis specification. Development. 2013;140:2334–2344. doi: 10.1242/dev.094748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguero T., Jin Z., Chorghade S., Kalsotra A., King M., Yang J. Maternal Dead-end 1 promotes translation of nanos1 by binding the eIF3 complex. Development. 2017;144:3755–3765. doi: 10.1242/dev.152611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., Su Y.L., Li L.S., Yang Z., Chen S., Xiong J., Fu X.H., Peng X.N. Mouse dead end 1-β interacts with c-Jun and stimulates activator protein 1 transactivation. Mol. Med. Rep. 2015;11:1701–1707. doi: 10.3892/mmr.2014.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noguchi T., Noguchi M. A recessive mutation (ter) causing germ cell deficiency and a high incidence of congenital testicular teratomas in 129/Sv-ter mice. J. Natl. Cancer Inst. 1985;75:385–392. [PubMed] [Google Scholar]
- 11.Sakurai T., Katoh H., Moriwaki K., Noguchi T., Noguchi M. The ter primordial germ cell deficiency mutation maps near Grl-1 on mouse chromosome 18. Mamm. Genome. 1994;5:333–336. doi: 10.1007/BF00356550. [DOI] [PubMed] [Google Scholar]
- 12.Stevens L.C. A new inbred subline of mice (129-terSv) with a high incidence of spontaneous congenital testicular teratomas. J. Natl. Cancer Inst. 1973;50:235–242. doi: 10.1093/jnci/50.1.235. [DOI] [PubMed] [Google Scholar]
- 13.Noguchi T., Stevens L.C. Primordial germ cell proliferation in fetal testes in mouse strains with high and low incidences of congenital testicular teratomas. J. Natl. Cancer Inst. 1982;69:907–913. [PubMed] [Google Scholar]
- 14.Asada Y., Varnum D.S., Frankel W.N., Nadeau J.H. A mutation in the Ter gene causing increased susceptibility to testicular teratomas maps to mouse chromosome 18. Nat. Genet. 1994;6:363–368. doi: 10.1038/ng0494-363. [DOI] [PubMed] [Google Scholar]
- 15.Youngren K.K., Coveney D., Peng X., Bhattacharya C., Schmidt L.S., Nickerson M.L., Lamb B.T., Deng J.M., Behringer R.R., Capel B., Rubin E.M., Nadeau J.H., Matin A. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Northrup E., Zschemisch N., Eisenblätter R., Glage S., Wedekind D., Cuppen E., Dorsch M., Hedrich H. The ter mutation in the rat Dnd1 gene initiates gonadal teratomas and infertility in both genders. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038001. 38010.31371/journal.pone.0038001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng F., Pan Y., Lu Y.M., Zhu L., Chen S. RNA-binding protein Dnd1 promotes breast cancer apoptosis by stabilizing the Bim mRNA in a miR-221 binding site. BioMed Res. Int. 2017:2017. doi: 10.1155/2017/9596152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu W., Gong F., Zhang T., Chi B., Wang J. RNA-binding protein Dnd1 inhibits epithelial–mesenchymal transition and cancer stem cell-related traits on hepatocellular carcinoma cells. Biotechnol. Lett. 2017;39:1359–1367. doi: 10.1007/s10529-017-2375-5. [DOI] [PubMed] [Google Scholar]
- 19.Liu X., Wang A., Heidbreder C.E., Jiang L., Yu J., Kolokythas A., Huang L., Dai Y., Zhou X. MicroRNA-24 targeting RNA-binding protein DND1 in tongue squamous cell carcinoma. FEBS Lett. 2010;584:4115–4120. doi: 10.1016/j.febslet.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhandari A., Gordon W., Dizon D., Hopkin A.S., Gordon E., Yu Z., Andersen B. The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene. 2013;32:1497–1507. doi: 10.1038/onc.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wampfler J., Federzoni E.A., Torbett B.E., Fey M.F., Tschan M.P. The RNA binding proteins RBM38 and DND1 are repressed in AML and have a novel function in APL differentiation. Leuk. Res. 2016;41:96–102. doi: 10.1016/j.leukres.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Zechel J., Doerner S., Lager A., Tesar P., Heaney J., Nadeau J. Contrasting effects of Deadend1 (Dnd1) gain and loss of function mutations on allelic inheritance, testicular cancer, and intestinal polyposis. BMC Genet. 2013;14:54. doi: 10.1186/1471-2156-1114-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q., Li W., Liu G.H., Tang W. MicroRNA-24 regulates the growth and chemosensitivity of the human colorectal cancer cells by targeting RNA-binding protein DND1. J. BUON. 2019;24(4):1476–1481. 1476-1481. [PubMed] [Google Scholar]
- 24.Cerami E., Gao J., Dogrusoz U., Gross B., Sumer S., Aksoy B., Jacobsen A., Byrne C., Heuer M., Larsson E., Antipin Y., Reva B., Goldberg A., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J., Aksoy B., Dogrusoz U., Dresdner G., Gross B., Sumer S., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269) doi: 10.1126/scisignal.2004088. PMID: 23550210; PMCID: PMC24160307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lakshmi S.P., Reddy A.T., Banno A., Reddy R.C. PPAR agonists for the prevention and treatment of lung cancer. PPAR Res. 2017;2017:8252796. doi: 10.1155/2017/8252796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsons R. Discovery of the PTEN tumor suppressor and its connection to the PI3K and AKT oncogenes. Cold Spring Harb Perspect. Med. 2020;10 doi: 10.1101/cshperspect.a036129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bar-Shavit R., Maoz M., Kancharla A., Nag J.K., Agranovich D., Grisaru-Granovsky S., Uziely B. G protein-coupled receptors in cancer. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17081320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saurer L., Zysset D., Rihs S., Mager L., Gusberti M., Simillion C., Lugli A., Zlobec I., Krebs P., Mueller C. TREM-1 promotes intestinal tumorigenesis. Sci. Rep. 2017;7:14870. doi: 10.1038/s41598-017-14516-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosco M.C., Raggi F., Varesio L. Therapeutic potential of targeting TREM-1 in inflammatory diseases and cancer. Curr. Pharmaceut. Des. 2016;22:6209–6233. doi: 10.2174/1381612822666160826110539. [DOI] [PubMed] [Google Scholar]
- 32.Wang M., Zhao J., Zhang L., Wei F., Lian Y., Wu Y., Gong Z., Zhang S., Zhou J., Cao K., Li X., Xiong W., Li G., Zeng Z., Guo C. Role of tumor microenvironment in tumorigenesis. J. Cancer. 2017;8:761–773. doi: 10.7150/jca.17648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J., Nefedova Y., Lei A., Gabrilovich D., Neutrophils, PMN-MDSC Their biological role and interaction with stromal cells. Semin. Immunol. 2018;35:19–28. doi: 10.1016/j.smim.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu Y., Liu S., Zeng S., Shen H. From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019;38:396. doi: 10.1186/s13046-019-1396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barriga V., Kuol N., Nurgali K., Apostolopoulos V. The complex interaction between the tumor micro-environment and immune checkpoints in breast cancer. Cancers. 2019;11 doi: 10.3390/cancers11081205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashton T.M., McKenna W.G., Kunz-Schughart L.A., Higgins G.S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. 2018;24:2482–2490. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- 37.Harding M.A., Theodorescu D. RhoGDI signaling provides targets for cancer therapy. Eur. J. Cancer. 2010;46:1252–1259. doi: 10.1016/j.ejca.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S., You S., Hwang D. Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat. Rev. Cancer. 2011;11:708–718. doi: 10.1038/nrc3124. [DOI] [PubMed] [Google Scholar]
- 39.Kwon N.H., Fox P.L., Kim S. Aminoacyl-tRNA synthetases as therapeutic targets. Nat. Rev. Drug Discov. 2019;18:629–650. doi: 10.1038/s41573-019-0026-3. [DOI] [PubMed] [Google Scholar]
- 40.Aras S., Zaidi M.R. TAMeless traitors: macrophages in cancer progression and metastasis. Br. J. Cancer. 2017;117:1583–1591. doi: 10.1038/bjc.2017.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu G., Yang X., Li C., Wang F., Cui J., Li B., Xiao H., Tang K., Cui Z. High DND1 level indicates a poor prognostic factor in prostate cancer. Dis. Markers. 2021;2021:9948241. doi: 10.1155/2021/9948241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.