Abstract
A total of 145 penicillin-nonsusceptible Streptococcus pneumoniae strains were isolated from young carriers in Greece and analyzed by antibiotic susceptibility testing, serotyping, restriction fragment end labeling (RFEL), and penicillin-binding protein (PBP) genotyping. The serotypes 23A and 23F (54%), 19A and 19F (25%), 9V (5%), 15A, 15B, and 15C (4%), 6A and 6B (4%), and 21 (4%) were most prevalent in this collection. Fifty-three distinct RFEL types were identified. Sixteen different RFEL clusters, harboring 2 to 32 strains each, accounted for 82% of all strains. Eight of these genetic clusters representing 60% of the strains were previously identified in other countries. A predominant lineage of 66 strains (46%) harboring five RFEL types and the serotypes 19F and 23F was closely related to the pandemic clone Spain23F-1 (genetic relatedness of ≥85%). Another lineage, representing 11 strains, showed close genetic relatedness to the pandemic clone France9V-3. Another lineage of 8 serotype 21 strains was Greece specific since the RFEL types were not observed in an international collection of 193 genotypes from 16 different countries. Characterization of the PBP genes pbp1a, pbp2b, and pbp2x revealed 20 distinct PBP genotypes of which PBP type 1-1-1, initially observed in the pandemic clones 23F and 9V, was predominantly present in 11 RFEL types in this Greek collection of penicillin-nonsusceptible strains (55%). Sixteen PBP types covering 52 strains (36%) were Greece specific. This study underlines the strong contribution of penicillin-resistant international clones to the prevalence and spread of penicillin-nonsusceptible pneumococci among young children in Greece.
Streptococcus pneumoniae is worldwide a common cause of invasive diseases such as meningitis, bacteremia, and pneumonia and of upper respiratory tract infections (1). Pneumococci are often part of the normal nasopharyngeal flora. Especially young children and elderly people are at risk of becoming colonized with S. pneumoniae. The colonization risk increases where crowding occurs, e.g., in day care centers, hospitals, and nursing homes (23, 30, 31). Although a positive correlation between colonization with pneumococci and acute otitis media has been found (12, 36), a relation between pneumococcal carriage and invasive disease has not yet been proven.
Since the late 1970s and 1980s, antibiotic resistance among pneumococci has become an emerging problem. Several (multi)drug-resistant clones have rapidly spread throughout various European countries, North and Latin America, and Asia (2, 18, 20). Some of these clones were initially discovered in Spain, where so far the prevalence of penicillin resistance has reached levels of up to 60% (4). The most striking example is the spread of the pandemic multidrug-resistant clone 6B from Spain to Iceland in the late 1980s, where until 1988 no drug resistance among S. pneumoniae isolates had been reported. From 1989 to 1992 penicillin resistance rose steeply from 2.3 to 17% (39). In 1993, the resistance level among pneumococcal isolates in Iceland reached 20%, of which the vast majority was associated with the serogroups 6, 19, and 23 (25). In 1991, Munoz et al. have reported the intercontinental spread of a multidrug resistant S. pneumoniae clone of serotype 23F from Spain to the United States (29). Since then this clone, recently designated pandemic clone Spain23F-1 (http://www.wits.ac.za/pmen/pmen.htm) has rapidly spread throughout the United States (27). Finally, Gasc et al. have described in 1995 the spread of a penicillin-resistant pneumococcal clone of serotype 9V (pandemic clone France9V-3) from Spain to France (14). Throughout the years these clones have been identified in many countries in different parts of the world (10, 20, 42). In addition, novel drug-resistant clones have been reported in France, former Czechoslovakia, Spain, Hungary, Japan, South Africa, the United States, Chile, England, etc. which tend to spread in a nationwide manner (6, 13, 16, 17, 19, 35, 37, 45). The risk for colonization with and spread of antibiotic-resistant strains is related to younger-age children, occurrence of refractory middle ear infections, previous antibiotic consumption, and day care attendance (9, 28, 32).
In Greece, the emergence of antibiotic resistance among pneumococcal isolates was recognized in the mid 1990s (41). During the period December 1995 through February 1996, 53% of the pneumococci isolated from healthy carriers attending day care centers appeared to be resistant to one or more antibiotics, while 29% of these isolates were penicillin nonsusceptible (41). In a recent study in which 2,448 infants and toddlers were screened during a 2-year period (1997 to 1999) for pneumococcal carriage, 16% of the pneumococci demonstrated reduced susceptibility to penicillin (40; G. Syrogiannopoulos, I. Grivea, G. Katopodis, and N. Beratis, unpublished data). The aim of the current study was to identify the molecular epidemiological nature of the penicillin-nonsusceptible pneumococci isolated in Greece. For this purpose, molecular analysis was performed on penicillin-nonsusceptible isolates of the two Greek studies. A total of 145 (multi)resistant pneumococcal isolates collected from both studies were characterized by drug susceptibility testing, serotyping, restriction fragment end labeling (RFEL), and penicillin-binding protein (PBP) genotyping.
MATERIALS AND METHODS
Bacteriology.
Penicillin-nonsusceptible S. pneumoniae strains were isolated from the nasopharynx of 338 children attending seven day care centers in the city of Patras, Southwestern Greece, from December 1995 to February 1996 and from 2,448 children under the age of 2 years visiting health care centers in Southern and Central Greece from February 1997 to February 1999. Bacteriological diagnosis and susceptibility testing were carried out at the Laboratory of the Division of Pediatric Infectious Disease of the University of Patras, Patras, Greece. The bacteriological methods and serotyping have been described previously (40, 41). Molecular analysis was performed on 92% of the penicillin-nonsusceptible isolates, i.e., 34 strains collected from seven day care centers in Patras and 111 strains collected from 12 different provinces in Central and Southern Greece. In addition, the Greek isolates were compared with an international collection of pneumococcal strains representing 193 distinct RFEL types originating from 16 different countries in America, Europe, Africa, and Asia (M. Sluijter, unpublished observations), in which the international clones pandemic clone Spain23F-1, pandemic clone France9V-3, and pandemic clone Spain6B-2 are present (http://www.wits.ac.za/pmen/pmen.htm).
RFEL analysis.
Typing of the 145 pneumococcal strains by RFEL analysis was performed as described by van Steenbergen et al. (44) and as adapted by Hermans et al. (22). Briefly, purified pneumococcal DNA was digested by the restriction enzyme EcoRI. The DNA restriction fragments were end labeled at 72°C with [α-32P]dATP using DNA polymerase (Goldstar; Eurogentec, Seraing, Belgium). The radiolabeled fragments were denatured and separated electrophoretically on a 6% polyacrylamide sequencing gel containing 8 M urea. Subsequently, the gel was transferred onto filter paper, vacuum dried (HBI, Saddlebrook, N.Y.), and exposed for various times at room temperature to ECL Hyperfilms (Amersham, Bucks, United Kingdom).
BOX PCR fingerprinting.
Typing of the 145 pneumococcal strains by BOX PCR fingerprinting was performed as described by van Belkum et al. (43). Briefly, 50 ng of pneumococcal DNA was amplified by PCR (4 min at 94°C [predenaturation]; 40 cycles of 1 min at 94°C, 1 min at 60°C, and 2 min at 74°C; and 2 min at 74°C [extension]), using primer BOX-A (5′-ATACTCTTCGAAAATCTCTTCAAAC), which was designed from the primary structure of the pneumococcal BOX repeat motif. The amplified products were separated on a 1.5% agarose gel. Gels were stained with ethidium bromide, and the banding patterns were evaluated visually.
PBP genotyping.
Genetic polymorphism of the penicillin resistance genes pbp1a, pbp2b, and pbp2x of the penicillin-nonsusceptible isolates was investigated by restriction fragment length polymorphism (RFLP) analysis as described previously (20). Briefly, we amplified the genes by PCR. The primers used to amplify the genes pbp1a, pbp2b, and pbp2x were described previously (7, 11). The amplification products (5 μl) were digested by restriction endonuclease HinfI and separated by agarose gel electrophoresis. Gels were scanned and analyzed by the Geldoc 2000 system (Bio-Rad). The different PBP genotypes received a three-number code (e.g., 6-12-34) referring to the RFLP patterns of the genes pbp1a (6), pbp2b (11), and pbp2x (33), respectively.
Computer-assisted analysis of the DNA banding patterns.
The RFEL types were analyzed using the Windows version of the Gelcompar software version 4 (Applied Maths, Kortrijk, Belgium) after imaging of the RFEL autoradiograms using the Image master DTS (Pharmacia Biotech, Uppsala, Sweden). To this end, the DNA fragments in the molecular size range of 160 to 400 bp were explored. The DNA banding patterns were normalized using pneumococcus-specific bands present in the RFEL banding patterns of all strains. Comparison of the banding patterns was performed by unweighted-pair-group method using arithmetic averages (34) and using the Jaccard similarity coefficient applied to peaks (38). Computer-assisted analysis and methods and algorithms used in this study were carried out according to the instructions of the manufacturer of Gelcompar. A tolerance of 1.2% in band position was applied during comparison of the DNA patterns. For evaluation of the genetic relatedness of the isolates, we used the following definitions: (i) strains of particular RFEL type are 100% identical by RFEL analysis; (ii) a RFEL cluster represents a group of RFEL types that differs in only one band (ca. >95% genetic relatedness); and (iii) an RFEL lineage represents a group of RFEL types that differs in <4 bands (ca. >85% genetic relatedness).
Statistical analysis.
For statistical analysis of the results, we used the Fisher exact test.
RESULTS
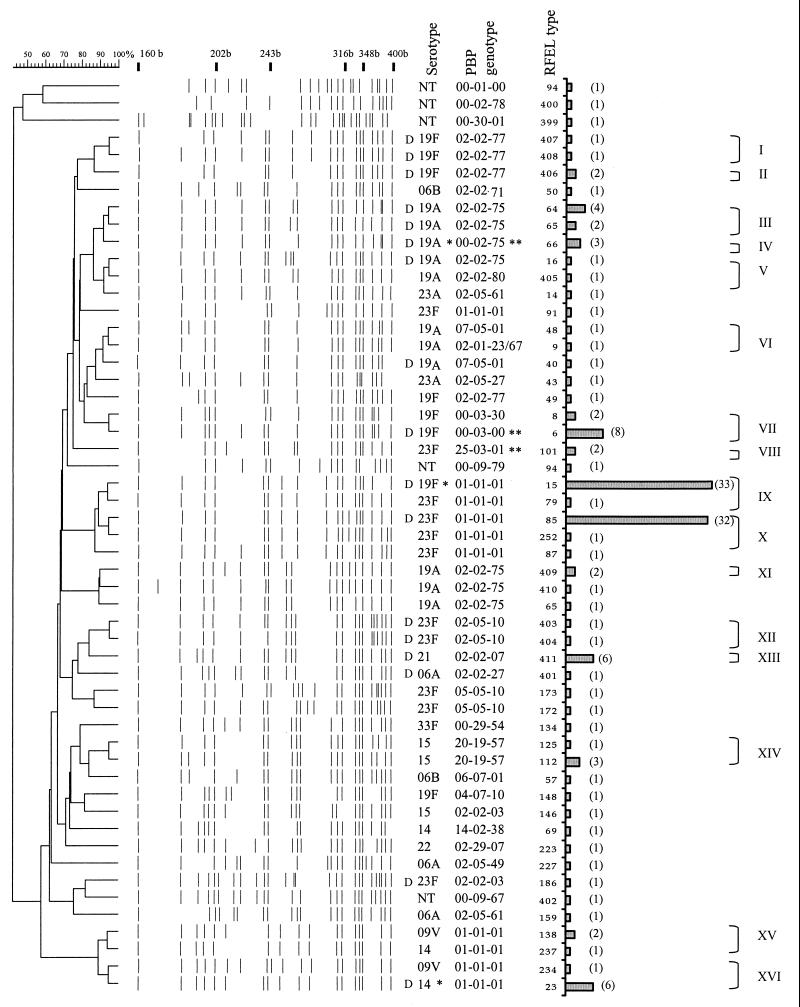
A total of 145 penicillin-nonsusceptible pneumococcal strains were analyzed using serotyping, RFEL, and PBP genotyping. Serotyping revealed 12 different serotypes, namely, 23F (52%); 23A (2%); 19F (15%); 19A (10%); 9V (5%); 15A, 15B, and 15C (4%); 6A (2%); 6B (1%); 21 (4%); 14 (2%); 22 (1%); and 33F (1%) (40, 41; Syrogiannopoulos et al., unpublished). RFEL analysis divided the strains into 53 distinct RFEL genotypes (Fig. 1). Sixteen genetic clusters were observed in this collection of strains, representing 82% of the strains and varying in size from 2 to 32 strains. The genetic relatedness within these clusters was confirmed by BOX PCR (43) (data not shown). Five of the sixteen clusters contained two serotypes, while one of the clusters harbored three different serotypes. We compared the 53 Greek RFEL types with our international library in which 193 RFEL genotypes of pneumococci from 16 different countries are present (M. Sluijter, unpublished observations). Six of the clusters representing 60% of the isolates were previously seen. To analyze the genetic heterogeneity in the penicillin resistance genes, we performed PBP genotyping. Twenty distinct PBP genotypes were observed (Table 1). PBP genotype 1-1-1, initially observed in the pandemic clones 23F and 9V, was most predominantly observed in this Greek collection; 81 strains representing 13 distinct RFEL types shared this penicillin resistance genotype.
FIG. 1.
Dendrogram of the 53 RFEL types observed among 145 penicillin-nonsusceptible pneumococcal isolates from the nasopharynxes of Greek children. Molecular sizes of reference bands are indicated in bases (b). Serotypes, PBP types, RFEL types, bars representing the number of isolates per RFEL type, clusters and cluster codes are also shown. NT, nontypeable; D, RFEL types observed at least once in day care centers. Code “00” refers to untypeable PBP genotypes. Major serotypes (∗) and PBP types (∗∗) within RFEL types are indicated (for details, see Table 1).
TABLE 1.
Clusters, serotypes, PBP genotypes, and resistance patterns of the 145 penicillin-resistant pneumococci isolated from the nasopharynx of young infants in Greece
| Clustera | No. of strains/no. of RFEL types | Serotype(s) (no. of strains) | PBP typeb (no. of strains) | Resistance patternc |
|---|---|---|---|---|
| I | 2/2 | 19F | 2-2-77 | P |
| II | 2/1 | 19F | P | |
| III | 6/2 | 19A and 19F | 2-2-75 | P |
| IV | 3/1 | 19A (2) | 2-2-75 | P |
| 9V (1) | 1-1-1 | P | ||
| V | 2/2 | 19A | 2-2-75 | P |
| 2-2-80 | P | |||
| 1/1 | 23A | 2-5-61 | PCTS | |
| VI | 2/2 | 19A | 7-5-1 | P(TE)S |
| 2-1-23 | P(TE)S | |||
| 1/1 | 19A | 7-5-1 | PCTS | |
| 1/1 | 23A | 2-5-27 | PCTS | |
| VII | 10/2 | 19F | 1(0)-3-30(0) | PTE(S) |
| VIII | 2/1 | 23F | 2-2-71 | PCT |
| 25-3-1 | PS | |||
| IX | 32/2 | 23F | 1-1-1 | PCTECLS |
| X | 33/2 | 23F (32) | PCTS | |
| 19F (1) | PCTS | |||
| 1/1 | 23F | PCTES | ||
| XI | 2/1 | 19A | 2-2-75 | PS |
| 2/2 | 19A | PS | ||
| XII | 2/2 | 23F | 2-5-10 | P |
| XIII | 6/1 | 21 (5) | 2-2-76 | P |
| 2/2 | 23F | 5-5-10 | P | |
| XIV | 4/2 | 15 | 2-19-57 (2) | P |
| 20-19-57 (2) | P | |||
| XV | 8/2 | 9V, 14, and 23A | 1-1-1 | P(CT)S |
| XVI | 3/2 | 23A and 14 | PS |
Genetic clustering according to RFEL typing exclusively matched with BOX PCR fingerprinting.
Code “(0)” refers to untypeable PBP genotypes.
The antibiotic resistance pattern was evaluated for penicillin (P), erythromycin (E), chloramphenicol (C), tetracycline (T), clindamycin (CL) and sulfamethoxazole-trimethoprim (S).
Clusters IX and X were the most predominantly observed clusters and consisted of 31 (21%) and 32 strains (22%), respectively (Fig. 1, Table 1). Both clusters belonged to one predominant lineage of 66 genetically related strains, representing five RFEL types and harboring the serotypes 23F and 19F. This lineage was closely related to the pandemic clone Spain23F-1 (genetic relatedness of ≥90%) which is widely spread all over the world (8, 26, 42). All strains belonging to this lineage showed resistance to penicillin, chloramphenicol, tetracycline, and sulfamethoxazole-trimethoprim. In addition, cluster I was also resistant to erythromycin and clindamycin. Similar to the characteristics of the pandemic clone Spain23F-1, this lineage invariably demonstrated PBP genotype 1-1-1.
The second lineage representing cluster VII contained 10 strains of serotypes 19F (Fig. 1, Table 1). Cluster VII was observed previously in The Netherlands, Thailand, and Vietnam and showed reduced susceptibility to penicillin, tetracycline, erythromycin and, in most cases, to sulfamethoxazole-trimethoprim. This cluster had another common feature since the genetic analysis of pbp1a by PBP genotyping of the vast majority of the strains was not applicable. Consequently, the PBP genotype of the majority of these strains was 0-3-30. Interestingly, this observation is in agreement with the genetically related strains present in the international data library.
The third lineage representing 12 strains and the clusters III, IV, and V displayed serotype 19A (Fig. 1, Table 1). These strains did not cluster with isolates from the international library and their PBP genotype did not match with any of the PBP genotypes present in the international library. All strains displayed the pbp1a and pbp2b genotype 2-2 that matched with the majority of the penicillin-susceptible pneumococci analyzed so far (Sluijter, unpublished). We observed alterations in the PBP profile of pbp2x, resulting in the genotypes 2-2-75 and 2-2-80. These PBP genotypes invariably corresponded to intermediate resistance to penicillin only.
The fourth lineage represented the clusters XV (two strains) and XVI (nine strains) and harbored the serotypes 9V, 14, and 23A (Fig. 1, Table 1). These clusters corresponded to the pandemic clone France9V-3. All strains showed the PBP genotype 1-1-1 and were invariably resistant to penicillin, sulfamethoxazole-trimethoprim and, in some strains, to chloramphenicol and tetracycline.
The fifth lineage represented the clusters XIII (serotype 21; six strains) and XII (serogroup 23F; two strains) (Fig. 1, Table 1). These clusters were Greece specific since they were not present in the international library. Interestingly, these clusters consisted of strains that were isolated in day care centers only. Cluster XIII was mostly observed in a single day care center, whereas cluster XII was exclusively observed in another day care center (Fig. 1). Cluster XIII displayed PBP type 2-2-76 corresponding to low penicillin resistance. Cluster XII showed a different PBP genotype 2-5-10 with alterations in pbp2b and pbp2x. None of the latter two PBP genotypes were present in the international library.
Both study groups were divided in day care center attendees and non-day care attendees to analyze the contribution of day care center attendance on clustering of pneumococcal strains. We observed that 86% of the strains from the day care center group containing 45 pneumococcal isolates belonged to a cluster. Within the non-day care center group representing 100 pneumococcal isolates, 80% of the strains demonstrated genetic clustering. There was no statistical difference between the two groups. We reanalyzed our data for the largest day care center in Patras, where 21 strains were isolated. We observed that 19 of these 21 strains (90%) clustered within four clusters. Although this percentage demonstrated a higher degree of genetic clustering in children attending day care centers, this difference was statistically not significant.
DISCUSSION
Serotyping of the penicillin-nonsusceptible isolates revealed that 79% of the Greek strains belonged to serotypes 23F, 23A, 19F, and 19A (40, 41; Syrogiannopoulos et al., unpublished). These data correspond to earlier findings in other European countries and in the Americas in which these serogroups also significantly contributed to the prevalence of penicillin-resistant S. pneumoniae among young carriers (2, 3, 15, 23, 33, 42).
RFEL genotyping of the 145 penicillin-nonsusceptible isolates showed that 82% of the strains matched within genetic clusters. These data suggest that penicillin-resistant pneumococci rapidly spread among children. This is in line with earlier findings by Hermans et al., who demonstrated that the degree of genetic clustering of penicillin-resistant strains in the Netherlands and Thailand is 70 and 74%, respectively, whereas the degree of genetic clustering among Dutch penicillin-susceptible isolates is 32% (21). Similar data have been reported in various other parts of the world, including Europe, the United States, and South America (8, 20, 26, 42).
The present study clearly demonstrates the significant contribution of the pandemic clone Spain23F-1 to the prevalence and spread of penicillin-nonsusceptible pneumococci among young children in Greece. This is in agreement with studies in other countries in which the predominance of this pandemic clone has also been observed (8, 26, 42). In addition, in the latter studies a second predominant clone, the pandemic clone France9V-3, was highly contributive to the penicillin-resistant pneumococcal population. This clone is also present in our Greek collection, but it does not play a predominant role.
Serogroup 19 represented the major serotype among five lineages. The lineages of cluster VI and cluster VII represented mainly multidrug-resistant isolates and were closely related to isolates found in The Netherlands, Thailand, and Vietnam. Multilocus sequence typing (MLST) has demonstrated that cluster VII also matches with multidrug-resistant serotype 19F isolates from Taiwan (B. Spratt, personal communication). The three other lineages, representing clusters I and II, clusters III, IV, and V, and cluster XI, respectively, showed intermediate resistance to penicillin only and displayed new PBP genotypes with alterations in pbp2x only. MLST has demonstrated that clusters III, IV, and V match with a drug-susceptible serotype 19A invasive isolate from the United Kingdom. In general, this study showed a relation between accumulation of alterations in the three PBP genes and level of penicillin resistance; high resistance levels were often associated with changes in the DNA banding patterns of all three PBP genes (18). In addition, there was also a correlation between multidrug resistance and cumulative alterations in all three PBP genes. This phenomenon is in agreement with the previous observations of Hermans et al. and is hypothesized to be the effect of frequent horizontal cotransfer of resistance genes other than PBP genes in pneumococci with high-level penicillin resistance (20).
Only one lineage, representing two RFEL clusters of serotype 21 and 23F, respectively, was found to be Greece specific since they were not present in the international data library. In addition, this lineage was found only in three day care centers. This is strongly suggestive for the dissemination of pneumococcal clones among day care center attendees within and between day care centers. MLST typing has recently demonstrated that this RFEL cluster of serotype 21 matches a drug-susceptible invasive isolate of serotype 21 in the United Kingdom (B. Spratt, personal communication).
In conclusion, our observations demonstrate a high degree of genetic clustering among penicillin-nonsusceptible, often multidrug-resistant pneumococci in young children in Greece, mainly caused by the spread of a restricted number of penicillin-resistant S. pneumoniae clones. Limiting antibiotic prescription and promoting compliance would probably contribute to the control this problem. A better alternative to prevent the spread of multidrug resistant clones in the near future, however, is large-scale vaccination using pneumococcal conjugate vaccines. Although the initial results of the conjugate vaccination trials look promising (5, 24; R. Dagan, N. Givon, P. Yagupsky, et al., Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. G552, 1998), the epidemiological consequences of such strategies need to be monitored in detail.
ACKNOWLEDGMENTS
We thank C. P. Elzenaar, A. J. Timmers-Reker, and M. Sluijter for technical support.
This work was sponsored by the Sophia Foundation for Medical Research (grant 268) and the NWO (grant SGO-Inf. 005), The Netherlands.
REFERENCES
- 1.Anonymous. Prevention of pneumococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP) Morb Mortal Wkly Rep. 1997;46:1–24. [PubMed] [Google Scholar]
- 2.Appelbaum P C, Gladkova C, Hryniewicz W, Kojouharov B, Kotulova D, Mihalcu F, Schindler J, Setchanova L, Semina N, Trupl J, Tyski S, Urbaskova P, Jacobs M R. Carriage of antibiotic-resistant Streptococcus pneumoniae by children in eastern and central Europe—a multicenter study with use of standardized methods. Clin Infect Dis. 1996;23:712–717. doi: 10.1093/clinids/23.4.712. [DOI] [PubMed] [Google Scholar]
- 3.Arnold K E, Leggiadro R J, Breiman R F, Lipman H B, Schwartz B, Appleton M A, Cleveland K O, Szeto H C, Hill B C, Tenover F C, Elliott J A, Facklam R R. Risk factors for carriage of drug-resistant Streptococcus pneumoniae among children in Memphis, Tennessee. J Pediatr. 1996;128:757–764. doi: 10.1016/s0022-3476(96)70326-8. [DOI] [PubMed] [Google Scholar]
- 4.Baquero F, Garcia-Rodriguez J A, Garcia de Lomas J, Aguilar L. Antimicrobial resistance of 1,113 Streptococcus pneumoniae isolates from patients with respiratory tract infections in Spain: results of a 1-year (1996–1997) multicenter surveillance study. The Spanish Surveillance Group for Respiratory Pathogens. Antimicrob Agents Chemother. 1999;43:357–359. doi: 10.1128/aac.43.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black S, Shinefield H, Fireman B, Lewis E, Ray P, Hansen J R, Elvin L, Ensor K M, Hackell J, Siber G, Malinoski F, Madore D, Chang I, Kohberger R, Watson W, Austrian R, Edwards K. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Northern California Kaiser Permanente Vaccine Study Center Group. Pediatr Infect Dis J. 2000;19:187–195. doi: 10.1097/00006454-200003000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Coffey T J, Berron S, Daniels M, Garcia-Leoni M E, Cercenado E, Bouza E, Fenoll A, Spratt B G. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988–1994): novel major clones of serotypes 14, 19F and 15F. Microbiology. 1996;142:2747–2757. doi: 10.1099/13500872-142-10-2747. [DOI] [PubMed] [Google Scholar]
- 7.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 8.Corso A, Severina E P, Petruk V F, Mauriz Y R, Tomasz A. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb Drug Resist. 1998;4:325–337. doi: 10.1089/mdr.1998.4.325. [DOI] [PubMed] [Google Scholar]
- 9.Dagan R, Leibovitz E, Greenberg D, Yagupsky P, Fliss D M, Leiberman A. Dynamics of pneumococcal nasopharyngeal colonization during the first days of antibiotic treatment in pediatric patients. Pediatr Infect Dis J. 1998;17:880–885. doi: 10.1097/00006454-199810000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Davies T, Goering R V, Lovgren M, Talbot J A, Jacobs M R, Appelbaum P C. Molecular epidemiological survey of penicillin-resistant Streptococcus pneumoniae from Asia, Europe, and North America. Diagn Microbiol Infect Dis. 1999;34:7–12. doi: 10.1016/s0732-8893(98)00156-4. [DOI] [PubMed] [Google Scholar]
- 11.Dowson C G, Hutchison A, Spratt B G. Extensive re-modelling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989;3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 12.Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik D, Tung Y. Relationship between nasopharyngeal colonization and the development of otitis media in children. Tonawanda/Williamsville Pediatrics J Infect Dis. 1997;175:1440–1445. doi: 10.1086/516477. [DOI] [PubMed] [Google Scholar]
- 13.Figueiredo A M, Austrian R, Urbaskova P, Teixeira L A, Tomasz A. Novel penicillin-resistant clones of Streptococcus pneumoniae in the Czech Republic and in Slovakia. Microb Drug Resist. 1995;1:71–78. doi: 10.1089/mdr.1995.1.71. [DOI] [PubMed] [Google Scholar]
- 14.Gasc A M, Geslin P, Sicard A M. Relatedness of penicillin-resistant Streptococcus pneumoniae serogroup 9 strains from France and Spain. Microbiology. 1995;141(Pt. 3):623–627. doi: 10.1099/13500872-141-3-623. [DOI] [PubMed] [Google Scholar]
- 15.Ghaffar F, Friedland I R, McCracken G H., Jr Dynamics of nasopharyngeal colonization by Streptococcus pneumoniae. Pediatr Infect Dis J. 1999;18:638–646. doi: 10.1097/00006454-199907000-00016. [DOI] [PubMed] [Google Scholar]
- 16.Gherardi G, Inostrozo J S, O'Ryan M, Prado V, Prieto S, Arellano C, Facklam R R, Beall B. Genotypic survey of recent beta-lactam-resistant pneumococcal nasopharyngeal isolates from asymptomatic children in Chile. J Clin Microbiol. 1999;37:3725–3730. doi: 10.1128/jcm.37.11.3725-3730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gherardi G, Whitney C G, Facklam R R, Beall B. Major related sets of antibiotic-resistant pneumococci in the United States as determined by pulsed-field gel electrophoresis and pbp1a-pbp2b-pbp2x-dhf restriction profiles. J Infect Dis. 2000;181:216–229. doi: 10.1086/315194. [DOI] [PubMed] [Google Scholar]
- 18.Hakenbeck R. β-Lactam-resistant streptococcus pneumoniae: epidemiology and evolutionary mechanism. Chemotherapy. 1999;45:83–94. doi: 10.1159/000007170. [DOI] [PubMed] [Google Scholar]
- 19.Hall L M, Whiley R A, Duke B, George R C, Efstratiou A. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J Clin Microbiol. 1996;34:853–859. doi: 10.1128/jcm.34.4.853-859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermans P W, Sluijter M, Dejsirilert S, Lemmens N, Elzenaar K, van Veen A, Goessens W H, de Groot R. Molecular epidemiology of drug-resistant pneumococci: toward an international approach. Microb Drug Resist. 1997;3:243–251. doi: 10.1089/mdr.1997.3.243. [DOI] [PubMed] [Google Scholar]
- 21.Hermans P W, Sluijter M, Elzenaar K, van Veen A, Schonkeren J J, Nooren F M, van Leeuwen W J, de Neeling A J, van Klingeren B, Verbrugh H A, de Groot R. Penicillin-resistant Streptococcus pneumoniae in the Netherlands: results of a 1-year molecular epidemiologic survey. J Infect Dis. 1997;175:1413–1422. doi: 10.1086/516474. [DOI] [PubMed] [Google Scholar]
- 22.Hermans P W, Sluijter M, Hoogenboezem T, Heersma H, van Belkum A, de Groot R. Comparative study of five different DNA fingerprint techniques for molecular typing of Streptococcus pneumoniae strains. J Clin Microbiol. 1995;33:1606–1612. doi: 10.1128/jcm.33.6.1606-1612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellner J D, Ford-Jones E L. Streptococcus pneumoniae carriage in children attending 59 Canadian child care centers. Toronto Child Care Centre Study Group. Arch Pediatr Adolesc Med. 1999;153:495–502. doi: 10.1001/archpedi.153.5.495. [DOI] [PubMed] [Google Scholar]
- 24.Korkeila M, Lehtonen H, Ahman H, Leroy O, Eskola J, Kayhty H. Salivary anti-capsular antibodies in infants and children immunised with Streptococcus pneumoniae capsular polysaccharides conjugated to diphtheria or tetanus toxoid. Vaccine. 2000;18:1218–1226. doi: 10.1016/s0264-410x(99)00393-x. [DOI] [PubMed] [Google Scholar]
- 25.Kristinsson K G. Epidemiology of penicillin resistant pneumococci in Iceland. Microb Drug Resist. 1995;1:121–125. doi: 10.1089/mdr.1995.1.121. [DOI] [PubMed] [Google Scholar]
- 26.Marchese A, Ramirez M, Schito G C, Tomasz A. Molecular epidemiology of penicillin-resistant Streptococcus pneumoniae isolates recovered in Italy from 1993 to 1996. J Clin Microbiol. 1998;36:2944–2949. doi: 10.1128/jcm.36.10.2944-2949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDougal L K, Facklam R, Reeves M, Hunter S, Swenson J M, Hill B C, Tenover F C. Analysis of multiply antimicrobial-resistant isolates of Streptococcus pneumoniae from the United States. Antimicrob Agents Chemother. 1992;36:2176–2184. doi: 10.1128/aac.36.10.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melander E, Molstad S, Persson K, Hansson H B, Soderstrom M, Ekdahl K. Previous antibiotic consumption and other risk factors for carriage of penicillin-resistant Streptococcus pneumoniae in children. Eur J Clin Microbiol Infect Dis. 1998;17:834–838. doi: 10.1007/s100960050202. [DOI] [PubMed] [Google Scholar]
- 29.Muñoz R, Coffey T J, Daniels M, Dawson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, et al. Intercontinental spread of a multiresistant clone of genotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 30.Nuorti J P, Butler J C, Crutcher J M, Guevara R, Welch D, Holder P, Elliott J A. An outbreak of multidrug-resistant pneumococcal pneumonia and bacteremia among unvaccinated nursing home residents. N Engl J Med. 1998;338:1861–1868. doi: 10.1056/NEJM199806253382601. [DOI] [PubMed] [Google Scholar]
- 31.Principi N, Marchisio P, Schito G C, Mannelli S. Risk factors for carriage of respiratory pathogens in the nasopharynx of healthy children. Ascanius Project Collaborative Group. Pediatr Infect Dis J. 1999;18:517–523. doi: 10.1097/00006454-199906000-00008. [DOI] [PubMed] [Google Scholar]
- 32.Reichler M R, Allphin A A, Breiman R F, Schreiber J R, Arnold J E, McDougal L K, Facklam R R, Boxerbaum B, May D, Walton R O, et al. The spread of multiply resistant Streptococcus pneumoniae at a day care center in Ohio. J Infect Dis. 1992;166:1346–1353. doi: 10.1093/infdis/166.6.1346. [DOI] [PubMed] [Google Scholar]
- 33.Reichler M R, Rakovsky J, Sobotova A, Slacikova M, Hlavacova B, Hill B, Krajcikova L, Tarina P, Facklam R R, Breiman R F. Multiple antimicrobial resistance of pneumococci in children with otitis media, bacteremia, and meningitis in Slovakia. J Infect Dis. 1995;171:1491–1496. doi: 10.1093/infdis/171.6.1491. [DOI] [PubMed] [Google Scholar]
- 34.Romesburg H. Cluster analysis for researchers. Malabar, Fla: Krieger; 1990. pp. 9–28. [Google Scholar]
- 35.Sibold C, Wang J, Henrichsen J, Hakenbeck R. Genetic relationships of penicillin-susceptible and -resistant Streptococcus pneumoniae strains isolated on different continents. Infect Immun. 1992;60:4119–4126. doi: 10.1128/iai.60.10.4119-4126.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sluijter M, Faden H, de Groot R, Lemmens N, Goessens W H, van Belkum A, Hermans P W. Molecular characterization of pneumococcal nasopharynx isolates collected from children during their first 2 years of life. J Clin Microbiol. 1998;36:2248–2253. doi: 10.1128/jcm.36.8.2248-2253.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith A M, Klugman K P. Three predominant clones identified within penicillin-resistant South African isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:385–389. doi: 10.1089/mdr.1997.3.385. [DOI] [PubMed] [Google Scholar]
- 38.Sneath P. Numerical taxonomy. San Francisco, Calif: W. H. Freeman; 1973. pp. 131–132. [Google Scholar]
- 39.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 40.Syrogiannopoulos G, Grivea I, Katopodis G, Jacobs G P, M, Beratis N. Carriage of antibiotic-resistant Streptococcus pneumoniae in Greek infants and toddlers. Eur J Clin Microbiol Infect Dis. 2000;19:288–293. doi: 10.1007/s100960050477. [DOI] [PubMed] [Google Scholar]
- 41.Syrogiannopoulos G A, Grivea I N, Beratis N G, Spiliopoulou A E, Fasola E L, Bajaksouzian S, Appelbaum P C, Jacobs M R. Resistance patterns of Streptococcus pneumoniae from carriers attending day-care centers in southwestern Greece. Clin Infect Dis. 1997;25:188–194. doi: 10.1086/514526. [DOI] [PubMed] [Google Scholar]
- 42.Tomasz A, Corso A, Severina E P, Echaniz-Aviles G, Brandileone M C, Camou T, Castaneda E, Figueroa O, Rossi A, Di Fabio J L. Molecular epidemiologic characterization of penicillin-resistant Streptococcus pneumoniae invasive pediatric isolates recovered in six Latin-American countries: an overview. PAHO/Rockefeller University Workshop. Pan American Health Organization. Microb Drug Resist. 1998;4:195–207. doi: 10.1089/mdr.1998.4.195. [DOI] [PubMed] [Google Scholar]
- 43.van Belkum A, Sluijuter M, de Groot R, Verbrugh H, Hermans P W. Novel BOX repeat PCR assay for high-resolution typing of Streptococcus pneumoniae strains. J Clin Microbiol. 1996;34:1176–1179. doi: 10.1128/jcm.34.5.1176-1179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Steenbergen T J, Colloms S D, Hermans P W, de Graaff J, Plasterk R H. Genomic DNA fingerprinting by restriction fragment end labeling. Proc Natl Acad Sci USA. 1995;92:5572–5576. doi: 10.1073/pnas.92.12.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida R, Hirakata Y, Kaku M, Tomono K, Maesaki S, Yamada Y, Kamihira S, Jacobs M R, Appelbaum P C, Kohno S. Genetic analysis of serotype 23F Streptococcus pneumoniae isolates from several countries by penicillin-binding protein gene fingerprinting and pulsed-field gel electrophoresis. Chemotherapy. 1999;45:158–165. doi: 10.1159/000007178. [DOI] [PubMed] [Google Scholar]