Abstract
Endemic human coronaviruses (hCoVs) 229E and OC43 cause respiratory disease with recurrent infections, while severe acute respiratory syndrome (SARS)-CoV-2 spreads across the world with impact on health and societies. Here, we report an image-based multicycle infection procedure with α-coronavirus hCoV-229E-eGFP in an arrayed chemical library screen of 5440 clinical and preclinical compounds. Toxicity counter selection and challenge with the β-coronaviruses OC43 and SARS-CoV-2 in tissue culture and human airway epithelial explant cultures (HAEEC) identified four FDA-approved compounds with oral availability. Methylene blue (MB, used for the treatment of methemoglobinemia), Mycophenolic acid (MPA, used in organ transplantation) and the anti-fungal agent Posaconazole (POS) had the broadest anti-CoV spectrum. They inhibited the shedding of SARS-CoV-2 and variants-of-concern (alpha, beta, gamma, delta) from HAEEC in either pre- or post exposure regimens at clinically relevant concentrations. Co-treatment of cultured cells with MB and the FDA-approved SARS-CoV-2 RNA-polymerase inhibitor Remdesivir reduced the effective anti-viral concentrations of MB by 2-fold, and Remdesivir by 4 to 10-fold, indicated by BLISS independence synergy modelling. Neither MB, nor MPA, nor POS affected the cell delivery of SARS-CoV-2 or OC43 (+)sense RNA, but blocked subsequent viral RNA accumulation in cells. Unlike Remdesivir, MB, MPA or POS did not reduce the release of viral RNA in post exposure regimen, thus indicating infection inhibition at a post-replicating step as well. In summary, the data emphasize the power of unbiased, full cycle compound screens to identify and repurpose broadly acting drugs against coronaviruses.
Keywords: Human coronavirus drug screen at single cell resolution in arrayed format, Human explant nasal and bronchial epithelial cells, Repurposing methylene blue, Mycophenolic acid, Posaconazole, Drug synergy in vitro, Post-entry coronavirus inhibition, SARS-CoV-2 variants of concern
Graphical abstract
1. Introduction
In December 2019, a local outbreak of pneumonia caused by an unknown agent was reported in Wuhan (Hubei, China), and soon after the agent was identified as SARS-CoV-2 causing Coronavirus disease 2019 (COVID-19) (Wu et al., 2020). As of Jan 18, 2022, there were more than 330 million PCR positive cases and 5.5 million deaths related to SARS-CoV-2 related to SAT (computed by the Johns Hopkins Coronavirus Research Center)(Home). Although vaccinations with mRNA and viral vectors strongly attenuate the severity of SARS-CoV-2 caused disease (Baden et al., 2021; Voysey et al., 2021), evidence emerges that vaccination will not suffice to contain the ongoing pandemics, and incompletely protects humans from severe COVID-19 (Planas et al., 2021; Hacisuleyman et al., 2021; Mlcochova et al., 2021; Kreer et al., 2020; Creech et al., 2021). Infection breakthrough is in part associated with emerging SARS-CoV-2 variants. Variants of concern are more contagious (alpha variant B.1.1.7, and delta variant B.1.617.2), and exhibit reduced susceptibility to antibodies from vaccinated or recovered individuals (beta variant B.1.351, gamma variant P.1 derived from B.1.1.28, delta variant B.1.617.2). Notably, many of the mutations of concern are located in the spike glycoprotein of the SARS-CoV-2 variants.
Severe COVID-19 is accompanied by respiratory failure, and patients require mechanical ventilation at intensive care units, and are at risk to develop acute respiratory distress syndromes, multiple organ dysfunction syndromes or failures and death (Cascella et al., 2021). Current treatment options are limited, and include supportive care, and administration of the viral RNA polymerase inhibitor Remdesivir together with corticosteroids (https://www.covid19treatmentguidelines.nih.gov/therapeutic-management/). Besides the mRNA vaccines, there are five FDA approved anti-COVID-19 therapies - the repurposed viral polymerase inhibitor Remdesivir (Beigel et al., 2020a), the janus kinase (JAK) 1 and 2 inhibitor Baricitinib blocking proinflammatory syndrome, the anti-S-protein IgG monoclonal antibody LY-CoV555, the anti-SARS-CoV-2 antibody cocktail Casirivimab/Imdevimab, and the monoclonal antibody Actemra blocking the interleukin 6 receptor (https://racetoacure.stanford.edu). Recently, the FDA emergency-use authorized Paxlovid, a SARS-CoV-2 3CL protease inhibitor, and Molnupiravir, an RNA mutagen for the virus. Currently, Remdesivir in combination with anti-inflammatory agents, such as the corticosteroid dexamethasone is a broadly used treatment to restrict severe lung inflammation and multiorgan failure upon marked immune cell dysregulation and cytokine storm syndrome (Neurath, 2021).
Drug repurposing allows for rapid identification of clinically approved or investigational compounds towards emerging indications. This approach offers a multitude of advantages over de novo drug development (reviewed in Ashburn and Thor, 2004). Most importantly, candidate compounds have a demonstrated safety record allowing for direct transition to human clinical trials. For example, the ReFrame library comprising some 12 000 compounds was used to screen single round SARS-CoV-2 infections, focussing on virus replication but omitting assembly and egress processes (Bakowski et al., 2021). This yielded compounds inhibiting genome replication and steps upstream of replication. Identified compounds included MK-4482 (Molnupiravir, nucleoside analog), Nafamostat mesylate (inhibitor of entry), hydroxycytidine (nucleoside analog), and nelfinavir, an HIV protease inhibitor also blocking human adenovirus infections (Georgi et al., 2020a). Other drugs proposed for repurposing against COVID-19 include the translation elongation inhibitor Aplidin (Plitidepsin) approved for the treatment of multiple myeloma (Riva et al., 2020; Losada et al., 2016; White et al., 2021), or the anti-viral anti-helmintic protonophore Niclosamide blocking endosomal low pH dependent viruses (Jurgeit et al., 2012), as well as the anti-malaria agents Amodiaquine and Artesunate, the HIV protease inhibitor combination Lopinavir/Ritonavir, the RNA polymerase inhibitors Favipiravir and Ribavirin used to treat influenza virus and respiratory syncytial virus infections, respectively, the nucleoside analogue EIDD-2801 (MK-4482 or Molnupiravir), the influenza virus neuraminidase inhibitor Oseltamivir, and the SARS-CoV-2 Nsp12 RNA-polymerase inhibitor AT-527 (Good et al., 2021).
Here, we present the results from a multicycle drug repurposing screen which uncovers a range of drug candidates for COVID-19 treatment. Unlike canonical drug screens, our assay sampled the entire replication cycle of a GFP expressing variant of the endemic α-coronavirus hCoV-229E in the human hepatoma cell line Huh7. We validated the initial hits by two hCoVs, OC43 and SARS-CoV-2 in nasal and bronchial human airway epithelial explant cultures (HAEEC) grown at air-liquid interface, and identified 9 systemically useable compounds with broad coronavirus effects. Four of them are approved for human use, namely Methylene blue (MB, also known as Methylthioninium chloride), Mycophenolic acid (MPA), Posaconazole (POS), and R788 (Fostamatinib), two are in phase II clinical trials (MLN4924, Ravuconazole), and three in preclinical tests (GPP-78, Ro 48–8071, SB-505124). One of the approved compounds (MPA) has anti-inflammatory effects in humans, and one (MB) has been widely used as a placebo control drug in clinical trials.
2. Methods
2.1. Viruses
hCoV-229E-GFP and SARS-CoV-2 “Wuhan” (TAR clone 3.3, München-1.1/2020/929) were obtained from Dr. Volker Thiel (University of Bern) (Cervantes-Barragan et al., 2010; Thao et al., 2020). SARS-CoV-2 Alpha (2 012 212 272 7D 15.01.21) and Beta (SDV4-SD1-4D 08/02/21) variants were obtained from Dr. Isabella Eckerle (University of Geneva). SARS-CoV-2 Gamma (NH-RIVM10915/2021) and Delta (NH-RIVM-27142/2021) variants were obtained from the RIVM (Netherlands) through European Virus Archive global. hCoV-229E-GFP was plaque purified and expanded on Huh7 cells for 48h. Supernatant was collected and cleared by centrifugation at 5000×g for 10 min. Human CoV-OC43 (ATCC-VR-1558) was obtained from LGC Standards Limited, (Teddington, United Kingdom) and expanded as described above. Virus titers were determined by FFU titration according to GFP or immunofluorescence signal, and by TCID50 titration according to the Spearman-Kärber method. SARS-CoV-2 variants were checked by routine Sanger sequencing of the Spike glycoprotein (S-ORF nucleotide position: 20–800).
2.2. Cell lines
Huh7 and Vero E6 cells were obtained from Dr. Volker Thiel (University of Bern, Switzerland). Polyclonal Huh7-ACE2, HeLa-ACE2 and A549-ACE2 were generated by stable transfection with a lentiviral vector (pLVX-ACE2-IRES-BSD). Parental HeLa and A549 cells were obtained from ATCC. The parental HeLa cell line additionally expressed an inducible eCas9 and a non-targeting sgRNA.
2.3. Cell culture
Huh7, Huh7-ACE2, HeLa-ACE2 and A549-ACE2 cells were maintained in Dulbecco's Modified Eagle Medium (D6429; Sigma-Aldrich, St. Louis, USA) supplemented with 10% fetal bovine serum (FBS, 10270; Invitrogen, Carlsbad, USA), non-essential amino acids (M7145; Sigma-Aldrich, St. Louis, USA) and subcultured by PBS washing and trypsinisation (C-41020; Trypsin-EDTA, Sigma-Aldrich, St. Louis, USA) twice weekly. Cell lines were kept at 37 °C, 5% CO2, 95% humidity. Infections with Huh7 cells were conducted with Minimal Essential Medium (MEM, Sigma-Alrich, St. Louis, USA) supplemented with 10% FBS.
2.4. Compound library
The Prestwick chemical library (PCL) was purchased from Prestwick Chemical (Illkirch, France). The remaining compounds were purchased from abcr GmbH, AK Scientific, Inc., Abcam plc., Acros Organics, AdipoGen Life Sciences, Inc., Advanced ChemBlocks Inc., Alinda Chemical Trade Company Ltd., Angene, Apollo Scientific Ltd., Axon Medchem, BIOTREND Chemikalien GmbH, BLD Pharmatech Ltd., Biosynth Carbosynth, Cayman Chemical Company, ChemBridge Corporation, ChemDiv Inc., Chemodex Ltd., Cohesion Biosciences Ltd., Combi-Blocks, Inc., Enamine Ltd., Enzo Life Sciences Inc., Fluorochem Ltd., Focus Biomolecules, InterBioScreen Ltd., J&K Scientific Ltd., Key Organics Ltd., LabSeeker, Inc, Life Chemicals Inc., Matrix Scientific, Maybridge Ltd., MedChemExpress, Merck KGaA, Otava Ltd., Pharmeks Ltd., Ramidus AB, SYNkinase, Santa Cruz Biotechnology, Selleck Chemicals LLC, Sigma-Aldrich, Specs, Target Molecule Corp., Tocris Bioscience, Tokyo Chemical Industry, Toronto Research Chemicals, UkrOrgSyntez Ltd., Vitas-M Laboratory Ltd. or Wuhan ChemFaces Biochemical Co., Ltd.. Compounds were dissolved in DMSO at a concentration of 10 mM.
2.5. High throughput compound screening
The screen was conducted in four completely independent biological replicates without technical replicates. For fluidics handling, a Matrix WellMate dispenser and Matrix WellMate tubing cartridges (Thermo Fisher Scientific, Waltham, USA), an Assist Plus pipetting robot (Integra Biosciences AG, Zizers, Switzerland) and an Echo acoustic dispenser (Labcyte Inc., Indiana, USA) were used. Compounds were spotted in random order at 1.67 μM final concentration in microscopy grade 384-well plates (Perkin-Elmer, Waltham, USA), and frozen. Plates were barcoded and shipped to University of Zurich without content lists to ensure blinding. Pre-spotted plates were thawed at room temperature 30 min prior to cell seeding. About 6000 Huh7 cells were seeded in 25 μl/well MEM +10% FCS and incubated at 37 °C, 5% CO2, 95% humidity overnight. Infection with hCoV-229E-GFP was performed by adding 50 focus forming units (FFU) per well in 5 μl and incubated at 33.5 °C, 5% CO2, 95% humidity for 48h. Finally, cells were fixed by adding 10 μl 16% paraformaldehyde (Sigma-Aldrich, St. Louis, USA) containing 0.4 μg/ml Hoechst 33342 (Sigma-Aldrich, St. Louis, USA). The fixation reaction was quenched in 25 mM NH4Cl in phosphate-buffered saline (PBS) for 30 min. Plates were washed three times with PBS. Finally, PBS was replaced with PBS + 0.02% N3. Plates were imaged with an automated inverted microscope (ImageXpress Micro Confocal High-Content Imaging System) at 4x magnification (Molecular Devices, San Jose, USA). For the validation experiments with hCoV-OC43 and SARS-CoV-2, compounds were spotted at 1.75 μM, cells seeded in 80 μl, incubated at 37 °C, 5% CO2 and 95% humidity overnight and infected by adding 120 FFU in 20 μl. After 68 h at 33.5 °C (OC43) or 24 h or 48 h at 37 °C (SARS-CoV-2), cells were fixed as described above. Subsequently, the experiments were subjected to immunostaining. Cells were permeabilized with 0.2% Triton-X-100 for 5 min, washed with PBS, incubated with primary antibodies against coronavirus nucleoprotein (Chemicon MAB 9013 for OC43, Rockland 200-401-A50 for SARS-CoV-2) in PBS + 1% bovine serum albumin (BSA) for 1 h, washed and incubated with a secondary antibody in PBS + 1% BSA for 1 h. Cells were then washed again, PBS was replaced with PBS + 0.02% N3 and finally, plates were imaged as described above. Only after the acquisition of images, the content lists were provided to UZH by EPFL to ensure the image analysis was conducted blinded.
2.6. Image analysis
Viral infection and cell health were analyzed using Plaque2.0 (Yakimovich et al., 2015a), with five parameters: number of nuclei, number of nuclei from infected cells, infection index, total intensity of the infected cells and number of plaques (infection foci).
2.7. Post-processing
Results obtained by image analysis with Plaque2.0 were annotated and filtered using R version 4.0.2 in RStudio Version 1.3.1056. Readout values were per-plate normalized by the mean values of the DMSO controls. Compounds were scored as toxic if the mean number of nuclei was lower than the mean number of nuclei of all negative control wells minus two times the standard deviation of the number of nuclei of all negative control (DMSO, no compound) wells. Compounds were considered hits if the mean GFP intensity signal value of all replicates falls below the mean of all negative control wells minus three times the standard deviation of all negative control wells for that specific readout. Hit lists were combined for all parameters. In the validation experiments, compounds were considered toxic if the mean number of nuclei was lower than the mean number of nuclei of all negative control wells divided by two.
2.8. Drug synergy
H1299 or Huh7 cells (about 20000 per well in a 96-well imaging plate) were treated with indicated concentrations of Remdesivir and MB at 37 °C, either alone and in combination resulting in a nine by nine matrix. After about 12h, the cells were infected with hCoV-229E-GFP virus at MOI 0.001 at 33.5 °C, fixed with 4% PFA and stained with Hoechst 33342 nuclear dye at 3 d pi. The experiment was performed in triplicate. The resulting plaques were quantified using Plaque 2.0 (Yakimovich et al., 2015b). The plaque number readout was normalized using the maximum number of plaques observed in the well of interest. The inhibitory effect of the drugs was represented as:
Synergy of the drugs was assessed using BLISS synergy score calculated and visualized by using the web tool Synergy Finder (http://www.synergyfinder.org) (Zheng et al., 2021).
2.9. RNA FISH with branched DNA signal amplification
Fifteen thousand Huh7 wt or Huh7-Ace2 cells were seeded in a 96-well plate in the presence of Remdesivir (1 μM), POS (10 μM), MB (2 μM) or MPA (2 μM). On the following day, cells were inoculated with hCoV-OC43 or Sars-CoV-2 at 33.5 °C and 37 °C, respectively, for 2h with input virus amounts that resulted in about 15 uncoated genomes in the cells. Inocula were washed away, and cells incubated with fresh infection medium containing the compounds for 2, 8 or 24 h pi, fixed with 3% PFA in PBS for 30 min at RT, washed twice with PBS, dehydrated by subsequent incubation with 50%, 70% and 100% ethanol for 2 min at RT, and stored at −20 °C until staining. Samples were rehydrated by incubation with 70%, 50% ethanol and PBS for 2 min at RT. Rehydrated samples were FISH-stained against either OC43 or SARS-CoV-2 ORF1a mRNA using ViewRNA mRNA FISH assay with type 1 probe Alexa Fluor 546, according to the manufacturer's instructions. Probes were custom made #6007025-01 and #6007037-01, respectively, directed against the ORF1a sequences between positions 2635–3663 and 401–1327 respectively, ThermoFisher). Subsequently, cells were incubated in PBS containing DAPI and succinimidyl ester AlexaFluor 647 (Thermo Fisher) for 10 min at RT, and imaged using an ImageXpress Micro confocal microscope (Molecular Devices, 60 μm pinhole, 15 stacks, 1.5 μm slice thickness) with a 40x NA 0.95 objective. For quantification, cells were segmented according to the DAPI and succinimidyl ester signals, and segmented OC43 or SARS-CoV-2 (+) RNA dots assigned to single cells using CellProfiler.
2.10. Nasal and bronchial HAEEC
Human nasal and bronchial airway tissue (MucilAir™, Epithelix SA, Geneva, Switzerland) cultured on transwell inserts (24-well plate) were maintained at air-liquid interface according to the supplier's instructions and cell culture medium (EP05MM). The samples were mycoplasma-free based on acetate kinase or carbamate kinase activity measurements of the supplier. The bronchial HAEEC were obtained from individual donors (Batch nr: MD0810) and the nasal HAEEC from a pool of fourteen healthy donors (Batch nr: MP0009, and MP010). All donors were non-smokers. Movies of cilia beating were recorded at room temperatures with an inverted light microscope (Axiovert 135) at 100X magnification using CellF software (Olympus Soft Imaging Solutions GmbH, version 3.0).
2.11. SARS-CoV-2 infection of MucilAir™ tissue and drug treatment
Three days prior infection, MucilAir™ tissue apical surface were washed with 200 μl of warmed MucilAir™ culture medium at 37 °C for 20 min to homogenize the amount of mucus between inserts. Inserts were inoculated on the apical side with 1000 FFU of SARS-CoV-2 in a final volume of 100 μl at 37 °C for 2 h. Then the SARS-CoV-2 inoculum was removed and the apical surfaces quickly washed two-times with PBS. SARS-CoV-2 produced at the apical surface was collected at different time point pi by 20 min apical wash and quantified in parallel by RT-qPCR and TCID50 titration with Vero E6 cells. Drug treatment was done at the indicated times pre and/or pi at indicated concentrations in fresh basolateral medium. Nasal and bronchial MucilAir™ tissue analysis was performed in duplicate.
2.12. Quantification of ciliary motion using optical flow
Transmitted light movies of ciliary beating were acquired at room temperature with an inverted light microscope (Axiovert 135) at 100x magnification using the cellF software (Olympus Soft Imaging Solutions GmbH, version 3.0) during 20 s. Ciliary motion was assessed by computing the median of the TV-L1 norm of the optical flow (Zach et al., 2007), using a scikit-image implementation with the default parameters (van der Walt et al., 2014).
2.13. SARS-CoV-2 RNA quantification by real-time RT-qPCR
SARS-CoV-2 RNAs were extracted from samples with the Quick-RNA™ MiniprepPlus Kit (Zymo, R1058) after TRIzol™ reagent (Invitrogen, 15596026) treatment, according to the manufacturer's instructions. The number of viral genome copies was evaluated by one-step real-time RT-qPCR of the SARS-CoV-2 M-gene with the Superscript TMIII Platinum One step Quantitative Kit (Invitrogen, 11732-020). Five μl of extracted RNAs were added to 15 μl of reaction mixture containing: reaction mix buffer (1x), 0.5 μl of MgSO4 buffer, reference dye ROX (50 nM), each primer (400 nM), Taqman probe (100 nM), and 0.4 μl SuperScript III RT/Platinum® TaqMix. The final volume was made up to 20 μl with nuclease-free water. Thermal cycling was performed in a QuantStudio 3 Real-Time PCR System thermocycler (Thermo Fisher Scientific) in MicroAmp® Optical 96-well reaction plates (Applied Biosystems, N8010560). Conditions were: reverse transcription at 50 °C for 20 min, Taq DNA polymerase activation for 2 min at 95 °C, and then 45 cycles of amplification consisting of DNA denaturation for 15 s at 95 °C, and combined annealing/extension for 1 min at 60 °C. Fluorescence data were collected at the end of each cycle. The RT-qPCR measurements were internally controlled by linear regression analyses of the Ct values from RNA standards against their respective copy numbers in the range of 103 to 106 per μl yielding R2 correlation coefficients of 0.9657 with inter-assay (n = 8) coefficients of variation ranging from 3.2 to 3.8%. The SARS-qRNAs were in vitro transcribed from a synthetic SARS-CoV-2 M-gene DNA template (Microsynth AG, Balgach Switzerland) under control of a T7 promoter by using the HiScribe™ T7 High Yield RNA Synthesis Kit (NEB, E2040). Then synthesized SARS-qRNAs were quantified with a NanoDrop spectrophotometer (Thermo Fisher Scientific). The oligonucleotides are presented in Table S1. We conducted PCR control measurements with comparable results using probes for either the M or the Pol gene (Nsp12), the latter only present in genomic but not subgenomic RNA (Kim et al., 2020), thereby ruling out the possibility that the M probe would measure preferentially subgenomic RNA fragments (Table S2).
2.14. Key resources table
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Primary antibodies | ||
| Mouse monoclonal anti-OC43_nucleoprotein | Merck | Cat #MAB9013 (clone542-7D) |
| Rabbit polyclonal anti-SARS-CoV2_nucleoprotein | Rockland | Cat #200-401-A50 LOT 45372 |
| Mouse monoclonal anti-dsRNA |
English & Scientific Consulting |
J2 https://doi.org/10.1016/0166-0934(94)90023-X |
|
Virus Strains | ||
| hCoV-229E-GFP | Kindly provided by Dr. Volker Thiel (University of Bern, Switzerland) Cervantes-Barragan et al. MBio. 2010 |
https://doi.org/10.1128/mBio.00171-10 |
| hCoV-OC43 (ATCC-VR-1558) | American Type Cell Culture | Cat #VR-1558 |
| SARS-CoV2 “wuhan” TAR clone 3.3, München-1.1/2020/929 | Kindly provided by Dr. Volker Thiel (University of Bern, Switzerland) Thi Nhu Thao et al. Nature. 2020 |
https://doi.org/10.1038/s41586-020-2294-9 |
| SARS-CoV-2 Alpha (2 012 212 272 7D 15.01.21) | Kindly provided by Dr. Isabella Eckerle (University of Geneva) | N/A |
| SARS-CoV2 Beta (SDV4-SD1-4D 08/02/21) | Kindly provided by Dr. Isabella Eckerle (University of Geneva) | N/A |
| SARS-CoV-2 Gamma (NH-RIVM10915/2021) | RIVM (through EVaG) | Not available anymore |
| SARS-CoV-2 Delta (NH-RIVM27142/2021) | RIVM (through EVaG) | N/A |
|
Chemicals, Peptides, and Recombinant Proteins | ||
|
Cell culture reagents |
/ |
/ |
| DMEM medium | Sigma-Aldrich | Cat #D6429 |
| Non-essential amino acids (NEAA) | Sigma-Aldrich | Cat #M7145 |
| Fetal Calf Serum (FBS) | Gibco | Cat #10270-106 |
| Penicillin-streptomycin | Sigma-Aldrich | Cat #P0781 |
| Trypsin-EDTA | Sigma-Aldrich | Cat #T3924 |
| PBS Buffer w/o Ca2+ and Mg2+ | Animated/Bioconcept | Cat #3-05P29-M |
| MucilAir medium |
Epithelix |
Cat #EP005 |
|
Drugs, Chemicals |
/ |
/ |
| DMSO | Sigma-Aldrich | Cat #D2650-5X |
| Remdesivir | Medchemexpress | Cat #HY-104077 LOT 46182 |
| Methylene Blue | Medchemexpress | Cat #HY-14536/ LOT 64303 |
| Mycophenolic acid | Medchemexpress | Cat #HY-B0421 LOT 15B95 |
| Posaconazole | Medchemexpress | Cat #HY-17373 LOT 12325 |
| Paraformaldehyd | Sigma-Aldrich | Cat #158127 |
| TRIzol® Reagent | Invitrogen | Cat #15596026 |
| Isopropanol |
Carlo Erba reagents |
Cat #415156 |
|
Molecular biology, RT-qPCR |
/ |
/ |
| Quick-RNATM MiniprepPlus Kit | Zymo | Cat #R1058 |
| Superscript TMIII Platinum One step Quantitative Kit | Invitrogen | Cat #11732-020 |
| SARS-CoV2 RNA Quantitative standards | This paper | N/A |
| Quantigene ViewRNA high content screening assay kit | ThermoFisher Scientific | QVP0011, QVP0201/LP1-550, QVP0201/LP4-488, custom-made probes |
|
| ||
|
Experimental Models: Cell Lines | ||
| Monkey: Vero E6 | Kindly provided by Dr. Volker Thiel (University of Bern, Switzerland) | |
| Human: Huh7 | Kindly provided by Dr. Volker Thiel (University of Bern, Switzerland) | |
| Human: Huh7-ACE2 | Laboratory-made by stable transfection with a lentivector (pLVX-ACE2-IRES-BSD) | |
| Human: Hela | American Type Cell Culture | |
| Human: Hela-ACE2 | Laboratory-made by stable transfection with a lentivector (pLVX-ACE2-IRES-BSD) | |
| Human: A549 | American Type Cell Culture | |
| Human: A549-ACE2 | Laboratory-made by stable transfection with a lentivector (pLVX-ACE2-IRES-BSD) | |
| Human: H1299 | Kindly provided by Dr. Silvio Hemmi (University of Zurich, Switzerland) | |
| human airway epithelium cells (hAEC), Nasal | Epithelix | MucilAir™ Cat #EP02 Pool of 14-donors; lot#MP009 |
| human airway epithelium cells (hAEC), Bronchial | Epithelix | MucilAir™ Cat #EP01 Single donor; lot# MD0810 |
|
| ||
|
Software and Algorithms | ||
| Graphpad Prism | GraphPad Software, Inc. La Jolla | Version 8 |
| Plaque2.0 | Greber Laboratory | https://doi.org/10.1371/journal.pone.0138760 |
| R version 4.0.2 in RStudio Version 1.3.1056 | ||
3. Replicates, statistics and randomization
Biological replicates of HAEEC experiments have been conducted with two independently prepared inserts followed by independent infection analyses. All drug inhibition experiments with HAEEC were repeated twice or three times with independent inserts at drug concentrations of 0.1, 1, 3.3, 5, 10 and 20 μM in post exposure regimen with essentially identical results. Randomization of donors for nasal HAEEC was achieved by pooling cells from 14 different donors, as described by the supplier (Epithelix SA, Geneva, Switzerland). Bronchial inserts were obtained from a single donor (Epithelix SA, Geneva, Switzerland). Further information about sample collection of biological data points and technical replicates is indicated in the corresponding figure legends.
4. Data sharing
The raw data of the screen are available under https://doi.org/10.17632/47xctms897.1.
5. Role of the funding source
The funders had no impact on the design of the study, the collection, analyses or interpretation of data, the writing of the report or the publication strategy.
6. Ethics statement
Nasal polyp epithelial cells and bronchial epithelial cells were purchased from Epithelix SARL (Geneva, Switzerland). Mucilair™ cells were reconstituted from patients undergoing surgical nasal polypectomy or bronchial biopsies, respectively. All samples from Epithelix SARL were obtained with informed consent, according to the declaration of Helsinki on biomedical research (Hong Kong amendment, 1989), and received approval from local ethics committee (commission cantonale d’éthique de la recherche CCER de Genève). Coronavirus storage and procedures have been registered and validated by the Swiss Federal Office for the Environment (Bundesamt für Umwelt, BAFU) and the Swiss Federal Office of Public Health (Bundesamt für Gesundheit, BAG); Ecogen A203032-00, registered the 01-Sep-2020 for a duration of 5-years for the University of Zurich (Department of Molecular Life Sciences).
7. Results
7.1. Compound identification by image-based full infection-cycle screening using hCoV-229E-GFP
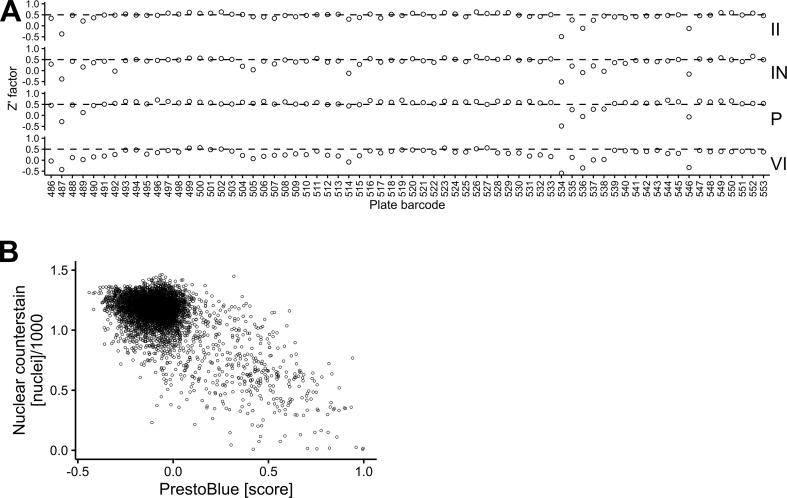
Host targeting with chemical compounds combined with full cycle image-based analyses is a powerful approach against viral infections, as recently demonstrated with the identification of the HIV protease inhibitor Nelfinavir blocking human adenovirus egress from infected cells (Georgi et al., 2020a, Georgi et al., 2020b). Here we used this approach to search for broadly effective coronavirus inhibitors. The screen with Huh7 cells infected with hCoV-229E-GFP was followed by high-throughput imaging as described (Yakimovich et al., 2015a; Georgi et al., 2020b). The infection assay scoring viral plaque formation was highly robust, as indicated by Z’ factors (Zhang et al., 1999) at or above 0.5, indicating excellent assay quality and consistent analysis pipelines (Fig. S1A).
We screened the EPFL BSF-repurposing compound collection, curated and corrected based on information made available by the Broad Institute (Corsello et al., 2017). The overall procedure is depicted in Fig. 1 . The library comprises 5440 mostly repurposable compounds, 39% are clinical compounds, 26% are in phase I – III trials, 34% in preclinical development and 1% have been withdrawn. The collection covers substantial chemical space with 5032 clusters, 4641 of which contain a single compound, as determined by sphere exclusion clustering analysis with a Tanimoto distance of 0.3 between centroids. One thousand two hundred eighty compounds were contained within the Prestwick Chemical Library (PCL) and purchased as such. The remaining 4160 were purchased from 34 different suppliers, including MedChem Express (59.5%), MolPort (35.5%) and others. All compounds were interrogated for chemical integrity and purity by LC-MS, and assessed for toxicity by PrestoBlue staining (Fig. S1B), a resazurin based assay for measuring overall energy levels of cell populations (Liu, 1981). These results are in good agreement with the number of segmented nuclei based on Hoechst 33342 staining of the screening plates (see Table S3).
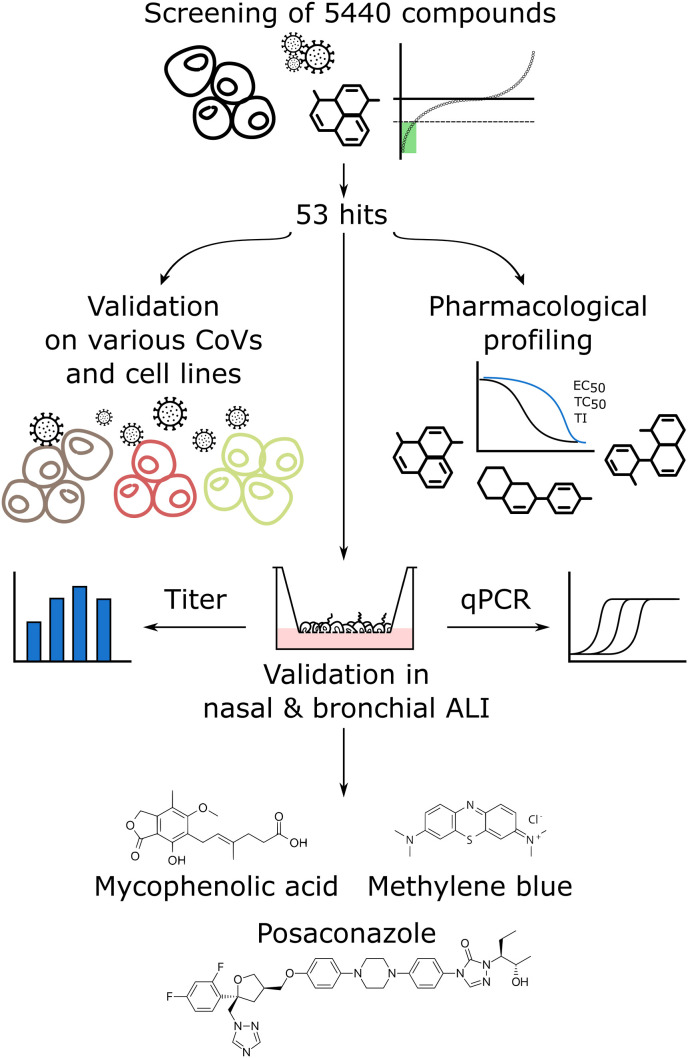
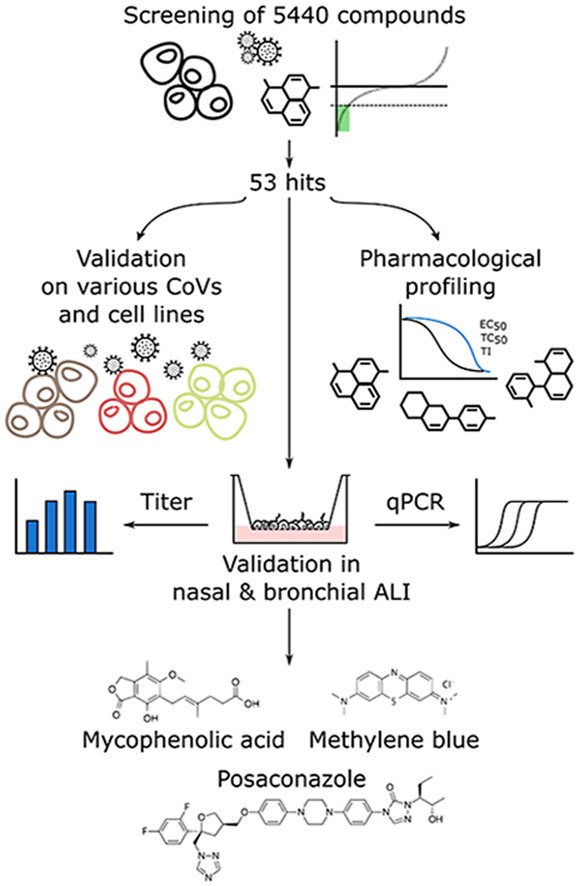
Fig. 1.
Workflow of the screening procedure Initially, we assessed a library of 5440 compounds for efficacy against hCoV-229E-GFP. This yielded 53 hits, which were tested for efficacy against hCoV-OC43 and SARS-CoV-2 in different cell lines, including Vero-E6, Huh7-ACE2, A549-ACE2 and HeLa-ACE2. In parallel, we determined the EC50, TC50 and the ratio TC50/EC50, i.e., the quasi therapeutic index (TI). A selection of hits in advanced clinical state (approved and systemically administrable) was tested for SARS-CoV-2 inhibition in differentiated nasal and bronchial airway epithelia grown at air-liquid interface. Methylene blue (MB) and mycophenolic acid (MPA) and posaconazole (POS) were the most potent inhibitors of SARS-CoV-2 infectious particle formation. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To conduct the infection screen, the library was arrayed in microscopy grade 384-well assay plates, followed by seeding of Huh7 cells, infection with hCoV-229E-GFP, and high-throughput imaging (Fig. 2 A). This allowed us to extract image analysis parameters, including total cell count, infected cell count, total GFP intensity and number of infection foci (plaques) (see also Fig. S1A). Two subsequent validation filters were applied with hCoV-OC43 infection of Huh7 cells, and SARS-CoV-2 infection of Vero E6, Huh7 expressing the angiotensin-converting enzyme 2 (ACE2), HeLa-ACE2 and A549-ACE2 cell lines. A set of selected drugs was then tested in nasal and bronchial HAEEC. These procedures identified MPA, MB and POS to be effective against 229E-GFP, OC-43 and SARS-CoV-2 infections, inhibiting virus accumulation and cell-cell transmission. These parameters were normalized per-plate to the mean value of all negative control wells. A compound was classified as hit inhibiting infection if a Z score of −3 for all of these normalized infection parameters was obtained, that is, if its value was less than the mean of the negative controls minus three standard deviations (Fig. 2B). Compounds were flagged toxic if the mean cell count as determined by nuclear counterstaining was below the mean of all negative control wells minus two standard deviations. Analysis and post-processing yielded a total of 53 compounds fulfilling these criteria, and was followed by validation for effective concentrations (EC50) and toxicity (TC50). The hit compounds were classified either as antiseptics, antifungals, antibacterials, anti-cancer, metabolism or anti-inflammatory agents (Fig. 2C). We selected MB, MPA and POS for further validation. MB and POS effectively reduced the number of infection foci. MPA reduced the total GFP intensity, while Remdesivir showed complete infection inhibition of hCoV-229E-GFP at 0.33 μM (Fig. 2D).
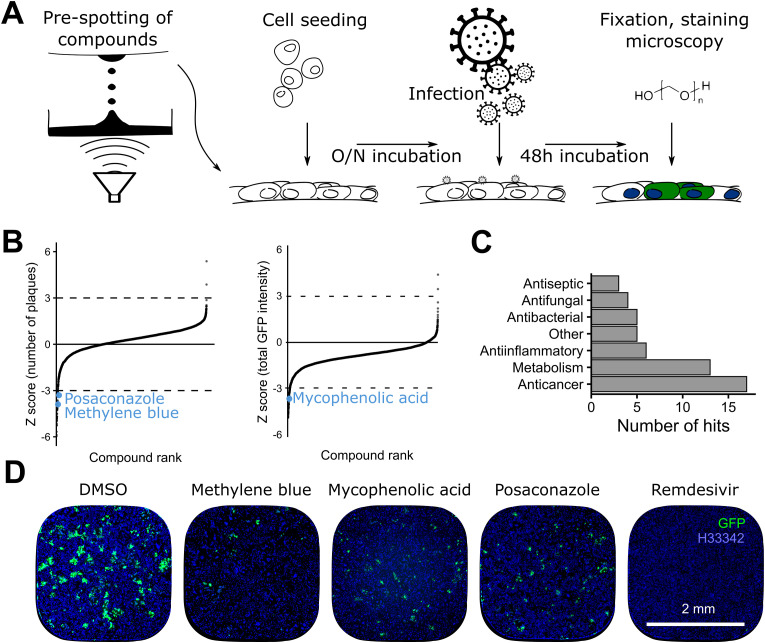
Fig. 2.
Overview of the arrayed 5440 compounds library screen A) Schematic representation of the screening procedure, at 1.67 μM per compound. B) Ranking of compounds by number of plaques and total GFP intensity. The cut-off value was set at a Z score of −3 (see Methods). MB, MPA and POS are highlighted in blue. Data points represent means of 4 independent biological replicates. C) Hit classification. Most of the hits classified as anticancer agents or modulators of metabolism. D) Example images of cells treated with the indicated compounds as described in A. In the negative control (DMSO), clusters of GFP positive cells represent infection foci (green). Nuclei are represented in blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
7.2. Dose-response validation and hit extension to hCoV-OC43 and SARS-CoV-2
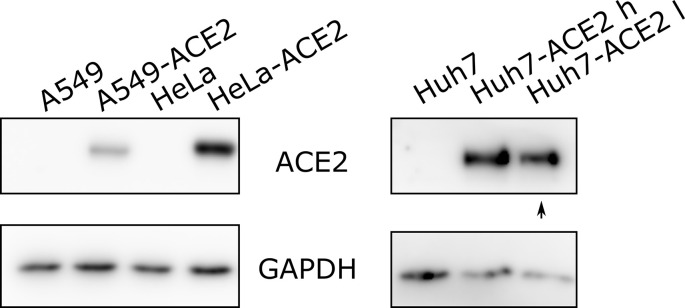
To assess the therapeutic potential of the 53 hit compounds EC50 and TC50 values were determined for hCoV-229E-GFP in Huh7 cells by conducting dose-response experiments with concentrations from 1.525 nM to 50 μM. The results are summarized in Table 1 and Table S3. Thirty nine compounds had an EC50 < 2 μM and a therapeutic index (TI) of > 2, as determined by fluorescent focus formation (FFF). The slight discrepancy to the results obtained in the initial screen at 1.67 μM was due to additional parameters used for hit scoring in the original screen, including number of GFP positive cells, total GFP intensity and infection index, in addition to the number of fluorescent foci. Notably, the dose-response analyses yielded LY2090314 as the most potent compound, where the lowest concentration tested (1.525 nM) resulted in 41% reduction of 229E-GFP infection foci. We conservatively estimated of an EC50 of about 2 nM, and a TI of 15830 based on an observed TC50 of 31.7 μM. MB had an EC50 of 1.43 μM, a TC50 of 8.71 μM (TI of 6.09), and the EC50 of MPA was estimated to 1.85 μM and TC50 45.8 μM, resulting in a TI of 24.8 (Fig. 3 A). POS had an EC50 of about 2.5 μM and showed essentially no cell loss up to 50 μM. In addition, 39 compounds used at 1.75 μM proved to be effective and non-toxic against hCoV-OC43 infection of Huh7 cells (Table 1). We next generated Huh7, human lung epithelial A549 and human cervical carcinoma HeLa cells expressing ACE2 by lentivirus transduction (Fig. S2). These ACE2 transduced cells as well as African green monkey kidney epithelial Vero E6 cells were susceptible to SARS-CoV-2 (Fig. 3B and Table 1). Eleven compounds were effective in two or more cell lines against SARS-CoV-2, and two of them in all cell lines, namely POS and Ro 48–8071. The preclinical compound Ro 48–8071 had the best hit profile regarding effectiveness in all four cell lines against SARS-CoV-2, and hCoV-OC43. Ro 48–8071 affects oxidosqualene cyclase in cholesterol biosynthesis (Maione et al., 2015a). In addition, the launched anti-fungal compound POS showed a similar profile, but did not meet the dose-response criterion with an EC50 of 4.20 μM against 229E-GFP infection (Fig. 3A). We did, however, not observe any toxicity effects for POS up to 50 μM. MPA, Thonzonium bromide and MLN4924 were effective against SARS-CoV-2 in three cell lines, and also inhibited hCoV-OC43. Notably, the number of compounds effective against both hCoV-229E-GFP and hCoV-OC43 was much higher than those effective against SARS-CoV-2. This effect did not seem to be cell-line dependent, as for example Huh7-ACE2 used for SARS-CoV-2 are derived from Huh7 cells used with hCoV-229E-GFP and hCoV-229E. It is unlikely though that temperature differences in infection (37 °C versus 33.5 °C) affected drug effectiveness against SARS-CoV-2. In summary, the cell line screens with the endemic coronaviruses 229E, OC43 and the pandemic SARS-CoV-2 uncovered six clinically launched compounds, MB, MPA, POS and R788-Fostamatinib, and the topical compounds Thonzonium bromide, a monocationic surfactant with bacteriocidal effects (Chan et al., 2012), and Cetylpyridinium used in mouth wash formulations (Pitten and Kramer, 2001), which inhibited all three viruses in the micromolar concentration range, similar to Remdesivir (Table 1, Table 2 , Table S3). In the following, we focussed on the systemic compounds.
Table 1.
Validation of the 53 top hits from the initial full cycle infection screen.
| Compound | 229E-GFP D-R Huh7 |
OC43 Huh7 |
SARS-CoV-2 Huh7-ACE2 | SARS-CoV-2 VeroE6 | SARS-CoV-2 HeLa-ACE2 | SARS-CoV-2 A549-ACE2 |
|---|---|---|---|---|---|---|
| A0001 | ✔ | |||||
| Fostamatinib | ✔ | ✔ | ✔ | ✔ | ||
| Methylene Blue | ✔ | ✔ | ✔ | |||
| MLN4924 | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Mycophenolic acid | ✔ | ✔ | ✔ | ✔ | ✔ | |
| OTSSP167 | ✔ | |||||
| Posaconazole | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Ro 48–8071 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Abafungin | ✔ | ✔ | ✔ | |||
| Acivicin | ✔ | ✔ | ||||
| Adarotene | ✔ | |||||
| Amuvatinib | ✔ | ✔ | ||||
| Anisomycin | ✔ | ✔ | ||||
| APY0201 | ✔ | ✔ | ||||
| AT9283 | ✔ | ✔ | ||||
| AZD2858 | ✔ | ✔ | ✔ | |||
| AZD-5438 | ✔ | ✔ | ||||
| Betulinic acid | ✔ | ✔ | ||||
| Cerdulatinib | ✔ | ✔ | ||||
| Cetalkonium chloride | ✔ | ✔ | ✔ | |||
| Cetylpyridinium | ✔ | ✔ | ✔ | |||
| CHIR-124 | ✔ | ✔ | ✔ | |||
| CHIR-98014 | ✔ | ✔ | ||||
| CUDC-907 | ||||||
| Cyclopiazonic-acid | ✔ | ✔ | ||||
| Eprinomectin | ✔ | |||||
| FH535 | ✔ | |||||
| Geldanamycin | ✔ | |||||
| GPP-78 | ✔ | ✔ | ✔ | ✔ | ||
| GSK 3 Inhibitor IX | ✔ | ✔ | ✔ | |||
| GZD824 | ✔ | ✔ | ✔ | |||
| Isavuconazole | ✔ | |||||
| JTE-013 | ✔ | ✔ | ||||
| LE-135 | ✔ | ✔ | ||||
| Lomefloxacin HCl | ||||||
| LY2090314 | ✔ | ✔ | ||||
| Mebendazole | ✔ | ✔ | ||||
| Oxaprozin | ||||||
| Pelitinib | ✔ | ✔ | ✔ | |||
| PF-04457845 | ✔ | ✔ | ||||
| PF-3845 | ✔ | ✔ | ||||
| PFK-015 | ✔ | |||||
| RAF265 | ✔ | ✔ | ✔ | |||
| Ravuconazole | ✔ | ✔ | ✔ | ✔ | ||
| SB225002 | ✔ | |||||
| SB-505124 | ✔ | ✔ | ||||
| Tanaproget | ✔ | ✔ | ||||
| TCS-21311 | ✔ | ✔ | ✔ | |||
| Thonzonium bromide | ✔ | ✔ | ✔ | ✔ | ✔ | |
| TRO 19622 | ||||||
| UK-5099 | ✔ | ✔ | ||||
| Verteporfin | ✔ | ✔ | ||||
| VU29 | ||||||
| Total | 39 | 39 | 7 | 9 | 10 | 16 |
All compounds identified as hits in the original screen are listed. The compounds highlighted in bold at the top of the table are clinically launched. Columns represent different validation experiments with the corresponding viruses and cell lines. Ticks mark compounds qualifying as hits in the respective experiment. There is a good agreement between the original screen, 229E-GFP dose-response (D-R) and hCoV-OC43 in Huh7 cells with fewer compounds also being effective against SARS-CoV-2 (Wuhan).
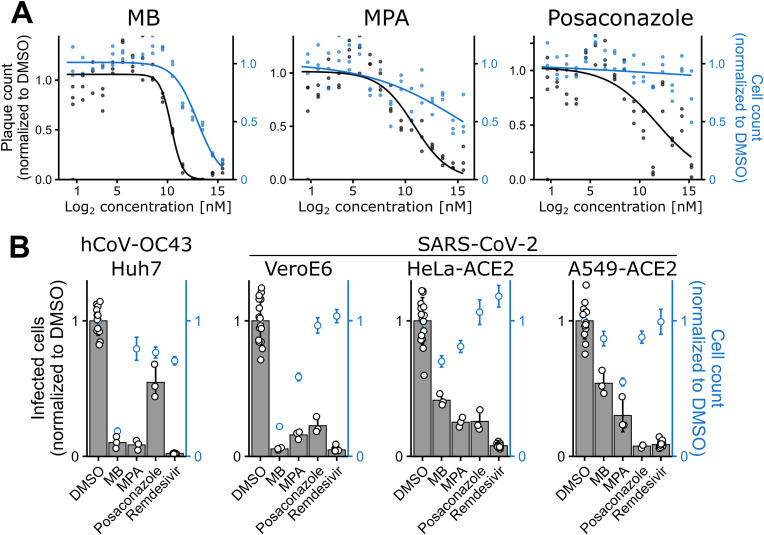
Fig. 3.
MB and MPA inhibit hCoV-229E-GFP, OC43 and SARS-CoV-2 infection A) Dose-response curves of MB, MPA and POS treated Huh7 cells infected with hCoV-229E-GFP at 50 FFU. After 48h of infection, cells were fixed and imaged. Curves were fitted through the data points using a 3-parameter log-logistic function with a lower limit of 0. Raw data from single wells are shown. Three technical replicates are shown. Note the concentration-dependent inhibition of viral plaque formation by MB, MPA and POS. B) Broad hCoV inhibition profiles of MB, MPA and POS at 1.67 μM. Indicated cell lines were infected with either hCoV-OC43 (68h) or SARS-CoV-2 (24 or 48h) in presence of MB, MPA or POS, and stained for newly synthesized viral nucleoprotein. The infection index was between 10 and 30%, except for Vero E6, where it was around 5%. Data points representing infected cell counts are from raw data of individual wells. Bar heights represent the mean of four technical replicates and whiskers represent the standard deviation. Blue data points representing cell counts show means ± standard deviation from four technical replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Efficacy of clinical and preclinical compounds against SARS-CoV-2 in cell cultures.
| Compound | Original indication | Clinical status | EC50 [μM] | TC50 [μM] | Susceptible cell lines (n.) |
|---|---|---|---|---|---|
| A0001 | Friedreich ataxia | Phase II | 0.497 | 8.4 | 0 |
| Methylene Blue | Methemoglobinemia | Launched | 1.426 | 8.709 | 2 |
| MLN4924 | Cancer | Phase II | 0.032 | 40.39 | 3 |
| Mycophenolic acid | Immunosuppressant | Launched | 1.85 | 45.823 | 3 |
| OTSSP167 | Cancer | Experimental | 0.041 | 0.766 | 0 |
| Posaconazole | Antifungal | Launched | 4.2 | >50 | 3 |
| R788-Fostamatinib | Immune thrombo-cytopenic purpura | Launched | 1.331 | 14.709 | 2 |
| Ro 48-8071 | Cancer | Experimental | 1.859 | 8.642 | 4 |
Repurposing compounds at different stages of clinical development with good separation of hCoV-229E-GFP inhibition and toxicity (columns 4 and 5) were selected, and tested for anti-viral efficacy against SARS-CoV-2 in nasal and bronchial HAEEC. Effects against SARS-CoV-2 in four different cell lines (Vero E6, Huh7-ACE2, A549-ACE2, and Hela-ACE2) are indicated in the last column.
7.3. MB and Remdesivir synergize against CoV infection and block the accumulation of viral (+)RNA in cells
We first explored if the top systemic compounds MB, MPA and POS had synergistic effects against hCoV-229E-GFP in the full cycle fluorescent focus formation assay. Since MPA and POS did not show synergistic effects with Remdesivir on hCoV-229E-GFP infection of Huh7 cells, we focussed on MB. Striking synergy effects occurred against hCoV-229E-GFP in human hepatoma Huh7 cells at 500 nM MB in combination with 50 nM Remdesivir, yielding a BLISS synergy score of 50 (Fig. 4 A and B). These effects were similar in human lung carcinoma cells (H1299) with MB at 5 μM and Remdesivir at 250 nM, yielding a BLISS synergy score of 31.3 (Fig. 4B). The results indicate that suitable concentrations of Remdesivir and MB enhance anti-coronavirus effects suggesting that Remdesivir and MB affect infection by independent mechanisms.
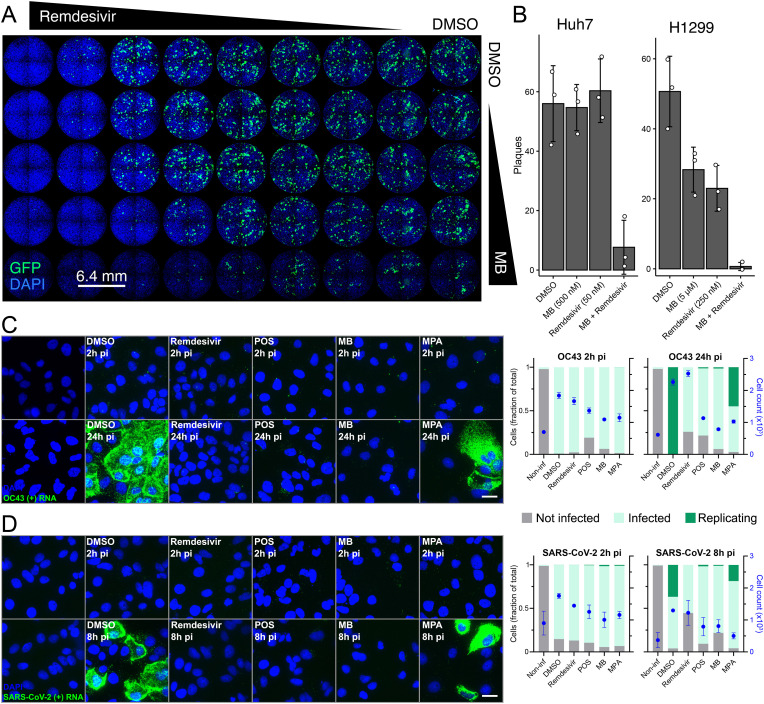
Fig. 4.
MB and remdesivir synergize against hCoV-229E infection, and independently reduce the intracellular load of replicated OC43 and SARS-CoV-2 plus strand RNA, alongside with POS and MPA A) Overview of a 96-well plate infected with hCoV-229E-GFP and treated with combinations of remdesivir (1 nM–250 nM) and MB (100 nM–1 μM) in 2-, 4-, 5-, 10-fold serial dilutions. B) Quantification of plaque counts in presence of the indicated concentrations of MB and remdesivir in Huh7 (left panel) and H1299 (right panel). C) Huh7 cells were infected with OC43 for 2 or 24h in presence of remdesivir (1 μM), POS (10 μM), MB (2 μM) or MPA (2 μM). Cells were fixed and stained against OC43 ORF1a (+)RNA using RNA FISH with branched DNA signal amplification. Nuclei were stained with DAPI. OC43 (+)RNA dots were segmented and quantified per cell. Infected cells contained at least one viral RNA per cell; replicating cells were defined as containing more than seventy-five viral RNAs per cell, i.e., the maximal detection limit for counting individual mRNA puncta in infected Huh7 cells (see similar numbers for A549 cells in, Suomalainen et al., 2020). Images are maximum projections. Scale bar, 10 μm. Bar graphs in panels C) and D) denote not infected cells (grey bars), infected cells containing individual (+)RNA puncta (light green) and cells containing replicated viral RNA (dark green). D) Huh7-ACE2 cells were infected with SARS-CoV-2 for 2 or 8h in presence of remdesivir (1 μM), POS (10 μM), MB (2 μM) or MPA (2 μM). Cells were fixed, stained against SARS-CoV-2 ORF1a (+)RNA and processed as described in (C). Images are maximum projections. Scale bar, 10 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To address the anti-viral mechanisms of MB, MPA and POS, we developed a procedure to score intracellular RNA of OC-43 and SARS-CoV-2 at MOI 1 in Huh7 and Huh7-ACE2 cells by fluorescence in situ hybridization using probes against the (+)RNA strand in ORF1a. The procedure was adapted from previous work with adenovirus and rhinovirus (Yakimovich et al., 2015b; Suomalainen et al., 2020; Suomalainen and Greber, 2021). The infection phenotype was scored in MB, MPA, POS and Remdesivir-treated cells by automated measurement of fluorescent puncta representing single viral RNA molecules. At 2 h pi, 80% or more of the cells contained at least one discernible fluorescent punctum of OC43 (Fig. 4C) or SARS-CoV-2 (Fig. 4D), akin to DMSO control treated cells. This suggested that none of compounds had a strong effect on virus uptake into cells. In contrast, strong differences to the control DMSO cells occurred upon MB, MPA, POS or Remdesivir treatment of SARS-CoV-2 and OC43 infections at 8 h and 24 h pi, respectively. Since OC43 infection kinetics is slower than that of SARS-CoV-2, different time points were used for the analyses. While control cells had abundant intracellular viral (+)RNA, MB, POS and Remdesivir-treated cells showed essentially no indications of increased viral RNA. This was contrasted by MPA-treated cells showing an intermediate phenotype, with some cells containing replicated viral RNA. The levels of viral RNA-free cells upon MB, POS or Remdesivir treatment remained constant or increased slightly compared to 2 h pi (Fig. 4C and D, grey bars), suggesting that these drugs lead to elimination of incoming viral RNA from cells over time. We conclude that MB, MPA and POS block viral (+)RNA enrichment in infected cells, and MPA has somewhat less pronounced effects.
7.4. MB, MPA and POS inhibit SARS-CoV-2 infection of HAEEC
Based on the broad antiviral effects of MB, MPA and POS in cell cultures, we tested these compounds together with another systemic launched compound hit R788-Fostamatinib, and four preclinical compounds in SARS-CoV-2 infections of well differentiated human nasal or bronchial airway cells grown at air-liquid interface (Epithelix, MucilAir™). Cells were either pre-treated or post-treated one day after apical inoculation of SARS-CoV-2 using low micromolar concentrations of compounds. The preclinical compounds comprised the broad kinase inhibitor OTSSP167 (Zhang et al., 2018), the vitamin E metabolite A0001 (a-tocopheryl-quinone) (Hawi et al., 2012), the NAE (Nedd8 activating enzyme) inhibitor MLN4924 (Pevonedistat) (Soucy et al., 2009a) and the oxidosqualene cyclase inhibitor Ro48-8071 involved in cholesterol biosynthesis (Maione et al., 2015b). However, none of the four preclinical compounds tested was considered further. OTSSP167 and A0001 lacked anti-viral activity, while MLN4924 and Ro48-8071 were toxic in HAEEC (Table S4). Remarkably, R788-Fostamatinib exhibited no consistent anti-viral effects on SARS-CoV-2 titer release in the apical milieu at 3.3 or 10 μM but gave rise to changes in HAEEC morphology, and therefore was not further considered.
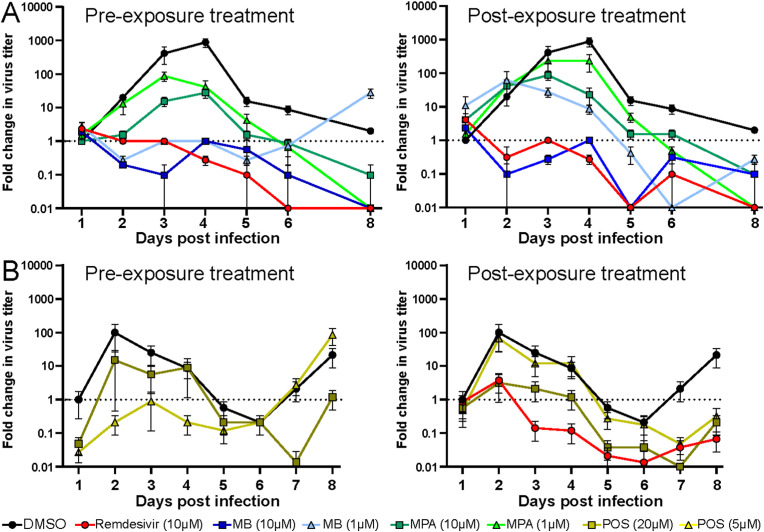
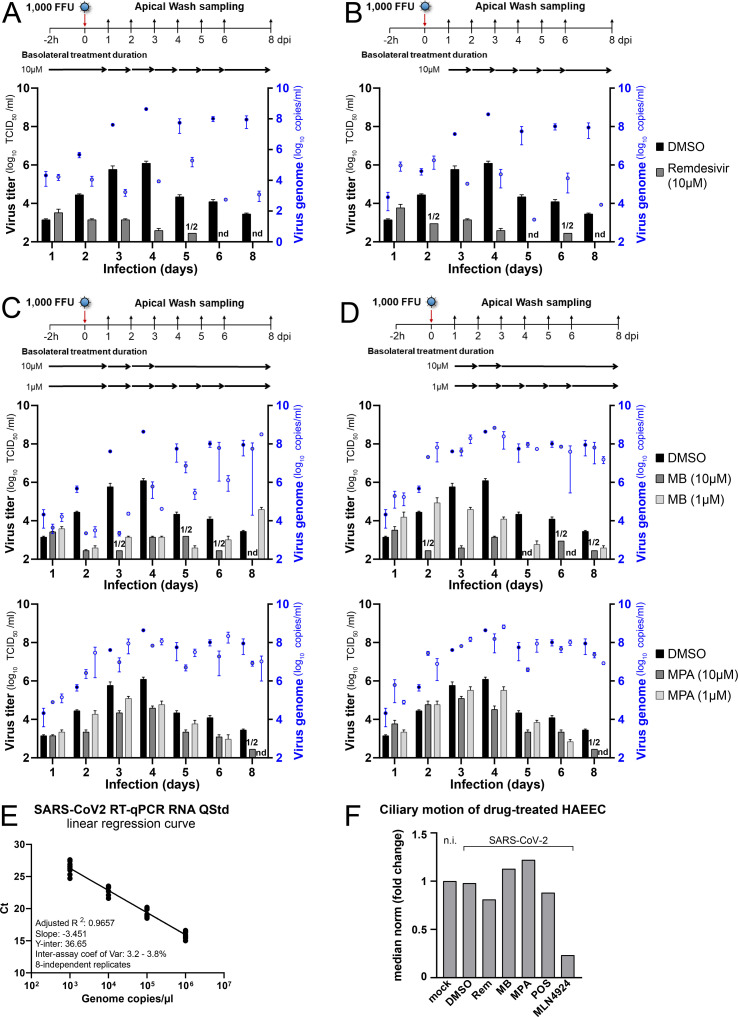
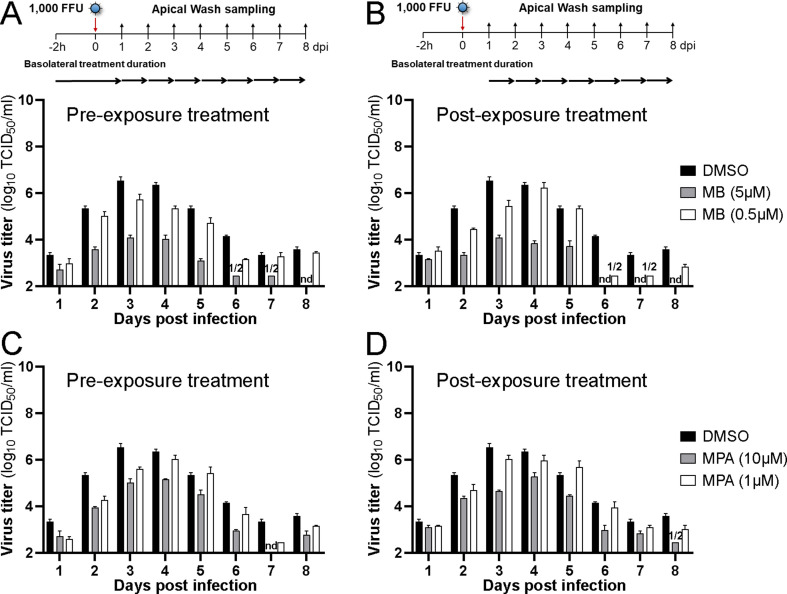
In contrast to Fostamatinib, MB, MPA, and POS-treated HAEEC remained morphologically unchanged in both pre- and post-exposure settings, akin to Remdesivir-treated cells, the latter as previously reported (Pizzorno et al., 2020). MB, MPA and POS treated inserts displayed robust ciliary beating akin to uninfected cells, whereas exposure to a toxic concentration of MLN4924 strongly impaired ciliary movement (Fig. S3F, MB at 5 μM and MPA 10 μM, Movies S1-5 at 6d pi, POS, 50 μM, Movies S6 and S7 at 5d pi). The pre-exposure treatment with MB at 1 or 10 μM blocked the release of infectious virus by about 3 logs at 3–4 d pi and virus levels were undetectable 8 d pi, akin to Remdesivir, while DMSO-treated cells released large amounts of virus titers peaking at 6.1 log10 TCID50/ml (Fig. 5 A, Figs. S3A–D). Although the pretreatment with MB reduced the viral genome load in the apical milieu by about 2 logs at 3–4 d pi, as indicated by RT-qPCR measurements, the genome load increased towards 8 d pi, and was largely unaffected in the post-exposure regimen, unlike Remdesivir-treated cells, which consistently showed 2–3 logs lower genome copies than the DMSO controls (Figs. S3A–D). MPA treatment had significant antiviral effects, at both 1 and 10 μM, albeit less pronounced than MB, and with delayed efficacy (Fig. 5A). Strikingly, the genome copy numbers in the apical supernatant were only mildly reduced by MPA in both pre- and post exposure regimen. Nevertheless, the antiviral effects of both MB and MPA were reproduced in bronchial HAEEC, thus confirming the results in nasal HAEEC (Figs. S4A–D). POS treatment (20 μM) also showed significant antiviral effects in pre- and post-exposure settings (Fig. 5B, Fig. S5). Intriguingly, the reduction of virus titer was more pronounced if POS was added post exposure.
Fig. 5.
MB, MPA and POS inhibit SARS-CoV-2 infection of nasal HAEEC. Antiviral effects of drug treatments represented as means ± SEM of two independent replicates. Baseline levels represent the apical means ± SEM of virus titer from the DMSO control sample at 1d pi. The same DMSO control was used for the pre- and post-exposure treatments. A) Nasal HAEEC grown at ALI were inoculated apically with 1000 FFU of SARS-CoV2 Wuhan (day 0), and treated with drugs in the basolateral medium in a pre-infection regimen, starting at 2h prior to virus inoculation (left) or in a post-infection regimen starting at 1 d pi (right). MB and MPA were administrated daily until day 3 at 10 μM or until day 6 at 1 μM. B) POS (5 and 20 μM) was administrated daily until day 8. Remdesivir (10 μM) and DMSO served respectively as positive and negative drug treatment control and were administrated daily until day 8. SARS-CoV-2 released to the apical side was collected daily by apical washing and quantified by virus TCID50 titration.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.crviro.2022.100019.
The following are the supplementary data related to this article:
Multimedia component 5Supplemental Movie 1: Ciliary beating in mock-treated, uninfected bronchial HAEEC. Bronchial HAEEC grown at ALI were mock-infected (day 0), and left untreated and uninfected. Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 6d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Multimedia component 6Supplemental Movie 2: Ciliary beating in DMSO-treated bronchial HAEEC. Bronchial HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 10 μM DMSO in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 6d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Multimedia component 7Supplemental Movie 3: Ciliary beating in Remdesivir-treated bronchial HAEEC. Bronchial HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 10 μM Remdesivir in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 6d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Multimedia component 8Supplemental Movie 4: Ciliary beating in MB-treated bronchial HAEEC. Bronchial HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 5 μM MB in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 6d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Multimedia component 9Supplemental Movie 5: Ciliary beating in MPA-treated bronchial HAEEC. Bronchial HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 10 μM MPA in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 6 d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Supplemental Movie 6: Ciliary beating in POS-treated nasal HAEEC. Nasal HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 50 μM POS in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 5d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Supplemental Movie 7: Ciliary beating in MLN4924-treated nasal HAEEC. Nasal HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 1 μM MLN4924 in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 5d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
7.5. MB, MPA and POS inhibit SARS-CoV-2 variant infections of HAEEC
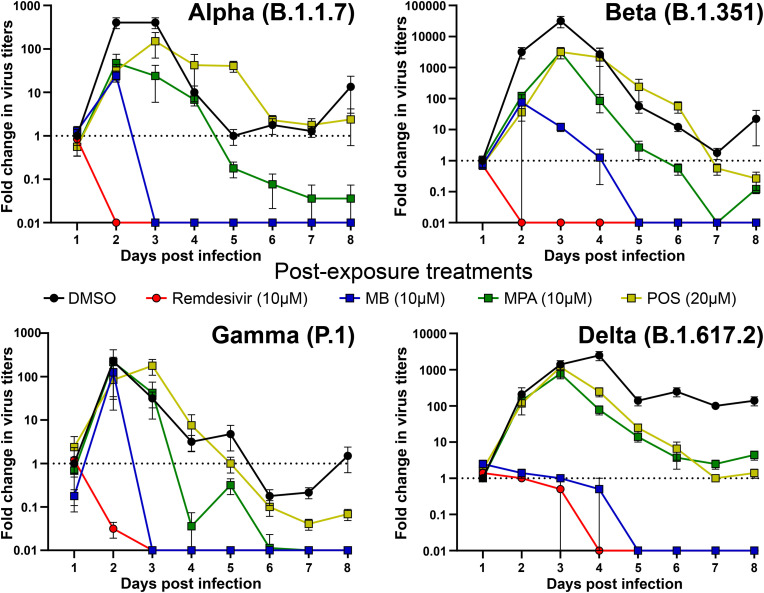
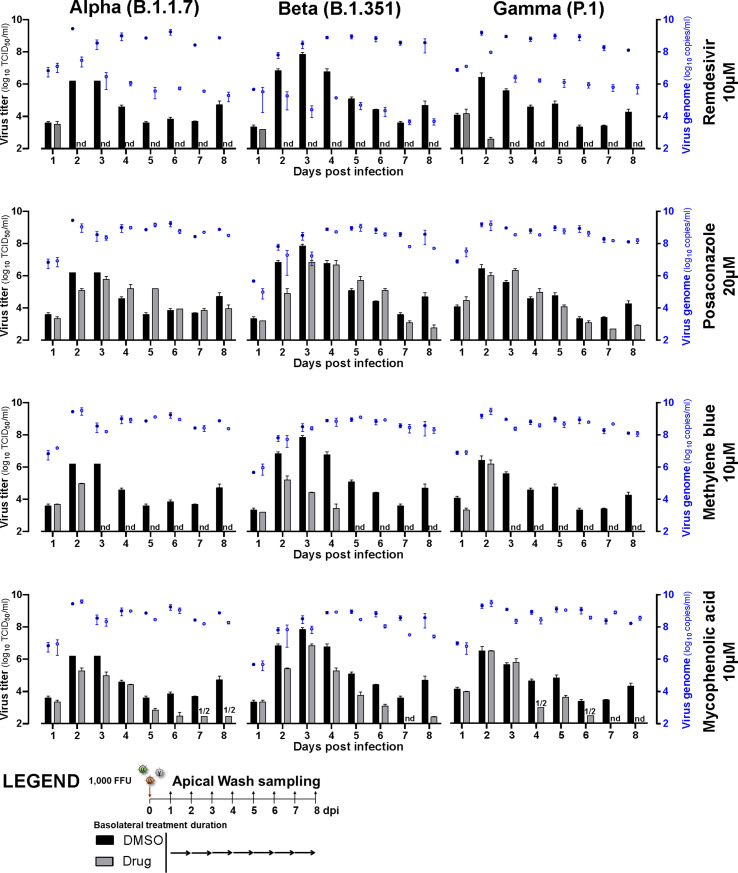
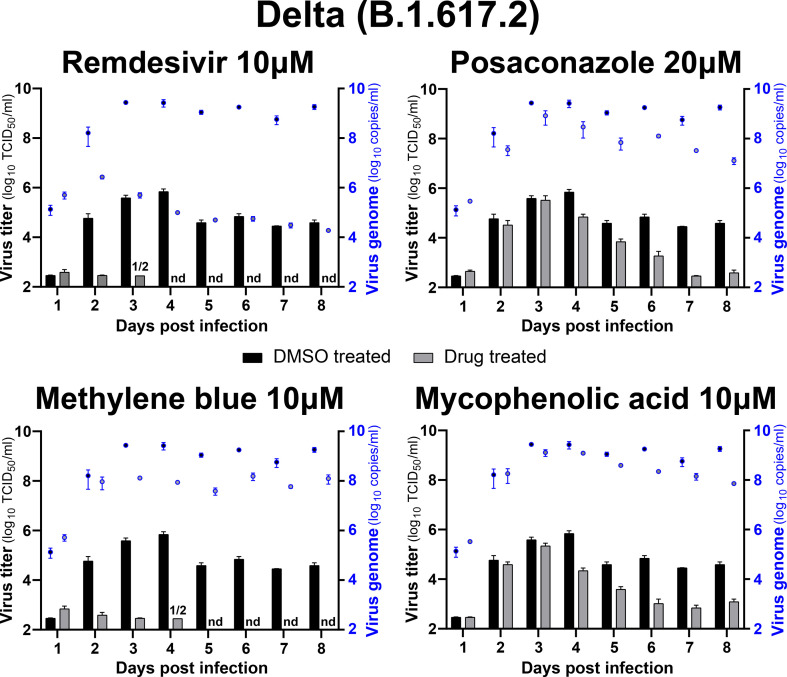
We next tested the effects of MB, MPA and POS on SARS-CoV-2 variants of concern. These variants have higher human to human transmissions and exhibit reduced susceptibility against antibodies from mRNA- and AdV vector-immunized vaccinees. While POS showed detectable albeit little efficacy against the alpha (B1.1.7), beta (B1.351), gamma (P.1) and delta (B.1.617.2) variants of SARS-CoV-2 in post exposure settings with nasal HAEEC, MB and MPA were strongly effective, similarly as against SARS-CoV-2 ‘Wuhan’ (Fig. 6 , Fig. S6, Fig. S7). Apical release of viral genomes was not reduced in post exposure treatments with either MB, MPA or POS, in contrast to Remdesivir, consistent with the findings in SARS-CoV-2 ‘Wuhan’ infections (Figs. S3B and D, Suppl Tab 4, Figs. S6 and S7). All the virus TCID50 titers and genome copy numbers are listed in Table S4. Together, our results demonstrate broad efficacy of MB and MPA and somewhat lesser effects by POS against seven human coronaviruses, the SARS-CoV-2 ‘Wuhan’ strain, the alpha, beta, gamma, and delta variants as well as the endemic CoV viruses 229E and OC43, by a mechanism distinct from Remdesivir.
Fig. 6.
MPA, MB, and POS inhibit SARS-CoV-2 alpha, beta, gamma and delta variant infections of nasal HAEEC Antiviral effects of drug treatment represented as means ± SEM of two independent replicates. Baseline levels represent the apical means ± SEM of virus titer from the DMSO control sample at 1d pi. Note that the official WHO nomenclature of the SARS-CoV2 variants (alpha, beta, gamma, delta) refer to the commonly used PANGO lineages (B.1.1.7, B.1.351, P.1, and B.1.617.2). Nasal HAEEC grown at ALI were inoculated apically with 1000 FFU of SARS-CoV2 alpha, beta, gamma or delta variants (day 0) and subjected to drug treatment in the basolateral medium, in a post-infection regimen starting at 1 d pi. MB (10 μM), MPA (10 μM), and POS (20 μM) were administered daily until day 8. Remdesivir (10 μM) and DMSO served as drug treatment controls. SARS-CoV-2 released to the apical side was collected daily by apical washing and quantified by virus TCID50 titration.
8. Discussion
Individuals suffering from COVID-19 are in need of acute medical treatment with broadly available safe and effective antivirals. In addition, the zoonotic and anthroponotic transmission of coronaviruses from animals to humans and from humans to animals (Corman et al., 2018; Oreshkova et al., 2020) may demand the treatment of livestock and pets with compounds against coronaviruses to shrink viral reservoirs. Hence, broad acting anti-virals at affordable cost will be required, a demand, which may be fullfilled by repurposing small chemical compounds targeting the host. Host targeting is likely less prone to raise viral resistance, unlike direct virus targeting, as in case of Remdesivir or the mutator nucleoside MK-4482/EIDD-2821 (Cox et al., 2021; Agostini et al., 2018; Lo et al., 2020; Sheahan et al., 2020). Notably, Remdesivir leads to stalling of the coronavirus RNA-dependent RNA polymerase and has a conserved mode of inhibition across different viruses, where a single point mutation in the Ebolavirus polymerase mediates drug resistance (Lo et al., 2020; Yin et al., 2020).
De novo development of antivirals is a lengthy and costly process and insufficient to resolve the current crisis in short terms (Ward et al., 2015). Our large scale drug repurposing screen identified broadly acting clinically approved small molecular weight compounds. We took advantage of the curated and mass spectrometry-validated BSF library composed of 5440 distinct chemical compounds, most of which have been approved for human use in nonviral indications or in clinical development. We harnessed the power of fluorescence imaging assessing the full virus replication cycle from entry to egress and spread to neighboring cells. This procedure previously identified Nelfinavir as an adenovirus egress inhibitor, notably in contrast to a previous publication earmarking it as ineffective against adenovirus in PCR-based virus replication assays (Georgi et al., 2020b; Gantt et al., 2011). To the best of our knowledge, none of the drug screening projects carried out by others has used our collection of compounds with a full cycle image-based assay.
The coronavirus screen developed here scored fluorescent focus formation of hCoV-229E-GFP. We validated the hits by toxicity assessments, counter screened with a second endemic coronavirus hCoV-OC43, and SARS-CoV-2 infection. It is unlikely that our full cycle screen discovers coronavirus entry inhibitors, as the entry pathways of endemic hCoVs are distinct from SARS-CoV-2 (Corman et al., 2018). For example, the hCoV-229E S-glycoprotein binds to aminopeptidase N, hCoV-NL63 lacking a furin cleavage site binds to the ACE2 receptor, HKU1 and OC43 binds to O-acetylated sialic acid (Yeager et al., 1992; Wu et al., 2009; Hulswit et al., 2019). SARS-CoV and SARS-CoV-2 bind to ACE2 (Yan et al., 2020). Upon furin cleavage of the S-glycoprotein, the latter contacts neuropilin for efficient entry (Cantuti-Castelvetri et al., 2020; Daly et al., 2020). Hence, the compounds that we identified likely target host functions in virus progeny formation, egress from infected cells or transmission to neighboring cells. We confirmed this notion for MB, MPA and POS, which did not block the uptake of viral RNA but reduced the levels of replicated viral genomes in the infected cells.
8.1. MB is a broad antiviral against coronavirus infections
Previous studies had suggested that MB inhibits SARS-CoV2 entry in Vero E6 cells (Gendrot et al., 2020), or SARS-CoV-2 S-protein pseudotyped viruses in HEK-ACE2 cells (Bojadzic et al., 2020). This contrasts our RNA FISH staining of the incoming viral RNA, which showed no effect of MB, MPA, POS or Remdesivir on the amount of intracellular virus genome for SARS-CoV-2 and OC43 at 2h pi. In addition, MB, MPA and Remdesivir did not inhibit the virus or genome equivalent release from HAEEC in pre-exposure treatments at 1dpi, although POS had an inhibitory effect. This strongly argues for post-entry mode-of-action of MB, MPA and Remdesivir, the latter being a defined RdRP inhibitor.
MB has long been used in humans against malaria and methemoglobinema (Walter-Sack et al., 2009; Schirmer et al., 2003; Patnaik et al., 2014). Many trials have employed it as a placebo agent with an extremely high safety profile. Yet, MB is not inert, it is an oxido-reductive thiazine dye cycling between the oxidated form (MB) and the reduced form leuco-MB (L-MB) (Buchholz et al., 2008). The reduction occurs through cellular flavo oxido-reductases, such as glutathione reductase, thioredoxin reductase or the mitochondrial membrane associated dihydrolipoamide dehydrogenase at the expense of nicotinamide adenine dinucleotide phosphate (NADPH). L-MB readily auto-oxidates to MB, a process which consumes molecular oxygen and yields hydrogen peroxide, a strong oxidizing agent. The oxidizing conditions trigger the expression of a range of response genes, most notably the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2). Nrf2 interlinks with a range of cellular stress and homeostasis pathways, such as the oxidative and xenobiotic stress response, mitochondrial respiration, the UPR in the endoplasmic reticulum and autophagy (Tonelli et al., 2018; Prasad and Greber, 2021). It is thus conceivable that the overexpression of Nrf2 has negative effects on SARS-CoV-2 infection. In support of this notion, Nrf2 mediated gene expression was found to be suppressed in biopsies obtained from COVID-19 patients (Olagnier et al., 2020). Moreover, another top performer in our screen, GPP-78 targets nicotinamide phosphoribosyltransferase, a crucial enzyme in the NAD salvage/recycling pathway (Khan et al., 2007), lending further support to the importance of redox control in coronavirus infection.
Intriguingly, our data reveal a discrepancy between infectious progeny virus production and viral genome copy numbers released from the HAEEC. In control cells, SARS-CoV-2 progeny peaked at 3–4 d pi and declined by 2–3 logs 8 d pi, while the genome copy numbers remained constant after day 4 of infection (Fig. S3). By using two distinct pairs of PCR primers, one for the RNA polymerase subunit Nsp12 and the other one for the M-gene, and by including the RNA polymerase inhibitor Remdesivir (which reduced the genome copy numbers), we exclude that this discrepancy is an artifact to subgenomic replicons known to comprise the M-gene. Importantly, the discrepancy was in perfect agreement with a previous study quantifying the N-gene RNA (Robinot et al., 2021). Since infected HAEEC cells treated with MB (and also MPA and to a lesser extent POS) strongly reduced the amounts of infectious virus but not the genome copy numbers, we suggest that MB and MPA block hCoV progeny formation and apical release of virions. Since MB, MPA and POS reduced the intracellular viral (+)RNA levels in Huh7 cells, these compounds may induce intracellular degradation or export rather than blocking synthesis of viral RNA. In contrast, the viral RNA polymerase inhibitor Remdesivir blocked the synthesis of viral RNA, as demonstrated by both intracellular and extracellular viral RNA measurements. This argues that MB, MPA and POS act by a distinct mode of action compared to Remedesivir. This contention is supported by our observation that MB and Remdesivir synergize against hCoV cell culture infection in the range of 0.5–5 μM MB and 50–500 nM Remdesivir.
These results encourage interventional trials against COVID-19 combining two FDA-approved drugs, MB and Remdesivir. The former is already given intravenous (i.v., or per oral) for the treatment of pediatric and adult patients with acquired methemoglobinemia, and the latter for treatment of early COVID-19, while it is largely ineffective at late stages of coronavirus disease (Ohl et al., 2021). Pharmacokinetics data from patients show that MB is well absorbed from the GI tract and reaches similar blood concentration levels as upon i.v. administration, although its organ distribution differs compared to i.v. administration in rats (Peter et al., 2000). For instance, 2 mg/kg dosage (recommended for treatment of methemoglobinemia) yields plasma levels of about 0.1 μM with a half life of 5–6h. Remarkably, a COVID-19 phase II clinical trial at 1 mg/kg per oral (every 8 h for two days, and then twice daily for the following 12 days) reported a 50% mortality reduction of severe COVID-19 cases, as well as increased blood oxygenation and respiratory rate (Hamidi-Alamdari et al., 2021). Clinical trials comparing regular as well as reduced dosages of MB and Remdesivir may be warranted (Beigel et al., 2020a; Hamidi-Alamdari et al., 2021). Endpoints may include survival and blood oxygenation levels, both of which are expected to be enhanced by the combination treatment due to the anti-viral effects of both agents and the potency of MB to treat methemoglobinemia in patients resistant to oxygen therapy.
8.2. A two-pronged action of MPA - anti-viral and anti-inflammatory
Our screen and follow-up experiments identified MPA as a broad anti-coronavirus inhibitor. MPA is a non-competitive inhibitor of inosine-5′-monophosphate dehydrogenase (IMPDH) involved in guanine biosynthesis (Hedstrom, 2009). IMPDH interacts with and supports SARS-CoV nonstructural protein 14 (Nsp14), a guanine-N7-methyl-transferase implicated in virus replication and transcription (Chen et al., 2009). In addition, MPA leads to a reduction in the levels of the furin protease in human pluripotent stem cell derived lung organoids, and inhibits the entry of S-glycoprotein pseudotyped vesicular stomatitis particles (Han et al., 2021). Furin has a key role in viral pathogenesis and dissemination, and is required for the limited proteolysis of the SARS-CoV-2 S-protein, and S-protein engagement with the neuropilin-1 receptor and virus infectivity, especially neuronal and heart cell infections (Cantuti-Castelvetri et al., 2020; Daly et al., 2020).
MPA is also a weak immunosuppressant used in transplant patients as Mycophenolate Mofetil (MMF, Myfortic®, Novartis) for reduction of transplant rejection risk (Han et al., 2021). MMF is readily converted to MPA by cellular esterases, and orally or intravenously broadly bioavailable at low side effects. And MPA inhibits SARS-CoV-2 infection of humanized mice and human lung organoids (Park, 2011). An initial clinical case reported that a COVID-19 patient affected by autoimmune blistering disease (ABD) during immunosuppressive treatment, i.e. azathioprine, was treated with MMF, and experienced only a mild form of COVID-19, without pulmonary complications or detrimental effects (Di Altobrando et al., 2020; Balestri et al., 2020). Notably, SARS-CoV-2 leads to a massive destruction of host mRNAs (Banerjee et al., 2020), except transcripts encoding the NFkB pathway, a situation, which leads to cytokine storms in COVID-19 patients (Mehta et al., 2020).
Given the immunosuppressive anti-inflammatory effects of MPA, we suggest a two-pronged modality of MPA towards treating COVID-19. One is an antiviral action at moderate concentration to reduce systemic disease, and the other one is anti-inflammatory. For clinical applications, we envision a low dosage modality of MPA/MMF in mild COVID-19 cases (0.1 g, twice daily), and a higher dosage in mid and severe COVID-19 cases (1.5 g, twice daily), resulting in plasma concentrations of 1–15 μg/ml MPA/MMF (approximately 2–30 μM) (Staatz and Tett, 2007). The precise dosage would have to be tuned individually as MPA/MMF exhibits considerable within-subject variability of 9–17 h half life. This regimen goes along with the notions that 1) moderate concentrations of MPA/MMF (1–3 μM) already exhibit noticeable anti-viral effects, and 2) higher doses (10 μM) have well described immunosuppressive effects in organ transplant patients at risk reduction of transplant rejection. Anti-inflammatory and immuno-suppressive agents may be beneficial to severe COVID-19 cases as shown with corticosteroid treatment (Neurath, 2021; Coperchini et al., 2020), albeit with variable outcomes and requiring close patient monitoring (Beigel et al., 2020b; Cain and Cidlowski, 2020).
In contrast to corticosteroids, MPA/MFF has well characterized distinct anti-inflammatory effects. For example, the reversible inhibition of IMPDH results in blockage of de novo guanosine synthesis, which suppresses the proliferation of T and B lymphocytes (Eugui et al., 1991). Hence, MPA/MFF may best be applied for days rather than weeks. B and T cells are largely devoid of salvage pathways for the synthesis of guanosine, and hence are particularly susceptible to MPA. MPA/MMF treatment leads to a reduction in mRNAs encoding pro-inflammatory cytokines, such as TNF-a, IL-6 & IL-1b (Lv et al., 2015; Huang et al., 2020). It furthermore inhibits the activity of 6-pyruvoyl tetrahydro-biopterin synthase involved in the biosynthesis of tetrahydrobiopterin from GTP, a cofactor of aromatic amino acid monooxygenases and nitric oxide synthase. This inhibition leads to a reduction of the proinflammatory metabolite peroxynitrite (Srinivas et al., 2003), and could contribute to the beneficial effects of MPA/MFF in COVID-19. Along the same vein, SB-505124 was a triple hit against hCoV-229E-GFP, hCoV-OC43 and SARS-CoV-2 in our human cell culture screens. It is a reversible ATP competitive inhibitor of the TGF-beta type I receptor serine/threonine kinase, also known as activin receptor-like kinase (ALK) 4, as well as ALK5 and 7 (Byfield et al., 2004). The TGF-beta receptor pathway controls differentiation, chemotaxis, proliferation, and activation of immune cells, and its inhibition by SB-505124 might contribute to a reduction in inflammation.
8.3. Azole-based anti-fungal agents and compounds interfering with cholesterol biosynthesis have broad effects against coronaviruses
The azole-based antifungal drugs POS and Ravuconazole were effective in cell culture and HAEEC against coronaviruses, including SARS-CoV-2 infection, along with the azole-based antifungals, Mebendazole, which was discarded because of autofluorescence in follow-up experiments. These agents target sterol 14α demethylase converts lanosterol to ergosterol in sterol biosynthesis (Mann et al., 2003; Lepesheva and Waterman, 2011; Hata et al., 1996). POS has been approved in the US, EU and Australia against invasive fungal infections in immunocompromised patients e.g. after organ or bone marrow transplants. It has been in clinical use since 2006 and generica are available, making it an interesting broadly available candidate for clincal trials against COVID-19. Intriguingly, another hit compound in the cholesterol pathway was Ro 48–8071 fumarate blocking cell culture coronavirus infections, with unknown anti-viral effects in HAEEC owing to toxicity (Table S4). Ro 48–8071 is a potent inhibitor of oxidosqualene cyclase (OSC) which catalyzes the conversion of monooxidosqualene to lanosterol (Morand et al., 1997), and could interfere with positive-strand RNA viruses, including hepatitis C virus (Romero-Brey et al., 2012), poliovirus (Belov et al., 2012), and rhinovirus (Roulin et al., 2014). Collectively, these data suggest that the cholesterol biosynthesis pathway is a potential drug target in SARS-CoV-2 infection.
8.4. Repurposing SARS-CoV-2 inhibitors in clinical development
One of our strongest hits in the cell culture screens was MLN4924 (Pevonedistat) with an excellent dose-response profile against hCoV-229E-GFP (EC50 30 nM; TC50 40.39 μM) and considerable inhibition of SARS-CoV-2. It is a promising candidate in retinoblastoma therapy (Aubry et al., 2020), but unfortunately exhibited toxicity in our HAEEC experiments at low micromolar concentrations (Table S4). Nonetheless, MLN4924 is an interesting tool compound in cell culture, where it targets the Nedd8 activating enzyme (NAE) with potential anti-neoplastic activity (Soucy et al., 2009b; Lan et al., 2016), and prevents the degradation of Nrf2 (Zhao et al., 2014). Possibly, MLN4924 and MB have a similar mode-of-action against SARS-CoV-2 by dysregulating the oxidative state of the cell and enhancement of Nrf2.
8.5. Other compounds
Our screen revealed several additional launched compounds already described for their potential to treat COVID-19, including Fostamatinib, Eprinomectin and Betulinic acid. Fostamatinib is an FDA approved Syk tyrosine kinase inhibitor for the treatment of chronic immune thrombocytopenia (Kost-Alimova et al., 2020), and is in a phase 3 clinical trial against COVID-19 (NCT04629703). Although Fostamatinib inhibited SARS-CoV-2 infection of cultured human cells and African green monkey Vero cells, we did not pursue this compound further due to morphological alterations in the HAEEC, including outgrowth of fibroblasts. Likewise, we did not follow up on Eprinomectin, a benzodiazepine receptor agonist with anti-parasitic activity but unknown mode-of-action, though approved for veterinary use in horses, cattle and cats, akin to other avermectins (El-Saber Batiha et al., 2020). We refrained from further investigating Betulinic acid, a pentacyclic triterpenoid with attributed anti-retroviral, anti-malarial, and anti-inflammatory properties but also anti-proliferative and apoptotic effects (Verma et al., 2020).
Intriguingly, our high-throughput screen with 229E-GFP revealed a cluster of five distinct compounds targeting glycogen synthase kinase 3 (GSK-3), namely LY2090314 (Atkinson et al., 2015), which was the most potent compound identified in our hCoV-229E-GFP (EC50 2 nM), GSK-3 inhibitor IX (Meijer et al., 2003), TCS-21311 (Thoma et al., 2011), AZD2858 (Marsell et al., 2012) and CHIR-98014 (Ring et al., 2003). GSK-3 phosphorylates a variety of proteins in glycogen metabolism, innate immunity and apoptosis (Jope and Johnson, 2004), and some of its inhibitors are approved to treat cancer, inflammation, Alzheimer's, and diabetes (Augello et al., 2020). GSK-3 inhibitors have also been proposed for SARS-CoV-2 treatment, based on the notion that GSK-3 is implicated in phosphorylation of SARS-CoV-2 N-protein (Liu et al., 2021). Unfortunately, none of the GSK-3 inhibitors identified in our screen translated antiviral effects to SARS-CoV-2 infections of HAEEC.
In conclusion, our image-based, high content, full cycle screen against human coronavirus infection provides a high quality hit list of clinical and preclinical compounds, including five clinically launched drugs, of which MB, MPA and POS were found to inhibit virus production in HAEEC. Two topical agents, thonzonium bromide, a monocationic surfactant with bacteriocidal effects (Chan et al., 2012), and the mouth wash component Cetylpyridinium scored in coronavirus cell culture infections, and may represent additional repurposing candidates for future analyses.
8.6. Perspective
Our top compounds are effective against SARS-CoV-2 Wuhan strain as well as four variants of concern, alpha, beta, gamma, and delta. These variants likely evolved in humans since December 2019, indicating that evolution and turnover of the circulating SARS-CoV-2 do not affect the efficacy of our inhibitors in human explant cell cultures. This is distinct from inhibitors targeting the virus directly, which the virus rapidly evades, for example favipiravir. Favipiravir is an RNA mutagen with antiviral effects (Delang et al., 2018) approved in Japan to treat pandemic influenza virus infections. It has anti-viral effects against SARS-CoV-2 in hamsters (Driouich et al., 2021), and is in clinical trials against COVID-19 (Joshi et al., 2021). A recent high-throughput computational protein design protocol identified mutational hotspots in the SARS-CoV-2 RNA polymerase confering favipiravir resistance, and remarkably some of these mutations already exist in SARS-CoV-2 genomes circulating in humans (Padhi et al., 2021; Greber, 2021). This implies that direct anti-viral agents readily face drug-resistant viruses, even before treatment, and likely have limited therapeutic effects. We surmise that host targeting anti-virals, such as the compounds identified in this study will have a higher fitness cost for virus to evade than direct anti-viral agents, akin to a recent study on chemical evolution of rhinovirus (Murer et al., 2021).
Author contributions
UFG initiated, conceived and supervised the project. LM, RV, NM, LY, DS, AGG, AP, DO, MB, FG, MS, FK, GT, UFG designed, carried out or analyzed experiments. LM, RV provided data visualization. VA, LM provided data curation and software. LM, RV, UFG wrote the paper. GT, UFG acquired funding. All authors read and approved the manuscript.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: UFG has been a consultant and stock owner in 3V-Biosciences (now Sagimet Biosciences), a consultant to F. Hoffmann-La Roche Ltd and to Union Therapeutics A/S. The authors filed a patent application on the use of MPA for the treatment of COVID-19 (EP20213904, European Patent Office, University of Zurich).
Acknowledgements
We acknowledge financial support from the Swiss National Science foundation (SNSF) to UFG and GT (31CA30_196177/1), the NCCR Chemical Biology supported by the SNSF for purchasing the BSF-EPFL repurposing collection, and a special Coronavirus Research grant from the University of Zurich to UFG. We are grateful to Dr. Adriano Aguzzi and Dr. Simone Hornemann for granting generous access to their BSL3 laboratory. We thank Dr. Volker Thiel and Dr. Nadine Ebert for their kind gifts of hCoV-229E-GFP, SARS-CoV-2 isolate Munich, cell lines, as well as discussions and hands-on information, as well as Dr. Daria Seiler, Jonathan Vesin, Antoine Gibelin and Julien Bortoli for their technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crviro.2022.100019.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplemental Table 1:Primers used for SARS-CoV-2 PCR reactions.
Supplemental Table 2: Threshold cycle comparison for SARS-CoV2 M and the Pol-gene RT-qPCR.
Supplemental Table 3: Overview of all compound hits. Chemical hit compounds from the screen in Huh7 cell infected with hCoV-229E-GFP are listed with their EC50 and TC50 concentrations, their clinical status and original treatment indication, effects in four cell lines against SARS-CoV-2 and their proposed mode of application.
Supplemental Table 4: Detailed presentation of compound effects on SARS-CoV-2 infection of HAEEC. The table presents all the virus titers as TCID50 per ml and genome copy numbers per ml in the apical supernatant from 610 distinct SARS-CoV2 infections of nasal and bronchial HAEEC treated with different compounds. Note, no measurable virus titer could be recorded in pilot experiments, and hence no basolateral measurements were conducted on a routine basis. The table also features the experiment ID, reference and the type of HAEEC, drug treatment (including concentrations and pre- or post-exposure regimen), the SARS-CoV2 genetic variants, and the sampling time pi. Virus titers and genomes numbers are expressed as log10 values. Data presented are annotated to the corresponding figure in the manuscript. Abbreviations: Yes (Y), No (N), ns (not show in the manuscript), NR (not realized).
figs1.
Z’ factors of each screening plate and comparison between nuclear counterstain and PrestoBlue. A) Each facet depicts the per-plate Z’ factors for the assessed parameters. II: infection index, IN: infected nuclei, P: plaques, VI: total virus intensity. The dashed line intersects the y-axis at 0.5. Mean Z’ factors are II = 0.432, IN = 0.378, P = 0.509, VI = 0.276. B) The number of nuclei was assessed based on the nuclear counterstain in the screening setup. In contrast, the PrestoBlue based assay was performed in absence of virus, but all the other parameters were kept the same. The score of the Prestoblue assay was normalized with the positive control set as 1, and the negative control as 0. As a result, there is a negative correlation between the parameters.
figs2.
ACE2 expression of lentivirus transduced cell lines. ACE2 expression was verified in cell lines transduced with lentivirus encoding human ACE2. An unspecific band was observed in both the untransduced and the transduced HeLa cells (not shown). The GAPDH loading control for the blot shown on the left side with A549 and HeLa cell lines was performed with the same lysates, but run on separate gels. The Huh7-ACE2 samples (h, l) shown on the right side are derived from separate cell populations, transduced with either a high or a low MOI of lentivirus. We used the low MOI transduced cells (small arrow), because ACE2 expression was sufficient to mediated SARS-CoV-2 infection.
figs3.
MB, MPA inhibit SARS-CoV-2 infection of nasal HAEEC. Viral titer (TCID50/ml) readout (left y-axis) are those shown in Fig. 5A, complemented here with the corresponding viral genome copy numbers (blue right y-axis). Data represent the mean+/-SEM of two independent replicates. Inserts treated in a pre-infection regimen, starting at 2h prior to virus inoculation (left) or in a post-infection regimen starting at 1d pi (right) with Remdesivir (A-B), MB (C), and with MPA (D). Not determined (nd) indicates virus titers below 2.4 log10 TCID50/ml in the two replicates, and (1/2) indicates virus titers below 2.4 log10 TCID50/ml in one replicate. E) The SARS-CoV-2 RT-qPCR inter-assay reproducibility is represented with the linear regression curve generated from the Ct values and the copy numbers (106 to 103 copies per μl) of the RNA quantitative standards in eight independent replicates. The coefficient of correlation (CV) was calculated for each quantitative point according to the formula: %CV = (SD / Ct mean)×100. F) Effect of drug treatment on ciliary motion of SARS-CoV-2 infected nasal HAEEC. Transmitted light movies were recorded at 5d pi (MLN4924, Posaconazole), 6d pi (DMSO, Remdesivir, MB, MPA), or 6d in culture in absence of virus or drug (mock). Ciliary motion was assessed by computing the median norm of the optical flow per frame. Fold changes are normalized to mock. Panel refers to Supplemental Movies 1-7.
figs4.
MB and MPA inhibit SARS-CoV-2 infection of bronchial HAEEC. Bronchial HAEEC grown at air-liquid interface were inoculated apically with 1,000 FFU of SARS-CoV2 (day 0), and subjected to drug treatment in the basolateral medium. Drugs were applied in a pre-infection regimen, starting at 2h prior to SARS-CoV2 inoculation (A, C) or in a post-infection regimen starting at 1d post inoculation (B, D). MB (5 and 0.5 μM) and MPA (10 and 1 μM) were administrated daily until day 8. DMSO was used as a negative control. SARS-CoV-2 released to the apical side was collected daily by apical washing and quantified by virus TCID50 titration. Data represent the mean±SEM of two independent replicates. Not determined (nd) indicates virus titers below 2.4 log10 TCID50/ml in the two replicates, and (1/2) indicates virus titers below 2.4 log10 TCID50/ml in one replicate.
figs5.
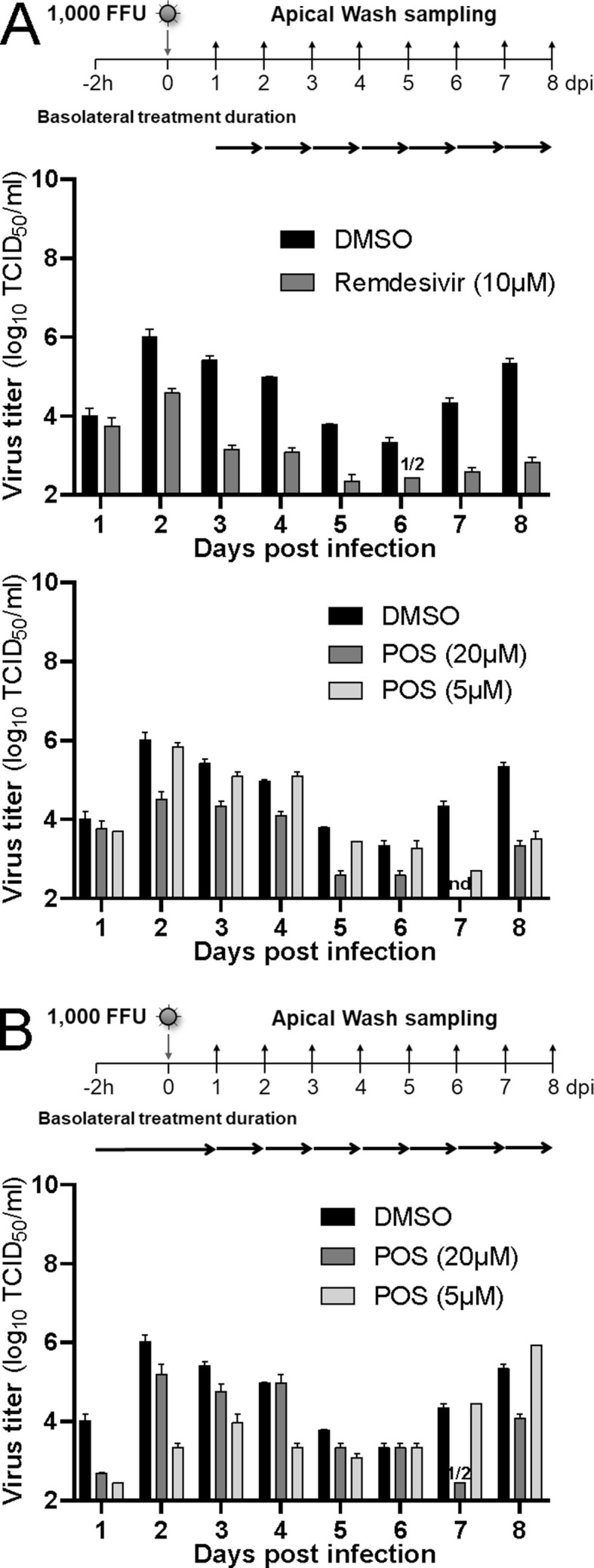
TCID50 data of POS treated nasal HAEEC infected with SARS-CoV-2. Viral titer (TCID50/ml) readout of the samples shown in Fig. 5B. Panel A) shows post-exposure treatment with 10 μM Remdesivir, and 5 and 20 μM POS. Panel B) depicts pre-exposure treatment with 5 and 20 μM POS. Data represent the mean±SEM of two independent replicates. Not determined (nd) indicates virus titers below 2.4 log10 TCID50/ml in the two replicates, and (1/2) indicates virus titers below 2.4 log10 TCID50/ml in one replicate.
figs6.
TCID50 and viral genome copy numbers of MB, MPA and POS treated nasal HAEEC inoculated with SARS-CoV-2 alpha, beta and gamma variants. Viral titer (TCID50/ml) readout (bars) of the samples shown in Fig. 6, and viral genome equivalents (copies/ml, blue dots). Data represent the means±SEM of two independent replicates. Not determined (nd) indicates virus titers below 2.4 log10 TCID50/ml in the two replicates, and (1/2) indicates virus titers below 2.4 log10 TCID50/ml in one replicate.
figs7.
TCID50 and viral genome copy numbers of MB, MPA and POS treated nasal HAEEC inoculated with SARS-CoV-2 delta variant of concern. Viral titer (TCID50/ml) readout (bars) of the samples shown in Fig. 6, and viral genome equivalents (copies/ml, blue dots). Data represent the means±SEM of two independent replicates. Not determined (nd) indicates virus titers below 2.4 log10 TCID50/ml in the two replicates, and (1/2) indicates virus titers below 2.4 log10 TCID50/ml in one replicate.
References
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2):e00221–18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- Atkinson J.M., Rank K.B., Zeng Y., Capen A., Yadav V., Manro J.R., et al. Activating the Wnt/β-catenin pathway for the treatment of melanoma–application of LY2090314, a novel selective inhibitor of glycogen synthase kinase-3. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0125028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry A., Yu T., Bremner R. Preclinical studies reveal MLN4924 is a promising new retinoblastoma therapy. Cell Death Discov. 2020;6(1):1–12. doi: 10.1038/s41420-020-0237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augello G., Emma M.R., Cusimano A., Azzolina A., Montalto G., McCubrey J.A., et al. The role of GSK-3 in cancer immunotherapy: GSK-3 inhibitors as a new frontier in cancer treatment. Cells. 2020;9(6):1427. doi: 10.3390/cells9061427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowski M.A., Beutler N., Wolff K.C., Kirkpatrick M.G., Chen E., Nguyen T.H., et al. Drug repurposing screens identify chemical entities for the development of COVID-19 interventions. Nat. Commun. 2021;12(1):3309. doi: 10.1038/s41467-021-23328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestri R., Rech G., Girardelli C.R. Occurrence of SARS-CoV-2 during mycophenolate mofetil treatment for pemphigus. J. Eur. Acad. Dermatol. Venereol. 2020;34(9):e435–e436. doi: 10.1111/jdv.16578. PMC7267403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183(5):1325–1339 e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., et al. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., et al. Remdesivir for the treatment of Covid-19. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov G.A., Nair V., Hansen B.T., Hoyt F.H., Fischer E.R., Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. J. Virol. 2012;86(1):302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojadzic D., Alcazar O., Buchwald P. Methylene blue inhibits the SARS-CoV-2 spike-ACE2 protein-protein interaction-a mechanism that can contribute to its antiviral activity against COVID-19. Front. Pharmacol. 2020;11:600372. doi: 10.3389/fphar.2020.600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz K., Schirmer R.H., Eubel J.K., Akoachere M.B., Dandekar T., Becker K., et al. Interactions of methylene blue with human disulfide reductases and their orthologues from Plasmodium falciparum. Antimicrob. Agents Chemother. 2008;52(1):183–191. doi: 10.1128/AAC.00773-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byfield S.D., Major C., Laping N.J., Roberts A.B. SB-505124 is a selective inhibitor of transforming growth factor-β type I receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2004;65(3):744–752. doi: 10.1124/mol.65.3.744. [DOI] [PubMed] [Google Scholar]
- Cain D.W., Cidlowski J.A. After 62 years of regulating immunity, dexamethasone meets COVID-19. Nat. Rev. Immunol. 2020;20(10):587–588. doi: 10.1038/s41577-020-00421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Aleem A., Dulebohn S.C., Di Napoli R. StatPearls Publishing; Treasure Island (FL): 2021. Features, Evaluation, and Treatment of Coronavirus (COVID-19). StatPearls. [PubMed] [Google Scholar]
- Cervantes-Barragan L., Züst R., Maier R., Sierro S., Janda J., Levy F., et al. Dendritic cell-specific antigen delivery by coronavirus vaccine vectors induces long-lasting protective antiviral and antitumor immunity. mBio. 2010;1(4):e00171–10. doi: 10.1128/mBio.00171-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.Y., Prudom C., Raines S.M., Charkhzarrin S., Melman S.D., De Haro L.P., et al. Inhibitors of V-ATPase proton transport reveal uncoupling functions of tether linking cytosolic and membrane domains of V0 subunit a (Vph1p) J. Biol. Chem. 2012;287(13):10236–10250. doi: 10.1074/jbc.M111.321133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai H., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. Unit. States Am. 2009;106(9):3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsello S.M., Bittker J.A., Liu Z., Gould J., McCarren P., Hirschman J.E., et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat. Med. 2017;23(4):405–408. doi: 10.1038/nm.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 2021;6(1):11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creech C.B., Walker S.C., Samuels R.J. SARS-CoV-2 vaccines. JAMA. 2021;325(13):1318–1320. doi: 10.1001/jama.2021.3199. [DOI] [PubMed] [Google Scholar]
- Daly J.L., Simonetti B., Klein K., Chen K.-E., Williamson M.K., Antón-Plágaro C., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370(6518):861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delang L., Abdelnabi R., Neyts J. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antivir. Res. 2018;153:85–94. doi: 10.1016/j.antiviral.2018.03.003. [DOI] [PubMed] [Google Scholar]
- Di Altobrando A., Patrizi A., Bardazzi F. Should SARS-CoV-2 influence immunosuppressive therapy for autoimmune blistering diseases? J. Eur. Acad. Dermatol. Venereol. 2020;34(7):e295–e297. doi: 10.1111/jdv.16491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich J.S., Cochin M., Lingas G., Moureau G., Touret F., Petit P.R., et al. Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model. Nat. Commun. 2021;12(1):1735. doi: 10.1038/s41467-021-21992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Saber Batiha G., Alqahtani A., Ilesanmi O.B., Saati A.A., El-Mleeh A., Hetta H.F., et al. Avermectin derivatives, pharmacokinetics, therapeutic and toxic dosages, mechanism of action, and their biological effects. Pharmaceuticals. 2020;13(8):196. doi: 10.3390/ph13080196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugui E.M., Mirkovich A., Allison A.C. Lymphocyte-selective antiproliferative and immunosuppressive effects of mycophenolic acid in mice. Scand. J. Immunol. 1991;33(2):175–183. doi: 10.1111/j.1365-3083.1991.tb03747.x. [DOI] [PubMed] [Google Scholar]
- Gantt S., Carlsson J., Ikoma M., Gachelet E., Gray M., Geballe A.P., et al. The HIV protease inhibitor nelfinavir inhibits Kaposi's sarcoma-associated herpesvirus replication in vitro. Antimicrob. Agents Chemother. 2011;55(6):2696–2703. doi: 10.1128/AAC.01295-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrot M., Andreani J., Duflot I., Boxberger M., Le Bideau M., Mosnier J., et al. Methylene blue inhibits replication of SARS-CoV-2 in vitro. Int. J. Antimicrob. Agents. 2020;56(6):106202. doi: 10.1016/j.ijantimicag.2020.106202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi F., Andriasyan V., Witte R., Murer L., Hemmi S., Yu L., et al. The FDA-approved drug Nelfinavir inhibits lytic cell-free, but not cell-associated non-lytic transmission of human adenovirus. Antimicrob. Agents Chemother. 2020;64(9):e01002–e01020. doi: 10.1128/AAC.01002-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi F., Kuttler F., Murer L., Andriasyan V., Witte R., Yakimovich A., et al. A high-content image-based drug screen of clinical compounds against cell transmission of adenovirus. Sci. Data. 2020;7(1):1–12. doi: 10.1038/s41597-020-00604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good S.S., Westover J., Jung K.H., Zhou X.J., Moussa A., La Colla P., et al. AT-527, a double prodrug of a guanosine nucleotide analog, is a potent inhibitor of SARS-CoV-2 in vitro and a promising oral antiviral for treatment of COVID-19. Antimicrob. Agents Chemother. 2021;65(4) doi: 10.1128/AAC.02479-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber U.F. Two years into COVID-19 – lessons in SARS-CoV-2 and a perspective from papers in FEBS Letters. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2021 doi: 10.1002/1873-3468.14226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E., Hale C., Saito Y., Blachere N.E., Bergh M., Conlon E.G., et al. Vaccine breakthrough infections with SARS-CoV-2 variants. N. Engl. J. Med. 2021;384(23):2212–2218. doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidi-Alamdari D., Hafizi-Lotfabadi S., Bagheri-Moghaddam A., Safari H., Mozdourian M., Javidarabshahi Z., et al. Methylene blue for treatment of hospitalized Covid-19 patients: a randomized, controlled, open-label clinical trial, phase 2. Rev. Invest. Clin. 2021;73(3):190–198. doi: 10.24875/RIC.21000028. [DOI] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589(7841):270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K., Kimura J., Miki H., Toyosawa T., Nakamura T., Katsu K. In vitro and in vivo antifungal activities of ER-30346, a novel oral triazole with a broad antifungal spectrum. Antimicrob. Agents Chemother. 1996;40(10):2237–2242. doi: 10.1128/aac.40.10.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawi A., Heald S., Sciascia T. Use of an adaptive study design in single ascending-dose pharmacokinetics of A0001 (α-Tocopherylquinone) in healthy male subjects. J. Clin. Pharmacol. 2012;52(1):65–77. doi: 10.1177/0091270010390807. [DOI] [PubMed] [Google Scholar]
- Hedstrom L. IMP dehydrogenase: structure, mechanism, and inhibition. Chem. Rev. 2009;109(7):2903–2928. doi: 10.1021/cr900021w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home. COVID-19 Map - Johns Hopkins Coronavirus Resource Center.
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R.J., Lang Y., Bakkers M.J., Li W., Li Z., Schouten A., et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116(7):2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jope R.S., Johnson G.V.W. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci. 2004;29(2):95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Joshi S., Parkar J., Ansari A., Vora A., Talwar D., Tiwaskar M., et al. Role of favipiravir in the treatment of COVID-19. Int. J. Infect. Dis. 2021;102:501–508. doi: 10.1016/j.ijid.2020.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgeit A., McDowell R., Moese S., Meldrum E., Schwendener R., Greber U.F. Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects. PLoS Pathog. 2012;8(10) doi: 10.1371/journal.ppat. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J.A., Forouhar F., Tao X., Tong L. Nicotinamide adenine dinucleotide metabolism as an attractive target for drug discovery. Expert Opin. Ther. Targets. 2007;11(5):695–705. doi: 10.1517/14728222.11.5.695. [DOI] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4) doi: 10.1016/j.cell.2020.04.011. 914-921 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost-Alimova M., Sidhom E.-H., Satyam A., Chamberlain B.T., Dvela-Levitt M., Melanson M., et al. A high-content screen for mucin-1-reducing compounds identifies fostamatinib as a candidate for rapid repurposing for acute lung injury. Cell Rep. Med. 2020;1(8):100137. doi: 10.1016/j.xcrm.2020.100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., et al. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 2020;182(4):843–854. doi: 10.1016/j.cell.2020.06.044. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan H., Tang Z., Jin H., Sun Y. Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci. Rep. 2016;6(1):1–12. doi: 10.1038/srep24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesheva G.I., Waterman M.R. Sterol 14alpha-demethylase (CYP51) as a therapeutic target for human trypanosomiasis and leishmaniasis. Curr. Top. Med. Chem. 2011;11(16):2060–2071. doi: 10.2174/156802611796575902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. Rapid biochemical test for measuring chemical toxicity. Bull. Environ. Contam. Toxicol. 1981;26(2) doi: 10.1007/BF01622068. [DOI] [PubMed] [Google Scholar]
- Liu X., Verma A., Ramage H., Garcia G., Myers R.L., Lucas A., et al. Targeting the coronavirus nucleocapsid protein through GSK-3 inhibition. Proc Natl Acad Sci U S A. 2021;118(42) doi: 10.1073/pnas.2113401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M.K., Albariño C.G., Perry J.K., Chang S., Tchesnokov E.P., Guerrero L., et al. Remdesivir targets a structurally analogous region of the Ebola virus and SARS-CoV-2 polymerases. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117(43):26946–26954. doi: 10.1073/pnas.2012294117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Muñoz-Alonso M.J., García C., Sánchez-Murcia P.A., Martínez-Leal J.F., Domínguez J.M., et al. Translation elongation factor eEF1A2 is a novel anticancer target for the marine natural product plitidepsin. Sci. Rep. 2016;6(1) doi: 10.1038/srep35100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q.-K., Liu J.-X., Li S.-N., Gao Y.-J., Lv Y., Xu Z.-P., et al. Mycophenolate mofetil modulates differentiation of Th1/Th2 and the secretion of cytokines in an active Crohn's disease mouse model. Int. J. Mol. Sci. 2015;16(11):26654–26666. doi: 10.3390/ijms161125985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maione F., Oliaro-Bosso S., Meda C., Di Nicolantonio F., Bussolino F., Balliano G., et al. The cholesterol biosynthesis enzyme oxidosqualene cyclase is a new target to impair tumour angiogenesis and metastasis dissemination. Sci. Rep. 2015;5(1):1–12. doi: 10.1038/srep09054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maione F., Oliaro-Bosso S., Meda C., Di Nicolantonio F., Bussolino F., Balliano G., et al. The cholesterol biosynthesis enzyme oxidosqualene cyclase is a new target to impair tumour angiogenesis and metastasis dissemination. Sci. Rep. 2015;5:9054. doi: 10.1038/srep09054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann P.A., Parmegiani R.M., Wei S.Q., Mendrick C.A., Li X., Loebenberg D., et al. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14alpha-demethylase. Antimicrob. Agents Chemother. 2003;47(2):577–581. doi: 10.1128/AAC.47.2.577-581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsell R., Sisask G., Nilsson Y., Sundgren-Andersson A.K., Andersson U., Larsson S., et al. GSK-3 inhibition by an orally active small molecule increases bone mass in rats. Bone. 2012;50(3):619–627. doi: 10.1016/j.bone.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer L., Skaltsounis A.-L., Magiatis P., Polychronopoulos P., Knockaert M., Leost M., et al. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem. Biol. 2003;10(12):1255–1266. doi: 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. PMC8566220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand O.H., Aebi J.D., Dehmlow H., Ji Y.-H., Gains N., Lengsfeld H., et al. Ro 48-8.071, a new 2, 3-oxidosqualene: lanosterol cyclase inhibitor lowering plasma cholesterol in hamsters, squirrel monkeys, and minipigs: comparison to simvastatin. J. Lipid Res. 1997;38(2):373–390. [PubMed] [Google Scholar]
- Murer L., Petkidis A., Vallet T., Vignuzzi M., Greber U.F. Chemical evolution of Rhinovirus identifies capsid-destabilizing mutations driving low pH-independent genome uncoating. J. Virol. 2021:JVI0106021. doi: 10.1128/JVI.01060-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath M.F. COVID-19: biologic and immunosuppressive therapy in gastroenterology and hepatology. Nat. Rev. Gastroenterol. Hepatol. 2021;18(10):705–715. doi: 10.1038/s41575-021-00480-y. PMC8239481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohl M.E., Miller D.R., Lund B.C., Kobayashi T., Richardson Miell K., Beck B.F., et al. Association of remdesivir treatment with survival and length of hospital stay among US Veterans hospitalized with COVID-19. JAMA Netw. Open. 2021;4(7) doi: 10.1001/jamanetworkopen.2021.14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olagnier D., Farahani E., Thyrsted J., Blay-Cadanet J., Herengt A., Idorn M., et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020;11(1):1–12. doi: 10.1038/s41467-020-18764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreshkova N., Molenaar R.J., Vreman S., Harders F., Munnink B.B.O., Hakze-van Der Honing R.W., et al. SARS-CoV-2 infection in farmed minks, The Netherlands, April and May 2020. Euro Surveill. 2020;25(23):2001005. doi: 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhi A.K., Dandapat J., Saudagar P., Uversky V.N., Tripathi T. Interface-based design of the favipiravir-binding site in SARS-CoV-2 RNA-dependent RNA polymerase reveals mutations conferring resistance to chain termination. FEBS Lett. 2021;595(18):2366–2382. doi: 10.1002/1873-3468.14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. The emergence of mycophenolate mofetilin dermatology: from its roots in the world of organ transplantation to its versatile role in the dermatology treatment room. J. Clin. Aesthetic Dermatol. 2011;4(1):18. [PMC free article] [PubMed] [Google Scholar]
- Patnaik S., Natarajan M.M., James E.J., Ebenezer K. Methylene blue unresponsive methemoglobinemia. Indian J. Crit. Care Med.: peer-reviewed official publication of Indian Society of Critical Care Medicine. 2014;18(4):253. doi: 10.4103/0972-5229.130582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter C., Hongwan D., Kupfer A., Lauterburg B.H. Pharmacokinetics and organ distribution of intravenous and oral methylene blue. Eur. J. Clin. Pharmacol. 2000;56(3):247–250. doi: 10.1007/s002280000124. [DOI] [PubMed] [Google Scholar]
- Pitten F.-A., Kramer A. Efficacy of cetylpyridinium chloride used as oropharyngeal antiseptic. Arzneimittelforschung. 2001;51(7):588–595. doi: 10.1055/s-0031-1300084. [DOI] [PubMed] [Google Scholar]
- Pizzorno A., Padey B., Julien T., Trouillet-Assant S., Traversier A., Errazuriz-Cerda E., et al. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell. Rep. Med. 2020;1(4):100059. doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Prasad V., Greber U.F. The endoplasmic reticulum unfolded protein response–homeostasis, cell death and evolution in virus infections. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2021;45(5) doi: 10.1093/femsre/fuab016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring D.B., Johnson K.W., Henriksen E.J., Nuss J.M., Goff D., Kinnick T.R., et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 2003;52(3):588–595. doi: 10.2337/diabetes.52.3.588. [DOI] [PubMed] [Google Scholar]
- Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586(7827):113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinot R., Hubert M., de Melo G.D., Lazarini F., Bruel T., Smith N., et al. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat. Commun. 2021;12(1):4354. doi: 10.1038/s41467-021-24521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I., Merz A., Chiramel A., Lee J.-Y., Chlanda P., Haselman U., et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8(12) doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin P.S., Lötzerich M., Torta F., Tanner L.B., van Kuppeveld F.J.M., Wenk M.R., et al. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe. 2014;16(5):677–690. doi: 10.1016/j.chom.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Schirmer R.H., Coulibaly B., Stich A., Scheiwein M., Merkle H., Eubel J., et al. Methylene blue as an antimalarial agent. Redox Rep. 2003;8(5):272–275. doi: 10.1179/135100003225002899. [DOI] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12(541) doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy T.A., Smith P.G., Milhollen M.A., Berger A.J., Gavin J.M., Adhikari S., et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Soucy T.A., Smith P.G., Milhollen M.A., Berger A.J., Gavin J.M., Adhikari S., et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Srinivas T.R., Kaplan B., Meier-Kriesche H.-U. Mycophenolate mofetil in solid-organ transplantation. Expet Opin. Pharmacother. 2003;4(12):2325–2345. doi: 10.1517/14656566.4.12.2325. [DOI] [PubMed] [Google Scholar]
- Staatz C.E., Tett S.E. Clinical pharmacokinetics and pharmacodynamics of mycophenolate in solid organ transplant recipients. Clin. Pharmacokinet. 2007;46(1):13–58. doi: 10.2165/00003088-200746010-00002. [DOI] [PubMed] [Google Scholar]
- Suomalainen M., Greber U.F. Virus infection variability by single-cell profiling. Viruses. 2021;13(8):1568. doi: 10.3390/v13081568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M., Prasad V., Kannan A., Greber U.F. Cell-to-cell and genome-to-genome variability of adenovirus transcription tuned by the cell cycle. J. Cell Sci. 2020;134(5) doi: 10.1242/jcs.252544. jcs.252544. [DOI] [PubMed] [Google Scholar]
- Thao T.T.N., Labroussaa F., Ebert N., V’kovski P., Stalder H., Portmann J., et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020;582(7813):561–565. doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- Thoma G., Nuninger F., Falchetto R., Hermes E., Tavares G.A., Vangrevelinghe E., et al. Identification of a potent Janus kinase 3 inhibitor with high selectivity within the Janus kinase family. J. Med. Chem. 2011;54(1):284–288. doi: 10.1021/jm101157q. [DOI] [PubMed] [Google Scholar]
- Tonelli C., Chio I.I.C., Tuveson D.A. Transcriptional regulation by Nrf2. Antioxidants Redox Signal. 2018;29(17):1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt S., Schonberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., et al. scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Twilley D., Esmear T., Oosthuizen C.B., Reid A.-M., Nel M., et al. Anti-SARS-CoV natural products with the potential to inhibit SARS-CoV-2 (COVID-19) Front. Pharmacol. 2020;11:1514. doi: 10.3389/fphar.2020.561334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter-Sack I., Rengelshausen J., Oberwittler H., Burhenne J., Mueller O., Meissner P., et al. High absolute bioavailability of methylene blue given as an aqueous oral formulation. Eur. J. Clin. Pharmacol. 2009;65(2):179–189. doi: 10.1007/s00228-008-0563-x. [DOI] [PubMed] [Google Scholar]
- Ward D.J., Hammond E., Linden-Phillips L., Stevens A.J. Trends in clinical development timeframes for antiviral drugs launched in the UK, 1981–2014: a retrospective observational study. BMJ Open. 2015;5(11) doi: 10.1136/bmjopen-2015-009333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K.M., Rosales R., Yildiz S., Kehrer T., Miorin L., Moreno E., et al. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science. 2021;371(6532):926–931. doi: 10.1126/science.abf4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Li W., Peng G., Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. Unit. States Am. 2009;106(47):19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimovich A., Andriasyan V., Witte R., Wang I.-H., Prasad V., Suomalainen M., et al. Plaque2. 0—a high-throughput analysis framework to score virus-cell transmission and clonal cell expansion. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0138760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimovich A., Andriasyan V., Witte R., Wang I.H., Prasad V., Suomalainen M., et al. Plaque2.0-A high-throughput analysis framework to score virus-cell transmission and clonal cell expansion. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0138760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., et al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357(6377):420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zach C., Pock T., Bischof H., editors. A Duality Based Approach for Realtime TV-L1 Optical Flow. Springer Berlin Heidelberg; Berlin, Heidelberg: 2007. [Google Scholar]
- Zhang J.-H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhou X., Li Y., Xu Y., Lu K., Li P., et al. Inhibition of maternal embryonic leucine zipper kinase with OTSSP167 displays potent anti-leukemic effects in chronic lymphocytic leukemia. Oncogene. 2018;37(41):5520–5533. doi: 10.1038/s41388-018-0333-x. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Morgan M.A., Sun Y. Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxidants Redox Signal. 2014;21(17):2383–2400. doi: 10.1089/ars.2013.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Wang W., Aldahdooh J., Malyutina A., Shadbahr T., Pessia A., et al. SynergyFinder Plus: towards a better interpretation and annotation of drug combination screening datasets. bioRxiv. 2021 doi: 10.1101/2021.06.01.446564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 5Supplemental Movie 1: Ciliary beating in mock-treated, uninfected bronchial HAEEC. Bronchial HAEEC grown at ALI were mock-infected (day 0), and left untreated and uninfected. Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 6d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Multimedia component 6Supplemental Movie 2: Ciliary beating in DMSO-treated bronchial HAEEC. Bronchial HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 10 μM DMSO in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 6d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Multimedia component 7Supplemental Movie 3: Ciliary beating in Remdesivir-treated bronchial HAEEC. Bronchial HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 10 μM Remdesivir in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 6d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Multimedia component 8Supplemental Movie 4: Ciliary beating in MB-treated bronchial HAEEC. Bronchial HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 5 μM MB in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 6d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Multimedia component 9Supplemental Movie 5: Ciliary beating in MPA-treated bronchial HAEEC. Bronchial HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 10 μM MPA in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 6 d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Supplemental Movie 6: Ciliary beating in POS-treated nasal HAEEC. Nasal HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 50 μM POS in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 5d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Supplemental Movie 7: Ciliary beating in MLN4924-treated nasal HAEEC. Nasal HAEEC grown at ALI were inoculated apically with 1,000 FFU of SARS-CoV2 ‘Wuhan’ (day 0), and treated daily with 1 μM MLN4924 in the basolateral medium using a post-infection regimen (1d pi). Prior to imaging, the apical surface was washed with 200 μl of pre-warmed culture medium at 37°C for 20 min to remove mucus. Cilia beating was recorded 5d pi, in live mode at room temperature with an inverted microscope (Axiovert 135, Zeiss) at 50× magnification. Time span: 20 sec at 20 Hz.
Supplemental Table 1:Primers used for SARS-CoV-2 PCR reactions.
Supplemental Table 2: Threshold cycle comparison for SARS-CoV2 M and the Pol-gene RT-qPCR.
Supplemental Table 3: Overview of all compound hits. Chemical hit compounds from the screen in Huh7 cell infected with hCoV-229E-GFP are listed with their EC50 and TC50 concentrations, their clinical status and original treatment indication, effects in four cell lines against SARS-CoV-2 and their proposed mode of application.
Supplemental Table 4: Detailed presentation of compound effects on SARS-CoV-2 infection of HAEEC. The table presents all the virus titers as TCID50 per ml and genome copy numbers per ml in the apical supernatant from 610 distinct SARS-CoV2 infections of nasal and bronchial HAEEC treated with different compounds. Note, no measurable virus titer could be recorded in pilot experiments, and hence no basolateral measurements were conducted on a routine basis. The table also features the experiment ID, reference and the type of HAEEC, drug treatment (including concentrations and pre- or post-exposure regimen), the SARS-CoV2 genetic variants, and the sampling time pi. Virus titers and genomes numbers are expressed as log10 values. Data presented are annotated to the corresponding figure in the manuscript. Abbreviations: Yes (Y), No (N), ns (not show in the manuscript), NR (not realized).