Abstract
Few data are available on the molecular subtypes of all penicillin-nonsusceptible Streptococcus pneumoniae (PNSP) from a defined population base. Pulsed-field gel electrophoresis (PFGE), serotyping, and antibiotic susceptibility testing were performed for all available invasive PNSP isolates for which the penicillin (MIC) was ≥0.1 μg/ml from Baltimore, Md., during 1995–1996 (n = 143). The dendrogram analysis of PFGE patterns included 32 distinct clonal groups. Six major clonal groups included two-thirds of the PNSP strains. Major clonal groups 2, 3, 4, and 6 strains were genetically related to four previously described international clones and were all multidrug resistant. Major clonal group 3 was genetically related to the Tennessee23F-4 clone and contained all four strains for which the penicillin MIC was 8 μg/ml. Most of the clonal group 1 and 5 strains had intermediate susceptibility to penicillin and were rarely multidrug resistant. The latter clonal groups represent two previously undescribed penicillin-intermediate pneumococcal clones. Clonal group homogeneity was greater for serotype 9V, 19A, and 23F strains than for serotype 6A, 6B, 14, and 19F strains. The classification of PNSP strains into clonal groups is essential for future population-based epidemiologic studies of PNSP.
Streptococcus pneumoniae is responsible for an estimated 50,000 cases of bacteremia, 3,000 cases of meningitis, 7 million cases of otitis media, and several hundred thousand cases of pneumonia in the United States each year (4, 20, 32, 34). The overall yearly incidence of pneumococcal bacteremia is estimated to be 15 to 35 cases per 100,000 population (2, 3, 14, 17, 18).
Penicillin-nonsusceptible S. pneumoniae (PNSP) was uncommon in the United States until the 1990s, when the rates of antibiotic resistance rapidly increased. Although 90 serotypes of S. pneumoniae exist (15), a small number of serotypes account for the majority of PNSP (17). The emergence of PNSP in the United States appears to be partially related to the dissemination of multidrug-resistant (MDR) international pneumococcal clones (6–8, 10, 13, 19, 21–23, 26, 30).
Recent molecular studies of PNSP in the United States have shown that the majority of strains can be subtyped into less than 10 clonal groups by pulsed-field gel electrophoresis (PFGE) (9, 12). These studies included a sample of isolates for which the penicillin MIC is ≥1 μg/ml from various areas of the United States.
The purpose of the present study was to characterize the phenotypic characteristics of PNSP isolates associated with invasive disease in a defined population base to facilitate our ongoing studies of the epidemiology of PNSP.
MATERIALS AND METHODS
Active surveillance for invasive pneumococcal infection was initiated in the Baltimore Metropolitan Area (BMA) on 1 January 1995 as part of the Maryland Bacterial Invasive Disease Surveillance project (BIDS) (14). BIDS is the Active Bacterial Core Surveillance component of the multistate Emerging Infections Program Network that is coordinated by the Centers for Disease Control and Prevention (CDC). BMA, with a population of 2.5 million, comprises Baltimore City and Baltimore, Anne Arundel, Carroll, Harford, and Howard Counties. The surveillance case definition is the isolation of S. pneumoniae from a normally sterile body fluid from a BMA resident of any age. All laboratories based in acute-care hospitals in BMA participate, as do other microbiology laboratories that process blood cultures. For each eligible patient, the hospital infection control professional completes a one-page case report form, which includes demographic (e.g., gender, age, and race) and brief clinical information, and the bacterial isolate is submitted for species confirmation and MIC testing. Biweekly telephone calls are made to hospital infection control practitioners to ascertain cases not reported spontaneously. Periodic laboratory audits are performed to identify unreported cases.
Bacterial isolates and antibiotic susceptibility.
Available BIDS pneumococcal isolates for which the penicillin MIC is ≥0.1 μg/ml isolated during 1995 and 1996 were included in the present study. The MICs of penicillin, cefotaxime, erythromycin, tetracycline, trimethoprim-sulfamethoxazole (TMP-SXZ), clindamycin, ofloxocin, and vancomycin were determined by broth microdilution testing by a CDC contract laboratory by methods recommended by the National Committee for Clinical Laboratory Standards (27). Pneumococcal serotypes were determined by the latex agglutination test and were confirmed by the Quellung reaction with type-specific antiserum prepared at CDC. Eleven PNSP international clones obtained from the American Type Culture Collection were included for comparison: Spain23F-1 (strain Sp264, ATCC 700669) (8, 21), Spain6B-2 (strain GM17, ATCC 700670) (6), France9V-3 (strain TL7, ATCC 700671) (22), Tennessee23F-4 (strain SP196, ATCC 51916) (7, 23), Spain14-5 (strain VH14, ATCC 700672) (6), Hungary19A-6 (strain HUN663, ATCC 700673) (10, 26), South Africa19A-7 (strain 17619, ATCC 700674) (30), South Africa6B-8 (strain 50803, ATCC 700675) (30), England14-9 (strain PN93/872/B, ATCC 700676) (13), Slovakia14-10 (strain 29055, ATCC 700677) (19), and Slovakia19A-11 (strain 6571, ATCC 700678) (10).
Isolates for which the penicillin MIC was between 0.1 and 1 μg/ml were defined as penicillin intermediate (Peni), and those for which the penicillin MIC was ≥2.0 μg/ml were defined as penicillin resistant (Penr). The Peni and Penr strains were collectively defined as PNSP. A strain was defined as MDR if it was nonsusceptible to at least two of the following antibiotics: penicillin and/or cefotaxime, erythromycin, TMP-SXZ, tetracycline, ofloxacin, and chloramphenicol.
PFGE.
Two PFGE protocols that resulted in identical banding patterns were used (24, 25). A simplified protocol deleted the lysis step, eliminated proteinase K, and required shorter incubation times (24). Equal amounts of bacterial suspension and 2% low-melting-temperature agarose (Sea Plaque; FMC Bioproducts, Rockland, Maine) were mixed and pipetted into 100-μl plug molds. After solidification on ice for 10 min, the plugs were incubated in lysis enzymes and buffer: 2 ml of buffer (1 M NaCl, 100 mM EDTA, 6 mM Tris-HCl, 0.5% Brij 58, 0.5% deoxycholate, 0.5% N-lauroyl sarcosine [pH 7.6]) supplemented with 1 mg of lysozyme per ml and 50 μg of RNase A per ml for 3 h at 37°C. Each plug was incubated with 2 ml of ES buffer (0.5 M EDTA, 1% N-lauroyl sarcosine [pH 8.5 to 9.3]) and 100 μg of proteinase K per ml for 6 to 18 h at 50°C. The plugs were washed three times with 10 ml of TE buffer containing 10 mM Tris-HCl and 1 mM EDTA (pH 7.6) at 37°C for 15 to 30 min. After preincubation of a plug (2 by 10 mm) in buffer (NE #4; New England BioLabs, Inc., Beverly, Mass.) for 20 min, the DNA was digested with buffer (NE #4) mixed with 30 U of SmaI and 200 μg of bovine serum albumin per ml at room temperature for 3 to 18 h. Each section of plug (2 by 5 mm) was loaded into a 1% agarose gel. PFGE was performed with a contour-clamped electrophoretic field gel apparatus (DRIII) under the following conditions: pulse times, 1 to 30 s for 18 h and 5 to 9 s for 8 h; 198 V; flow rate, 1 liter/min; temperature, 14°C. After the gel was stained with ethidium bromide, the image was digitized on a Gel Doc 2000 System (Bio-Rad, Hercules, Calif.).
Statistical analysis.
Data were analyzed with Epi Info, version 6.04, software (CDC). The χ2 and Fisher exact tests were used for the analysis of dichotomous variables. The genetic relatedness of strains was determined by analyzing the PFGE patterns with Molecular Analyst/Multi-Analyst computer programs (Bio-Rad). Dendrograms were created by use of the unweighted pair group method with arithmetic averages, the Dice coefficient, and a position tolerance of 1.5%. The Molecular Analyst cophenetic correlation was calculated for all dendrograms. The cophenetic correlation indicates the degree of correlation between the computer-derived degree of genetic relatedness and the visual display by a dendrogram. A minimum 70% correlation is necessary to ensure that the dendrogram faithfully represents the actual degree of genetic relatedness between strains. A PFGE-based clonal group was defined as a group of isolates with genetically related PFGE patterns. The clonal groups were determined by use of the criteria of Tenover et al. (33) and the dendrogram-derived degree of genetic relatedness. In general, the PFGE patterns of strains categorized within a clonal group had six or fewer differences from each other (33) and ≥80% genetic relatedness on the dendrogram. Strains were defined as genetically related to an international clone if their PFGE patterns differed by six or fewer bands from the PFGE patterns of the respective clones. Six clonal groups had five or more strains per group; these were defined as major clonal groups and were enumerated as groups 1 to 6. Twenty-six remaining clonal groups had less than five strains per group; these were defined as minor clonal groups and were enumerated as groups 7 to 32.
To determine the degree of clonal group homogeneity for each serotype, we summarized the distribution of the number of clonal groups for each individual serotype using the formula (X12 + X22 + … Xn2)/N2, where X denotes the number of isolates for each clonal group and N represents the total number of isolates. This formula was derived by weighting the frequency distribution among the clonal groups according to the number of isolates within each clonal group by use of the formula X1(X1/N) + X2(X2/N) + … Xn(Xn/N) and then adjusting for the sample size by dividing by the total number of isolates (N) for that serotype.
To determine whether differences in antimicrobial susceptibility patterns were independent of serotype, we controlled for serotype in the analysis. For the analysis of the antimicrobial susceptibilities of the six major clonal groups, clonal group 1 was compared to clonal group 2 and clonal group 3 was compared to clonal group 4. The majority of the strains in clonal groups 1 and 2 were serotype 9V, and all of the strains in major clonal groups 3 and 4 were serotype 23F. All of the clonal group 5 and 6 strains were serotype 19A and serotype 6B, respectively. No other major clonal group contained primarily serotype 19A or 6B strains. Therefore, the strains in clonal groups 5 and 6 were compared to nonclonal group 5 serotype 19A and nonclonal group 6 serotype 6B isolates, respectively.
RESULTS
Study isolates.
A total of 1,412 patients with invasive pneumococcal infection were reported from 1 January 1995 to 31 December 1996. The pneumococcal isolates were available for 1,136 (80.5%) of these patients, of which 86 (7.6%) and 81 (7.1%) were found to be Peni and Penr, respectively. Of the 167 PNSP isolates, 143 (86%) were available for subtyping.
Of the 143 PNSP isolates, 140 (98%) were from blood and <1% each were from pleural fluid, joint fluid, and cerebrospinal fluid. Blacks between the ages of 5 and 64 years with PNSP infections had a lower proportion of Penr isolates (38.5%) than whites of the same age (64.3%) (P = 0.04). Twenty-eight (40.6%) of the isolates from patients from Baltimore City were resistant to penicillin whereas 44 (59.5%) of the isolates from patients from the remaining BMA counties were resistant to penicillin (P = 0.02) (Table 1).
TABLE 1.
Patient characteristics and serotype distribution of isolates from patients with invasive pneumococcal infection due to Peni and Penr strains
| Characteristic | No. (row %) of patients
|
P value | |
|---|---|---|---|
| Intermediate (n = 71) | Resistant (n = 72) | ||
| Age (yr) | |||
| 0–4 | |||
| White | 5 (33.3) | 10 (66.7) | 0.55 |
| Black | 7 (43.8) | 9 (56.3) | |
| 5–64 | |||
| White | 10 (35.7) | 18 (64.3) | 0.04 |
| Black | 24 (61.5) | 15 (38.5) | |
| ≥65 yrs | |||
| White | 21 (56.8) | 16 (43.2) | 0.76 |
| Black | 3 (50) | 3 (50) | |
| Gender | |||
| Male | 46 (52.3) | 42 (47.7) | 0.43 |
| Female | 25 (45.5) | 30 (54.5) | |
| Place of residence | |||
| Baltimore City | 41 (59.4) | 28 (40.6) | 0.02 |
| Rest of BMA | 30 (40.5) | 44 (59.5) | |
| Serotype | |||
| 6A | 2 (28.6) | 5 (71.4) | <0.01 |
| 6B | 7 (50) | 7 (50) | |
| 9V | 15 (38.5) | 24 (61.5) | |
| 14 | 4 (36.4) | 7 (63.6) | |
| 19A | 25 (92.6) | 2 (7.4) | |
| 19F | 4 (57.1) | 3 (42.9) | |
| 23F | 9 (28.1) | 23 (71.9) | |
| Othera | 5 (83.3) | 1 (16.7) | |
Serotypes include three nontypeable isolates one isolate each of serotypes 9N, 12F, and 35B. All except the serotype 35B isolates were Peni.
Clonal groups.
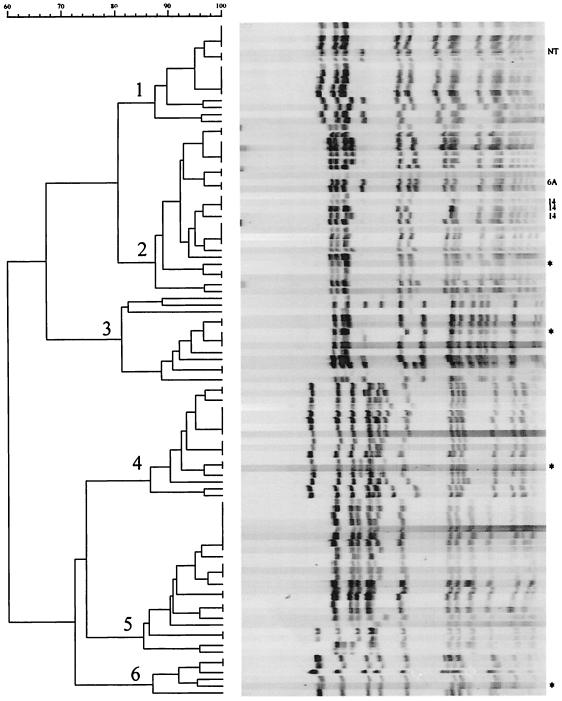
The 143 PNSP strains were classified into 32 clonal groups (Table 2). The six major clonal groups (Fig. 1) accounted for 95 (66.4%) of the PNSP isolates. The remaining 48 (33.6%) strains were categorized into 26 minor clonal groups.
TABLE 2.
Distribution of clonal groups of PNSP strains in Baltimore
| Clonal group | No. (%) of isolates per clonal group:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6A | 6B | 9N | 9V | 12F | 14 | 19A | 19F | 23F | 35B | NTe | Total | |
| 1 | 14 | 1 | 15 (10.5) | |||||||||
| 2 | 1 | 20a | 3 | 24 (16.8) | ||||||||
| 3 | 13b | 13 (9.1) | ||||||||||
| 4 | 16c | 16 (11.2) | ||||||||||
| 5 | 22 | 22 (15.4) | ||||||||||
| 6 | 5d | 5 (3.5) | ||||||||||
| 7 | 2 | 2 (1.4) | ||||||||||
| 8 | 1 | 1 | 2 (1.4) | |||||||||
| 9 | 4 | 4 (2.8) | ||||||||||
| 10 | 1 | 2 | 3 (2.1) | |||||||||
| 11 | 1 | 1 (0.7) | ||||||||||
| 12 | 1 | 1 (0.7) | ||||||||||
| 13 | 1 | 1 (0.7) | ||||||||||
| 14 | 3 | 3 (2.1) | ||||||||||
| 15 | 2 | 2 (1.4) | ||||||||||
| 16 | 1 | 1 | 2 (1.4) | |||||||||
| 17 | 2 | 2 (1.4) | ||||||||||
| 18 | 4 | 4 (2.8) | ||||||||||
| 19 | 2 | 2 (1.4) | ||||||||||
| 20 | 2 | 2 (1.4) | ||||||||||
| 21 | 1 | 1 (0.7) | ||||||||||
| 22 | 1 | 1 (0.7) | ||||||||||
| 23 | 1 | 1 (0.7) | ||||||||||
| 24 | 1 | 1 (0.7) | ||||||||||
| 25 | 1 | 1 (0.7) | ||||||||||
| 26 | 1 | 1 (0.7) | ||||||||||
| 27 | 1 | 1 (0.7) | ||||||||||
| 28 | 1 | 1 (0.7) | ||||||||||
| 29 | 1 | 1 (0.7) | ||||||||||
| 30 | 3 | 3 (2.1) | ||||||||||
| 31 | 2 | 2 (1.4) | ||||||||||
| 32 | 3 | 3 (2.1) | ||||||||||
| Total | 7 (4.8) | 14 (9.8) | 1 (0.7) | 39 (27.2) | 1 (0.7) | 11 (7.7) | 27 (18.9) | 7 (4.8) | 32 (22.4) | 1 (0.7) | 3 (2.1) | 143 |
| Clonal group homogeneity | 0.18 | 0.26 | 1.00 | 0.40 | 1.00 | 0.26 | 0.68 | 0.22 | 0.42 | 1.00 | 0.33 | |
PFGE pattern related to the France9V-3 strain.
PFGE pattern related to the Tennessee23F-4 strain.
PFGE pattern related to the Spain23F-1 strain.
PFGE pattern related to the Spain6B-2 strain.
NT, nontypeable.
FIG. 1.
Dendrogram analysis of 104 PNSP strains in the six major clonal groups; the cophenetic correlation is 89.4%. Asterisks indicate the PFGE patterns for the international clones. All strains within a major clonal group have the same serotype except as indicated. NT, nontypeable.
Four of the major clonal groups were genetically related to an international pneumococcal clone and accounted for 58 (40.6%) of all PNSP isolates; clonal group 2 was related to the France9V-3 clone, clonal group 3 was related to the Tennessee23F-4 clone, clonal group 4 was related to the Spain23F-1 clone, and clonal group 6 was related to the Spain6B-2 clone. Most of the clonal group 2 and 5 strains were Peni serotype 9V and 19A strains, respectively. These isolates accounted for 35 (49.3%) of the Peni strains.
In general, strains categorized into a major clonal group were of the same serotype. Clonal groups 1 and 2 consisted of 87.2% serotype 9V strains, clonal groups 3 and 4 consisted of 100% serotype 23F strains, and clonal groups 5 and 6 consisted of 100% 19A and 6B strains (Table 2). The cophenetic correlation for the dendrogram was 89.4% (Fig. 1).
The majority of strains of serotypes 6A, 14, and 19F were present in multiple minor clonal groups. One major clonal group and multiple minor clonal groups contained serotype 6B and 19A strains. Accordingly, the measure of clonal group homogeneity was higher for serotypes 9V, 19A, and 23F (range, 0.40 to 0.68) than for serotypes 6A, 6B, 14, and 19F (range, 0.18 to 0.26) (Table 2).
Antibiotic susceptibility.
The penicillin MICs for the study isolates were distributed as follows: for 62 (43.4%) isolates the penicillin MIC was 0.1 to 0.5 μg/ml, for 9 (6.3%) isolates the penicillin MIC was 1.0 μg/ml, for 43 (30%) isolates the penicillin MIC was 2.0 μg/ml, for 25 (17.5%) isolates the penicillin MIC was 4.0 μg/ml; and for 4 (2.8%) isolates the penicillin MIC was 8.0 μg/ml. Of all 143 PNSP strains, 101 (79%) were nonsusceptible to TMP-SXZ, 35 (24.5%) were nonsusceptible to erythromycin, 68 (47.6%) were nonsusceptible to cefotaxime, 30 (21%) were nonsusceptible to tetracycline, 29 (20.3%) were nonsusceptible to chloramphenicol, 12 (8.4%) were nonsusceptible to clindamycin, and 9 (6.3%) were nonsusceptible to ofloxocin. None of the isolates were nonsusceptible to vancomycin.
Serotype was strongly associated with the level of penicillin resistance. For example, 23 (71.9%) of the serotype 23F isolates but only 2 (7.4%) of the serotype 19A isolates were Penr (P < 0.001) (Table 1). Serotype 19A was less likely to be associated with MDR strains and serotype 23F was more likely to be associated with MDR strains compared to the associations for the remainder of the serotypes.
Major clonal groups 1 and 2 differentiated Peni from Penr strains among the serotype 9V strains (Table 3). The major clonal groups differed substantially by antibiotic susceptibility pattern. The susceptibility patterns of clonal group 2 to 4 and 6 strains mirrored the susceptibility patterns of the respective international clones with a few exceptions. Specifically, the erythromycin susceptibility patterns of the Spain23F-1 and Spain6B-2 clones and the penicillin susceptibility pattern of the Tennessee23F-4 clone differed from those of most of the strains in these clonal groups. The clonal groups which were genetically related to an international clone were more likely than the remainder of the clonal groups to contain MDR strains (Table 3).
TABLE 3.
Distribution of Penr and MDR pneumococcal strains by clonal group
| Clonal group | No. (row %) of Penr strains (n = 72) | P value | No. (row %) of MDR strains (n = 109) | P value |
|---|---|---|---|---|
| Clonal group | ||||
| 1 | 1 (6.7) | <0.01 | 3 (11.1) | <0.01 |
| 2a | 21 (87.5) | 24 (100) | ||
| Clonal group | ||||
| 3b | 7 (53.8) | <0.01 | 13 (100) | 1.00 |
| 4c | 16 (100) | 16 (100) | ||
| Clonal group 5 | 1 (4.5) | 0.34 | 15 (68.2) | 1.00 |
| Non-clonal group 5, serotype 19A | 1 (20) | 3 (60) | ||
| Clonal group 6d | 5 (100) | 0.02 | 5 (100) | 0.23 |
| Non-clonal group 6, serotype 6B | 2 (22.2) | 6 (66.7) | ||
| Clonal groups 2 to 4 and 6 | 49 (84.5) | <0.01 | 58 (100) | <0.01 |
| Remainder of isolates | 23 (27.1) | 51 (60) |
PFGE pattern related to France9V-3 strain.
PFGE pattern related to the Tennessee23F-4 strain.
PFGE pattern related to the Spain23F-1 strain.
PFGE pattern related to the Spain6B-2 strain.
Major clonal group 3, which was genetically related to the Tennessee23F-4 clone, included two subsets of unique MDR strains. The only four isolates for which the penicillin MIC was 8 μg/ml and the only two isolates that were both Peni and cefotaxime resistant were present in this clonal group. All six of these strains were also resistant to cefotaxime and TMP-SXZ. Six (46%) of the clonal group 3 strains were Peni, whereas none of the major clonal group 4 strains were Peni (Table 3).
DISCUSSION
This is the first study that has described the molecular epidemiology of all invasive PNSP isolates from an entire metropolitan area. This approach provides a complete representation of the genetic diversity of invasive PNSP isolates for a defined population base. Other studies which have described the molecular epidemiology of PNSP in the United States have focused on a select sample of PNSP isolates for which the penicillin MIC is ≥1 μg/ml (9, 12).
In this study, we found that two-thirds of invasive PNSP isolates in Baltimore could be characterized into six major clonal groups and that nearly half of the PNSP isolates were genetically related to one of four recognized international clones. Furthermore, antibiotic susceptibility patterns differed substantially between major clonal groups, even among isolates of the same serotype. PNSP strains which were genetically related to an international clone were MDR.
To the best of our knowledge, two of the major clonal groups that we identified (clonal groups 1 and 5) have not been previously described as international clones or identified in other studies of the molecular epidemiology of PNSP. This is most likely because previous studies have focused on Penr isolates, whereas these clonal groups largely consisted of Peni strains (9, 12). Nearly all of the clonal group 1 and 5 strains were serotype 9V or 19A, and for 95% of these strains the penicillin MICs were <1 μg/ml. As would be expected, the majority of the strains were not MDR.
We also found that the degree of genetic diversity varied by serotype. Each of the six major clonal groups largely comprised strains of one serotype. Serotypes 9V, 19A, and 23F were associated with more clonal group homogeneity than serotypes 6A, 6B, 14, and 19F. Whether these differences in genetic diversity by serotype are due to different rates of recombination, different lineages of S. pneumoniae, or some other mechanism is not known.
As has been described for other geographic areas, the majority of PNSP strains in BMA were of serotypes 6A, 6B, 9V, 14, 19A, and 23F (17). The present data are in accordance with those from two recent studies which found that 70 to 93% of the strains for which the penicillin MIC was ≥1.0 μg/ml could be subtyped into 1 of 10 PFGE types (9, 12).
PFGE is a standardized method for the subtyping of bacteria and has been used effectively to track the worldwide spread of major international clones. The dendrogram has become an essential component of the analysis of the genetic relatedness of a large number of isolates. Unfortunately, no standardized method for the grouping of isolates into clonal groups has been developed. To complicate matters, computer-based analyses are operator dependent (11) and the genetic relatedness of isolates can be substantially altered by modifying the band sensitivity parameters. We defined genetic relatedness by a combination of dendrogram and visual inspection. These two methods resulted in a classification which was supported by the phenotypic characteristics of the clones and a dendrogram with a high degree of cophenetic correlation. Strains within a major clonal group had a ≥80% correlation on the dendrogram. In addition, our clonal groups generally had at least seven-band differences between groups and less than seven-band differences within groups. For clonal groups which contained an international clone, all strains were genetically related to the clone by the criteria of Tenover et al. (33) and often had the susceptibility pattern of the respective clone. The notable exceptions were the strains of major clonal groups 1 and 2. Some of the clonal group 1 strains had differences of five to six bands compared to the pattern for the France9V-3 clone of major clonal group 2. However, the dendrogram categorized these strains into two clonal groups which were phenotypically distinct. Our high degree of cophenetic correlation confirmed that the dendrogram faithfully represented the computer-derived degree of genetic relatedness between strains.
The use of only one molecular subtyping technique and one restriction enzyme is a limitation of this study that could have led to an underestimation of the genetic diversity of our isolates. PFGE provides discriminatory power equal to that of BOX fingerprinting or restriction fragment end labeling and discriminatory power higher than those of PCR and ribotyping (16). However, PNSP strains with a different penicillin-binding protein gene (dhf) can have identical PFGE patterns (12).
Despite the substantial degree of MDR among isolates in Baltimore, all PNSP isolates were susceptible to vancomycin and there was nearly complete susceptibility to ofloxacin (1, 28). However, fluoroquinolone-nonsusceptible pneumococci are increasing in frequency in Canada, most likely due to the increasing use of fluoroquinolones (5). In addition, the emergence of vancomycin-nonsusceptible Staphylococcus aureus raises the possibility that vancomycin-resistant S. pneumoniae may develop in the future (29, 31). The changing epidemiology of drug resistance in the United States underscores the need to use these antibiotics prudently and to continue to monitor resistance rates to ensure the development of appropriate antibiotic therapy guidelines. Since geographic variations in the penicillin susceptibilities of S. pneumonia isolates have been detected, these data may not be able to be generalized to other areas of the United States.
In summary, we defined the clonal groups and phenotypic characteristics of PNSP isolates responsible for invasive infection in BMA. The classification that we used to define PFGE-based clonal groups resulted in groups with distinctive phenotypic features that were characteristic of those of the previously reported international clones and were often serotype specific. The identification of two previously undescribed clonal groups was most likely due to the inclusion of S. pneumoniae isolates for which penicillin MICs were ≥0.1 and <1 μg/ml. Future studies will determine whether new clonal groups have emerged or whether the frequencies of the present clonal groups have changed since 1996. The definition of these clonal groups will be crucial for our ongoing studies to understand the demographic, geographic, and seasonal differences in PNSP (B. A. Albanese, Z. H. Reed, J. C. Roche, M. A. Pass, C. G. Whitney, and L. H. Harrison, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1047, p. 152, 1999).
ACKNOWLEDGMENTS
We thank the participating hospital infection control practitioners and microbiology laboratory personnel in BMA for identifying patients with pneumococcal infections and providing the bacterial isolates; Yvonne Dean-Hibbert for assistance in conducting surveillance; Kim Holmes for assistance with data collection; and Althea Glenn, Laboratories Administration, Maryland Department of Health and Mental Hygiene, for processing the isolates. We gratefully acknowledge Victor Yu for expertise and David McDevitt for susceptibility testing of selected isolates. We thank Terry Thompson and Lashondra Shealey for assistance with the serotype testing of selected isolates, Linda McDougal for providing the susceptibility patterns of the international pneumococcal clones, and Susan Hunter for guidance with the dendrogram analysis. We also thank the Emerging Infections Project in Maryland, especially the staff of the Epidemiology and Disease Control Program of the Maryland Department of Health and Mental Hygiene, for support.
This work was supported by the National Foundation for Infectious Diseases (to M.C.M.) and the National Vaccine Program, Emerging Infections Program Network, National Center for Infectious Diseases, CDC (to L.H.H).
REFERENCES
- 1.Bartlett J G, Breiman R F, Mandell L A, File T M., Jr Community-acquired pneumonia in adults: guidelines for management. The Infectious Diseases Society of America. Clin Infect Dis. 1998;26:811–838. doi: 10.1086/513953. [DOI] [PubMed] [Google Scholar]
- 2.Bennett N M, Buffington J, LaForce F M. Pneumococcal bacteremia in Monroe County, New York. Am J Public Health. 1992;82:1513–1516. doi: 10.2105/ajph.82.11.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breiman R F, Spika J S, Navarro V J, Darden P M, Darby C P. Pneumococcal bacteremia in Charleston County, South Carolina: a decade later. Arch Intern Med. 1990;150:1401–1405. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Prevention of pneumococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP) Morb Mortal Wkly Rep. 1997;46(RR-8):1–24. [PubMed] [Google Scholar]
- 5.Chen D K, McGeer A, de Azavedo J C, Low D E. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. Canadian Bacterial Surveillance Network. N Engl J Med. 1999;341:233–239. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 6.Coffey T J, Berron S, Daniels M. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988–1994): novel major clones of serotypes 14, 19F and 15F. Microbiology. 1996;142:2747–2757. doi: 10.1099/13500872-142-10-2747. [DOI] [PubMed] [Google Scholar]
- 7.Coffey T J, Daniels M, McDougal L K, Dowson C G, Tenover F C, Spratt B G. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 1995;39:1306–1313. doi: 10.1128/aac.39.6.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 9.Doern G V, Brueggemann A B, Blocker M, Dunne M, Holley P, Kehl K S, Duval J, Kugler K, Putnam S, Rauch A, Pfaller M. Clonal relationships among high-level penicillin-resistant Streptococcus pneumoniae in the United States. Clin Infect Dis. 1998;27:757–761. doi: 10.1086/514937. [DOI] [PubMed] [Google Scholar]
- 10.Figueiredo A M, Austrian R, Urbaskova P, Teixeira L A, Tomasz A. Novel penicillin-resistant clones of Streptococcus pneumoniae in the Czech Republic and in Slovakia. Microb Drug Resist. 1995;1:71–78. doi: 10.1089/mdr.1995.1.71. [DOI] [PubMed] [Google Scholar]
- 11.Gerner-Smidt P, Graves L M, Hunter S, Swaminathan B. Computerized analysis of restriction fragment length polymorphism patterns: comparative evaluation of two commercial software packages. J Clin Microbiol. 1998;36:1318–1323. doi: 10.1128/jcm.36.5.1318-1323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gherardi G, Whitney C G, Facklam R R, Beall B. Major related sets of antibiotic-resistant pneumococci in the United States as determined by pulsed-field gel electrophoresis and pbp1a-pbp2b-pbp2x-dhf restriction profiles. J Infect Dis. 2000;181:216–229. doi: 10.1086/315194. [DOI] [PubMed] [Google Scholar]
- 13.Hall L M, Whiley R A, Duke B, George R C, Efstratiou A. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J Clin Microbiol. 1996;34:853–859. doi: 10.1128/jcm.34.4.853-859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison L H, Dwyer D M, Billmann L, Kolczak M S, Schuchat A. Invasive pneumococcal infection in Baltimore, Md: implications for immunization policy. Arch Intern Med. 2000;160:89–94. doi: 10.1001/archinte.160.1.89. [DOI] [PubMed] [Google Scholar]
- 15.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermans P W M, Sluijter M, Hoogenboezem T, Heersma H, van Belkum A, de Groot R. Comparative study of five different DNA fingerprint techniques for molecular typing of Streptococcus pneumoniae strains. J Clin Microbiol. 1995;33:1606–1612. doi: 10.1128/jcm.33.6.1606-1612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann J, Cetron M S, Farley M M, Baughman W S, Facklam R R, Elliott J A, Deaver K A, Breiman R. The prevalence of drug-resistant Streptococcus pneumoniae in Atlanta. N Engl J Med. 1995;333:471–476. doi: 10.1056/NEJM199508243330803. [DOI] [PubMed] [Google Scholar]
- 18.Istre G R, Tarpay M, Anderson M, Pryor A, Welch D. Invasive disease due to Streptococcus pneumoniae in an area with a high rate of relative penicillin resistance. J Infect Dis. 1987;156:732–735. doi: 10.1093/infdis/156.5.732. [DOI] [PubMed] [Google Scholar]
- 19.Jabes D, Nachman S, Tomasz A. Penicillin-binding protein families: evidence for the clonal nature of penicillin resistance in clinical isolates of pneumococci. J Infect Dis. 1989;159:16–25. doi: 10.1093/infdis/159.1.16. [DOI] [PubMed] [Google Scholar]
- 20.Jernigan D B, Cetron M S, Breiman R F. Minimizing the impact of drug-resistant Streptococcus pneumoniae: a strategy from the DRSP working group. JAMA. 1996;275:206–209. [PubMed] [Google Scholar]
- 21.Klugman K. Pneumococcal molecular epidemiology network. ASM News. 1998;64:371. [Google Scholar]
- 22.Lefevre J C, Bertrand M A, Faucon G. Molecular analysis by pulsed-field gel electrophoresis of penicillin-resistant Streptococcus pneumoniae from Toulouse, France. Eur J Clin Microbiol Infect Dis. 1995;14:491–497. doi: 10.1007/BF02113426. [DOI] [PubMed] [Google Scholar]
- 23.McDougal L K, Rasheed J K, Biddle J W, Tenover F C. Identification of multiple clones of extended-spectrum cephalosporin-resistant Streptococcus pneumoniae isolates in the United States. Antimicrob Agents Chemother. 1995;39:2282–2288. doi: 10.1128/aac.39.10.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McEllistrem M C, Stout J E, Harrison L H. Simplified protocol for pulsed-field gel electrophoresis analysis of Streptococcus pneumoniae. J Clin Microbiol. 2000;38:351–353. doi: 10.1128/jcm.38.1.351-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreno F, Crisp C, Jorgensen J, Patterson J. The clinical and molecular epidemiology of bacteremias at a university hospital caused by pneumococci not susceptible to penicillin. J Infect Dis. 1995;172:427–432. doi: 10.1093/infdis/172.2.427. [DOI] [PubMed] [Google Scholar]
- 26.Munoz R, Musser J M, Crain M, Briles D E, Marton A, Parkinson A J, Sorensen U, Tomasz A. Geographic distribution of penicillin-resistant clones of Streptococcus pneumoniae: characterization by penicillin-binding protein profile, surface protein A typing, and multilocus enzyme analysis. Clin Infect Dis. 1992;15:112–118. doi: 10.1093/clinids/15.1.112. [DOI] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS document M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 28.Quagliarello V J, Scheld W M. Drug therapy: treatment of bacterial meningitis. N Engl J Med. 1997;336:708–716. doi: 10.1056/NEJM199703063361007. [DOI] [PubMed] [Google Scholar]
- 29.Sieradzki K, Roberts R B, Haber S W, Tomasz A. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N Engl J Med. 1999;340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 30.Smith A M, Klugman K P. Three predominant clones identified within penicillin-resistant South African isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:385–389. doi: 10.1089/mdr.1997.3.385. [DOI] [PubMed] [Google Scholar]
- 31.Smith T L, Pearson M L, Wilcox K R, Cruz C, Lancaster M V, Robinson-Dunn B, Tenover F C, Zervos M J, Band J D, White E, Jarvis W R. Emergence of vancomycin resistance in Staphylococcus aureus. Glycopeptide-Intermediate Staphylococcus aureus Working Group. N Engl J Med. 1999;340:493–501. doi: 10.1056/NEJM199902183400701. [DOI] [PubMed] [Google Scholar]
- 32.Stool S E, Field M J. The impact of otitis media. Pediatr Infect Dis J. 1989;8:S11–S14. [PubMed] [Google Scholar]
- 33.Tenover F C, Arbeit R D, Goering R V, Mickelson P A, Murray B E, Persing D H, Swaminathan B. Guest commentary: interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams W W, Hickson M A, Kane M A, Kendal A P, Spika J S, Hinman A R. Immunization policies and vaccine coverage among adults: the risk for missed opportunities. Ann Intern Med. 1988;108:616–625. doi: 10.7326/0003-4819-108-4-616. [DOI] [PubMed] [Google Scholar]