Abstract
Radiotherapy constitutes a major therapeutic modality for early management of breast cancer. Despite the high efficacy in treating breast cancer (BC), radiation resistance and tumor recurrence are major hurdles in breast cancer radiotherapy. Herein, stable isotope labeling by amino acids in cell culture (SILAC) was employed, along with the parallel-reaction monitoring (PRM)-based targeted quantitative proteomic method, to examine the differences in kinase protein expression in MCF-7 and MDA-MB-231 breast cancer cells and their corresponding radioresistant C6 and C5 clones. We quantified the relative protein expression levels of 300 and 281 kinases in C5/MDA-MB-231 and C6/MCF-7 pairs of breast cancer cells, respectively. We also showed that TAF9, which was one of the differentially expressed kinases, enhances radiation resistance in breast cancer cells. Moreover, a correlation analysis of gene expression suggested TAF9’s role in upregulating the expression of genes involved with radioresistance. Overall, our study uncovered a large number of differentially expressed kinases accompanied with the acquisition of radioresistance and revealed a role of TAF9 in promoting radioresistance in breast cancer.
Keywords: quantitative proteomics, parallel-reaction monitoring, SILAC, kinase, kinome, breast cancer, radioresistance, TAF9
Graphical Abstract
INTRODUCTION
Breast cancer is among the most frequent malignancy in women.1 The global incidence of breast cancer has an annual growth rate of 3.1%, where the total number of cases were 641 000, 1.6 million, and almost 2 million in 1980, 2010, and 2017, respectively.2,3 In addition, breast cancer represents the fifth most common cause of cancer death in women.4
Multiple approaches have been employed for treating breast cancer, where the majority of early-stage patients undergo breast-conserving surgery with radiotherapy or mastectomy.5 Radiotherapy is a treatment method wherein high-energy γ rays introduce DNA damage and kill rapidly proliferating cancer cells. It has been evolving to improve precision in targeting the diseased tissue while minimizing radiation delivery to nearby vital organs like the heart and lungs.6 Despite the advances in radiation therapy, uniform radiation doses are typically applied without considering the variations among different subtypes of breast cancer.6 As a result, the benefits of this therapy may not be uniform across all patients owing to individual variations in sensitivity toward radiation therapy.6 Moreover, some patients may experience recurrences and develop radiation resistance, which confers poor prognosis and diminished quality of life.6 Thus, radioresistance presents a major obstacle to the current breast cancer radiotherapy,7 and the discovery of novel molecular targets that modulate radiosensitivity may provide a venue for improving therapeutic efficacy and for developing personalized radiotherapy.
Recent studies showed that dysregulations in expression and activity of kinases are associated with the acquisition of resistance toward breast cancer radiation therapy. For example, abnormal expression of maternal embryonic leucine zipper kinase (MELK)8 and ataxia-telangiectasia mutated (ATM)9 are known to be accompanied by breast cancer that are refractory toward radiation treatment. In addition, Guo et al.10 reported the application of a targeted proteomic method, relying on the utilization of isotope-labeled desthiobiotin-ATP acyl phosphate probe together with liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis in multiple-reaction monitoring (MRM) mode to reveal the roles of kinases in the acquisition of resistance toward radiation in MCF-7 breast cancer cells. However, the results from the ATP-affinity probe enrichment may be affected by not only the protein expression level but also the activity of a kinase, rendering it difficult to differentiate their individual contributions. In addition, the earlier version of the kinome profiling method only allowed for the quantifications of 120 unique kinases;10 thus, some low-abundance kinases associated with radioresistance may not be detected. Therefore, there is a need for in-depth profiling of differential expression of protein kinases that may contribute to the development of resistance to breast cancer radiotherapy.
Herein, we employed our previously developed parallel-reaction monitoring (PRM)-based LC-MS/MS method11–16 to examine the differential expression of kinase proteins in two pairs of breast cancer cell lines, i.e., the MDA-MB-231 and MCF-7 breast cancer cells and their corresponding radio-resistant subclones. Our results facilitated the measurements of the relative expression levels of 300 and 281 kinase proteins in the MDA-MB-231 and MCF-7 pairs of breast cancer cells, respectively. We also revealed a role of TAF9 in promoting acquired radiation resistance in breast cancer cells.
MATERIALS AND METHODS
Cell Culture
The radioresistant clone (clone 6 or C6) of MCF-7 cells was produced by treating the parental cells with a total of 60 Gy of γ rays and further exposed to an additional term of 30 fractions of γ rays.17,18 Similarly, MDA-MB-231 cells were exposed to a combined dose of 30 Gy of ionizing radiation to yield the radioresistant C5 clone.18,19
The details for cell culture conditions are provided in the Supporting Information. Approximately 5 × 106 cells were harvested by trypsinization, washed twice with cold phosphate-buffered saline (PBS), and lysed. The details are provided in the Supporting Information.
Tryptic Digestion of Protein Lysates
The whole cell lysates prepared from MCF-7 or MDA-MB-231 cells were combined with those of their radioresistant counter-parts at a mass ratio of 1:1 (determined by Bradford assay), denatured with 8 M urea, and subsequently treated with dithiothreitol and iodoacetamide for cysteine reduction and alkylation, respectively. The proteins were then digested overnight, in 50 mM NH4HCO3 (pH 8.5) and at 37 °C, with modified MS-grade trypsin (Pierce), where the enzyme/substrate ratio was 1:100. The resulting peptide mixture was dried in a Speed-vac, desalted using OMIX C18 pipette tips (Agilent Technologies), and analyzed by LC-MS/MS in the PRM mode.
Liquid Chromatography-Parallel Reaction Monitoring (LC-PRM) Analysis
All LC-PRM experiments were performed on a Q Exactive Plus quadrupole-Orbitrap mass spectrometer, where an EASY-nLC 1000 system (Thermo Fishier Scientific) was employed for the separation. Up to four most abundant unique peptides identified from shotgun proteomic analysis were included as precursor ions for each kinase protein. For each of these peptides, the six most abundant y ions from MS/MS for each peptide were chosen for peptide identification and quantification. A mass accuracy of 20 ppm or better for fragment ions was applied as a criterion during peptide identification in the Skyline platform. The resulting LC-MS/MS data were processed with Skyline (version 3.5) for extracted-ion chromatogram generation and peak area integration.20 The Supporting Information again provides the detailed experimental conditions.
Western Blot
The sample preparation procedures for Western blot analysis are described in the Supporting Information. The primary antibodies included human CHK1 (Cell Signaling Technology, 2360S, 1:2000 dilution) and TAF9 (Proteintech, 10544-1-AP, 1:2500 dilution). The secondary antibodies were horseradish peroxidase-conjugated antirabbit IgG and IRDye 680LT goat antimouse IgG. Membranes were also probed with anti-actin (Cell Signaling no. 4967, 1:10 000 dilution) or anti-GAPDH (Santa Cruz Biotechnology, sc-32233, 1:5000 dilution) antibody to verify equal loading of proteins.
TCGA, UALCAN, and GEPIA Data Analysis
The mRNA expression profiles and clinical data for the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC)21 cohort were retrieved using cBioPortal (http://www.cbioportal.org/data_sets.jsp).22 A total of 1137 patients in the METABRIC cohort with radiation therapy was employed for TCGA analysis. Median mRNA expression levels of genes encoding kinases were further employed as cutoff for categorizing breast cancer patients into low and high expression groups (568 and 569 patients, respectively). The log-rank test was employed to determine p-values, and a p < 0.05 was considered significant.
Box plots for kinase mRNA expression were generated using the gene expression data for cell lines derived from 114 normal breast tissues and 1097 breast cancer tissues using UALCAN (http://ualcan.path.uab.edu/index.html)23 The t-test was conducted with a PERL script in Comprehensive Perl Archive Network (CPAN) module “Statistics::TTest” (http://search.cpan.org/~yunfang/Statistics-TTest-1.1.0/TTest.pm).23 Correlation analysis of gene expression was conducted using GEPIA,24 where the expression of genes of interest in 1085 breast cancer tissues was analyzed. Pearson’s correlation coefficient was employed to calculate the distance metric.
Plasmid Construction and Clonogenic Survival Assay
The coding region of human TAF9 gene was amplified from a cDNA library prepared from mRNAs of HEK293T cells and cloned into the pRK7 plasmid after restriction digestion using XbaI and BamHI. The resulting pRK7-TAF9 plasmid was confirmed by Sanger sequencing.
MCF-7 and MDA-MB-231 cells were transfected with a control pRK7 vector or pRK7-TAF9 using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommended procedures. At 24 h following the transfection, the cells were seeded in triplicate in six-well plates at a concentration of 900 cells per well and incubated overnight. On the following day, the cells were exposed to 0, 1, 2, 3.5, or 5 Gy of X-rays, delivered by a Rad Source RS-2000 cabinet irradiator (Rad Source Technologies, Buford, GA). Following a 10 day incubation, cell colonies were fixed with a 6% glutaraldehyde solution for 1 h and subsequently stained for half an hour with 0.5% crystal violet. Those colonies with at least 50 cells were counted.
RESULTS AND DISCUSSION
Quantitative Interrogation of Differences in Expression of Kinase Proteins in Two Matched Pairs of Radioresistant/Parental Breast Cancer Cells
To unveil potential functions of kinases in acquired radioresistance, we employed a PRM-based targeted proteomic method,11–16 in combination with stable isotope labeling by amino acids in cell culture (SILAC),25 to screen for differentially expressed kinase proteins in two matched pairs of breast cancer cells, i.e., parental (WT) MCF-7 and MDA-MB-231 cells, as well as their corresponding radioresistant C6 and C5 clones (Figure 1a).
Figure 1.
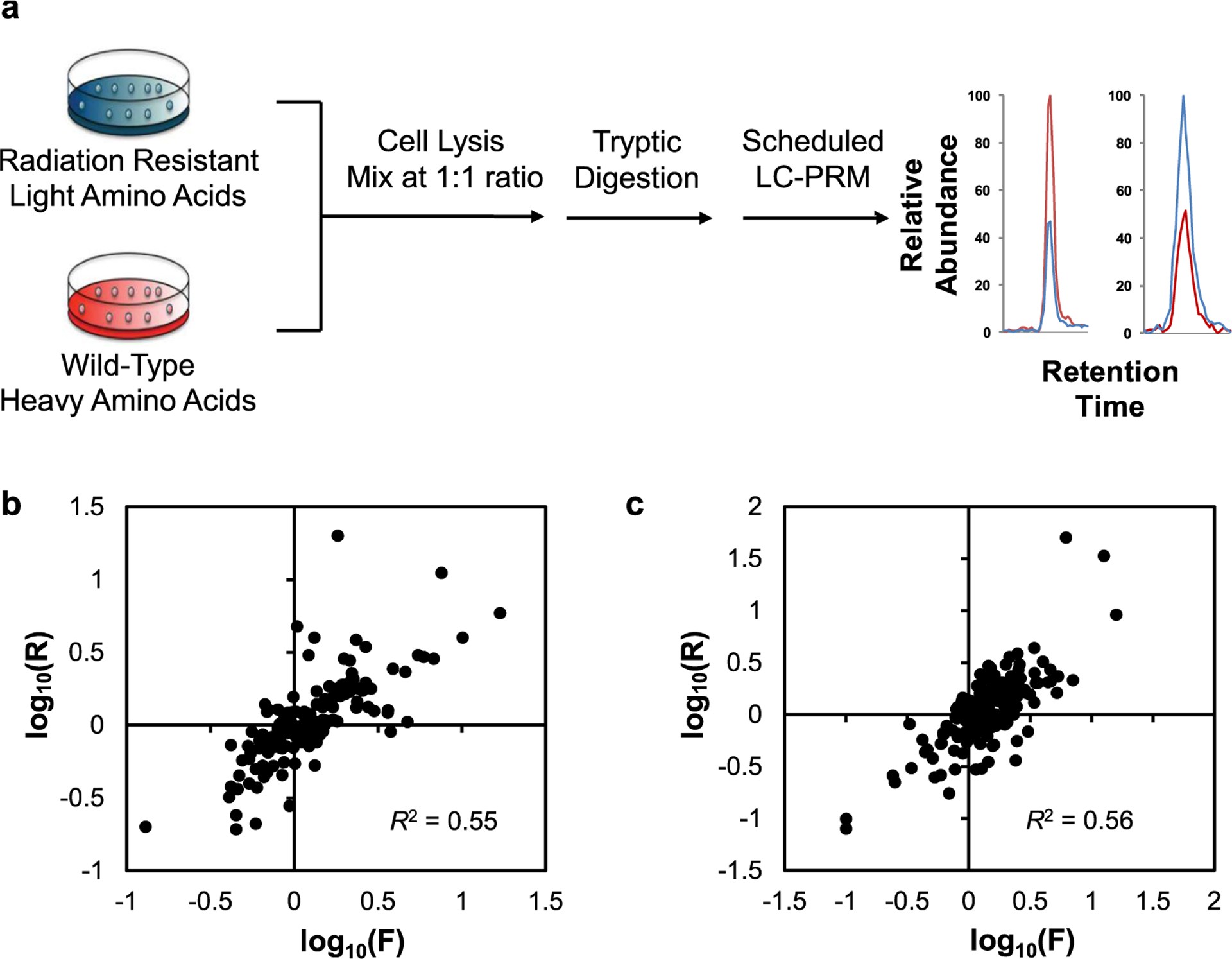
PRM-based targeted proteomic method for quantitative analyses of differences in kinase protein expression in two pairs of radioresistant/parental breast cancer cells. (a) A schematic diagram illustrating the workflow for the PRM method. (b, c) Scatter plots displaying the correlations between the ratios of kinase protein expression in C6/MCF-7 (b) and C5/MDA-MB-231 (c) pairs of breast cancer cells derived from LC-MS/MS analyses in conjunction with forward and reverse SILAC experiments.
The targeted proteomic analysis enabled the quantifications of 281 and 300 unique kinases in the MCF-7 WT/C6 and MDA-MB-231 WT/C5 pairs of breast cancer cells, respectively (Figures 1b,c and 2, and Table S1). All four to six PRM transitions selected for each peptide derived from kinases showed the same elution time and exhibited a dot product (dotp) value26 over 0.7. Additionally, over 90% of all of the quantified kinase proteins were detected in both forward and reverse SILAC experiments (Figure 1b,c and Table S1). Furthermore, the peptide ratios derived from forward and reverse SILAC labeling experiments showed linear fits with R2 of 0.55 and 0.56 (Figure 1b,c), where the variations may arise from biological heterogeneities and variabilities in sample preparation procedures. These results, therefore, revealed the robustness and reasonably good reproducibility of the PRM method.
Figure 2.
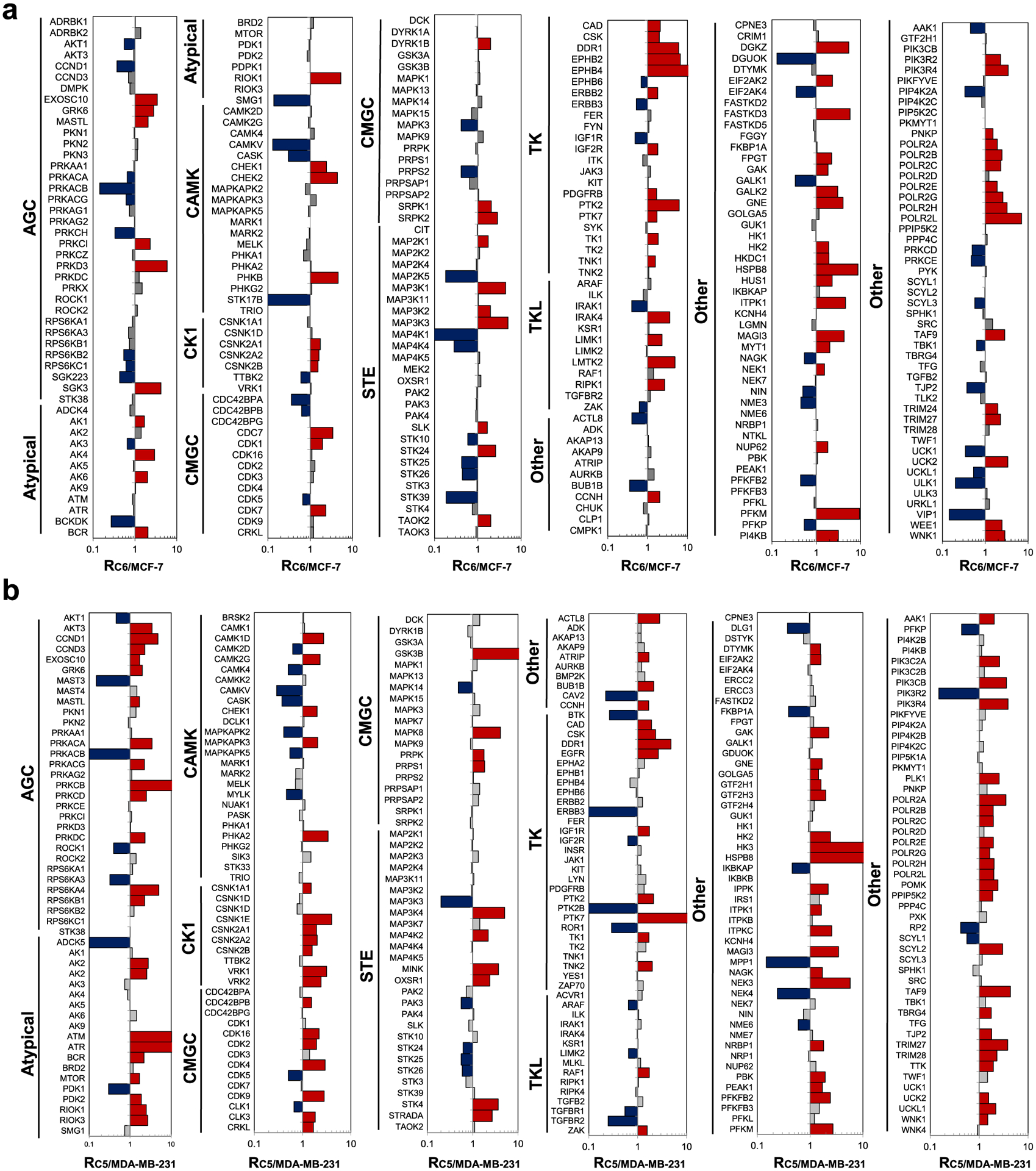
Differences in the expression of kinase proteins in C6/MCF-7 (a) and C5/MDA-MB-231 (b) pairs of breast cancer cells. The kinase protein expression data represent the average of results from a total four replicate experiments (two forward and two reverse SILAC experiments each). The kinase protein expression ratios measured from individual SILAC experiments are listed in Table S1.
Previously, Guo et al.10 employed desthiobiotin-ATP probe together with LC-MRM analysis to reveal the roles of kinases in the acquisition of radioresistance in MCF-7 cells. We next compared the reported MRM and our PRM data to assess the changes in protein expression levels and ATP-binding affinities of kinases upon acquisition of radioresistance in MCF-7 cells. We found that 87 kinases were commonly quantified in the two data sets (Figure 3a and Table S2). Kinase ratios (C6/MCF-7) derived from PRM and MRM analyses lacked apparent correlation (Figure 3b), suggesting that many kinases were with altered ATP-binding affinities during the acquisition of radioresistance. The comparison of the two data sets revealed that the perturbations of some kinases (e.g., CHK1 and CDK1) are due to protein expression alone, whereas the ATP-binding affinities of MAP4K4 and MAPK13 are induced profoundly during the acquisition of radioresistance (Figure 3b). This agrees with a recent study showing the role of MAP4K4 in regulating DNA double-strand break (DSB) repair.27 Along this line, we uncovered enhanced ATP-binding affinities of 10 kinases and diminished affinities of 15 kinases, which could be important in the acquisition of radioresistance in MCF-7 cells (Table S2).
Figure 3.
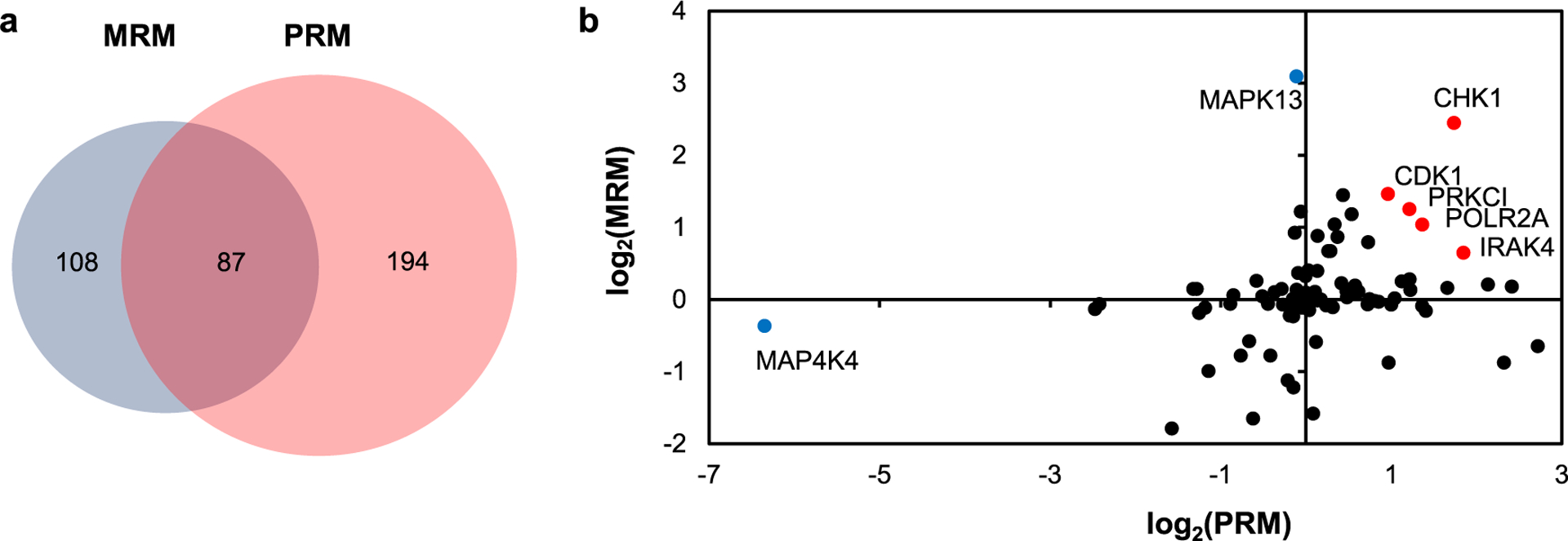
Comparison of quantification results acquired from PRM and MRM analyses. (a) Venn diagram displaying the number of overlapped and unique kinases quantified in the MRM and PRM analyses for the C6/MCF-7 pair of breast cancer cells. (b) Scatter plot displaying the correlation between the ratios of kinases obtained from MRM and PRM analysis in the C6/MCF-7 pair of breast cancer cells.
Altered Expressions of Kinase Proteins upon the Acquisition of Radioresistance in Breast Cancer Cells
The PRM method facilitated the quantifications of the relative expressions of 227 common kinases in the two pairs of breast cancer cells, which exhibited similar changes in kinome profiles upon the development of radioresistance (Figure 4a,b). In particular, 45 kinases were commonly altered by at least 1.5-fold in both lines of radioresistant breast cancer cells when compared to the corresponding parental lines (Figure 4c and Table 1). In this vein, it is of note that MDA-MB-231 and MCF-7 cells carry mutant and wild-type p53 gene, respectively;28 hence, we deduce that the acquired radioresistance-elicited changes in expression levels of these kinases are independent of p53 status in breast cancer cells. Moreover, several of these kinases were previously characterized to confer radiation resistance in cancer cells. For example, CHK1, which is involved in DNA-damage-checkpoint responses, is known to be associated with the acquisition of radioresistance in cancer cells.29,30 In addition, upregulation of HK2 was found to elicit radioresistance by increasing the glycolysis rate.31 Moreover, DDR1, a receptor tyrosine kinase, is able to regulate autophagy and ultimately modulate radiosensitivity in glioblastoma.32
Figure 4.
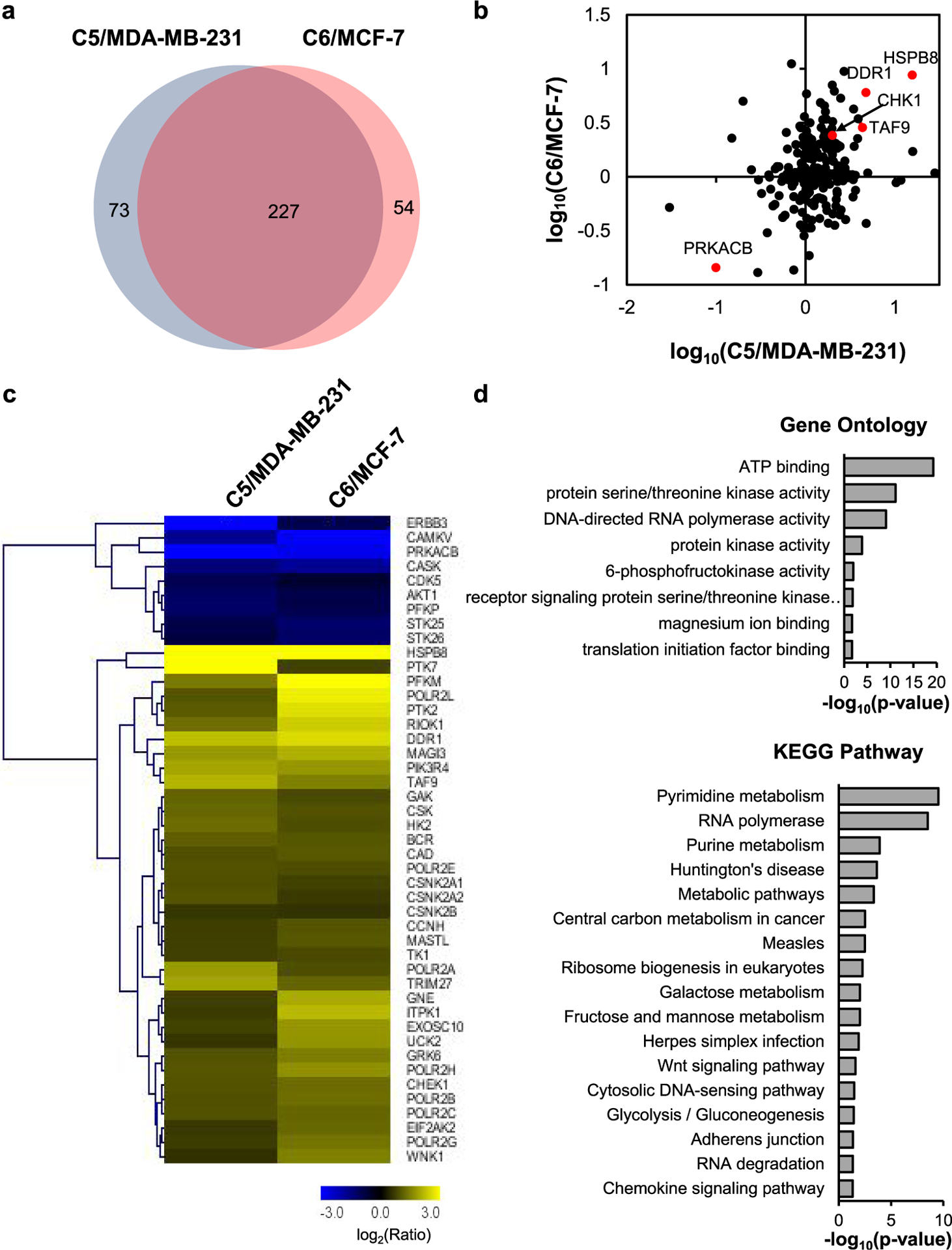
Comparison of the quantified kinases in the two pairs of breast cancer cells. (a) Venn diagram showing the overlap between quantified kinases from C5/MDA-MB-231 and C6/MCF-7 pairs of breast cancer cells. (b) Scatter plot displaying the correlation between the expression ratios of kinases obtained for the two pairs of radioresistant/parental (WT) cells. (c) Heatmap displaying the variations in the expression of the commonly altered kinases in the two pairs of radioresistant/WT breast cancer cell lines. Genes are clustered with Euclidean distance. The data represent the average values of the results obtained from two forward and two reverse SILAC experiments (see Table 1 for detailed ratios). (d) GO and KEGG pathway analyses of commonly perturbed kinases in the two matched pairs of radioresistant/parental breast cancer cell lines.
Table 1.
Relative Protein Expression Levels of Those Kinases that Are Altered by At Least 1.5-Fold in Both MCF-7 and MDA-MB-231 Pairs of Radioresistant/Parental Cellsa
| protein | C5/MDA-MB-231 | C6/MCF-7 |
|---|---|---|
| AKT1 | 0.46 ± 0.21 | 0.55 ± 0.14 |
| BCR | 2.17 | 2.05 ± 0.30 |
| CAD | 1.92 ± 0.39 | 2.08 ± 0.14 |
| CAMKV | 0.29 ± 0.03 | 0.13 |
| CASK | 0.37 | 0.30 ± 0.17 |
| CCNH | 1.67 ± 0.63 | 2.03 ± 0.21 |
| CDK5 | 0.51 ± 0.16 | 0.67 ± 0.11 |
| CHEK1 | 2.00 ± 0.14 | 2.42 ± 0.62 |
| CSK | 2.33 ± 0.84 | 1.96 ± 0.41 |
| CSNK2A1 | 1.95 ± 1.03 | 1.72 ± 0.08 |
| CSNK2A2 | 2.01 ± 0.20 | 1.61 ± 0.10 |
| CSNK2B | 1.58 ± 0.14 | 1.53 ± 0.26 |
| DDR1 | 4.76 | 6.01 ± 3.53 |
| EIF2AK2 | 1.60 ± 0.17 | 2.34 ± 0.52 |
| ERBB3 | 0.03 | 0.52 |
| EXOSC10 | 1.72 ± 0.35 | 3.41 ± 1.38 |
| GAK | 2.27 ± 0.55 | 1.84 ± 0.28 |
| GNE | 1.68 ± 0.6 | 4.00 |
| GRK6 | 1.98 | 2.80 ± 1.82 |
| HK2 | 2.44 ± 0.65 | 1.91 ± 0.34 |
| HSPB8 | 15.71 ± 11.95 | 8.75 ± 8.66 |
| ITPK1 | 1.64 ± 0.98 | 4.54 ± 1.80 |
| MAGI3 | 3.46 | 4.23 |
| MASTL | 1.70 ± 0.38 | 2.06 ± 0.27 |
| PFKM | 2.72 ± 0.69 | 9.45 ± 5.44 |
| PFKP | 0.43 ± 0.01 | 0.54 ± 0.05 |
| PIK3R4 | 3.90 ± 3.51 | 3.45 |
| POLR2A | 3.53 ± 1.58 | 1.92 ± 0.18 |
| POLR2B | 1.94 ± 0.68 | 2.46 ± 0.44 |
| POLR2C | 1.96 ± 0.10 | 2.30 ± 1.25 |
| POLR2E | 1.96 ± 0.39 | 1.90 ± 0.40 |
| POLR2G | 1.65 ± 0.07 | 2.58 ± 1.16 |
| POLR2H | 2.02 ± 0.90 | 3.24 ± 0.75 |
| POLR2L | 2.00 ± 0.46 | 7.07 ± 4.33 |
| PRKACB | 0.10 ± 0.00 | 0.14 |
| PTK2 | 2.11 ± 0.90 | 6.21 |
| PTK7 | 15.93 ± 13.90 | 1.71 ± 0.10 |
| RIOK1 | 2.48 ± 0.69 | 5.33 ± 2.00 |
| STK25 | 0.56 | 0.42 |
| STK26 | 0.59 ± 0.16 | 0.44 ± 0.20 |
| TAF9 | 4.35 | 2.85 |
| TK1 | 1.71 ± 0.64 | 1.84 ± 0.20 |
| TRIM27 | 3.85 | 2.26 ± 1.31 |
| UCK2 | 1.56 ± 0.43 | 3.38 ± 1.14 |
| WNK1 | 1.51 ± 0.33 | 2.85 ± 1.66 |
The values represent the mean ratio ± S.D. Some kinases were quantified in only one replicate; thus, no S.D. was reported for these kinases.
To understand further the molecular mechanisms through which the differentially expressed kinases may contribute to radioresistance, we carried out Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses (Figure 4d). Interestingly, RNA polymerases stand out in both GO and KEGG pathway analyses, while seven subunits of RNA polymerase II complex were commonly upregulated in the two radioresistant lines over the respective parental lines. Furthermore, analysis of The Cancer Genome Atlas (TCGA) data among those breast cancer patients with radiotherapy revealed that increased expression of one out of seven RNA polymerase II subunits, i.e., POLR2H, was associated positively with poorer survival of patients with breast cancer (Figure 5a and Table S3). Therefore, our results suggest that RNA polymerase II may promote the acquisition of radioresistance.
Figure 5.
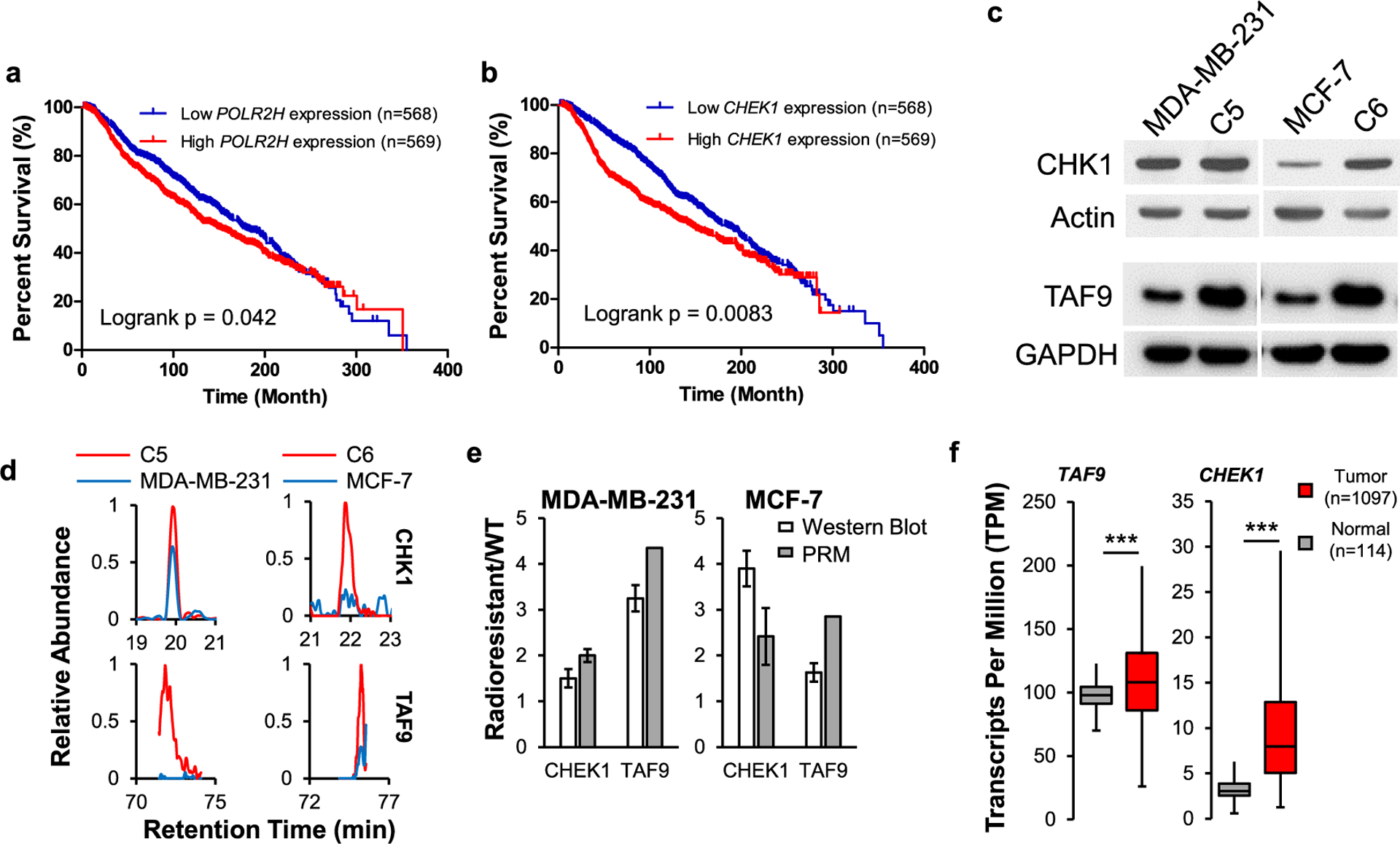
Differential expressions of CHK1 and TAF9 in acquired radioresistance. Patient survival correlates with the mRNA expression levels of POLR2H (a) and CHEK1 (b) genes in the METABRIC cohort with radiation therapy. The p-value was calculated using the log-rank test. (c) Western blot for verifying the relative expression levels of CHK1 and TAF9 proteins in the paired breast cancer cells. (d) PRM traces for quantifying CHK1 and TAF9 proteins. (e) Quantitative comparison of the ratios of CHK1 and TAF9 in the paired breast cancer cells obtained from PRM and Western blot analysis. The mean ± S.D. of the quantification results (n = 3) are shown. (f) Box-and-whisker plot illustrating the comparison of mRNA levels of CHEK1 and TAF9 genes in normal breast tissues and primary breast cancer tissues. Data were retrieved from 114 normal breast tissues and 1097 breast cancer tissues from UALCAN. Box plots represent the interquartile range (IQR), including minimum, 25th percentile, median, 75th percentile, and maximum values, with outliers being excluded from the plot.
TAF9 Drives the Acquisition of Radioresistance in Breast Cancer Cells
To uncover those differentially expressed kinases that modulate the acquisition of radioresistance in breast cancer cells, we compared our kinome quantification results with the mRNA levels of kinases in primary breast tumor tissues and normal tissues (Table S3). We found that the differential mRNA expressions of 25 out of 45 perturbed kinases were positively correlated with their protein expressions, including CHK1 and TAF9, while the expression of CHK1 was positively correlated with poorer survival of breast cancer patients (Figure 5b). Consistent with the PRM data, our Western blot results revealed increased expressions of CHK1 and TAF9 in the radioresistant breast cancer cells over the parental breast cancer cells (Figure 5c–e). These results also support the quantification accuracy of the PRM method.
We also observed higher mRNA expression levels of CHEK1, which encodes CHK1 kinase, and TAF9 genes in the breast tumors compared to normal tissues (Figure 5f). Since CHK1 is known to be involved in the DNA damage response signaling and correlated with radiosensitivity of cancer cells,29,30 we focused on examining the function of TAF9 in acquired radioresistance in breast cancer cells.
TAF9 is a component of multiple protein complexes, including the transcription factor IID (TFIID) complex and the TBP-free TAFII complex (TFTC).33 Because both the TFIID complex and TFTC are required for the transcriptional regulation modulated by RNA polymerase II, TAF9 may regulate RNA polymerase II-catalyzed transcription. As noted above, the protein expression levels of seven out of eight quantified subunits of RNA polymerase II were elevated in both radioresistant lines of breast cancer cells. Thus, TAF9 may assume a central role in the acquisition of radioresistance in breast cancer cells by regulating RNA polymerase II-based transcription.
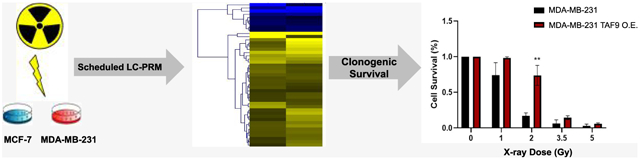
To assess directly the role of TAF9 in the acquisition of radioresistance, we overexpressed TAF9 gene in MCF-7 and MDA-MB-231 cells, and assessed its impact on the survival of breast cancer cells following exposure to X-rays. Results from our clonogenic survival assay showed increased survival of MCF-7 and MDA-MB-231 cells upon ectopic overexpression of TAF9 compared to transfection with empty vector control, with the most pronounced decrease being observed for cells exposed with 2 Gy of X-rays (Figure 6a,b).
Figure 6.
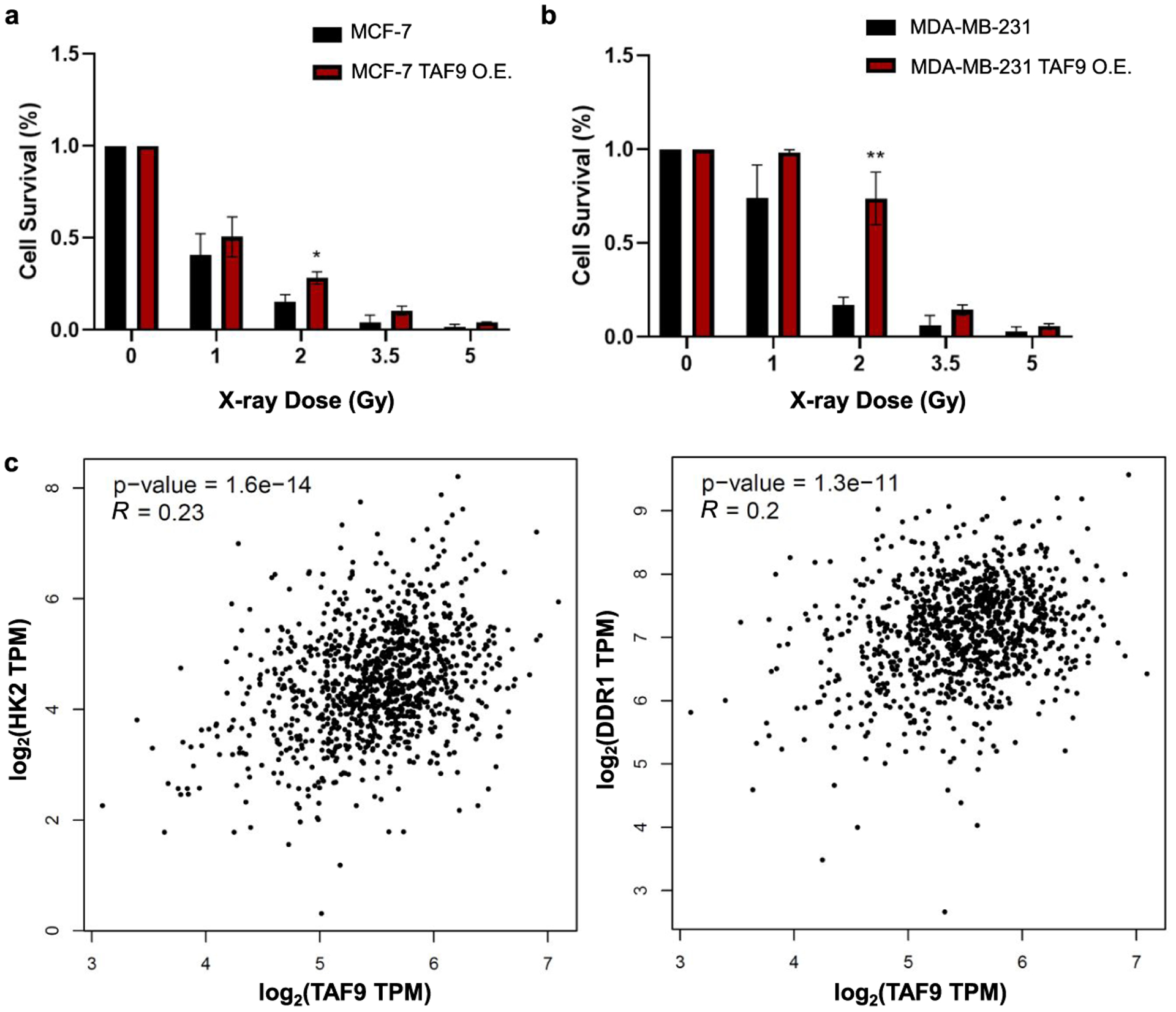
TAF9 modulates the acquisition of radiation resistance in breast cancer cells. (a-b) Relative cell survival of MCF-7 (a) and MDA-MB-231 (b) breast cancer cells with exposure to different doses of X-rays after ectopic overexpression TAF9 or transfection with an empty vector (control). Unpaired, two-tailed Student’s t-test was used to determine the p-values: *, 0.01 ≤ p < 0.05; **, 0.001 ≤ p < 0.01. (c) Correlations between mRNA expression levels of TAF9 and those of HK2 or DDR1 gene obtained from 1085 breast cancer patients.
As TAF9 binds directly to several transcription factors such as p5334,35 and VP16,36 thereby regulating gene transcription, we also asked if TAF9 regulates the transcription of radioresistant-related genes. For this purpose, we compared the mRNA expression levels of TAF9 and drivers of radioresistance in 1085 breast cancer tissues. HK2 and DDR1 are two known drivers that we identified to be upregulated in both radioresistant cell lines, as described above. The mRNA expression levels of these two genes are positively correlated with the mRNA level of TAF9 gene in breast cancer patients, indicating that TAF9 may assume a potential role in the upregulation of those drivers for radiation resistance (Figure 6c).
CONCLUSIONS
By applying a PRM-based targeted proteomic method, we investigated the alterations in kinase protein expression that are accompanied with acquired radioresistance in two pairs of breast cancer cells. With this method, we quantified 300 and 281 kinases in MDA-MB-231/C5 and MCF-7/C6 pairs of breast cancer cells, respectively. Among these kinases, 45 were commonly altered by at least 1.5-fold in both pairs of breast cancer cell lines. We also validated that one of upregulated kinases in the radioresistant breast cancer cells, TAF9, promotes the acquisition of radioresistance in cultured breast cancer cells. Moreover, gene correlation analysis suggested TAF9’s role in positively regulating the expression of other genes involved in radioresistance. Overall, our study revealed a new role of TAF9 in promoting the acquisition of radioresistance in breast cancer and uncovered a number of other kinases with potential functions in radioresistance.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R01 CA210072), and the authors would like to thank Prof. Jian-Jian Li for providing radioresistant breast cancer cells employed in this study.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.1c00075.
Uncropped Western blot images; experimental conditions for cell culture, LC-PRM analysis, and Western blot analysis (PDF)
Relative expression levels of kinases in MCF-7 and MDA-MB-231 pairs of radioresistant/parental breast cancer cells (Table S1) (XLSX)
Comparison between the ratios obtained from MRM and PRM analysis in C6/MCF-7 paired breast cancer cells (Table S2) (XLSX)
Correlation between the expressions of kinases and in MCF-7 and MDA-MB-231 radioresistant/parental cells and the survival among the patients with radiotherapy or mRNA expression in breast cancer tissues versus normal breast tissues (Table S3) (XLSX)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jproteome.1c00075
The authors declare no competing financial interest.
All of the raw files for LC-PRM analyses of kinases for the two paired radioresistant breast cancer cells were deposited into PeptideAtlas with the identifier number of PASS01644 (http://www.peptideatlas.org/PASS/PASS01644).
Contributor Information
Weili Miao, Department of Chemistry, University of California, Riverside, California 92521-0403, United States.
David Bade, Environmental Toxicology Graduate Program, University of California, Riverside, California 92521-0403, United States.
Yinsheng Wang, Department of Chemistry and Environmental Toxicology Graduate Program, University of California, Riverside, California 92521-0403, United States.
REFERENCES
- (1).Harbeck N; Penault-Llorca F; Cortes J; Gnant M; Houssami N; Poortmans P; Ruddy K; Tsang J; Cardoso F Breast cancer. Nat. Rev. Dis. Primers 2019, 5, No. 66. [DOI] [PubMed] [Google Scholar]
- (2).Bray F; Ferlay J; Laversanne M; Brewster DH; Gombe Mbalawa C; Kohler B; Piñeros M; Steliarova-Foucher E; Swaminathan R; Antoni S; Soerjomataram I; Forman D Cancer incidence in five continents: inclusion criteria, highlights from volume X and the global status of cancer registration. Int. J. Cancer 2015, 137, 2060–2071. [DOI] [PubMed] [Google Scholar]
- (3).Li N; Deng Y; Zhou L; Tian T; Yang S; Wu Y; Zheng Y; Zhai Z; Hao Q; Song D; Zhang D; Kang H; Dai Z Global burden of breast cancer and attributable risk factors in 195 countries and territories, from 1990 to 2017: results from the global burden of disease study 2017. J. Hematol. Oncol 2019, 12, No. 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Sullivan R; Peppercorn J; Sikora K; Zalcberg J; Meropol NJ; Amir E; Khayat D; Boyle P; Autier P; Tannock IF; Fojo T; Siderov J; Williamson S; Camporesi S; McVie JG; Purushotham AD; Naredi P; Eggermont A; Brennan MF; Steinberg ML; De Ridder M; McCloskey SA; Verellen D; Roberts T; Storme G; Hicks RJ; Ell PJ; Hirsch BR; Carbone DP; Schulman KA; Catchpole P; Taylor D; Geissler J; Brinker NG; Meltzer D; Kerr D; Aapro M Delivering affordable cancer care in high-income countries. Lancet Oncol. 2011, 12, 933–980. [DOI] [PubMed] [Google Scholar]
- (5).Moo T-A; Sanford R; Dang C; Morrow M Overview of breast cancer therapy. PET Clin. 2018, 13, 339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Choi J; Yoon YN; Kim N; Park CS; Seol H; Park I-C; Kim H-A; Noh WC; Kim J-S; Seong M-K Predicting radiation resistance in breast cancer with expression status of phosphorylated S6K1. Sci. Rep 2020, 10, No. 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yadav P; Shankar BS Radio resistance in breast cancer cells is mediated through TGF-β signalling, hybrid epithelial-mesenchymal phenotype and cancer stem cells. Biomed. Pharmacother 2019, 111, 119–130. [DOI] [PubMed] [Google Scholar]
- (8).Speers C; Zhao SG; Kothari V; Santola A; Liu M; Wilder-Romans K; Evans J; Batra N; Bartelink H; Hayes DF; Lawrence TS; Brown PH; Pierce LJ; Feng FY Maternal embryonic leucine zipper kinase (MELK) as a novel mediator and biomarker of radioresistance in human breast cancer. Clin. Cancer Res 2016, 22, 5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bian L; Meng Y; Zhang M; Guo Z; Liu F; Zhang W; Ke X; Su Y; Wang M; Yao Y; Wu L; Li D ATM expression is elevated in established radiation-resistant breast cancer cells and improves DNA repair efficiency. Int. J. Biol. Sci 2020, 16, 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Guo L; Xiao Y; Fan M; Li JJ; Wang Y Profiling global kinome signatures of the radioresistant MCF-7/C6 breast cancer cells using MRM-based targeted proteomics. J. Proteome Res 2015, 14, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Miao W; Guo L; Wang Y Imatinib-induced changes in protein expression and ATP-binding affinities of kinases in chronic myelocytic leukemia cells. Anal. Chem 2019, 91, 3209–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Miao W; Li L; Liu X; Qi TF; Guo L; Huang M; Wang Y A targeted quantitative proteomic method revealed a substantial reprogramming of kinome during melanoma metastasis. Sci. Rep 2020, 10, No. 2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Miao W; Li L; Wang Y High-throughput targeted quantitative analysis of the interaction between HSP90 and kinases. Anal. Chem 2019, 91, 11507–11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Miao W; Wang Y Targeted quantitative kinome analysis identifies PRPS2 as a promoter for colorectal cancer metastasis. J. Proteome Res 2019, 18, 2279–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Miao W; Wang Y Quantitative interrogation of the human kinome perturbed by two BRAF inhibitors. J. Proteome Res 2019, 18, 2624–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Miao W; Yuan J; Li L; Wang Y Parallel-reaction monitoring-based proteome-wide profiling of differential kinase protein expression during prostate cancer metastasis in vitro. Anal. Chem 2019, 91, 9893–9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ahmed KM; Dong S; Fan M; Li JJ Nuclear factor-κB p65 inhibits mitogen-activated protein kinase signaling pathway in radioresistant breast cancer cells. Mol. Cancer Res 2006, 4, No. 945. [DOI] [PubMed] [Google Scholar]
- (18).Miao W; Fan M; Huang M; Li JJ; Wang Y Targeted profiling of heat shock proteome in radioresistant breast cancer cells. Chem. Res. Toxicol 2019, 32, 326–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cao N; Li S; Wang Z; Ahmed KM; Degnan ME; Fan M; Dynlacht JR; Li JJ NF-κB-mediated HER2 overexpression in radiation-adaptive resistance. Radiat. Res 2009, 171, 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).MacLean B; Tomazela DM; Shulman N; Chambers M; Finney GL; Frewen B; Kern R; Tabb DL; Liebler DC; MacCoss MJ Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Curtis C; Shah SP; Chin S-F; Turashvili G; Rueda OM; Dunning MJ; Speed D; Lynch AG; Samarajiwa S; Yuan Y; Gräf S; Ha G; Haffari G; Bashashati A; Russell R; McKinney S; METABRIC Group; Langerød A; Green A; Provenzano E; Wishart G; Pinder S; Watson P; Markowetz F; Murphy L; Ellis I; Purushotham A; Børresen-Dale A-L; Brenton JD; Tavaré S; Caldas C; Aparicio S The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Gao J; Aksoy BA; Dogrusoz U; Dresdner G; Gross B; Sumer SO; Sun Y; Jacobsen A; Sinha R; Larsson E; Cerami E; Sander C; Schultz N Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signaling 2013, 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Chandrashekar DS; Bashel B; Balasubramanya SAH; Creighton CJ; Ponce-Rodriguez I; Chakravarthi BVSK; Varambally S UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Tang Z; Li C; Kang B; Gao G; Li C; Zhang Z GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ong S-E; Blagoev B; Kratchmarova I; Kristensen DB; Steen H; Pandey A; Mann M Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 2002, 1, 376–386. [DOI] [PubMed] [Google Scholar]
- (26).de Graaf EL; Altelaar AF; van Breukelen B; Mohammed S; Heck AJ Improving SRM assay development: a global comparison between triple quadrupole, ion trap, and higher energy CID peptide fragmentation spectra. J. Proteome Res 2011, 10, 4334–4341. [DOI] [PubMed] [Google Scholar]
- (27).Deng M; Lin J; Nowsheen S; Liu T; Zhao Y; Villalta PW; Sicard D; Tschumperlin DJ; Lee S; Kim J; Lou Z Extracellular matrix stiffness determines DNA repair efficiency and cellular sensitivity to genotoxic agents. Sci. Adv 2020, 6, No. eabb2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Olivier M; Eeles R; Hollstein M; Khan MA; Harris CC; Hainaut P The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat 2002, 19, 607–614. [DOI] [PubMed] [Google Scholar]
- (29).Biau J; Chautard E; Verrelle P; Dutreix M Altering DNA repair to improve radiation therapy: specific and multiple pathway targeting. Front. Oncol 2019, 9, No. 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wang W-J; Wu S-P; Liu J-B; Shi Y-S; Huang X; Zhang Q-B; Yao K-T MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells. Cancer Res. 2013, 73, 1219. [DOI] [PubMed] [Google Scholar]
- (31).Zhong J-T; Zhou S-H Warburg effect, hexokinase-II, and radioresistance of laryngeal carcinoma. Oncotarget 2017, 8, 14133–14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Vehlow A; Klapproth E; Jin S; Hannen R; Hauswald M; Bartsch J-W; Nimsky C; Temme A; Leitinger B; Cordes N Interaction of discoidin domain receptor 1 with a 14-3-3-beclin-1-Akt1 complex modulates glioblastoma therapy sensitivity. Cell Rep. 2019, 26, 3672.e7–3683.e7. [DOI] [PubMed] [Google Scholar]
- (33).Frontini M; Soutoglou E; Argentini M; Bole-Feysot C; Jost B; Scheer E; Tora L TAF9b (formerly TAF9L) is a bona fide TAF that has unique and overlapping roles with TAF9. Mol. Cell. Biol 2005, 25, 4638–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Uesugi M; Verdine GL The α-helical FXXΦΦ motif in p53: TAF interaction and discrimination by MDM2. Proc. Natl. Acad. Sci. U.S.A 1999, 96, 14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Yoon JW; Lamm M; Iannaccone S; Higashiyama N; Leong KF; Iannaccone P; Walterhouse D p53 modulates the activity of the GLI1 oncogene through interactions with the shared coactivator TAF9. DNA Repair 2015, 34, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Uesugi M; Nyanguile O; Lu H; Levine AJ; Verdine GL Induced α helix in the VP16 activation domain upon binding to a human TAF. Science 1997, 277, 1310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


